Summary
The most efficient means of generating cellular energy is through aerobic respiration. Under anaerobic conditions, several prokaryotes can replace oxygen by nitrate as final electron acceptor. During denitrification, nitrate is reduced via nitrite, NO and N2O to molecular nitrogen (N2) by four membrane‐localized reductases with the simultaneous formation of an ion gradient for ATP synthesis. These four multisubunit enzyme complexes are coupled in four electron transport chains to electron donating primary dehydrogenases and intermediate electron transfer proteins. Many components require membrane transport and insertion, complex assembly and cofactor incorporation. All these processes are mediated by fine‐tuned stable and transient protein–protein interactions. Recently, an interactomic approach was used to determine the exact protein–protein interactions involved in the assembly of the denitrification apparatus of Pseudomonas aeruginosa. Both subunits of the NO reductase NorBC, combined with the flavoprotein NosR, serve as a membrane‐localized assembly platform for the attachment of the nitrate reductase NarGHI, the periplasmic nitrite reductase NirS via its maturation factor NirF and the N2O reductase NosZ through NosR. A nitrate transporter (NarK2), the corresponding regulatory system NarXL, various nitrite (NirEJMNQ) and N2O reductase (NosFL) maturation proteins are also part of the complex. Primary dehydrogenases, ATP synthase, most enzymes of the TCA cycle, and the SEC protein export system, as well as a number of other proteins, were found to interact with the denitrification complex. Finally, a protein complex composed of the flagella protein FliC, nitrite reductase NirS and the chaperone DnaK required for flagella formation was found in the periplasm of P. aeruginosa. This work demonstrated that the interactomic approach allows for the identification and characterization of stable and transient protein–protein complexes and interactions involved in the assembly and function of multi‐enzyme complexes.
Introduction
Most organisms, with the exception of a few fermentative bacteria, utilize membrane‐associated respiratory processes for cellular energy generation. During respiration, electrons are transported along a chain of redox‐active cofactors fixed to large, membrane‐associated enzyme complexes driven by the corresponding redox potentials (Borrero‐de Acuna et al., 2016). The linked release of energy is employed for the formation of a chemo‐osmotically active proton/sodium ion gradient, which in turn allows for ATP formation via proton/sodium ion‐driven ATP synthases (Jahn and Jahn, 2012). Multiple electron donors and acceptors can be employed by a wealth of different enzyme complexes carrying diverse redox cofactors (Marreiros et al., 2016). However, all these components have to be localized in the appropriate membrane, assembled with their cofactors, and possess the appropriate contacts required for productive electron transfer. It has been shown that stacking of the different complexes facilitates correct channelling and transport of electrons through the respiratory chains (Guo et al., 2016). The loss of energy is thereby minimized, and undesired side reactions, such as the formation of free reactive oxygen species (ROS), are reduced. There are two different hypotheses regarding the nature of the interactions that might occur between complexes I–V of the mitochondrial respiratory chain, the ‘fluid‐state’ and the ‘solid‐state’ models (Lapuente‐Brun et al., 2013). According to the fluid‐state model, the diverse respiratory complexes are able to diffuse without restraints along the inner mitochondrial membrane. In this case, electrons are transferred from one complex to the succeeding one when both entities randomly collide (Porras and Bai, 2015). In contrast, the solid model assumes that respiratory complexes are built in a highly organized and rigid manner (Enriquez, 2016). However, there is experimental evidence supporting co‐occurrence of elements of both models. Accordingly, it has been presumed that in a natural system, the dynamic interchange between both states is the most effective mode. This leads to the ‘plasticity’ or ‘dynamic aggregate’ model, in which the complexes can freely switch from one state to the other (Acin‐Perez and Enriquez, 2014). In this model, complexes I, III and IV strongly interact with each other creating the respirasome supercomplex, which oxidizes NADH. The respirasome in turn interacts with complex II (succinate dehydrogenase) which accepts electrons from FADH2. The coenzyme Q and cytochrome c, normally found in pools, are able to diffuse along the membrane and associate with these complexes, transferring the electrons from one complex to another (Alcazar‐Fabra et al., 2016). The formation of tight respirasome complexes either in eukaryotes or in prokaryotes has been demonstrated by different methods. Many higher‐ordered complexes were discovered and analysed by Blue‐native gel electrophoresis, like the respirasomes from yeast (Schagger and Pfeiffer, 2000), mouse fibroblast (Lapuente‐Brun et al., 2013), spinach (Krause et al., 2004) and potato (Bultema et al., 2009). Supercomplexes from bovine mitochondria were visualized with single particle cryoelectron tomography (Dudkina et al., 2011). Immunochemical and proteomics methods were used for the elucidation of complex composition (Borrero‐de Acuna et al., 2016). Similarly, several respiratory supercomplexes were analysed in bacteria like the sulfide oxidase–oxygen reductase from Aquifex aeolicus (Prunetti et al., 2010) and the respirasome from Paracoccus denitrificans (Stroh et al., 2004). The dynamics of oxidative phosphorylation complexes in Eschericha coli was recently discussed (Magalon et al., 2012; Magalon and Alberge, 2016). However, knowledge of the assembly, composition, function and degradation of all these complexes is limited.
The denitrification machinery of Pseudomonas aeruginosa
The ubiquitously found, metabolically highly versatile bacterium Pseudomonas aeruginosa is proliferates in diverse environments, such as soil, water and even on the surfaces of hospital equipment (Talwalkar and Murray, 2016). It is also an important opportunistic pathogen and one of the most predominant pathogens causing acute and chronic lung infections in immunocompromised hosts (Oliver et al., 2000; Driscoll et al., 2007; Auerbach et al., 2015). P. aeruginosa is a facultative anaerobe able to respire nitrate or nitrite and to ferment pyruvate and arginine when oxygen becomes exhausted (Vander Wauven et al., 1984; Eschbach et al., 2004). The capability of thriving at low oxygen partial pressure facilitates the invasion of the mucus of cystic fibrosis patients (Alvarez‐Ortega and Harwood, 2007). Worldwide, there are over 70.000 cases of P. aeruginosa infections of patients with this ion‐channel‐defect genetic disease (Aloush et al., 2006). P. aeruginosa replaces molecular oxygen as terminal electron acceptor for the respiratory chain with different N‐oxides during anaerobiosis. An electron transport with the terminal enzyme nitrate reductase converts nitrate into nitrite, generates a proton/sodium ion gradient and synthesizes ATP. Similarly, nitrite is reduced to NO, N2O and further to molecular nitrogen by 3 further respiratory chains with the terminal enzymes nitrite, NO and N2O reductase (Zumft, 1997). Two of these reductases (nitrite reductase and N2O reductase) are localized in the periplasm, and the other two (nitrate reductase and NO reductase) reside in the inner membrane (Schobert and Jahn, 2010). Three regulatory systems control the onset of denitrification (Schreiber et al., 2007; Trunk et al., 2010). The Fnr/Crp‐type regulator Anr (PA1544) senses the absence of oxygen, while the two‐component regulatory system NarXL (PA3878; PA3879) monitors nitrate, and a second Fnr/Crp‐type regulator Dnr (PA0527) detects the formation of NO (Pessi and Haas, 2000; Rinaldo et al., 2005). In a regulatory cascade reaction, Anr, Dnr and NarXL gradually induce the denitrification operon (Van Alst et al., 2007). Many of the denitrification proteins produced are exported via the Sec protein secretion system into the periplasm (Denks et al., 2014). The incorporation of essential cofactors (Fe‐S‐clusters, molybdenum cofactor, haem, metals) requires their synthesis, transport and insertion by specialized proteins (Blasco et al., 2001; Magalon and Mendel, 2015; Dailey et al., 2017). To elucidate the protein–protein interaction network involved in the formation of the denitrification apparatus, a proteomics‐based interactomic approach was combined with electron microscopy (Borrero‐de Acuna et al., 2016).
The proteomics‐based interactomic approach
First, bait proteins were selected on the basis of results from physiological experiments. For example, mutants in the genes for NO reductase subunits B (PA0524) and C (PA0523); (NorBC) and the flavoprotein NosR (PA3391) revealed a nitrate reductase‐deficient growth phenotype (Borrero‐de Acuna et al., 2016). However, an intact nitrate reductase was detected in these mutants using appropriate antibodies. Thus, NorBC and NosR were presumed to functionally interact with nitrate reductase to form a higher‐ordered protein complex anchored to the inner membrane (Vaccaro et al., 2015; Cutruzzola and Frankenberg‐Dinkel, 2016; Zhang et al., 2017). NorBC and NosR were therefore selected as bait proteins for the interactomic experiments. Another interesting physiological observation was the failure of P. aeruginosa nitrite reductase (NirS) gene mutants to build an intact flagellum resulting in the loss of swimming motility (Borrero‐de Acuna et al., 2015). The periplasmic NirS was therefore selected as a bait protein to identify its interaction partners, using affinity chromatography purification coupled with mass spectrometry (Borrero‐de Acuna et al., 2015).
The workflow of affinity copurification of prey proteins with the corresponding bait proteins, coupled to prey protein identification by LC‐MS/MS, is shown in Fig. 1A. Denitrifying conditions were achieved by anaerobically incubating nitrate‐supplemented cultures until the late exponential phase (Galimand et al., 1991). At this point, protein cross‐linking through addition of formaldehyde was carried out to stabilize scarce and transient protein–protein interactions within the protein complexes. The new peptides, i.e. peptides not found in the native proteins, created by cross‐linking and released by trypsin treatment, are shown in Fig. 1B. LC‐MS/MS analyses readily identified non‐cross‐linked peptides, whereas inter‐ and trans‐peptides posed problems for identification because the corresponding m/z ratios were shifted (Fig. 1B). This observation was used for the identification of the interacting domains of two proteins (NirS and FliC) as outlined below. Due to the chemical nature of this cross‐linker, its penetration through cellular membranes is rather fast (20 min). Furthermore, formaldehyde cross‐linking preserves the native structures of the proteins in the formed complexes. Cross‐linking was quenched by adding an amino acid such as glycine. Formed formaldehyde‐based cross‐links were disrupted again by subjecting the samples to high temperatures (95°C) for short periods of about 20 min (Sutherland et al., 2008), thereby allowing recovery of analysable peptides for mass spectrometry analysis. However, this method does not distinguish between directly bait‐bound proteins and proteins indirectly bound via other bait‐bound polypeptides.
Figure 1.
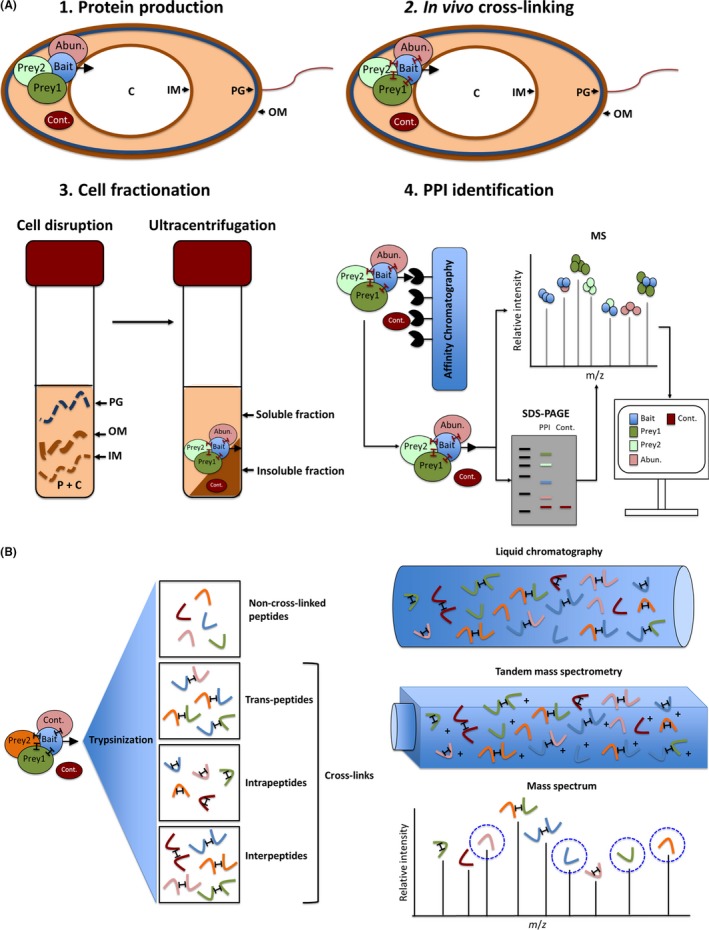
General interactomic workflow. The illustration depicts step by step the protein–protein interaction elucidation pathway via affinity chromatography copurification coupled with mass spectrometry.
A. Shows: (1) Induction of the bacterial host for bait–protein production by the use of anaerobic growth condition in the presence of nitrate; (2) the in vivo cross‐linking by the addition of diffusible cross‐linkers; (3) the cell fractionation and separation of in‐ and soluble fraction by centrifugation; and in (4) the affinity tag‐based purification of the formed and cross‐linked protein complexes, their visualization on an SDS–PAGE gel, the mass spectrometry analyses of corresponding peptides and the computational identification of bait interaction partner candidates.
B. shows the downstream processing of cross‐linked peptides. There are four possible varieties of resulting peptides after trypsin digestion: non‐cross‐linked peptides, interpeptides (cross‐links between diverse peptides of the same protein), intrapeptides (cross‐links within the same peptide of a certain protein) and trans‐peptides (cross‐links between peptides of different proteins). The LC‐MS/MS analysis is depicted in detail. Abun., Abundant proteins that unspecifically attach to the bait–prey complex (identified by the analysis of their protein abundance prior to affinity purification enrichment. Thus, proteins enriched after purification are considered as potential interaction partners); PPI, protein–protein interactions; Cont., contaminant with affinity to the column material (detected with a control strain lacking the expression plasmid); MS, mass spectrometry; PG, peptidoglycan; C, cytoplasm; IM, inner membrane; OM, outer membrane.
Due to the different cellular locations of our bait proteins, NirS in the periplasm and NorBC/NosR in the membrane, different preparation procedures for the isolation of the cross‐linked complexes and the corresponding background control samples were established. Periplasmic proteins were released by an osmotic shock treatment by addition of 300 mM sucrose, avoiding complete cell disruption and minimizing release of cytoplasmic proteins, followed by ultracentrifugation (100 000 × g) (Nicke et al., 2013). Membrane proteins were obtained by French Press disruption, followed by ultracentrifugation to separate soluble from insoluble proteins. Membrane proteins were subsequently solubilized by addition of Triton X‐100 for further purification. Afterwards, all isolated protein fractions were purified by affinity chromatography with stringent washing to eliminate contaminants (see Fig. 1) (Makowski et al., 2016). Contaminants consist of proteins interacting with the column material. Periplasmic and membrane protein fractions prepared in parallel from the parental strain without bait served as background controls.
Affinity‐purified protein complexes were subject to quantitative LC/MS‐MS‐based proteomics. If a prey was measured in larger amounts in the protein complex in comparison with the natural appearance in the proteomic analysis of the disrupted P. aeruginosa, prior to affinity chromatography purification (control), its interaction with the bait was considered to be specific. For this purpose, the abundance of a specific protein was measured by elucidating the average area of its three most prominent mass spectrometry peaks in the sample before and after affinity purification. A semi‐quantitative value was given to each polypeptide according to this average area that directly reflects protein abundance. The abundance of each protein prior and subsequent to affinity purification was then compared (Ong and Mann, 2005; Kaake et al., 2010; Howden et al., 2013). The software Proteome Discoverer was employed for m/z analysis and identification of interaction partner candidates (Al Shweiki et al., 2017).
Tandem affinity purification has proven useful for precise protein interaction partner determination, avoiding the presence of contaminants to a high extent (Burckstummer et al., 2006). Here, the use of just one tag combined with the application of stringent washing steps led to highly accurate results. In our approach, different types of affinity tags, i.e. His6‐tag as well as the Strep‐tag II, were employed. The gene encoding the periplasmic protein NirS was genetically fused to Strep‐tagII, whereas membrane protein encoding genes norC, norB, nosR were fused to the His6‐tag. Expression of all of these genes was driven by the cognate native promoters and thereby controlled by the intrinsic transcriptional mechanisms of P. aeruginosa. The identification of potential N‐terminal signal peptides of the bait proteins is crucial to properly locate the selected tag (Dalbey and Kuhn, 2012). C‐terminal tags should not interfere with protein translocation. The tagged bait proteins were produced in mutant backgrounds eliminating the non‐tagged counterpart of the protein of interest (i.e. norC‐His6‐tag in a transposon norC mutant).
The antibody‐based interactomic approach
Identified protein–protein interactions were confirmed by a second, technically independent, approach. In this case, antibodies against the potential interaction partners were obtained and used for in vivo colocalization experiments by electron microscopy. Polyclonal antibodies against the different baits used (NorB, NorC, NosR, NirS) and preys detected (NarH, DnaK, FliC) were raised. DnaK and NosZ antibodies were synthesized using the pET14b plasmid and E. coli BL21. Specific antibodies against NarH, NosR, NirS, NorC and FliC were produced by immunization using peptides representing specific soluble domains of the proteins of interest, namely peptides spanning amino acid residues 50 to 61, 102 to 116, 118 to 132 and 106 to 130, for NorC; 190 to 203 and 404 to 418 for NosR; 1 to 276, 277 to 392, and 418 to 513, for NarH; 379 to 392, 526 to 540, and 541 to 555, for NirS; and the first 174 amino acids, for FliC. In the case of membrane proteins NarH, NosR and NorC, the chosen peptides were hydrophilic representing loops of the protein protruding into the cytoplasm or periplasm. Based on the known crystal structures of the soluble proteins NirS and FliC, peptides known to be exposed at the protein surface were preferred (Cutruzzola et al., 2001; Sun et al., 2002; Song and Yoon, 2014); (PDB: 1GJQ and PDB: 4NX9).
Obtained antibodies were conjugated to immunogold particles of different sizes (10 and 15 nm) and used to visualize by transmission electron microscopy potential colocalization of protein targets in individual cells. Both the cellular locations of individual target proteins, and their interaction partners, were thereby visualized at the single cell level (Masoumi et al., 2008; Borrero‐de Acuna et al., 2016).
NorCB and NosR are the membrane‐based assembly platform for the denitrification Supercomplex
Pseudomonas aeruginosa was grown under anaerobic, denitrifying conditions. Protein interaction partners of NorBC and NosR were determined using the outlined comparative proteomics approach. Selected protein–protein interactions were visualized using the antibody‐based electron microscopy approach as described above. The robustness of our approach was demonstrated by the strong enrichment of NorB by the tagged NorC bait and vice versa. NosR was encountered as strong interaction partner for both proteins (Fig 2A).
Figure 2.
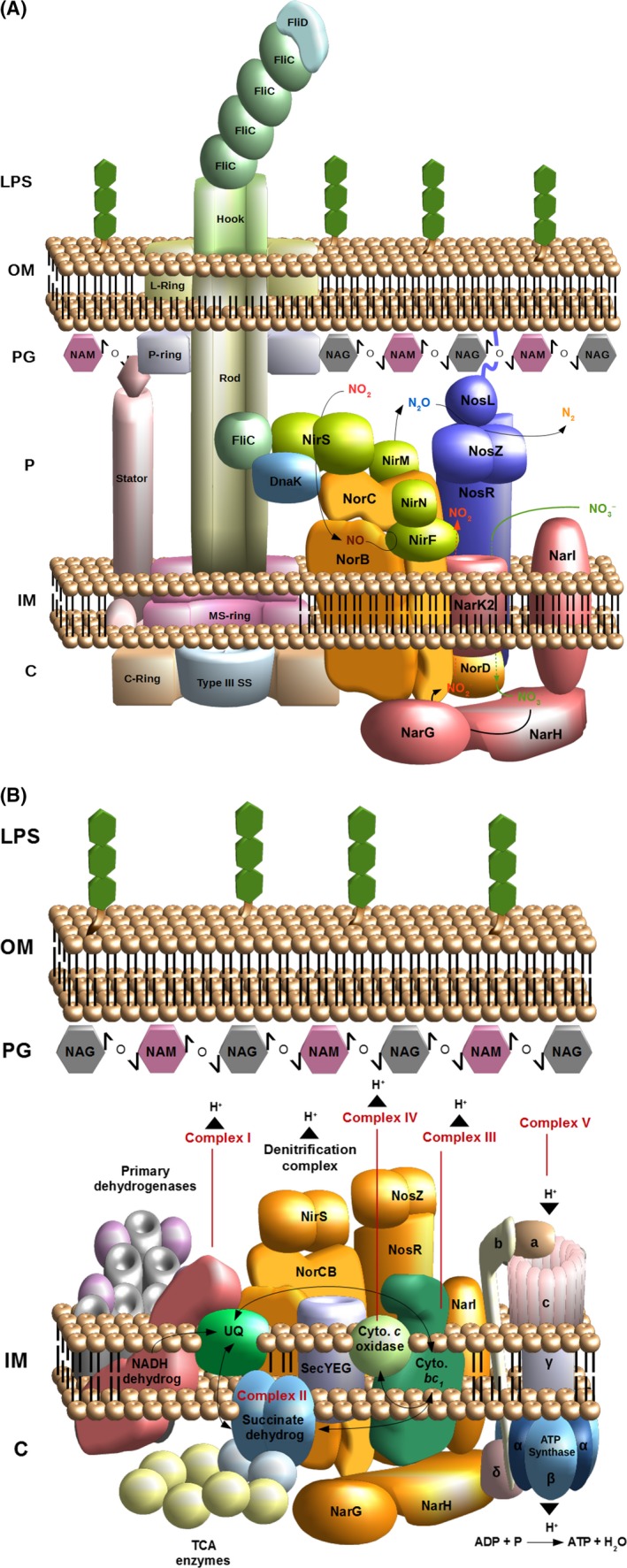
Illustration of the detected denitrification supercomplexes.
A. The protein interaction partners for NirS, NosR, NorC and NorB that are related to denitrification or flagellum assembly (FliC and DnaK) are shown. The four reduction steps → ; → NO; NO → N2O and N2O → N2 are represented. The flagellum structure is drafted based on previous works on Salmonella enterica and E. coli.
B. The respirasome of Pseudomonas aeruginosa encompassing, among others, primary dehydrogenases, respiratory chain complexes I‐V, ubiquinol, cytochrome c and the SecAYEG translocon. The electron flow is specified with black arrows. The proton translocation associated with ATP generation is also depicted. NAM, N‐acetylglucosamine; NAG, N‐acetylmuramic acid; LPS, lipopolysaccharide; OM, outer membrane; PG, peptidoglycan; P, periplasm; IM, inner membrane; C, cytoplasm; type III SS, type III secretion system; TCA enzymes, enzymes involved in the tricarboxylic acid (TCA) cycle.
The nitrate reductase NarGH (PA3875; PA3874) subunits firmly interacted with NorCB, whereas membrane‐spanning NarI (PA3872) was not found to be involved in the interaction. The nitrate/nitrite antiporter NarK2 (PA3876), responsible for nitrate import into the cytoplasm, and the parallel export of formed nitrite, interacted with NorC. This transporter was found to be essential for denitrification by P. aeruginosa (Sharma et al., 2006). A second known nitrate transporter, NarK1 (PA3877), was not found in the protein complex. The next step of denitrification is the reduction of nitrite to NO by the nitrite reductase NirS. NirF (PA0516), a protein involved in nitrite reductase (NirS) maturation via the incorporation of haem d 1 into the enzyme (Bali et al., 2010), interacted strongly with NorCB. Furthermore, the protein NirN (PA0509) was also part of NorBC interactome (Adamczack et al., 2014). Finally, NirS was found in complex with NorB. These findings are consistent with the previous observation that NirS forms a maturation triad, a stable triple complex, with NirF and NirN (Fig. 2A) (Nicke et al., 2013). Strong interactions between NirM (PA0518), a c‐type cytochrome donating electrons to NirS, and NorC were also observed (Hasegawa et al., 2001). Furthermore, azurin (PA4292), also known as cytochrome c 551, which transfers electrons from the cytochrome bc 1 complex to terminal reductases of denitrification (Williams et al., 2007; Santini et al., 2014), was incorporated into this protein complex by its contact to NorC. Finally, NorC exhibited significant affinity to NirQ (PA0520), a regulatory protein required for formation of the denitrification machinery (Hayashi et al., 1998).
NosR is an iron–sulfur protein harbouring a flavin cofactor facing the periplasm and redox centres located on the cytoplasmic side of the inner membrane (Wunsch and Zumft, 2005) and, as indicated above, is tightly bound to the respirasome via NorC (Fig. 2A). NosR was, itself, found to interact with NarG, NarH, NirE (PA0510), NirQ, NirS and NirM. Importantly, the N2O reductase NosZ (PA3392) exhibited a strong interaction with NosR. The thus far not well understood NosR function seems to be linked to NosZ maturation (Cuypers et al., 1992; Arai et al., 2003; Wunsch et al., 2003). NosR also associates with NosL (PA3396), a lipoprotein attached to the outer membrane described as a copper‐binding protein. This protein is probably responsible for the insertion and coordination of the multicopper centre within NosZ (McGuirl et al., 2001; Taubner et al., 2004). Therefore, the N2O reduction machinery appears to interact with the rest of the denitrification complex principally via the NosR membrane protein and not via NorCB (Fig. 2A).
Denitrification protein colocalization studies were carried out with pairs of antibodies, raised against NarH, NirS, NorC, NosZ an NosR and subsequently conjugated to 15‐ or 10‐nm gold particles (Rohde et al., 2003) that were applied to ultrathin sections of P. aeruginosa and visualized by electron microscopy. When the observed distance between two different sized gold particles was < 25 nm, it was assumed that the two corresponding proteins had been colocalized, and hence interacted with one another (Elamin et al., 2011). The results of these colocalization experiments were highly consistent with the conclusions of the proteomics experiments. An important control for the colocalization approach was that when two proteins were known to be separated by the inner membrane, as is the case for cytoplasmic NarH and periplasmic NirS or NosZ, colocalization was not observed.
These observations explain results of our previous physiological experiments, in which norB mutants exhibit impaired nitrate and nitrite reduction in vitro and in vivo, namely that the absence of NorBC results in destabilization of the respirasome in such a way that NarGHI and NirS are unable to conduct their enzymatic functions. While NarH can still be detected in corresponding mutant bacteria, NirS cannot and hence must be prone to proteolytic degradation.
The denitrification supercomplex as platform for the attachment of the corresponding electron transport chains
Several primary dehydrogenases were found tightly bound to the NorCB‐NosR platform. The most significant interaction partners detected were NADH dehydrogenase Nuo (Platt et al., 2008) (PA2638 to PA2644), proline dehydrogenase PutA (PA0782) (Nakada et al., 2002), l‐lactate dehydrogenase (PA4771) (Eschbach et al., 2004) and the D‐amino acid dehydratase (PA3357) (Ikeno et al., 2006). Diverse subunits of the FoF1 ATP synthase (PA5553 to 5560) (Cook et al., 2014) were also linked to the respiration complex. Surprisingly, several enzymes mediating the TCA cycle, like malate: quinone oxidoreductase (PA3452), succinate dehydrogenase (PA1582 to PA1584), isocitrate dehydrogenase (PA2624), citrate synthase (PA1580), succinyl‐coenzyme A (succinyl‐CoA) synthetase (PA1588) and 2‐oxoglutarate dehydrogenase (PA1585), were also found to be part of the supercomplex, as were enzymes related to the pyruvate metabolism, such as various subunits of pyruvate dehydrogenase (PA5015), pyruvate kinase (PA4329), phosphopyruvate hydratase (PA3035), acetyl‐CoA carboxylase (PA3112, PA3639 and PA4848) and phosphoenolpyruvate synthase (PA1770) (Ornston, 1971; Meylan et al., 2017) (Fig. 2B).
The denitrification supercomplex as a platform for corresponding transport and maturation factors
The respirasome included several enzymes involved in haem biosynthesis, like the haem biosynthesis‐associated protein (PA5257), the potential enzyme of haem biosynthesis HemX (PA5258), coproporphyrinogen III dehydrogenase HemN (PA1546), coproporphyrinogen III oxidase HemF (PA0024) and porphobilinogen synthase HemB (PA5243) (Dailey et al., 2017). Furthermore, the haem d1 biosynthesis proteins NirF (PA0516), NirJ (PA0511), NirL (PA0514) and NirE (PA0510) were found attached (Layer et al., 2010). Haem d1 is a cofactor of nitrite reductase NirS. The electron donor systems for NirS, the c‐type cytochromes NirM (PA0518) and NirN (PA0509) were also associated with the complex, as were cytochrome c 1 (PA4429), cytochrome c 5 CycB (PA5300), cytochrome c 4 precursor Cc4 (PA5490), cytochrome c oxidase of the cbb3‐type (PA1552‐4), and the cytochrome c‐type biogenesis proteins CcmE (PA1479) and CycH (PA1483) (Williams et al., 2007). Proteins involved in the biogenesis of iron–sulfur clusters, like NfuA (PA1847) (Roche et al., 2013), of the molybdenum cofactor, like MoaB1 (PA3915) (Andreae et al., 2014; Kasaragod and Schindelin, 2016; Fernandez‐Barat et al., 2017), and of ubiquinone, like UbiE (PA5063) (Jacewicz et al., 2013; Aussel et al., 2014), were also found in the complex. Finally, multiple proteins constituting the SecAYEG translocon, like YajC (PA3822), SecA (PA4403), SecD (PA3821), SecF (PA3820) and YidC (PA5568), were identified as interaction partners (Dalbey and Kuhn, 2012; Kudva et al., 2013). This is consistent with the need to secrete most of the proteins involved in denitrification to the periplasm.
The periplasmic NirS‐DnaK‐FliC complex is essential for flagellum formation
During elucidation of interaction partners for NirS, a triad constituted of NirS‐DnaK‐FliC was found in the periplasm of P. aeruginosa by the proteomics approach and subsequently confirmed by colocalization analyses (Borrero‐de Acuna et al., 2015), suggesting a role of the respirasome in motility. Consistent with this suggestion, the nirS transposon mutant exhibited swimming impairment in swimming motility assays, and electron microscopic examination of the mutant revealed defective flagellum formation (Fig. 3A). This finding raised the question of whether the role of NirS in motility is structural or enzymatic, i.e. whether or not the nitrite reductase activity of NirS is essential for flagellum formation and motility. We therefore examined the swimming ability of a nirF mutant, because NirF is required for NirS maturation, and a nirF mutant produces an intact NirS protein lacking nitrite reductase activity (Adamczack et al., 2014). This experiment revealed that the nirF mutant exhibited a normal flagellum and mobility, indicating a structural role of NirS in flagellum formation (Borrero‐de Acuna et al., 2015). As crystal structures of FliC and NirF are available, their contact surfaces in the triad could be identified by means of the proteomics approach. Detection of cross‐linked FliC‐NirS peptides, and comparison with their un‐cross‐linked counterparts revealed that NirS and FliC interacted via surfaces involving the AAEQYQGAASAVDPTHVVR, CAGCHGVLRK and GQQYLEALITYGTPLGMPNWGSSGELSK peptides of NirS (Fig. 3B) and the NQVLQQAGT, AILAQANQLPQAVLSLLR and LGITASINDK peptides of FliC (Fig. 3B). Although the presence of the DnaK in the triad and in the periplasm was unambiguously documented by electron microscopy (Borrero‐de Acuna et al., 2015), its role in the triad remains to be determined.
Figure 3.
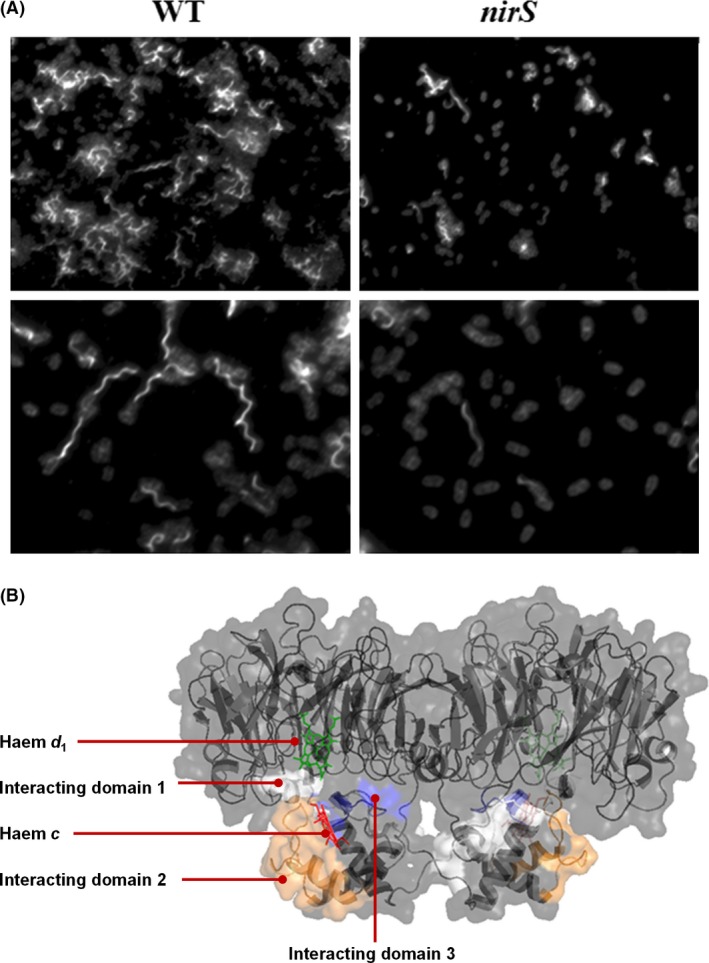
Visualization of impaired flagellar formation in the Pseudomonas aeruginosa nirS mutant and determination of the interacting domains of the NirS and FliC proteins by LC‐MS/MS.
A. The flagellar formation in the wild type and nirS strains (grown under anaerobic conditions and 20 mM arginine) is shown by fluorescence microscopy. Antibodies against DnaK and FliC were raised and employed for detection. Goat anti‐rabbit Alexa 488 (for FliC) or goat anti‐rabbit Alexa 568 (for DnaK) were used as fluorescently labelled secondary antibodies. DAPI dye was utilized for DNA staining.
B. The interacting domains between NirS and FliC elucidated by mass spectrometry by determination of the cross‐linked peptides are shown for NirS: AAEQYQGAASAVDPTHVVR (white), CAGCHGVLRK (blue) and GQQYLEALITYGTPLGMPNWGSSGELSK (orange). The haem d 1 (green) and haem c (red) are also highlighted in the NirS structure.
Conclusion and outlook
A dynamic supercomplex for anaerobic denitrification is formed at a membrane‐localized platform via transient and stable protein–protein interactions ensuring efficient electron transfer for ATP generation. Essential complex maturation and control proteins are intrinsic to the complex. Moreover, enzymes of this supercomplex also serve for unexpected structural purposes during flagellum assembly. It seems that we presently only have a glimpse of a novel protein–protein interaction world coordinating the structural assembly and function of protein complexes serving central cellular processes at the cytoplasmic membrane. The question of the mechanisms of complex disassembly and component breakdown or recycling during adaptation to new environmental conditions, as occurring during oxygen respiration, will be a research focus of the future. How are membrane‐associated transport and signal perception processes included in the observed protein complex dynamics? How is this membrane activity coordinated with cytoplasmic activities? Clearly, we have to think complex.
Conflict of Interest
None declared.
Acknowledgements
This work was supported by ERC grant IPBSL, awarded to K.N.T. and Ricardo Amils and the funding by the Deutsche Forschungsgemeinschaft (Forschergruppe PROTRAIN) granted to M.J. and D.J.
Microbial Biotechnology (2017) 10(6), 1523–1534
Funding information
This work was supported by ERC grant (ERC 250350‐IPBSL), awarded to K.N.T. and Ricardo Amils and the funding by the Deutsche Forschungsgemeinschaft
References
- Acin‐Perez, R. , and Enriquez, J.A. (2014) The function of the respiratory supercomplexes: the plasticity model. Biochim Biophys Acta 1837: 444–450. [DOI] [PubMed] [Google Scholar]
- Adamczack, J. , Hoffmann, M. , Papke, U. , Haufschildt, K. , Nicke, T. , Broring, M. , et al (2014) NirN protein from Pseudomonas aeruginosa is a novel electron‐bifurcating dehydrogenase catalyzing the last step of heme d 1 biosynthesis. J Biol Chem 289: 30753–30762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Shweiki, M.R. , Monchgesang, S. , Majovsky, P. , Thieme, D. , Trutschel, D. , and Hoehenwarter, W. (2017) Assessment of label‐free quantification in discovery proteomics and impact of technological factors and natural variability of protein abundance. J Proteome Res 16: 1410–1424. [DOI] [PubMed] [Google Scholar]
- Alcazar‐Fabra, M. , Navas, P. , and Brea‐Calvo, G. (2016) Coenzyme Q biosynthesis and its role in the respiratory chain structure. Biochim Biophys Acta 1857: 1073–1078. [DOI] [PubMed] [Google Scholar]
- Aloush, V. , Navon‐Venezia, S. , Seigman‐Igra, Y. , Cabili, S. , and Carmeli, Y. (2006) Multidrug‐resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob Agents Chemother 50: 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez‐Ortega, C. , and Harwood, C.S. (2007) Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol Microbiol 65: 153–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreae, C.A. , Titball, R.W. , and Butler, C.S. (2014) Influence of the molybdenum cofactor biosynthesis on anaerobic respiration, biofilm formation and motility in Burkholderia thailandensis . Res Microbiol 165: 41–49. [DOI] [PubMed] [Google Scholar]
- Arai, H. , Mizutani, M. , and Igarashi, Y. (2003) Transcriptional regulation of the nos genes for nitrous oxide reductase in Pseudomonas aeruginosa . Microbiology 149: 29–36. [DOI] [PubMed] [Google Scholar]
- Auerbach, A. , Kerem, E. , Assous, M.V. , Picard, E. , and Bar‐Meir, M. (2015) Is infection with hypermutable Pseudomonas aeruginosa clinically significant? J Cyst Fibros 14: 347–352. [DOI] [PubMed] [Google Scholar]
- Aussel, L. , Pierrel, F. , Loiseau, L. , Lombard, M. , Fontecave, M. , and Barras, F. (2014) Biosynthesis and physiology of coenzyme Q in bacteria. Biochim Biophys Acta 1837: 1004–1011. [DOI] [PubMed] [Google Scholar]
- Bali, S. , Warren, M.J. , and Ferguson, S.J. (2010) NirF is a periplasmic protein that binds d 1 heme as part of its essential role in d 1 heme biogenesis. FEBS J 277: 4944–4955. [DOI] [PubMed] [Google Scholar]
- Blasco, F. , Guigliarelli, B. , Magalon, A. , Asso, M. , Giordano, G. , and Rothery, R.A. (2001) The coordination and function of the redox centres of the membrane‐bound nitrate reductases. Cell Mol Life Sci 58: 179–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrero‐de Acuna, J.M. , Molinari, G. , Rohde, M. , Dammeyer, T. , Wissing, J. , Jansch, L. , et al (2015) A periplasmic complex of the nitrite reductase NirS, the chaperone DnaK and the flagellum protein FliC is essential for flagellum assembly and motility in Pseudomonas aeruginosa . J Bacteriol 197: 3066–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrero‐de Acuna, J.M. , Rohde, M. , Wissing, J. , Jansch, L. , Schobert, M. , Molinari, G. , et al (2016) Protein network of the Pseudomonas aeruginosa denitrification apparatus. J Bacteriol 198: 1401–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultema, J.B. , Braun, H.P. , Boekema, E.J. , and Kouril, R. (2009) Megacomplex organization of the oxidative phosphorylation system by structural analysis of respiratory supercomplexes from potato. Biochim Biophys Acta 1787: 60–67. [DOI] [PubMed] [Google Scholar]
- Burckstummer, T. , Bennett, K.L. , Preradovic, A. , Schutze, G. , Hantschel, O. , Superti‐Furga, G. , and Bauch, A. (2006) An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods 3: 1013–1019. [DOI] [PubMed] [Google Scholar]
- Cook, G.M. , Greening, C. , Hards, K. , and Berney, M. (2014) Energetics of pathogenic bacteria and opportunities for drug development. Adv Microb Physiol 65: 1–62. [DOI] [PubMed] [Google Scholar]
- Cutruzzola, F. , and Frankenberg‐Dinkel, N. (2016) Origin and impact of nitric oxide in Pseudomonas aeruginosa biofilms. J Bacteriol 198: 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutruzzola, F. , Brown, K. , Wilson, E.K. , Bellelli, A. , Arese, M. , Tegoni, M. , et al (2001) The nitrite reductase from Pseudomonas aeruginosa: essential role of two active‐site histidines in the catalytic and structural properties. Proc Natl Acad Sci USA 98: 2232–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuypers, H. , Viebrock‐Sambale, A. , and Zumft, W.G. (1992) NosR, a membrane‐bound regulatory component necessary for expression of nitrous oxide reductase in denitrifying Pseudomonas stutzeri . J Bacteriol 174: 5332–5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey, H.A. , Dailey, T.A. , Gerdes, S. , Jahn, D. , Jahn, M. , O'Brian, M.R. and Warren, M.J. (2017) Prokaryotic heme biosynthesis: multiple pathways to a common essential product. Microbiol Mol Biol Rev 81: e00048–16. https://doi.org/10.1128./MMBR.00048.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbey, R.E. , and Kuhn, A. (2012) Protein traffic in Gram‐negative bacteria–how exported and secreted proteins find their way. FEMS Microbiol Rev 36: 1023–1045. [DOI] [PubMed] [Google Scholar]
- Denks, K. , Vogt, A. , Sachelaru, I. , Petriman, N.A. , Kudva, R. , and Koch, H.G. (2014) The Sec translocon mediated protein transport in prokaryotes and eukaryotes. Mol Membr Biol 31: 58–84. [DOI] [PubMed] [Google Scholar]
- Driscoll, J.A. , Brody, S.L. , and Kollef, M.H. (2007) The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67: 351–368. [DOI] [PubMed] [Google Scholar]
- Dudkina, N.V. , Kudryashev, M. , Stahlberg, H. , and Boekema, E.J. (2011) Interaction of complexes I, III, and IV within the bovine respirasome by single particle cryoelectron tomography. Proc Natl Acad Sci USA 108: 15196–15200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elamin, A.A. , Stehr, M. , Spallek, R. , Rohde, M. , and Singh, M. (2011) The Mycobacterium tuberculosis Ag85A is a novel diacylglycerol acyltransferase involved in lipid body formation. Mol Microbiol 81: 1577–1592. [DOI] [PubMed] [Google Scholar]
- Enriquez, J.A. (2016) Supramolecular organization of respiratory complexes. Annu Rev Physiol 78: 533–561. [DOI] [PubMed] [Google Scholar]
- Eschbach, M. , Schreiber, K. , Trunk, K. , Buer, J. , Jahn, D. , and Schobert, M. (2004) Long‐term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J Bacteriol 186: 4596–4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez‐Barat, L. , Ciofu, O. , Kragh, K.N. , Pressler, T. , Johansen, U. , Motos, A. , et al (2017) Phenotypic shift in Pseudomonas aeruginosa populations from cystic fibrosis lungs after 2‐week antipseudomonal treatment. J Cyst Fibros 16: 222–229. [DOI] [PubMed] [Google Scholar]
- Galimand, M. , Gamper, M. , Zimmermann, A. , and Haas, D. (1991) Positive FNR‐like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa . J Bacteriol 173: 1598–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, R. , Gu, J. , Wu, M. , and Yang, M. (2016) Amazing structure of respirasome: unveiling the secrets of cell respiration. Protein Cell 7: 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa, N. , Arai, H. , and Igarashi, Y. (2001) Two c‐type cytochromes, NirM and NirC, encoded in the nir gene cluster of Pseudomonas aeruginosa act as electron donors for nitrite reductase. Biochem Biophys Res Commun 288: 1223–1230. [DOI] [PubMed] [Google Scholar]
- Hayashi, N.R. , Arai, H. , Kodama, T. , and Igarashi, Y. (1998) The nirQ gene, which is required for denitrification of Pseudomonas aeruginosa, can activate the RubisCO from Pseudomonas hydrogenothermophila . Biochim Biophys Acta 1381: 347–350. [DOI] [PubMed] [Google Scholar]
- Howden, A.J. , Geoghegan, V. , Katsch, K. , Efstathiou, G. , Bhushan, B. , Boutureira, O. , et al (2013) QuaNCAT: quantitating proteome dynamics in primary cells. Nat Methods 10: 343–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeno, S. , Aoki, D. , Hamada, M. , Hori, M. , and Tsuchiya, K.S. (2006) DNA sequencing and transcriptional analysis of the kasugamycin biosynthetic gene cluster from Streptomyces kasugaensis M338‐M1. J Antibiot (Tokyo) 59: 18–28. [DOI] [PubMed] [Google Scholar]
- Jacewicz, A. , Izumi, A. , Brunner, K. , Schnell, R. , and Schneider, G. (2013) Structural insights into the UbiD protein family from the crystal structure of PA0254 from Pseudomonas aeruginosa . PLoS ONE 8: e63161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn, M. and Jahn, D. (2012) Electron Transfer Reactions and Oxidative Phosphorylation. Biochemical Pathways: An Atlas of Biochemistry and Molecular Biology, 2nd edn New York, NY: Wiley. [Google Scholar]
- Kaake, R.M. , Wang, X. , and Huang, L. (2010) Profiling of protein interaction networks of protein complexes using affinity purification and quantitative mass spectrometry. Mol Cell Proteomics 9: 1650–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasaragod, V.B. , and Schindelin, H. (2016) Structural Framework for Metal Incorporation during Molybdenum Cofactor Biosynthesis. Structure 24: 782–788. [DOI] [PubMed] [Google Scholar]
- Krause, F. , Reifschneider, N.H. , Vocke, D. , Seelert, H. , Rexroth, S. , and Dencher, N.A. (2004) “Respirasome”‐like supercomplexes in green leaf mitochondria of spinach. J Biol Chem 279: 48369–48375. [DOI] [PubMed] [Google Scholar]
- Kudva, R. , Denks, K. , Kuhn, P. , Vogt, A. , Muller, M. , and Koch, H.G. (2013) Protein translocation across the inner membrane of Gram‐negative bacteria: the Sec and Tat dependent protein transport pathways. Res Microbiol 164: 505–534. [DOI] [PubMed] [Google Scholar]
- Lapuente‐Brun, E. , Moreno‐Loshuertos, R. , Acin‐Perez, R. , Latorre‐Pellicer, A. , Colas, C. , Balsa, E. , et al (2013) Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 340: 1567–1570. [DOI] [PubMed] [Google Scholar]
- Layer, G. , Reichelt, J. , Jahn, D. , and Heinz, D.W. (2010) Structure and function of enzymes in heme biosynthesis. Protein Sci 19: 1137–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalon, A. , and Alberge, F. (2016) Distribution and dynamics of OXPHOS complexes in the bacterial cytoplasmic membrane. Biochim Biophys Acta 1857: 198–213. [DOI] [PubMed] [Google Scholar]
- Magalon, A. , and Mendel, R.R. (2015) Biosynthesis and insertion of the molybdenum cofactor. EcoSal Plus 6 [In press] doi: 10.1128/ecosalplus.ESP‐0006‐2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalon, A. , Arias‐Cartin, R. , and Walburger, A. (2012) Supramolecular organization in prokaryotic respiratory systems. Adv Microb Physiol 61: 217–266. [DOI] [PubMed] [Google Scholar]
- Makowski, M.M. , Willems, E. , Jansen, P.W. , and Vermeulen, M. (2016) Cross‐linking immunoprecipitation‐MS (xIP‐MS): topological analysis of chromatin‐associated protein complexes using single affinity purification. Mol Cell Proteomics 15: 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marreiros, B.C. , Calisto, F. , Castro, P.J. , Duarte, A.M. , Sena, F.V. , Silva, A.F. , et al (2016) Exploring membrane respiratory chains. Biochim Biophys Acta 1857: 1039–1067. [DOI] [PubMed] [Google Scholar]
- Masoumi, A. , Heinemann, I.U. , Rohde, M. , Koch, M. , Jahn, M. , and Jahn, D. (2008) Complex formation between protoporphyrinogen IX oxidase and ferrochelatase during haem biosynthesis in Thermosynechococcus elongatus . Microbiology 154: 3707–3714. [DOI] [PubMed] [Google Scholar]
- McGuirl, M.A. , Bollinger, J.A. , Cosper, N. , Scott, R.A. , and Dooley, D.M. (2001) Expression, purification, and characterization of NosL, a novel Cu(I) protein of the nitrous oxide reductase (nos) gene cluster. J Biol Inorg Chem 6: 189–195. [DOI] [PubMed] [Google Scholar]
- Meylan, S. , Porter, C.B. , Yang, J.H. , Belenky, P. , Gutierrez, A. , Lobritz, M.A. , et al (2017) Carbon sources tune antibiotic susceptibility in Pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chem Biol 24: 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada, Y. , Nishijyo, T. , and Itoh, Y. (2002) Divergent structure and regulatory mechanism of proline catabolic systems: characterization of the putAP proline catabolic operon of Pseudomonas aeruginosa PAO1 and its regulation by PruR, an AraC/XylS family protein. J Bacteriol 184: 5633–5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicke, T. , Schnitzer, T. , Munch, K. , Adamczack, J. , Haufschildt, K. , Buchmeier, S. , et al (2013) Maturation of the cytochrome cd 1 nitrite reductase NirS from Pseudomonas aeruginosa requires transient interactions between the three proteins NirS, NirN and NirF. Biosci Rep 33: 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, A. , Canton, R. , Campo, P. , Baquero, F. , and Blazquez, J. (2000) High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288: 1251–1254. [DOI] [PubMed] [Google Scholar]
- Ong, S.E. , and Mann, M. (2005) Mass spectrometry‐based proteomics turns quantitative. Nat Chem Biol 1: 252–262. [DOI] [PubMed] [Google Scholar]
- Ornston, L.N. (1971) Regulation of catabolic pathways in Pseudomonas . Bacteriol Rev 35: 87–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessi, G. , and Haas, D. (2000) Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum‐sensing regulators LasR and RhlR in Pseudomonas aeruginosa . J Bacteriol 182: 6940–6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt, M.D. , Schurr, M.J. , Sauer, K. , Vazquez, G. , Kukavica‐Ibrulj, I. , Potvin, E. , et al (2008) Proteomic, microarray, and signature‐tagged mutagenesis analyses of anaerobic Pseudomonas aeruginosa at pH 6.5, likely representing chronic, late‐stage cystic fibrosis airway conditions. J Bacteriol 190: 2739–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porras, C.A. , and Bai, Y. (2015) Respiratory supercomplexes: plasticity and implications. Front Biosci (Landmark Ed) 20: 621–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunetti, L. , Infossi, P. , Brugna, M. , Ebel, C. , Giudici‐Orticoni, M.T. , and Guiral, M. (2010) New functional sulfide oxidase‐oxygen reductase supercomplex in the membrane of the hyperthermophilic bacterium Aquifex aeolicus . J Biol Chem 285: 41815–41826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldo, S. , Giardina, G. , Brunori, M. , and Cutruzzola, F. (2005) N‐oxide sensing in Pseudomonas aeruginosa: expression and preliminary characterization of DNR, an FNR‐CRP type transcriptional regulator. Biochem Soc Trans 33: 184–186. [DOI] [PubMed] [Google Scholar]
- Roche, B. , Aussel, L. , Ezraty, B. , Mandin, P. , Py, B. , and Barras, F. (2013) Reprint of: iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim Biophys Acta 1827: 923–937. [DOI] [PubMed] [Google Scholar]
- Rohde, M. , Puls, J. , Buhrdorf, R. , Fischer, W. , and Haas, R. (2003) A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol Microbiol 49: 219–234. [DOI] [PubMed] [Google Scholar]
- Santini, S. , Bizzarri, A.R. , Yamada, T. , Beattie, C.W. , and Cannistraro, S. (2014) Binding of azurin to cytochrome c 551 as investigated by surface plasmon resonance and fluorescence. J Mol Recognit 27: 124–130. [DOI] [PubMed] [Google Scholar]
- Schagger, H. , and Pfeiffer, K. (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J 19: 1777–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schobert, M. , and Jahn, D. (2010) Anaerobic physiology of Pseudomonas aeruginosa in the cystic fibrosis lung. Int J Med Microbiol 300: 549–556. [DOI] [PubMed] [Google Scholar]
- Schreiber, K. , Krieger, R. , Benkert, B. , Eschbach, M. , Arai, H. , Schobert, M. , and Jahn, D. (2007) The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J Bacteriol 189: 4310–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, V. , Noriega, C.E. , and Rowe, J.J. (2006) Involvement of NarK1 and NarK2 proteins in transport of nitrate and nitrite in the denitrifying bacterium Pseudomonas aeruginosa PAO1. Appl Environ Microbiol 72: 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W.S. , and Yoon, S.I. (2014) Crystal structure of FliC flagellin from Pseudomonas aeruginosa and its implication in TLR5 binding and formation of the flagellar filament. Biochem Biophys Res Commun 444: 109–115. [DOI] [PubMed] [Google Scholar]
- Stroh, A. , Anderka, O. , Pfeiffer, K. , Yagi, T. , Finel, M. , Ludwig, B. , and Schagger, H. (2004) Assembly of respiratory complexes I, III, and IV into NADH oxidase supercomplex stabilizes complex I in Paracoccus denitrificans . J Biol Chem 279: 5000–5007. [DOI] [PubMed] [Google Scholar]
- Sun, W. , Arese, M. , Brunori, M. , Nurizzo, D. , Brown, K. , Cambillau, C. , et al (2002) Cyanide binding to cd 1 nitrite reductase from Pseudomonas aeruginosa: role of the active‐site His369 in ligand stabilization. Biochem Biophys Res Commun 291: 1–7. [DOI] [PubMed] [Google Scholar]
- Sutherland, B.W. , Toews, J. , and Kast, J. (2008) Utility of formaldehyde cross‐linking and mass spectrometry in the study of protein‐protein interactions. J Mass Spectrom 43: 699–715. [DOI] [PubMed] [Google Scholar]
- Talwalkar, J.S. , and Murray, T.S. (2016) The approach to Pseudomonas aeruginosa in cystic fibrosis. Clin Chest Med 37: 69–81. [DOI] [PubMed] [Google Scholar]
- Taubner, L.M. , McGuirl, M.A. , Dooley, D.M. , and Copie, V. (2004) 1H, 13C, 15N backbone and sidechain resonance assignments of apo‐NosL, a novel copper(I) binding protein from the nitrous oxide reductase gene cluster of Achromobacter cycloclastes . J Biomol NMR 29: 211–212. [DOI] [PubMed] [Google Scholar]
- Trunk, K. , Benkert, B. , Quack, N. , Munch, R. , Scheer, M. , Garbe, J. , et al (2010) Anaerobic adaptation in Pseudomonas aeruginosa: definition of the Anr and Dnr regulons. Environ Microbiol 12: 1719–1733. [DOI] [PubMed] [Google Scholar]
- Vaccaro, B.J. , Thorgersen, M.P. , Lancaster, W.A. , Price, M.N. , Wetmore, K.M. , Poole, F.L. 2nd , et al (2015) Determining roles of accessory genes in denitrification by mutant fitness analyses. Appl Environ Microbiol 82: 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Alst, N.E. , Picardo, K.F. , Iglewski, B.H. , and Haidaris, C.G. (2007) Nitrate sensing and metabolism modulate motility, biofilm formation, and virulence in Pseudomonas aeruginosa . Infect Immun 75: 3780–3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Wauven, C. , Pierard, A. , Kley‐Raymann, M. , and Haas, D. (1984) Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four‐gene cluster encoding the arginine deiminase pathway. J Bacteriol 160: 928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, H.D. , Zlosnik, J.E. , and Ryall, B. (2007) Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa . Adv Microb Physiol 52: 1–71. [DOI] [PubMed] [Google Scholar]
- Wunsch, P. , and Zumft, W.G. (2005) Functional domains of NosR, a novel transmembrane iron‐sulfur flavoprotein necessary for nitrous oxide respiration. J Bacteriol 187: 1992–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunsch, P. , Herb, M. , Wieland, H. , Schiek, U.M. , and Zumft, W.G. (2003) Requirements for CuA and Cu‐S center assembly of nitrous oxide reductase deduced from complete periplasmic enzyme maturation in the nondenitrifier Pseudomonas putida . J Bacteriol 185: 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Trncik, C. , Andrade, S.L. , and Einsle, O. (2017) The flavinyl transferase ApbE of Pseudomonas stutzeri matures the NosR protein required for nitrous oxide reduction. Biochim Biophys Acta 1858: 95–102. [DOI] [PubMed] [Google Scholar]
- Zumft, W.G. (1997) Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61: 533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]


