Summary
Metaldehyde is a common molluscicide, used to control slugs in agriculture and horticulture. It is resistant to breakdown by current water treatment processes, and its accumulation in drinking water sources leads to regular regulatory failures in drinking water quality. To address this problem, we isolated metaldehyde‐degrading microbes from domestic soils. Two distinct bacterial isolates were cultured, that were able to grow prototrophically using metaldehyde as sole carbon and energy source. One isolate belonged to the genus Acinetobacter (strain designation E1) and the other isolate belonged to the genus Variovorax (strain designation E3). Acinetobacter E1 was able to degrade metaldehyde to a residual concentration < 1 nM, whereas closely related Acinetobacter strains were completely unable to degrade metaldehyde. Variovorax E3 grew and degraded metaldehyde more slowly than Acinetobacter E1, and residual metaldehyde remained at the end of growth of the Variovorax E3 strain. Biological degradation of metaldehyde using these bacterial strains or approaches that allow in situ amplification of metaldehyde‐degrading bacteria may represent a way forward for dealing with metaldehyde contamination in soils and water.
Introduction
Metaldehyde (CH₃CHO)₄ is an ether, formed from a cyclic tetramerization of acetaldehyde (Fig. 1A) (Kekulé and Zincke, 1872). Metaldehyde was initially used as a solid fuel firelighter ‘Meta‐fuel’ (Miller, 1928), but its major contemporary use is as a molluscicide in agriculture and horticulture. Its application in controlling slugs was known as early as 1934 (Gimingham, 1940), and it is now widely used in both agricultural fields and domestic gardens. It is applied as a pelleted bran bait that inhibits slug feeding after exposure (Wedgwood and Bailey, 1988), causing effects such as the distention and disintegration of the Golgi apparatus and endoplasmic reticulum in the mucus cells of slugs (Triebskorn et al., 1998).
Figure 1.
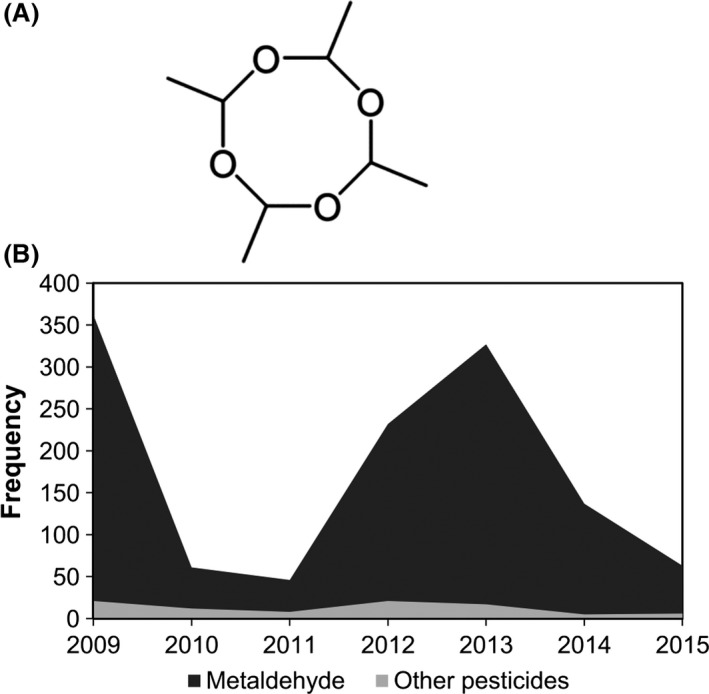
A. Skeletal structure of metaldehyde.
B. Frequency of water quality failures per year in the UK due to metaldehyde or all other pesticides. Compiled from the Drinking Water Inspectorate annual regional reports, available from http://www.dwi.gov.uk/about/annual-report.
In 2014, Metaldehyde accounted for 87% of all recorded molluscicide applications on agricultural fields in the UK (Garthwaite et al., 2015). 112 tonnes were applied over 920 thousand hectares (21% of surveyed arable land used to grow crops) in Britain in 2014; primarily on wheat, oilseed rape and potato crops (Garthwaite et al., 2015). The vast majority of failures in drinking water quality in the UK, due to pesticide contamination, are caused by metaldehyde exceeding the regulatory limit of 0.1 μg l−1 (≡ 0.6 nM) (European Union Council Directive 98/83/EC) (Fig. 1B).
The recalcitrance of metaldehyde to degradation at ambient temperature (Fleischmann et al., 2000) is problematic for water treatment, as metaldehyde is not removed by conventional water treatment processes (Kay and Grayson, 2014). Researchers are pursuing a variety of chemical and physical approaches to deal with the problem of metaldehyde contamination (Autin et al., 2013; Doria et al., 2013; Tao and Fletcher, 2013, 2014). But currently, no economical method exists to degrade or remove metaldehyde from water.
It has been shown that the xenobiotic metaldehyde can be quickly degraded in soils (Zhang et al., 2011) and is oxidized to carbon dioxide under aerobic conditions in unsterilized soils (EFSA, 2010). This strongly suggests the involvement of microbes in its degradation, although no microorganisms have been isolated to date that degrade metaldehyde. The degradation of metaldehyde to CO2 is strongly exothermic [heat of combustion 3370 kJ mol−1 (Fleischmann et al., 2000)], suggesting that it has the potential to be a carbon and energy source to support microbial growth. Soils are home to a vast array of microbes and represent a source of metabolic activities that may be of use in industrial and medicinal applications (Delmont et al., 2011). Here, we enriched microbes from soils and report the first isolation and identification of microbial isolates capable of using metaldehyde as a sole source of energy and carbon for growth.
Results and Discussion
Two distinct metaldehyde‐degrading strains were isolated from domestic soils
Metaldehyde‐degrading bacteria were selected in a mineral medium consisting of salts Na2HPO4 (55 mM), KH2PO4 (11 mM), NH4Cl (6 mM) and MgSO4 (0.4 mM) (pH 7). This was supplemented with 2 ml l−1 of a trace elements solution (Vishniac and Santer, 1957). Metaldehyde was provided as sole carbon source and control cultures lacked metaldehyde. Ability to grow using metaldehyde was tested in both liquid enrichment cultures and on solid media, containing 1.5% agarose. 100 ml liquid cultures were inoculated with 1 g of soil obtained from domestic gardens in York, UK. Cultures were incubated at 30°C for 3 days, 1 ml of enrichment media was subcultured into fresh media and incubated for a further 3 days and subsequently samples were spread onto agarose plates containing 2800 μM (500 mg l−1) metaldehyde. Fifty to 200 colonies were obtained on plates when the enrichments were carried out in liquid culture in the presence of 570 μM (100 mg l−1) metaldehyde, but not following control enrichments in the absence of metaldehyde. 1 g samples of the same domestic soils were re‐suspended in 10 ml of sterile water and 100 μl aliquots spread directly onto agarose plates containing metaldehyde. Two to five colonies grew on these plates. The morphology of all the colonies was white, round and glossy. Ten isolates were picked for further analysis and named E1‐E6 and M1‐M4, to designate the source soils used. Soil E had a recent history of metaldehyde utilization, whereas soil M had not been treated with metaldehyde for at least 5 years. In each case, the isolated strains grew on agarose plates supplemented with metaldehyde, but not in its absence, suggesting they were utilizing metaldehyde as a carbon and energy source.
On subculturing the metaldehyde‐degrading strains, each strain appeared to be a pure culture, except strain E4 which yielded two distinct colony morphologies, and was subsequently subdivided into E4a and E4b. Colonies from strains E1, E3, E4a, E4b, E5, M1 and M4 were used for amplification of 16S rDNA as described previously with primers U8F and U1492R (Eden et al., 1991). Amplification was achieved using GoTaq polymerase (Promega) with a standard programme of: 98°C for 30 s; 35 cycles of 98°C for 10 s, 50°C for 30 s, 72°C for 60 s; 72°C for 10 min. PCR products were purified using QIAquick PCR purification kit (Qiagen, Manchester, UK) following the manufacturer's instructions. For restriction fragment length polymorphism (RFLP) analysis, 1 μg of purified DNA was digested for 1 or 3 h at 37°C using restriction enzyme HhaI. RFLP revealed two distinctly different ribotypes (see Supporting Information). Two examples of each ribotype were sequenced. Sanger sequencing was used to obtain the nucleotide sequences of the U8F‐U1492R amplicons of E1, M1, E3 and E4a using U8F as sequencing primer. Sequences from E1 and M1 were aligned using ClustalX V2.1 and found to be identical across the > 900 base region where the base sequence could be confidently assigned. Similarly, the sequences from E3 and E4a were found to be identical across a > 900 base region.
Subsequent investigation focused on the strains E1 and E3. The sequences of E1 and E3 (see Supporting Information) type strains of A. pittii, A. oleivorans and A. seifertii also had 99% identity to E1. The E3 sequence has 99% identity to type strains of Variovorax boronicumulans, V. paradoxus, V. guangxiensis, V. ginsengisoli. Based on these analyses, the isolates have been assigned genera and designated Acinetobacter E1 and Variovorax E3.
The disappearance of metaldehyde from minimal media is proportional to the growth of Acinetobacter E1 and Variovorax E3 in pure cultures
Triplicate cultures of Acinetobacter E1 and Variovorax E3 were grown in minimal media with 850 μM (150 mg l−1) metaldehyde, incubated at 30°C with shaking at 200 rpm. An additional three flasks of media were not inoculated. Periodic samples were taken from each culture, and an uninoculated media flask and OD600 measurements were made. Contemporaneously, cellular material was removed from samples by centrifugation at 5000 × g, the supernatant aspirated and stored at −20°C for later analysis of metaldehyde content. Growth curves are shown in Fig. 2A. During the exponential growth phase, Acinetobacter E1 had a doubling time of 8.5 h, and Variovorax E3 had a doubling time of c. 22 h. There was no increase in optical density in the uninoculated control culture. Metaldehyde concentration of culture media samples was quantified by liquid chromatography‐mass spectrometry (for method, see Supporting Information). Metaldehyde disappeared over a similar timescale to the growth of the E1 and E3 isolates (Fig. 2B). The disappearance of metaldehyde from the cultures was correlated with the growth of the isolates (Fig. 2C and D). As the sole carbon and energy source present in the culture medium, it can be concluded that the strains were catabolizing metaldehyde for growth. Variovorax E3 catabolizes metaldehyde more slowly and has a longer lag time, lower maximum optical density, longer doubling time and higher final concentration of residual metaldehyde compared to Acinetobacter E1.
Figure 2.
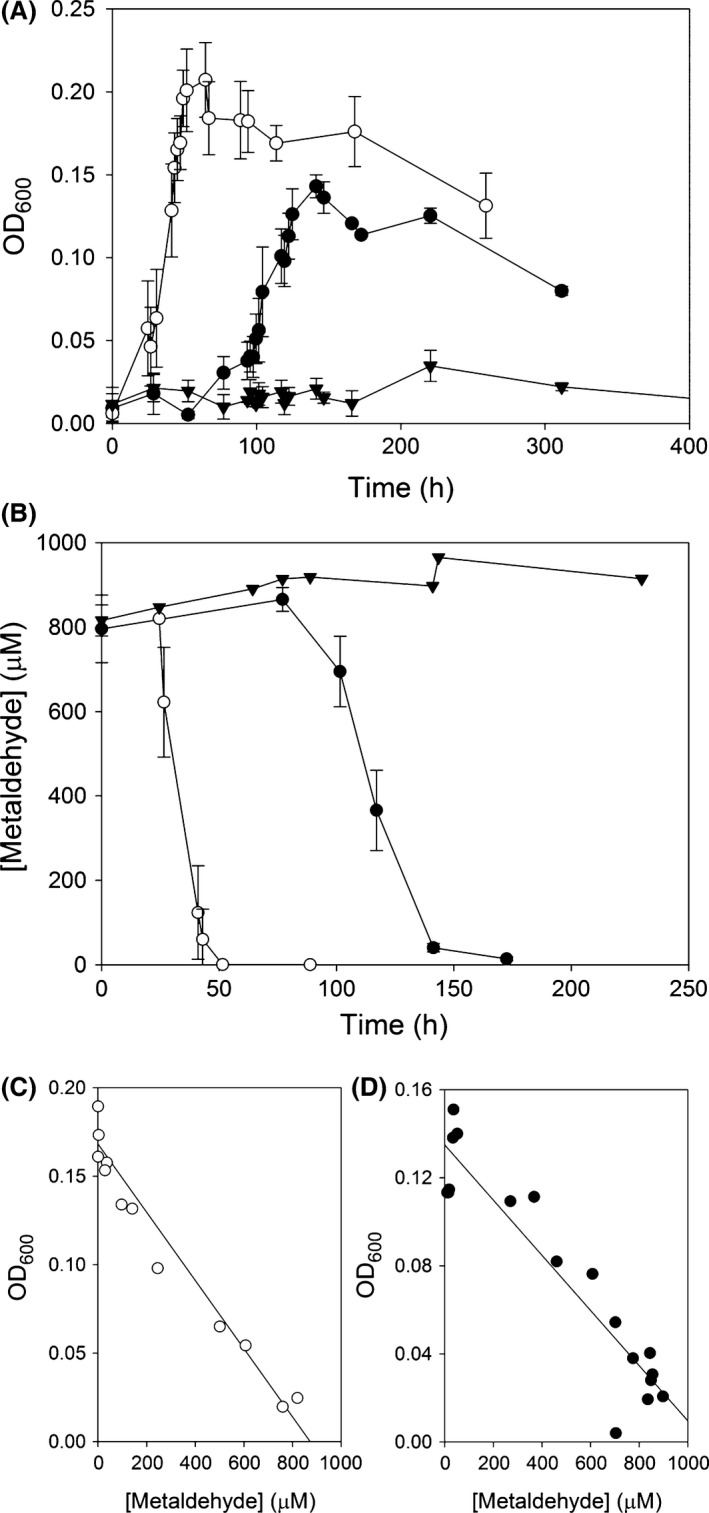
Growth and metaldehyde utilization by Acinetobacter E1 and Variovorax E3.
A. Mean OD 600 (measured using a Jenway 6300 spectrophotometer) in liquid culture with 850 μM metaldehyde as sole carbon and energy source, inoculated with single colonies of Acinetobacter E1 (open circles) and Variovorax E3 (filled circles), or not inoculated (filled triangles). Error bars give SD of triplicate independent cultures.
B. Mean [metaldehyde] in culture media during growth of Acinetobacter E1 (open circles) and Variovorax E3 (filled circles), or not inoculated (filled triangles). Error bars give SD of triplicate independent cultures. Correlation between culture optical density and residual metaldehyde concentration during growth of (C) Acinetobacter E1 (R 2 = 0.94) and (D) Variovorax E3 (R 2 = 0.88) in media containing metaldehyde as the sole energy and carbon source.
Utilization of metaldehyde by Acinetobacter E1 is a property not shared by other Acinetobacter
The remainder of the work focused on Acinetobacter E1 which has faster growth kinetics, and a more rapid and complete utilization of metaldehyde, compared to Variovorax E3. Acinetobacter E1 was unable to grow using glucose, fructose, arabinose or glycerol as alternative carbon substrates.
It was desirable to identify other strains related to Acinetobacter E1 for comparative purposes. A. calcoaceticus RUH 2202 (Nemec et al., 2011) was purchased from the Belgian Coordinated Collection of Microorganisms, A. calcoaceticus ANC3678 (Nemec et al., 2011), A. calcoaceticus NIPH1 (Nemec et al., 1999), A. pittii ANC3678 (Nemec et al., 2011) A. pittii 70.29 (Seifert et al., 1994) and A. baylyi DSM14961 (Carr et al., 2003) from the CIP culture collection (Pasteur Institute, Paris). The ability of these Acinetobacter to use metaldehyde was assessed by streaking colonies from an LB plate onto a MSM + metaldehyde plate and inoculating into liquid media containing 850 μM metaldehyde. There were no signs of growth in either media after 4 days’ incubation at 30°C. Acinetobacter E1, unlike strain RUH 2202, was able to grow on phenol, whereas A. calcoaceticus RUH 2202 grew on 1% ethanol as a carbon source, but strain E1 could not grow with ethanol. Both Acinetobacter strains E1 and RUH 2202 grew on acetate as a carbon source, which allowed for comparative analysis of metaldehyde utilization under the same growth conditions. Following growth on acetate as sole carbon source, Acinetobacter E1 utilized 40 μM metaldehyde over a 30 min period, whereas there was no loss of metaldehyde in cultures of A. calcoaceticus RUH 2202 (Fig. 3A).
Figure 3.
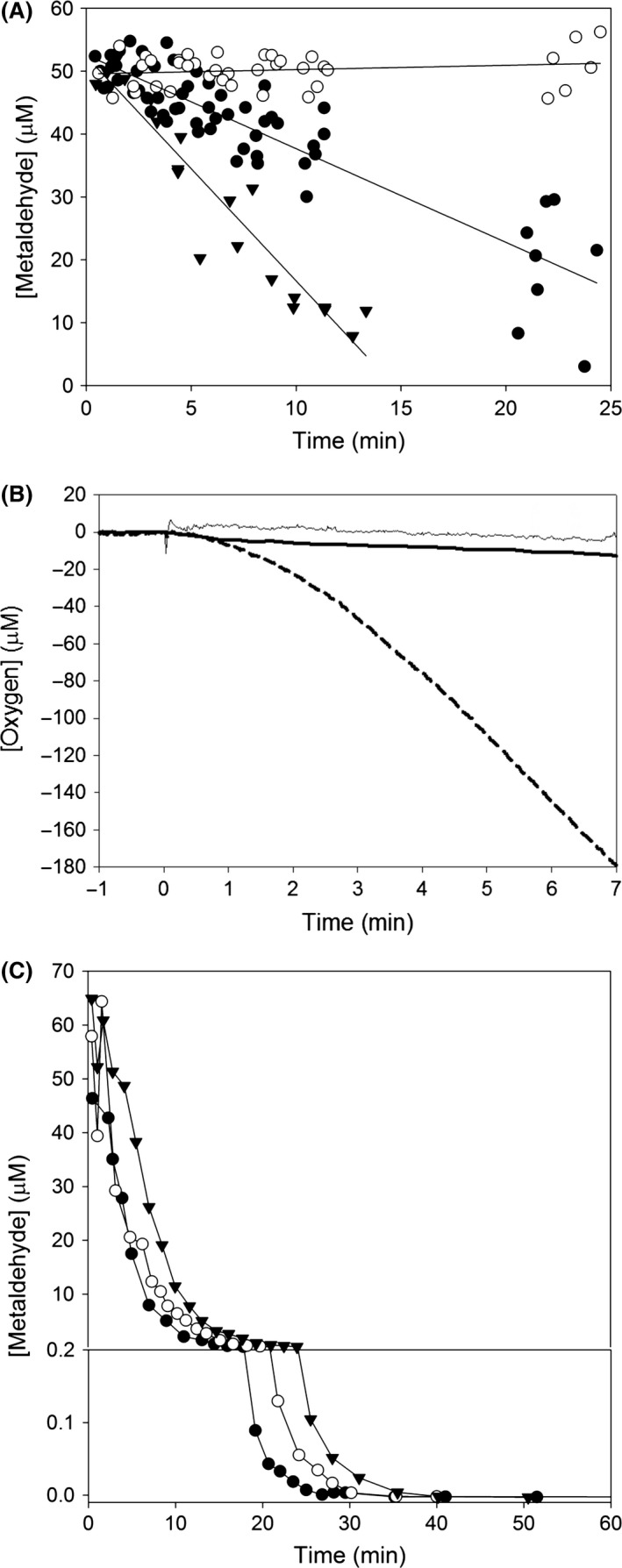
Metaldehyde utilization and metaldehyde‐dependent oxygen utilization.
A. Metaldehyde utilization in samples of washed Acinetobacter cells resuspended to an OD 600 = 1.0 treated with 53 μM metaldehyde following culture of Acinetobacter E1 in acetate (filled circles; rate of metaldehyde utilization = 1.5 ± 0.1 μM min−1) or in metaldehyde (filled triangles; rate of metaldehyde utilization = 3.8 ± 0.3 μM min−1), or strain RUH 2202 grown with acetate (open circles; rate of metaldehyde utilization = −0.1 ± 0.1 μM min−1) as sole carbon source.
B. Metaldehyde‐dependent oxygen utilization in samples of washed Acinetobacter cells resuspended to an OD 600 = 1.0 treated with 53 μM metaldehyde added at time zero. A. calcoaceticus RUH2202 (cultured in acetate) (solid thin line; rate of O2 utilization = 1.6 ± 0.4 μM min−1), Acinetobacter E1 cultured in acetate (solid thick line; rate of O2 utilization = 2.7 ± 1.1 μM min−1) or in metaldehyde (dashed line; rate of O2 utilization = 24.5 ± 3.8 μM min−1). Data are representative of at least three replicates.
C. Three time‐courses of metaldehyde degradation following culture of Acinetobacter E1 with metaldehyde as sole carbon source. Metaldehyde axis is split to show rate of disappearance between 0–0.2 μM, and 0.2–50 μM metaldehyde.
Acinetobacter E1 degrades metaldehyde to completion, and this degradation is followed by oxygen consumption
Following growth on metaldehyde, Acinetobacter E1 utilized 40 μM metaldehyde over a 12 min period (Fig. 3A). This suggests a c. twofold increase in activity of the metaldehyde‐degrading enzyme following culturing with metaldehyde. Furthermore, suspensions of Acinetobacter E1 utilize oxygen in a metaldehyde‐dependent manner after growth on metaldehyde, but not after growth on acetate (Fig. 3B). This oxygen consumption is delayed compared to metaldehyde disappearance, indicating that the metaldehyde catabolism involves metaldehyde degradation, followed by an oxygen‐dependent metabolic step. The apparent KM of cell suspensions of Acinetobacter E1 for metaldehyde was c. 50 μM, and it is noted that metaldehyde was degraded to below the limit of detection in these experiments (< 1 nM metaldehyde) in 30 min (Fig. 3C), which suggests that this or similar strains may have value in future bioremediation strategies.
Metaldehyde is a xenobiotic (i.e. only in existence due to human activity via chemical synthesis) that has been in widespread use for about 100 years. The metaldehyde‐degrading strains Acinetobacter E1 and Variovorax E3 share evolutionary heritage with other bacteria with versatile metabolism (Fewson, 1967; Willems et al., 1991) and a demonstrated ability to degrade xenobiotics (Mirgain et al., 1993; Greene et al., 2000; Sorensen et al., 2005; Wang and Gu, 2006; Bruland et al., 2009; Carbajal‐Rodriguez et al., 2011; Zhang et al., 2012; Rajoo et al., 2013; Murdoch and Hay, 2015) and other potentially recalcitrant chemicals (Reisfeld et al., 1972; Abbott et al., 1973; Koh et al., 1985; Hwang and Draughon, 1994; Singh and Lin, 2008; Zhao et al., 2009). The metabolic versatility of Acinetobacter and Variovorax isolates varies between isolates, presumably due to horizontal acquisition of genetic traits, selected in particular environments. Future work will focus on identifying the mechanistic basis for metaldehyde degradation.
To conclude, here we have demonstrated the first isolation of bacteria capable of degrading the commonly used molluscicide metaldehyde. Metaldehyde is a stable polymer of acetaldehyde which consists of a ring structure in which the bonds are aliphatic C‐C single bonds and C‐O ethers. Biological degradation of metaldehyde via the metabolic processes in bacteria such as Acinetobacter E1 and Variovorax E3 may prove valuable in dealing with metaldehyde contamination in natural environments and drinking water sources.
Conflict of interest
None declared.
Supporting information
Fig. S1. RFLP analysis of metaldehyde‐degrading bacterial isolates.
Acknowledgements
JCT was supported by a Biotechnology and Biological Sciences Research Council (BBSRC) studentship.
Microbial Biotechnology (2017) 10(6), 1824–1829
References
- Abbott, B.J. , Laskin, A.I. , and McCoy, C.J. (1973) Growth of Acinetobacter calcoaceticus on ethanol. Appl Microbiol 25: 787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autin, O. , Hart, J. , Jarvis, P. , MacAdam, J. , Parsons, S.A. , and Jefferson, B. (2013) The impact of background organic matter and alkalinity on the degradation of the pesticide metaldehyde by two advanced oxidation processes: UV/H(2)O(2) and UV/TiO(2). Water Res 47: 2041–2049. [DOI] [PubMed] [Google Scholar]
- Bruland, N. , Wubbeler, J.H. , and Steinbuchel, A. (2009) 3‐mercaptopropionate dioxygenase, a cysteine dioxygenase homologue, catalyzes the initial step of 3‐mercaptopropionate catabolism in the 3,3‐thiodipropionic acid‐degrading bacterium Variovorax paradoxus . J Biol Chem 284: 660–672. [DOI] [PubMed] [Google Scholar]
- Carbajal‐Rodriguez, I. , Stoveken, N. , Satola, B. , Wubbeler, J.H. , and Steinbuchel, A. (2011) Aerobic degradation of mercaptosuccinate by the Gram negative bacterium Variovorax paradoxus Strain B4. J Bacteriol 193: 527–539.21075928 [Google Scholar]
- Carr, E.L. , Kampfer, P. , Patel, B.K. , Gurtler, V. , and Seviour, R.J. (2003) Seven novel species of Acinetobacter isolated from activated sludge. Int J Syst Evol Microbiol 53: 953–963. [DOI] [PubMed] [Google Scholar]
- Delmont, T.O. , Malandain, C. , Prestat, E. , Larose, C. , Monier, J.M. , Simonet, P. , and Vogel, T.M. (2011) Metagenomic mining for microbiologists. ISME J 5: 1837–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria, F.C. , Borges, A. , Kim, J. , Nathan, A. , Joo, J. , and Campos, L. (2013) Removal of metaldehyde through photocatalytic reactions using nano‐sized zinc oxide composites. Water Air Soil Pollut 224: 1–9. [Google Scholar]
- Eden, P.A. , Schmidt, T.M. , Blakemore, R.P. , and Pace, N.R. (1991) Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction‐amplified 16S rRNA‐specific DNA. Int J Syst Bacteriol 41: 324–325. [DOI] [PubMed] [Google Scholar]
- EFSA (2010) Conclusion on the peer review of the pesticide risk assessment of the active substance metaldehyde. EFSA Journal. DOI: 10.2903/j.efsa.2010.1856. [Google Scholar]
- Fewson, C.A. (1967) The identity of Gram negative bacterium NCIB 8250 (Vibrio 01). J Gen Microbiol 48: 107–110. [DOI] [PubMed] [Google Scholar]
- Fleischmann, G. , Jira, R. , Bolt, H.M. and Golka, K. (2000) Acetaldehyde In Ullmann's Encyclopedia of Industrial Chemistry. Hoboken, New Jersey, USA: Wiley‐VCH Verlag GmbH & Co. KGaA. [Google Scholar]
- Garthwaite, D. , Barker, I. , Laybourn, R. , Huntly, A. , Parrish, G.P. , Hudson, S. and Thygesesn, H. (2015). In Pesticide Usage Survey Report 263 ‐Arable crops in the UK. Department for Environment. London: Defra. [Google Scholar]
- Gimingham, C. (1940) Some recent contributions by English workers to the development of methods of insect control. Ann Appl Biol 27: 161–175. [Google Scholar]
- Greene, E.A. , Beatty, P.H. , and Fedorak, P.M. (2000) Sulfolane degradation by mixed cultures and a bacterial isolate identified as a Variovorax sp. Arch Microbiol 174: 111–119. [DOI] [PubMed] [Google Scholar]
- Hwang, C.A. , and Draughon, F.A. (1994) Degradation of Ochratoxin A by Acinetobacter calcoaceticus . J Food Protect 57: 410–414. [DOI] [PubMed] [Google Scholar]
- Kay, P. , and Grayson, R. (2014) Using water industry data to assess the metaldehyde pollution problem. Water Environ J 28: 410–417. [Google Scholar]
- Kekulé, A. , and Zincke, T. (1872) Ueber das sogenannte chloraceten und die polymeren modificationen des aldehyds. Justus Liebigs Annalen der Chemie 162: 125–150. [Google Scholar]
- Koh, J.S. , Yamakawa, T. , Kodama, T. , and Minoda, Y. (1985) Rapid and dense culture of Acinetobacter calcoaceticus on palm oil. Agr Biol Chem Tokyo 49: 1411–1416. [Google Scholar]
- Miller, R. (1928) Poisoning by “Meta Fuel” tablets (metacetaldehyde). Arch Dis Child 3: 292–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirgain, I. , Green, G.A. , and Monteil, H. (1993) Degradation of atrazine in laboratory microcosms: isolation and identification of the biodegrading bacteria. Environ Toxicol Chem 12: 1627–1634. [Google Scholar]
- Murdoch, R.W. , and Hay, A.G. (2015) The biotransformation of ibuprofen to trihydroxyibuprofen in activated sludge and by Variovorax Ibu‐1. Biodegradation 26: 105–113. [DOI] [PubMed] [Google Scholar]
- Nemec, A. , Janda, L. , Melter, O. , and Dijkshoorn, L. (1999) Genotypic and phenotypic similarity of multiresistant Acinetobacter baumannii isolates in the Czech Republic. J Med Microbiol 48: 287–296. [DOI] [PubMed] [Google Scholar]
- Nemec, A. , Krizova, L. , Maixnerova, M. , van der Reijden, T.J. , Deschaght, P. , Passet, V. , et al (2011) Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus‐Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res Microbiol 162: 393–404. [DOI] [PubMed] [Google Scholar]
- Rajoo, S. , Ahn, J.O. , Lee, H.W. , and Jung, J.K. (2013) Isolation and characterization of a novel epsilon‐caprolactam‐degrading microbe. Acinetobacter calcoaceticus, from industrial wastewater by chemostat enrichment. Biotechnol Lett 35: 2069–2072. [DOI] [PubMed] [Google Scholar]
- Reisfeld, A. , Rosenber, E. and Gutnick, D. (1972) Microbial degradation of crude oil: factors affecting dispersion in sea water by mixed and pure cultures. Appl Microbiol 24, 363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert, H. , Schulze, A. , Baginski, R. , and Pulverer, G. (1994) Comparison of four different methods for epidemiologic typing of Acinetobacter baumannii . J Clin Microbiol 32: 1816–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, C. , and Lin, J. (2008) Isolation and characterization of diesel oil degrading indigenous microrganisms in Kwazulu‐Natal, South Africa. Afr J Biotechnol 7: 1927–1932. [Google Scholar]
- Sorensen, S.R. , Rasmussen, J. , Jacobsen, C.S. , Jacobsen, O.S. , Juhler, R.K. , and Aamand, J. (2005) Elucidating the key member of a linuron‐mineralizing bacterial community by PCR and reverse transcription‐PCR denaturing gradient gel electrophoresis 16S rRNA gene fingerprinting and cultivation. Appl Environ Microb 71: 4144–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, B. , and Fletcher, A.J. (2013) Metaldehyde removal from aqueous solution by adsorption and ion exchange mechanisms onto activated carbon and polymeric sorbents. J Hazard Mater 244–245: 240–250. [DOI] [PubMed] [Google Scholar]
- Tao, B. , and Fletcher, A.J. (2014) Catalytic degradation and adsorption of metaldehyde from drinking water by functionalized mesoporous silicas and ion‐exchange resin. Sep Purif Technol 124: 195–200. [Google Scholar]
- Triebskorn, R. , Christensen, K. , and Heim, G. (1998) Effects of orally and dermally applied metaldehyde on mucus cells of slugs (Deroceras reticulatum) depending on temperature and duration of exposure. J Mollus Stud 64: 467–487. [Google Scholar]
- Vishniac, W. , and Santer, M. (1957) The thiobacilli. Bacteriol Rev 21: 195–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.P. , and Gu, J.D. (2006) Degradability of dimethyl terephthalate by Variovorax paradoxus T4 and Sphingomonas yanoikuyae DOS01 isolated from deep‐ocean sediments. Ecotoxicol 15: 549–557. [DOI] [PubMed] [Google Scholar]
- Wedgwood, M.A. , and Bailey, S.E. (1988) The inhibitory effects of the molluscicide metaldehyde on feeding, locomotion and faecal elimination of three pest species of terrestrial slug. Ann Appl Biol 112: 439–457. [Google Scholar]
- Willems, A. , De Ley, J. , Gillis, M. , and Kersters, K. (1991) Comamonadaceae, a New Family Encompassing the Acidovorans ribosomal RNA complex, including Variovorax paradoxus gen. nov., comb. nov., for Alcaligenes paradoxus (Davis 1969). Int J Syst Bacteriol 41: 445–450. [Google Scholar]
- Zhang, H.‐Y. , Wang, C. , Lu, H.‐Z. , Guan, W.‐B. , and Ma, Y.‐Q. (2011) Residues and dissipation dynamics of molluscicide metaldehyde in cabbage and soil. Ecotox Environ Safe 74: 1653–1658. [DOI] [PubMed] [Google Scholar]
- Zhang, H.J. , Zhou, Q.W. , Zhou, G.C. , Cao, Y.M. , Dai, Y.J. , Ji, W.W. , et al (2012) Biotransformation of the neonicotinoid insecticide Thiacloprid by the bacterium Variovorax boronicumulans Strain J1 and mediation of the major metabolic pathway by nitrile hydratase. J Agr Food Chem 60: 153–159. [DOI] [PubMed] [Google Scholar]
- Zhao, X.H. , He, X. , Wang, J.N. , Song, Y.M. , Geng, G.X. , and Wang, J.H. (2009) Biodegradation of Swainsonine by Acinetobacter calcoaceticus strain YLZZ‐1 and its isolation and identification. Biodegradation 20: 331–338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. RFLP analysis of metaldehyde‐degrading bacterial isolates.


