Summary
In this study, we show the proof of concept for the production of defined oligo‐isoprenoids with terminal functional groups that can be used as starting materials for various purposes including the synthesis of isoprenoid‐based plastics. To this end, we used three types of rubber oxygenases for the enzymatic cleavage of rubber [poly(cis‐1,4‐isoprene)]. Two enzymes, rubber oxygenase RoxAX sp and rubber oxygenase RoxBX sp, originate from Xanthomonas sp. 35Y; the third rubber oxygenase, latex‐clearing protein (LcpK30), is derived from Gram‐positive rubber degraders such as Streptomyces sp. K30. Emulsions of polyisoprene (latex) were treated with RoxAX sp, RoxBX sp, LcpK30 or with combinations of the three proteins. The cleavage products were purified by solvent extraction and FPLC separation. All products had the same general structure with terminal functions (CHO‐CH 2‐ and ‐CH 2‐COCH 3) but differed in the number of intact isoprene units in between. The composition and m/z values of oligo‐isoprenoid products were determined by HPLC‐MS analysis. Our results provide a method for the preparation of reactive oligo‐isoprenoids that can likely be used to convert polyisoprene latex or rubber waste materials into value‐added molecules, biofuels, polyurethanes or other polymers.
Introduction
Natural rubber has been produced in huge amounts for more than a century by cultivating the rubber tree (Hevea brasiliensis), and the material is used for a variety of applications, as an example for the production of rubbers, tyres, sealings, latex gloves and many other items. The main component of rubber is the hydrocarbon poly(cis‐1,4‐isoprene). For most of today's applications of rubber, an important material property is the molecular weight of the polymer that – when high – gives rise to superior material properties that are necessary for example for the production of tyres. However, no attention has been given so far to the use of rubber for the biotechnological preparation of low molecular fine chemicals (Förster‐Fromme and Jendrossek, 2010; Kamm, 2014; Akhlaghi et al., 2015; Schrader and Bohlmann, 2015). In this contribution, we describe the proof of concept for the use of rubber oxygenases to cleave polyisoprene‐containing (waste) materials to low molecular products and to produce functionalized oligo‐isoprenoids with defined structure. The generated products can be used either directly as biofuels or value‐added materials which can be obtained by conversion of oligo‐isoprenoids to new products such as polyurethanes and related isoprene‐containing polymers.
Only two major types of rubber‐cleaving enzymes have been described so far. One is the rubber oxygenase RoxA that was first isolated from Xanthomonas sp. 35Y (Tsuchii and Takeda, 1990; Braaz et al., 2004) and has been found only in Gram‐negative rubber‐degrading bacteria (Birke et al., 2013). The genome sequence of Xanthomonas sp. 35Y has been determined (Sharma, V., Siedenburg, G., Birke, J., Mobeen, F., Jendrossek, D., Srivastava, T.P. unpubl. data). RoxA of Xanthomonas sp. 35Y (RoxAXsp) is a c‐type dihaem dioxygenase (≈70 kDa, Fig. 1A) and cleaves poly(cis‐1,4‐isoprene) into 2‐oxo‐4,8‐dimethyl‐trideca‐4,8‐diene‐1‐al (ODTD), a C15 compound with a terminal keto and aldehyde group as the main product (Fig. 1B) (Braaz et al., 2005; Schmitt et al., 2010). The structure of RoxAXsp has been solved (Seidel et al., 2013), and molecular insights in the active site of RoxAXsp as well as the cleavage mechanism have been obtained by the construction and biochemical characterization of RoxAXsp muteins (Birke et al., 2012).
Figure 1.
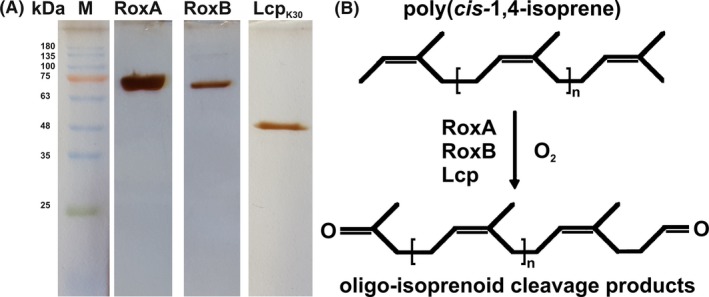
SDS‐PAGE of purified rubber oxygenases. RoxAX sp and RoxBX sp were purified from filter‐concentrated supernatants of L‐rhamnose/LB‐medium‐grown ∆roxA Xanthomonas sp. 35Y cells with genome‐integrated rox AX sp or rox BX sp gene respectively. LcpK30 was purified from soluble French‐press extracts of L‐rhamnose/LB‐medium‐grown E. coli (p4782.1::strep‐lcp K30) via Strep‐Tactin HC gravity flow affinity chromatography.
A. Purified proteins were separated by SDS‐PAGE and stained with silver. A molecular mass standard (M) with kDa values indicated is shown.
B. Oxidative cleavage of rubber. Poly(cis‐1,4‐isoprene) (100 < n < ≈ 10 000) is oxidatively cleaved by rubber oxygenases to oligo‐isoprenoids with terminal keto‐ and aldehyde groups. The methanol‐soluble products differ in the number of intact isoprene units (n) with 1 ≤ n < ≈ 12.
The second type of rubber oxygenase is a protein designated as latex‐clearing protein (Lcp) (Rose et al., 2005; Hiessl et al., 2012; Yikmis and Steinbüchel, 2012). Lcps (≈40 kDa, Fig. 1A) are widespread in or even specific for Gram‐positive rubber‐degrading bacteria, such as Streptomyces sp. K30 (LcpK30) (Rose et al., 2005), Gordonia polyisoprenivorans, Gordonia westfalica (Arenskötter et al., 2001; Bröker et al., 2008), and were recently isolated from Gordonia polyisoprenivorans VH2 (Hiessl et al., 2014) Streptomyces sp. K30 (Birke et al., 2015; Röther et al., 2016) and from Rhodococcus rhodochrous RPK1 (Watcharakul et al., 2016). The amino acid sequences of RoxAs and Lcps are not related although both enzymes catalyse the oxidative cleavage of the double bonds in poly(cis‐1,4‐isoprene) and both cleave polyisoprene to products with terminal keto and aldehyde groups (Fig. 1B). In contrast to RoxAs that cleave rubber to only one major end‐product (ODTD), Lcps produce a mixture of oligo‐isoprenoids (C20, C25, C30 and higher oligo‐isoprenoids, Fig. 2) (Ibrahim et al., 2006; Birke and Jendrossek, 2014). Lcps are b‐type cytochromes and share a common domain of unknown function (DUF2236) (Hiessl et al., 2014; Birke et al., 2015). Recently, the importance of several strictly conserved residues within the DUF2236 domain for stability and activity was determined (Röther et al., 2016).
Figure 2.
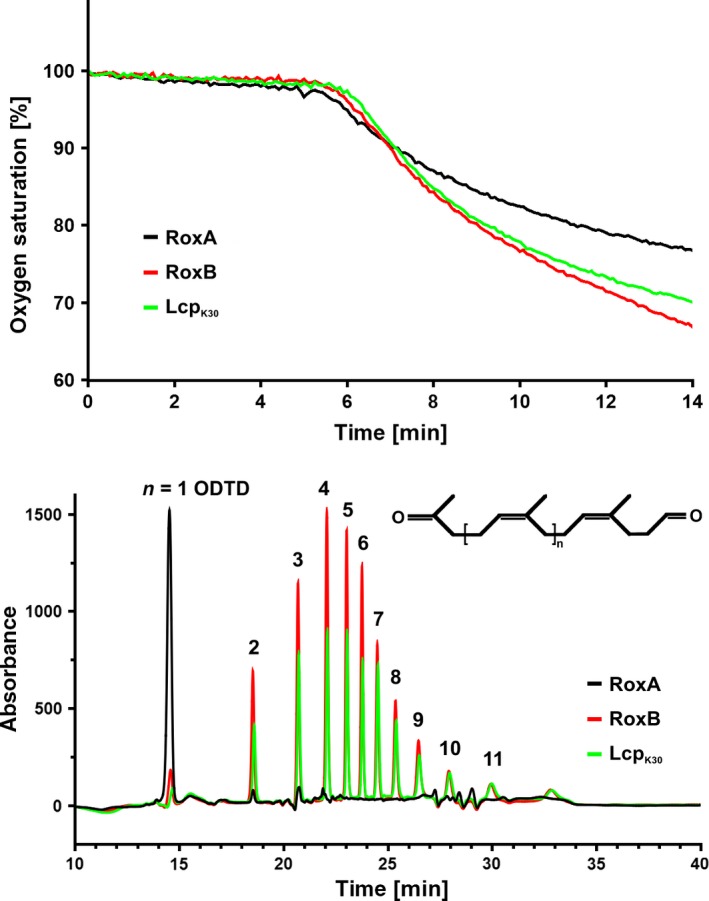
Activities and product analysis of rubber oxygenases. Activities of purified rubber oxygenases (LcpK30, RoxAX sp and RoxBX sp) were determined by following the consumption of dissolved oxygen at 37°C in a Oxy4 V2 apparatus, Presens, Regensburg, Germany, as described recently (Röther et al., 2017) (top). 4 μg each of LcpK30, RoxAX sp or RoxBX sp was added to 1 ml of an emulsion of polyisoprene latex in potassium phosphate buffer (100 mM, pH 7) at ≈5.5 min. The initial slopes correspond to specific activities of 6.2, 2.6, 6.4 U mg−1 for LcpK30, RoxAX sp or RoxBX sp respectively. One unit corresponds to the consumption of one molecule of dioxygen per minute. The products of polyisoprene cleavage were determined by HPLC‐based analysis of the ethylacetate‐extracted cleavage products (bottom). For LcpK30 and RoxBX sp, a typical pattern of oligo‐isoprenoids varying in the number of subunits (n = 2–11) was observed. For RoxA, 12‐oxo‐4,8‐dimethyltrideca‐4,8‐diene‐1‐al (ODTD, n = 1) was detected as the major cleavage product.
Very recently, a third type of rubber oxygenase, RoxB, was discovered (Birke et al., 2017). The coding sequence is provided under the accession No KY 498024. RoxBXsp was identified as a RoxAXsp homologue in Xanthomonas sp. 35Y and shared some properties with RoxAs: RoxBXsp is also a c‐type dihaem protein with an apparent molecular weight of around 70 kDa), but it has only a low sequence similarity to RoxAXsp (38%). However, RoxBXsp differs from RoxAs in cleaving polyisoprene to a mixture of oligo‐isoprenoids (C20, C25, C30 and higher oligo‐isoprenoids, Fig. 2B). This has previously been described only for Lcps. Therefore, RoxBXsp combines properties of RoxAs and Lcps (Birke et al., 2017). RoxB is related in amino acid sequence to the latA gene product of Rhizobacter gummiphilus (83%) (Kasai et al., 2017). The latA gene was recently discovered to code for a protein that is responsible for the cleavage of polyisoprene in R. gummiphilus. However, no information on the properties of the expressed LatA protein is yet available.
Recombinant overexpression of rubber oxygenases
Despite the fact that all so far described rubber oxygenases must be post‐translationary modified to incorporate the haem cofactor, overexpression of highly active rubber oxygenases is surprisingly easy: RoxAXsp can be expressed extracellularly in quantities of ≈15 mg l−1 from recombinant Xanthomonas sp. 35Y strains which harbour a roxA Xsp gene on the chromosome under the control of an rhamnose‐inducible promoter (Hambsch et al., 2010; Birke et al., 2012). We assume that the amount of produced rubber oxygenase can be increased by a combination of medium optimization, inducer concentration and the time point of addition and harvest. Lcps have been successfully overexpressed intracellularly in recombinant E. coli using conventional induction by rhamnose (Birke et al., 2015; Watcharakul et al., 2016) or via autoinduction (Andler and Steinbüchel, 2017). Secretion of the mature Lcps via the TAT secretion pathway in E. coli (Yikmis et al., 2008) or Bacillus subtilis (van Dijl and Hecker, 2013) should be possible. However, the secretion pathways used for RoxA and RoxB proteins have not yet been identified. If pure proteins are necessary, tagged versions of Lcps can be purified in high yields using a one step affinity chromatography (≈ 15 mg LcpK30 l−1 culture for Strep‐tagged Lcp). The tag also offers the opportunity for enzyme immobilization. Furthermore, over‐production of haem containing rubber oxygenases might be limited by the intracellular availability of the cofactor. An increase in the efficiency of haem biosynthesis, e g., by the expression of gamma‐aminolevulinic acid synthase and gamma‐aminolevulinic acid dehydratase could be used to overcome this limitation (Doss and Philipp‐Dormston, 1975).
Purification of rubber oxygenases
We purified each one representative of the three types of rubber oxygenase (RoxAXsp, RoxBXsp and LcpK30, Fig. 1) and used the purified proteins alone or in combination for the production of oligo‐isoprenoids from polyisoprene latex. Produced oligo‐isoprenoids were purified by HPLC and FPLC, and the identity of the isolated products was confirmed by ESI‐MS analysis.
Untagged RoxAXsp and RoxBXsp were purified from the culture fluid of recombinant ∆roxA Xanthomonas sp. 35Y strains which harboured either the roxA Xsp or the roxB Xsp gene integrated into the chromosome under the control of an L‐rhamnose‐inducible promoter using a two‐step purification procedure as described recently (Birke et al., 2012, 2017). LcpK30 was expressed intracellularly in form of an N‐terminal Strep‐tagged protein and was purified from recombinant E. coli as described previously (Röther et al., 2016). Fig. 1A shows that all three proteins were of high purity and activity determinations confirmed high specific activities of 2.6 U mg−1 (RoxAXsp), 6.2 U mg−1 (LcpK30) and 6.4 U mg−1 (RoxBXsp) at 37°C for the three purified rubber oxygenases (Fig. 2 top). HPLC analysis of the solvent‐extracted products confirmed the cleavage of polyisoprene to ODTD (C15 oligo‐isoprenoid) as major product by RoxAXsp and the formation of a mixture of C20 and higher oligo‐isoprenoids in case of RoxBXsp and LcpK30 (Fig. 2 bottom). ODTD was present only in minor amounts in the products obtained from RoxBXsp and LcpK30.
The finding of only one cleavage product (C15 oligo‐isoprenoid ODTD) for the RoxAXsp‐catalysed reaction and the identification of multiple cleavage products (C20 and higher oligo‐isoprenoids) in case of the RoxBXsp‐ or LcpK30‐cleaved polyisoprene suggested that RoxAXsp on the one side and RoxBXsp and LcpK30 on the other side employ different cleavage mechanisms. We assume that RoxAXsp has a ‘molecular ruler’ and uses an exo‐type mechanism to cleave the polyisoprene chain (Seidel et al., 2013). This explains the formation of only one main cleavage product of a defined length (ODTD). In contrast, in case of RoxBXsp and LcpK30, the formation of multiple products of different length suggests that these rubber oxygenases do not have such a molecular ruler and cleave the polyisoprene chain randomly in an endo‐type mechanism resulting in the observed mixture of oligo‐isoprenoids of different lengths.
Synergistic effect of RoxB and of Lcp on polyisoprene cleavage by RoxA
The generation of oligo‐isoprenoids by endo‐cleavage of polyisoprene molecules (with RoxBXsp or LcpK30) increases the number of free polyisoprene chains. A higher concentration of polyisoprenoid ends should enhance the efficiency of polyisoprene cleavage by rubber oxygenases with an endo‐type cleavage such as RoxAXsp. We therefore determined whether the amount of ODTD produced by RoxA could be increased by the presence of trace amounts of RoxBXsp or LcpK30. The presence of 0.2 μg ml−1 purified RoxBXsp or LcpK30 in the assay mixture did not lead to the formation of substantial amounts of ODTD (factor being < 0.02 relative to 1.0 by 2 μg of RoxAXsp, Fig. 3). However, when combined, 2 μg ml−1 RoxAXsp and 0.2 μg ml−1 purified RoxBXsp or LcpK30 increased the amount of produced ODTD by a factor of 1.4 or 1.5, respectively, in comparison with the values obtained with 2 μg RoxAXsp or LcpK30 alone (Fig. 3). Furthermore, the synergistic effect was investigated with respect to a kinetic effect enhancing the speed of the cleavage reaction, representing a major factor to be considered upon industrial employment of the reaction. To this end, the oxygen consumption rates by LcpK30 (0.4 μg) and RoxAXsp (4 μg) alone were determined, combined (added) in silico and were then compared to an experiment in which both enzymes were simultaneously present. As evident from Fig. 4, the simultaneous presence of low amounts of LcpK30 increased the specific oxygen consumption by a factor of 1.4 (2.6 U mg−1) relative to the in silico combined oxygen consumption rates (1.8 U mg−1). These results also showed that the presence of terminal aldehyde and keto groups did not inhibit the cleavage of these oligo‐isoprenoids to ODTD by RoxAXsp. Furthermore, the efficiency of rubber degradation was enhanced when each an endo‐ and exo‐type rubber oxygenase were simultaneously present. These data provide a plausible explanation for the presence of the roxA and roxB gene in Xanthomonas sp. 35Y due to a synergistic effect; in the presence of both gene products, ODTD is the only observed cleavage product for the facilitated uptake into the cells and use as a source of carbon and energy.
Figure 3.
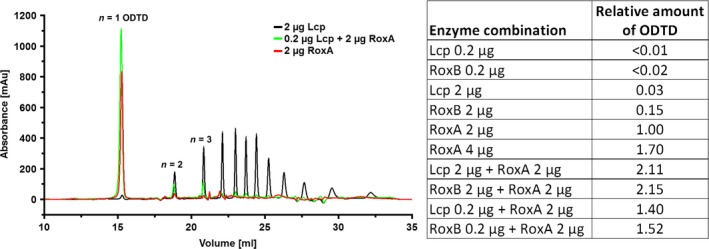
Synergistic effect during rubber cleavage. Polyisoprene latex was cleaved by different amounts and combinations of rubber oxygenases as indicated (left). The amounts of produced ODTD (Table on the right) were determined from the ODTD areas in HPLC chromatograms (exemplary shown in the image on the left). The ODTD‐specific area obtained for 2 μg of RoxAX sp was set as 1.0. The addition of only 0.2 μg Lcp increased ODTD formation by 2 μg of RoxAX sp by a factor of 1.4 and only trace amounts of higher oligo‐isoprenoids (n = 2 and 3; n indicates the number of intact isoprene units, see structure shown in Fig. 1B) were determined. A similar effect with 1.5‐fold higher amount of produced ODTD was observed for a combination of 2 μg RoxAX sp and 0.2 μg RoxBX sp.
Figure 4.
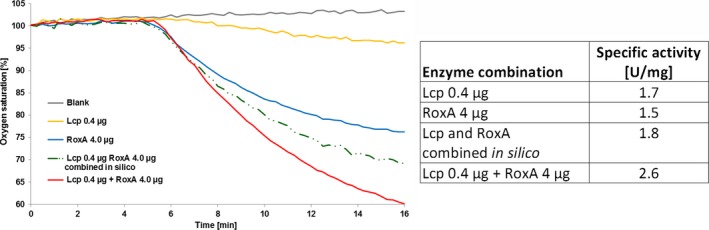
Synergistic effect of the presence of LcpK30 on the specific activity of RoxAX sp. The oxygen consumption rates of 0.4 μg LcpK30, of 4 μg of RoxAX sp and of a mixture of 0.4 μg LcpK30 and of 4 μg of RoxAX sp were recorded. The values for 0.4 μg LcpK30 and 4 μg RoxAXsp were combined in silico and the slope of the resulting curve was calculated to determine a theoretical specific activity. Note that the specific activities of the reaction in the presence of both enzymes were 1.4‐fold higher (2.6 U mg−1) compared to the in silico combined values of the two reactions with the single enzymes (1.8 U mg−1).
Separation and purification of oligo‐isoprenoids
As shown in Fig 2B, the cleavage of polyisoprene by RoxBXsp or by LcpK30 yielded a mixture of oligo‐isoprenoids (C20 and higher oligo‐isoprenoids). For the application of these compounds as fine chemicals or as building blocks for (polymer) plastic synthesis in organic chemistry, the preparation of large amounts of pure oligo‐isoprenoids is preferable. To demonstrate the isolation of isoprenoids at a higher scale, we increased the volume of polyisoprene latex and replaced the HPLC‐based separation of oligo‐isoprenoids by an FPLC separation system because FPLC systems can be up‐scaled more easily than HPLC‐based separations. As a proof of principle, we treated 1 litre of 5% (wt/vol) polyisoprene latex in 100 mM potassium phosphate buffer, pH 7 with 4 mg of purified LcpK30 and incubated the assay mixture for 24 h at room temperature while stirring at 200 rpm. The produced oligo‐isoprenoids were solvent‐extracted with 100 ml ethylacetate. The solvent was evaporated, and the products (≈100 mg) were dissolved in 5 ml methanol. 200 μl of the dissolved products was then applied to a PEP RPC HR5/5‐FPLC column that had been equilibrated with 50% methanol: water and eluted by the application of an increasing step gradient to 100% methanol at a constant flow rate of 1.5 ml min−1. Peaks were automatically fractioned (≈2 ml per peak) by monitoring the absorbance at 210 nm. As shown in Fig. 5 left, the same eleven individual peaks were identified that had been detected on the analytical HPLC column (Fig. 2). The compound of each of the separated peaks was collected individually, concentrated by evaporation and dissolved in 100 μl of methanol. When each of the isolated compounds was separately run on the analytical HPLC column, the successful isolation of each oligo‐isoprenoid was demonstrated by the appearance of one homogeneous peak (Fig. 5, right). The m/z values of the isolated oligo‐isoprenoids were determined by HPLC‐MS and were in agreement with the structural formulas and the theoretical values for the individual oligo‐isoprenoids (Table 1).
Figure 5.
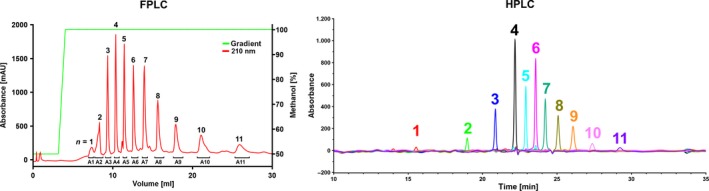
Separation of oligo‐isoprenoid mixtures by FPLC, HPLC and HPLC‐MS. 200 μl oligo‐isoprenoid solution in methanol (prepared by digestion of polyisoprene with LcpK30 as described in the main text) was applied to a reversed‐phase FPLC column (Pep RPC HR 5/5, 1 ml bed volume) and separated by a step gradient from 50% water: methanol to 100% methanol (green line) (left image). Absorption at 210 nm (red line) was used to fractionate peaks representing different oligo‐isoprenoids (A1‐A11, corresponding to n = 1–11). Aliquots of each separately collected fraction (A1 to A11) were applied to analysis via HPLC. An overlay image consisting of all eleven HPLC chromatograms is shown on the right. The superposition of the chromatograms shows the high resolution power of the used FPLC column. The masses (m/z values) of each isolated compound were confirmed by HPLC‐ESI‐MS and are provided in Table 1.
Table 1.
Oligo‐isoprenoids produced by enzymatic cleavage of polyisoprene using purified rubber oxygenases
| No of intact isoprene units [n] | Elemental formula | m/z | m/z [M+H]+ | m/z [M+Na]+ | m/z [M+Na+CH3OH]+ | FPLC peak area [%] |
|---|---|---|---|---|---|---|
| 1 | C15H24O2 (ODTD) 12‐oxo‐4,8‐dimethyl‐trideca‐4,8‐diene‐1‐al | 236.178 | 237.185 | 259.167 | 291.193 | 2.9 |
| 2 | C20H32O2 | 304.240 | 305.248 | 327.230 | 359.256 | 6.8 |
| 3 | C25H40O2 | 372.303 | 373.310 | 395.292 | 427.318 | 9.6 |
| 4 | C30H48O2 | 440.365 | 441.373 | 463.355 | 495.381 | 10.5 |
| 5 | C35H56O2 | 508.428 | 509.435 | 531.417 | 563.444 | 10.3 |
| 6 | C40H64O2 | 576.491 | 577.498 | 599.480 | 631.506 | 10.4 |
| 7 | C45H72O2 | 644.553 | 645.561 | 667.543 | 699.569 | 12.4 |
| 8 | C50H80O2 | 712.616 | 713.623 | 735.605 | 767.631 | 10.1 |
| 9 | C55H88O2 | 780.678 | 781.686 | 803.668 | 835.694 | 8.7 |
| 10 | C60H96O2 | 848.741 | 849.748 | 871.730 | 903.757 | 9.5 |
| 11 | C65H104O2 | 916.804 | 917.811 | 939.793 | 971.819 | 9.0 |
Polyisoprene latex was treated with purified rubber oxygenase (LcpK30), and cleavage products were extracted with ethylacetate and dissolved in methanol. Products were analysed by HPLC‐ESI‐MS analysis before and after purification of individual peaks by FPLC (Fig. 4). For each compound the theoretical m/z values and the values corresponding to the protonated ([M+H]+), the sodium ion adduct ([M+Na]+) and for the sodium ion+ methanol adduct forms ([M+Na+CH3OH]+) are indicated. The relative amounts (in %) of each prepared oligo‐isoprenoid are also provided.
Conclusions and outlook
Polyisoprene in form of natural rubber latex is a cheap bulk compound and is available in the ton‐scale. Cleavage of polyisoprene by rubber oxygenases and separation of produced oligo‐isoprenoids is fairly possible. In this study, eleven oligo‐isoprenoids of the ‘ODTD‐family’ with one to eleven central isoprene units (n) between the terminal aldehyde and keto functional groups could be separately prepared. The highest yields were obtained for ODTD (RoxAXsp alone) and for the C30 to C50 compounds (LcpK30 or RoxBXsp alone). Purification of oligo‐isoprenoids by FPLC can be easily up‐scaled for the mass production of oligo‐isoprenoids. The use of tyres and other materials containing vulcanized rubbers as substrates for enzymatic degradation by different rubber oxygenases is also possible; however, the presence of sulfur bridges and other components complicates the efficiency of enzymatic cleavage of vulcanized rubber waste and therefore limit – at present – the use of rubber oxygenases to the cleavage of unprocessed natural rubber latex. Mechanical, chemical and/or physical pre‐treatments of rubber wastes (e.g. grinding, solvent extraction, desulphurization) might help to make processed rubber wastes also accessible for enzymatic cleavage. Isoprenoids derived from rubber can be used for the production of fragrances, hormones and pharmaceuticals, creating interest in cheap synthesis pathways see (Förster‐Fromme and Jendrossek, 2010; Schewe et al., 2015). Furthermore, they can be also used in chemical or enzymatic cyclization reactions (Siedenburg et al., 2012, 2013) for the production of cyclic compounds or can be used as biofuels (Mewalal et al., 2017). This study provides purified, reactive oligo‐isoprenoids that can likely be used to convert rubber waste, e.g., from tires into precursors for the synthesis of value‐added compounds. The reactivity of the aldehydes might be directly used to form covalent bonds with other molecules (e.g. with amines). Alternatively, the keto groups of the oligo‐isoprenoids can be chemically or enzymatically reduced to the corresponding mono‐ or di‐alcohols. The reduction in the C15 compound ODTD to the corresponding alcohol by enzymatic reduction has been previously demonstrated (Braaz et al., 2005). Enzymatic generation of isoprenoid‐diols can help to provide precursors for the production of polymers from sustainably produced monomers, e.g., for the production of polyurethanes and might be an alternative to chemical methods for the conversion of polyisoprenes to polyurethanes (Anancharoenwong, 2011).
Conflict of Interest
None declared.
Acknowledgements
We thank Weber and Schaer (Hamburg) for providing polyisoprene and IBA Life sciences (Göttingen) for providing Strep‐Tactin columns and cooperation during up‐scaling of rubber oxygenase purification. This work was supported by the Graduiertenkolleg GRK1708 to the University of Tübingen and by a grant of the Deutsche Forschungsgemeinschaft to D.J.
Microbial Biotechnology (2017) 10(6), 1426–1433
Funding Information
This work was supported by the Graduiertenkolleg GRK1708 to the University of Tübingen and by a grant of the Deutsche Forschungsgemeinschaft to D.J.
References
- Akhlaghi, S. , Gedde, U.W. , Hedenqvist, M.S. , Brana, M.T.C. , and Bellander, M. (2015) Deterioration of automotive rubbers in liquid biofuels: a review. Renew Sustain Energy Rev 43: 1238–1248. [Google Scholar]
- Anancharoenwong, E. , 2011. Synthesis and characterization of cis‐1, 4‐polyisoprene‐based polyurethane coatings; study of their adhesive properties on metal surface. Université du Maine, English. NNT: 2011LEMA1009.
- Andler, R. , and Steinbüchel, A. (2017) A simple, rapid and cost‐effective process for production of latex clearing protein to produce oligopolyisoprene molecules. J Biotechnol 241: 184–192. [DOI] [PubMed] [Google Scholar]
- Arenskötter, M. , Baumeister, D. , Berekaa, M.M. , Pötter, G. , Kroppenstedt, R.M. , Linos, A. , and Steinbüchel, A. (2001) Taxonomic characterization of two rubber degrading bacteria belonging to the species Gordonia polyisoprenivorans and analysis of hyper variable regions of 16S rDNA sequences. FEMS Microbiol Lett 205: 277–282. [DOI] [PubMed] [Google Scholar]
- Birke, J. , and Jendrossek, D. (2014) Rubber oxygenase and latex clearing protein cleave rubber to different products and use different cleavage mechanisms. Appl Environ Microbiol 80: 5012–5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birke, J. , Hambsch, N. , Schmitt, G. , Altenbuchner, J. , and Jendrossek, D. (2012) Phe317 is essential for rubber oxygenase RoxA activity. Appl Environ Microbiol 78: 7876–7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birke, J. , Röther, W. , Schmitt, G. , and Jendrossek, D. (2013) Functional identification of rubber oxygenase (RoxA) in soil and marine myxobacteria. Appl Environ Microbiol 79: 6391–6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birke, J. , Röther, W. , and Jendrossek, D. (2015) Latex clearing protein (Lcp) of Streptomyces sp. strain K30 Is a b‐type cytochrome and differs from rubber oxygenase A (RoxA) in its biophysical properties. Appl Environ Microbiol 81: 3793–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birke, J. , Röther, W. and Jendrossek, D. (2017) RoxB is a novel type of rubber oxygenase that combines properties of rubber oxygenase RoxA and latex clearing protein (Lcp). Appl Environ Microbiol 83, in press, https://doi.org/10.1128/aem.00721-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braaz, R. , Fischer, P. , and Jendrossek, D. (2004) Novel type of heme‐dependent oxygenase catalyzes oxidative cleavage of rubber (poly‐cis‐1,4‐isoprene). Appl Environ Microbiol 70: 7388–7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braaz, R. , Armbruster, W. , and Jendrossek, D. (2005) Heme‐dependent rubber oxygenase RoxA of Xanthomonas sp. cleaves the carbon backbone of poly(cis‐1,4‐Isoprene) by a dioxygenase mechanism. Appl Environ Microbiol 71: 2473–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröker, D. , Dietz, D. , Arenskötter, M. , and Steinbüchel, A. (2008) The genomes of the non‐clearing‐zone‐forming and natural‐rubber‐ degrading species Gordonia polyisoprenivorans and Gordonia westfalica harbor genes expressing Lcp activity in Streptomyces strains. Appl Environ Microbiol 74: 2288–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijl, J.M. , and Hecker, M. (2013) Bacillus subtilis: from soil bacterium to super‐secreting cell factory. Microb Cell Fact 12: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doss, M. , and Philipp‐Dormston, W.K. (1975) Over‐production of porphyrins and heme in heterotrophic bacteria. Z Naturforsch C Biosci 30: 425–426. [DOI] [PubMed] [Google Scholar]
- Förster‐Fromme, K. , and Jendrossek, D. (2010) Catabolism of citronellol and related acyclic terpenoids in pseudomonads. Appl Microbiol Biotechnol 87: 859–869. [DOI] [PubMed] [Google Scholar]
- Hambsch, N. , Schmitt, G. , and Jendrossek, D. (2010) Development of a homologous expression system for rubber oxygenase RoxA from Xanthomonas sp. J Appl Microbiol 109: 1067–1075. [DOI] [PubMed] [Google Scholar]
- Hiessl, S. , Schuldes, J. , Thuermer, A. , Halbsguth, T. , Broeker, D. , Angelov, A. , et al (2012) Involvement of two latex‐clearing proteins during rubber degradation and insights into the subsequent degradation pathway revealed by the genome sequence of Gordonia polyisoprenivorans strain VH2. Appl Environ Microbiol 78: 2874–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiessl, S. , Boese, D. , Oetermann, S. , Eggers, J. , Pietruszka, J. , and Steinbüchel, A. (2014) Latex clearing protein‐an oxygenase cleaving Poly(cis‐1,4‐Isoprene) rubber at the cis double bonds. Appl Environ Microbiol 80: 5231–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim, E. , Arenskötter, M. , Luftmann, H. , and Steinbüchel, A. (2006) Identification of poly(cis‐1,4‐isoprene) degradation intermediates during growth of moderately thermophilic actinomycetes on rubber and cloning of a functional lcp homologue from Nocardia farcinica strain E1. Appl Environ Microbiol 72: 3375–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai, D. , Imai, S. , Asano, S. , Tabata, M. , Iijima, S. , Kamimura, N. , et al (2017) Identification of natural rubber degradation gene in Rhizobacter gummiphilus NS21. Biosci Biotechnol Biochem 81: 614–620. [DOI] [PubMed] [Google Scholar]
- Mewalal, R. , Rai, D.K. , Kainer, D. , Chen, F. , Külheim, C. , Peter, G.F. , and Tuskan, G.A. (2017) Plant‐derived terpenes: a feedstock for specialty biofuels. Trends Biotechnol 35: 227–240. [DOI] [PubMed] [Google Scholar]
- Rose, K. , Tenberge, K.B. , and Steinbüchel, A. (2005) Identification and characterization of genes from Streptomyces sp strain K30 responsible for clear zone formation on natural rubber latex and poly(cis‐1,4‐isoprene) rubber degradation. Biomacromolecules 6: 180–188. [DOI] [PubMed] [Google Scholar]
- Röther, W. , Austen, S. , Birke, J. , and Jendrossek, D. (2016) Molecular insights in the cleavage of rubber by the latex‐clearing‐protein (Lcp) of Streptomyces sp. strain K30. Appl Environ Microbiol 82: 6593–6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röther, W. , Birke, J. , and Jendrossek, D. (2017) Assays for the detection of rubber oxygenase activities. Bio‐protocol 7: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schewe, H. , Mirata, M.A. , and Schrader, J. (2015) Bioprocess engineering for microbial synthesis and conversion of isoprenoids. Adv Biochem Eng Biotechnol 148: 251–286. [DOI] [PubMed] [Google Scholar]
- Schmitt, G. , Seiffert, G. , Kroneck, P.M.H. , Braaz, R. and Jendrossek, D. ., 2010. Spectroscopic properties of rubber oxygenase RoxA from Xanthomonas sp., a new type of dihaem dioxygenase. Microbiology (Reading, Engl.) 156: 2537–2548. [DOI] [PubMed] [Google Scholar]
- Schrader, J. and Bohlmann, J. , 2015. Biotechnology of Isoprenoids. Cham: Springer; https://doi.org/10.1007/978-3-319-20107-8 [Google Scholar]
- Seidel, J. , Schmitt, G. , Hoffmann, M. , Jendrossek, D. , and Einsle, O. (2013) Structure of the processive rubber oxygenase RoxA from Xanthomonas sp. Proc Natl Acad Sci USA 110: 13833–13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedenburg, G. , Jendrossek, D. , Breuer, M. , Juhl, B. , Pleiss, J. , Seitz, M. , et al (2012) Activation‐independent cyclization of monoterpenoids. Appl Environ Microbiol 78: 1055–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedenburg, G. , Breuer, M. , and Jendrossek, D. (2013) Prokaryotic squalene‐hopene cyclases can be converted to citronellal cyclases by single amino acid exchange. Appl Microbiol Biotechnol 97: 1571–1580. [DOI] [PubMed] [Google Scholar]
- Tsuchii, A. , and Takeda, K. (1990) Rubber‐degrading enzyme from a bacterial culture. Appl Environ Microbiol 56: 269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers, C.E. , Behrendorff, J.B.Y.H. , Bongers, M. , Brennan, T.C.R. , Bruschi, M. , and Nielsen, L.K. (2015) Production of industrially relevant isoprenoid compounds in engineered microbes. Microbiol Monogr 26: 303–334. [Google Scholar]
- Watcharakul, S. , Röther, W. , Birke, J. , Umsakul, K. , Hodgson, B. , and Jendrossek, D. (2016) Biochemical and spectroscopic characterization of purified Latex Clearing Protein (Lcp) from newly isolated rubber degrading Rhodococcus rhodochrous strain RPK1 reveals novel properties of Lcp. BMC Microbiol 16: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yikmis, M. , and Steinbüchel, A. (2012) Historical and recent achievements in the field of microbial degradation of natural and synthetic rubber. Appl Environ Microbiol 78: 4543–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yikmis, M. , Arenskoetter, M. , Rose, K. , Lange, N. , Wernsmann, H. , Wiefel, L. , and Steinbüchel, A. (2008) Secretion and transcriptional regulation of the latex‐clearing protein, Lcp, by the rubber‐degrading bacterium Streptomyces sp strain K30. Appl Environ Microbiol 74: 5373–5382. [DOI] [PMC free article] [PubMed] [Google Scholar]


