Summary
Lignocellulosic biomass, which mainly consists of cellulose, hemicellulose and lignin, is the most abundant renewable source for production of biofuel and biorefinery products. The industrial use of plant biomass involves mechanical milling or chipping, followed by chemical or physicochemical pretreatment steps to make the material more susceptible to enzymatic hydrolysis. Thereby the cost of enzyme production still presents the major bottleneck, mostly because some of the produced enzymes have low catalytic activity under industrial conditions and/or because the rate of hydrolysis of some enzymes in the secreted enzyme mixture is limiting. Almost all of the lignocellulolytic enzyme cocktails needed for the hydrolysis step are produced by fermentation of the ascomycete Trichoderma reesei (Hypocreales). For this reason, the structure and mechanism of the enzymes involved, the regulation of their expression and the pathways of their formation and secretion have been investigated in T. reesei in considerable details. Several of the findings thereby obtained have been used to improve the formation of the T. reesei cellulases and their properties. In this article, we will review the achievements that have already been made and also show promising fields for further progress.
Introduction
Plant biomass is the most abundant renewable source for conversion to biofuel and biorefinery products. It consists of lignocellulose primary made of various polysaccharides and the aromatic polymer lignin, which provide mechanical strength to the plants and render them highly resistant to attack from pathogens (Houston et al., 2016). The main polysaccharides in lignocellulose comprise the glucohomopolysaccharide cellulose (20–50%, w/w) and hemicelluloses (15–35%, w/w), which – depending on the plant–are primarily made up of a xylan, glucuronoxylan, xyloglucan, glucomannan and arabinoxylan backbones with heterogeneous side‐chains (Kubicek, 2013; Álvarez et al., 2016).
The use of the monosaccharides that constitute the plant biomass polymers implies their efficient hydrolysis, which – because of its recalcitrance and heterogeneity – is still a major technical challenge. To achieve this, the plant debris have first to be reduced in size by milling or chipping, followed by a mild chemical or physicochemical pretreatment step to make the biomass more susceptible to hydrolysis by the enzymes. Finally, enzymatic hydrolysis depolymerizes cellulose to d‐glucose and the still present hemicelluloses to monosaccharides such as d‐glucose, d‐xylose and l‐arabinose. These two steps are known to be the cost‐intensive part of biomass conversion (Brethauer and Studer, 2015; Payne et al., 2015; Gupta et al., 2016a; Kubicek and Kubicek, 2016). The current situation and advances in the pretreatment technology have been subject to several excellent recent reviews and shall therefore not be covered here (Xu and Huang, 2014; Silveira et al., 2015; Capolupo and Faraco, 2016; Rabemanolontsoa and Saka, 2016). As for the enzymatic hydrolysis step, some of the enzymes in the cellulase cocktail have too low catalytic activity, which impacts the amount needed for the saccharification step. In addition, the composition of the secreted enzyme mixture may not be optimal for certain applications because of the presence of some enzymes in limiting amounts (see below for details). Preparation and use of these enzymes are therefore a major cost factor (Chundawat et al., 2011; Klein‐Marcuschamer et al., 2012; Gupta et al., 2016a; Kubicek and Kubicek, 2016). Enzymatic hydrolysis is nevertheless considered the best available procedure because – in contrast to chemical hydrolysis – it does not produce compounds that inhibit the further conversion of the hydrolysate to biofuels and platform chemicals by fermentation (Payne et al., 2015).
To remove the bottleneck at the enzymatic hydrolysis step, a significant amount of work has been performed to understand the performance of the respective enzymes (Beckham et al., 2014; Harris et al., 2014; Payne et al., 2015; Kubicek and Kubicek, 2016). As cellulose represents the most recalcitrant material of the plant biomass, and yields just d‐glucose, the preferred monosaccharide for further use, the majority of work has been performed on cellulolytic enzymes. In the microbial world, there are two fundamentally different strategies for the hydrolysis of cellulose (Payne et al., 2015): the ‘bound enzyme paradigm’ (cellulosome) that is present in anaerobic microorganisms (Bae et al., 2013), and the ‘free enzyme paradigm’ that is used by aerobic microorganisms (Gupta et al., 2016a). The latter is responsible for the vast majority of plant cell wall degradation in nature (Kubicek, 2013). Among the organisms capable of doing this, the ascomycete Trichoderma reesei (Hypocreales) was the first microorganism selected for closer investigation because of its potential to produce an efficient cellulose‐hydrolysing enzyme mixture (for review on the history of the detection and use of T. reesei see Druzhinina and Kubicek, 2016). Despite the fact that other fungi also produce powerful cellulolytic mixtures, which in some cases even exhibit properties superior to those produced by T. reesei (see e.g. Gusakov, 2011; Wang et al., 2012 for review), T. reesei is still almost exclusively used for cellulase production by industry because technologies for its use and handling are based on a seventy years of experience (Bischof et al., 2016).
For a long time, the canonical view of action of fungal cellulases was that endoglucanases (EGs; ‘nonprocessive cellulases’) act by cleaving cellulose chains in amorphous regions within the polymers chain, and cellobiohydrolases (CBHs; processive cellulases) hydrolsze cellulose chains at the end and release the disaccharide cellobiose (‘exo‐endo’ model). This disaccharide is then hydrolysed by β‐glucosidases to glucose. According to their structure, these enzymes are grouped into various glycoside hydrolase (GH) families, which are archived in the CAZyme (carbohydrate active enzymes) database (Lombard et al., 2014). Cellulases produced by T. reesei belong to five GH families (Table 1): endo‐β‐1,4‐d‐glucanases are found in GH5, GH7, GH12 and GH45, and CBHs in GH6 and GH7. GH7 is the only family that contains both CBHs (CEL7A; previously named CBH1) and endo‐β‐1,4‐d‐glucanases (CEL7B, previously named EGL1).
Table 1.
Trichoderma reesei cellulolytic enzymes
| Enzyme type | EC number | GH family | Abbreviated namea | Full name | Previous name | Protein IDb |
|---|---|---|---|---|---|---|
| Cellobiohydrolases | EC 3.2.1.91 | GH6 | CEL6A | Cellobiohydrolase II | CBHII | Trire2:72567 |
| EC 3.2.1.91 | GH7 | CEL7A | Cellobiohydrolase I | CBHI | Trire2:123989 | |
| Endo‐β‐1,4‐d‐glucanases | EC 3.2.1.4 | GH5 | CEL5A | Endoglucanase II | EGL2 | Trire2:120312 |
| EC 3.2.1.4 | GH5 | NN | Endoglucanase | Trire2:53731 | ||
| EC 3.2.1.4 | GH5 | NN | Endoglucanase | Trire2:82616 | ||
| EC 3.2.1.4 | GH7 | CEL7B | Endoglucanase I | EGLI | Trire2:122081 | |
| EC 3.2.1.4 | GH12 | CEL12A | Endoglucanase III | EGL3 | Trire2:123232 | |
| EC 3.2.1.4 | GH12 | NN | Trire2:77284 | |||
| EC 3.2.1.4 | GH45 | CEL45A | Endoglucanase V | EGL5 | Trire2:49976 | |
| β‐d‐glucosidases | EC 3.2.1.21 | GH3 | CEL3A | β‐glucosidase I | BGL1 | Trire2:76672 |
| EC 3.2.1.21 | GH3 | CEL3B | β‐glucosidase | Trire2:121735 | ||
| EC 3.2.1.21 | GH3 | CEL3C | β‐glucosidase | Trire2:82227 | ||
| EC 3.2.1.21 | GH3 | CEL3E | β‐glucosidase | Trire2:76227 | ||
| EC 3.2.1.21 | GH3 | CEL3F | β‐glucosidase | Trire2:104797 | ||
| EC 3.2.1.21 | GH3 | CEL3H | β‐glucosidase | Trire2:108671 | ||
| EC 3.2.1.21 | GH3 | CEL3J | β‐glucosidase | Trire2:66832 |
NN, no specific name given yet; the GH3 03B2‐glucosidases CEL3D and CEL3G are not listed because they are intracellular enzymes (Guo et al., 2016).
Refers to the T. reesei genome database (http://genome.jgi.doe.gov/Trire2/Trire2.home.html).
GH5 cellulases are most abundant in fungi (Li and Walton, 2017), and also three members of this family are present in T. reesei. GH7 enzymes are common, and orthologues of CEL7A are the most prevalent cellulolytic enzymes in the secretomes of biomass‐degrading fungi. Because of its processive mode of catalysis (like also CEL6A, which is however usually present in much smaller concentrations), it depolymerizes cellulose most rapidly and is also responsible for the majority of hydrolytic turnover. The GH6 family is currently the only known family that comprises cellulases that act from the nonreducing end of the cellulose chain. CEL7A and CEL6A therefore act in synergism and are considered as primary components in biomass degradation cocktails.
GH12 enzymes are typically characterized by a low molecular weight (25 kDa), and – in contrast to most other cellulases – lack a cellulose‐binding domain (CBM1) and glycosylation. They can therefore diffuse deeper into cellulosic material, which made them preferred candidates for the laundry industry. GH45 cellulases are also generally small but have a broader substrate specificity compared to GH5 and GH7 endo‐β‐1, 4‐d‐glucanases. Interestingly, GH45 enzymes are structurally and evolutionarily related to plant expansins.
Trichoderma reesei β‐glucosidases are found in the GH1 and GH3. Those belonging to GH1 are exclusively intracellular enzymes, whereas seven of the nine GH3 β‐glucosidases are secreted into the medium (Guo et al., 2016). CEL3A (previously named BGL1) accounts for most of the β‐glucosidase secreted activity.
The ‘exo/endo’ model (for review see Kubicek, 2013) has been revised by Stahlberg et al. (1993) and Kurašin and Väljamäe (2011) who showed that CEL7A is also able to conduct ‘endo‐initiation’ and is therefore not a true exocellulase. However, neither the EGs nor the CBHs from fungi can cause massive cellulose decomposition. They rather ‘peel one layer at a time’ (Payne et al., 2015). Elwyn T. Reese and co‐workers had therefore in the fifties already proposed that a decrystallizing protein must exist, which does not hydrolyse the β‐glycosidic linkage but possesses the ability to swell or disrupt cellulose (the so called ‘C1 factor’; Reese et al., 1950). The nature of this C1 protein was enigmatic for a long time. The recently detected lytic polysaccharide monooxygenases (LPMO; originally classified as endoglucanases GH61), attack the highly crystalline regions of cellulose and cleave cellulose by oxidation. They could therefore represent the earlier postulated C1 factor (Morgenstern et al., 2014; Johansen, 2016a,b). Another T. reesei protein, SWO1 which bears an expansin‐like domain has also been considered to fulfill a function in preparing cellulase or hydrolytic attack. But recently published results with SWO1 that had been produced in a cellulase‐negative T. reesei strain failed to show a synergy with cellulases in the degradation of pretreated lignocellulose (Eibinger et al., 2016). The authors therefore concluded that SWO1 ‘is not a C1 factor of degradation of pure cellulose’. A summary of our current understanding of enzymatic cellulose hydrolysis by ‘free cellulases’ is given in Fig. 1).
Figure 1.
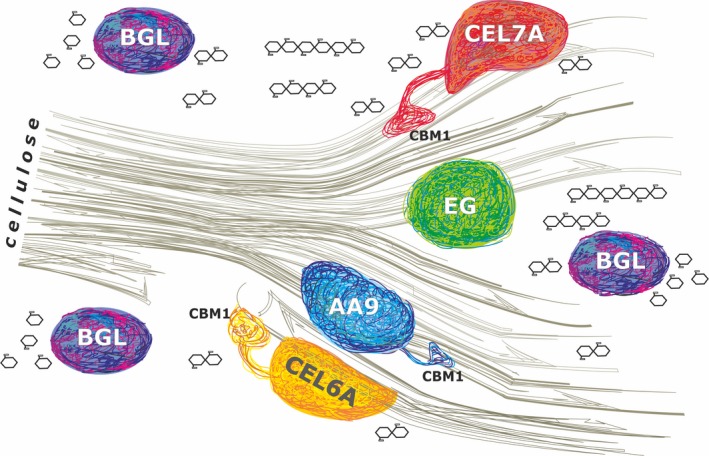
Cartoon summarizing the current knowledge about the Trichoderma reesei enzymes that attack and hydrolyse cellulose. Abbreviations: CEL7A cellobiohydrolase CBH1; CEL6A cellobiohydrolase CBH2; CBM1 cellulose‐binding domain, if present; EG, endoglucanase; AA9, lytic polysaccharide monooxygenase; BGL ‐ β‐glucosidase. CBH1 and CBH2 cleave at the (reducing and non‐reducing, respectively) ends of the cellulose chain. EG cleaves in amorphous cellulose regions, AA9 can act on both crystalline and less‐crystalline regions. The oligosaccharides are further hydrolysed to d‐glucose by β‐glucosidase (BGL, EC 3.2.1.21).
Not surprisingly, T. reesei has been the subject of intensive investigation towards the improvement of its cellulases and reducing the costs for their production and use. This involved attempts to increase their intrinsic activity, facilitating their production, and the reinforcement of existing cellulase preparations by auxiliary proteins and enzymes from other organisms (Wilson, 2009; Horn et al., 2012; Peterson and Nevalainen, 2012; Hu et al., 2015; Müller et al., 2015; Payne et al., 2015; Kubicek and Kubicek, 2016). In this review, we will summarize the progress in these fields, thereby focusing exclusively on cellulose hydrolysis by T. reesei.
Protein engineering of T. reesei cellulase
Despite the progress that has been made in molecular understanding of the reaction mechanism of several cellulases and their binding to cellulose (for extensive review see Payne et al., 2015), these findings have (at least in the published literature) not yet led to strategies for improvement of cellulase activities of T. reesei by genetic engineering. Rather, the focus has been put on the stability of the enzymes at increased temperatures, activity in broader pH ranges and the relief from inhibition by components present in the saccharification mixture.
Engineering high‐temperature tolerance of cellulases
Thermostable cellulases are a major goal in the lignocellulose degradation technology. Performing lignocellulose saccharification at elevated temperatures would provide several benefits such as increased specific activity and stability, prevention of growth of contaminants and increased mass transfer rate due to lower fluid viscosity at high substrate concentrations (Viikari et al., 2007; Blumer‐Schuette et al., 2014). In addition, the biomass saccharification rate is inhibited by lignin that is still present, but this inhibition is less pronounced in thermostable enzymes (Rahikainen et al., 2013). Trichoderma reesei is mesophilic and its enzymes are consequently only moderately tolerant to temperatures above 50 °C (Chokhawala et al., 2015). While the incorporation of respective cellulase genes from thermotolerant fungi such as Acremonium thermophilum, Thermoascus aurantiacus, Chaetomium thermophilum, Myceliophthora thermophila, or Thielavia terrestris is an option (Viikari et al., 2007; Voutilainen et al., 2008), there have also been several attempts to improve the thermostability of T. reesei cellulases by protein engineering: Sandgren et al. (2003) pioneered this area by comparing the amino acid sequence of CEL12A from T. reesei and its thermally much less stable orthologue from T. citrinoviride. The two proteins differed only in 14 amino acids, and exchanging each of them in T. reesei CEL12A by those from T. citrinoviride CEL12A identified an A35S substitution that most strongly decreases the thermal stability. Interestingly, a highly thermostable CEL12A from an unidentified Streptomyces sp. displayed an A35V substitution, and the introduction of this A35V mutation into T. reesei CEL12A resulted in a Tm that was 7.7 °C higher than that of the native enzyme. Mutations of neither A35, S35 nor V35 had an influence on the overall protein structure or hydration of the protein. However, the presence of a hydrophobic amino acid such as A or V at position 35 caused tighter van der Waals interactions with its three neighbouring amino acid side‐chains and closed the entry to the β‐sheet sandwich core. This behaviour, known as cavity‐filling mutation, has been observed in thermal stabilizing mutations in other GHs too (Karshikoff et al., 2015).
Lantz et al. (2010) demonstrated that the two CBHs CEL6A and CEL7A of T. reesei are more thermolabile than the other enzymes involved in cellulose breakdown (like endo‐β‐1,4‐glucanase CEL5A and the β‐glucosidase CEL3A). They are therefore likely rate limiting to the performance of the whole cellulase mixture at saccharification temperatures over 50 °C. Consequently Day et al. (2007) compared the amino acid sequence and structure of 42 CEL7A family members from various fungi and identified 19 sites that could be involved in increased thermostability. Using site saturation mutagenesis, they indeed showed that mutations in 18 of these sites enhanced temperature stability, and the introduction of all 18 sites into CEL7A produced a variant, which exhibited an increase in T m of 14.8 °C (T m 76.0 °C; Lantz et al., 2010).
Arnold and colleagues from the California Institute of Technology have made substantial contributions to GH7 engineering primarily on the basis of computational prediction tools such as structure‐guided recombination. They used non‐contiguous recombination (NCR), a method that identifies pieces of structure that can be swapped among homologous proteins to create new chimeric proteins (Smith et al., 2013a). By this means they designed a library of chimeric enzymes, in which blocks of the structure from T. reesei CEL7A and the two thermostable CEL7A homologues from Talaromyces emersonii and C. thermophilum, respectively, were shuffled to create 531,438 possible variants (Smith et al., 2013b). Selecting a maximally informative subset of 35 chimeras for analysis, they found that these blocks contributed additively to the stability of a chimera. Two highly stabilizing blocks displayed the same two mutations (T360A and F362M), and they increased the thermal stability of CEL6A by 1 and 3°C, respectively (Smith et al., 2013b).
Engineering cellulases for resistance towards ionic liquids
As explained above, the recalcitrant nature of native lignocellulose makes a pretreatment by chemical, physical or biological means necessary. Classical pretreatment procedures, however, are performed in a separate step before hydrolysis, and so necessitate extensive washing (for review see Brethauer and Studer, 2015). Consequently, pretreatment and hydrolysis in a one‐pot reaction would improve the process from an economic point of view. Among the various pretreatment methods, the use of ionic liquids (ILs) would be most appropriate because it does not involve harsh conditions and does not lead to the formation of toxic by‐products (Uju et al., 2012). However, the strongly nucleophilic ILs interact with the positively charged residues on the surface of the cellulases and inactivate them (Li et al., 2015). A possibility for overcoming this is a chemical modification of the primary amino groups in T. reesei CEL7A by succinylation and acetylation, which results in a doubling of the rate of cellulose hydrolysis in 15% (v/v) of the IL 1‐butyl‐3‐methylimidazolium chloride (BMIM‐Cl) (Xu et al., 2016). ILs can also strongly bind to the cellulose‐binding tunnel in the catalytic domain of CEL7A (Li et al., 2015), and interrupt binding of cellulose to the hydrophobic amino acids that are located on the flat face of CBM1 (Wahlstrom et al., 2014). Li et al. (2015) identified six amino acids near the active site that strongly bind the 1‐butyl‐3‐methylimidazolium cation (BMIM]+) and provided in silico evidence that mutation of these residues reduced [BMIM]+ binding and enhanced the tolerance to ILs. However, the production of such a mutated enzyme by T. reesei has not yet been documented.
Manipulation of the enzyme composition
The oldest tool to manipulate the enzyme spectrum produced by T. reesei is the introduction or deletion of one or more cellulase or hemicellulase genes. Soon after the development of the first transformation systems for T. reesei (Penttilä et al., 1987; Gruber et al., 1990; Smith et al., 1991), first successful attempts in this direction had been published (Harkki et al., 1991; Kubicek‐Pranz et al., 1991). Manipulation or elimination of the production of certain cellulase components is particularly important for the laundry and textile industry, where they can damage tissues (Galante et al., 1998).
Another limitation of the T. reesei cellulase system is its low activity of β‐glucosidase, which leads to accumulation of cellobiose during biomass hydrolysis. This in turn inhibits cellobiohydrolase and endo‐β‐1,4‐glucanase activities and so reduces the saccharification rate (Gruno et al., 2004). This low β‐glucosidase activity is not due to too low expression of the respective genes but because the main extracellular β‐glucosidase – CEL1B – remains trapped to a high percentage within the fungus’ cell wall and only part of it slowly released into the medium during autolysis (Kubicek, 1981). Although the cell wall components responsible for this trapping have been identified (Rath et al., 1995), there have been no attempts to use this information for producing strains that secrete more CEL1B into the medium. Instead, this β‐glucosidase deficiency was compensated by the introduction of β‐glucosidase genes from other fungi into T. reesei. Various donors have been reported, such as Penicillium decumbens (Ma et al., 2011), Aspergillus aculeatus (Nakazawa et al., 2012; Treebupachatsakul et al., 2015), a Periconia sp. (Dashtban and Qin, 2012), Rasamsonia emersonii (Ellilä et al., 2017), Neosartorya fischeri (Xue et al., 2016) and Chaetomium atrobrunneum (Colabardini et al., 2016). The enzymes from the last three thermotolerant species also are more stable at higher temperatures, thereby removing the limitation of saccharification by the low‐temperature stability of T. reesei CEL1B during hydrolysis (Viikari et al., 2007). To this end, Xue et al. (2016) reported that a fusion of the N. fischeri NfBgl3A gene to the cbh1 structural gene resulted in β‐glucosidase activities that were up to 175‐fold higher than those of the parent strain.
Cellobiose dehydrogenase (CBD, EC 1.1.99.18), a member of the enzyme arsenal auxiliary to glycoside hydrolases (family AA3; Levasseur et al., 2013), is an extracellular enzyme produced by various wood‐degrading fungi (Henriksson et al., 2000). It oxidizes the reducing ends of cellobiose and cellooligosaccharides to 1, 5‐lactones, which are subsequently hydrolysed to the corresponding carboxylic acids. CBD orthologues’ have also been found in several Pezizomycotina genomes (Table S1). Although T. reesei contains several members of the AA3 family too (Druzhinina and Kubicek, 2016) – a CBD orthologue is absent. Ayers et al. (1978) first suggested that CBD (named ‘cellobiose oxidase’ then) from Phanerochaete chrysosporium could be involved in cellulose biodegradation. Bey et al. (2011) demonstrated that the addition of CBD from the basidiomycete Pycnoporus cinnabarinus synergistically stimulated the saccharification of wheat straw by a T. reesei commercial cellulase cocktail. This stimulation is due to relieving CBH1 from the competitive inhibition by cellobiose, which is oxidized by CBD to the non‐inhibitory cellobionolactone. Consequently, Wang and Lu (2016) overexpressed a CBD‐encoding gene from P. chrysosporium under the cbh2 promoter in T. reesei, and obtained transformants with up to fourfold increased filter paper activity.
The identification of LPMOs as a group of enzymes that accelerate the breakdown of carbohydrate polymers like cellulose, chitin and starch by oxidative cleavage has been a breakthrough in lignocellulose conversion research (Johansen, 2016a,b). LPMOs belonging to the auxiliary enzyme family AA9 degrade cellulose nanofibrils on the surface into shorter fragments. They therefore assist cellulases to attack otherwise highly resistant crystalline substrate areas, which results in a faster and more complete surface degradation (Eibinger et al., 2014). Supplementation of the commercial cellulase cocktail Cellic CTec1 (Novozymes) with AA9 enzymes improved the hydrolysis of lignocellulose (Sun et al., 2015), which led to the new commercial cellulase preparations Cellic CTec2 and Cellic CTec3.
Because of the role of CBD in assisting the catalysis by AA9, it is possible that this stimulatory action of CBD (vide supra) is due to the stimulation of AA9. Trichoderma reesei has three AA9‐encoding genes, of which one is strongly induced during growth under cellulase inducing conditions. The enzyme has been overproduced in T. reesei (Karlsson et al., 2001), but the resulting cellulose‐hydrolysing activity of the secreted cellulolytic enzymes has not been identified (at that time, the enzyme was considered to be an endo‐β‐1,4‐glucanase, CEL61A). The successful overexpression of AA9 – together with CBD – in T. reesei may directly lead to strains with improved cellulase performance.
Finally, T. reesei lacks invertases that hydrolyse sucrose (Bergès et al., 1993), and therefore cannot grow on some cheap technical substrates such as sugarcane molasses. Ellilä et al. (2017) showed that this deficiency could be overcome by constitutively expressing an invertase gene from Aspergillus niger in T. reesei.
Engineering cellulase gene expression
Various cellular levels have been identified which influence the expression of cellulases (for overview see Fig. 2), which will be described below.
Figure 2.
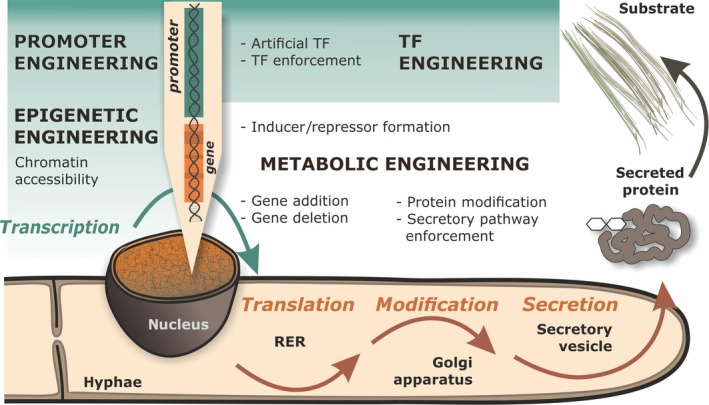
An overview of genetic engineering strategies that offer new perspectives for the further improvement of Trichoderma reesei cellulase formation. Abbreviations: TF – transcription factor, RER – rough endoplasmic reticulum.
Genetic engineering of transcriptional regulators
In T. reesei, the expression of all cellulases and of the majority of hemicellulases is induced in a coordinated manner by growth on carbon sources such as cellulosic substrates, or some disaccharides such as lactose or sophorose, respectively. The expression of the enzymes can therefore be manipulated by engineering a small number of regulators that are responsible for the coordinated expression of cellulases. In T. reesei, unlike in Neurospora crassa, Fusarium graminearum or Aspergillus spp., the transcriptional activator XYR1 (Stricker et al., 2006) is the major transcriptional activator of both cellulase and xylanase gene expression. It belongs to the fungal specific class of transcription factors that contain a domain that binds Zn2+ ions by six cysteine residues (Zn2Cys6). XYR1 binds to a 5′‐GGCW4‐3′ DNA motif (Furukawa et al., 2008). Deletion of this gene eliminates cellulase induction by all known inducers, whereas xyr1 overexpression enhances it (for review see Bischof et al., 2016; Gupta et al., 2016b; Druzhinina and Kubicek, 2016). Several strategies for manipulation of XYR1 (and of some other transcriptional regulators that will be described below) to enhance cellulase production or render its expression constitutive have therefore been presented (Table 2). Recently, da Silva Delabona et al. (2017) showed that a 26‐fold constitutive overexpression of xyr1 in another Trichoderma spp. (T. cf. harzianum) resulted in the production of an enzymatic complex that exhibited a 25% enhanced hydrolysis rate of sugarcane bagasse during the first 24 h of saccharification. Also, Ellilä et al. (2017) constitutively expressed a XYR1 (V821F) mutant (which leads to reduced glucose repression; Derntl et al., 2013) in T. reesei and obtained increased cellulase and xylanase production on sugarcane bagasse.
Table 2.
Published examples of improvement of cellulase production in Trichoderma reesei by genetic engineering
| Gene | Manipulation method | Promoter | References |
|---|---|---|---|
| xyr1 | Constitutive overexpression | pki1 | Seiboth, Karimi et al. (2012) |
| Constitutive overexpression | tcu1 | Lv et al. (2015) | |
| Constitutive overexpression, and ace1 downregulation | pdc1, downregulation by antisense with the same promoter | Wang et al. (2014) | |
| cbh1 | Promoter engineering | CRE1 binding sites exchanged against ACE2 and HAP2/3/5 binding sites | Zou et al. (2012) |
| cre1 | Deletion | Nakari‐Setälä et al. (2009) | |
| Truncation | Nakari‐Setälä et al. (2009); Mello‐de‐Sousa et al. (2014) | ||
| ace1 | Deletion | Aro et al. (2003) | |
| ace3 | Introduction of multiple gene copies | Häkkinen et al. (2014) | |
| lae1 | Constitutive overexpression | gpd1 | Seiboth‐Karimi et al. (2012) |
| vel1 | Constitutive overexpression | tef1 | Karimi Aghcheh et al. (2014) |
Three other proteins or protein complexes (i.e. the Zn2Cys6 transcriptional activators ACE2 and ACE3, and the CCAAT‐binding protein complex (HAP2/HAP3/HAP5) are also involved in the regulation of cellulase gene expression in T. reesei. So far, only the overexpression of ace3 has been shown to result in enhanced cellulase formation (Häkkinen et al., 2014). Other regulators may still be detected: introduction of multiple copies of the gene for the still uncharacterized Zn2Cys6 transcription factor Trire2:80291 into T. reesei was also shown to increase CBH1 formation twofold (Häkkinen et al., 2014). In addition, cellulase gene expression in N. crassa involves several other transcription factors, of which at least CLR2 and VIB1 (Coradetti et al., 2012; Xiong et al., 2014) have orthologues in T. reesei (Trire2:26163 and Trire2:54675, respectively; see genome annotation in Druzhinina et al., 2016). However, it is not known yet whether they also regulate cellulase gene expression in T. reesei, and – if so – whether their manipulation could be used to improve cellulase production.
Two interesting alternative perspectives for manipulation of cellulase gene expression have recently been presented: Zhang et al. (2017) constructed a hybrid cellulase regulator, which consisted of the DNA‐binding domain of the glucose‐repressor CRE1 (see below) fused to the XYR1 effector binding domains. Its overexpression in T. reesei led to an approximately 30‐fold increase of constitutive cellulase and hemicellulase production on glucose. Zhang et al. (2016b) constructed an artificial zinc finger protein library and expressed it in T. reesei. One of the respective transformants showed a 55% rise in cellulase activity against filter paper and an 8.1‐fold increased β‐glucosidase activity.
Cellulase gene expression has also been shown to be repressed by rapidly assimilated carbohydrates, which is due to the action of carbon catabolite repressor protein CRE1 (Nakari‐Setälä et al., 2009). CRE1 interferes both with constitutive as well as induced cellulase and hemicellulase gene expression, but the degree of this repression varies for different cellulase and hemicellulase genes (see Druzhinina and Kubicek, 2016 for review). As an example, only the constitutive but not the inductive expression of the cellobiohydrolase gene cel6A and of the xylanase gene xyn1 is repressed by CRE1 (Mach et al., 1996; Zeilinger et al., 2003). In contrast, CRE1 represses both constitutive and induced expression of the cellobiohydrolase 1 gene cel7a (Nakari‐Setälä et al., 2009). Elimination of the function of CRE1 had already been obtained by classical mutagenesis and resulted in a strain with increased cellulase formation in the presence of increased concentration of inducing carbon sources (cellulose or lactose; Eveleigh and Montenecourt, 1979). This strain – known as T. reesei RUT C30 – bears a truncated version of the cre1 gene that expresses only the zinc finger but not the transactivating domains (CRE1‐96; Ilmén et al., 1996). It is noteworthy that some strains used for the production of cellulases and hemicellulases on the industrial scale are descendants of RUT C30. Recombinant strains of T. reesei with cre1 loss of function have been analysed too, but – in contrast to RUT C30 – they have a pleomorphic phenotype (Nakari‐Setälä et al., 2009) and their use in cellulase production is therefore limited.
Another Zn2Cys6 transcription factor – ACE1 – acts as a partial repressor of cellulase and xylanase gene expression, and strains carrying a deleted allele displayed enhanced cellulase formation (Aro et al., 2003). The physiological conditions that trigger ACE1 repression are not understood yet. ACE1 is an orthologue of Aspergillus nidulans StzA/SltA (Aro et al., 2003) that plays a role in Ca2+ homoeostasis (Spielvogel et al., 2008) and has further been implicated in nitrogen control (Chilton et al., 2008). It is however intriguing that the ACE1 orthologue of Colletotrichum gloeosporioides is mainly involved in appressorium formation (Dubey et al., 2016). An elucidation of the function of ACE1 in T. reesei will be essential before ace1 can be used for science‐based strain engineering.
After this review had been submitted, Cao et al. (2017) reported on the identification of a further repressor of cellulase gene expression, RCE1. Disruption of its gene enhanced the induced expression of cellulase genes and led to a significant delay in termination of induction. RCE1 did not participate in CRE1‐mediated catabolite repression, but antagonized the binding of XYR1 to the cellulase promoters.
Promoter engineering
Manipulation of the binding sites for transcriptional regulators in the cellulase genes is another strategy to modify their expression. Given the number of cellulases produced by T. reesei, this can be a time‐consuming process. Nevertheless, replacing the CRE1 binding sites within the cbh1 promoter by the binding sites for the transcription activators ACE2 and the HAP2/HAP3/HAP5 complex enhanced transcription of a test gene (green fluorescent protein) under cellulase inducing conditions sevenfold (Zou et al., 2012). This approach may have its merit when the expression of only a few genes is targeted. On the other hand, engineering the xyr1 promoter (XYR1 also needs to be induced for cellulase and hemicellulase formation; Lichius et al., 2014) – should provide a much more straightforward approach towards enhancement or modulation of cellulase production. Yet no such experiments have been published up to date.
Epigenetic engineering
Epigenetic engineering has recently been reviewed as a potentially new tool for industrial improvement of fungi (Aghcheh and Kubicek, 2015). The term has been coined by Waddington (1942) for heritable changes in gene expression that do not involve changes in the underlying DNA sequences. In the current understanding, two major levels of epigenetic regulation are important: DNA methylation; and chromatin remodelling by histone modification. While evidence for a regulatory role of DNA‐methylation in T. reesei is not available, some aspects of chromatin remodelling have been demonstrated.
Gupta et al. (2016b) have recently discussed the remodelling of chromatin by transcriptional activators and emphasized it as an emerging approach for improvement of cellulase formation. Under cellulase inducing conditions, the chromatin packing around the cellulase‐encoding genes cbh1 and cbh2 opens, but this does not occur in a xyr1‐deletion strain (Mello‐de‐Sousa et al., 2015, 2016). Consequently, Mello‐de‐Sousa et al. (2015) identified several genes encoding chromatin remodelling enzymes, including histone acetyltransferases, whose expression is significantly different in the ∆xyr1 and its parental strain. Whether these genes could be used to improve cellulase production has not been tested yet. Another histone acetyltransferase (GCN5), however, has been shown to be necessary for cellulase gene expression, and the acetylations of K9 and K14 of histone H3 in the cbh1 promoter were strongly reduced in the T. reesei Δgcn5 strain (Xin et al., 2013).
The carbon catabolite repressor CRE1 has also been implicated in chromatin remodelling: strains expressing a non‐functional CRE1 exhibit a loss of positioned nucleosomes within the cbh1 and cbh2 genes under repressing conditions only (Zeilinger et al., 2003; Ries et al., 2014). Interestingly, the truncated version of CRE1 (CRE1‐96) of RUT C30 seems to trigger chromatin opening (Mello‐de‐Sousa et al., 2014), likely by regulating the expression of snf2/htf1 (Portnoy et al., 2011), a putative helicase that might participate in an ATP‐dependent chromatin remodelling complex (Mello‐de‐Sousa et al., 2014). Identification and engineering of chromatin remodelling proteins are obviously a promising possibility for strain development.
In addition, there is another group of fungal genes that participate in chromatin modification and whose manipulation has been shown to modulate cellulase and hemicellulase production: the Velvet‐LaeA/LAE1 complex (Seiboth, Karimi et al., 2012; Karimi Aghcheh et al., 2014; Liu et al., 2016). LaeA, originally identified in A. nidulans as a regulator of secondary metabolite gene expression, has characteristics of an S‐adenosyl‐l‐methionine arginine protein methyltransferase (Sarikaya‐Bayram et al., 2015). However, LaeA appears not to methylate histones. Rather it seems to function by interaction with the transcription factors of the Velvet protein complex. Constitutive overexpression of both lae1 as well as vel1 in T. reesei has been shown to enhance cellulase expression and secretion (Seiboth, Karimi et al., 2012; Karimi Aghcheh et al., 2014).
Metabolic engineering
A significant body of work has been dedicated to the characterization of the metabolic steps that are involved in the signalling of cellulase gene expression. The current state of knowledge in this area has recently been reviewed (Druzhinina and Kubicek, 2016; Gupta et al., 2016b). Despite a fair amount of progress, an essential improvement of cellulase production by the manipulation of individual genes involved in this process has not yet occurred. This is in part due to the fact that manipulation of genes involved in central metabolism and particularly in signal transduction often produce pleiotropic effects that are difficult to use for strain improvement. Whole‐genome dedicated strategies, such as the use of whole‐cell metabolic models, likely offer the best available strategy for the identification of the limiting steps for cellulase production. To this end, Castillo et al. (2016) applied the CoReCo (comparative metabolic reconstruction framework) pipeline (Pitkänen et al., 2014) for the construction of a high‐quality metabolic model of T. reesei (BIOMODELS database). The model contains a biomass equation, reaction boundaries and uptake/export reactions, which make it ready for the simulation of protein production processes. It was applied by Pakula et al. (2016) for analysis of the flux balances of its metabolism, using a transcriptomic analysis of protein production by T. reesei in chemostat cultivations. The study showed that high protein (cellulase) production might be limited at the level of biosynthesis of sulfur‐containing amino acids, which identifies a clear target for metabolic engineering that was so far not detected by other means.
The engineering toolbox of T. reesei
A suitable molecular tool box that enables easy exchange and manipulation of genes is a prerequisite for engineering T. reesei or individual components of its cellulolytic cocktail, as reviewed above. This topic has been subject of several exhaustive recent reviews (Steiger, 2013; Bischof and Seiboth, 2014; Bischof et al., 2016; Gupta et al., 2016b), and we shall therefore discuss it only very briefly. The low efficiency of gene targeting has for a long time been a major bottleneck in this regard, but was finally – like in several other fungi – solved by inactivating components of the non‐homologous end joining pathway of DNA repair such as tku70 or tmus53 (Guangtao et al., 2009; Steiger et al., 2011; Schuster et al., 2012). More recently, Ouedraogo et al. (2015, 2016) demonstrated that an I‐SceI mediated double strand break in a T. reesei tku70 gene improved transformation efficiencies and increased homologous integration up to 90–100%. Also the CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 system (D'Agostino and D'Aniello, 2017) has been adapted for use with T. reesei (Liu et al., 2015). It depends only on the Cas9 (CRISPR‐associated) nuclease which uses a single chimeric guide RNA for targeting, and so introduces specific DNA double strand breaks that stimulate gene targeting.
Further, a number of new approaches were offered that allowed the insertion of expression cassettes at defined genomic regions. Derntl et al. (2015) constructed T. reesei strains bearing truncated versions of several genes resulting in auxotrophic phenotypes, and tested the rescue of prototrophy by transformation with a complementary fragment of the same gene. Among them, they used the ade2 locus (encoding a phosphoribosylaminoimidazole carboxylase necessary for purine biosynthesis) because integration into this locus destroys ade2, the Δade2 disruptants accumulate polymerized 5‐aminoimidazole ribonucleotide (Ugolini and Bruschi, 1996) and can therefore easily be identified by the red colour. Derntl et al. (2015) also successfully demonstrated the applicability of the asl1 (encoding argininosuccinate lyase of arginine biosynthesis), the hah1 (encoding homoaconitate dehydratase involved in lysine biosynthesis) and the pyr4 genes (encoding the orotidine‐5‐phosphate decarboxylase of the uridine biosynthesis pathway) for this approach to obtain site‐directed integration in T. reesei.
Even if the transformation frequency is high, identification of the transformants that express the target protein at desired levels can still be a tedious task because the expression levels between transformants can be very variable. Subramanian et al. (2017) have therefore adapted the 2A peptide system from the foot‐and‐mouth disease virus to T. reesei. This tool allows multiple independent genes to be transcribed as a single mRNA. Upon translation, the 2A peptide sequence causes a ‘ribosomal skip’ between its two C‐terminal amino acids and generates two independent gene products. A target protein can therefore be cotranslated with a readily testable marker protein, and colonies with desired expression levels can so be identified. The authors tested this by coexpression of CEL7A and an enhanced green fluorescent protein as a marker and obtained similar levels of expression of both proteins. This system therefore offers an efficient strategy to test the expression of heterologous proteins in T. reesei, and provides a novel platform for multi‐protein‐expression in approximately equimolar ratios using a single polycistronic gene expression cassette.
A further essential tool needed for strain engineering is promoters that allow high constitutive expression without dependence on special physiological conditions. An important issue is thereby that the transcriptions factors that bind to the respective regulatory elements in these promoters are present in sufficient quantities in the cells because their use would otherwise be limiting the expression by titration effects. Table 3 summarizes such promoters and their use in T. reesei.
Table 3.
Trichoderma reesei promoters used
| Promoter | Condition | ||
|---|---|---|---|
| Constitutive expression | Glycolytic genes (pki1, gpd1, pgd1, eno1, pdc1 | Li et al. (2012); Seiboth, Karimi et al., (2012); Wang et al. (2014); Linger et al. (2015) | |
| cre1:xyr1 hybrid | Zhang et al. (2017) | ||
| cDNA1 | Uzbas et al. (2012) | ||
| tef1 | Seiboth, Karimi et al. (2012) | ||
| Inducible expressiona | tcu1 copper transporter | Copper deficiency | Lv et al. (2015) |
| TauD‐dioxigenase | Methionine deficiency | Bischof et al. (2015) | |
Promoters of cellulase and xylanase genes are not listed.
Also, there is a demand for promoters that could be temporarily turned either on or off. Traditionally, the T. reesei cbh1 and xyn1 promoters (of the genes encoding the cellobiohydrolase CEL7A and the endo‐β‐1,4‐xylanase XYN1, respectively) have been used for this purpose, but the respective inducers or repressors also act on other cellulases and xylanases and are therefore not sufficiently specific (for review see Bischof and Seiboth, 2014). To solve this problem, Bischof et al. (2015) introduced the promoter of a gene encoding Trire2:123979 (a putative dioxygenase of unknown function) which in T. reesei can be strongly repressed by addition of 0.1 mM l‐methionine. An alternative strategy was presented by Lv et al. (2015), using the promoter of the gene encoding the copper transporter TCU1 (Trire2:52315), which can be repressed by Cu2+ ions of > 0.2 μM. This system was recently used to manipulate the expression of the histone acetyltransferase GCN5 (Trire2:64680) in T. reesei (Zheng et al., 2016).
Towards the development of an inducible expression system, Zhang et al. (2016a) constructed a modular synthetic regulator by fusing the gene fragments encoding the DNA‐binding domains of yeast Gal4, the light responsive domain of N. crassa Vivid, and the Herpex simplex VP16 transactivation domains. The fusion gene was put under the control of a synthetic promoter carrying five copies of the Gal4 consensus binding site. It was successfully expressed in T. reesei in the presence of light. However, the operation of this system at an industrial scale with high‐density suspensions of cellulosic substrates and fungal mycelium has not been tested yet.
Conclusions
The potential of lignocellulosic biomass as renewable feedstock for substituting a significant fraction of today's fossil fuel consumption has been an attractive theory for many years. In recent years, the first industrial‐scale cellulosic ethanol plants have started to operate. Yet competition with technologies based on fossil resources is still hampered by the high costs for the pretreatment and enzymatic saccharification steps due to the natural recalcitrance of lignocellulosic biomass. In this article, we have focused on saccharification, and discussed how the production and properties of the cellulases from the major industrial producer T. reesei could be changed towards a better process economy. In this regard, it is interesting to note that – despite the amount of work dedicated to the understanding of the catalytic mechanisms of some of the major cellulases – comparatively little work has been published that shows whether the introduction of activity increasing mutations indeed lead to strains that produce better enzyme preparations. Thereby it should also be borne in mind that cellulases function at a solid–liquid interface of a physically and chemically heterogeneous substrate, i.e. native or pretreated plant cell walls, which clearly impact the activity of the enzyme, and makes the enzyme dependent on other enzyme partners. Finally, it is still an open question whether (and if so why) the multiplicity of some cellulases (e.g. the endo‐β‐1,4‐glucanases) and the LPMOs are essential for the overall hydrolysis of commercially used lignocellulose substrates. Answers to these questions are needed for genetic engineering of T. reesei strains to produce more active cellulase mixtures.
Engineering of cellulase production is a related, yet different issue: industrial strains producing extremely high cellulase concentrations (over 120 g l−1 total protein) have been reported (Gupta et al., 2016a), and it is worth of discussion whether a further increase (or shortening of the fermentation time needed to achieve this) is feasible and required. Smaller companies or newcomers may, however, want to construct their own producer strain, and can now select from a variety of conditions for this purpose. Also, the carbon source for production of the enzymes needs not to be the same that is used for saccharification, and the producer strain, therefore, may have to be tailored to form cellulases on media not containing lignocellulose or containing a mixture of lignocellulose and other carbon sources. Unfortunately, most of the alterations achieved by metabolic engineering, promoter or transcription factor engineering and epigenetic engineering have been tested only under laboratory conditions, frequently using minimal media, and by only measuring the relative abundance of cellulase mRNA. The presence of a heterogenous polymer and the chemical changes that occur in the medium during its hydrolysis will likely have a strong impact on the function of these engineered strains. It is therefore difficult to predict whether the introduced mutation would indeed by beneficial for the industrial process. We strongly recommend that such engineered strains should always also be tested on proper lignocellulose substrates. In addition, an in vitro saccharification experiment with the enzymes produced in these cultivations should also be included.
Conflict of interest
None declared.
Supporting information
Table S1. 100 best hits in a blastp analysis* using the H. insolens cellobiose dehydrogenase as a query.
Acknowledgements
The authors acknowledge support from the Austrian Science Fund FWF to CPK (I‐1249) and ISD (P25613‐B20).
Microbial Biotechnology (2017) 10(6), 1485–1499
Funding information
Austrian Science Foundation (Grant/Award Number: ‘I‐1214’,‘P25613‐B2’).
References
- Aghcheh, R.K. , and Kubicek, C.P. (2015) Epigenetics as an emerging tool for improvement of fungal strains used in biotechnology. Appl Microbiol Biotechnol 99: 6167–6181. [DOI] [PubMed] [Google Scholar]
- Álvarez, C. , Reyes‐Sosa, F.M. , and Díez, B. (2016) Enzymatic hydrolysis of biomass from wood. Microb Biotechnol 9: 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro, N. , Ilmén, M. , Saloheimo, A. , and Penttilä, M. (2003) ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl Environ Microbiol 69: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers, A.R. , Ayers, S.B. , and Eriksson, K.E. (1978) Cellobiose oxidase, purification and partial characterization of a hemoprotein from Sporotrichum pulverulentum . Eur J Biochem 90: 171–181. [DOI] [PubMed] [Google Scholar]
- Bae, J. , Morisaka, H. , Kuroda, K. , and Ueda, M. (2013) Cellulosome complexes: natural biocatalysts as arming microcompartments of enzymes. J Mol Microbiol Biotechnol 23: 370–378. [DOI] [PubMed] [Google Scholar]
- Beckham, G.T. , Ståhlberg, J. , Knott, B.C. , Himmel, M.E. , Crowley, M.F. , Sandgren, M. , et al (2014) Towards a molecular‐level theory of carbohydrate processivity in glycoside hydrolases. Curr Opin Biotechnol 27: 96–106. [DOI] [PubMed] [Google Scholar]
- Bergès, T. , Barreau, C. , Peberdy, J.F. , and Boddy, L.M. (1993) Cloning of an Aspergillus niger invertase gene by expression in Trichoderma reesei . Curr Genet 24: 53–59. [DOI] [PubMed] [Google Scholar]
- Bey, M. , Berrin, J.G. , Poidevin, L. , and Sigoillot, J.C. (2011) Heterologous expression of Pycnoporus cinnabarinus cellobiose dehydrogenase in Pichia pastoris and involvement in saccharification processes. Microb Cell Fact 10: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof, R. , and Seiboth, B. (2014) Molecular tools for strain improvement of Trichoderma spp In Biotechnology and Biology of Trichoderma. Gupta V.G., Schmoll M., Herrera‐Estrella A., Upadhyay R.S., Druzhinina I., and Tuohy M. (eds). Oxford: Elsevier, pp. 179–191. [Google Scholar]
- Bischof, R.H. , Horejs, J. , Metz, B. , Gamauf, C. , Kubicek, C.P. , and Seiboth, B. (2015) L‐Methionine repressible promoters for tuneable gene expression in Trichoderma reesei . Microb Cell Fact 14: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof, R.H. , Ramoni, J. , and Seiboth, B. (2016) Cellulases and beyond: the first 70 years of the enzyme producer Trichoderma reesei . Microb Cell Fact 15: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer‐Schuette, S.E. , Brown, S.D. , Sander, K.B. , Bayer, E.A. , Kataeva, I. , Zurawski, J.V. , et al (2014) Thermophilic lignocellulose deconstruction. FEMS Microbiol Rev 38: 393–448. [DOI] [PubMed] [Google Scholar]
- Brethauer, S. , and Studer, M.H. (2015) Biochemical conversion processes of lignocellulosic biomass to fuels and chemicals – a review. Chimia (Aarau) 69: 572–581. [DOI] [PubMed] [Google Scholar]
- Cao, Y. , Zheng, F. , Wang, L. , Zhao, G. , Chen, G. , Zhang, W. , and Liu, W. (2017) Rce1, a novel transcriptional repressor, regulates cellulase gene expression by antagonizing the transactivator Xyr1 in Trichoderma reesei . Mol Microbiol. doi: 10.1111/mmi.13685 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Capolupo, L. , and Faraco, V. (2016) Green methods of lignocellulose pretreatment for biorefinery development. Appl Microbiol Biotechnol 100: 9451–9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo, S. , Barth, D. , Arvas, M. , Pakula, T.M. , Pitkänen, E. , Blomberg, P. , et al (2016) Whole‐genome metabolic model of Trichoderma reesei built by comparative reconstruction. Biotechnol Biofuels 9: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilton, I.J. , Delaney, C.E. , Barham‐Morris, J. , Fincham, D.A. , Hooley, P. , and Whitehead, M.P. (2008) The Aspergillus nidulans stress response transcription factor StzA is ascomycete‐specific and shows species‐specific polymorphisms in the C‐terminal region. Mycol Res 112: 1435–1446. [DOI] [PubMed] [Google Scholar]
- Chokhawala, H.A. , Roche, C.M. , Kim, T.‐W. , Atreya, M.E. , Vegesna, N. , Dana, C.M. , et al (2015) Mutagenesis of Trichoderma reesei endoglucanase I: impact of expression host on activity and stability at elevated temperatures. BMC Biotechnol 15: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chundawat, S.P. , Beckham, G.T. , Himmel, M.E. , and Dale, B.E. (2011) Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu Rev Chem Biomol Eng 2: 121–145. [DOI] [PubMed] [Google Scholar]
- Colabardini, A.C. , Valkonen, M. , Huuskonen, A. , Siika‐Aho, M. , Koivula, A. , Goldman, G.H. , and Saloheimo, M. (2016) Expression of two novel β‐glucosidases from Chaetomium atrobrunneum in Trichoderma reesei and characterization of the heterologous protein products. Mol Biotechnol 58: 821–831. [DOI] [PubMed] [Google Scholar]
- Coradetti, S.T. , Craig, J.P. , Xiong, Y. , Shock, T. , Tian, C. , and Glass, N.L. (2012) Conserved and essential transcription factors for cellulase gene expression in ascomycete fungi. Proc Natl Acad Sci USA 109: 7397–7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino, Y. and D'Aniello, S. (2017) Molecular basis, applications and challenges of CRISPR/Cas9: a continuously evolving tool for genome editing. Brief Funct Genomics. doi: 10.1093/bfgp/elw038 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Dashtban, M. , and Qin, W. (2012) Overexpression of an exotic thermotolerant β‐glucosidase in Trichoderma reesei and its significant increase in cellulolytic activity and saccharification of barley straw. Microb Cell Fact 11: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, A. , Goedegebuur, F. , Gualfetti, P. , Mitchinson, C. , Neefe, P. , Sandgren, M. , et al (2007) Novel variant Hypocrea jecorina CBH1 cellulases. US patent 2007/0173431
- Derntl, C. , Gudynaite‐Savitch, L. , Calixte, S. , White, T. , Mach, R.L. , and Mach‐Aigner, A.R. (2013) Mutation of the Xylanase regulator 1 causes a glucose blind hydrolase expressing phenotype in industrially used Trichoderma strains. Biotechnol Biofuels 6: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derntl, C. , Kiesenhofer, D.P. , Mach, R.L. , and Mach‐Aigner, A.R. (2015) Novel strategies for genomic manipulation of Trichoderma reesei with the purpose of strain engineering. Appl Environ Microbiol 81: 6314–6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druzhinina, I.S. , Kopchinskiy, A.G. , Kubicek, E.M. , and Kubicek, C.P. (2016) A complete annotation of the chromosomes of the cellulase producer Trichoderma reesei provides insights in gene clusters, their expression and reveals genes required for fitness. Biotechnol Biofuels 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druzhinina, I. S. , and Kubicek, C. P. (2016) Familiar stranger: ecologicagenomics of the model saprotroph and industrial enzyme producer Trichoderma reesei breaks the stereotypes. Adv Appl Microbiol 95: 69–147. [DOI] [PubMed] [Google Scholar]
- Dubey, A.K. , Barad, S. , Luria, N. , Kumar, D. , Espeso, E.A. , and Prusky, D.B. (2016) Cation‐stress‐responsive transcription factors SltA and CrzA regulate morphogenetic processes and pathogenicity of Colletotrichum gloeosporioides . PLoS ONE 11: e0168561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eibinger, M. , Ganner, T. , Bubner, P. , Rosker, S. , Kracher, D. , Haltrich, D. , et al (2014) Cellulose surface degradation by a lytic polysaccharide monooxygenase and its effect on cellulase hydrolytic efficiency. J Biol Chem 289: 35929–35938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eibinger, M. , Sigl, K. , Sattelkow, J. , Ganner, T. , Ramoni, J. , Seiboth, B. , et al (2016) Functional characterization of the native swollenin from Trichoderma reesei: study of its possible role as C1 factor of enzymatic lignocellulose conversion. Biotechnol Biofuels 9: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellilä, S. , Fonseca, L. , Uchima, C. , Cota, J. , Goldman, G.H. , Saloheimo, M. , et al (2017) Development of a low‐cost cellulase production process using Trichoderma reesei for Brazilian biorefineries. Biotechnol Biofuels 10: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eveleigh, D.E. , and Montenecourt, B.S. (1979) Increasing yields of extracellular enzymes. Adv Appl Microbiol 25: 57–74. [DOI] [PubMed] [Google Scholar]
- Furukawa, T. , Shida, Y. , Kitagami, N. , Ota, Y. , Adachi, M. , Nakagawa, S. , Shimada, R. , Kato, M. , Kobayashi, T. , Okada, H. , Ogasawara, W. , and Morikawa, Y. (2008) Identification of the cis‐acting elements involved in regulation of xylanase III gene expression in Trichoderma reesei PC‐3‐7. Fungal Genet Biol 45: 1094–1102. [DOI] [PubMed] [Google Scholar]
- Galante, Y.M. , De Conti, A. , and Monteverdi, R. (1998) Application of Trichoderma enzymes in the textile industry In Trichoderma and Gliocladium: Enzymes, Biological Control and Commercial Applications. Harman G.E., and Kubicek C.P. (eds). London, UK: Taylor and Francis Ltd., pp. 311–325. [Google Scholar]
- Gruber, F. , Visser, J. , Kubicek, C.P. , and De Graaff, L. (1990) Cloning of the Trichoderma reesei pyrG gene and its use as a homologous marker for a high‐frequency transformation system. Curr Genet 18: 451–456. [DOI] [PubMed] [Google Scholar]
- Gruno, M. , Valjamae, P. , Pettersson, G. , and Johansson, G. (2004) Inhibition of the Trichoderma reesei cellulases by cellobiose is strongly dependent on the nature of the substrate. Biotechnol Bioeng 86: 503–511. [DOI] [PubMed] [Google Scholar]
- Guangtao, Z. , Hartl, L. , Schuster, A. , Polak, S. , Schmoll, M. , Wang, T. , et al (2009) Gene targeting in a nonhomologous end joining deficient Hypocrea jecorina . J Biotechnol 139: 146–151. [DOI] [PubMed] [Google Scholar]
- Guo, B. , Sato, N. , Biely, P. , Amano, Y. , and Nozaki, K. (2016) Comparison of catalytic properties of multiple β‐glucosidases of Trichoderma reesei . Appl Microbiol Biotechnol 100: 4959–4968. [DOI] [PubMed] [Google Scholar]
- Gupta, V.K. , Kubicek, C.P. , Berrin, J.G. , Wilson, D.W. , Couturier, M. , Berlin, A. , et al (2016a) Fungal enzymes for bio‐products from sustainable and waste biomass. Trends Biochem Sci 41: 633–645. [DOI] [PubMed] [Google Scholar]
- Gupta, V.K. , Steindorff, A.S. , de Paula, R.G. , Silva‐Rocha, R. , Mach‐Aigner, A.R. , Mach, R.L. , and Silva, R.N. (2016b) The post‐genomic era of Trichoderma reesei: What's next? Trends Biotechnol 34: 970–982. [DOI] [PubMed] [Google Scholar]
- Gusakov, A.V. (2011) Alternatives to Trichoderma reesei in biofuel production. Trends Biotechnol 29: 419–425. [DOI] [PubMed] [Google Scholar]
- Häkkinen, M. , Valkonen, M.J. , Westerholm‐Parvinen, A. , Aro, N. , Arvas, M. , Vitikainen, M. , et al (2014) Screening of candidate regulators for cellulase and hemicellulase production in Trichoderma reesei and identification of a factor essential for cellulase production. Biotechnol Biofuels 7: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkki, A. , Mäntylä, A. , Penttilä, M. , Muttilainen, S. , Bühler, R. , Suominen, P. , et al (1991) Genetic engineering of Trichoderma to produce strains with novel cellulase profiles. Enzyme Microb Technol 13: 227–233. [DOI] [PubMed] [Google Scholar]
- Harris, P.V. , Xu, F. , Kreel, N.E. , Kang, C. , and Fukuyama, S. (2014) New enzyme insights drive advances in commercial ethanol production. Curr Opin Chem Biol 19: 162–170. [DOI] [PubMed] [Google Scholar]
- Henriksson, G. , Johansson, G. , and Pettersson, G. (2000) A critical review of cellobiose dehydrogenases. J Biotechnol 78: 93–113. [DOI] [PubMed] [Google Scholar]
- Horn, S.J. , Vaaje‐Kolstad, G. , Westereng, B. , and Eijsink, V.G. (2012) Novel enzymes for the degradation of cellulose. Biotechnol Biofuels 5: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston, K. , Tucker, M.R. , Chowdhury, J. , Shirley, N. , and Little, A. (2016) The plant cell wall: a complex and dynamic structure as revealed by the responses of genes under stress conditions. Front Plant Sci 7: 984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J. , Chandra, R. , Arantes, V. , Gourlay, K. , van Dyk, J.S. , and Saddler, J.N. (2015) The addition of accessory enzymes enhances the hydrolytic performance of cellulase enzymes at high solid loadings. Bioresour Technol 186: 149–153. [DOI] [PubMed] [Google Scholar]
- Ilmén, M. , Thrane, C. , and Penttilä, M. (1996) The glucose repressor gene cre1 of Trichoderma: isolation and expression of a full‐length and a truncated mutant form. Mol Gen Genet 251: 451–460. [DOI] [PubMed] [Google Scholar]
- Johansen, K.S. (2016a) Lytic polysaccharide monooxygenases: the microbial power tool for lignocellulose degradation. Trends Plant Sci 21: 926–936. [DOI] [PubMed] [Google Scholar]
- Johansen, K.S. (2016b) Discovery and industrial applications of lytic polysaccharide mono‐oxygenases. Biochem Soc Trans 44: 143–149. [DOI] [PubMed] [Google Scholar]
- Karimi Aghcheh, R. , Németh, Z. , Atanasova, L. , Fekete, E. , Paholcsek, M. , Sandor, E. , et al (2014) The VELVET A orthologue VEL1 of Trichoderma reesei regulates fungal development and is essential for cellulase gene expression. PLoS ONE 9: e112799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson, J. , Saloheimo, M. , Siika‐Aho, M. , Tenkanen, M. , Penttilä, M. , and Tjerneld, F. (2001) Homologous expression and characterization of Cel61A (EG IV) of Trichoderma reesei . Eur J Biochem 268: 6498–6507. [DOI] [PubMed] [Google Scholar]
- Karshikoff, A. , Nilsson, L. , and Ladenstein, R. (2015) Rigidity versus flexibility: the dilemma of understanding protein thermal stability. FEBS J 282: 3899–3917. [DOI] [PubMed] [Google Scholar]
- Klein‐Marcuschamer, D. , Oleskowicz‐Popiel, P. , Simmons, B.A. , and Blanch, H.W. (2012) The challenge of enzyme cost in the production of lignocellulosic biofuels. Biotechnol Bioeng 109: 1083–1087. [DOI] [PubMed] [Google Scholar]
- Kubicek, C.P. (1981) Release of carboxymethyl‐cellulase and β‐glucosidase from the cell walls of Trichoderma reesei . Eur J Appl Microbiol Biotechnol 13: 226–231. [Google Scholar]
- Kubicek, C.P. (2013) Fungi and Lignocellulosic Biomass. Hoboken, NJ, USA: John Wiley & Sons, Inc. [Google Scholar]
- Kubicek, C.P. , and Kubicek, E.M. (2016) Enzymatic deconstruction of plant biomass by fungal enzymes. Curr Opin Chem Biol 35: 51–57. [DOI] [PubMed] [Google Scholar]
- Kubicek‐Pranz, E.M. , Gruber, F. , and Kubicek, C.P. (1991) Transformation of Trichoderma reesei with the cellobiohydrolase II gene as a means for obtaining strains with increased cellulase production and specific activity. J Biotechnol 20: 83–94. [Google Scholar]
- Kurašin, M. , and Väljamäe, P. (2011) Processivity of cellobiohydrolases is limited by the substrate. J Biol Chem 286: 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz, S. E. , Goedegebuur, F. , Hommes, R. , Kaper, T. , Kelemen, B. R. , Mitchinson, C. , Wallace, L. , Ståhlberg, J. , and Larenas, E. A. (2010) Hypocrea jecorina CEL6A protein engineering. Biotechnol Biofuels 3: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levasseur, A. , Drula, E. , Lombard, V. , Coutinho, P.M. , and Henrissat, B. (2013) Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels 6: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B. and Walton, J.D. (2017) Functional diversity for biomass deconstruction in family 5 subfamily 5 (GH5_5) of fungal endo‐β1,4‐glucanases. Appl Microbiol Biotechnol 101: 4093–4101. [DOI] [PubMed] [Google Scholar]
- Li, J. , Wang, J. , Wang, S. , Xing, M. , Yu, S. , and Liu, G. (2012) Achieving efficient protein expression in Trichoderma reesei by using strong constitutive promoters. Microb Cell Fact 11: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Wang, L. , Zhou, R. , and Mu, Y. (2015) Ionic liquid induced inactivation of cellobiohydrolase I from Trichoderma reesei . Green Chem 17: 1618–1625. [Google Scholar]
- Lichius, A. , Seidl‐Seiboth, V. , Seiboth, B. , and Kubicek, C.P. (2014) Nucleo‐cytoplasmic shuttling dynamics of the transcriptional regulators XYR1 and CRE1 under conditions of cellulase and xylanase gene expression in Trichoderma reesei . Mol Microbiol 94: 1162–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linger, J.G. , Taylor, L.E. 2nd , Baker, J.O. , Vander Wall, T. , Hobdey, S.E. , Podkaminer, K. , et al (2015) A constitutive expression system for glycosyl hydrolase family 7 cellobiohydrolases in Hypocrea jecorina . Biotechnol Biofuels 8: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, R. , Chen, L. , Jiang, Y. , Zhou, Z. , and Zou, G. (2015) Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov 1: 15007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, K. , Dong, Y. , Wang, F. , Jiang, B. , Wang, M. , and Fang, X. (2016) Regulation of cellulase expression, sporulation, and morphogenesis by velvet family proteins in Trichoderma reesei . Appl Microbiol Biotechnol 100: 769–779. [DOI] [PubMed] [Google Scholar]
- Lombard, V. , Golaconda Ramulu, H. , Drula, E. , Coutinho, P.M. and Henrissat, B. (2014) The carbohydrate‐active enzymes database (CAZy) in 2013. Nucleic Acids Res 42: D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, X. , Zheng, F. , Li, C. , Zhang, W. , Chen, G. , and Liu, W. (2015) Characterization of a copper responsive promoter and its mediated overexpression of the xylanase regulator 1 results in an induction‐independent production of cellulases in Trichoderma reesei . Biotechnol Biofuels 8: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L. , Zhang, J. , Zou, G. , Wang, C. , and Zhou, Z. (2011) Improvement of cellulase activity in Trichoderma reesei by heterologous expression of a beta‐glucosidase gene from Penicillium decumbens . Enzyme Microb Technol 49: 366–371. [DOI] [PubMed] [Google Scholar]
- Mach, R.L. , Strauss, J. , Zeilinger, S. , Schindler, M. , and Kubicek, C.P. (1996) Carbon catabolite repression of xylanase I (xyn1) gene expression in Trichoderma reesei . Mol Microbiol 21: 1273–1281. [DOI] [PubMed] [Google Scholar]
- Mello‐de‐Sousa, T.M. , Gorsche, R. , Rassinger, A. , Poças‐Fonseca, M.J. , Mach, R.L. , and Mach‐Aigner, A.R. (2014) A truncated form of the carbon catabolite repressor 1 increases cellulase production in Trichoderma reesei . Biotechnol Biofuels 7: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello‐de‐Sousa, T.M. , Rassinger, A. , Pucher, M.E. , dos Santos Castro, L. , Persinoti, G.F. , Silva‐Rocha, R. , et al (2015) The impact of chromatin remodelling on cellulase expression in Trichoderma reesei . BMC Genom 16: 588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello‐de‐Sousa, T.M. , Rassinger, A. , Derntl, C. , Poças‐Fonseca, M.J. , Mach, R.L. , and Mach‐Aigner, R.A. (2016) The relation between promoter chromatin status, XYR1 and cellulase expression in Trichoderma reesei . Curr Genomics 17: 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern, I. , Powlowski, J. , and Tsang, A. (2014) Fungal cellulose degradation by oxidative enzymes: from dysfunctional GH61 family to powerful lytic polysaccharide monooxygenase family. Brief Funct Genomics 13: 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, G. , Várnai, A. , Johansen, K.S. , Eijsink, V.G.H. , and Horn, S.J. (2015) Harnessing the potential of LPMO‐containing cellulase cocktails poses new demands on processing conditions. Biotechnol Biofuels 8: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakari‐Setälä, T. , Paloheimo, M. , Kallio, J. , Vehmaanperä, J. , Penttilä, M. , and Saloheimo, M. (2009) Genetic modification of carbon catabolite repression in Trichoderma reesei for improved protein production. Appl Environ Microbiol 75: 4853–4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa, H. , Kawai, T. , Ida, N. , Shida, Y. , Kobayashi, Y. , Okada, H. , et al (2012) Construction of a recombinant Trichoderma reesei strain expressing Aspergillus aculeatus β‐glucosidase 1 for efficient biomass conversion. Biotechnol Bioeng 109: 92–99. [DOI] [PubMed] [Google Scholar]
- Ouedraogo, J.P. , Arentshorst, M. , Nikolaev, I. , Barends, S. , and Ram, A.F. (2015) I‐SceI‐mediated double‐strand DNA breaks stimulate efficient gene targeting in the industrial fungus Trichoderma reesei . Appl Microbiol Biotechnol 99: 10083–10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouedraogo, J.P. , Arentshorst, M. , Nikolaev, I. , Barends, S. , and Ram, A.F. (2016) I‐SceI enzyme mediated integration (SEMI) for fast and efficient gene targeting in Trichoderma reesei . J Biotechnol 222: 25–28. [DOI] [PubMed] [Google Scholar]
- Pakula, T.M. , Nygren, H. , Barth, D. , Heinonen, M. , Castillo, S. , Penttilä, M. , and Arvas, M. (2016) Genome wide analysis of protein production load in Trichoderma reesei . Biotechnol Biofuels 9: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne, C.M. , Knott, B.C. , Mayes, H.B. , Hansson, H. , Himmel, M.E. , Sandgren, M. , et al (2015) Fungal cellulases. Chem Rev 115: 1308–1448. [DOI] [PubMed] [Google Scholar]
- Penttilä, M. , Nevalainen, H. , Rättö, M. , Salminen, E. , and Knowles, J. (1987) A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei . Gene 61: 155–164. [DOI] [PubMed] [Google Scholar]
- Peterson, R. , and Nevalainen, H. (2012) Trichoderma reesei RUT‐C30—thirty years of strain improvement. Microbiology 158: 58–68. [DOI] [PubMed] [Google Scholar]
- Pitkänen, E. , Jouhten, P. , Hou, J. , Syed, M.F. , Blomberg, P. , Kludas, J. , et al (2014) Comparative genome‐scale reconstruction of gapless metabolic networks for present and ancestral species. PLoS Comput Biol 10: 1003465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy, T. , Margeot, A. , Linke, R. , Atanasova, L. , Fekete, E. , Sandor, E. , et al (2011) The CRE1 carbon catabolite repressor of the fungus Trichoderma reesei: a master regulator of carbon assimilation. BMC Genom 12: 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabemanolontsoa, H. , and Saka, S. (2016) Various pretreatments of lignocellulosics. Bioresour Technol 199: 83–91. [DOI] [PubMed] [Google Scholar]
- Rahikainen, J. , Moilanen, U. , Nurmi‐Rantala, S. , Lappas, A. , Koivula, A. , Viikarim, L. , and Kruus, M. (2013) Effect of temperature on lignin‐derived inhibition studied with three structurally different cellobiohydrolases. Bioresour Technol 146: 118–125. [DOI] [PubMed] [Google Scholar]
- Rath, H. , Messner, R. , Kosma, P. , Altmann, F. , März, L. , et al (1995) The α‐D‐mannan core of a complex cell‐wall heteroglycan of Trichoderma reesei is responsible for β‐glucosidase activation. Arch Microbiol 164: 414–419. [PubMed] [Google Scholar]
- Reese, E.T. , Siu, R.G. , and Levinson, H.S. (1950) The biological degradation of soluble cellulose derivatives and its relationship to the mechanism of cellulose hydrolysis. J Bacteriol 59: 485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries, L. , Belshaw, N.J. , Ilmén, M. , Penttilä, M.E. , Alapuranen, M. , and Archer, D.B. (2014) The role of CRE1 in nucleosome positioning within the cbh1 promoter and coding regions of Trichoderma reesei . Appl Microbiol Biotechnol 98: 749–762. [DOI] [PubMed] [Google Scholar]
- Sandgren, M. , Gualfetti, P.J. , Shaw, A. , Gross, L.S. , Saldajeno, M. , Day, A.G. , et al (2003) Comparison of family 12 glycoside hydrolases and recruited substitutions important for thermal stability. Protein Sci 12: 848–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarikaya‐Bayram, Ö. , Palmer, J.M. , Keller, N. , Braus, G.H. , and Bayram, Ö. (2015) One Juliet and four Romeos: VeA and its methyltransferases. Front Microbiol 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster, A. , Bruno, K.S. , Collett, J.R. , Baker, S.E. , Seiboth, B. , Kubicek, C.P. , and Schmoll, M. (2012) A versatile toolkit for high throughput functional genomics with Trichoderma reesei . Biotechnol Biofuels 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiboth, B. , Karimi, R. A. , Phatale, P. A. , Linke, R. , Hartl, L. , Sauer, D. G. , Smith, K. M. , Baker, S. E. , Freitag, M. , and Kubicek, C. P. (2012) The putative protein methyltransferase LAE1 controls cellulase gene expression in Trichoderma reesei . Mol Microbiol 84: 1150–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Delabona, P. , Rodrigues, G.N. , Zubieta, M.P. , Ramoni, J. , Codima, C.A. , Lima, D.J. , et al (2017) The relation between xyr1 overexpression in Trichoderma harzianum and sugarcane bagasse saccharification performance. J Biotechnol 246, 24–32. [DOI] [PubMed] [Google Scholar]
- Silveira, M.H. , Morais, A.R. , da Costa Lopes, A.M. , Olekszyszen, D.N. , Bogel‐Łukasik, R. , Andreaus, J. , and Pereira‐Ramos, I. (2015) Current pretreatment technologies for the development of cellulosic ethanol and biorefineries. Chemsuschem 8: 3366–3390. [DOI] [PubMed] [Google Scholar]
- Smith, J.L. , Bayliss, F.T. , and Ward, M. (1991) Sequence of the cloned pyr4 gene of Trichoderma reesei and its use as a homologous selectable marker for transformation. Curr Genet 19: 27–33. [DOI] [PubMed] [Google Scholar]
- Smith, M.A. , Romero, P.A. , Wu, T. , Brustad, E.M. , and Arnold, F.H. (2013a) Chimeragenesis of distantly‐related proteins by noncontiguous recombination. Protein Sci 22: 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M.A. , Bedbrook, C.N. , Wu, T. , and Arnold, F.H. (2013b) Hypocrea jecorina cellobiohydrolase I stabilizing mutations identified using noncontiguous recombination. ACS Synth Biol 2: 690–696. [DOI] [PubMed] [Google Scholar]
- Spielvogel, A. , Findon, H. , Arst, H.N. , Araújo‐Bazán, L. , Hernández‐Ortíz, P. , Stahl, U. , et al (2008) Two zinc finger transcription factors, CrzA and SltA, are involved in cation homoeostasis and detoxification in Aspergillus nidulans . Biochem J 414: 419–429. [DOI] [PubMed] [Google Scholar]
- Stahlberg, J. , Johansson, G. , and Pettersson, G. (1993) Trichoderma reesei has no true exo‐cellulase: all intact and truncated cellulases produce new reducing end groups on cellulose. Biochim Biophys Acta 1157: 107–113. [DOI] [PubMed] [Google Scholar]
- Steiger, M.G. (2013) Molecular tools in Trichoderma genetic studies In Trichoderma: Biology and Applications. Mukherjee P.K., Horwitz B.A., Singh U.S., Mukherjee M., and Schmoll M. (eds). Oxfordshire: CABI, pp. 128–143. [Google Scholar]
- Steiger, M.G. , Vitikainen, M. , Uskonen, P. , Brunner, K. , Adam, G. , Pakula, T. , et al (2011) Transformation system for Hypocrea jecorina (Trichoderma reesei) that favours homologous integration and employs reusable bidirectionally selectable markers. Appl Environ Microbiol 77: 114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker, A. R. , Grosstessner‐Hain, K. , Würleitner, E. , and Mach, R. L. (2006) Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and D‐xylose metabolism in Hypocrea jecorina . Eukaryot Cell 5: 2128–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, V. , Schuster, L.A. , Moore, K.T. , Taylor, L.E. 2nd , Baker, J.O. , Vander Wall, T.A. , et al (2017) Linger JG, A versatile 2A peptide‐based bicistronic protein expressing platform for the industrial cellulase producing fungus, Trichoderma reesei . Biotechnol Biofuels 10: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, F.F. , Hong, J. , Hu, J. , Saddler, J.N. , Fang, X. , Zhang, Z. , and Shen, S. (2015) Accessory enzymes influence cellulase hydrolysis of the model substrate and the realistic lignocellulosic biomass. Enzyme Microb Technol 79–80: 42–48. [DOI] [PubMed] [Google Scholar]
- Treebupachatsakul, T. , Shioya, K. , Nakazawa, H. , Kawaguchi, T. , Morikawa, Y. , Shida, Y. , et al (2015) Utilization of recombinant Trichoderma reesei expressing Aspergillus aculeatus β‐glucosidase I (JN11) for a more economical production of ethanol from lignocellulosic biomass. J Biosci Bioeng 120: 657–665. [DOI] [PubMed] [Google Scholar]
- Ugolini, S. , and Bruschi, C.V. (1996) The red/white colony color assay in the yeast Saccharomyces cerevisiae: epistatic growth advantage of white ade8‐18, ade2 cells over red ade2 cells. Curr Genet 30: 485–492. [DOI] [PubMed] [Google Scholar]
- Uju, S.Y. , Nakamoto, A. , Goto, M. , Tokuhara, W. , Noritake, Y. , Katahira, S. , et al (2012) Short time ionic liquids pretreatment on lignocellulosic biomass to enhance enzymatic saccharification. Bioresour Technol 103: 446–452. [DOI] [PubMed] [Google Scholar]
- Uzbas, F. , Sezerman, U. , Hartl, L. , Kubicek, C.P. , and Seiboth, B. (2012) A homologous production system for Trichoderma reesei secreted proteins in a cellulase‐free background. Appl Microbiol Biotechnol 93: 1601–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viikari, L. , Alapuranen, M. , Puranen, T. , Vehmaanperä, J. , and Siika‐Aho, M. (2007) Thermostable enzymes in lignocellulose hydrolysis. Adv Biochem Eng Biotechnol 108: 121–145. [DOI] [PubMed] [Google Scholar]
- Voutilainen, S.P. , Puranen, T. , Siika‐Aho, M. , Lappalainen, A. , Alapuranen, M. , Kallio, J. , et al (2008) Cloning, expression, and characterization of novel thermostable family 7 cellobiohydrolases. Biotechnol Bioeng 101: 515–528. [DOI] [PubMed] [Google Scholar]
- Waddington, C.H. (1942) The epigenotype. Int J Epidemiol 41: 10–13. [DOI] [PubMed] [Google Scholar]
- Wahlstrom, R. , Rahikainen, J. , Kruus, K. , and Suurnakki, A. (2014) Cellulose hydrolysis and binding with Trichoderma reesei Cel5A and Cel7A and their core domains in ionic liquid solutions. Biotechnol Bioeng 111: 726–733. [DOI] [PubMed] [Google Scholar]
- Wang, M. , and Lu, X. (2016) Exploring the synergy between cellobiose dehydrogenase from Phanerochaete chrysosporium and cellulase from Trichoderma reesei . Front Microbiol 7: 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Li, Z. , Fang, X. , Wang, L. , and Qu, Y. (2012) Cellulolytic enzyme production and enzymatic hydrolysis for second‐generation bioethanol production. Adv Biochem Eng Biotechnol 128: 1–24. [DOI] [PubMed] [Google Scholar]
- Wang, W. , Meng, F. , Liu, P. , Yang, S. , and Wie, D. (2014) Construction of a promoter collection for genes co‐expression in filamentous fungus Trichoderma reesei . J Ind Microbiol Biotechnol 41: 1709–1718. [DOI] [PubMed] [Google Scholar]
- Wilson, D.B. (2009) Cellulases and biofuels. Curr Opin Biotechnol 20: 295–299. [DOI] [PubMed] [Google Scholar]
- Xin, Q. , Gong, Y. , Lv, X. , Chen, G. , and Liu, W. (2013) Trichoderma reesei histone acetyltransferase Gcn5 regulates fungal growth, conidiation, and cellulase gene expression. Curr Microbiol 67: 580–589. [DOI] [PubMed] [Google Scholar]
- Xiong, Y. , Sun, J. , and Glass, N.L. (2014) VIB1, a link between glucose signaling and carbon catabolite repression, is essential for plant cell wall degradation by Neurospora crassa . PLoS Genet 10: e1004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z. , and Huang, F. (2014) Pretreatment methods for bioethanol production. Appl Biochem Biotechnol 174: 43–62. [DOI] [PubMed] [Google Scholar]
- Xu, J. , Xiong, P. , and He, B. (2016) Advances in improving the performance of cellulase in ionic liquids for lignocellulose biorefinery. Bioresour Technol 200: 961–970. [DOI] [PubMed] [Google Scholar]
- Xue, X. , Wu, Y. , Qin, X. , Ma, R. , Luo, H. , Su, X. , and Yao, B. (2016) Revisiting overexpression of a heterologous β‐glucosidase in Trichoderma reesei: fusion expression of the Neosartorya fischeri Bgl3A to cbh1 enhances the overall as well as individual cellulase activities. Microb Cell Fact 15: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilinger, S. , Schmoll, M. , Pail, M. , Mach, R.L. , and Kubicek, C.P. (2003) Nucleosome transactions on the Hypocrea jecorina (Trichoderma reesei) cellulase promoter cbh2 associated with cellulase induction. Mol Genet Genomics 270: 46–55. [DOI] [PubMed] [Google Scholar]
- Zhang, G. , Liu, P. , Wie, W. , Wang, X. , Wei, D. , and Wang, W. (2016a) A light‐switchable bidirectional expression system in filamentous fungus Trichoderma reesei . J Biotechnol 240: 85–93. [DOI] [PubMed] [Google Scholar]
- Zhang, F. , Bai, F. , and Zhao, X. (2016b) Enhanced cellulase production from Trichoderma reesei Rut‐C30 by engineering with an artificial zinc finger protein library. Biotechnol J 11: 1282–1290. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Li, Y. , Zhao, X. , and Bai, F. (2017) Constitutive cellulase production from glucose using the recombinant Trichoderma reesei strain overexpressing an artificial transcription activator. Bioresour Technol 223: 317–322. [DOI] [PubMed] [Google Scholar]
- Zheng, F. , Cao, Y. , Lv, X. , Wang, L. , Li, C. , Zhang, W. , et al (2016) A copper‐responsive promoter replacement system to investigate gene functions in Trichoderma reesei: a case study in characterizing SAGA genes. Appl Microbiol Biotechnol 101, 2067–2078. [DOI] [PubMed] [Google Scholar]
- Zou, G. , Shi, S. , Jiang, Y. , van den Brink, J. , de Vries, R.P. , Chen, L. , et al (2012) Construction of a cellulase hyper‐expression system in Trichoderma reesei by promoter and enzyme engineering. Microb Cell Fact 11: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. 100 best hits in a blastp analysis* using the H. insolens cellobiose dehydrogenase as a query.


