Summary
Petroleum‐based plastics have replaced many natural materials in their former applications. With their excellent properties, they have found widespread uses in almost every area of human life. However, the high recalcitrance of many synthetic plastics results in their long persistence in the environment, and the growing amount of plastic waste ending up in landfills and in the oceans has become a global concern. In recent years, a number of microbial enzymes capable of modifying or degrading recalcitrant synthetic polymers have been identified. They are emerging as candidates for the development of biocatalytic plastic recycling processes, by which valuable raw materials can be recovered in an environmentally sustainable way. This review is focused on microbial biocatalysts involved in the degradation of the synthetic plastics polyethylene, polystyrene, polyurethane and polyethylene terephthalate (PET). Recent progress in the application of polyester hydrolases for the recovery of PET building blocks and challenges for the application of these enzymes in alternative plastic waste recycling processes will be discussed.
Introduction
Plastics can be easily moulded into different shapes and forms (Andrady, 2015b,d). Due to their low weight, durability and low production cost, they can be readily manufactured to an expanding range of products used for a variety of civil and industrial applications (Andrady and Neal, 2009; Thompson et al., 2009; Andrady, 2015d). In many areas, they have substituted natural materials as well as paper and glass in most of their former uses (Andrady and Neal, 2009). As a result, plastics have become omnipresent in our daily life. Over the last 50 years, the global production of plastics has continuously increased and reached 322 million tons in 2015 (PlasticsEurope, 2016). In Europe, packaging (39.9%), building and construction (19.7%) and automotive (8.9%) are the leading application sectors for the plastic industry (PlasticsEurope, 2016). The majority of plastics are made from fossil‐based feedstocks (Hopewell et al., 2009; Andrady, 2015d). Polyethylene (PE), polypropylene (PP), polystyrene (PS), polyvinyl chloride (PVC), polyethylene terephthalate (PET) and polyurethane (PUR) are the main types of plastics, which correspond to over 80% of the total demand in Europe (PlasticsEurope, 2016; Fig. 1). It has been estimated that nearly 8% of the total global fossil fuel production is utilized as raw materials or to provide energy for the manufacturing of plastics (Hopewell et al., 2009; Andrady, 2015d).
Figure 1.
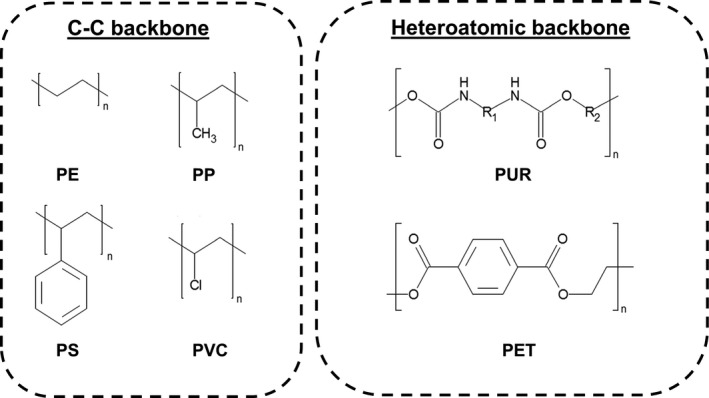
Backbone structural formula of widely used petroleum‐based plastics (PlasticsEurope, 2016).
As a result of the widespread use and consumption of plastic products, up to 25.8 million tons of post‐consumer plastic wastes is annually generated in Europe alone (PlasticsEurope, 2016). Packaging materials or other short‐lived disposable plastic items that are discarded within a year of manufacturing make up nearly 60% of the total plastic waste in Europe (WRAP, 2016). In 2014, 69% of post‐consumer plastic waste in Europe was recovered by recycling and energy generation processes, whereas 31% still ended up in landfills (PlasticsEurope, 2016).
Most of the petroleum‐based plastics have been considered as notably resistant to microbial degradation (Andrady, 1994; Zheng et al., 2005; Mueller, 2006; Tokiwa et al., 2009). The majority of plastics manufactured today are therefore estimated to persist in the environment for a very long time (Hopewell et al., 2009; Andrady, 2015a). In addition, the careless disposal of plastics waste, especially in developing countries, is aggravating the associated environmental problems (Andrady, 1994, 2015a; Rigamonti et al., 2014; Jambeck et al., 2015). The degradation process of plastic waste causing serious environmental damages can be expected to be considerably different in landfills, terrestrial and marine environments (Kyrikou and Briassoulis, 2007; Andrady, 2015a; Gewert et al., 2015). It has been shown that hazardous chemicals released from plastic waste in landfills can contaminate the groundwater (North and Halden, 2013). An increasing amount of plastic waste is also entering marine environments. A calculated up to 12 million tons of plastics produced by coastal countries worldwide has entered the oceans in 2010 and this amount is expected to grow steadily (Jambeck et al., 2015). Plastic pollution has deadly effects on marine mammals from ingestion or getting entangled in plastic debris (Derraik, 2002; Andrady, 2015e; Wilcox et al., 2015; Nelms et al., 2016). Microplastics, partially degraded plastic debris of less than 5 mm in diameter, have been shown to pose an even more serious impact on marine ecosystems by concentrating persistent organic pollutants. These are often hydrophobic compounds with a high affinity to microplastics, thereby entering the food chains when microplastics are ingested by marine wildlife (Andrady, 2011, 2015e; Van Cauwenberghe et al., 2013; Law and Thompson, 2014).
The development of biodegradable plastics is providing a promising alternative to their counterparts made from petrochemicals (Andrady, 2015d; Iwata, 2015). Due to their lower durability and lack of compatibility with existing equipment and end‐of‐life management systems, the scale of production and use of biodegradable plastics are, however, still very limited (Gourmelon, 2015). In addition, not all of the so‐called bioplastics derived from renewable resources are readily biodegradable (Tokiwa et al., 2009; Soroudi and Jakubowicz, 2013; Yates and Barlow, 2013; Andrady, 2015d; Prieto, 2016). They may also persist in the environment for a considerable long time depending on local abiotic factors that facilitate their breakdown and subsequent biodegradation (Swift, 2015; Hopewell et al., 2009; Andrady, 2015a). The enzymatic degradation of biodegradable plastics has been reviewed elsewhere (Tokiwa et al., 2009; Bhardwaj et al., 2013; Banerjee et al., 2014).
The biodegradation of recalcitrant plastics has become a focus of research (for recent reviews, see Eubeler et al., 2010; Gu, 2003; Krueger et al., 2015; Lucas et al., 2008; Shah et al., 2008b; Sivan, 2011; Zheng et al., 2005). The complex process of their biodegradation in the environment has been considered as the result of a combination of many abiotic and biotic factors (Mueller, 2006; Lucas et al., 2008; Sivan, 2011). As schematically shown in Fig. 2, following a deterioration cooperatively accomplished by abiotic factors and microorganisms, the bulk polymer becomes fragmented with more exposed surfaces available for biological attack. Inducible extracellular enzymes play a crucial role in the further depolymerization process of the synthetic polymers (Lucas et al., 2008; Sivan, 2011; Bhardwaj et al., 2013). Plant polymers are the natural substrates for key enzymes capable of attacking the polymer backbones of synthetic plastics. For example, cutinases can hydrolyse cutin, an aliphatic polyester found in the plant cuticle (Kolattukudy, 1981; Heredia, 2003). These enzymes are also able to hydrolyse the ester bonds in PET and PUR (Chen et al., 2013; Wei et al., 2014c; Schmidt et al., 2017). Several enzymes involved in the metabolism of plant lignin are also involved in the degradation of the thermoplastic polyolefin PE (Sivan, 2011; Restrepo‐Flórez et al., 2014).
Figure 2.
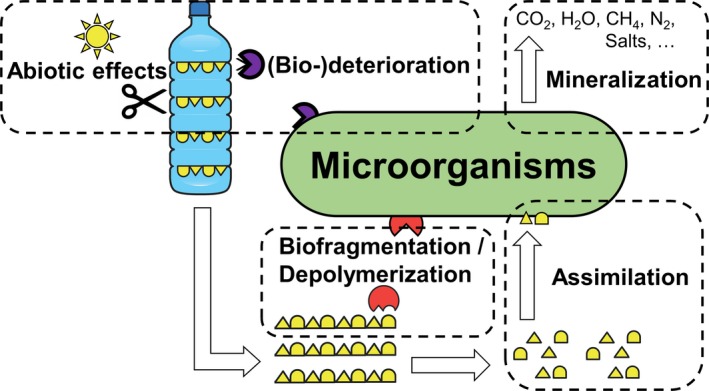
Schematic illustration of plastic biodegradation (Lucas et al., 2008).
Once the molecular size of the synthetic polymers has been reduced to a range of 10–50 carbon atoms, the degradation products can be taken up into the cell for further metabolization (Lucas et al., 2008; Restrepo‐Flórez et al., 2014). The intracellular enzymatic processes involved are beyond the scope of this review.
We will briefly summarize recent advances in the enzymatic degradation of the widely used recalcitrant petroleum‐based plastics PE, PS, PUR and PET and discuss the challenges for the development of efficient plastic‐degrading enzymes and their potential use in alternative recycling processes.
Physiochemical properties of synthetic plastics as obstacles for their enzymatic degradation
Synthetic plastics show a high resistance to many physical, chemical and biological factors (Andrady and Neal, 2009; Thompson et al., 2009). However, this durability also results in their extremely slow degradation in the environment. The hydrophobicity, degree of crystallinity, surface topography and molecular size of the synthetic polymers are important factors restricting their biodegradability (Tokiwa et al., 2009; Webb et al., 2013; Restrepo‐Flórez et al., 2014). Polymers with hydrolysable chemical bonds in their backbone such as PET (Webb et al., 2013) and PUR (Cregut et al., 2013) are more susceptible to biodegradation than PE, PS, PP and PVC (Zheng et al., 2005; Tokiwa et al., 2009; Krueger et al., 2015; Fig. 1). Their highly stable carbon‐carbon (C‐C) bonds have to be oxidized first prior to their further depolymerization (Zheng et al., 2005; Restrepo‐Flórez et al., 2014). Abiotic factors such as UV irradiation, oxygen, temperature, as well as the presence of chemical oxidants, therefore play a crucial role in the degradation of PE and PP in the environment (Bonhomme et al., 2003; Jakubowicz, 2003; Hakkarainen and Albertsson, 2004; Koutny et al., 2006a,b; Arkatkar et al., 2010; Fig. 3).
Figure 3.
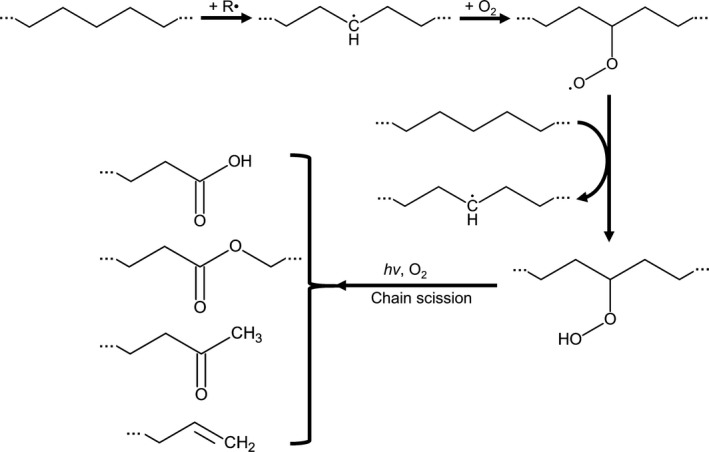
Simplified scheme of the abiotic degradation of polyethylene by oxygen and light. Radicals (R·) can be generated by photo‐oxidation mediated by chemical oxidants (modified based on Koutny et al., 2006b).
The high molecular weight of synthetic polymers with hydrophobic repeating units determines their insolubility in water prohibiting a rapid assimilation by microorganisms (Zheng et al., 2005; Arutchelvi et al., 2008; Restrepo‐Flórez et al., 2014; Krueger et al., 2015). Their degradation by enzymes can be considered as a surface erosion process that is strictly depending on the surface properties of the polymers (Mueller, 2006; Lucas et al., 2008; Restrepo‐Flórez et al., 2014). A high degree of hydrophobicity, a low specific surface area and a smooth surface topography restrict the formation of a biofilm by polymer‐degrading microorganisms (Lucas et al., 2008; Loredo‐Trevino et al., 2012; Cregut et al., 2013; Restrepo‐Flórez et al., 2014; Wei et al., 2014a). The hydrophobic polymer surface has also been shown to prohibit an effective adsorption and catalytic performance of polymer‐degrading enzymes (Espino‐Rammer et al., 2013; Ribitsch et al., 2015, 2013; Sammond et al., 2014). The microbial degradation is furthermore restricted by the low surface‐to‐volume ratio of the plastic debris. A micronization of different PET materials to obtain particle sizes between 0.25 and 0.5 mm was shown to markedly improve their subsequent degradation by a bacterial polyester hydrolase by increasing the accessible surface area for the enzyme (Gamerith et al., 2017). A pretreatment of plastic waste may therefore be a prerequisite in biocatalytic recycling, thereby resulting in further process costs.
Most petroleum‐based plastics are semi‐crystalline polymers containing both crystalline and amorphous regions, the latter being more susceptible to microbial attacks (Sarkar et al., 2006; Tokiwa et al., 2009; Loredo‐Trevino et al., 2012; Webb et al., 2013; Restrepo‐Flórez et al., 2014; Fig. 4). The degree of crystallinity of the polymers has therefore a strong influence on their biodegradability (Urgun‐Demirtas et al., 2007; Brueckner et al., 2008; Jenkins and Harrison, 2008; Eberl et al., 2009; Ronkvist et al., 2009; Horn et al., 2012; Webb et al., 2013; Restrepo‐Flórez et al., 2014; Wei et al., 2014a). However, the crystalline parts of synthetic plastics can also be degraded enzymatically. For example, the crystalline fraction of a thermally pretreated PE sample was also degraded following the complete consumption of the amorphous parts (Manzur et al., 1997; Restrepo‐Flórez et al., 2014). The complete decomposition of low crystalline PET films by a fungal polyester hydrolase has been shown to occur at an almost linear rate, indicating that the crystalline parts were also attacked by the enzyme (Ronkvist et al., 2009).
Figure 4.
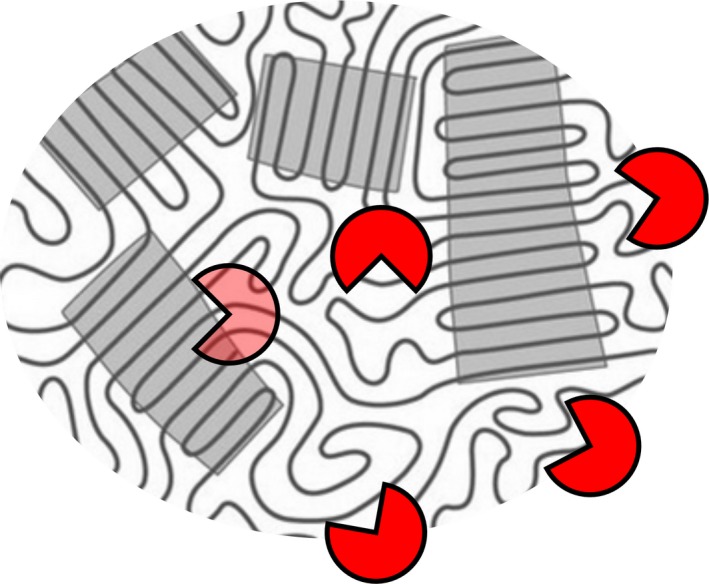
Schematic illustration of a semi‐crystalline polymer containing both amorphous and crystalline regions (grey areas). The amorphous parts are more susceptible to enzymatic attacks.
Enzymatic degradation of plastics with carbon‐carbon backbones
Biodegradation of PE, PP, PS and PVC is hampered by the lack of hydrolysable functional groups in their backbones (Tokiwa et al., 2009; Restrepo‐Flórez et al., 2014; Krueger et al., 2015). The initial breakdown of the polymers in the environment and the observed reduction in their molecular weight have been mainly attributed to a synergistic action of biotic and abiotic factors (Bonhomme et al., 2003; Hakkarainen and Albertsson, 2004; Eubeler et al., 2010; Restrepo‐Flórez et al., 2014). The carbonyl groups formed as a result of UV irradiation or oxidizing agents have been considered as more accessible for a subsequent microbial attack (Albertsson et al., 1987; Karlsson et al., 1988; Albertsson and Karlsson, 1990; Koutny et al., 2006a,b; Fontanella et al., 2010; Fig. 3). Biodegradation studies of plastics with C‐C backbones have therefore mostly been carried out with preoxidized or thermally pretreated substrates under laboratory conditions (Motta et al., 2009; Ojeda et al., 2009; Jeyakumar et al., 2013; Restrepo‐Flórez et al., 2014).
Polyethylene is the most common plastic with a C‐C backbone. Various types of PE have been subjected to biodegradation studies in the last decades (Restrepo‐Flórez et al., 2014; Sen and Raut, 2015). Microbial enzymes capable of degrading lignin, a heterogeneous cross‐linked phenolic polymer with oxidizable C‐C bonds found in plant cell walls (Freudenberg and Neish, 1968; Suhas et al., 2007), have been reported to be involved in the biodegradation of PE (Restrepo‐Flórez et al., 2014; Krueger et al., 2015). These include laccases (EC 1.10.3.2.), manganese peroxidase (MnP, EC 1.11.1.13) and lignin peroxidases (LiP, EC 1.11.1.14). As the redox potential required for the breakdown of lignin is considerably lower than for the homogenous C‐C backbone of PE, an efficient degradation of PE by these enzymes can, however, not be expected (Krueger et al., 2015).
A thermostable laccase from Rhodococcus ruber C208 degraded UV‐irradiated PE films both in culture supernatants and in cell‐free extracts in the presence of copper (Santo et al., 2013). As a result of oxidations and polymer chain scissions mainly within the amorphous part of the PE films, an increased amount of carbonyl groups and a reduction in the molecular weight were observed after 2 weeks of incubation with the enzyme at 37°C. A laccase from Trametes versicolor also strongly reduced the molecular weight of a PE membrane in the presence of 1‐hydroxybenzotriazole, which mediated the oxidation of non‐phenolic substrates by the enzyme (Fujisawa et al., 2001). MnP from the white‐rot fungi Phanerochaete chrysosporium ME‐446 and the isolate IZU‐154 have been described as key enzymes for the degradation of a high molecular weight PE membrane (Iiyoshi et al., 1998). Surfactants including Tween 80, Tween 20 and CHAPSO have been shown to promote the degradation of PE by the partially purified MnP (Iiyoshi et al., 1998; Ehara et al., 2000). The genes encoding the most active MnP from IZU‐154 have been identified and further characterized, however only with respect to the oxidation of 2,6‐dimethoxyphenol (Matsubara et al., 1996) and the degradation of nylon‐66 (Deguchi et al., 1998). Bacillus cereus also degraded UV‐irradiated PE associated with a pronounced extracellular production of both laccases and MnP (Sowmya et al., 2014). However, the incubation of similarly pretreated PE with a partially purified laccase and a MnP from Penicillium simplicissimum resulted only in a negligible weight loss of less than 1% (Sowmya et al., 2015). The concentrated culture supernatants of lignocellulose‐degrading Streptomyces species containing LiP activity were reported to degrade the PE fraction of a heat‐treated plastic blend (Pometto et al., 1992). Similarly, the extracellular LiP and MnP of Phanerochaete chrysosporium MTCC‐787 were reported to degrade 70% of a preoxidized high molecular weight PE sample within 15 days of incubation (Mukherjee and Kundu, 2014).
Alkane hydroxylases (AH) of the AlkB family (EC 1.14.15.3) can catalyse the degradation of hydrocarbon oligomers by terminal or subterminal oxidation (Rojo, 2010). A recombinant AH from Pseudomonas sp. E4 expressed in Escherichia coli BL21 converted 20% of the low molecular weight PE sample to CO2 after incubation for 80 days at 37°C (Yoon et al., 2012). A recombinant E. coli strain simultaneously expressing the complete AH system from Pseudomonas aeruginosa E7 including an alkane monooxygenase, rubredoxin and rubredoxin reductase degraded about 30% of this PE sample (Jeon and Kim, 2015).
A purified hydroquinone peroxidase (EC 1.11.1.7) of the lignin‐decolorizing Azotobacter beijerinckii HM121 degraded PS, an aromatic thermoplastic with a C‐C backbone (Fig. 1), in a two‐phase system consisting of dichloromethane and water. PS in the organic phase was rapidly converted to small water‐soluble products within 5 min of reaction at 30°C in the presence of hydrogen peroxide and tetramethylhydroquinone (Nakamiya et al., 1997). However, this two‐phase process has apparently not been further developed for a recycling process of PS waste. While the degradation of PE and PS by novel bacterial strains isolated from the guts of insect larva has been reported recently, the corresponding enzymes involved have not been identified yet (Yang et al., 2014, 2015).
The above‐mentioned studies used only culture supernatants or partially purified enzyme preparations and required long incubation times. For PVC, another important plastic with a C‐C backbone, enzymes directly involved in its degradation have not been reported yet.
The use of whole cells rather than isolated enzymes has been proposed as a potentially better approach for the biodegradation of plastics with C‐C backbones. Recently, the utilization of microbial communities or mixed cultures with defined microbial strains has shown an improved performance in the degradation of PS and PE compared with the use of single microorganisms (Roy et al., 2008; Esmaeili et al., 2013; Yang et al., 2015; Mukherjee et al., 2016).
Enzymatic degradation of PUR
Polyurethane is a polymer composed of di‐ or polyisocyanate and polyols linked by carbamate (urethane) bonds (Seymour and Kauffman, 1992; Fig. 1). The urethane bond connects the crystalline rigid segments consisting of isocyanate and the chain extender with the amorphous parts composed of a polyester or polyether (Nomura et al., 1998; Ruiz et al., 1999; Urgun‐Demirtas et al., 2007). Depending on the polyols used for the polycondensation reaction, polyether and polyester PUR with different characteristic properties can be manufactured (Seymour and Kauffman, 1992). The presence of aromatic esters and the extent of the crystalline fraction of the polymer are important factors influencing the biodegradation of PUR (Urgun‐Demirtas et al., 2007; Cregut et al., 2013). PUR can be depolymerized by microbial ureases, esterases and proteases hydrolysing the urethane and ester bonds of the plastic (Howard, 2012a; Loredo‐Trevino et al., 2012; Cregut et al., 2013; Fig. 5). Enzymes degrading polyester PUR from bacteria (Nakajima‐Kambe et al., 1995; Howard and Blake, 1998; Stern and Howard, 2000; Howard et al., 2001, 2012b; Schmidt et al., 2017; Shah et al., 2008a) and fungi (Pathirana and Seal, 1984a,b; Crabbe et al., 1994; Russell et al., 2011) have been described. It has been postulated that proteases hydrolyse the amide and urethane bonds, while ureases attack the urea linkages (Labow et al., 1996; Ruiz et al., 1999; Matsumiya et al., 2010). Esterases and proteases also hydrolyse the ester bonds in polyester PUR as a major mechanism for its enzymatic depolymerization (Nakajima‐Kambe et al., 1999; Tang et al., 2001a,b; Howard, 2002). Although a cleavage of urethane bonds in polyether PUR by bacterial (Akutsu‐Shigeno et al., 2006) and fungal (Owen et al., 1996) hydrolases has been reported, this type of PUR is much more recalcitrant to enzymatic attack compared with polyester PUR (Nakajima‐Kambe et al., 1999; Christenson et al., 2006). However, a weight loss of a polyether PUR of more than 60% following incubation with fungal isolates has been reported recently without yet a characterization of the enzymes involved (Álvarez‐Barragán et al., 2016).
Figure 5.
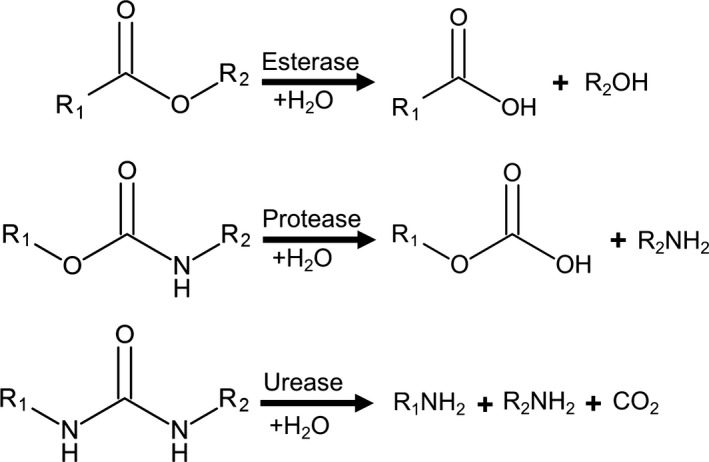
Cleavage of linkages in polyurethane by esterases, proteases and ureases (modified from Phua et al., 1987 and Loredo‐Trevino et al., 2012).
The enzymatic hydrolysis of insoluble PUR polymer is a surface erosion process depending on the efficient adsorption of the biocatalysts on the polymer surface prior to the polymer breakdown (Akutsu et al., 1998). Compared with the wild‐type enzyme, an increased yield in degradation products was observed following the incubation of a solid polyester PUR with a polyamidase from Nocardia farcinica fused to the hydrophobic polymer binding module of the polyhydroxyalkanoate depolymerase from Alcaligenes faecalis (Gamerith et al., 2016). In contrast, the enzyme with the fused binding module did not show a better performance than the wild‐type enzyme in the hydrolysis of soluble PUR substrates, confirming the importance of the initial enzyme adsorption process in the degradation of solid PUR.
Two types of PUR esterases with apparent synergistic activities in the degradation of PUR have been reported (Nakajima‐Kambe et al., 1995, 1999; Akutsu et al., 1998; Howard and Hilliard, 1999). A membrane‐bound esterase of Delftia acidovorans was shown to be essential for the adhesion of the microorganism on the polymer surface as well as its initial hydrolysis. A second secreted PUR esterase benefitting from the close vicinity to the substrate catalysed the subsequent depolymerization of PUR, thereby increasing the surface area for further cell adhesion mediated by the membrane‐bound esterase (Nakajima‐Kambe et al., 1995, 1999; Cregut et al., 2013).
While previously isolated PUR‐degrading enzymes showed only low catalytic efficiencies, whole‐cell catalysis has been suggested as a more promising approach for a biotechnical recycling process of PUR waste materials (Cregut et al., 2013). PUR foam particles and fragments, which represent the most abundant PUR waste materials (Seymour and Kauffman, 1992), will be difficult to attack by microbial catalysts in an aqueous recycling system. A mechanical size reduction in the PUR foam and the use of immobilized biomass have been suggested to overcome the low bioavailability of this PUR waste material (Cregut et al., 2013). However, an efficient biocatalytic degradation approach of PUR with application potential in a biotechnical recycling process has not yet been demonstrated.
Enzymatic degradation of PET
Polyethylene terephthalate is a polymer of terephthalic acid and ethylene glycol linked by ester bonds (Webb et al., 2013). Owing to its widespread uses in packaging materials, beverage bottles and the textile industry, the global PET production has exceeded 41.6 million tons in 2014 (Research and Markets, 2015). The durability and the resulting low biodegradability of PET are due to the presence of repeating aromatic terephthalate units in its backbone and the corresponding limited mobility of the polymer chains (Marten et al., 2003, 2005). The semi‐crystalline PET polymer also contains both amorphous and crystalline fractions with a strong effect on its biodegradability (see Physiochemical properties of synthetic plastics as obstacles for their enzymatic degradation, Fig. 4). At industrial processing conditions such as fibre spinning, injection moulding and film blowing, PET materials with different degrees of crystallinity are obtained as a result of flow‐induced crystallization which determines the final structure and properties of the semi‐crystalline polymer (Lamberti, 2014; Wang et al., 2016). PET beverage bottles with a crystallinity of over 30% (Liu et al., 2004) and PET fibres up to 40% (Lee et al., 2013) are common PET products which pose a high recalcitrance to enzymatic degradation.
At a temperature close to the glass transition temperature (T g) of PET above 65°C (Alves et al., 2002), the amorphous parts of the polymer become flexible and more accessible to an enzymatic attack (Parikh et al., 1993; Ronkvist et al., 2009). Consequently, the high T g of PET requires enzymes with a high stability in this temperature range (Ronkvist et al., 2009; Sulaiman et al., 2014; Wei et al., 2014b; Then et al., 2016). Recently, an enzymatic hydrolysis of amorphous PET at a reaction temperature of 30°C has also been reported. However, only very low degradation rates could be observed (Yoshida et al., 2016). A number of lipases (Vertommen et al., 2005; Eberl et al., 2009; Ronkvist et al., 2009), esterases (Liebminger et al., 2007) and cutinases (Müller et al., 2005; Alisch‐Mark et al., 2006; Araujo et al., 2007; Nimchua et al., 2007; Ronkvist et al., 2009; Herrero Acero et al., 2011; Chen et al., 2013; Wei et al., 2014c) from fungal and actinomycete species hydrolyse amorphous PET and modify the surface of PET films and fibres (Zimmermann and Billig, 2011). Carboxylesterases from Bacillus licheniformis, Bacillus subtilis and Thermobifida fusca also partially hydrolysed PET fibres and showed a high activity against PET oligomers (Billig et al., 2010; Oeser et al., 2010; Ribitsch et al., 2011; Lülsdorf et al., 2015; Barth et al., 2016). Lipases display low activity against PET due to their lid structure covering the buried hydrophobic catalytic centre and the resulting limited accessibility for polymeric substrates (Guebitz and Cavaco‐Paulo, 2008; Eberl et al., 2009; Zimmermann and Billig, 2011). In contrast, cutinases lacking a lid structure were able to cause significant weight losses from amorphous PET films (Ronkvist et al., 2009; Sulaiman et al., 2014; Wei et al., 2016). Structural analyses of both fungal (Martinez et al., 1992; Longhi et al., 1997) and bacterial cutinases (Kitadokoro et al., 2012; Roth et al., 2014; Sulaiman et al., 2014; Miyakawa et al., 2015) revealed an exposed active site close to the surface of the enzymes, which is essential for the recognition and interaction with polymeric substrates (Guebitz and Cavaco‐Paulo, 2008; Kitadokoro et al., 2012).
The thermostable cutinase HiC from Thermomyces (formerly Humicola) insolens is the most active fungal polyester hydrolase reported so far (Ronkvist et al., 2009). After a reaction time of 96 h at 70°C, HiC hydrolysed a low crystalline (7%) PET film almost completely, suggesting that the crystalline part of the PET film was also degraded at this reaction temperature. The thermostable bacterial LC‐cutinase hydrolysed approximately 25% of a low crystalline PET film for 24 h at the same reaction temperature (Sulaiman et al., 2014). The gene encoding this enzyme is homologous to the polyester hydrolases of Thermobifida species (Herrero Acero et al., 2011) and has been isolated from a plant compost metagenome (Sulaiman et al., 2012). Bivalent metal ions such as Ca2+ and Mg2+ enhanced the thermostability of several polyester hydrolases from actinomycetes and resulted in an increased hydrolytic activity against PET near the T g of PET (Thumarat et al., 2012; Kawai et al., 2014; Sulaiman et al., 2014; Miyakawa et al., 2015; Then et al., 2015, 2016; Wei et al., 2016). By substitution of the metal binding site with a salt bridge or a disulfide bridge, variants of the polyester hydrolase TfCut2 from Thermobifida fusca KW3 also readily degraded amorphous PET films at 70°C in the absence of metal ions (Then et al., 2015, 2016). The stabilizing effect of phosphate anions at a concentration of up to 1 M (Jensen et al., 1995; Park et al., 2001) also promoted the activity of these enzymes against PET (Schmidt et al., 2016).
Although some polyester hydrolases displayed high activity against amorphous PET materials at reaction temperatures above 50°C, crystalline and biaxially oriented PET was degraded to a much lesser extent at the same reaction conditions (Eberl et al., 2009; Ronkvist et al., 2009; Wei et al., 2016; Gamerith et al., 2017; Fig. 6). It is therefore presently not possible to completely degrade PET fibres and beverage bottles with a higher percentage of crystallinity (Liu et al., 2004; Lee et al., 2013) or biaxially oriented PET within short reaction times with these enzymes (Zhang et al., 2004; Zimmermann and Billig, 2011; Carniel et al., 2016; Yoshida et al., 2016; Gamerith et al., 2017).
Figure 6.
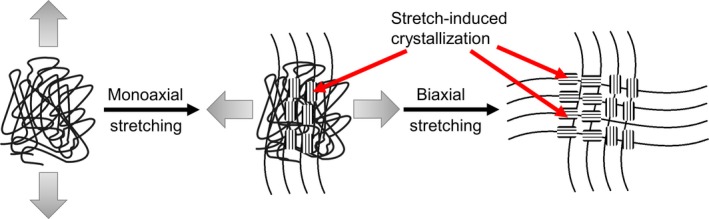
Schematic illustration of the biaxial stretching of an amorphous polymer and the stretch‐induced crystallization (Zhang et al., 2016). Due to the small crystallite size below the wavelength of visible light, the resulting material remains transparent after the biaxial stretching (Jariyasakoolroj et al., 2015).
The enzymatic hydrolysis of PET is also a surface erosion process (Zhang et al., 2004; Müller et al., 2005; Mueller, 2006; Wei et al., 2014a). The hydrophobic nature of PET represents a barrier which hampers the effective adsorption of enzymes to the polymer surface for the hydrolytic reaction (Atthoff and Hilborn, 2007). Unlike enzymes hydrolysing natural polymers such as polyhydroxyalkanoates (Knoll et al., 2009) or cellulose (Atthoff and Hilborn, 2007), specific binding domains responsible for substrate adsorption are absent in cutinases (Chen et al., 2013; Wei et al., 2014c). Their initial adsorption to the surface of PET is presumably mediated by hydrophobic regions surrounding the catalytic site (Herrero Acero et al., 2011). By site‐directed mutagenesis of selected amino acids in these regions, variants with enhanced activity against PET could be obtained (Wei, 2011; Herrero Acero et al., 2013). The fusion of polymer and cellulose binding domains (Ribitsch et al., 2013) or hydrophobins (Ribitsch et al., 2015) also enhanced the adsorption of cutinases to the surface of PET and resulted in higher yields of hydrolysis products. A truncation of 71 N‐terminal residues of an esterase from Clostridium botulinum exposed a hydrophobic patch, which facilitated its adsorption to PET and improved its hydrolytic activity (Biundo et al., 2016).
Selected amino acid residues in close vicinity to the catalytic centre were suggested to be important for the recognition and interaction with the polymeric substrate by various polyester hydrolases (Guebitz and Cavaco‐Paulo, 2008; Kitadokoro et al., 2012). By modification of the size and the hydrophobicity of these residues, the hydrolytic activity against PET of a polyester hydrolase from Fusarium solani (Araujo et al., 2007) and Thermobifida fusca (Silva et al., 2011) could be increased.
In addition to ethylene glycol, terephthalate, mono‐(2‐hydroxyethyl) terephthalate (MHET) and bis‐(2‐hydroxyethyl) terephthalate (BHET) are the main water‐soluble products obtained by the enzymatic hydrolysis of PET (Vertommen et al., 2005; Wei et al., 2012; Fig. 7). The polyester hydrolase TfCut2 is strongly inhibited by MHET and BHET (Barth et al., 2015a). By performing the hydrolysis of amorphous PET films in an enzyme reactor fitted with an ultrafiltration membrane, the product inhibition could be avoided by the continuous removal of MHET and BHT (Barth et al., 2015b). As a result, a 1.7‐fold higher amount of hydrolysis products were obtained from amorphous PET films after a reaction time of 24 h. With a dual enzyme reaction system composed of a polyester hydrolase and the immobilized carboxylesterase TfCa from Thermobifida fusca KW3, a twofold higher yield of degradation products could be obtained compared with those without TfCa (Barth et al., 2016). In this one‐pot process, TfCa prevented the inhibition of TfCut2 by specifically binding and hydrolysing MHET and BHET in the reaction medium. Similarly, a reaction system composed of the fungal polyester hydrolase HiC and the lipase CalB from Candida antarctica also showed a 7.7‐fold increase in the yield of terephthalate obtained due to the concomitant degradation of MHET catalysed by CalB (Carniel et al., 2016). A recently described enzyme from Ideonella sakaiensis 201‐F6, which catalysed specifically the hydrolysis of MHET (Yoshida et al., 2016), could be a further candidate for a one‐pot system to promote PET hydrolysis in an enzyme reactor. The susceptibility of TfCut2 to product inhibition could also be mitigated by modifying a key amino acid residue involved in the interaction with a low molecular weight PET model compound (Wei et al., 2016). As a result, a 2.7‐fold higher weight loss of an amorphous PET film was obtained with this variant after a reaction time of 50 h compared with the wild‐type enzyme.
Figure 7.
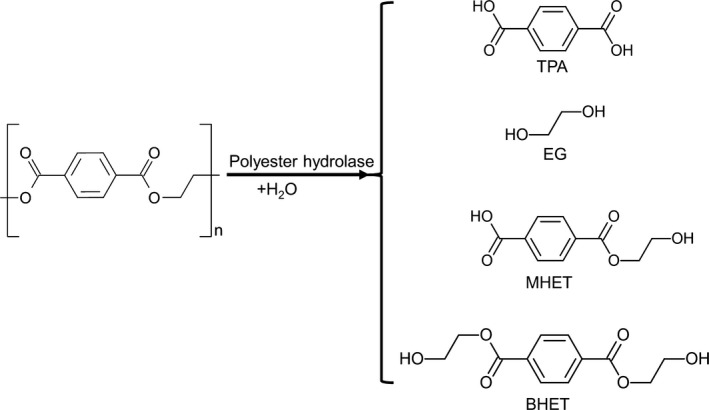
Water‐soluble products obtained from PET films degraded by a polyester hydrolase (Barth et al., 2015a). TPA, terephthalic acid; EG, ethylene glycol; MHET, mono‐(2‐hydroxyethyl) terephthalate; BHET, bis‐(2‐hydroxyethyl) terephthalate.
Prospects for a biocatalytic recycling of recalcitrant plastic waste
The recovery of chemical feedstocks that can be used for the production of virgin polymers in a closed‐loop recycling process is considered as the most sustainable option to solve the plastic waste problem (Al‐Salem et al., 2009; Andrady, 2015c). A drastic increase in the amount of plastic waste being recycled instead of discarded will therefore be necessary in the future (World Economic Forum, 2016). Increasing knowledge on microbial enzymes able to degrade petroleum‐based recalcitrant plastics will promote the further development of environmentally friendly plastic recycling processes (Müller et al., 2005; Barth et al., 2015b, 2016). Although several redox enzymes have been found to contribute to the degradation of PE, a complete biocatalytic degradation of plastics with C‐C backbones has not been demonstrated yet. The identification of suitable enzymes and a better understanding of the degradation mechanism will be necessary before an application of enzymes for a recycling of PE waste could be envisaged. Instead, whole‐cell catalysis with single microorganisms or even microbial communities might provide an alternative strategy for a biotechnical PE recycling.
Synthetic polyesters such as PET and also polyester PUR have been shown to be susceptible to enzymatic degradation by microbial polyester hydrolases. A range of fungal and bacterial enzymes has been described recently able to modify and degrade PET films and fibres. As a proof of principle, the conversion of amorphous PET in an enzyme reactor to its monomers has been demonstrated recently (Barth et al., 2015b). Although the crystalline parts of PET can also be degraded by the enzymes, this process is still too slow to be applied for a biocatalytic recycling of PET beverage bottles or textile fibres. Metagenomic approaches have already shown to facilitate the access to novel polyester hydrolases from the environment (Sulaiman et al., 2012). Directed evolution strategies can be applied for the identification of mutation hot spots in polyester hydrolases complementing enzyme optimization strategies by semi‐rational re‐design (Thumarat et al., 2012; Kawai et al., 2014). In the search for improved biocatalysts, high‐throughput screening methods specifically designed to monitor polyester hydrolase activities enable a rapid identification of these enzymes and their variants (Wei et al., 2012). The discovery of novel microbial polyester hydrolases and the construction of highly active variants therefore remain a key challenge for the development of a viable biocatalytic recycling process for post‐consumer PET waste.
Conflict of interest
The authors have declared no conflict of interest.
Acknowledgements
Support by grants from the European Union's Horizon 2020 Research and Innovation Program under Agreement No. 633962 and from the German Federal Ministry of Education and Research (Project No. 031A227E) is acknowledged.
Microbial Biotechnology (2017) 10(6), 1308–1322
Funding Information Bundesministerium für Bildung und Forschung (031A227E); Horizon 2020 (633962).
Contributor Information
Ren Wei, Email: wei@uni-leipzig.de.
Wolfgang Zimmermann, Email: wolfgang.zimmermann@uni-leipzig.de.
References
- Akutsu, Y. , Nakajima‐Kambe, T. , Nomura, N. , and Nakahara, T. (1998) Purification and properties of a polyester polyurethane‐degrading enzyme from Comamonas acidovorans TB‐35. Appl Environ Microbiol 64: 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akutsu‐Shigeno, Y. , Adachi, Y. , Yamada, C. , Toyoshima, K. , Nomura, N. , Uchiyama, H. , and Nakajima‐Kambe, T. (2006) Isolation of a bacterium that degrades urethane compounds and characterization of its urethane hydrolase. Appl Microbiol Biotechnol 70: 422–429. [DOI] [PubMed] [Google Scholar]
- Albertsson, A.‐C. , and Karlsson, S. (1990) The influence of biotic and abiotic environments on the degradation of polyethylene. Prog Polym Sci 15: 177–192. [Google Scholar]
- Albertsson, A.‐C. , Andersson, S.O. , and Karlsson, S. (1987) The mechanism of biodegradation of polyethylene. Polym Degrad Stabil 18: 73–87. [Google Scholar]
- Alisch‐Mark, M. , Herrmann, A. , and Zimmermann, W. (2006) Increase of the hydrophilicity of polyethylene terephthalate fibres by hydrolases from Thermomonospora fusca and Fusarium solani f. sp. pisi . Biotechnol Lett 28: 681–685. [DOI] [PubMed] [Google Scholar]
- Al‐Salem, S.M. , Lettieri, P. , and Baeyens, J. (2009) Recycling and recovery routes of plastic solid waste (psw): a review. Waste Manage 29: 2625–2643. [DOI] [PubMed] [Google Scholar]
- Álvarez‐Barragán, J. , Domínguez‐Malfavón, L. , Vargas‐Suárez, M. , González‐Hernández, R. , Aguilar‐Osorio, G. , and Loza‐Tavera, H. (2016) Biodegradative activities of selected environmental fungi on a polyester polyurethane varnish and polyether polyurethane foams. Appl Environ Microbiol 82: 5225–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves, N.M. , Mano, J.F. , Balaguer, E. , Meseguer Duenas, J.M. , and Gomez Ribelles, J.L. (2002) Glass transition and structural relaxation in semi‐crystalline poly(ethylene terephthalate): a DSC study. Polymer 43: 4111–4122. [Google Scholar]
- Andrady, A.L. (1994) Assessment of environmental biodegradation of synthetic polymers. J Macromol Sci C 34: 25–76. [Google Scholar]
- Andrady, A.L. (2011) Microplastics in the marine environment. Mar Pollut Bull 62: 1596–1605. [DOI] [PubMed] [Google Scholar]
- Andrady, A.L. (2015a) Degradation of plastics in the environment In Plastics and Environmental Sustainability. Hoboken, New Jersey: John Wiley & Sons, pp. 145–184. [Google Scholar]
- Andrady, A.L. (2015b) An introduction to plastics In Plastics and Environmental Sustainability. Hoboken, New Jersey: John Wiley & Sons, pp. 55–82. [Google Scholar]
- Andrady, A.L. (2015c) Managing plastic waste In Plastics and Environmental Sustainability. Hoboken, New Jersey: John Wiley & Sons, pp. 255–293. [Google Scholar]
- Andrady, A.L. (2015d) Plastic products In Plastics and Environmental Sustainability. Hoboken, New Jersey: John Wiley & Sons, pp. 83–119. [Google Scholar]
- Andrady, A.L. (2015e) Plastics in the oceans In Plastics and Environmental Sustainability. Hoboken, New Jersey: John Wiley & Sons, pp. 295–318. [Google Scholar]
- Andrady, A.L. , and Neal, M.A. (2009) Applications and societal benefits of plastics. Philos T R Soc B 364: 1977–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo, R. , Silva, C. , O'Neill, A. , Micaelo, N. , Guebitz, G. , Soares, C.M. , et al (2007) Tailoring cutinase activity towards polyethylene terephthalate and polyamide 6,6 fibers. J Biotechnol 128: 849–885. [DOI] [PubMed] [Google Scholar]
- Arkatkar, A. , Juwarkar, A.A. , Bhaduri, S. , Uppara, P.V. , and Doble, M. (2010) Growth of Pseudomonas and Bacillus biofilms on pretreated polypropylene surface. Int Biodeterior Biodegradation 64: 530–536. [Google Scholar]
- Arutchelvi, J. , Sudhakar, M. , Arkatkar, A. , Doble, M. , Bhaduri, S. , and Uppara, P. (2008) Biodegradation of polyethylene and polypropylene. Ind J Biotechnol 7: 9–22. [Google Scholar]
- Atthoff, B. , and Hilborn, J. (2007) Protein adsorption onto polyester surfaces: is there a need for surface activation? J Biomed Mat Res B‐Appl Biomat 80B: 121–130. [DOI] [PubMed] [Google Scholar]
- Banerjee, A. , Chatterjee, K. , and Madras, G. (2014) Enzymatic degradation of polymers: a brief review. Mat Sci Technol 30: 567–573. [Google Scholar]
- Barth, M. , Oeser, T. , Wei, R. , Then, J. , Schmidt, J. , and Zimmermann, W. (2015a) Effect of hydrolysis products on the enzymatic degradation of polyethylene terephthalate nanoparticles by a polyester hydrolase from Thermobifida fusca . Biochem Eng J 93: 222–228. [Google Scholar]
- Barth, M. , Wei, R. , Oeser, T. , Then, J. , Schmidt, J. , Wohlgemuth, F. , and Zimmermann, W. (2015b) Enzymatic hydrolysis of polyethylene terephthalate films in an ultrafiltration membrane reactor. J Mem Sci 494: 182–187. [Google Scholar]
- Barth, M. , Honak, A. , Oeser, T. , Wei, R. , Belisário‐Ferrari, M.R. , Then, J. , et al (2016) A dual enzyme system composed of a polyester hydrolase and a carboxylesterase enhances the biocatalytic degradation of polyethylene terephthalate films. Biotechnol J 11: 1082–1087. [DOI] [PubMed] [Google Scholar]
- Bhardwaj, H. , Gupta, R. , and Tiwari, A. (2013) Communities of microbial enzymes associated with biodegradation of plastics. J Polym Environ 21: 575–579. [Google Scholar]
- Billig, S. , Oeser, T. , Birkemeyer, C. , and Zimmermann, W. (2010) Hydrolysis of cyclic poly(ethylene terephthalate) trimers by a carboxylesterase from Thermobifida fusca KW3. Appl Microbiol Biotechnol 87: 1753–1764. [DOI] [PubMed] [Google Scholar]
- Biundo, A. , Ribitsch, D. , Steinkellner, G. , Gruber, K. , and Guebitz, G.M. (2016) Polyester hydrolysis is enhanced by a truncated esterase: less is more. Biotechnol J In Press doi:10.1002/biot.201600450. [DOI] [PubMed] [Google Scholar]
- Bonhomme, S. , Cuer, A. , Delort, A.M. , Lemaire, J. , Sancelme, M. , and Scott, G. (2003) Environmental biodegradation of polyethylene. Polym Degrad Stabil 81: 441–452. [Google Scholar]
- Brueckner, T. , Eberl, A. , Heumann, S. , Rabe, M. , and Guebitz, G.M. (2008) Enzymatic and chemical hydrolysis of poly(ethylene terephthalate) fabrics. J Polym Sci A‐Polym Chem 46: 6435–6443. [Google Scholar]
- Carniel, A. , Valoni, E. , Nicomedes, J. , Gomes, A.D.C. , and Castro, A.M.D. (2016) Lipase from Candida antarctica (CALB) and cutinase from Humicola insolens act synergistically for PET hydrolysis to terephthalic acid. Proc Biochem In Press doi:10.1016/j.procbio.2016.07.023. [Google Scholar]
- Chen, S. , Su, L. , Chen, J. , and Wu, J. (2013) Cutinase: characteristics, preparation, and application. Biotechnol Adv 31: 1754–1767. [DOI] [PubMed] [Google Scholar]
- Christenson, E.M. , Patel, S. , Anderson, J.M. , and Hiltner, A. (2006) Enzymatic degradation of poly(ether urethane) and poly(carbonate urethane) by cholesterol esterase. Biomaterials 27: 3920–3926. [DOI] [PubMed] [Google Scholar]
- Crabbe, J.R. , Campbell, J.R. , Thompson, L. , Walz, S.L. , and Schultz, W.W. (1994) Biodegradation of a colloidal ester‐based polyurethane by soil fungi. Int Biodeterior Biodegradation 33: 103–113. [Google Scholar]
- Cregut, M. , Bedas, M. , Durand, M.J. , and Thouand, G. (2013) New insights into polyurethane biodegradation and realistic prospects for the development of a sustainable waste recycling process. Biotechnol Adv 31: 1634–1647. [DOI] [PubMed] [Google Scholar]
- Deguchi, T. , Kitaoka, Y. , Kakezawa, M. , and Nishida, T. (1998) Purification and characterization of a nylon‐degrading enzyme. App Environ Microb 64: 1366–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derraik, J.G.B. (2002) The pollution of the marine environment by plastic debris: a review. Mar Pollut Bull 44: 842–852. [DOI] [PubMed] [Google Scholar]
- Eberl, A. , Heumann, S. , Bruckner, T. , Araujo, R. , Cavaco‐Paulo, A. , Kaufmann, F. , et al (2009) Enzymatic surface hydrolysis of poly(ethylene terephthalate) and bis(benzoyloxyethyl) terephthalate by lipase and cutinase in the presence of surface active molecules. J Biotechnol 143: 207–212. [DOI] [PubMed] [Google Scholar]
- Ehara, K. , Iiyoshi, Y. , Tsutsumi, Y. , and Nishida, T. (2000) Polyethylene degradation by manganese peroxidase in the absence of hydrogen peroxide. J Wood Sci 46: 180–183. [Google Scholar]
- Esmaeili, A. , Pourbabaee, A.A. , Alikhani, H.A. , Shabani, F. , and Esmaeili, E. (2013) Biodegradation of low‐density polyethylene (LDPE) by mixed culture of Lysinibacillus xylanilyticus and Aspergillus niger in soil. PLoS ONE 8: e71720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espino‐Rammer, L. , Ribitsch, D. , Przylucka, A. , Marold, A. , Greimel, K.J. , Herrero Acero, E. , et al (2013) Two novel class II hydrophobins from Trichoderma spp. stimulate enzymatic hydrolysis of poly(ethylene terephthalate) when expressed as fusion proteins. Appl Environ Microbiol 79: 4230–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubeler, J.P. , Bernhard, M. , and Knepper, T.P. (2010) Environmental biodegradation of synthetic polymers ii. Biodegradation of different polymer groups. TrAC Trends Analy Chem 29: 84–100. [Google Scholar]
- Fontanella, S. , Bonhomme, S. , Koutny, M. , Husarova, L. , Brusson, J.‐M. , Courdavault, J.‐P. , et al (2010) Comparison of the biodegradability of various polyethylene films containing pro‐oxidant additives. Polym Degrad Stabil 95: 1011–1021. [Google Scholar]
- Freudenberg, K. , and Neish, A.C. (1968) Constitution and Biosynthesis of Lignin. Berlin, Heidelberg: Springer‐Verlag, pp. 1–132. [Google Scholar]
- Fujisawa, M. , Hirai, H. , and Nishida, T. (2001) Degradation of polyethylene and nylon‐66 by the laccase‐mediator system. J Polym Environ 9: 103–108. [Google Scholar]
- Gamerith, C. , Herrero Acero, E. , Pellis, A. , Ortner, A. , Vielnascher, R. , Luschnig, D. , et al (2016) Improving enzymatic polyurethane hydrolysis by tuning enzyme sorption. Polym Degrad Stabil 132: 69–77. [Google Scholar]
- Gamerith, C. , Zartl, B. , Pellis, A. , Guillamot, F. , Marty, A. , Acero, E.H. , and Guebitz, G.M. (2017) Enzymatic recovery of polyester building blocks from polymer blends. Proc Biochem In Press doi:10.1016/j.procbio.2017.01.004. [Google Scholar]
- Gewert, B. , Plassmann, M.M. , and MacLeod, M. (2015) Pathways for degradation of plastic polymers floating in the marine environment. Environ Sci Proc Imp 17: 1513–1521. [DOI] [PubMed] [Google Scholar]
- Gourmelon, G. (2015) Global plastic production rises, recycling lags In Vital Signs, vol 22. Starke L. (ed). Washington: Worldwatch Institute, pp. 91–95. http://vitalsigns.worldwatch.org/sites/default/files/vital_signs_trend_plastic_full_pdf.pdf [Google Scholar]
- Gu, J.‐D. (2003) Microbiological deterioration and degradation of synthetic polymeric materials: recent research advances. Int Biodeterior Biodegadationr 52: 69–91. [Google Scholar]
- Guebitz, G.M. , and Cavaco‐Paulo, A. (2008) Enzymes go big: surface hydrolysis and functionalization of synthetic polymers. Trends Biotechnol 26: 32–38. [DOI] [PubMed] [Google Scholar]
- Hakkarainen, M. , and Albertsson, A.‐C. (2004) Environmental degradation of polyethylene In Long Term Properties of Polyolefins. Albertsson A.‐C. (ed). Berlin Heidelberg: Springer‐Verlag, pp. 177–200. [Google Scholar]
- Heredia, A. (2003) Biophysical and biochemical characteristics of cutin, a plant barrier biopolymer. Biochim Biophys Acta 1620: 1–7. [DOI] [PubMed] [Google Scholar]
- Herrero Acero, E. , Ribitsch, D. , Steinkellner, G. , Gruber, K. , Greimel, K. , Eiteljoerg, I. , et al (2011) Enzymatic surface hydrolysis of pet: effect of structural diversity on kinetic properties of cutinases from Thermobifida . Macromolecules 44: 4632–4640. [Google Scholar]
- Herrero Acero, E. , Ribitsch, D. , Dellacher, A. , Zitzenbacher, S. , Marold, A. , Steinkellner, G. , et al (2013) Surface engineering of a cutinase from Thermobifida cellulosilytica for improved polyester hydrolysis. Biotechnol Bioeng 110: 2581–2590. [DOI] [PubMed] [Google Scholar]
- Hopewell, J. , Dvorak, R. , and Kosior, E. (2009) Plastics recycling: challenges and opportunities. Philos T R Soc B 364: 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, S.J. , Vaaje‐Kolstad, G. , Westereng, B. , and Eijsink, V. (2012) Novel enzymes for the degradation of cellulose. Biotechnol Biofuels 5: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, G.T. (2002) Biodegradation of polyurethane: a review. Int Biodeterior Biodegradation 49: 245–252. [Google Scholar]
- Howard, G.T. (2012a) Polyurethane biodegradation In Microbial Degradation of Xenobiotics. Singh S.N. (ed). Berlin, Heidelberg: Springer‐Verlag, pp. 371–394. [Google Scholar]
- Howard, G.T. , and Blake, R.C. (1998) Growth of Pseudomonas fluorescens on a polyester–polyurethane and the purification and characterization of a polyurethanase–protease enzyme. Int Biodeterior Biodegradation 42: 213–220. [Google Scholar]
- Howard, G.T. , and Hilliard, N.P. (1999) Use of coomassie blue‐polyurethane interaction inscreening of polyurethanase proteins and polyurethanolytic bacteria. Int Biodeterior Biodegradation 43: 23–30. [Google Scholar]
- Howard, G.T. , Crother, B. , and Vicknair, J. (2001) Cloning, nucleotide sequencing and characterization of a polyurethanase gene (pueB) from Pseudomonas chlororaphis . Int Biodeterior Biodegradation 47: 141–149. [Google Scholar]
- Howard, G.T. , Norton, W.N. , and Burks, T. (2012b) Growth of Acinetobacter gerneri P7 on polyurethane and the purification and characterization of a polyurethanase enzyme. Biodegradation 23: 561–573. [DOI] [PubMed] [Google Scholar]
- Iiyoshi, Y. , Tsutsumi, Y. , and Nishida, T. (1998) Polyethylene degradation by lignin‐degrading fungi and manganese peroxidase. J Wood Sci 44: 222. [Google Scholar]
- Iwata, T. (2015) Biodegradable and bio‐based polymers: future prospects of eco‐friendly plastics. Angew Chem Int Edit 54: 3210–3215. [DOI] [PubMed] [Google Scholar]
- Jakubowicz, I. (2003) Evaluation of degradability of biodegradable polyethylene (PE). Polym Degrad Stabil 80: 39–43. [Google Scholar]
- Jambeck, J.R. , Geyer, R. , Wilcox, C. , Siegler, T.R. , Perryman, M. , Andrady, A. , et al (2015) Plastic waste inputs from land into the ocean. Science 347: 768–771. [DOI] [PubMed] [Google Scholar]
- Jariyasakoolroj, P. , Tashiro, K. , Wang, H. , Yamamoto, H. , Chinsirikul, W. , Kerddonfag, N. , and Chirachanchai, S. (2015) Isotropically small crystalline lamellae induced by high biaxial‐stretching rate as a key microstructure for super‐tough polylactide film. Polymer 68: 234–245. [Google Scholar]
- Jenkins, M.J. , and Harrison, K.L. (2008) The effect of crystalline morphology on the degradation of polycaprolactone in a solution of phosphate buffer and lipase. Polym Adv Technol 19: 1901–1906. [Google Scholar]
- Jensen, W.A. , Armstrong, J.M. , de Giorgio, J. , and Hearn, M.T.W. (1995) Stability studies on maize leaf phosphoenolpyruvate carboxylase. Biochemistry 34: 472–480. [DOI] [PubMed] [Google Scholar]
- Jeon, H.J. , and Kim, M.N. (2015) Functional analysis of alkane hydroxylase system derived from Pseudomonas aeruginosa E7 for low molecular weight polyethylene biodegradation. Int Biodeterior Biodegradation 103: 141–146. [Google Scholar]
- Jeyakumar, D. , Chirsteen, J. , and Doble, M. (2013) Synergistic effects of pretreatment and blending on fungi mediated biodegradation of polypropylenes. Biores Technol 148: 78–85. [DOI] [PubMed] [Google Scholar]
- Karlsson, S. , Ljungquist, O. , and Albertsson, A.‐C. (1988) Biodegradation of polyethylene and the influence of surfactants. Polym Degrad Stabil 21: 237–250. [Google Scholar]
- Kawai, F. , Oda, M. , Tamashiro, T. , Waku, T. , Tanaka, N. , Yamamoto, M. , et al (2014) A novel Ca2+‐activated, thermostabilized polyesterase capable of hydrolyzing polyethylene terephthalate from Saccharomonospora viridis AHK190. Appl Microbiol Biotechnol 98: 10053–10064. [DOI] [PubMed] [Google Scholar]
- Kitadokoro, K. , Thumarat, U. , Nakamura, R. , Nishimura, K. , Karatani, H. , Suzuki, H. , and Kawai, F. (2012) Crystal structure of cutinase est119 from Thermobifida alba AHK119 that can degrade modified polyethylene terephthalate at 1.76 A resolution. Polym Degrad Stabil 97: 771–775. [Google Scholar]
- Knoll, M. , Hamm, T.M. , Wagner, F. , Martinez, V. , and Pleiss, J. (2009) The PHA depolymerase engineering database: a systematic analysis tool for the diverse family of polyhydroxyalkanoate (PHA) depolymerases. BMC Bioinformatics 10: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolattukudy, P.E. (1981) Structure, biosynthesis, and biodegradation of cutin and suberin. Annual Rev Plant Physiol 32: 539–567. [Google Scholar]
- Koutny, M. , Sancelme, M. , Dabin, C. , Pichon, N. , Delort, A.‐M. , and Lemaire, J. (2006a) Acquired biodegradability of polyethylenes containing pro‐oxidant additives. Polym Degrad Stabil 91: 1495–1503. [Google Scholar]
- Koutny, M. , Lemaire, J. , and Delort, A.‐M. (2006b) Biodegradation of polyethylene films with prooxidant additives. Chemosphere 64: 1243–1252. [DOI] [PubMed] [Google Scholar]
- Krueger, M.C. , Harms, H. , and Schlosser, D. (2015) Prospects for microbiological solutions to environmental pollution with plastics. Appl Microbiol Biotechnol 99: 8857–8874. [DOI] [PubMed] [Google Scholar]
- Kyrikou, I. , and Briassoulis, D. (2007) Biodegradation of agricultural plastic films: a critical review. J Polym Environ 15: 125–150. [Google Scholar]
- Labow, R.S. , Erfle, D.J. , and Santerre, J.P. (1996) Elastase‐induced hydrolysis of synthetic solid substrates: poly(ester‐urea‐urethane) and poly(ether‐urea‐urethane). Biomaterials 17: 2381–2388. [DOI] [PubMed] [Google Scholar]
- Lamberti, G. (2014) Flow induced crystallisation of polymers. Chem Soc Rev 43: 2240–2252. [DOI] [PubMed] [Google Scholar]
- Law, K.L. , and Thompson, R.C. (2014) Microplastics in the seas. Science 345: 144–145. [DOI] [PubMed] [Google Scholar]
- Lee, J.H. , Lim, K.S. , Hahm, W.G. , and Kim, S.H. (2013) Properties of recycled and virgin poly(ethylene terephthalate) blend fibers. J Appl Polym Sci 128: 1250–1256. [Google Scholar]
- Liebminger, S. , Eberl, A. , Sousa, F. , Heumann, S. , Fischer‐Colbrie, G. , Cavaco‐Paulo, A. , and Guebitz, G.M. (2007) Hydrolysis of pet and bis‐(benzoyloxyethyl) terephthalate with a new polyesterase from Penicillium citrinum . Biocatal Biotrans 25: 171–177. [Google Scholar]
- Liu, R.Y.F. , Hu, Y.S. , Schiraldi, D.A. , Hiltner, A. , and Baer, E. (2004) Crystallinity and oxygen transport properties of PET bottle walls. J Appl Polym Sci 94: 671–677. [Google Scholar]
- Longhi, S. , Czjzek, M. , Lamzin, V. , Nicolas, A. , and Cambillau, C. (1997) Atomic resolution (1.0 Å) crystal structure of Fusarium solani cutinase: stereochemical analysis. J Mol Biol 268: 779–799. [DOI] [PubMed] [Google Scholar]
- Loredo‐Trevino, A. , Gutierrez‐Sanchez, G. , Rodruguez‐Herrera, R. , and Aguilar, C.N. (2012) Microbial enzymes involved in polyurethane biodegradation: a review. J Polym Environ 20: 258–265. [Google Scholar]
- Lucas, N. , Bienaime, C. , Belloy, C. , Queneudec, M. , Silvestre, F. , and Nava‐Saucedo, J.‐E. (2008) Polymer biodegradation: mechanisms and estimation techniques ‐ a review. Chemosphere 73: 429–442. [DOI] [PubMed] [Google Scholar]
- Lülsdorf, N. , Vojcic, L. , Hellmuth, H. , Weber, T.T. , Mussmann, N. , Martinez, R. , and Schwaneberg, U. (2015) A first continuous 4‐aminoantipyrine (4‐aap)‐based screening system for directed esterase evolution. Appl Microbiol Biotechnol 99: 5237–5246. [DOI] [PubMed] [Google Scholar]
- Manzur, A. , Cuamatzi, F. , and Favela, E. (1997) Effect of the growth of Phanerochaete chrysosporium in a blend of low density polyethylene and sugar cane bagasse. J Appl Polym Sci 66: 105–111. [Google Scholar]
- Marten, E. , Müller, R.‐J. , and Deckwer, W.‐D. (2003) Studies on the enzymatic hydrolysis of polyesters i. Low molecular mass model esters and aliphatic polyesters. Polym Degrad Stabil 80: 485–501. [Google Scholar]
- Marten, E. , Müller, R.‐J. , and Deckwer, W.‐D. (2005) Studies on the enzymatic hydrolysis of polyesters. II. Aliphatic‐aromatic copolyesters. Polym Degrad Stabil 88: 371–381. [Google Scholar]
- Martinez, C. , De Geus, P. , Lauwereys, M. , Matthyssens, G. , and Cambillau, C. (1992) Fusarium solani cutinase is a lipolytic enzyme with a catalytic serine accessible to solvent. Nature 356: 615–618. [DOI] [PubMed] [Google Scholar]
- Matsubara, M. , Suzuki, J. , Deguchi, T. , Miura, M. , and Kitaoka, Y. (1996) Characterization of manganese peroxidases from the hyperlignolytic fungus IZU‐154. Appl Environ Microbiol 62: 4066–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumiya, Y. , Murata, N. , Tanabe, E. , Kubota, K. , and Kubo, M. (2010) Isolation and characterization of an ether‐type polyurethane‐degrading micro‐organism and analysis of degradation mechanism by Alternaria sp. J Appl Microb 108: 1946–1953. [DOI] [PubMed] [Google Scholar]
- Miyakawa, T. , Mizushima, H. , Ohtsuka, J. , Oda, M. , Kawai, F. , and Tanokura, M. (2015) Structural basis for the Ca2+‐enhanced thermostability and activity of pet‐degrading cutinase‐like enzyme from Saccharomonospora viridis ahk190. Appl Microbiol Biotechnol 99: 4297–4307. [DOI] [PubMed] [Google Scholar]
- Motta, O. , Proto, A. , De Carlo, F. , De Caro, F. , Santoro, E. , Brunetti, L. , and Capunzo, M. (2009) Utilization of chemically oxidized polystyrene as co‐substrate by filamentous fungi. Int J Hyg Environ Health 212: 61–66. [DOI] [PubMed] [Google Scholar]
- Mueller, R.‐J. (2006) Biological degradation of synthetic polyesters–enzymes as potential catalysts for polyester recycling. Process Biochem 41: 2124–2128. [Google Scholar]
- Mukherjee, S. , and Kundu, P.P. (2014) Alkaline fungal degradation of oxidized polyethylene in black liquor: studies on the effect of lignin peroxidases and manganese peroxidases. J Appl Polym Sci 131: 40738. [Google Scholar]
- Mukherjee, S. , Roy Chowdhuri, U. , and Kundu, P.P. (2016) Bio‐degradation of polyethylene waste by simultaneous use of two bacteria: Bacillus licheniformis for production of bio‐surfactant and Lysinibacillus fusiformis for bio‐degradation. RSC Advances 6: 2982–2992. [Google Scholar]
- Müller, R.‐J. , Schrader, H. , Profe, J. , Dresler, K. , and Deckwer, W.‐D. (2005) Enzymatic degradation of poly(ethylene terephthalate): rapid hydrolyse using a hydrolase from T. fusca . Macro Rap Comm 26: 1400–1405. [Google Scholar]
- Nakajima‐Kambe, T. , Onuma, F. , Kimpara, N. , and Nakahara, T. (1995) Isolation and characterization of a bacterium which utilizes polyester polyurethane as a sole carbon and nitrogen source. FEMS Microbiol L 129: 39–42. [DOI] [PubMed] [Google Scholar]
- Nakajima‐Kambe, T. , Shigeno‐Akutsu, Y. , Nomura, N. , Onuma, F. , and Nakahara, T. (1999) Microbial degradation of polyurethane, polyester polyurethanes and polyether polyurethanes. Appl Microbiol Biotechnol 51: 134–140. [DOI] [PubMed] [Google Scholar]
- Nakamiya, K. , Sakasita, G. , Ooi, T. , and Kinoshita, S. (1997) Enzymatic degradation of polystyrene by hydroquinone peroxidase of Azotobacter beijerinckii HM121. J Ferm Bioeng 84: 480–482. [Google Scholar]
- Nelms, S.E. , Duncan, E.M. , Broderick, A.C. , Galloway, T.S. , Godfrey, M.H. , Hamann, M. , et al (2016) Plastic and marine turtles: a review and call for research. ICES J Mar Sci 73: 165–181. [Google Scholar]
- Nimchua, T. , Punnapayak, H. , and Zimmermann, W. (2007) Comparison of the hydrolysis of polyethylene terephthalate fibers by a hydrolase from Fusarium oxysporum and Fusarium solani . Biotechnol J 2: 361–364. [DOI] [PubMed] [Google Scholar]
- Nomura, N. , Shigeno‐Akutsu, Y. , Nakajima‐Kambe, T. , and Nakahara, T. (1998) Cloning and sequence analysis of a polyurethane esterase of Comamonas acidovorans TB‐35. J Ferm Bioeng 86: 339–345. [Google Scholar]
- North, E.J. , and Halden, R.U. (2013) Plastics and environmental health: the road ahead. Rev Environ Health 28: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeser, T. , Wei, R. , Baumgarten, T. , Billig, S. , Föllner, C. , and Zimmermann, W. (2010) High level expression of a hydrophobic poly(ethylene terephthalate)‐hydrolyzing carboxylesterase from Thermobifida fusca KW3 in Escherichia coli bl21(de3). J Biotechnol 146: 100–104. [DOI] [PubMed] [Google Scholar]
- Ojeda, T. , Freitas, A. , Dalmolin, E. , Pizzol, M.D. , Vignol, L. , Melnik, J. , et al (2009) Abiotic and biotic degradation of oxo‐biodegradable foamed polystyrene. Polym Degrad Stabil 94: 2128–2133. [Google Scholar]
- Owen, S. , Otani, T. , Masaoka, S. , and Ohe, T. (1996) The biodegradation of low‐molecular‐weight urethane compounds by a strain of Exophiala jeanselmei . Biosci Biotechnol Biochem 60: 244–248. [DOI] [PubMed] [Google Scholar]
- Parikh, M. , Gross, R.A. , and McCarthy, S.P. (1993) The effect of crystalline morphology on enzymatic degradation kinetics In Biodegradable Polymers and Packaging. Ching C., Kaplan D.L., and Thomas E.L. (eds). Lancaster‐Basel: Technomic, pp. 407–415. [Google Scholar]
- Park, M.T. , Lee, M.S. , Choi, J.Y. , Kim, S.C. , and Lee, G.M. (2001) Orthophosphate anion enhances the stability and activity of endoxylanase from Bacillus sp . Biotechnol Bioeng 72: 434–440. [DOI] [PubMed] [Google Scholar]
- Pathirana, R.A. , and Seal, K.J. (1984a) Studies on polyurethane deteriorating fungi. I: isolation and characterization of the test fungi employed. Int Biodeterior 20: 163–168. [Google Scholar]
- Pathirana, R.A. , and Seal, K.J. (1984b) Studies on polyurethane deteriorating fungi. II: an examination of their enzyme activities. Int Biodeterior 20: 229–235. [Google Scholar]
- Phua, S.K. , Castillo, E. , Anderson, J.M. , and Hiltner, A. (1987) Biodegradation of a polyurethane in vitro. J Biomed Mat Res 21: 231–246. [DOI] [PubMed] [Google Scholar]
- PlasticsEurope (2016) Plastics – The Facts 2016. Brussels, Belgium: PlasticsEurope; http://www.plasticseurope.org/Document/plastics-the-facts-2016-15787.aspx?Page=DOCUMENT&FolID=2 [Google Scholar]
- Pometto, A.L. , Lee, B.T. , and Johnson, K.E. (1992) Production of an extracellular polyethylene‐degrading enzyme(s) by streptomyces species. Appl Environ Microbiol 58: 731–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto, A. (2016) To be, or not to be biodegradable… that is the question for the bio‐based plastics. Microb Biotech 9: 652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Research and Markets (2015) Global polyethylene terephthalate Market (PET resin) ‐ by end‐use industries, products, and regions ‐ market size, demand forecasts, industry trends and updates (2014–2020). http://www.researchandmarkets.com/reports/3505772/global-polyethylene-terephtalate-market-pet
- Restrepo‐Flórez, J.‐M. , Bassi, A. , and Thompson, M.R. (2014) Microbial degradation and deterioration of polyethylene – a review. Int Biodeterior Biodegradation 88: 83–90. [Google Scholar]
- Ribitsch, D. , Heumann, S. , Trotscha, E. , Herrero Acero, E. , Greimel, K. , Leber, R. , et al (2011) Hydrolysis of polyethyleneterephthalate by p‐nitrobenzylesterase from Bacillus subtilis . Biotechnol Prog 27: 951–960. [DOI] [PubMed] [Google Scholar]
- Ribitsch, D. , Yebra, A.O. , Zitzenbacher, S. , Wu, J. , Nowitsch, S. , Steinkellner, G. , et al (2013) Fusion of binding domains to Thermobifida cellulosilytica cutinase to tune sorption characteristics and enhancing pet hydrolysis. Biomacromol 14: 1769–1776. [DOI] [PubMed] [Google Scholar]
- Ribitsch, D. , Herrero Acero, E. , Przylucka, A. , Zitzenbacher, S. , Marold, A. , Gamerith, C. , et al (2015) Enhanced cutinase‐catalyzed hydrolysis of polyethylene terephthalate by covalent fusion to hydrophobins. Appl Environ Microbiol 81: 3586–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigamonti, L. , Grosso, M. , Mueller, J. , Martinez Sanchez, V. , Magnani, S. , and Christensen, T.H. (2014) Environmental evaluation of plastic waste management scenarios. Resour Conserv Recy 85: 42–53. [Google Scholar]
- Rojo, F. (2010) Enzymes for aerobic degradation of alkanes In Handbook of Hydrocarbon and Lipid Microbiology. Timmis K.N. (ed). Berlin, Heidelberg: Springer‐Verlag, pp. 781–797. [Google Scholar]
- Ronkvist, Ã.S.M. , Xie, W. , Lu, W. , and Gross, R.A. (2009) Cutinase‐catalyzed hydrolysis of poly(ethylene terephthalate). Macromolecules 42: 5128–5138. [Google Scholar]
- Roth, C. , Wei, R. , Oeser, T. , Then, J. , Foellner, C. , Zimmermann, W. , and Sträter, N. (2014) Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca . Appl Microbiol Biotechnol 98: 7815–7823. [DOI] [PubMed] [Google Scholar]
- Roy, P.K. , Titus, S. , Surekha, P. , Tulsi, E. , Deshmukh, C. , and Rajagopal, C. (2008) Degradation of abiotically aged LDPE films containing pro‐oxidant by bacterial consortium. Polym Degrad Stab 93: 1917–1922. [Google Scholar]
- Ruiz, C. , Main, T. , Hilliard, N.P. , and Howard, G.T. (1999) Purification and characterization of two polyurethanase enzymes from Pseudomonas chlororaphis . Int Biodeterior Biodegradation 43: 43–47. [Google Scholar]
- Russell, J.R. , Huang, J. , Anand, P. , Kucera, K. , Sandoval, A.G. , Dantzler, K.W. , et al (2011) Biodegradation of polyester polyurethane by endophytic fungi. Appl Environ Microbiol 77: 6076–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammond, D.W. , Yarbrough, J.M. , Mansfield, E. , Bomble, Y.J. , Hobdey, S.E. , Decker, S.R. , et al (2014) Predicting enzyme adsorption to lignin films by calculating enzyme surface hydrophobicity. J Biolog Chem 289: 20960–20969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santo, M. , Weitsman, R. , and Sivan, A. (2013) The role of the copper‐binding enzyme ‐ laccase ‐ in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber . Int Biodeterior Biodegradation 84: 204–210. [Google Scholar]
- Sarkar, S. , Singha, P.K. , Dey, S. , Mohanty, M. , and Adhikari, B. (2006) Synthesis, characterization, and cytotoxicity analysis of a biodegradable polyurethane Mater Manuf Process 21: 291–296. [Google Scholar]
- Schmidt, J. , Wei, R. , Oeser, T. , Belisário‐Ferrari, M.R. , Barth, M. , Then, J. , and Zimmermann, W. (2016) Effect of Tris, MOPS, and phosphate buffers on the hydrolysis of polyethylene terephthalate films by polyester hydrolases. FEBS Open Bio 6: 919–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, J. , Wei, R. , Oeser, T. , Dedavid e Silva, L.A. , Breite, D. , Schulze, A. , and Zimmermann, W. (2017) Degradation of polyester polyurethane by bacterial polyester hydrolases. Polymers 9: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen, S.K. , and Raut, S. (2015) Microbial degradation of low density polyethylene (LDPE): a review. J Environ Chem Eng 3: 462–473. [Google Scholar]
- Seymour, R.B. , and Kauffman, G.B. (1992) Polyurethanes: a class of modern versatile materials. J Chem Educ 69: 909. [Google Scholar]
- Shah, A.A. , Hasan, F. , Akhter, J.I. , Hameed, A. , and Ahmed, S. (2008a) Degradation of polyurethane by novel bacterial consortium isolated from soil. Ann Microbiol 58: 381. [Google Scholar]
- Shah, A.A. , Hasan, F. , Hameed, A. , and Ahmed, S. (2008b) Biological degradation of plastics: a comprehensive review. Biotech Adv 26: 246–265. [DOI] [PubMed] [Google Scholar]
- Silva, C. , Da, S. , Silva, N. , Matama, T. , Araujo, R. , Martins, M. , et al (2011) Engineered Thermobifida fusca cutinase with increased activity on polyester substrates. Biotechnol J 6: 1230–1239. [DOI] [PubMed] [Google Scholar]
- Sivan, A. (2011) New perspectives in plastic biodegradation. Curr Opin Biotech 22: 422–426. [DOI] [PubMed] [Google Scholar]
- Soroudi, A. , and Jakubowicz, I. (2013) Recycling of bioplastics, their blends and biocomposites: a review. Euro Polym J 49: 2839–2858. [Google Scholar]
- Sowmya, H.V. , Ramalingappa, M.K. , and Thippeswamy, B. (2014) Biodegradation of polyethylene by Bacillus cereus . Adv Polym Sci Technol‐ Int J 4: 28–32. [Google Scholar]
- Sowmya, H.V. , Ramalingappa, M.K. , and Thippeswamy, B. (2015) Degradation of polyethylene by Penicillium simplicissimum isolated from local dumpsite of shivamogga district. Environ Develop Sustain 17: 731–745. [Google Scholar]
- Stern, R.V. , and Howard, G.T. (2000) The polyester polyurethanase gene (pueA) from Pseudomonas chlororaphis encodes a lipase. FEMS Microbiol Lett 185: 163–168. [DOI] [PubMed] [Google Scholar]
- Suhas, Carrott, P.J.M. , and Ribeiro Carrott, M.M.L. (2007) Lignin – from natural adsorbent to activated carbon: a review. Biores Tech 98: 2301–2312. [DOI] [PubMed] [Google Scholar]
- Sulaiman, S. , Yamato, S. , Kanaya, E. , Kim, J.J. , Koga, Y. , Takano, K. , and Kanaya, S. (2012) Isolation of a novel cutinase homolog with polyethylene terephthalate‐degrading activity from leaf‐branch compost by using a metagenomic approach. Appl Environ Microbiol 78: 1556–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulaiman, S. , You, D.J. , Kanaya, E. , Koga, Y. , and Kanaya, S. (2014) Crystal structure and thermodynamic and kinetic stability of metagenome‐derived lc‐cutinase. Biochemistry 53: 1858–1869. [DOI] [PubMed] [Google Scholar]
- Swift, G. (2015) Degradable polymers and plastics in landfill sites In Encyclopedia of Polymer Science and Technology. Mark F.M. (ed). Hoboken: John Wiley & Sons, pp. 1–13. [Google Scholar]
- Tang, Y.W. , Labow, R.S. , and Santerre, J.P. (2001a) Enzyme‐induced biodegradation of polycarbonate‐polyurethanes: dependence on hard‐segment chemistry. J Biomed Mat Res 57: 597–611. [DOI] [PubMed] [Google Scholar]
- Tang, Y.W. , Labow, R.S. , and Santerre, J.P. (2001b) Enzyme‐induced biodegradation of polycarbonate polyurethanes: dependence on hard‐segment concentration. J Biomed Mat Res 56: 516–528. [DOI] [PubMed] [Google Scholar]
- Then, J. , Wei, R. , Oeser, T. , Barth, M. , Belisário‐Ferrari, M.R. , Schmidt, J. , and Zimmermann, W. (2015) Ca2+ and Mg2+ binding site engineering increases the degradation of polyethylene terephthalate films by polyester hydrolases from Thermobifida fusca . Biotechnol J 10: 592–598. [DOI] [PubMed] [Google Scholar]
- Then, J. , Wei, R. , Oeser, T. , Gerdts, A. , Schmidt, J. , Barth, M. , and Zimmermann, W. (2016) A disulfide bridge in the calcium binding site of a polyester hydrolase increases its thermal stability and activity against polyethylene terephthalate. FEBS Open Bio 6: 425–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, R.C. , Swan, S.H. , Moore, C.J. , and vom Saal, F.S. (2009) Our plastic age. Phil Trans R Soc B 364: 1973–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumarat, U. , Nakamura, R. , Kawabata, T. , Suzuki, H. , and Kawai, F. (2012) Biochemical and genetic analysis of a cutinase‐type polyesterase from a thermophilic Thermobifida alba AHK119. Appl Microbiol Biotechnol 95: 419–430. [DOI] [PubMed] [Google Scholar]
- Tokiwa, Y. , Calabia, B. , Ugwu, C. , and Aiba, S. (2009) Biodegradability of plastics. Int J Mol Sci 10: 3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urgun‐Demirtas, M. , Singh, D. , and Pagilla, K. (2007) Laboratory investigation of biodegradability of a polyurethane foam under anaerobic conditions. Polym Degrad Stabil 92: 1599–1610. [Google Scholar]
- Van Cauwenberghe, L. , Vanreusel, A. , Mees, J. , and Janssen, C.R. (2013) Microplastic pollution in deep‐sea sediments. Environ Poll 182: 495–499. [DOI] [PubMed] [Google Scholar]
- Vertommen, M.A. , Nierstrasz, V.A. , Veer, M. , and Warmoeskerken, M.M. (2005) Enzymatic surface modification of poly(ethylene terephthalate). J Biotechnol 120: 376–386. [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Ma, Z. , and Li, L. (2016) Flow‐induced crystallization of polymers: molecular and thermodynamic considerations. Macromolecules 49: 1505–1517. [Google Scholar]
- Webb, H. , Arnott, J. , Crawford, R. , and Ivanova, E. (2013) Plastic degradation and its environmental implications with special reference to poly(ethylene terephthalate). Polymers 5: 1. [Google Scholar]
- Wei, R. (2011) Hydrolysis of polyethylene terephthalate by cutinases from Thermobifida fusca KW3. PhD Thesis. Leipzig: Universität Leipzig. [Google Scholar]
- Wei, R. , Oeser, T. , Billig, S. , and Zimmermann, W. (2012) A high‐throughput assay for enzymatic polyester hydrolysis activity by fluorimetric detection. Biotechnol J 7: 1517–1521. [DOI] [PubMed] [Google Scholar]
- Wei, R. , Oeser, T. , Barth, M. , Weigl, N. , Lübs, A. , Schulz‐Siegmund, M. , et al (2014a) Turbidimetric analysis of the enzymatic hydrolysis of polyethylene terephthalate nanoparticles. J Mol Catal B‐Enzym 103: 72–78. [Google Scholar]
- Wei, R. , Oeser, T. , Then, J. , Kühn, N. , Barth, M. , and Zimmermann, W. (2014b) Functional characterization and structural modeling of synthetic polyester‐degrading hydrolases from Thermomonospora curvata . AMB Express 4: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, R. , Oeser, T. , and Zimmermann, W. (2014c) Synthetic polyester‐hydrolyzing enzymes from thermophilic actinomycetes. Adv Appl Microbiol 89: 267–305. [DOI] [PubMed] [Google Scholar]
- Wei, R. , Oeser, T. , Schmidt, J. , Meier, R. , Barth, M. , Then, J. , and Zimmermann, W. (2016) Engineered bacterial polyester hydrolases efficiently degrade polyethylene terephthalate due to relieved product inhibition. Biotechnol Bioeng 113: 1658–1665. [DOI] [PubMed] [Google Scholar]
- Wilcox, C. , Van Sebille, E. , and Hardesty, B.D. (2015) Threat of plastic pollution to seabirds is global, pervasive, and increasing. Proc Natl Acad Sci USA 112: 11899–11904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Economic Forum (2016) The new plastics economy: Rethinking the future of plastics. http://www3.weforum.org/docs/WEF_The_New_Plastics_Economy.pdf
- WRAP (2016) Plastics market situation report Waste & Resources Action Programme, Banbury, UK. http://www.wrap.org.uk/sites/files/wrap/Plastics_Market_Situation_Report.pdf
- Yang, J. , Yang, Y. , Wu, W.‐M. , Zhao, J. , and Jiang, L. (2014) Evidence of polyethylene biodegradation by bacterial strains from the guts of plastic‐eating waxworms. Environ Sci Technol 48: 13776–13784. [DOI] [PubMed] [Google Scholar]
- Yang, Y. , Yang, J. , Wu, W.‐M. , Zhao, J. , Song, Y. , Gao, L. , et al (2015) Biodegradation and mineralization of polystyrene by plastic‐eating mealworms: part 2. Role of gut microorganisms. Environm Sci Technol 49: 12087–12093. [DOI] [PubMed] [Google Scholar]
- Yates, M.R. , and Barlow, C.Y. (2013) Life cycle assessments of biodegradable, commercial biopolymers ‐ a critical review. Resour Conser Recycl 78: 54–66. [Google Scholar]
- Yoon, M. , Jeon, H. , and Kim, M. (2012) Biodegradation of polyethylene by a soil bacterium and AlkB cloned recombinant cell. J Bioremed Biodegrad 3: 145. [Google Scholar]
- Yoshida, S. , Hiraga, K. , Takehana, T. , Taniguchi, I. , Yamaji, H. , Maeda, Y. , et al (2016) A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351: 1196–1199. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Wang, X. , Gong, J. , and Gu, Z. (2004) A study on the biodegradability of polyethylene terephthalate fiber and diethylene glycol terephthalate. J Appl Polym Sci 93: 1089–1096. [Google Scholar]
- Zhang, Q. , Zhang, R. , Meng, L. , Lin, Y. , Chen, X. , Li, X. , et al (2016) Biaxial stretch‐induced crystallization of poly(ethylene terephthalate) above glass transition temperature: the necessary of chain mobility. Polymer 101: 15–23. [Google Scholar]
- Zheng, Y. , Yanful, E.K. , and Bassi, A.S. (2005) A review of plastic waste biodegradation. Crit Rev Biotechnol 25: 243–250. [DOI] [PubMed] [Google Scholar]
- Zimmermann, W. , and Billig, S. (2011) Enzymes for the biofunctionalization of poly(ethylene terephthalate). Adv Biochem Eng Biot 125: 97–120. [DOI] [PubMed] [Google Scholar]


