Abstract
Autism spectrum disorders (ASDs) are neurodevelopmental disorders caused by various genetic and environmental factors that result in synaptic abnormalities. ASD development is suggested to involve microglia, which have a role in synaptic refinement during development. Autophagy and related pathways are also suggested to be involved in ASDs. However, the precise roles of microglial autophagy in synapses and ASDs are unknown. Here, we show that microglial autophagy is involved in synaptic refinement and neurobehavior regulation. We found that deletion of atg7, which is vital for autophagy, from myeloid cell-specific lysozyme M-Cre mice resulted in social behavioral defects and repetitive behaviors, characteristic features of ASDs. These mice also had increases in dendritic spines and synaptic markers and altered connectivity between brain regions, indicating defects in synaptic refinement. Synaptosome degradation was impaired in atg7-deficient microglia and immature dendritic filopodia were increased in neurons co-cultured with atg7-deficient microglia. To our knowledge, our results are the first to show the role of microglial autophagy in the regulation of the synapse and neurobehaviors. We anticipate our results to be a starting point for more comprehensive studies of microglial autophagy in ASDs and the development of putative therapeutics.
Introduction
Autism spectrum disorders (ASDs) are neurodevelopmental disorders characterized by impaired social interaction, communication deficits, repetitive behaviors, and narrow and intense interests.1 Various genetic studies suggest the association of ASD with abnormalities in cellular pathways related to postsynaptic glutamatergic synapses.2, 3 Increased dendritic spine density has been found in ASD brains4 and abnormal synaptic structures were observed in ASD model mice.5
Postnatal synaptic development is dynamically regulated by concurrent synapse formation and elimination in the mammalian cerebral cortex.6, 7 A surplus of synapses are formed early in development, more than what are usually maintained in the mature brain. The extra and unnecessary synapses are subsequently eliminated and a subset of synapses is maintained and strengthened.8, 9 Hence, precise regulation of synapse formation and elimination is important for the normal development of the brain, and reduced elimination of synapses, resulting in an excess, is thought to be associated with neurodevelopmental disorders such as ASD.10, 11
Increasing evidence suggests a central role for immune dysregulation in ASDs. Several ASD-risk genes are associated with the immune system and maternal immune system-related risk factors are related to ASDs.12 Microglia, a representative immune cell in the brain, have an important role in synaptic refinement10, 13, 14, 15 and are also thought to be involved in the pathogenesis of ASDs.12, 16, 17, 18, 19, 20 Many reports also show alterations in the activation, amount and distribution of microglia in ASD brains.17, 18, 19, 21
Various studies have suggested that autophagy pathways are involved in the pathogenesis of ASDs.22 ASD-associated exonic copy-number variants have been reported in the genes coding for proteins involved in autophagy pathways.23 Autophagy is the catabolic process that sequesters cytoplasm, including aberrant organelles and macromolecules, into double-membrane vesicles and delivers it to lysosomes for degradation and eventual recycling of the resulting macromolecules.24 Mammalian target of rapamycin (mTOR) is one of the important inhibitory regulators of autophagy induction.25, 26 Mice with PTEN mutation-mediated mTOR disinhibition, which inhibits autophagy, display autistic behaviors and abnormal neuronal arborization, suggesting that autophagy is deregulated in ASD.27 Autism-like phenotypes were observed in female mice lacking ambra-1, which is a positive regulator of beclin-1, a principal player in autophagosome formation.28 Inhibition of mTOR by rapamycin, which activates autophagy, restores autism-like symptoms and improves abnormal neuro-anatomical structures in PTEN mutant mice.29, 30 Recently, hyperactivated mTOR and impaired autophagy were observed in the post mortem temporal cortex of ASD patients.11
Accordingly, we speculated that the autophagic processes of microglia might be involved in synaptic pruning and wondered if microglial autophagy is also important in autistic behaviors. Although glia are much more abundant in the brain, and were recently identified as having a more important role in the regulation of synaptic activity and maintenance of homeostasis of the brain than previously thought,19, 23 the relationship between microglial autophagy and ASD remains to be elucidated.
Materials and methods
Animals
To generate mice lacking the atg7 gene in cells of the myeloid lineage, including microglia, we crossed Lyz2-Cre mice (stock number 4781; Jackson Laboratories, Bar Harbor, ME, USA) with Atg7fl/fl mice (provided by Masaaki Komatsu of the Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan). Littermates were used in all experiments. For direct visualization of Cre expression, ROSA26-tdTomato (stock number 7914; Jackson Laboratories) was used. PCR genotyping was conducted with the following primers: 5′-CCCAGAAATG CCAGATTACG-3′ for wild-type, 5′-CTTGGGCTGC CAGAATTTCTC-3′ for common and 5′-TTACAGTCGG CCAGGCTGAC-3′ for Cre in Lyz2-Cre mice; and 5′-CCACTGGCCC ATCAGTGAGCATG-3′ for wild-type, 5′-CATCTTGTAGCACCTGCTGACCTGC-3′ for common and 5′-GCGGATCCTCGTATAATGTATGCTATACGAAGTTAT-3′ for loxP in Atg7fl/fl mice. Animals were bred at the Laboratory Animal Facility in the Asan Institute for Life Sciences under specific pathogen-free conditions and maintained at a constant ambient temperature (22±1 °C) with a 12:12-h light:dark cycle. Animals were housed 3–5 per cage with ad libitum access to food and water. All experiments were designed to minimize the number of animals used, and all procedures were carried out in accordance with the Institute of Laboratory Animal Resources (ILAR) ‘Guide for the Care and Use of Laboratory Animals’. This study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the Asan Institute for Life Science (approval number 2014-02-022).
Mouse behavior tests
Detailed materials and methods are provided in the Supplementary Information.
Cell cultures
Microglia were isolated from mixed primary glial cultures that were obtained from the cerebral cortices of 3-day-old mice as previously described.31 Primary cultures of mouse cortical neurons were prepared from the brains of embryonic day 16 pups as previously described.32 Detailed materials and methods are provided in the Supplementary Information.
Western blot and other experiments
Western blots were performed according to standard methods under denaturing and reducing conditions as previously described.33 Detailed materials and methods are provided in the Supplementary Information.
Results
Generation of mice with Atg7-deficient microglia
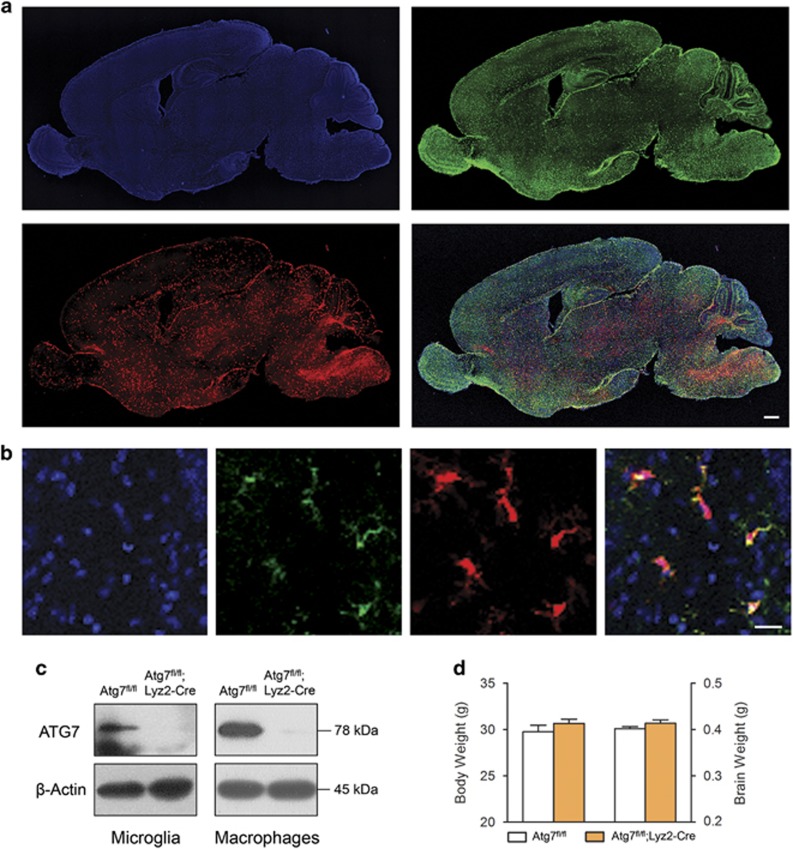
To investigate the contribution of microglial autophagy to synaptic refinement and ASD, we produced Atg7fl/fl;Lyz2-Cre mice. In these mice, Atg7, which is vital for autophagy, is deleted in myeloid lineage-specific lysozyme M-expressing cells as previously described.34 By crossing Atg7fl/fl;Lyz2-Cre mouse with ROSA26-tdTomato reporter mice harboring loxP-STOP-loxP sequences before the tdTomato gene, we visualized the Cre-expressing microglia. As shown in the sagittal plane of Atg7fl/+;Lyz2Cre/+;ROSA26-tdTomato mouse brain at postnatal day 7, Tomato-expressing microglia were detected in all brain regions (Figure 1a) and colocalized with Iba-1-positive microglia (Figure 1b). The protein level of Atg7 in primary cultured microglia as well as in peritoneal macrophages collected from Atg7fl/fl;Lyz2-Cre mice was either not detectable or very low in western blot analysis (Figure 1c). Atg7fl/fl;Lyz2-Cre mice were developmentally normal and fertile. Adults did not show any differences in body or brain weight compared with controls (Figure 1d).
Figure 1.
Phenotypes of mice with Atg7-deficient microglia. (a) Sagittal images of Atg7fl/+;Lyz2Cre/+;ROSA26-tdTomato mouse brain at postnatal day 7 stained with an antibody against Iba-1 (green) and Hoechst 33342 (blue). Tomato expression (red) was detected in all brain regions. Scale bar=500 μm. (b) Most of the Tomato expression (red) was co-localized with Iba-1-positive microglia (green). Scale bar=20 μm. (c) Atg7 protein was not detected in the lysate of primary microglia cultures or in peritoneal macrophages collected from Atg7fl/fl;Lyz2-Cre mouse. (d) Body and brain weights of adult Atg7fl/fl;Lyz2-Cre mice were similar to those of controls (n=12 for each group).
Mice with Atg7-deficient microglia show impaired social interaction and repetitive behavior
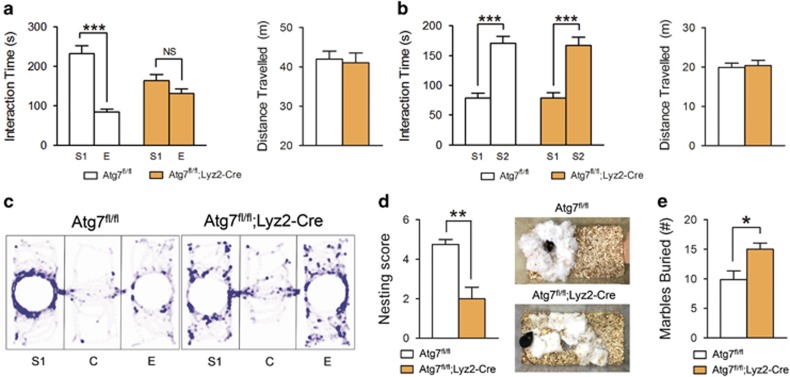
Autism is defined by impaired social interaction and communication, and repetitive behaviors. First, we examined whether Atg7fl/fl;Lyz2-Cre mice showed abnormal social interaction. In the three-chamber social interaction assay, Atg7fl/fl;Lyz2-Cre male mice did not show any preference for stranger 1, whereas control male mice spent significantly more time with stranger 1 than with the empty cup, indicating that Atg7fl/fl;Lyz2-Cre mice displayed autistic-like impairment of social interaction (Figure 2a). When the empty cup was occupied with another novel mouse (stranger 2), both control and Atg7fl/fl;Lyz2-Cre mice preferred to explore around stranger 2 (Figure 2b). There were no differences between the groups in the total distance travelled during each test (Figures 2a–c). Similar to male mice, female mice also showed deficient sociability (Supplementary Figure S1). These results show that mice with autophagy-deficient microglia have impaired sociability but normal social recognition. Next, we assessed the marble-burying test and nest-building behaviors. Atg7fl/fl;Lyz2-Cre mice buried more marbles than the controls (Figure 2d), reflecting increased repetitive behavior, which is one of the features of ASD. Atg7fl/fl;Lyz2-Cre mice also had worse nest-building scores than controls (Figure 2e).
Figure 2.
Autistic behaviors in mice with Atg7-deficient microglia. (a) In the three-chamber sociability test, Atg7fl/fl;Lyz2-Cre mice did not show any preference for stranger 1 (S1), whereas Atg7fl/fl mice spent more time in contact with S1 than an empty cup (E). No difference was observed in the total distance travelled between groups for 10 min. n=12 for each group. (b) Both Atg7fl/fl and Atg7fl/fl;Lyz2-Cre mice interacted with stranger 2 (S2) more than with S1 during a social novelty test. n=12 for each group. (c) Representative tracks during the sociability test are displayed. (d) In the nest-building test, Atg7fl/fl;Lyz2-Cre mice had lower scores than controls. n=7 for Atg7fl/fl, n=8 for Atg7fl/fl;Lyz2-Cre. (e) Atg7fl/fl;Lyz2-Cre mice buried more marbles than control mice in a 30-min period in the marble-burying test. n=7 for each group. *P<0.05, **P<0.01, ***P<0.001. NS, not significant (two-tailed Student’s t-test).
Increased dendritic spine density in mice with Atg7-deficient microglia
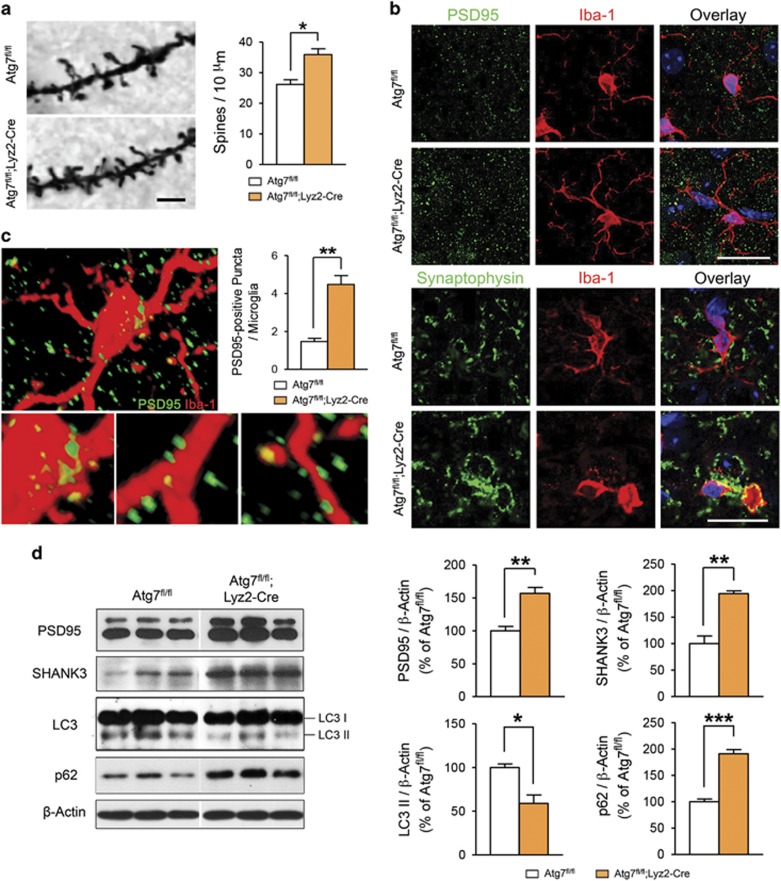
We hypothesized that the autistic-like behavior observed in Atg7fl/fl;Lyz2-Cre mice was owing to dysfunctional synaptic pruning by microglia during early brain development. Microglia have a crucial role in the development of functional brain connectivity by eliminating unnecessary synapses through phagolysosomes during the first 3 weeks of life.15 To test our hypothesis, we used a Golgi-Cox staining method to measure the numbers of dendritic spines in the primary auditory cortex (A1) and secondary somatosensory cortex (S2), which are considered the temporal cortices of mice.11, 35 At postnatal day 15, when synaptic pruning is frequent, dendritic spine numbers in basal segments were significantly increased in Atg7fl/fl;Lyz2-Cre mice compared with control animals (Figure 3a). In parallel, the protein level of PSD95 was significantly higher in Atg7fl/fl;Lyz2-Cre mice than in control mice (Figures 3d and e). Furthermore, we also found a higher protein level of SHANK3, a member of the SHANK family of postsynaptic density proteins, than in control mice (Figure 3d). Because SHANK3 is a key protein in synaptic scaffolding and imbalanced synaptic signaling may be involved in ASD,36 it is possible that the imbalance in SHANK proteins and the resulting signaling cascades induced the autistic-like behaviors observed in Atg7fl/fl;Lyz2-Cre mice.
Figure 3.
Increased dendritic spine density in mice with Atg7-deficient microglia. (a) Representative images of dendrites in the somatosensory 2 (S2) brain region from Atg7fl/fl and Atg7fl/fl;Lyz2-Cre mouse after Golgi-Cox staining. The number of dendritic spines in Atg7fl/fl;Lyz2-Cre mice was significantly increased compared with control mice at postnatal day 15. n=150 segments for Atg7fl/fl, n=60 segments for Atg7fl/fl;Lyz2-Cre. Scale bar=2 μm. (b) PSD95 and synaptophysin immunoreactivity was more frequently co-localized with Iba-1 immunoreactivity in the brain of Atg7fl/fl;Lyz2-Cre mice than that of Atg7fl/fl mice. Representative images from each group are shown. Scale bar=20 μm. (c) Three-dimensional confocal image, constructed from serial confocal z-stack images, of Iba-1-positive microglia (red) from an Atg7fl/fl;Lyz2-Cre mouse. High-magnitude images of PSD95-positive puncta (green) engulfed by Iba-1-positive microglia are shown in the lower panel. The number of PSD95-positive puncta per microglia (yellow) was significantly increased in Atg7fl/fl;Lyz2-Cre mice compared with controls at postnatal day 12. n=40 cells for each group. (d) Western blot images of synaptic markers (PSD95 and SHANK3) and autophagy-related proteins (LC3-II and p62). Intensities of both PSD95 and SHANK3 were significantly higher in Atg7fl/fl;Lyz2-Cre mice than in the controls. In Atg7fl/fl;Lyz2-Cre mice, the LC3-II intensity was decreased and that of p62 is increased compared with Atg7fl/fl mice. n=5 for each group. *P<0.05, **P<0.01, ***P<0.001 (two-tailed Student’s t-test).
Again, we confirmed higher PSD95 immunoreactivity in the brain sections of Atg7fl/fl;Lyz2-Cre mice than in the controls (Figure 3b). By double-labeling with PSD95 antibody and Iba-1 antibody, we found abundant co-localizations of PSD95- and Iba-1-positive immunoreactivities in Atg7fl/fl;Lyz2-Cre mice compared with control animals. When stained with an antibody against synaptophysin, a presynaptic marker, Iba-1 and synaptophysin co-localizations were also more frequently detected in Atg7fl/fl;Lyz2-Cre mice than in controls (Figure 3b). At postnatal day 12, we quantified the number of engulfed PSD95-positive puncta per microglia in the cortical area (Figure 3c). Engulfed PSD95-positive puncta were quantified in high-resolution three-dimensional projections of each microglia constructed from a confocal z-stack. The numbers of PSD95-positive puncta in Atg7fl/fl;Lyz2-Cre mice were significantly higher than in controls (Figure 3c).
In Atg7fl/fl;Lyz2-Cre mice, the protein levels of autophagy markers including LC3 (light chain-3)-II and p62 were significantly different from those of controls (Figure 3d). In western blot analysis, LC3-II, an autophagosome marker, was significantly decreased in Atg7fl/fl;Lyz2-cre mice and p62, a protein substrate for autophagy used for monitoring autophagic turnover,37 was significantly increased, reflecting the impaired autophagy in microglia. Taken together, we speculated that autophagy deficiency in microglia during early development results in increased dendritic spines because of reduced microglial autophagy-mediated elimination.
Impaired degradation of synapses and increased numbers of immature synapses owing to atg7-deficient microglia
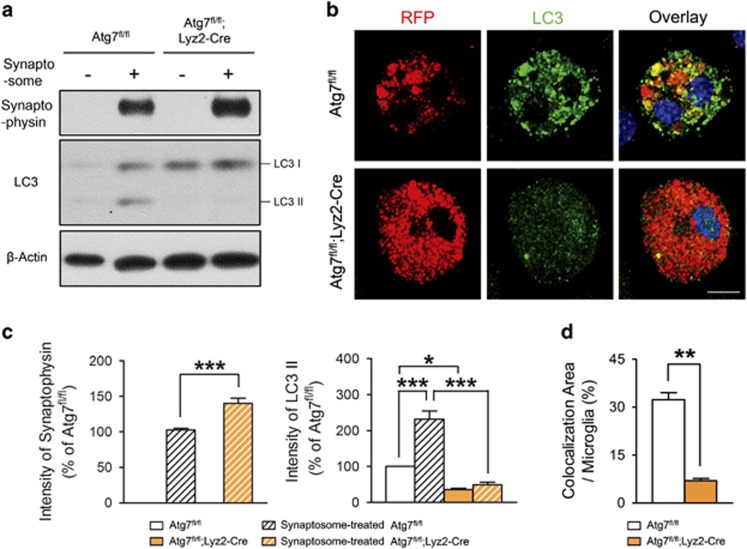
To investigate synaptic pruning by microglial autophagy in more detail, primary microglia were cultured from Atg7fl/fl or Atg7fl/fl;Lyz2-Cre mouse pups. We then treated microglial cultures with synaptosomes isolated from red fluorescent protein (RFP)-expressing mice to observe the degradation capacity of wild-type and atg7-deficient microglia (Figure 4). Western blots from cell lysates of cultures incubated for 12 h with synaptosomes showed that synaptophysin was increased in atg7-deficient microglia, indicating that synaptosome degradation was inefficient and delayed in atg7-deficient microglia compared with wild-type microglia (Figure 4a). Microtubule-associated protein LC3-II, an autophagosome marker, was decreased in atg7-deficient microglia, showing that autophagosome formation was impaired in atg7-deficient microglia (Figure 4a). RFP from synaptosomes accumulated more in atg7-deficient microglia than in wild-type microglia (Figure 4b), similar to the western blot results (Figure 4a). Immunocytochemistry with LC3 showed that the punctate forms of LC3, which represent autophagosomes, were increased in wild-type microglia and co-localized with RFP to a certain extent (Figure 4b), suggesting that autophagic processes are involved in the degradation of synapses. However, atg7-deficient microglia show few LC3 puncta and its scarce co-localization with RFP.
Figure 4.
Impaired degradation of the synaptosome by atg7-deficient microglia. (a, c) Primary microglia from Atg7fl/fl or Atg7fl/fl;Lyz2-Cre mice were cultured and isolated synaptosomes from red fluorescent protein (RFP)-expressing mouse brain were added to the microglial cultures. Western blots showed that synaptophysin is increased and LC3-II is decreased in atg7-deficient microglia compared with wild-type microglia. One-way analysis of variance with Tukey’s post hoc test. F(3,11)=355.40, P<0.001 (synaptophysin). F(3,11)=57.19, P<0.001 (LC3-II). n=3 for each group. (b, d) Immunocytochemistry of microglia showing that punctated forms of LC3 (green) were increased in wild-type microglia, with co-localization (yellow) of RFP-positive synaptosomes (red). LC3 signals were diffuse and the RFP signal was increased in atg7-deficient microglia compared with wild-type microglia. Scale bar=20 μm. *P<0.05, **P<0.01, ***P<0.001 (two-tailed Student’s t-test).
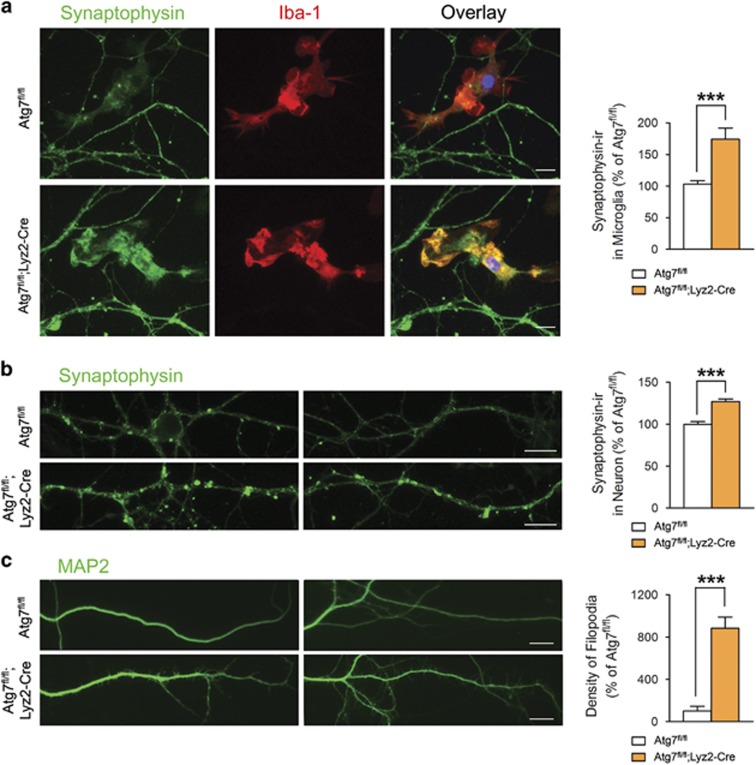
We next co-cultured primary mouse neurons with microglia from Atg7fl/fl or Atg7fl/fl;Lyz2-Cre mice to investigate the effects of microglial autophagy on the development of neurons. Similar to previous results (Figures 3 and 4), synaptophysin accumulation was more prominent in atg7-deficient microglia (Figure 5a), further confirming the impaired synapse degradation in autophagy-deficient microglia. Synaptophysin puncta were increased in neurons co-cultured with atg7-deficient microglia (Figure 5b), similar to the in vivo results (Figure 3). MAP2-positive dendritic filopodia were also increased in neurons co-cultured with atg7-deficient microglia (Figure 5c), suggesting that immature synapses were increased by autophagy-deficient microglia. Neurons co-cultured with atg7-deficient microglia also showed increased neurite thickness, formation of neurite fascicles and fragmented neurites (Supplementary Figure S2). Interestingly, these changes were not always accompanied by direct contact with microglia, suggesting that there are additional humoral factors affecting synapse development beyond the direct effect of microglial autophagy on synapse degradation.
Figure 5.
Neuronal changes after their co-culture with atg7-deficient microglia. (a) Primary mouse cortical neurons (18 days in vitro) were co-cultured with primary microglia from Atg7fl/fl or Atg7fl/fl;Lyz2-Cre mice for 2 weeks and then immunostained with antibodies against synaptophysin (green) and Iba-1 (red). Greater accumulation of synaptophysin was seen in atg7-deficient Iba-1-positive microglia than in wild-type microglia. Scale bar=20 μm. (b) Synaptophysin puncta were increased in neurons co-cultured with atg7-deficient microglia. Scale bar=20 μm. (c) MAP2-positive dendrites showed increased filopodia in neurons co-cultured with atg7-deficient microglia. Scale bar=20 μm. ***P<0.001 (two-tailed Student’s t-test).
Alteration of anatomical and functional brain connectivity in mice with autophagy-deficient microglia
Because deficient neuron-microglia signaling results in impaired functional brain connectivity,10 and neuritic and synaptic changes were observed in the neurons and brains of mice with atg7-deficient microglia (Figures 3, 4, 5 and Supplementary Figure S2), we explored brain connectivity using diffusion tensor imaging and resting-state functional magnetic resonance imaging (fMRI). In the brains of Atg7fl/fl;Lyz2-Cre mice, mean diffusivity was increased in S2 (Supplementary Figure S3a). This suggests that there are microstructural anomalies in several brain regions of Atg7fl/fl;Lyz2-Cre mice, which may affect anatomical and functional brain connectivity and may reflect the neuritic changes observed upon co-culture of neurons with atg7-deficient microglia (Supplementary Figure S2). Correlation matrices calculated from global fMRI BOLD signal analysis showed a trend toward altered fMRI functional connectivity from S2 to other cortical areas. The functional connectivity was increased from S2 to the anterior cingulate cortex, and decreased from S2 to the prefrontal cortex in Atg7fl/fl;Lyz2-cre mice compared with control mice (Supplementary Figure S3b). This further suggests that impaired microglial autophagy alters the microstructure and functional connectivity of brains, which may contribute to ASD pathogenesis.
Discussion
In the present study, we show for we believe the first time that loss of autophagy in early developmental microglia impairs synaptic pruning and results in increased dendritic spine density, and the abnormal social interaction and repetitive behaviors indicative of ASD. Hence, autophagy in microglia has an important role in regulating synaptic homeostasis and neuropsychological behaviors (Supplementary Figure S5).
Autophagy has been suggested to be involved in ASD pathogenesis.22, 23 Tang et al.11 recently discovered that autophagy is impaired in post mortem ASD temporal cortices by showing that the autophagosome marker LC3-II is decreased, and that the autophagy substrate p62 is increased. In addition, the phosphorylation of mTOR and its substrate is increased, indicating that mTOR is activated and able to inhibit autophagy. They further observed defects in synaptic pruning and ASD-like behaviors in the neuronal autophagy-deficient mice, showing that neuronal autophagy is important for regulating synaptic refinement and normal brain development. In addition, there may be another mechanism for synaptic pruning, namely, that microglia eliminates weak and inappropriate synapses by phagocytosis during postnatal development.13, 15 Results showing impaired autophagy in the post mortem ASD brain11 could be due to impaired autophagy not only in neurons, but also in glia, including microglia, because glia are more abundant than neurons. We showed here that microglial autophagy is also important in synaptic refinement and normal brain development (Figures 3, 4, 5). Impaired microglial autophagy may contribute to the overall autophagy impairment in the brains of ASD patients.11
Microglia are the resident macrophages in the brain that constantly survey the neural environment for pathogens, foreign materials and apoptotic cells; accordingly, microglia are considered the principal immunoeffector phagocytes in the brain.38, 39 Microglia not only degrade toxic materials, but also, once activated, secrete various inflammatory cytokines.31, 40 In addition to this phagocytic and inflammatory role, microglia have been recently revealed to have important regulatory roles in the normal brain. Microglia heve a major role in synaptic refinement by pruning weak and inappropriate synapses during postnatal development.13, 15 Compared with control mice, Atg7fl/fl;Lyz2-Cre mice showed increased dendritic spine density and synaptic markers, such as PSD95, SHANK3 and synaptophysin (Figure 3). Hence, autophagy in microglia is important for refining synapses during development, with impaired autophagy reducing synaptic pruning and increasing unnecessary synapses. Microglia can also secrete some factors such as insulin-like growth factor 1 to regulate neurogenesis and support neuronal survival during development.41, 42 Increased synaptic markers and dendritic filopodia, which reflect an inability of filopodia to properly mature into functional spines, as in ASD,43 in the co-culture of neurons with autophagy-deficient microglia, even in the absence of direct contact with microglia (Figure 5 and Supplementary Figure S2), suggests that the microglial secretion of certain factors affecting neurites and synapses is regulated by autophagy.
Autophagy is a catabolic process that degrades a cell’s own components, such as organelles or proteins, by capturing debris in double-membrane autophagosomes and fusing them with lysosomes. Notably, autophagic processes have recently been found to be involved in the degradation of not only a cell’s own components, but also extracellular materials such as pathogens and dead cells.31, 44, 45 Microglia deficient in atg7 show impaired degradation of synaptosomal fractions (Figure 4), which may reflect a disturbance in synaptic pruning by microglia. Hence, the extracellular synapses engulfed by microglia during development can be degraded by autophagic processes in microglia. Autophagy is important not only for degrading materials, but also for regulating inflammation.31, 44, 45 Macrophages are classified as classically activated (M1) or alternatively activated (M2) macrophages, which secrete pro-inflammatory and anti-inflammatory cytokines, respectively, and microglia can also be similarly classified.46, 47 Autophagy in macrophages is important for regulating the M1/M2 phenotype and its impairment increases pro-inflammatory M1 polarization and decreases anti-inflammatory M2 polarization.48, 49, 50, 51 Increased inflammation and immune dysregulation is associated with ASD52, 53, 54 and microglial activation is reported in ASD brains.17, 18, 19, 21 Microglial autophagy is important for regulating the inflammatory status in the brain31 and its impairment increases the expression of pro-inflammatory cytokines after LPS treatment (Supplementary Figure S4). Hence, under some inflammatory conditions that may increase the possibility of ASD,55, 56 impairment of microglial autophagy may aggravate the inflammatory condition, which may further affect ASD pathogenesis. Interestingly, autophagy, microglial activation and synaptic refinement by complement are also suggested to be involved in the pathogenesis of schizophrenia,57, 58, 59 which would be an interesting issue to address in the future.
The prevalence of ASD differs between males and females.60 Heterozygous ambra-1 deficiency results in autistic behaviors only in female mice, not in male mice. Activity-dependent neuroprotective protein (ADNP) is mutated in ASD patients and most male children with mutations, which to date, show more severe intellectual disability than females.61, 62, 63 ADNP haploinsufficiency results in a severe cognitive impairment in male mice, whereas female mice are partially spared.64, 65 Interestingly, ADNP is also expressed in microglia and is suggested to have compensatory protective functions against stressful conditions.66 These properties might also be impaired by ASD mutations. Males have more microglia in certain brain areas, including the preoptic area, hippocampus, parietal cortex and amygdala, suggesting that microglia have a role in the sexual differentiation of the brain.67, 68, 69 Although our experimental conditions revealed decreased sociability in both male and female mice with autophagy-deficient microglia (Figure 2 and Supplementary Figure 1), there could be differences between males and females in the severity or susceptibility of autistic behaviors and synapse regulations according to context and environment, which should be addressed in the future.
There is a wide degree of variation in the way ASD affects people. Regarding cognitive functions, autism can exist with any level of intelligence.70 Although intellectual disabilities are frequently accompanied by ASD, the proportion of ASD children who have IQs above 70 has increased.71 Thus, it will be interesting to understand the neurobiological background of the difference between ASD with low and high cognitive functions. Mice with autophagy-deficient microglia showed ASD phenotypes such as decreased sociability and repetitive behaviors, but intact cognitive function such as normal social novelty (Figure 2b). This feature may be a characteristic of the type of ASD caused by autophagy-deficient microglia, which will be an interesting topic to address in future studies.
Various MRI studies have revealed abnormalities in anatomical and functional connections in the brains of ASD patients.72, 73, 74 Defects in synaptic pruning by microglia during development result in reduced functional connectivity between brain regions and impaired social interaction.10 We also observed that microstructural connectivity is altered and that functional connectivity is decreased in the brains of Atg7fl/fl;Lyz2-cre mice in diffusion tensor imaging and fMRI analysis, which may be related to synaptic alterations caused by autophagy-deficient microglia (Supplementary Figure S3). Thus, autophagy in microglia may affect the functional connectivity between brain regions and the impairment of microglial autophagy results in reduced functional connectivity and ASD-like behaviors.
In summary, our findings reveal a role for microglial autophagy in synaptic pruning and maturation of neuronal connections during development and the formation of normal social behaviors. Increased dendritic spines and immature filopodia in neurons with autophagy-deficient microglia may contribute to ASD pathogenesis (Supplementary Figure S5). Hence, regulation of microglial autophagy could be a strategy for ASD therapy and prevention.
Acknowledgments
We thank Professor Masaaki Komatsu for providing the Atg7fl/fl mouse, Eun-Ju Chang for providing the RFP mouse brain and Eun-Young Choi for helping with the FACS sorting. This work was supported by the Medical Research Center Program through the National Research Foundation of Korea, funded by the Ministry of Science, Information and Communications Technology, and Future Planning (2008-0062286); the Korean Health Technology Research and Development Project, Ministry of Health & Welfare, Republic of Korea (HI13C1630); and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (2015R1A2A1A10053683).
Author contributions
H-JK performed the animal behavioral experiments; M-HC performed western blots and cell experiments; H-JK and M-HC performed immunohistochemistry and analyzed the data; WHS and JKK analyzed the fMRI and DTI data; E-YJ supported in performing the experiments; H-JK, M-HC, D-HK and S-YY wrote the manuscript; D-HK supervised the experiments; S-YY made the hypothesis, and designed and supervised the overall experiments.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
The authors declare no conflict of interest.
Supplementary Material
References
- King BH, Navot N, Bernier R, Webb SJ. Update on diagnostic classification in autism. Curr Opin Psychiatry 2014; 27: 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeron T. A synaptic trek to autism. Curr Opin Neurobiol 2009; 19: 231–234. [DOI] [PubMed] [Google Scholar]
- Peca J, Feng G. Cellular and synaptic network defects in autism. Curr Opin Neurobiol 2012; 22: 866–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutsler JJ, Zhang H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res 2010; 1309: 83–94. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol 2012; 4: a009886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purves D, Lichtman JW. Elimination of synapses in the developing nervous system. Science 1980; 210: 153–157. [DOI] [PubMed] [Google Scholar]
- Rakic P, Bourgeois JP, Eckenhoff MF, Zecevic N, Goldman-Rakic PS. Concurrent overproduction of synapses in diverse regions of the primate cerebral cortex. Science 1986; 232: 232–235. [DOI] [PubMed] [Google Scholar]
- Hua JY, Smith SJ. Neural activity and the dynamics of central nervous system development. Nat Neurosci 2004; 7: 327–332. [DOI] [PubMed] [Google Scholar]
- Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science 1996; 274: 1133–1138. [DOI] [PubMed] [Google Scholar]
- Zhan Y, Paolicelli RC, Sforazzini F, Weinhard L, Bolasco G, Pagani F et al. Deficient neuron-microglia signaling results in impaired functional brain connectivity and social behavior. Nat Neurosci 2014; 17: 400–406. [DOI] [PubMed] [Google Scholar]
- Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron 2014; 83: 1131–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes ML, McAllister AK. Immune mediators in the brain and peripheral tissues in autism spectrum disorder. Nat Rev Neurosci 2015; 16: 469–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 2012; 74: 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama R, Ikegaya Y. Microglia in the pathogenesis of autism spectrum disorders. Neurosci Res 2015; 100: 1–5. [DOI] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P et al. Synaptic pruning by microglia is necessary for normal brain development. Science 2011; 333: 1456–1458. [DOI] [PubMed] [Google Scholar]
- Rodriguez JI, Kern JK. Evidence of microglial activation in autism and its possible role in brain underconnectivity. Neuron Glia Biol 2011; 7: 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JT, Chana G, Pardo CA, Achim C, Semendeferi K, Buckwalter J et al. Microglial activation and increased microglial density observed in the dorsolateral prefrontal cortex in autism. Biol Psychiatry 2010; 68: 368–376. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Sugihara G, Ouchi Y, Nakamura K, Futatsubashi M, Takebayashi K et al. Microglial activation in young adults with autism spectrum disorder. JAMA Psychiatry 2013; 70: 49–58. [DOI] [PubMed] [Google Scholar]
- Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, Pardo CA. Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann Neurol 2005; 57: 67–81. [DOI] [PubMed] [Google Scholar]
- Gupta S, Ellis SE, Ashar FN, Moes A, Bader JS, Zhan J et al. Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat Commun 2014; 5: 5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetreault NA, Hakeem AY, Jiang S, Williams BA, Allman E, Wold BJ et al. Microglia in the cerebral cortex in autism. J Autism Dev Disord 2012; 42: 2569–2584. [DOI] [PubMed] [Google Scholar]
- Lee KM, Hwang SK, Lee JA. Neuronal autophagy and neurodevelopmental disorders. Exp Neurobiol 2013; 22: 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poultney CS, Goldberg AP, Drapeau E, Kou Y, Harony-Nicolas H, Kajiwara Y et al. Identification of small exonic CNV from whole-exome sequence data and application to autism spectrum disorder. Am J Hum Genet 2013; 93: 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci 2010; 13: 805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8: 445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SY, Choi JE, Kweon HS, Choe H, Kim SW, Hwang O et al. Okadaic acid increases autophagosomes in rat neurons: implications for Alzheimer's disease. J Neurosci Res 2008; 86: 3230–3239. [DOI] [PubMed] [Google Scholar]
- Kwon CH, Luikart BW, Powell CM, Zhou J, Matheny SA, Zhang W et al. Pten regulates neuronal arborization and social interaction in mice. Neuron 2006; 50: 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere E, Dahm L, Lu D, Hammerschmidt K, Ju A, Tantra M et al. Heterozygous ambra1 deficiency in mice: a genetic trait with autism-like behavior restricted to the female gender. Front Behav Neurosci 2014; 8: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Blundell J, Ogawa S, Kwon CH, Zhang W, Sinton C et al. Pharmacological inhibition of mTORC1 suppresses anatomical, cellular, and behavioral abnormalities in neural-specific Pten knock-out mice. J Neurosci 2009; 29: 1773–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CH, Zhu X, Zhang J, Baker SJ. mTor is required for hypertrophy of Pten-deficient neuronal soma in vivo. Proc Natl Acad Sci USA 2003; 100: 12923–12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MH, Cho K, Kang HJ, Jeon EY, Kim HS, Kwon HJ et al. Autophagy in microglia degrades extracellular beta-amyloid fibrils and regulates the NLRP3 inflammasome. Autophagy 2014; 10: 1761–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HL, Shim S, Kim DH, Won SH, Joo S, Kim S et al. beta-Amyloid is transmitted via neuronal connections along axonal membranes. Ann Neurol 2014; 75: 88–97. [DOI] [PubMed] [Google Scholar]
- Park JS, Ji IJ, An HJ, Kang MJ, Kang SW, Kim DH et al. Disease-associated mutations of TREM2 alter the processing of N-linked oligosaccharides in the Golgi apparatus. Traffic 2015; 16: 510–518. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006; 441: 880–884. [DOI] [PubMed] [Google Scholar]
- Benavides-Piccione R, Ballesteros-Yanez I, DeFelipe J, Yuste R. Cortical area and species differences in dendritic spine morphology. J Neurocytol 2002; 31: 337–346. [DOI] [PubMed] [Google Scholar]
- Lee EJ, Choi SY, Kim E. NMDA receptor dysfunction in autism spectrum disorders. Curr Opin Pharmacol 2015; 20: 8–13. [DOI] [PubMed] [Google Scholar]
- Ha YE, Kong KH, Cho MH, Kim DH, Song YS, Yoon SY. Vancomycin blocks autophagy and induces interleukin-1beta release in macrophages. J Antibiot (Tokyo) 2015; 68: 76–80. [DOI] [PubMed] [Google Scholar]
- Kreisel T, Frank MG, Licht T, Reshef R, Ben-Menachem-Zidon O, Baratta MV et al. Dynamic microglial alterations underlie stress-induced depressive-like behavior and suppressed neurogenesis. Mol Psychiatry 2014; 19: 699–709. [DOI] [PubMed] [Google Scholar]
- Streit WJ, Mrak RE, Griffin WS. Microglia and neuroinflammation: a pathological perspective. J Neuroinflam 2004; 1: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halle A, Hornung V, Petzold GC, Stewart CR, Monks BG, Reinheckel T et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol 2008; 9: 857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O, Ziv Y, Schwartz A, Landa G, Talpalar AE, Pluchino S et al. Microglia activated by IL-4 or IFN-gamma differentially induce neurogenesis and oligodendrogenesis from adult stem/progenitor cells. Mol Cell Neurosci 2006; 31: 149–160. [DOI] [PubMed] [Google Scholar]
- Ueno M, Fujita Y, Tanaka T, Nakamura Y, Kikuta J, Ishii M et al. Layer V cortical neurons require microglial support for survival during postnatal development. Nat Neurosci 2013; 16: 543–551. [DOI] [PubMed] [Google Scholar]
- Durand CM, Perroy J, Loll F, Perrais D, Fagni L, Bourgeron T et al. SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Mol Psychiatry 2012; 17: 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature 2011; 469: 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol 2013; 13: 722–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Le W. Differential roles of M1 and M2 microglia in neurodegenerative diseases. Mol Neurobiol 2015; 53: 1181–1194. [DOI] [PubMed] [Google Scholar]
- Orihuela R, McPherson CA, Harry GJ. Microglial M1/M2 polarization and metabolic states. Br J Pharmacol 2015; 173: 649–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CS, Kim JJ, Lee HM, Jin HS, Lee SH, Park JH et al. The AMPK-PPARGC1A pathway is required for antimicrobial host defense through activation of autophagy. Autophagy 2014; 10: 785–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouimet M. Autophagy in obesity and atherosclerosis: interrelationships between cholesterol homeostasis, lipoprotein metabolism and autophagy in macrophages and other systems. Biochim Biophys Acta 2013; 1831: 1124–1133. [DOI] [PubMed] [Google Scholar]
- Liao X, Sluimer JC, Wang Y, Subramanian M, Brown K, Pattison JS et al. Macrophage autophagy plays a protective role in advanced atherosclerosis. Cell Metab 2012; 15: 545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Zhao E, Ilyas G, Lalazar G, Lin Y, Haseeb M et al. Impaired macrophage autophagy increases the immune response in obese mice by promoting proinflammatory macrophage polarization. Autophagy 2015; 11: 271–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theoharides TC, Athanassiou M, Panagiotidou S, Doyle R. Dysregulated brain immunity and neurotrophin signaling in Rett syndrome and autism spectrum disorders. J Neuroimmunol 2015; 279: 33–38. [DOI] [PubMed] [Google Scholar]
- Rossignol DA, Frye RE. Evidence linking oxidative stress, mitochondrial dysfunction, and inflammation in the brain of individuals with autism. Front Physiol 2014; 5: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reus GZ, Fries GR, Stertz L, Badawy M, Passos IC, Barichello T et al. The role of inflammation and microglial activation in the pathophysiology of psychiatric disorders. Neuroscience 2015; 300: 141–154. [DOI] [PubMed] [Google Scholar]
- Ornoy A, Weinstein-Fudim L, Ergaz Z. Prenatal factors associated with autism spectrum disorder (ASD). Reprod Toxicol 2015; 56: 155–169. [DOI] [PubMed] [Google Scholar]
- Le Belle JE, Sperry J, Ngo A, Ghochani Y, Laks DR, Lopez-Aranda M et al. Maternal inflammation contributes to brain overgrowth and autism-associated behaviors through altered redox signaling in stem and progenitor cells. Stem Cell Rep 2014; 3: 725–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merenlender-Wagner A, Malishkevich A, Shemer Z, Udawela M, Gibbons A, Scarr E et al. Autophagy has a key role in the pathophysiology of schizophrenia. Mol Psychiatry 2015; 20: 126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N et al. Schizophrenia risk from complex variation of complement component 4. Nature 2016; 530: 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller N, Weidinger E, Leitner B, Schwarz MJ. The role of inflammation in schizophrenia. Front Neurosci 2015; 9: 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkovski M, Enticott PG, Fitzgerald PB. A review of the role of female gender in autism spectrum disorders. J Autism Dev Disord 2013; 43: 2584–2603. [DOI] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 2012; 485: 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 2012; 338: 1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsmoortel C, Vulto-van Silfhout AT, Coe BP, Vandeweyer G, Rooms L, van den Ende J et al. A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat Genet 2014; 46: 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amram N, Hacohen-Kleiman G, Sragovich S, Malishkevich A, Katz J, Touloumi O et al. Sexual divergence in microtubule function: the novel intranasal microtubule targeting SKIP normalizes axonal transport and enhances memory. Mol Psychiatry (e-pub ahead of print 19 January 2016; doi:10.1038/mp.2015.208). [DOI] [PubMed]
- Malishkevich A, Amram N, Hacohen-Kleiman G, Magen I, Giladi E, Gozes I. Activity-dependent neuroprotective protein (ADNP) exhibits striking sexual dichotomy impacting on autistic and Alzheimer's pathologies. Transl Psychiatry 2015; 5: e501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozes I, Zaltzman R, Hauser J, Brenneman DE, Shohami E, Hill JM. The expression of activity-dependent neuroprotective protein (ADNP) is regulated by brain damage and treatment of mice with the ADNP derived peptide, NAP, reduces the severity of traumatic head injury. Curr Alzheimer Res 2005; 2: 149–153. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Sholar PW, Bilbo SD. Sex differences in microglial colonization of the developing rat brain. J Neurochem 2012; 120: 948–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz KM, McCarthy MM. A starring role for microglia in brain sex differences. Neuroscientist 2015; 21: 306–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz KM, Nugent BM, Haliyur R, McCarthy MM. Microglia are essential to masculinization of brain and behavior. J Neurosci 2013; 33: 2761–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien G, Pearson J. Autism and learning disability. Autism 2004; 8: 125–140. [DOI] [PubMed] [Google Scholar]
- Investigators. AaDDMNSYP, (CDC). CfDCaP. Prevalence of autism spectrum disorders–Autism and Developmental Disabilities Monitoring Network, United States, 2006. MMWR Surveill Summ 2009; 58: 1–20. [PubMed] [Google Scholar]
- Sato JR, Balardin J, Vidal MC, Fujita A. Identification of segregated regions in the functional brain connectome of autistic patients by a combination of fuzzy spectral clustering and entropy analysis. J Psychiatry Neurosci 2015; 41: 140364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roine U, Roine T, Salmi J, Nieminen-von Wendt T, Tani P, Leppamaki S et al. Abnormal wiring of the connectome in adults with high-functioning autism spectrum disorder. Mol Autism 2015; 6: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahamy A, Behrmann M, Malach R. The idiosyncratic brain: distortion of spontaneous connectivity patterns in autism spectrum disorder. Nat Neurosci 2015; 18: 302–309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.