Abstract
Tumor suppressor TP53 is frequently mutated in colorectal cancer (CRC), and most mutations are missense type. Although gain-of-functions by mutant p53 have been demonstrated experimentally, the precise mechanism for malignant progression in in vivo tumors remains unsolved. We generated ApcΔ716 Trp53LSL•R270H villin-CreER compound mice, in which mutant p53R270H was expressed in the intestinal epithelia upon tamoxifen treatment, and examined the intestinal tumor phenotypes and tumor-derived organoids. Mutant Trp53R270H, but not Trp53-null mutation accelerated submucosal invasion with generation of desmoplastic microenvironment. The nuclear accumulation of p53 was evident in ApcΔ716 Trp53R270H/R270H homozygous tumors like human CRC. Although p53 was distributed to the cytoplasm in ApcΔ716 Trp53+/R270H heterozygous tumors, it accumulated in the nuclei at the invasion front, suggesting a regulation mechanism for p53 localization by the microenvironment. Importantly, mutant p53 induced drastic morphological changes in the tumor organoids to complex glandular structures, which was associated with the acquisition of invasiveness. Consistently, the branching scores of human CRC that carry TP53 mutations at codon 273 significantly increased in comparison with those of TP53 wild-type tumors. Moreover, allografted ApcΔ716 Trp53R270H/R270H organoid tumors showed a malignant histology with an increased number of myofibroblasts in the stroma. These results indicate that nuclear-accumulated mutant p53R270H induces malignant progression of intestinal tumors through complex tumor gland formation and acquisition of invasiveness. Furthermore, RNA sequencing analyses revealed global gene upregulation by mutant p53R270H, which was associated with the activation of inflammatory and innate immune pathways. Accordingly, it is possible that mutant p53R270H induces CRC progression, not only by a cell intrinsic mechanism, but also by the generation or activation of the microenvironment, which may synergistically contribute to the acceleration of submucosal invasion. Therefore, the present study indicates that nuclear-accumulated mutant p53R270H is a potential therapeutic target for the treatment of advanced CRCs.
Introduction
Molecular genetic studies have revealed that genetic alterations in driver genes induce the development of colorectal cancer (CRC) through an adenoma carcinoma sequence.1, 2, 3 It has recently been demonstrated that cumulative mutations in APC, KRAS, SMAD4 and TP53 in human intestinal cell-derived organoids are associated with the development of transplanted tumors in immunodeficient mice.4, 5 However, despite these findings, the precise functional role of each driver gene mutation in the malignant progression is still not fully understood. Moreover, precisely how the respective genetic alterations in tumor cells contribute to the generation of the microenvironment, which is an important factor in cancer progression, remains to be elucidated. In this regard, mouse genetic studies remain a powerful complementary approach that enables detailed in vivo experimentation and observations for delineating the mechanistic basis of tumorigenesis.
The TP53 is one of the most frequently mutated genes in cancer.6, 7 The target of p53 regulates many processes that prevent tumorigenesis, including cell cycle arrest, DNA repair and senescence.8 Notably, 74% of p53 mutations are missense mutations, which results in the formation of a mutant protein.9, 10 Mouse genetic studies have indicated that missense mutations in Trp53 at codon 172 and 270 cause adenocarcinomas in the lung and intestine, which are not developed in Trp53-null mutant mice, indicating that mutant p53 induces tumors in epithelial organs by a gain-of-function mechanism.11, 12 Furthermore, it has been shown that mutant p53 contributes to the invasion and metastasis of rhabdomyosarcoma,13 pancreatic cancer14 and sporadic and chemically induced colitis-associated intestinal tumors.15, 16 Mechanistically, mutant p53 promotes tumorigenesis by the activation of hepatocyte growth factor and platelet-derived growth factor receptor β signaling.17, 18 Moreover, mutant p53 alters gene expression at a global level through epigenetic mechanisms, namely by recruiting the SWI/SNF complex to remodel the chromatin of gene promoters,19 and through the induction of MLL1, MLL2 and MOZ to modify histone methylation and acetylation.20 However, the contribution of mutant p53 to tumor invasion and metastasis during in vivo tumorigenesis and the underlying mechanisms remains unclear.
In the present study, we generated ApcΔ716 and Trp53R270H compound mutant mice to examine the effect of mutant p53R270H on Apc mutation-induced intestinal tumors. ApcΔ716 mice develop benign intestinal tumors through β-catenin stabilization and subsequent Wnt signaling activation.21 Notably, mutant p53 strongly accumulates in the nuclei of tumor cells at the invasion front where desmoplastic microenvironment is generated. We observed that mutant p53 induces drastic morphological changes in tumor organoids with the acquisition of invasiveness, which is related to the increased branching of in vivo mouse tumors and human CRC. Mutant p53 also induced a marked shift in the transcriptome, which caused a significant activation of inflammatory and innate immune pathways together with Wnt/β-catenin signaling. Therefore, the present study indicates that mutant p53R270H induces intestinal tumor invasion through the acquisition of invasiveness with a complex tumor gland architecture, and mutant p53-induced activation of the inflammatory pathways and Wnt signaling may cooperatively contribute to these malignant phenotypes.
Results
Induction of submucosal invasion of intestinal tumors by Trp53 mutation
We generated ApcΔ716 Trp53+/LSL•R270H villin-CreER and ApcΔ716 Trp53LSL•R270H/LSL•R270H villin-CreER compound mice (hereafter, ApcΔ716 Trp53+/flox and ApcΔ716 Trp53flox/flox). The Trp53flox allele is functionally Trp53-null, and treatment of ApcΔ716 Trp53+/flox and ApcΔ716 Trp53flox/flox mice with tamoxifen results in the expression of mutant p53R270H in intestinal tumor cells (hereafter, ApcΔ716 Trp53+/R270H and ApcΔ716 Trp53R270H/R270H).
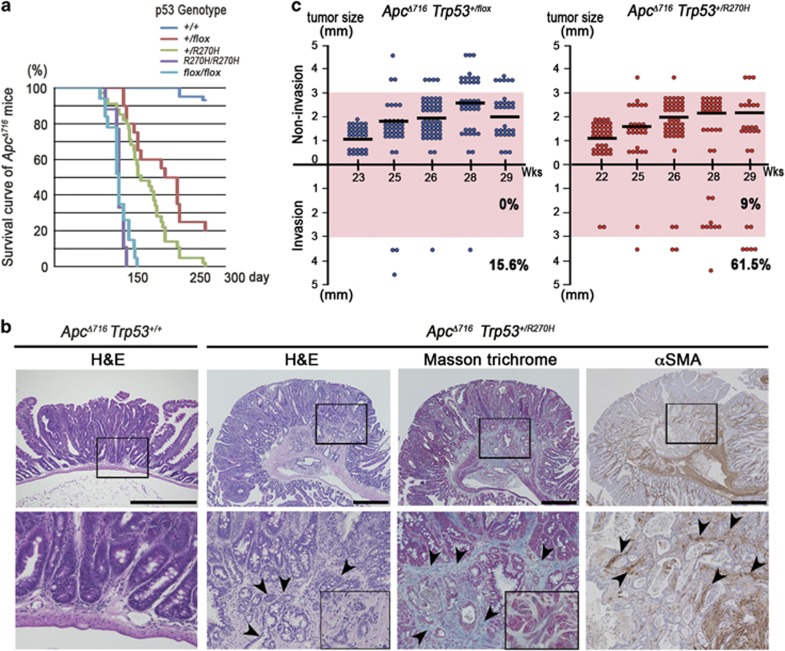
The mean lifespan of ApcΔ716 Trp53R270H/R270H mice was comparable to that of ApcΔ716 Trp53flox/flox mice, as they developed lymphomas owing to the loss of wild-type Trp53 (Figure 1a). ApcΔ716 Trp53+/R270H heterozygous mice survived longer than ApcΔ716 Trp53R270H/R270H mice, which allowed them to develop intestinal adenocarcinomas with submucosal invasion with desmoplasia consisting of collagen fibers and α-smooth muscle actin (αSMA)-positive myofibroblasts (Figure 1b, arrowheads). The mean polyp size in the ApcΔ716 Trp53+/flox and ApcΔ716 Trp53+/R270H mice (five mice from 22 to 29 weeks of ages for each genotype) was 1.97±0.82 mm and 1.95±0.74 mm, respectively, with no significant difference. However, the incidence of invasive tumors was markedly increased in ApcΔ716 Trp53+/R270H mice compared with ApcΔ716 Trp53+/flox mice (Figure 1c), which coincided with the shorter lifespan (161 days vs 202 days in ApcΔ716 Trp53+/R270H and ApcΔ716 Trp53+/flox, respectively, by the log-rank test, P=0.0162). Immunohistochemical analysis revealed that the expression of mesothelin, an invading CRC cell marker,22 and snail2, an epithelial-mesenchymal transition marker,23, 24 was induced at the submucosal invasive area of ApcΔ716 Trp53+/R270H adenocarcinomas, but it was not detected in the invasive area of ApcΔ716 Trp53+/flox tumors (Supplementary Figure 1a). In addition, the expression of the stem cell marker Cd44 was significantly increased in the tumor cells of invasive areas (Supplementary Figure 1b). Accordingly, it is possible that the mutant p53R270H causes intestinal tumor invasion through the induction of invasiveness and the undifferentiated characteristics of the tumor cells by a gain-of-function mechanism. In contrast, there was essentially no difference in the Ki67 labeling indices among ApcΔ716 Trp53+/+, ApcΔ716 Trp53+/R270H and ApcΔ716 Trp53R270H/R270H tumors. Likewise, apoptotic cells were rarely found in the tumors of either genotype (Supplementary Figure 1c and d).
Figure 1.
Submucosal invasion of ApcΔ716 intestinal polyps by mutant p53R270H. (a) The survival curves of each type of ApcΔ716 Trp53 compound mice are shown. (b) Representative photographs ApcΔ716 Trp53+/+ mouse intestinal adenomas (left, H&E) and ApcΔ716 Trp53+/R270H mouse invasive adenocarcinomas (right, H&E, Masson trichrome and αSMA immunostaining). The bottom photographs are enlarged images of the top (boxed area). The inset in the Masson trichrome staining image (bottom) shows a magnified view of dysplastic tumor cells. The arrowheads indicate invading tumor cells (H&E), collagen fibers (Masson Trichrome) and myofibroblasts (αSMA). Bars, 500 μm. (c) Size classification of intestinal tumors of ApcΔ716 Trp53+/flox mice (left) and ApcΔ716 Trp53+/R270Hmice (right) at indicated weeks of ages. The numbers of invasive and non-invasive tumors were separately scored. Each dot indicates an individual tumor. Horizontal bars indicate the mean polyp sizes. The percentages of invasive tumors are indicated for tumors <3 mm (pink area) and ⩾3 mm in diameter (outside of pink area). Wks, weeks.
Nuclear accumulation of p53 in the tumor cells of invasive area
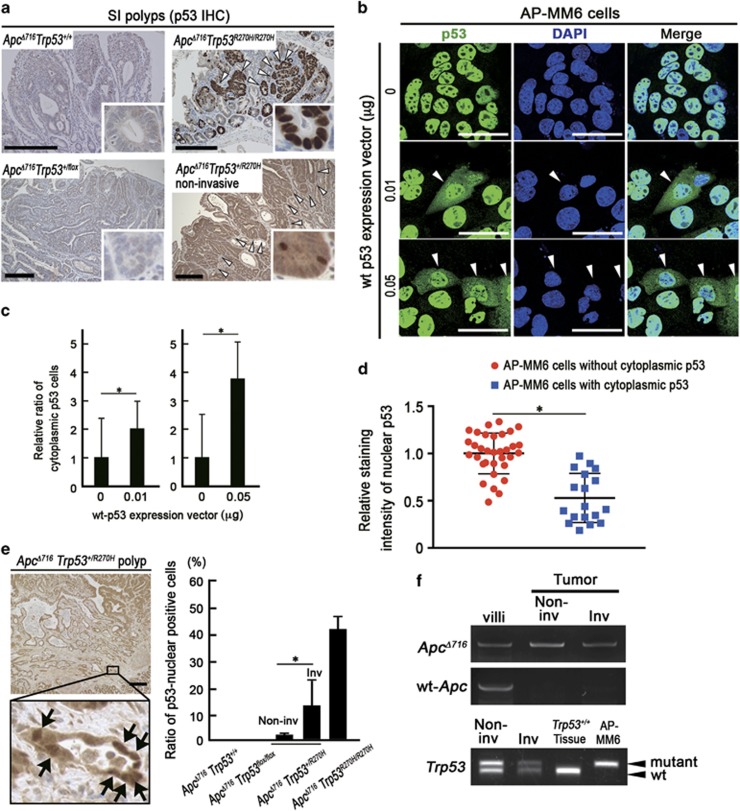
A striking nuclear accumulation of p53 was observed in the ApcΔ716 Trp53R270H/R270H mouse tumors like human CRC with TP53 mutations, whereas p53 was not detected in the ApcΔ716 Trp53+/+ and ApcΔ716 Trp53+/flox mouse tumors (Figure 2a and Supplementary Figure 2). Notably, p53 was stabilized and distributed to cytoplasm in ApcΔ716 Trp53+/R270H heterozygous mouse tumors, suggesting that wild-type p53 suppresses the nuclear accumulation of mutant p53, possibly through a dominant-negative mechanism. To assess this possibility, we established a cell line AP-MM6 from ApcΔ716 Trp53R270H/R270H mouse intestinal tumors. As anticipated, stabilized mutant p53 was accumulated in the nuclei of most AP-MM6 cells (Figure 2b). Of note, the expression of exogenous wild-type Trp53 in AP-MM6 cells caused a vector DNA amount-dependent increase of cells with cytoplasmic p53 distribution (Figures 2b and c). Moreover, in the AP-MM6 cells with cytoplasmic p53, the intensity of nuclear p53 significantly decreased (Figure 2d). These results support the idea that wild-type p53 suppresses the nuclear accumulation of mutant p53, although further studies are needed for validation.
Figure 2.
Nuclear localization of p53 in the invasive intestinal tumor cells. (a) Representative photographs of immunohistochemistry for p53 in small intestinal (SI) polyps of the indicated genotype mice. The insets show enlarged images. The arrowheads indicate p53 positive tumor cells. Bars, 200 μm. (b) Representative photographs of fluorescence immunocytochemistry for p53 (green), nuclear counterstaining with DAPI (blue) and merged images (right) of AP-MM6 cells transfected with the wild-type (wt) p53 expression vector (middle and bottom) and control (top). The arrowheads indicate AP-MM6 cells showing both nuclear and cytoplasmic p53 localization. Bars, 50 μm. (c) The ratio of AP-MM6 cells with cytoplasmic p53 distribution after transfection of 0.01 μg (left) or 0.05 μg (right) of wild-type p53 expression vector relative to the control level (mean±s.d.). Asterisks, P<0.05. (d) Relative staining intensity of nuclear p53 in AP-MM6 cells with or without cytoplasmic distribution of p53 after wt p53 vector transfection (blue and red, respectively). Each dot indicates a single cell. Asterisk, P<0.05. (e) Representative photographs of immunohistochemistry for p53 in the invasive region of ApcΔ716 Trp53+/R270H mouse intestinal tumors with low-power magnification (left top) and an enlarged image (left bottom). The arrows indicate tumor cells with nuclear-accumulated p53. Bars, 200 μm. The relative ratio of tumor cells with nuclear-accumulated p53 in the indicated genotypes is shown (mean±s.d.) (right). Asterisk, P<0.05. (f) An LOH analysis for Apc and Trp53 by LMD-based genomic PCR in the non-invasive (Non-inv) and invasive (Inv) areas of ApcΔ716 Trp53+/R270H intestinal tumors and normal villi as the controls. The mutant Trp53 and wild-type Trp53-specific bands are indicated as ‘mutant’ and ‘wt’, respectively. Genomic DNA of Trp53 wild-type mouse tissue and AP-MM6 were used for positive control of wt and mutant Trp53, respectively. Note that wild-type (wt) Apc was lost in both non-invasive and invasive tumor cells, whereas wild-type Trp53 is remained in these cells.
Interestingly, however, we found the nuclear accumulation of p53 in the invasion front of ApcΔ716 Trp53+/R270H mouse tumors where the number of stromal cells increased (Figure 2e). Moreover, wild-type Trp53 alleles were detected in these cells by laser microdissection-coupled genomic polymerase chain reaction (PCR), excluding the possibility of the lost of wild-type Trp53 by the loss of heterozygosity in the invasive area (Figure 2f). In contrast, wild-type Apc is lost in ApcΔ716 tumor cells, which is consistent with the findings from a previous report.21 Accordingly, it is possible that the subcellular distribution of mutant p53 in tumor cells is affected by the microenvironment of the invasive area.
Induction of the complex glandular structure of tumors by mutant p53R270H
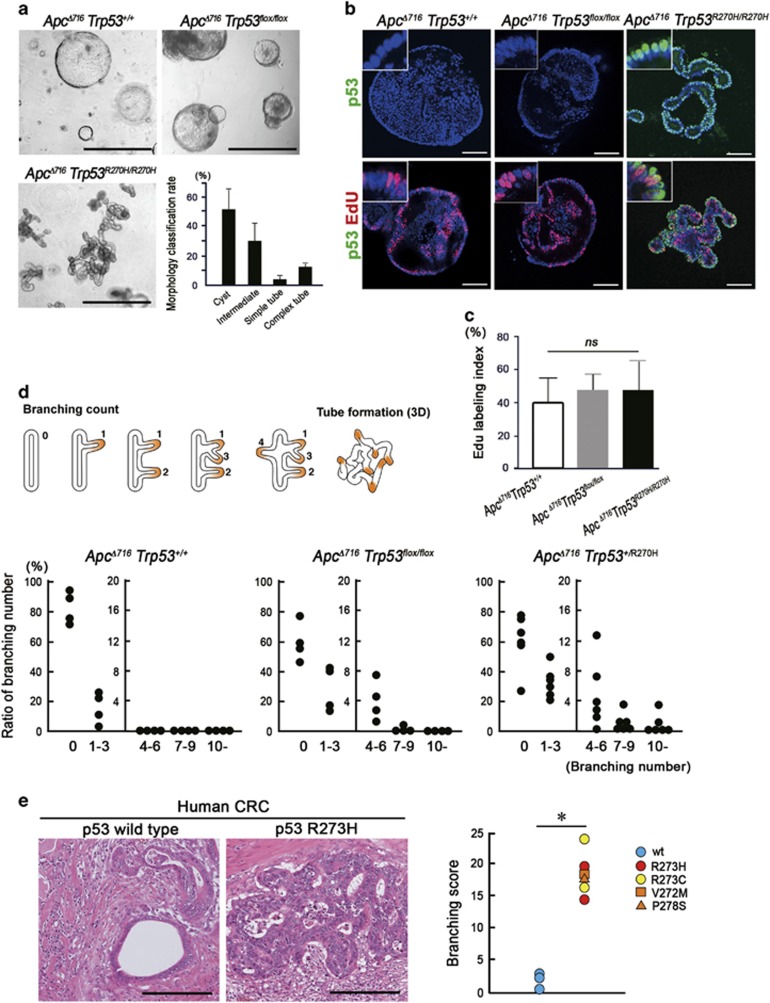
To investigate the role of nuclear-accumulated p53R270H in submucosal invasion, we examined intestinal tumor-derived organoids. In this study, we analyzed ApcΔ716 Trp53R270H/R270H tumor organoids because immunocytochemical staining showed a predominant p53 localization in the nucleus, whereas the nuclear accumulation was limited and cytoplasmic stabilization of p53 was not detected in the ApcΔ716 Trp53+/R270H organoids (Supplementary Figure 3a). Morphologically, the organoids derived from ApcΔ716 Trp53+/+ and ApcΔ716 Trp53flox/flox tumors showed round cystic structures, reflecting the uniform undifferentiated state caused by Wnt signaling activation in all cells (Figure 3a and Supplementary Figure 3b).25 ApcΔ716 KrasG12D and ApcΔ716 Tgfbr2ΔIEC tumor organoids also showed similar cystic morphologies. In contrast, ApcΔ716 Trp53R270H/R270H tumor organoids frequently formed simple as well as complex tube structures from Day 2 after each passage (Figure 3a and Supplementary Figure 4a), indicating that Trp53R270H mutation induces morphological changes by a gain-of-function. EdU (5-ethynyl-2’-deoxyuridine) labeling experiments showed no significant difference in the proliferation rate among ApcΔ716 Trp53+/+, ApcΔ716 Trp53flox/flox and ApcΔ716 Trp53R270H/R270H tumor organoids (Figures 3b and c).
Figure 3.
Induction of morphological changes of the tumors by mutant p53R270H. (a) Representative photographs of the indicated genotype organoids. Bars, 1 mm. The ratios of morphological classifications of ApcΔ716 Trp53R270H/R270H organoids are shown as a bar graph (mean±s.d.) (bottom right). (b) Representative confocal microscopy images of ApcΔ716 Trp53+/+ (left), ApcΔ716 Trp53flox/flox (center) and ApcΔ716 Trp53R270H/R270H tumor organoids (right). Fluorescence immunostaining for p53 (green, top) and double immunostaining for p53 and EdU (green and red, respectively, bottom) with DAPI staining (blue). The insets show enlarged images. Bars, 100 μm. (c) The EdU labeling index of tumor organoids is shown (mean±s.d.). ns, not significant. (d) A schematic diagram of the representative patterns of intestinal tumor branching leading to tube formation (top). The ratio of the branching number classification of ApcΔ716 Trp53+/+, ApcΔ716 Trp53flox/flox and ApcΔ716 Trp53+/R270H intestinal tumors are shown (bottom). Each dot represents the average rate (%) of the branching number in the individual mouse tumors. (e) Representative micrographs of human CRC with TP53 wild-type and R273H mutation (left, H&E). Bars, 200μm. Branching scores of human CRC with TP53 mutations around codon 273 (red, yellow and orange) compared with TP53 wild-type cases (blue) are shown (right). Asterisk, P<0.05.
To address the relationship between the complex glandular architecture of ApcΔ716 Trp53R270H/R270H organoids and the distinct histology of the in vivo tumors, we scored the branching of the intestinal tumor glands of each genotype mouse. Notably, the branching rate in ApcΔ716 Trp53+/R270H mouse tumors was markedly higher than that in ApcΔ716 Trp53+/+ or ApcΔ716 Trp53flox/flox counterparts (Figure 3d). Although the tumor size was small in ApcΔ716 Trp53R270H/R270H mice because of their short lifespan (Figure 1a), we confirmed that the branching rate tended to be higher in the ApcΔ716 Trp53R270H/R270H mouse tumors (polyp diameter 1–2 mm) than in size-matched ApcΔ716 Trp53flox/flox tumors (Supplementary Figure 3c). We next examined the branching frequency in human CRC carrying TP53 mutations around codon 273 (corresponding to codon 270 in mice). Importantly, CRC with TP53 mutations showed a significantly increased branching score compared with that in TP53 wild-type (Figure 3e). These results suggest that mutant p53 can promote structural atypia of intestinal tumors through the construction of a complex glandular structure.
Acquisition of invasiveness of tumor glands expressing p53R270H
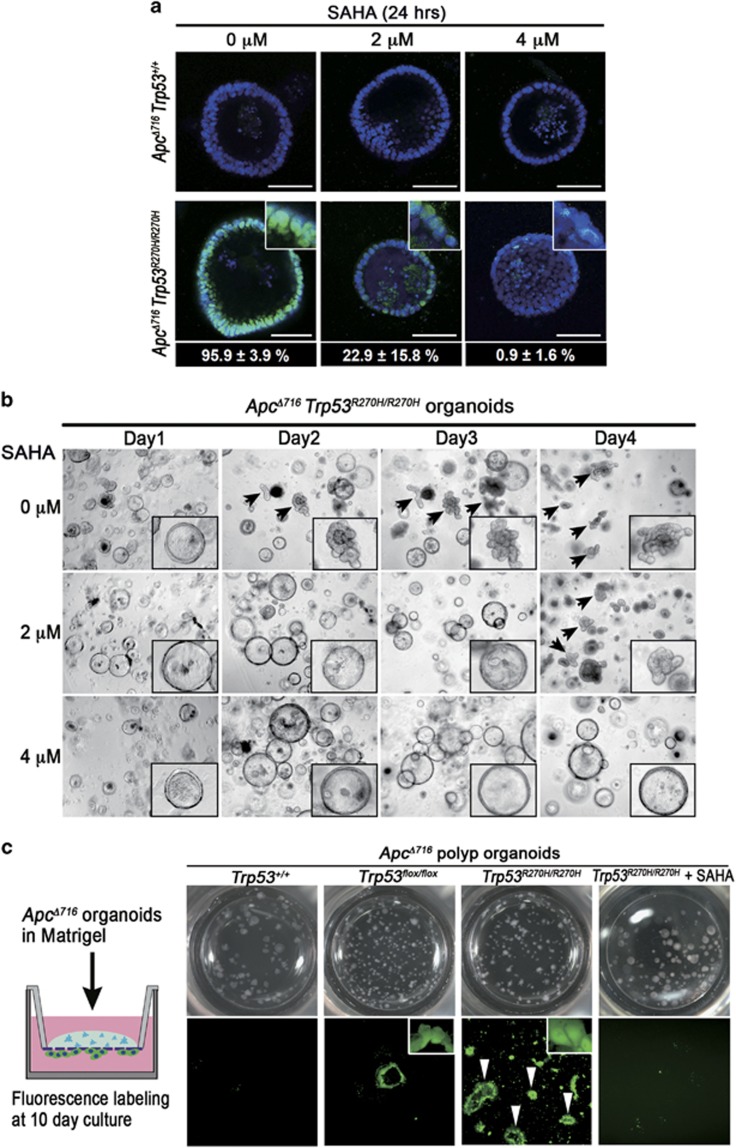
It has been reported that mutant p53 proteins are stabilized by interaction with the HSP90 chaperone mechanism, which inhibits MDM2 activity.26, 27 As HSP90 is positively regulated by histone deacetylase 6, targeting histone deacetylase 6 using suberoylanilide hydroxamic acid (SAHA) can destabilize the mutant p53 proteins.28 We confirmed that SAHA treatment abolished the nuclear accumulation of p53R270H in ApcΔ716 Trp53R270H/R270H organoids in a dose-dependent manner (Figure 4a). Importantly, SAHA treatment blocked the formation of complex glandular structures and reverted the morphology of ApcΔ716 Trp53R270H/R270H organoids to the round cystic typical of ApcΔ716 mouse tumor organoids (Figure 4b and Supplementary Figure 4a). These results suggest that p53R270H is required to maintain the complex tubular morphology of the organoids.
Figure 4.
Complex tube structure formation and the acquisition of invasiveness by mutant p53R270H. (a) Representative confocal microscopy images of fluorescence immunostaining for p53 (green) with nuclear counterstaining DAPI (blue) of ApcΔ716 Trp53+/+ (top) and ApcΔ716 Trp53R270H/R270H organoids (bottom) treated with the indicated concentration of SAHA. Insets indicate enlarged images. Bars, 100 μm. Ratios of the cells with nuclear-accumulated p53 in the ApcΔ716 Trp53R270H/R270H organoids treated with SAHA at each concentration are indicated in the bottom (mean%±s.d.%). (b) Representative dissecting microscopy photographs of ApcΔ716 Trp53R270H/R270H organoids at Days 1–4 after SAHA treatment with the indicated concentration. The arrows indicate organoids with complex tubular structures. Insets indicate enlarged images. (c) An illustration of the organoid invasion assay (left). Representative dissecting microscopy photographs of upper wells (top) and fluorescence microscopy images of the bottom surface of transwells (bottom) of the indicated genotype organoids and SAHA-treated ApcΔ716 Trp53R270H/R270H organoids. The arrowheads indicate invading tumor cells forming a glandular structure.
We next investigated the invasive capacity of organoids of different Trp53 genotypes using a transwell invasion assay. The results showed that ApcΔ716 Trp53+/+ organoids have no invasive capacity, whereas ApcΔ716 Trp53flox/flox organoids displayed only limited invasive properties (Figure 4c). In contrast, significant invasion was observed for the ApcΔ716 Trp53R270H/R270H organoids that retained their tubular structure during invasion (Figure 4c, arrowheads). Importantly, treatment of ApcΔ716 Trp53R270H/R270H organoids with SAHA effectively inhibited invasion (Figure 4c and Supplementary Figure 4b). These results indicate that p53R270H-induced complex gland formation is accompanied by the acquisition of invasive properties, although the possibility of a p53 degradation-independent mechanism by SAHA remains to be investigated.
Malignant tumor development by ApcΔ716 Trp53R270H/R270H tumor organoid transplantation
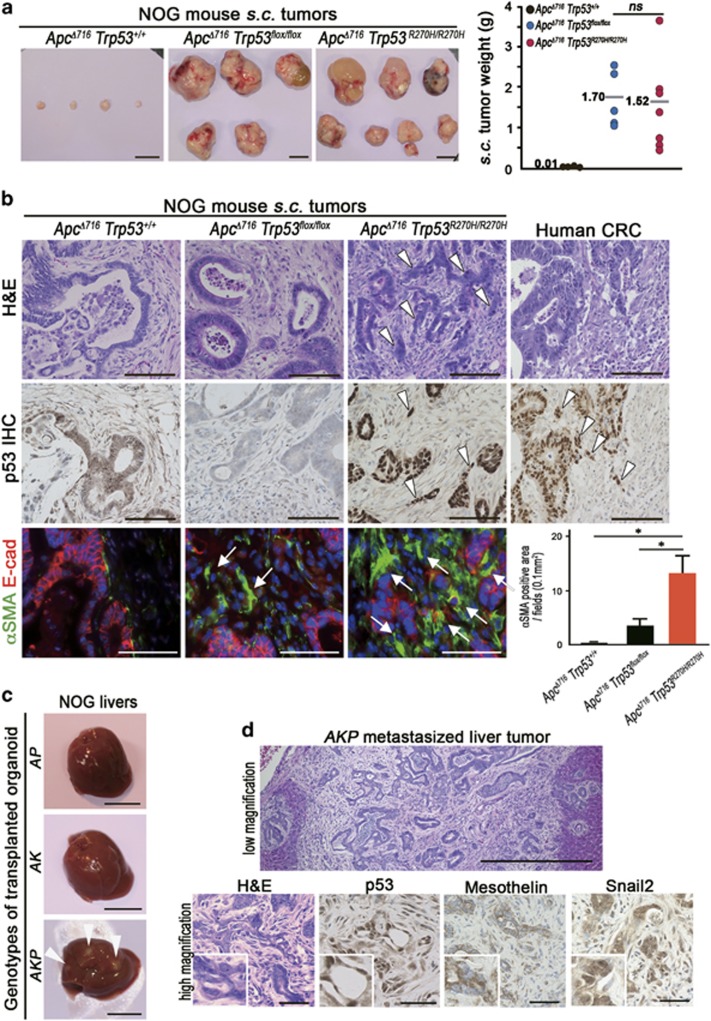
Given that ApcΔ716 Trp53R270H/R270H mice succumb prior to the development of malignant intestinal adenocarcinoma, we transplanted ApcΔ716 Trp53R270H/R270H tumor organoids subcutaneously (s.c.) into immunodeficient NOG mice to further examine their tumorigenicity. Control ApcΔ716 Trp53+/+ tumor organoids survived for 4 months after transplantation without forming palpable tumors (Figure 5a). In contrast, both ApcΔ716 Trp53flox/flox and ApcΔ716 Trp53R270H/R270H organoids formed large tumors of comparable sizes. Histologically, residual ApcΔ716 Trp53+/+ allografts and ApcΔ716 Trp53flox/flox organoid tumors showed mostly cystic structures (Figure 5b). In contrast, ApcΔ716 Trp53R270H/R270H organoid tumors presented a malignant histology with nuclear p53 accumulation, such as extensive branching of the tumor glands or occasional cell clusters in sheets (Figure 5b, arrowheads). The mean ratios of the cell clusters without gland formation in ApcΔ716 Trp53flox/flox and ApcΔ716 Trp53R270H/R270H tumors were 3.1% and 68.8%, respectively. Notably, the αSMA-positive area was increased significantly in the ApcΔ716 Trp53R270H/R270H s.c. tumor stroma (Figure 5b, arrows and bar graph), suggesting that the microenvironment is activated with increased myofibroblasts by the Trp53 mutation in cancer cells. These morphological characteristics of ApcΔ716 Trp53R270H/R270H tumors closely mimic those of moderately differentiated human colon cancer (Figure 5b).29
Figure 5.
Malignant tumor formation in NOG mice by p53R270H organoids. (a) Representative macroscopic photographs of the organoid-derived s.c. tumors in NOG mice at 4 months after transplantation (left, three photographs). Bars, 1 cm. The weights of s.c. tumors for each genotype with the average are plotted in a dot graph (right). ns, not significant. (b) Representative histology photographs of organoid-derived s.c. tumors and human CRC (stage IIIB). H&E (top) and immunohistochemistory for p53 (middle) and double fluorescence immunostaining for αSMA (green) and E-cadherin (red) (bottom). The arrowheads indicate the sheets of tumor cells in ApcΔ716 Trp53R270H/R270H s.c. tumors and human CRC. The arrows indicate αSMA-positive myofibroblasts. Bars, 100 μm. The αSMA-positive areas for each tumor genotype determined using fluorescence immunohistochemistry are shown as a bar graph (mean±s.d.). Asterisks, P<0.05. (c) The macroscopic photographs of NOG mouse livers at 1 month after injection of the organoids into the spleen. The arrowheads indicate metastasized tumors. AP, ApcΔ716 Trp53R270H/R270H; AK, ApcΔ716 KrasG12D; AKP, ApcΔ716 KrasG12D Trp53+/R270H. Bars, 1 cm. (d) Representative photographs of metastasized AKP organoid-derived tumors in NOG mouse livers. H&E staining (top and bottom left) and immunohistochemistry for p53, mesothelin and Snail2 (bottom, left to right). Bars, 500 μm (top) and 50 μm (bottom).
Despite their malignant histology, ApcΔ716 Trp53R270H/R270H organoid s.c. tumors did not metastasize to distant organs. To assess the metastatic potential of Trp53R270H cancer cells, we injected tumor-derived organoids into NOG mouse spleens to test their liver metastasis. In this experiment, we used organoids derived from ApcΔ716 Trp53R270H/R270H, ApcΔ716 KrasG12D and ApcΔ716 KrasG12D Trp53+/R270H mouse tumors. Among these genotypes, only ApcΔ716 KrasG12D Trp53+/R270H triple mutant organoids developed metastasis foci in the liver (Figure 5c). A histological analysis showed the moderately to poorly differentiated features of the metastasized tumors with p53 nuclear accumulation and the expression of mesothelin and Snail2 (Figure 5d). Collectively, these results indicate that the combination of p53 mutation with Kras activation can induce metastasis of intestinal tumors.
Expression of a wide range of genes induced by nuclear-accumulated p53R270H
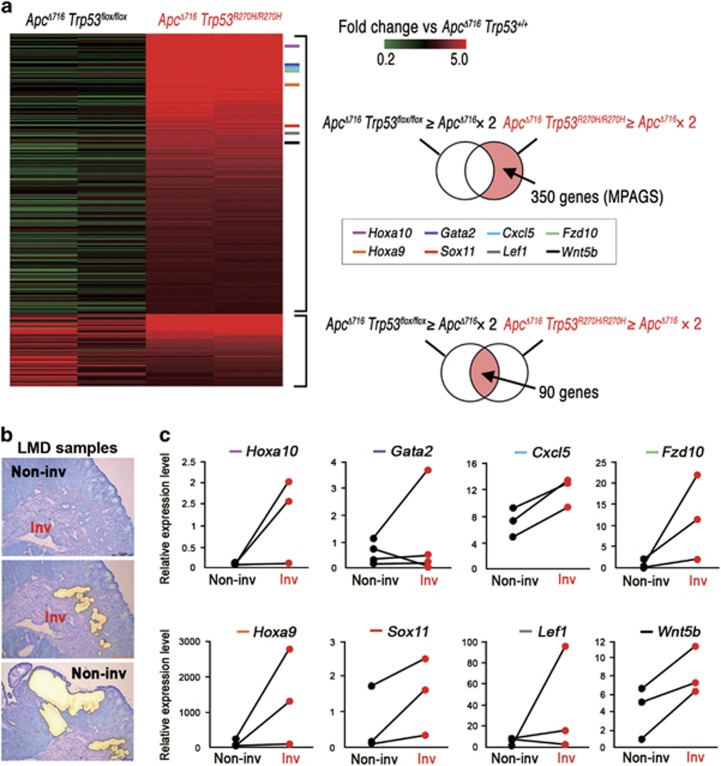
To examine the molecular mechanism through which p53R270H promotes malignant progression, we performed RNA Sequencing of the respective Trp53 genotype organoids. A clustering analysis showed a gene cluster specifically upregulated in the ApcΔ716 Trp53R270H/R270H tumor organoids (Supplementary Figure 5a, asterisk). Of the 440 genes elevated more than twofold in the ApcΔ716 Trp53R270H/R270H organoids relative to ApcΔ716 Trp53+/+, 350 genes were not upregulated in ApcΔ716 Trp53flox/flox organoids, and were thus designated as ‘a mutant p53-activated gene set (MPAGS)’ (Figure 6a and Supplementary Table 1). We selected eight genes (Hoxa10, Gata2, Cxcl5, Fzd10, Hoxa9, Sox11, Lef1 and Wnt5b) from MPAGS for the validation of a sequencing analysis by real-time reverse transcribed (RT)-PCR and confirmed the upregulation of these genes in ApcΔ716 Trp53R270H/R270H organoids compared with ApcΔ716 Trp53+/+ and ApcΔ716 Trp53flox/flox (Supplementary Figure 5b). As the nuclear accumulation of p53 is restricted to the submucosal invasive area of ApcΔ716 Trp53+/R270H tumors (Figure 2e), we separately collected tumor cells from the invasive and non-invasive areas of ApcΔ716 Trp53+/R270H mouse tumors by laser microdissection (Figure 6b), and analyzed the expression of the eight validated genes by real-time RT-PCR (Figure 6c). Notably, the expression of these genes was elevated in the invasive areas compared with their non-invasive counterparts, indicating that the nuclear localization of p53R270H causes a transcriptomic shift in tumor cells.
Figure 6.
Expression of a wide range of genes by nuclear-accumulated p53R270H. (a) A heat map of 440 genes that are upregulated in ApcΔ716 Trp53R270H/R270H organoids more than twofold compared with ApcΔ716 Trp53+/+ organoids. Of 440 genes, 350 were upregulated in ApcΔ716 Trp53R270H/R270H organoids, but not in ApcΔ716 Trp53flox/flox organoids, and were designated as the mutant p53-activated gene set (MPAGS), whereas the other 90 genes were upregulated in both ApcΔ716 Trp53R270H/R270H and ApcΔ716 Trp53flox/flox organoids. (b) Representative photographs of an intestinal tumor after the collection of invasive tumor cells (middle) and non-invasive tumor cells (bottom) by LMD. (c) Relative mRNA expression of eight selected genes from the MPAGS in non-invasive area (Non-inv) and invasive area (Inv) of ApcΔ716 Trp53+/R270H mouse tumors. The lines connect the real-time RT-PCR results of the non-invasion (closed) and invasion (red) areas of the same tumors.
Recent studies show that mutant p53 induces the epigenetic modification of chromatin.19, 20 We thus analyzed the accessibilities of the promoter regions of validated MPAGS genes. The data showed that the promoter regions of Hoxa9, Hoxa10, Gata2 and Lef1 genes were substantially more sensitive to the nuclease treatment in ApcΔ716 Trp53R270H/R270H tumor cells than in ApcΔ716 Trp53+/+ and ApcΔ716 Trp53flox/flox (Supplementary Figure 5c). These results indicate that mutant p53 induces a more opened chromatin structure, which rendered gene promoters more accessible to transcription factors, at least in a portion of MPAGS.
Activation of inflammatory and innate immune signaling in Apc Δ716 Trp53 R270H/R270H organoids
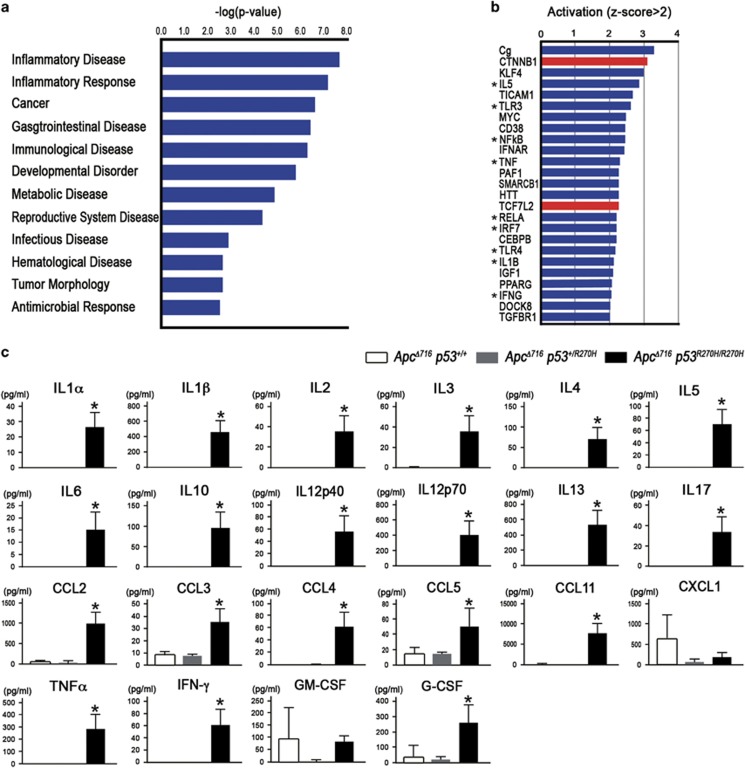
A gene ontology term analysis for the disease category using MPAGS indicated a significant activation of ‘Inflammatory Disease’ and ‘Inflammatory Response’ in addition to ‘Cancer’ in the ApcΔ716 Trp53R270H/R270H tumor cells (Figure 7a). Consistently, Ingenuity Pathway Analysis indicated that inflammatory and innate immune signaling pathways were significantly activated in ApcΔ716 Trp53R270H/R270H tumor organoid cells (Figure 7b, asterisks). We thus examined medium concentrations of cytokines and chemokines in the respective genotype organoid cultures. Consistently, the levels of most cytokines and chemokines were significantly elevated in ApcΔ716 Trp53R270H/R270H organoid culture media (Figure 7c). It is possible that Trp53 mutation in cancer cells triggers the generation of a microenvironment through the secretion of inflammatory factors (Supplementary Figure 6).
Figure 7.
Activation of inflammatory and innate immune pathways in tumor cells by p53R270H. (a) The results of a gene ontology (GO) analysis using MPAGS showing significantly activated disease category in ApcΔ716 Trp53R270H/R270H organoids aligned by P-values. (b) The results of an ingenuity pathway analysis (IPA) showing significantly upregulated pathways in ApcΔ716 Trp53R270H/R270H organoids aligned by P-values. Asterisks, inflammatory and innate immune pathways. Red bars, Wnt/β-catenin signaling. (c) Medium concentration of cytokines and chemokines in the organoid cultures for the indicated genotypes are shown as bar graphs (mean±s.d.). Asterisks, P<0.05.
Moreover, we found that the Wnt/β-catenin signaling pathway was also activated in p53R270H tumor organoids (Figure 7b, red bars). It has been shown that the concurrent activation of NF-κB and Wnt signaling pathways promotes a stem cell-like state.30 Accordingly, it is also conceivable that the Trp53 mutation induces stemness or an undifferentiated status of tumor cells through the simultaneous activation of inflammatory pathways and Wnt signaling (Supplementary Figure 6).
Discussion
The accumulation of driver gene mutations promotes cancer development and malignant progression.2 Recent studies have shown that the transformation of human normal colonic stem cells can be achieved by the accumulation of APC, KRAS, SMAD4 and TP53-null mutations.4, 5 However, these transformed organoids failed to show a fully metastatic malignant behavior of human CRC.4, 31 In contrast, we herein show that triple mutations in ApcΔ716, KrasG12D and Trp53R270H showed liver metastasis after being injected into the spleen of NOG mice. Accordingly, it is possible that Trp53R270H but not Trp53-null mutation promotes the metastatic potential of cancer cells.
In the present study, we also demonstrate that the nuclear localization of p53R270H is required for the induction of MPAGS expression, which subsequently induces the malignant phenotypes of intestinal tumors. Moreover, recent studies have shown that a p53 gain-of-function induces a global shift in gene expression through chromatin modification,19, 20 and we also consistently found that the promoter accessibility of MPAGS increased. It is therefore conceivable that the remodeling of chromatin in gene promoters by nuclear localized mutant p53 causes hyper-activation of transcription factors, leading to malignant phenotypes. Because >94% of human CRCs carry genetic alterations in the Wnt signaling pathway, such as in APC and β-catenin,3 the TCF/β-catenin complex is one of the possible transcription factors that are activated by mutant p53 in colon cancer cells. Indeed, our study demonstrates that the canonical Wnt pathway that had already been activated in Apc mutant tumors was further augmented by p53R270H, likely through an increased accessibility of Wnt-target genes to the β-catenin/TCF complex, or via the induction of cofactors that enhances the transcription of Wnt-target genes. It has been shown that an increased Wnt/β-catenin activity in CRC cells correlates with an invasive phenotype and cancer stem cell property.32, 33 Thus, it is possible that the hyper-activation of the Wnt/β-catenin signaling may be a mechanism that drives the malignant progression induced by p53R270H.
It has been shown in 3D cultures that the activation of Wnt/β-catenin signaling together with the epidermal growth factor pathway can induce the tube-like structures of intestinal epithelial cells.34 Moreover, such a mechanism may increase the invasive growth of tumor glands into stromal tissues.35 We herein observed that p53R270H induced marked morphological changes of tumor organoids to complex glandular structures that correlated with increased invasiveness. Moreover, we also demonstrated increased branching of the tumor glands in human CRC with TP53 mutations. These data indicate that such morphological and invasive features are driven by p53 mutations, fueled by a hyper-activated Wnt/β-catenin pathway to promote invasion and metastasis (Supplementary Figure 6).
We have previously demonstrated that inflammatory responses have a role in the malignant progression of intestinal tumors.36 It has been reported that mutant p53 prolongs TNF-α-induced NF-κB activation, which results in highly prone to inflammation-associated colon cancer,16 and that mutant p53 facilitates TNF-α signaling.37 These results, taken together, suggest that mutant p53 has a role in malignant progression through the induction of the inflammatory pathway. We show here that inflammatory and innate immune signaling pathways are significantly activated in tumor cells by mutant p53R270H expression, which may be caused by the activation of NF-κB through mutant p53-induced promoter accessibility. Accordingly, TP53/Trp53 mutation in cancer cells can activate the inflammatory microenvironment, which contributes to malignant progression.
Importantly, it has been shown that inflammatory and innate immune pathways through NF-κB and TLR2 together with Wnt/β-catenin signaling are important for the acquisition of stem cell properties.30, 38 Accordingly, the present results suggest that mutant p53R270H can induce stem cell properties in intestinal tumor cells through the activation of both NF-κB pathway and Wnt signaling (Figure 7b and Supplementary Figure 6). Moreover, it is possible that the cytokines expressed by cancer cells further activate inflammatory signaling in tumor cells by some positive feedback mechanism, which then accelerates mutant p53R270H-induced malignant progression.
In conclusion, we herein showed that Trp53R270H mutation promotes the invasion and metastasis of intestinal tumors. Nuclear-accumulated mutant p53R270H induces the acquisition of invasiveness associated with complex glandular structures of tumor organoids, which drives the submucosal invasion and distant metastasis of CRC. The activation of the inflammatory pathway together with Wnt signaling may be important factors to accelerate these mutant p53-induced malignant phenotypes. Therefore, the dynamic regulation of nuclear accumulation for mutant p53 represents an attractive treatment strategy against the malignant progression of CRC and thus warrants further investigation.
Materials and methods
Mouse experiments
ApcΔ716 and villin-CreER mice were previously described.21, 39 Tgfbr2flox/flox, Trp53LSL·R270H and KrasLSL•G12D mice were obtained from the Mouse Repository (NCI-Frederick, Frederick, MD, USA).11, 40, 41 The genetic background of all strains used in this study is C57BL/6. All animal experiments were performed with the protocol approved by the Committee on Animal Experimentation of Kanazawa University. The generation of the compound mice used in this study is provided in the Supplementary Materials and Methods. For the survival rate analysis, mice were observed until 264 days of age, and the mice were euthanized when they showed a moribund phenotype. The total numbers of polyps in both the small intestine and colon were scored and examined histologically at 22–29 weeks of age (n=5 each genotype).
Human CRC samples
The human CRC tissues were collected in National Cancer Center Hospital East, Japan with informed consent and nine samples were selected after examination of the TP53 mutation status (n=2 each for R273H, and R273C, n=1 each for V272M and P278S, n=3 for wild type). The four of the nine cases were collected for BREAC trial (Yuki et al., ASCO, 2015; Yamazaki et al., ASCO, 2015). The genetic and clinicopathological information for the human samples are presented in Supplementary Table 2. This study was approved by the institutional review board of the National Cancer Center East (registration #2005-043), and carried out according to the ethical principles of the Declaration of Helsinki.
Histology and immunohistochemistry
The tissue specimens were fixed in 4% paraformaldehyde, paraffin-embedded and sectioned at 4-μm thickness. The sections were stained with Haemotoxylin and Eosin or Masson’s trichrome stain. For immunohistochemistry, antibodies against E-cadherin (R&D Systems, Minneapolis, MN, USA), α-smooth muscle actin (Sigma, St Louis, MO, USA), p53 (CM5) (Leica Biosystems, Wetzlar, Germany), Snail2 (NOVUS Biologicals, Littleton, CO, USA), mesothelin (IBL, Fujioka, Japan), and Ki67 (Life Technologies, Grand Island, NY, USA) were used. Staining signals were visualized using the Vectastain Elite Kit (Vector Laboratories, Burlingame, CA, USA). Human CRC histology sections were also purchased (US Biomax, Rockville, MD, USA).
Scoring tumor gland branching
The branching frequency of the mouse tumor glands was scored using Haemotoxylin and Eosin sections (n=4~6 mice for each genotype). The branching numbers of the individual continuous glands found on the sections were scored blindly in two randomly selected microscopy fields, and three polyps were examined per mouse. The ratio of the numbers of branches was then calculated. The branching frequency of human CRC was scored blindly using Haemotoxylin and Eosin sections. The number of gland branching was counted in three individual glands at the invasion front, and the mean values were calculated.
Cell culture experiments
The mouse intestinal tumor cell line (AP-MM6) was established using ApcΔ716 Trp53R270H/R270H intestinal tumor-derived organoids by subcloning in a 96-well plate (deposited to Riken BioResource Center, Tsukuba, Japan). p53 expression vectors were transfected to AP-MM6 cells using Lipofectamine 2000 (Thermo Fisher Scientific, Rockford, IL, USA), and examined by immunocytochemistry using antibody for p53 (Leica Biosystems), E-cadherin (R&D Systems) and anti-rabbit IgG Alexa 488 as a secondary antibody (Molecular Probes, Eugene, OR, USA). The constructions of the p53 expression vectors are provided in the Supplementary Materials and Methods. The ratio of cells with cytoplasmic p53 was calculated by counting>4 × 103 cells, and experiments for each transfection condition were performed three times independently. The staining intensity of nuclear p53 was measured using the NIH image software program (NIH, Bethesda, MD, USA).
Organoid culture experiments
The organoid cultures were prepared from small intestinal tumors, as previously described.42 Organoid cell proliferation was examined using the Click-iT EdU Imaging System (Invitrogen, Carlsbad, CA, USA), and the EdU labeling index was calculated by counting more than 200 cells each for three independent organoids. The organoids were immunostained using anti-p53 antibody (Leica Biosystems) and Alexa Fluor 488-conjugated antibody (Molecular probes, Grand Island, NY, USA). For the inhibition of p53R270H, organoids were treated with pan-histone deacetylase inhibitor suberoylanilide hydroxamic acid (Sigma). The organoid morphologies were categorized as cyst, simple tube or complex tube structures based on their morphologies (Supplementary Figure 4a).
For the invasion assay, the organoids were dissociated with Accutase (Innovative Cell Technologies, San Diego, CA, USA), and 3 × 104 cells in Matrigel were seeded in Fluoroblock inserts with 8-μm pores (Corning, Corning, NY, USA). The culture media of the upper and bottom wells were the same as the organoid culture media. At 10 days after seeding, the cells invading through the pores of the inserts were stained with fluorescent dye Calcein AM (Corning) and examined with a fluorescence microscope. When invading cells were found, the well was judged to be ‘invasion-positive’, and the total number of ‘invasion-positive’ wells was scored for each genotype.
For the suspension array analysis, 1.5 × 105 cells of organoids were cultured for 4 days and the culture medium was collected and used for suspension array analyses according to the manufacturer’s instructions of the Bio-Plex Mouse Cytokine Assay kit (Bio-Rad, Hercules, CA, USA).
Organoid transplantation experiments
NOD/Shi-scid Il2rg−/− mice (NOG mice) were purchased (CIEA, Kawasaki, Japan). The organoids were mechanically dissociated, and 1 × 105 organoid cells were injected s.c. with Matrigel to NOG mice (n=4–7 dependent on genotypes). At 4 months after transplantation, the tumors were examined histologically. For the liver metastasis analysis, 1 × 105 organoid cells were injected with Matrigel into the NOG mouse spleens (n=2–4 dependent on genotypes). At 1 month after injection, the liver metastases were examined histologically.
Real-time RT-PCR
For real-time RT-PCR, tumor cells from the invasion and non-invasion areas of intestinal tumors of mice were isolated from frozen sections on a LMD7000 laser microdissection system (Leica Microsystems, Wetzlar, Germany). Total RNA was extracted from laser microdissection samples using an RNeasy Plus Micro extraction kit (Qiagen GmbH, Hilden, Germany), reverse transcribed using a PrimeScript RT reagent kit (Takara, Tokyo, Japan) and amplified using ExTaqII SYBR Premix (Takara) on a Stratagene Mx3000P real-time thermocycler (Agilent Technologies, Santa Clara, CA, Japan). The primer sequences are described in the Supplementary Materials and Methods.
Next-generation RNA sequencing
Poly A(+) mRNA extracted from the organoids (n=2 for each genotype) was used for the sequencing library construction by a TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA, USA) and an Agilent XT Auto System (Agilent Technologies). The libraries were sequenced by an Illumina HiSeq 2500 (Illumina). The raw reads were mapped to the mouse reference genome (mm10), and the reads per kilobase per million mapped reads (rpkm) values of the genes were calculated using the StrandNGS software package (Strand Genomics, San Francisco, CA, USA). The sequencing data were deposited in the Gene Expression Omnibus (accession code GSE81441).
Upstream pathway and gene ontology analyses
The MPAGS was analyzed for putative upstream regulators and processes using the Ingenuity Pathway Analysis and gene ontology Analysis software package (Ingenuity Systems; www.ingenuity.com). The activation z-scores were calculated as a measurement of the functional and translational activation in the upstream regulator analysis. Pathways with z-scores of >2 and P-values of <0.05 were designated as activated with statistical significance.
Statistical analysis
The data were analyzed using an unpaired t-test and are presented as the means±s.d. A value of P<0.05 was considered to be statistically significant.
Acknowledgments
We thank Yoshie Jomen, Ayako Tsuda and Manami Watanabe for technical assistance. Computations were partially performed on the NIG supercomputer at ROIS National Institute of Genetics. This work was supported by AMED-CREST, AMED, Japan Agency for Medical Research and Development, Japan, Grants-in-Aid for Scientific Research (A) (#15H02362) tand (C) (26430110) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell 1996; 87: 159–170. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscape. Science 2013; 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med 2015; 21: 256–262. [DOI] [PubMed] [Google Scholar]
- Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 2015; 521: 43–47. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature 2000; 408: 307–310. [DOI] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C et al. Mutational landscape and significance across 12 major cancer types. Nature 2013; 502: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer 2002; 2: 594–604. [DOI] [PubMed] [Google Scholar]
- Brosh R, Rotter V. When mutant gain new powers: news from the mutant p53 field. Nat Rev Cancer 2009; 9: 701–713. [DOI] [PubMed] [Google Scholar]
- Muller PAJ, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 2014; 25: 304–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 2004; 119: 847–860. [DOI] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM et al. Gain of function of p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 2004; 119: 861–872. [DOI] [PubMed] [Google Scholar]
- Doyle B, Morton JP, Delaney DW, Ridgway RA, Wilkins JA, Sansom OJ. p53 mutation and loss have different effects on tumorigenesis in a novel mouse model of pleomorphic rhabdomyosarcoma. J Pathol 2010; 222: 129–137. [DOI] [PubMed] [Google Scholar]
- Morton JP, Timpson P, Karim SA, Ridgway RA, Athineos D, Doyle B et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci USA 2010; 107: 246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PAJ, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S et al. Mutant p53 drives invasion by promoting integrin recycling. Cell 2009; 139: 1327–1341. [DOI] [PubMed] [Google Scholar]
- Cooks T, Pateras IS, Tarcic O, Solomon H, Schetter AJ, Wilder S et al. Mutant p53 prolongs NF-κB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell 2013; 23: 634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PAJ, Trinidad AG, Timpson P, Morton JP, Zanivan S, van den Berghe PVE et al. Mutant p53 enhances MET trafficking and signaling to drive cell scattering and invasion. Oncogene 2013; 32: 1252–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmueller S, Manchado E, Saborowski M, Morris JP IV, Wagenblast E, Davis CA et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor β signaling. Cell 2014; 157: 382–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister NT, Fomin V, Regunath K, Zhou JY, Zhou W, Silwal-Pandit L et al. Mutant p53 cooperates with the SWI/SNF chromatin remodeling complex to regulate VEGF2 in breast cancer cells. Genes Dev 2015; 29: 1298–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Sammons MA, Donahue G, Dou Z, Vedadi M, Getlik M et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature 2015; 525: 206–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima M, Oshima H, Kitagawa K, Kobayashi M, Itakura C, Taketo M. Loss of Apc heterozygosity and abnormal tissue building in nascent intestinal polyps in mice carrying a truncated Apc gene. Proc Natl Acad Sci USA 1995; 92: 4482–4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebig B, Brabletz T, Staege MS, Wulfänger J, Riemann D, Burdach S et al. Forced expression of ΔN-TCF-1B in colon cancer derived cell lines in accompanied by the induction of CEACAM5/6 and mesothelin. Cancer Lett 2005; 223: 159–167. [DOI] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 2012; 148: 1015–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoyel M. Gut stem cells, a story of snails, flies and mice. EMBO J 2015; 34: 1287–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011; 469: 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Marchenko ND, Schulz R, Fischer V, Velasco-Hernandez T, Talos F et al. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol Cancer Res 2011; 9: 577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Marchenko ND, Moll UM. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ 2011; 18: 1904–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrova EM, Yallowitz AR, Li D, Xu S, Schulz R, Proia DA et al. Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature 2015; 523: 352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming M, Ravula S, Tatishchev SF, Wang HL. Colorectal carcinoma: pathologic aspects. J Gastrointest Oncol 2012; 3: 153–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Göktuna SI, Ziegler PK et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 2013; 152: 25–38. [DOI] [PubMed] [Google Scholar]
- Salahudeen AA, Kuo CJ. Toward recreating colon cancer in human organoids. Nat Med 2015; 21: 215–216. [DOI] [PubMed] [Google Scholar]
- Fodde R, Brabletz T. Wnt/β-catenin signaling in cancer stemness and malignant behavior. Curr Opin Cell Biol 2007; 19: 150–158. [DOI] [PubMed] [Google Scholar]
- Vermeulen L, De Sousa E, Melo F, van der Heijden M, Cameron K, de Jong JH et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol 2010; 12: 468–476. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Fujii S, Sato A, Ibuka S, Kagawa Y, Ishii M et al. A combination of Wnt and growth factor signaling induces Arl4c expression to form epithelial tubular structures. EMBO J 2014; 33: 702–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Matsumoto S, Nojima S, Morii E, Kikuchi A. Arl4c expression in colorectal and lung cancers promotes tumorigenesis and may represent a novel therapeutic target. Oncogene 2015; 34: 4838–4844. [DOI] [PubMed] [Google Scholar]
- Oshima H, Nakayama M, Han TS, Naoi K, Ju X, Maeda Y et al. Suppressing TGFβ signaling in regenerating epithelia in an inflammatory microenvironment is sufficient to cause invasive intestinal cancer. Cancer Res 2015; 75: 766–776. [DOI] [PubMed] [Google Scholar]
- Di Minin G, Bellazzo A, Dal Ferro M, Chiaruttini G, Nuzzo S, Bicciato S et al. Mutant p53 reprograms TNF signaling in cancer cells through interaction with the tumor suppressor DAB2IP. Mol Cell 2014; 56: 617–629. [DOI] [PubMed] [Google Scholar]
- Scheeren FA, Kuo AH, van Weele LJ, Cai S, Glykofridis I, Sikandar SS et al. A cell-intrinsic role for TLR2-MYD88 in intestinal and breast epithelia and oncogenesis. Nat Cell Biol 2014; 16: 1238–1248. [DOI] [PubMed] [Google Scholar]
- el Marjou F, Janssen KP, Chang BH, Li M, Chan L, Louvard D et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 2004; 39: 186–193. [DOI] [PubMed] [Google Scholar]
- Chytil A, Magnunson MA, Wright CV, Moses HL. Conditional inactivation of the TGF-β type II receptor using Cre:Lox. Genesis 2002; 32: 73–75. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 2001; 15: 3243–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009; 459: 262–265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.