Main Text
Determining how conformational properties of unfolded proteins vary as a function of denaturant concentration has important implications for understanding the early stages of protein folding, and for our understanding of protein-solvent interactions. Upon dilution from a high concentration of denaturant into native conditions, does an unfolded protein undergo collapse first followed by folding, or does the chain remain expanded before folding and collapse occur concomitantly? The global dimensions of a protein are captured by its radius of gyration (Rg), meaning that, in principle, this question should be simple to answer by measuring Rg as a function of denaturant concentration. However, for several proteins, Rg, as directly quantified by small-angle x-ray scattering (SAXS) experiments, shows minimal changes as a function of denaturant concentration (1, 2). In contrast, for the same set of proteins, inferences drawn from single-molecule Förster resonance energy transfer (smFRET) experiments suggest Rg undergoes a significant and continuous collapse as denaturant concentration is reduced (2, 3, 4). These results have led to questions as to whether the observed discrepancy is due to the high protein concentrations needed for SAXS experiments, the presence of the dyes in smFRET experiments, or some additional inherent shortcoming of either or both experimental methods (1, 2, 5, 6, 7). In this issue, Song et al. (8) demonstrate that the observed discrepancy is not necessarily due to an inherent problem with either SAXS or smFRET. Instead, the discrepancy emerges due to the use of the conventional inference strategy utilized to convert mean FRET transfer efficiencies (〈E〉) measured in smFRET experiments into estimates of Rg that can then be directly compared to Rg measured from SAXS.
What is the conventional inference strategy, and why does it lead to this discrepancy? The conventional inference strategy consists of two parts. First, a homogenous polymer model is utilized to infer the end-to-end distance, REE, from the measured 〈E〉. Second, a one-to-one mapping between REE and Rg is used to extrapolate Rg from REE (4). Both of these inferences rely on the assumption that the underlying conformational ensemble expands or contracts uniformly—an assumption that ignores the chemical heterogeneity associated with polypeptides. Such an assumption is likely reasonable at high denaturant concentrations where the solvent quality is such that proteins behave like homopolymeric self-avoiding random coils, and thus explains why inferences regarding conformational properties of proteins often agree between SAXS and smFRET measurements under these conditions (1, 2, 3, 4, 8, 9). However, as denaturant concentration is decreased, there is a reduction in the effective solvent quality. This leads to the formation of sequence-dependent interactions that introduces conformational heterogeneity not captured by mean-field homopolymer models. Thus, as the concentration of denaturant decreases, the conventional inference strategy becomes increasingly inappropriate for inferring Rg from 〈E〉 due to the inherent decoupling of Rg and REE.
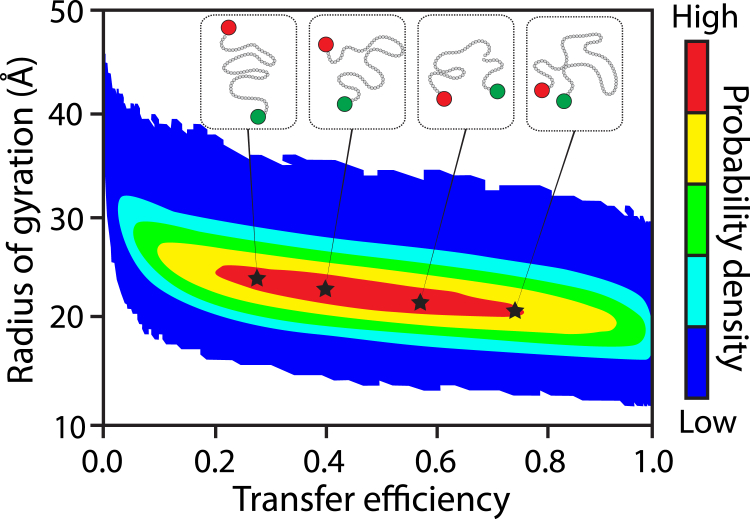
Song et al. (8) demonstrate the decoupling of Rg and REE by extracting conditional Rg distributions from minimal Cα resolution, explicit-chain simulations given a specific experimental 〈E〉. The use of explicit-chain simulations allows for the construction of a large ensemble of physically realizable conformations. Here, conformational heterogeneity is introduced by extracting subensembles that are consistent with a given E value. The authors show that by accounting for heterogeneity in the underlying ensembles, large changes in E can be consistent with minimal changes in Rg. This decoupling of Rg and E, and thus consequently REE, is shown in Fig. 1, which reproduces the 2D distribution of E and Rg. For unconstrained simulations, this 2D space illustrates how relatively similar Rg values can be consistent with a wide range of E values. Focusing on the archetypal Protein L, Song et al. (8) calculated Rg values from subensembles that are consistent with experimentally derived 〈E〉 values for a range of denaturant conditions. Using these values, the authors demonstrate that Rg remains relatively constant as a function of denaturant concentration, consistent with SAXS results.
Figure 1.
2D distribution of transfer efficiency (E) and radius of gyration (Rg) demonstrates the decoupling of E and Rg. Insets depict cartoon representations of conformations with similar Rg values but different E values. The terminal residues are depicted by green and red circles. This figure was adapted from Fig. 3 from Song et al. (8). To see this figure in color, go online.
Importantly, Song et al. (8) are clear that their strategy only provides a general rationale for the SAXS-smFRET discrepancy, but cannot yield insights on the sequence-dependent source of the conformational heterogeneity for a given protein. The extent of conformational heterogeneity, and thus whether the conformational properties of an unfolded protein can be reasonably approximated using homopolymer models, will be sequence dependent and is typically unknown a priori. As a result, understanding the extent and type of sequence-dependent interactions that lead to deviations from homopolymer models represents an important next step in interpreting smFRET data.
The lack of a one-to-one mapping between fluctuations at the ends of chains, as quantified by 〈E〉, and global size, as quantified by Rg, for heterogeneous ensembles (i.e., unfolded proteins in low denaturant concentrations) suggests that caution must be taken when converting between conformational descriptors that interrogate a specific length scale (〈E〉) and conformational descriptors that average over all interresidue distances (Rg). SAXS can provide an accurate description of an unfolded protein’s global size (Rg) and shape, but cannot necessarily offer information regarding a specific length scale (e.g., REE) without the application of some kind of model. Similarly, smFRET can yield an accurate description regarding the distance between two residues of an unfolded protein. However, the conversion of interresidue distances into a globally averaged conformational descriptor (e.g., Rg) requires extrapolation using some model. Thus, if both length-scale-specific and globally averaged conformational descriptions are desired, then additional strategies should be pursued. For SAXS, methods that quantify intramolecular distances, such as anomalous SAXS combined with gold labels, can be deployed to determine length-scale specific properties (10). Equivalently, for smFRET, multiple interdye distance measurements can be used to infer globally averaged properties (11). Additionally, as demonstrated by Song et al. and others, combining SAXS and/or smFRET measurements with explicit-chain simulations can yield further insights regarding the conformational behaviors of unfolded proteins (2, 5, 6, 8).
Altogether, these results suggest that theories designed for homopolymers may be inadequate to simultaneously provide accurate globally averaged and length-scale-specific conformational descriptions of unfolded proteins using either SAXS-derived Rg or smFRET-derived transfer efficiencies alone. Instead, to develop complete conformational descriptions of heterogeneous ensembles, a combination of experimental measurements designed to extract distinct conformational properties, as well as the aid of computational methods, are needed.
Editor: Rohit Pappu.
References
- 1.Yoo T.Y., Meisburger S.P., Plaxco K. Small-angle x-ray scattering and single-molecule FRET spectroscopy produce highly divergent views of the low-denaturant unfolded state. J. Mol. Biol. 2012;418:226–236. doi: 10.1016/j.jmb.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuertes G., Banterle N., Lemke E.A. Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements. Proc. Natl. Acad. Sci. USA. 2017;114:E6342–E6351. doi: 10.1073/pnas.1704692114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sherman E., Haran G. Coil-globule transition in the denatured state of a small protein. Proc. Natl. Acad. Sci. USA. 2006;103:11539–11543. doi: 10.1073/pnas.0601395103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merchant K.A., Best R.B., Eaton W.A. Characterizing the unfolded states of proteins using single-molecule FRET spectroscopy and molecular simulations. Proc. Natl. Acad. Sci. USA. 2007;104:1528–1533. doi: 10.1073/pnas.0607097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng W., Borgia A., Best R.B. Probing the action of chemical denaturant on an intrinsically disordered protein by simulation and experiment. J. Am. Chem. Soc. 2016;138:11702–11713. doi: 10.1021/jacs.6b05443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borgia A., Zheng W., Schuler B. Consistent view of polypeptide chain expansion in chemical denaturants from multiple experimental methods. J. Am. Chem. Soc. 2016;138:11714–11726. doi: 10.1021/jacs.6b05917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maity H., Reddy G. Folding of protein L with implications for collapse in the denatured state ensemble. J. Am. Chem. Soc. 2016;138:2609–2616. doi: 10.1021/jacs.5b11300. [DOI] [PubMed] [Google Scholar]
- 8.Song J., Gomes G.-N., Chan H.S. Conformational heterogeneity and FRET data interpretation for dimensions of unfolded proteins. Biophys. J. 2017;113:1012–1024. doi: 10.1016/j.bpj.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohn J.E., Millett I.S., Plaxco K.W. Random-coil behavior and the dimensions of chemically unfolded proteins. Proc. Natl. Acad. Sci. USA. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zettl T., Mathew R.S., Lipfert J. Absolute intramolecular distance measurements with angstrom-resolution using anomalous small-angle x-ray scattering. Nano Lett. 2016;16:5353–5357. doi: 10.1021/acs.nanolett.6b01160. [DOI] [PubMed] [Google Scholar]
- 11.Aznauryan M., Delgado L., Schuler B. Comprehensive structural and dynamical view of an unfolded protein from the combination of single-molecule FRET, NMR, and SAXS. Proc. Natl. Acad. Sci. USA. 2016;113:E5389–E5398. doi: 10.1073/pnas.1607193113. [DOI] [PMC free article] [PubMed] [Google Scholar]