Abstract
Background
Ferrous iron (Fe) and zinc (Zn) at high concentration in the soil cause heavy metal toxicity and greatly affect rice yield and quality. To improve rice production, understanding the genetic and molecular resistance mechanisms to excess Fe and Zn in rice is essential. Genome-wide association study (GWAS) is an effective way to identify loci and favorable alleles governing Fe and Zn toxicty as well as dissect the genetic relationship between them in a genetically diverse population.
Results
A total of 29 and 31 putative QTL affecting shoot height (SH), root length (RL), shoot fresh weight (SFW), shoot dry weight (SDW), root dry weight (RDW), shoot water content (SWC) and shoot ion concentrations (SFe or SZn) were identified at seedling stage in Fe and Zn experiments, respectively. Five toxicity tolerance QTL (qSdw3a, qSdw3b, qSdw12 and qSFe5 / qSZn5) were detected in the same genomic regions under the two stress conditions and 22 candidate genes for 10 important QTL regions were also determined by haplotype analyses.
Conclusion
Rice plants share partial genetic overlaps of Fe and Zn toxicity tolerance at seedling stage. Candidate genes putatively affecting Fe and Zn toxicity tolerance identified in this study provide valuable information for future functional characterization and improvement of rice tolerance to Fe and Zn toxicity by marker-assisted selection or designed QTL pyramiding.
Electronic supplementary material
The online version of this article (10.1186/s12864-017-4221-5) contains supplementary material, which is available to authorized users.
Keywords: Rice, Fe toxicity, Zn toxicity, GWAS, QTL
Background
Ferrous iron (Fe) and zinc (Zn) are essential trace elements for rice, as they are involved in numerous physiological and biochemical processes. In trace amounts, these two elements serve as pivotal cofactors for many enzymes and key structural motifs for transcriptional regulatory proteins. However, due to natural and industrial processes, Fe and Zn occurring in high quantities cause heavy metal toxicity that greatly affect rice growth and quality [1]. Fe toxicity is a serious constraint to the production of lowland rice grown in acid flooded soils [2]. Reported yield losses in fields usually range between 15 and 30%, but can also cause complete crop failure [3]. Zn toxicity can occur in acid soil and is extremely harmed to the growth of rice plants [4]. Moreover, high concentration of Fe and Zn in the soil may lead nutrient imbalance by limiting the absorption of other nutrients. Many researchers have studied the toxic effect of Fe [2, 5] and Zn [6, 7] on rice. Fe and Zn toxicities have similar phenotypic effects and both can result in oxidative cell damage accompanied by the induction of antioxidative defense mechanisms. The typical visual symptom caused by Fe toxicity is small brown-purple spots, appearing from the tips towards the base of the older leaves, commonly known as “bronzing” [8]. Zn toxicity symptoms are first characterized by leaf discoloration and bronzing of older leaves that then spread to the whole plant [9]. Plants have developed several resistance mechanisms such as restricted uptake, extrusion, chelation, trafficking, and storage to maintain essential trace element homeostasis and alleviate heavy metal toxicity [10].
Breeding resistant varieties is an economically sustainable solution to improve rice production under Fe and Zn stress conditions. The genetic variation for Fe and Zn toxicity tolerance in rice is controlled by several genes and its expression largely depends on the environment [8, 11]. Several genes responsible for the uptake, transport and accumulation of Fe and Zn have been identified in rice, which belong to five known protein families (OsNRAMPs, OsFROs, OsZIPs, OsFERs and OsYSLs) [12]. Many QTL for Fe or Zn toxicity tolerance in rice have been identified and mapped using DNA molecular markers in the populations derived from two parents [8, 9, 13–17]. These QTL are associated with some easily measurable traits like leaf bronzing index, shoot height, root length, shoot and root dry weight, tiller number under toxicity stress, because physiological process traits are difficult to measure and map at the whole population level [18]. Considering that biparental populations in linkage mapping only evaluate two alleles and provide limited insight into the analysis of complex traits unless the population is very large [19], association mapping is becoming an alternative method for dissecting complex traits controlled by multiple QTL and can evaluate a greater number of alleles in a broader population [20]. Due to strong interaction effects of genotype and environment in the field, studies of Fe and Zn tolerance, selection or testing of tolerance in hydroponics solution is a quicker and more efficient way to determine tolerant and sensitive lines in rice while controlling other environmental effects. To our best knowledge, few reports have conducted GWAS for dissecting complex traits related to Fe and Zn toxicity tolerance in rice so far [21].
Understanding of genetic and molecular mechanisms underlying Fe and Zn tolerance in rice is essential to accelerate the development of Fe and Zn tolerant varieties. The objectives of this study were the following: 1) to screen for Fe and Zn toxicity tolerance of a world-wide Oryza sativa indica subspecies; and 2) to identify QTL related to Fe and Zn toxicity tolerance.
Methods
Plant materials
A total of 222 indica rice accessions were introduced from IRRI, which were collected from 31 countries in Asia, Africa and Latin America. It was a subset taken from core collection established at the International Rice Research Institute (IRRI) and represents a well selected panel of phenotypically diverse lines for many agronomic traits. As previously indicated [22], two subgroups of the 222 associations were found by PCA 3 dimension plot (Additional file 1). Given the strong population differentiation, 11 accessions were removed and the remaining 211 accessions were used for the following analysis in this study.
Phenotypic evaluation
The experiments were carried out in greenhouse of Institute of Crop Sciences, Chinese Academy of Agricultural Sciences in Beijing. Zn toxicity experiment was conducted from May to June and Fe toxicity experiment was from August to September in 2016. Randomized complete block design (RCBD) consisting of 211 accessions (10 plants per accession) with 2 replicates were applied for both control and stress conditions. For zinc toxicity experiment, the temperature was around 28/20 °C (day/night) and the relative humidity was ~60%. For ferrous iron toxicity experiment, the temperature was around 32/25 °C (day/night) and the relative humidity was ~75%. The growth condition in greenhouse was regulated by using shade net and mechanical ventilation along with the cooling pad.
The seeds were surface sterilized with 5% sodium hypochlorite solution for 20 min and rinsed well with distilled water. Then seeds were soaked in distilled water in the dark at 30 °C for 48 h. Finally, 10 uniformly germinated seeds of each accession were directly sown in holes of perforated styrofoam sheets (10 lines × 13 rows) with a nylon net bottom in a plastic container. The styrofoam sheets were allowed to float on water up to five days and then transferred to Yoshida solution [23] for five days. At three-leaf stage, the Fe in the form of FeSO4 • 7H2O at the concentration of 300 mg L−1 (5.36 mM) (2.0 mg L−1 for control) or the Zn in the form of ZnSO4 • 7H2O at the concentration of 200 mg L−1(3.06 mM) (0.01 mg L−1 for control) were applied. The pH of the solution was adjusted to 5.0 at alternative day by 1 M NaOH / HCl. The solution was renewed every five days. After 21 days of treatment, the shoot height (SH), maximum root length (RL), and shoot fresh weight (SFW) of each plant was measured. Then the samples were kept in oven for 72 h under 50 °C. Finally shoot dry weight (SDW) and root dry weight (RDW) were recorded, and shoot water content (SWC) was determined by (SFW– SDW) / SFW × 100. For each trait, its stress tolerance index was calculated by trait ratio of stress to control condition. The average value of two replicates was taken for all traits.
Measurement of plant Fe and Zn concentrations
The concentration of Fe or Zn in shoot (SFe or SZn) samples under stress conditions were determined by atomic absorption spectrometry (AAS, Series2, Thermo Electron Corporation) with wet digestion method (GB/T 14609–2008). About 1 g dried shoot samples from each lines was digested with 5 ml mix acid (HNO3:HClO4 = 4:1, V/V) using a graphite liquation furnace. The heating process was as follows: 80 °C for 15 min, 120 °C for 20 min, 150 °C for 30 min and 180 °C for 60 min. Finally, the colorless or slightly yellow transparent liquid was diluted in 100 ml volumetric flask with distilled water. For Fe and Zn determinations, calibration standard solutions were prepared by diluting 1000 μg ml−1 standard solution (NCS, China). Two replicates were performed per sample and the average value was taken.
Genotyping
The genotyping data used in this study was from a high-density rice array (HDRA) composed of 700,000 single-nucleotide polymorphisms (SNPs). The HDRA was developed as an Affymetrix Custom GeneChip Array from a SNP discovery dataset consisting of ~16 millions SNPs (generated by re-sequencing 128 rice samples at ~7X genome coverage). Methods for the development of the HDRA, including SNP discovery and selection, probe design, genotype calling and quality control were described by McCouch [24]. The SNP with minor allele frequency (MAF) less than 0.05 were removed, and finally 395,553 SNP markers were selected for GWAS.
Data analysis and QTL mapping
The Spearman’s rank correlation coefficients between each trait pair were calculated by the SAS8.1 PROC CORR (SAS Institute, 1996). To estimate the effects of replication, genotype and conditions, analysis of variance was carried out by SAS8.1 PROC GLM.
We performed a genome wide association study (GWAS) to detect the trait-SNP associations for all measured traits using 395,553 SNPs and the mean trait values of the 211 accessions from each of the environments. Marker-trait associations were conducted by compressed mixed linear model implemented in the Genome Association and Prediction Integrated Tool (GAPIT), a package of R software [25]. The critical p-value for declaring significant marker-trait association was set to 1.0 × 10−4. Adjacent significant SNP with distances less than 200 kb were merged into single QTL.
Candidate gene identification for QTL affecting Fe and Zn toxicity tolerance
Here, QTL regions simultaneously meeting any of two items below were considered as important for candidate genes analysis of Fe or Zn toxicity tolerance: (1) affecting stress tolerance index for two traits; (2) consistently identified in both stress and control conditions with similar magnitude and same direction of gene effect; (3) close to previously cloned genes or mapped QTL. Besides, all QTL regions affecting concentrations of Fe and Zn in shoot were also analyzed for candidate genes. The following steps were conducted to identify candidate genes for important QTL identified. Firstly, for each important QTL region, the SNPs whose –log10 (p) located inside the interval of 1 unit of the peak SNP were regarded as significant. Secondly, all significant SNPs were used to check non-synonymous mutation in the coding sequence (CDS) region for all the genes located in the interval of each important QTL from the Rice Genome Annotation Project (RGAP). Thirdly, if more than two significant SNPs distributed in one gene, haplotype analysis was carried out for each of the candidate genes in each important QTL region using all non-synonymous SNPs located inside of the gene CDS region. Finally, candidate genes were determined by testing the significant differences among major haplotypes (containing more than 8 samples) for each important QTL through ANOVA.
Results
Phenotypic variation and trait correlation
Wide variation of all investigated traits in both control and stress conditions were observed in the current association panel. For both Fe and Zn toxicity experiments, significant differences between control and stress conditions were observed for all traits. Compared with control, the mean values of all traits under stress conditions were considerably decreased except RDW under Fe stress condition (Fig. 1). Based on criteria of lowest SFe or SZn and highest stress tolerance index of SH, SFW and SDW, seven accessions (CC127, CC123, CC31, CC155, CC139, CC141 and CC55) were selected as Fe toxicity tolerance while eight accessions (CC101, CC120, CC55, CC109, CC83, CC218, CC123 and CC155) selected as Zn toxicity tolerance (Data not shown), and three accessions (CC55, CC123, CC155) were tolerant to both Fe and Zn toxicity stresses. ANOVA results showed that differences among genotypes and environments were highly significant for all measured traits. Genotype explained an average of 85.7% of phenotypic variance ranging from 55.9% for FeSWC to 97.3% for FeSH, and environment explained an average of 84.5% of all measured traits, ranging from 68.8% for SWC to 94.7% for SDW in Fe toxicity experiment (Additional file 2). Genotype accounted for an average of 76.9% of phenotypic variance ranging from 58.5% of Zn/CKSDW to 95.0% for CKSH, and environment explained an average of 84.6% of all traits, ranging from 75.1% for RL to 94.6% for SH in Zn toxicity experiment (Additional file 3). The trait pair-wise correlations were similar under both Fe and Zn stress conditions. The same traits among control and stress conditions had high positive correlations, suggesting the effect of iron and zinc toxicity on these traits was similar across all lines. The aboveground traits such as SH, SFW, SDW had significantly positive correlations under stress vs. control conditions, indicating that genotypic patterns of plant biomass were similar under different environments. RL under Fe stress condition had no correlation with other traits except for SFe. The SFe (SZn) had significant negative correlation with SFW, SDW, RDW and SWC under stress condition, whereby the correlation coefficients (r) were −0.61 (−0.47), −0.59 (−0.40), −0.70 (−0.57) and −0.43 (−0.40), respectively (Additional file 4).
Fig. 1.
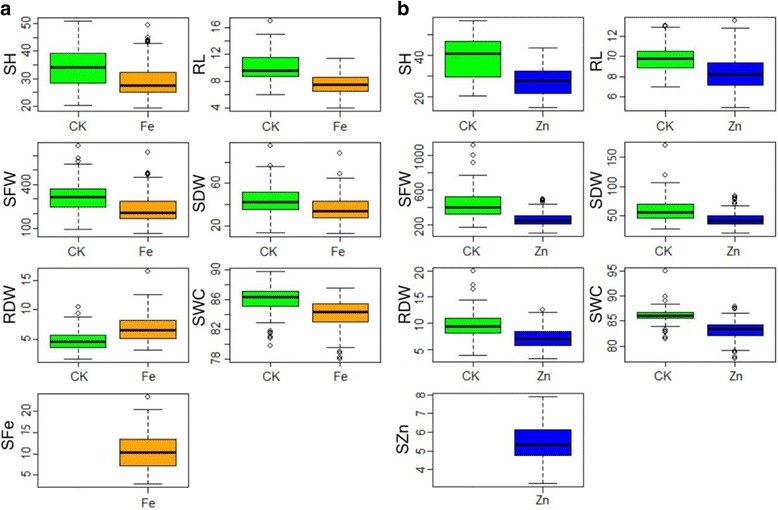
Box plot of seven measured traits in two Fe (a) and Zn (b) experiments. CK, Control condition; Fe, Ferrous iron toxicity stress condition; Zn, Zinc toxicity stress condition; SH, Shoot height; RL, Root length; SFW, Shoot fresh weight; SDW, Shoot dry weight; RDW, Root dry weight; SWC, Shoot water content; SFe, Fe concentration in shoot; SZn, Zn concentration in shoot
SNP markers
The 395,553 high quality SNP markers were evenly distributed across the 12 chromosomes covering an average 97.6% (372.96 Mb) of the rice genome published by International Rice Genome Sequencing Project [26], ranging from 94.1% (29.02 Mb) of chromosome 11 to 99.7% (28.44 Mb) of chromosome 8. The number of markers per chromosome ranged from 23,532 on chromosome 10 to 47,895 on chromosome 1. The average marker spacing was 943 bp across the whole genome ranging from 788 bp on chromosome 11 to 1041 bp on chromosome 5 (Table 1). Of these SNP markers, 23.1% were located in genes CDS region and 15.2% are non-synonymous SNPs.
Table 1.
Distributions of markers on 12 chromosomes
| Chr | Start (bp) | End (bp) | Size (Mb) | Count | Spacing | Length (Mb)a | Coverage (%) |
|---|---|---|---|---|---|---|---|
| 1 | 1579 | 43,256,417 | 43.25 | 47,895 | 903 | 45.06 | 96.0 |
| 2 | 2057 | 35,935,335 | 35.93 | 40,309 | 891 | 36.82 | 97.6 |
| 3 | 20,925 | 36,413,109 | 36.39 | 36,812 | 989 | 37.26 | 97.7 |
| 4 | 2212 | 35,462,406 | 35.46 | 35,119 | 1010 | 35.86 | 98.9 |
| 5 | 10,557 | 29,907,310 | 29.90 | 28,729 | 1041 | 30.04 | 99.5 |
| 6 | 2922 | 31,246,064 | 31.24 | 31,993 | 977 | 32.12 | 97.3 |
| 7 | 1638 | 29,691,817 | 29.69 | 28,919 | 1027 | 30.36 | 97.8 |
| 8 | 4149 | 28,441,872 | 28.44 | 30,496 | 933 | 28.53 | 99.7 |
| 9 | 38,389 | 22,939,999 | 22.90 | 25,045 | 914 | 23.84 | 96.1 |
| 10 | 3835 | 23,205,372 | 23.20 | 23,532 | 986 | 23.66 | 98.1 |
| 11 | 2041 | 29,020,003 | 29.02 | 36,841 | 788 | 30.83 | 94.1 |
| 12 | 2372 | 27,530,630 | 27.53 | 29,863 | 922 | 27.76 | 99.2 |
| Total | 372.96 | 395,553 | 943 | 382.15 | 97.7 |
aThe length of each chromosome and whole genome published by International Rice Genome Sequencing Project
Identification of QTL associated with differentiated responses to Fe stress
A total of 29 QTL were identified in Fe stress experiment (Additional file 5), including four detected only in control condition, 12 detected only under Fe stress condition, and 13 commonly detected under both control and Fe stress conditions. Among them, six QTL (qSh2, qRl2, qSfw2, qSdw6, qSwc2 and qSwc11) were identified for the trait ratio of stress to control conditions (Fig. 2, Table 2).
Fig. 2.
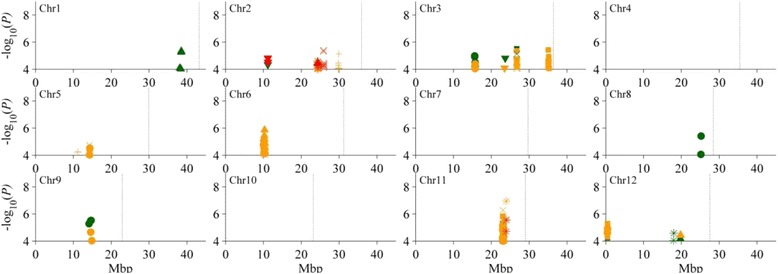
Manhattan plots of iron toxicity tolerance QTL in the whole genome. Significant SNPs from different conditions are displayed in different colors: control is green, ferrous iron stress is yellow, the ratio value between ferrous iron stress and control is red. The associated traits are represented by different symbols: shoot height = triangle up, root length = triangle down, shoot fresh weight = ×, shoot dry weight = square, root dry weight = circle, shoot water content = star, ion concentration = +
Table 2.
QTL identified with significant association to ferrous iron toxicity tolerance related traits
| Traita | QTL | Chr | Interval (Mb) | Control | Fe stress | Ratio of stress to control | Previously reported QTL or gene | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P valueb | Effectc | R 2(%)d | P value | Effect | R 2(%) | P value | Effect | R 2(%) | |||||
| SH | qSh1 | 1 | 38.11–38.54 | 4.62E-06 | −3.1 | 5.9 | |||||||
| qSh2 | 2 | 24.35–24.56 | 3.07E-05 | 3.0 | 4.8 | 9.03E-05 | 2.2 | 6.1 | 3.79E-05 | −0.04 | 7.3 | ||
| qSh6 | 6 | 10.17–10.38 | 5.69E-06 | 3.1 | 8.3 | 1.38E-06 | 3.3 | 9.5 | Qsh6a [22] | ||||
| qSh12 | 12 | 19.73–19.81 | 6.63E-05 | 3.2 | 4.4 | 3.49E-05 | 2.0 | 6.9 | |||||
| RL | qRl2 | 2 | 11.17–11.19 | 4.59E-05 | −0.7 | 8.3 | 1.51E-05 | 0.07 | 9.5 | ||||
| qRl3 | 3 | 23.50–23.68 | 1.56E-05 | 1.3 | 9.4 | 8.16E-05 | −0.8 | 7.8 | |||||
| SFW | qSfw2 | 2 | 25.87–25.97 | 5.32E-06 | −30.1 | 9.7 | 4.21E-06 | −0.05 | 10.2 | ||||
| qSfw3a | 3 | 26.68–26.79 | 7.39E-06 | 66.1 | 7.0 | 2.82E-05 | 42.1 | 5.5 | |||||
| qSfw5b | 5 | 14.23–14.33 | 1.83E-05 | 58.3 | 8.6 | ||||||||
| qSfw6 | 6 | 10.03–10.38 | 1.10E-05 | 40.5 | 9.0 | ||||||||
| qSfw11 | 11 | 23.03–23.42 | 8.06E-05 | 47.0 | 7.4 | 5.41E-07 | 53.5 | 11.9 | |||||
| SDW | qSdw3a | 3 | 26.68–26.79 | 3.16E-06 | 13.6 | 8.3 | 4.49E-06 | 12.7 | 9.3 | OsIRT1 [34] | |||
| qSdw3b | 3 | 35.01–35.29 | 2.60E-05 | 11.2 | 5.9 | 3.80E-06 | 13.6 | 8.8 | qSdw3 [10] | ||||
| qSdw6 | 6 | 10.12–10.38 | 1.03E-05 | 5.4 | 9.2 | 6.35E-05 | 0.06 | 7.9 | qSdw [24] | ||||
| qSdw11 | 11 | 23.03–23.19 | 5.35E-06 | 7.1 | 10.3 | 1.51E-06 | 6.8 | 11.0 | qLBI11 [10] | ||||
| qSdw12 | 12 | 0.48–0.49 | 5.82E-05 | 7.3 | 7.9 | 5.24E-06 | 7.5 | 9.8 | |||||
| RDW | qRdw3 | 3 | 15.60–15.70 | 1.04E-05 | −0.7 | 8.5 | 4.18E-05 | −0.9 | 7.8 | qFRRDW3 [25] | |||
| qRdw5b | 5 | 14.24–14.33 | 3.05E-05 | 1.1 | 8.0 | ||||||||
| qRdw6 | 6 | 10.03–10.38 | 1.78E-05 | 1.0 | 8.5 | ||||||||
| qRdw8 | 8 | 25.15–25.22 | 3.91E-06 | 1.0 | 9.4 | ||||||||
| qRdw9 | 9 | 14.56–14.84 | 2.97E-06 | 1.1 | 9.7 | 2.19E-05 | 1.2 | 8.4 | qFRRDW9–2 [25] | ||||
| qRdw11 | 11 | 23.00–23.18 | 6.87E-05 | 0.8 | 6.9 | 6.62E-06 | 1.2 | 9.5 | |||||
| qRdw12 | 12 | 0.42–0.48 | 1.19E-05 | 1.3 | 8.9 | ||||||||
| SWC | qSwc2 | 2 | 24.17–24.41 | 2.38E-05 | −0.8 | 8.4 | 2.79E-05 | −0.01 | 8.6 | ||||
| qSwc11 | 11 | ~23.95 | 1.14E-07 | −1.5 | 13.5 | 2.88E-06 | −0.02 | 10.9 | qSWC [13] | ||||
| qSwc12 | 12 | 17.94–17.99 | 2.63E-05 | −1.4 | 9.0 | ||||||||
| SFe | qSFe2 | 2 | 29.81–29.93 | 7.43E-06 | 1.7 | 9.4 | |||||||
| qSFe5 | 5 | 11.18–11.19 | 5.66E-05 | 2.3 | 7.5 | ||||||||
| qSFe6 | 6 | ~10.38 | 2.62E-05 | 1.8 | 8.2 | ||||||||
aSame as in Fig. 1
bthe peak value in the chromosome region
cAllele effect with respect to the minor allele
dPhenotypic variance explained
For SH, four QTL were identified on chromosomes 1, 2, 6 and 12. qSh1 was only detected under the control condition and explained 5.9% of phenotypic variance. The remaining three QTL, qSh2, qSh6 and qSh12 were commonly identified under both the control and stress conditions and explained 4.4% (6.1%) to 8.3% (9.5%) of phenotypic variance in control (stress) condition. And qSh2 also affected stress tolerance index of SH with 7.3% of phenotypic variance (Table 2).
Two QTL (qRl2 and qRl3) for RL were detected on chromosomes 2 and 3. qRl2 was identified under control condition with 8.3% of phenotypic variance, and it also affected stress tolerance index of RL and accounted for 9.5% of phenotypic variance. qRl3 was identified under both control and stress conditions, accounting for 9.4% and 7.8% of phenotypic variance, respectively. Interestingly, minor allele at qRl3 increased RL under control condition but reduced RL under stress condition (Table 2).
For SFW, five QTL were detected on chromosomes 2, 3, 5, 6 and 11. qSfw2, qSfw5b and qSfw6 were identified under stress condition, explaining 8.6% to 9.7% of phenotypic variance. And qSfw2 simultaneously affected stress tolerance index of SFW with 10.2% of phenotypic variance. qSfw3a and qSfw11 were identified under both control and stress conditions and accounted for 7.0% (5.5%) and 7.4% (11.9%) of phenotypic variance in control (stress) conditions, respectively (Table 2).
Five QTL governing SDW were identified on chromosomes 3, 6, 11 and 12. qSdw6 was detected only under stress condition and accounted for 9.2% of phenotypic variance. It also affected stress tolerance index of SDW with 7.9% of phenotypic variance. The other four QTL (qSdw3a, qSdw3b, qSdw11 and qSdw12) were identified under both control and stress conditions and explained phenotypic variance ranging from 5.9% to 11.0% (Table 2).
Seven QTL affecting for RDW were identified on chromosomes 3, 5, 6, 8, 9, 11 and 12. qRdw3, qRdw9 and qRdw11 were identified under both control and stress conditions with the phenotypic variance ranging from 6.9% to 9.5%. qRdw5b, qRdw6 and qRdw12 were detected only under stress condition and explained phenotypic variance ranging from 8.0% to 8.9%. qRdw8 was detected only under control condition with 9.4% of phenotypic variance (Table 2).
For SWC, three QTL were identified on chromosomes 2, 11 and 12. qSwc12 was detected only under control condition and accounted for 9.0% of phenotypic variance. qSwc2 and qSwc11 were detected only under stress condition and explained 8.4% and 13.5% of phenotypic variance, respectively, and they also affected stress tolerance index of SWC with 8.6% and 10.9% of phenotypic variance, respectively (Table 2).
Three QTL (qSFe2, qSFe5 and qSFe6) governing SFe were detected only under stress condition and accounted for 9.4%, 7.5% and 8.2% of phenotypic variance, respectively (Table 2).
Identification of QTL associated with differentiated responses to Zn stress
A total of 31 QTL were identified in Zn stress experiment (Additional file 6), including three only under control condition, 19 detected only under Zn stress condition, and nine commonly detected under both stress and control conditions. Among the latter, 10 QTL (qSh4a, qRl2, qRl8, qSfw5a, qSfw10, qRdw6, qRdw12, qSwc5, qSwc9 and qSwc10) had significant effects on stress tolerance index of their corresponding traits (Fig. 3, Table 3).
Fig. 3.
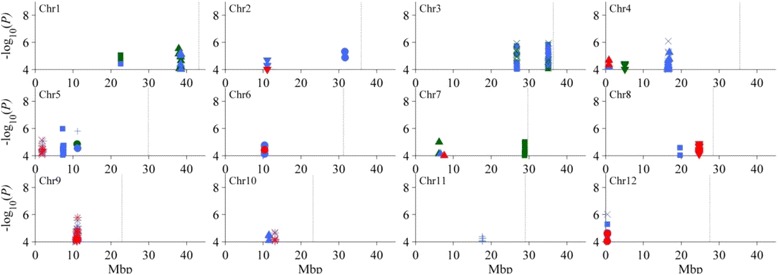
Manhattan plots of zinc toxicity tolerance QTL in the whole genome. Significant SNPs from different conditions are displayed in different colors: control is green, zinc stress is blue, the ratio between zinc stress and control is red. The symbols of associated traits are same as Fig. 2
Table 3.
QTL identified with significant association to zinc toxicity tolerance related traits
| Trait | QTL | Chr | Interval (Mb) | Control | Zn stress | Ratio of stress to control | Previously reported | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P valuea | Effectb | R 2(%)c | P value | Effect | R 2(%) | P value | Effect | R 2(%) | QTL | ||||
| SH | qSh1 | 1 | 37.93–38.54 | 2.85E-06 | −3.4 | 3.7 | 6.39E-06 | −1.8 | 4.7 | ||||
| qSh4a | 4 | 0.87–0.89 | 4.24E-05 | 1.7 | 3.8 | 2.06E-05 | 0.03 | 9.0 | |||||
| qSh4b | 4 | 16.38–16.87 | 5.42E-05 | 3.0 | 2.7 | 1.70E-05 | 2.3 | 4.2 | |||||
| qSh7 | 7 | 6.19–6.27 | 1.00E-05 | 2.6 | 3.3 | ||||||||
| qSh10 | 10 | 11.54–11.55 | 3.22E-05 | 2.2 | 3.9 | ||||||||
| RL | qRl2 | 2 | 11.10–11.11 | 1.99E-05 | −0.6 | 8.0 | 9.73E-05 | −0.05 | 6.9 | ||||
| qRl4 | 4 | 5.10–5.11 | 3.69E-05 | 0.4 | 6.8 | ||||||||
| qRl8 | 8 | 24.66–24.77 | 6.27E-05 | −0.9 | 7.0 | 1.31E-05 | −0.07 | 8.7 | |||||
| SFW | qSfw3a | 3 | 26.69–26.84 | 3.17E-06 | 87.7 | 6.7 | 3.91E-06 | 51.1 | 8.2 | ||||
| qSfw3b | 3 | 35.03–35.16 | 1.15E-06 | 95.9 | 7.3 | 1.70E-06 | 61.1 | 8.8 | qZNT-3 [11] | ||||
| qSfw4 | 4 | 16.41–16.58 | 8.09E-07 | 47.1 | 9.4 | ||||||||
| qSfw5a | 5 | 1.76–1.86 | 7.04E-05 | −31.2 | 6.0 | 2.63E-05 | −0.07 | 7.9 | |||||
| qSfw10 | 10 | 13.05–13.09 | 3.85E-05 | 24.6 | 6.4 | 2.05E-05 | 0.06 | 8.1 | |||||
| qSfw12 | 12 | 0.46–0.48 | 9.63E-07 | 64.9 | 9.3 | ||||||||
| SDW | qSdw1 | 1 | 22.54–22.59 | 8.77E-06 | 8.7 | 6.4 | 3.80E-05 | 8.2 | 6.3 | ||||
| qSdw3a | 3 | 26.69–26.84 | 6.20E-05 | 9.9 | 5.2 | 1.85E-06 | 8.3 | 4.4 | |||||
| qSdw3b | 3 | 35.01–35.22 | 3.12E-05 | 9.0 | 5.6 | 1.47E-06 | 8.6 | 10.5 | qZNT-3 [11] | ||||
| qSdw4 | 4 | 16.46–16.58 | 4.13E-05 | 5.5 | 6.2 | ||||||||
| qSdw5 | 5 | 7.24–7.55 | 1.04E-06 | 6.9 | 9.0 | ||||||||
| qSdw7 | 7 | 28.83–28.87 | 9.88E-06 | −7.7 | 6.3 | ||||||||
| qSdw8 | 8 | 19.65–19.68 | 2.55E-05 | 4.1 | 6.6 | ||||||||
| qSdw12 | 12 | 0.47–0.48 | 5.52E-05 | 8.6 | 5.2 | 5.11E-06 | 8.6 | 7.8 | |||||
| RDW | qRdw2 | 2 | 31.59–31.63 | 4.77E-06 | 0.8 | 9.8 | |||||||
| qRdw5a | 5 | 11.07–11.19 | 1.40E-05 | −1.3 | 8.7 | 2.80E-05 | −1.2 | 8.1 | |||||
| qRdw6 | 6 | ~10.38 | 1.68E-05 | −0.9 | 8.6 | 3.67E-05 | −0.06 | 8.4 | |||||
| qRdw12 | 12 | 0.46–0.48 | 9.22E-05 | −0.8 | 7.1 | 2.50E-05 | −0.05 | 8.8 | |||||
| SWC | qSwc5 | 5 | 1.83–2.03 | 8.47E-06 | −1.02 | 9.4 | 3.15E-05 | −0.01 | 7.3 | ||||
| qSwc9 | 9 | 10.81–11.19 | 2.65E-06 | 0.64 | 10.5 | 1.67E-06 | 0.01 | 8.6 | |||||
| qSwc10 | 10 | 13.05–13.12 | 2.31E-05 | 0.58 | 8.4 | 6.68E-05 | 0.01 | 6.7 | |||||
| SZn | qSZn5 | 5 | 11.18–11.19 | 1.57E-06 | 0.72 | 11.8 | |||||||
| qSZn11 | 11 | 17.63–17.83 | 4.10E-05 | −0.48 | 8.5 | ||||||||
aSame as in Fig. 1
bthe peak value in the chromosome region
cAllele effect with respect to the minor allele
dPhenotypic variance explained
Five QTL affecting SH were identified on chromosomes 1, 4, 7 and 10. qSh7 was identified only under control condition, accounting for 3.3% of phenotypic variance. qSh4a and qSh10 were detected only under stress condition and explained 3.8 and 3.9% of phenotypic variance, respectively. And qSh4a also affected stress tolerance index of SH with 9.0% of phenotypic variance. qSh1 and qSh4b were detected under both stress and control conditions and explained 3.7% (4.7%) and 2.7% (4.2%) of phenotypic variance in control (stress) conditions, respectively (Table 3).
Three QTL affecting RL were identified on chromosomes 2, 4 and 8. qRl2 and qRl8 were detected under Zn stress condition and accounted for 8.0% and 7.0% of phenotypic variance, respectively. What’s more, the two QTL also affected stress tolerance index of RL with 6.9% and 8.7% of phenotypic variance, respectively. qRl4 was identified only under control condition with 6.8% of phenotypic variance (Table 3).
For SFW, 6 QTL were detected on chromosomes 3, 4, 5, 10 and 12. Four QTL (qSfw4, qSfw12, qSfw5a and qSfw10) were identified under stress condition with phenotypic variance ranging from 6.0 to 9.4%. And the latter two QTL also affected stress tolerance index of SFW and accounted for 7.9 and 8.1%, respectively. qSfw3a and qSfw3b were detected under both control and stress conditions and explained phenotypic variation ranging from 6.7% to 8.8% (Table 3).
Eight QTL governing SDW were identified on chromosomes 1, 3, 4, 5, 7, 8 and 12. qSdw7 was detected only under control condition, accounting for 6.3% of phenotypic variance. qSdw4, qSdw5 and qSdw8 were identified under stress condition and explained 6.2%, 9.0% and 6.6% of phenotypic variance, respectively. Four QTL (qSdw1, qSdw3a, qSdw3b and qSdw12) were detected under both control and stress conditions with phenotypic variance ranging from 4.4% to 10.5% (Table 3).
For RDW, 4 QTL were detected on chromosomes 2, 5, 6 and 12. qRdw5a was identified under both control and stress conditions and explained 8.7% and 8.1% of phenotypic variance, respectively. Three QTL (qRdw2, qRdw6 and qRdw12) were detected under Zn stress condition with phenotypic variance ranging from 7.1 to 9.8%. Moreover, qRdw6 and qRdw12 also affected stress tolerance index of RDW with 8.4 and 8.8% of phenotypic variance, respectively (Table 3).
Three QTL (qSwc5, qSwc9 and qSwc10) affecting SWC were identified under stress condition, accounting for 9.4%, 10.5 and 8.4% of phenotypic variance, respectively. These QTL all had significant effects on stress tolerance index of SWC and explained 7.3%, 8.6 and 6.7% of phenotypic variance, respectively (Table 3).
For SZn, two QTL (qSZn5 and qSZn11) were detected under stress condition, accounting for 11.8% and 8.5% of phenotypic variance, respectively (Table 3).
Overlap of QTL for toxicity tolerance related traits across different stress conditions
In comparison of 29 QTL detected under Fe toxicity experiment and 31 QTL detected under Zn toxicity experiment, 8 QTL (qSh1, qRl2, qSfw3a, qSdw3a, qSdw3b, qSdw12, qRdw6 and qRdw12) were commonly identified in both experiments, including qSh1 detected under control condition, qRdw6 and qRdw12 detected under the two stress conditions, qSfw3a, qSdw3a, qSdw3b and qSdw12 detected under both control and the two stress conditions, and qRl2 detected by stress tolerance index. In addition, qSFe5 and qSZn5 were colocated in the region of 11.18–11.19 Mb on chromosome 5 affecting both Fe and Zn concentrations in shoots under Fe and Zn stresses.
Candidate genes for important QTL
A total of 22 candidate genes for 10 important QTL regions were found (Table 4), and only 11 candidate genes had non-synonymous SNPs within the CDS region and were performed for haplotype analysis (Fig. 4). There were 1 to 6 candidate genes for each important QTL region.
Table 4.
List of 22 candidate genes for 10 important QTL associated with Fe or Zn toxicity tolerance
| QTL | Candidate gene | Annotation |
|---|---|---|
| qSh6 | LOC_Os06g17690 | hypothetical protein |
| qSh6 | LOC_Os06g17800 | retrotransposon protein, putative, Ty3-gypsy subclass, expressed |
| qSh6 | LOC_Os06g17880 | NBS-LRR disease resistance protein, putative, expressed |
| qSdw3a | LOC_Os03g47149 | expressed protein |
| qSdw3a | LOC_Os03g47240 | Conserved hypothetical protein. |
| qSdw3a | LOC_Os03g47310 | transposon protein, putative, CACTA, En/Spm sub-class, expressed |
| qSdw3a | LOC_Os03g47330 | transposon protein, putative, CACTA, En/Spm sub-class |
| qSdw3a | LOC_Os03g47360 | Similar to F-box domain containing protein. |
| qSdw3a | LOC_Os03g47370 | LTPL95 - Protease inhibitor/seed storage/LTP family protein precursor, putative, expressed |
| qSdw3b | LOC_Os03g62050 | conserved hypothetical protein |
| qSdw3b | LOC_Os03g62060 | Similar to IAA-amino acid hydrolase 1 |
| qSdw3b | LOC_Os03g62170 | cyclase/dehydrase family protein, expressed |
| qSdw6 | LOC_Os06g17690 | hypothetical protein |
| qSdw6 | LOC_Os06g17880 | NBS-LRR disease resistance protein, putative, expressed |
| qSdw11 | LOC_Os11g38890 | retrotransposon protein, putative, unclassified, expressed |
| qSdw11 | LOC_Os11g38930 | tRNA-splicing endonuclease subunit Sen2, putative, expressed |
| qSdw11 | LOC_Os11g38959 | 40S ribosomal protein S9–2, putative, expressed |
| qRdw9 | LOC_Os09g24700 | Ribosomal protein L34e domain containing protein |
| qSwc5 | LOC_Os05g04410 | peroxidase precursor, putative, expressed |
| qSwc10 | LOC_Os10g25320 | initiation factor 2 subunit family domain containing protein, expressed |
| qSFe2 | LOC_Os02g48940 | expressed protein |
| qSFe2 | LOC_Os02g48950 | ubiquitin-conjugating enzyme, putative, expressed |
| qSZn11 | LOC_Os11g30370 | OsSPL19 - SBP-box gene family member, expressed |
| qSZn11 | LOC_Os11g30400 | expressed protein |
Fig. 4.
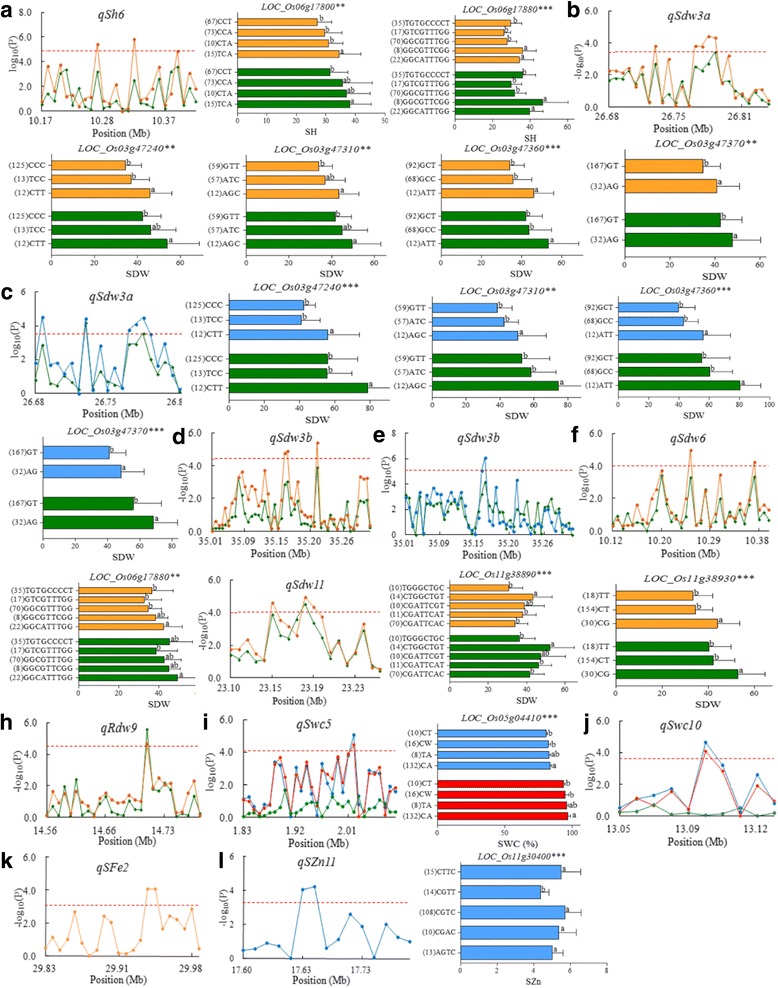
Manhattan plot of important QTL and haplotype analysis of candidate genes related to QTL including qSh6 (a), qSdw3a (b and c), qSdw3b (d and e), qSdw6 (f), qSdw11 (g), qRdw9 (h), qSwc5 (i), qSwc10 (j), qSFe2 (k) and qSZn11(l). Each point was a gene in the region of the QTL. Line and histogram in different colors indicated different conditions: green is control condition, yellow is ferrous iron stress condition, blue is zinc stress condition and red is the ratio of zinc stress to control condition. Dash line showed the threshold to determine candidate genes. The ** and *** suggested significance of ANOVA at p < 0.01and p < 0.001, respectively. The letter on histogram (a and b) indicated multiple comparisons result at the significant level 0.01. The value in bracket was the number of individuals for each haplotype
For qSh6 in the region 10.17–10.38 Mb on chromosome 6, 102 SNPs were identified in 26 genes. Among them, three candidate genes (LOC_Os06g17690, LOC_Os06g17800 and LOC_Os06g17880) were found for qSh6 (Fig. 4a). Significant differences in SH were detected between different haplotypes in two candidate genes (LOC_Os06g17800 and LOC_Os06g17880), and the major allele(s) in this two genes significantly decreased SH which had same effect as the peak SNP. No haplotype was found for LOC_Os06g17690.
For qSdw3a in the region 26.68–26.84 Mb on chromosome 3, 71 SNPs were identified in 25 genes, and 6 candidate genes (LOC_Os03g47149, LOC_Os03g47240, LOC_Os03g47310, LOC_Os03g47330, LOC_Os03g47360 and LOC_Os03g47370) were found. Among them, LOC_Os03g47149 was only detected under zinc stress condition (Fig. 4b and c). No haplotype was found in LOC_Os03g47149 and LOC_Os03g47330. Two haplotypes were found for LOC_Os03g47370, and three haplotypes found for the other three genes (LOC_Os03g47240, LOC_Os03g47310 and LOC_Os03g47360). The minor haplotypes significantly increased SDW under both Fe and Zn stress conditions.
For qSdw3b in the region from 35.01 to 35.29 Mb on chromosome 3, 163 SNPs were identified in 49 genes. Three candidate genes (LOC_Os03g62050, LOC_Os03g62060 and LOC_Os03g62170) were observed for qSdw3b, and only LOC_Os03g62170 was detected under Fe stress condition (Fig. 4d and e). No haplotype was found for all these genes.
For qSdw6 in the region from 10.12 to 10.38 Mb on chromosome 6, 163 SNPs were detected in 33 genes. Two candidate genes LOC_Os06g17690 and LOC_Os06g17880 were found for qSdw6 (Fig. 4f). Only LOC_Os06g17880 had haplotype and significant phenotypic differences for SDW were observed between the five haplotypes, indicating that LOC_Os06g17880 was the candidate gene for qSdw6.
For qSdw11 in the region from 23.03 to 23.19 Mb on chromosome 11, 100 SNPs were identified in 19 genes. Three candidate genes (LOC_Os11g38890, LOC_Os11g38930 and LOC_Os11g38959) were observed for qSdw11, but no haplotype was found for LOC_Os11g38959 (Fig. 4g). Haplotype analysis revealed that major haplotype significantly decreased SDW for the two genes (LOC_Os11g38890 and LOC_Os11g38930).
For qRdw9 in the region of 14.56–14.84 Mb on chromosome 9, 84 SNPs were detected in 27 genes. Only one candidate gene LOC_Os09g24700 was found for qRdw9 which was associated with RDW under both control and Fe stress conditions. No haplotype was found in this gene (Fig. 4h).
For qSwc5 in the region from 1.83 to 2.03 Mb on chromosome 5, 137 SNPs were identified in 30 genes. Only candidate gene LOC_Os05g04410 was observed for qSwc5. Haplotype analysis revealed that the major haplotype increased SWC both under Zn stress condition and the Zn stress tolerance index, indicating that it was the candidate gene for qSwc5 (Fig. 4i).
For qSwc10 located in the region 13.05–13.12 Mb on chromosome 10, 28 SNPs were detected in 10 genes. Only one candidate gene LOC_Os10g25320 was associated with significant SNP for qSwc10. No haplotype was found for the gene (Fig. 4j).
For qSFe2 in the region from 29.81 to 29.93 Mb on chromosome 2, 58 SNPs were detected in 22 genes. LOC_Os02g48940 and LOC_Os02g48950 were the only two candidate genes for qSFe2. No haplotype was found for these two genes (Fig. 4k).
For qSZn11 in the region 17.63–17.83 Mb on chromosome 11, 70 SNPs were identified in 15 genes. Two candidate genes LOC_Os11g30370 and LOC_Os11g30400 were observed for qSZn11. Five haplotypes were found for LOC_Os11g30400 but no haplotype for LOC_Os11g30370. Haplotype CGTT had significantly lower Zn concentration in shoot than the other four haplotypes (Fig. 4l).
Discussion
Differential QTL expressions and their association with ion toxicity tolerance in rice
Stress tolerance in crops can be characterized in morphological, physiological and biochemical levels [27]. Many morpho-physiological traits putatively contribute to stress tolerance, and each of these traits is typically controlled by multiple genes or QTL [28]. In consideration of crop tolerance to stress is influenced by environment to a great extent, it was proposed to improve stress tolerance by marker-assisted selection (MAS) for secondary traits such as morpho-physiological traits if genes/QTL affecting the secondary traits contributing to stress tolerance could be accurately mapped and characterized [29]. Secondary traits are plant characteristics that are associated with stress tolerance under stress and should be satisfied with following tests, (1) genetically correlated with stress tolerance in the stress conditions; (2) highly heritable in the screening system; (3) enough variation among lines for the trait; (4) possible to measure the trait rapidly and economically [29]. In our study, Fe and Zn tolerance-related morphological traits (SH, RL, SFW, SDW, RDW, SWC) in stress and control conditions and physiological traits (SFe and SZn) in stress condition were measured, and their derived traits such as ratio of stress to control were also calculated and all used for input data to detect QTL. Definitely, QTL affecting ratio traits were directly related to stress tolerance [30]. However, ratio traits may reduce trait differences or variations in the population, thus probably resulting in that some QTL were undetectable. So besides comparing QTL results using ratio traits, we also compared QTL detections between stress and control conditions as we dissected drought tolerance QTL in our previous reports [31, 32].
Most of QTL at seedling stage associated with SH, RL, SFW, SDW, RDW, SWC, SFe and SZn detected in this study were specific to either Fe or Zn stress condition. Of the 29 QTL identified in Fe stress experiments, four were detected only in control condition, and 12 were detected only under Fe stress condition (Table 2). In zinc stress experiment, three were identified only in normal condition, and 19 were detected only under Zn stress condition (Table 3). Of the 13 QTL detected under both control and Fe stress conditions, one (qRl3) had opposite phenotypic effect under control and Fe stress conditions, three (qSh12, qSfw3a and qSfw11) had effects that significantly differed in magnitude under control and Fe stress conditions, and nine (qSh2, qSh6, qSdw3a, qSdw3b, qSdw11, qSdw12, qRdw3, qRdw9 and qRdw11) behaved similarly under Fe stress and non-stress conditions. Of the 9 QTL detected under both control and Zn stress conditions, three (qSh1, qSfw3a and qSfw3b) had effects that differed significantly in magnitude under control and Zn stress conditions, and six (qSh4b, qSdw1, qSdw3a, qSdw3b, qSdw12 and qRdw5a) behaved similarly under Zn stress and control conditions. Among the above QTL, we believe that only two types of QTL really contributed to Fe or Zn toxicity tolerance. Type I was the QTL identified by the ratio of stress to control conditions, which can reduce trait difference between stress and non-stress conditions and thus contributes to toxicity tolerance. Type II was the QTL having the same orientation and similar magnitude of phenotypic effect under both control and stress conditions, suggesting they had stable expression under stress and non-stress conditions. Therefore, a total of 14 (48.3%) different QTL contributed to Fe toxicity tolerance under Fe stress, including six QTL (qSh2, qRl2, qSfw2, qSdw6, qSwc2 and qSwc11) belonged to type I and nine QTL (qSh2, qSh6, qSdw3a, qSdw3b, qSdw11, qSdw12, qRdw3, qRdw9 and qRdw11) belonged to type II. Similarly, 16 (51.6%) QTL were associated with Zn toxicity tolerance, including 10 QTL (qSh4a, qRl2, qRl8, qSfw5, qSfw10, qRdw6, qRdw12, qSwc5, qSwc9 and qSwc10) classified as type I and six (qSh4b, qSdw1, qSdw3a, qSdw3b, qSdw12 and qRdw5a) QTL classified into type II. Among above QTL contributing to Fe and Zn toxicity tolerance, six QTL, including qRl2 affecting RL, qSdw3a, qSdw3b and qSdw12 affecting SDW and qSFe5 (qSZn5) for concentrations of Fe (Zn) were detected in the same genomic regions under the both stress conditions. This indicates that there were probably some common genes or partially overlapping mechanisms for responses to Fe and Zn toxicity in rice.
Comparison of present QTL with previously reported QTL and cloned genes
Comparison of QTL affecting Fe and Zn toxicity tolerance detected in this study with previously reported QTL or cloned genes was performed within 1 Mb physical distance. Of the 14 QTL for Fe toxicity tolerance, 8 QTL were found to locate in the same or adjacent regions with previously reported QTL or cloned genes in rice (Table 2). For example, qSh6 affecting SH was mapped in the region 10.17–10.38 Mb on chromosome 6 which harbored previously reported qSh6a for SH under Fe stress condition [16]. qSfw3a for SFW and qSdw3a for SDW, detected in the region on chromosome 3 under both Fe and Zn stress conditions colocated with a previously mapped putative QTL for shoot dry weight detected by chromosomal segments substitution lines (CSSLs) [33] and OsIRT1 (LOC_Os03g46470), which is a functional metal transporter of Fe and actively engaged in Fe uptake from soils [34]. Its over-expression leads to increase Fe and Zn accumulation in rice [35]. qSdw3b and qSdw6 affecting SDW, located in the regions of 35.01–35.29 Mb on chromosome 3 and 10.12–10.38 Mb on chromosome 6, were colocated with qSDW3 [8] and qSdw [36] for SDW, respectively. qSdw11 affecting SDW on chromosome 11 colocated with qLBI11 which influences leaf bronzing index under ferrous iron stress toxicity condition [8]. qRdw3 and qRdw9 for RDW were colocated with qFRRDW3 and qFRRDW9–2 for FRRDW (Fe relative root dry weight), respectively [13]. qSwc11 affecting SWC colocated with qSWC for SWC [11]. Unlike QTL mapping of Fe toxicity tolerance, there are few reports on QTL mapping for Zn toxicity tolerance. Only two QTL, qSfw3b for SFW and qSdw3b for SDW in this study collocated with qZNT-3 affecting index score of Zn toxicity at rice seedling stage [9]. QTL regions for the Fe and Zn toxicity tolerance-related traits that were identified in different mapping populations and diverse environments could be beneficial for MAS-based development of Fe and Zn toxicity tolerant rice varieties.
Candidate gene identification for the important toxicity tolerance QTL
Although many studies reported QTL for different traits associated to Fe or Zn toxicity at seedling stage in rice [9, 13, 17, 18], no stable locus has been identified, fine-mapped or cloned so far [21]. The genetic mechanisms of rice tolerance to Fe and Zn toxicity seem to be complex. Using high density SNPs for GWAS and haplotype analysis of candidate genes, we found 22 candidate genes for 10 important QTL regions affecting the measured traits.
In the region 10.12–10.38 Mb on chromosome 6, both qSh6 and qSdw6 were located. Of the three candidate genes for qSh6 and two candidate genes for qSdw6, the most likely one was LOC_Os06g17880, which encodes an NBS-LRR protein involved in disease resistance, drought tolerance and salt tolerance [37–39]. In this research, qSdw3a and qSdw3b were identified under both control and stress conditions in Fe and Zn experiments. Of the six candidate genes for qSdw3a, LOC_Os03g47360 (similar to F-box domain containing protein) was the most likely one, as F-Box protein in rice was reported to be expressed under abiotic stress conditions [40]. Of the three candidate genes for qSdw3b, the most likely one was LOC_Os03g62060 (similar to IAA-amino acid hydrolase 1) even though no haplotype was found in it, because it is very important for plant growth [41]. The gene LOC_Os05g04410 is involved in peroxidase precursor whose POD-increasing activity reportedly is a defensive response to excess heavy metals in rice [42, 43]. Thus, LOC_Os05g04410 was the most likely candidate gene for qSwc5. For qSFe2, LOC_Os02g48950 (ubiquitin conjugating enzyme) was the most likely candidate gene, as ubiquitination plays important roles in plant abiotic stress response [44]. For qSZn11, LOC_Os11g30370 encodes OsSPL19 - SBP-box, a zinc finger protein involved in a variety of biological processes [45]. Due to low density of SNP, no haplotype was found for this gene. These two genes may be both the candidate genes of qSZn11.
Considerations for GWAS mapping in this study
GWAS used for association mapping of quantitative traits is becoming more and more feasible in recent years. In this study, a total of 395,553 SNPs remained after filtering low MAF (minor allele frequency) from the 700,000 SNPs, which roughly equates to 1 marker per 1 kb. It was sufficient to recognize genomic associations for Fe and Zn toxicity tolerance in rice and to identify significant SNP clusters for associated traits. The population used in this study was comparatively small and composed exclusively of indica accessions. Intra-subspecific analyses decrease the incidence of false-positive associations resulting from population structure. However, loci with low MAF were not detected in this study, thus some functional alleles probably escaped detection in this indica panel [46]. We selected significant SNPs in genes and performed haplotype analysis by non-synonymous SNP in CDS region for candidate genes analysis. There was uncertainty in candidate gene detection for some important QTL such as (qSdw3b, qSFe2 and qRdw9) due to absence of haplotype, resulting from low numbers of significant non-synonymous SNPs. Due to the low density of SNP in some QTL regions, it was also difficult to find non-synonymous significant SNPs in CDS region in genes located in the search interval as defined in this study. For instance, the region of 11.18–11.19 Mb on chromosome 5 is important, harboring qSFe5 and qSZn5 for concentrations of Fe and Zn in shoots under stress conditions. However, there was no non-synonymous significant SNP in the CDS region of the gene (LOC_Os05g19270) within the search interval. Therefore, high SNP density and large population size are important to identify candidate genes through GWAS.
Application in rice breeding for heavy metal toxicity tolerance
Higher Fe and Zn concentrations in the grains are desirable for human health. Ruengphayak found that the MuFRO1 mutant which tolerated Fe toxicity in the vegetative stage had 21–30% more grain Fe content than its wild type [47]. Therefore, breeding resistant rice varieties that tolerate high Fe and Zn concentrations with high content in the grains is an effective way to avoid soil pollution effects on agriculture [48]. Fe and Zn toxicity tolerance processes are difficult to define and measure. The secondary traits or symptoms associated with stress can help breeders make perform selection for stress tolerance [49]. In this study, SH, SFW and SDW were easily measured and more reliable than RL, RDW and SWC. And SFe (SZn) had significant a negative correlation with the aboveground physical traits. Consequently, favorable haplotypes or alleles of some toxicity tolerance QTL such as qSdw3a, qSdw3b, qSdw12, qSFe5 and qSZn5 may be useful for improving rice tolerance to Fe and Zn toxicity by marker-assisted selection (MAS) or QTL pyramiding. Three accessions (CC55, CC123 and CC155) with low concentrations of Fe and Zn and high tolerance index of aboveground traits under stress were identified to have strong Fe and Zn stress tolerance in this panel. At the four QTL regions mentioned above, these lines had the alleles for Fe and Zn toxicity tolerance (data not shown). So CC55, CC123 and CC155 could be used as donors in rice breeding for Fe and Zn toxicity tolerance by MAS.
Conclusions
Large genetic variations for eight Fe and Zn toxicity tolerance related traits existed in the panel of 211 indica accessions. Through GWAS, a total of 14 QTL for Fe toxicity tolerance and 16 QTL for Zn toxicity tolerance were identified, respectively. qSdw3a, qSdw3b, qSdw12 and qSFe5 / qSZn5 were detected in the same genomic regions under the two stress conditions, indicating that there are probably common genes and mechanisms governing Fe and Zn toxicity tolerance in rice. A total 22 candidate genes for 10 important QTL regions were determined by haplotype analyses. Five most likely candidates of five QTL (qSh6 / qSdw6, qSdw3a, qSdw3b, qSwc5, qSFe2) underlying aboveground traits under stress were inferred according to functional annotation. Three accessions (CC55, CC123 and CC155) having favorable alleles at the four loci showed strong Fe and Zn stress tolerance. The candidate genes affecting Fe and Zn toxicity tolerance and tolerant accessions identified in this study provide valuable information for future functional characterization and improvement of rice varieties for heavy metal toxicity tolerance by MAS.
Additional files
PCA 3D plot of the first three principal components (PC) in 222 accessions (a) and 211 accessions (b); Table S1. List of indica accessions used in the present study. (DOCX 201 kb)
ANOVA results of the measured traits under Fe toxicity for 211 indica accessions. (DOCX 25 kb)
ANOVA results for the measured traits under zinc toxicity for 211 indica accessions. (DOCX 26 kb)
Correlations between all measured traits under control (upper triangular) and stress (lower triangular) conditions in Fe (a) and Zn (b) experiments. (DOCX 275 kb)
Significant SNPs associated with measured traits in Fe experiment. (XLSX 34 kb)
Significant SNPs associated with measured traits in Zn experiment. (XLSX 40 kb)
Acknowledgements
We gratefully thank Dr. Susan R. McCouch for providing high-density rice array (HDRA) comprised of 700,000 SNPs data.
Funding
Introduction of rice germplasms was supported by the “948” Project to JX (2016-X16) from the Chinese Ministry of Agriculture (http://www.moa.gov.cn/) and the CAAS Innovative Team Awards to JX (http://www.caas.net.cn/), design of the study, data collection and experimental supplies were funded the “863” Key Project to JX (2014AA10A601) from the Chinese Ministry of Science and Technology (http://www.863.gov.cn/), data analysis and interpretation were funded by the Shenzhen Peacock Plan to ZL (http://www.szsti.gov.cn/) (#:20,130,415,095,710,361), and the publication costs of this article was provided by Bill and Melinda Gates Foundation project to ZL (#OPP1130530).
Availability of data and materials
The data sets supporting the results of this article are included within the article and its additional files.
Abbreviations
- CK
Control condition
- Fe/CK
Ratio of ferrous iron stress to control condition
- MAS
Marker-assisted selection
- RDW
Root dry weight
- RL
Root length
- SDW
Shoot dry weight
- SFe
Fe concentration in shoot;
- SFW
Shoot fresh weight
- SH
Shoot height
- SWC
Shoot water content
- SZn
Zn concentration in shoot;
- Zn/CK
Ratio of zinc stress to control condition
Authors’ contributions
JLX and ZKL designed the experiment; JZ, KC, SAN, XQZ, XQW, YW performed phenotypic evaluation; JZ and YLP performed analysis and interpretation of the data; JZ and LJX drafted the manuscript; MD and JP revised the MS. All authors approved the final version to be published.
Ethics approval and consent to participate
All rice germplasm resources used in this study were introduced and granted by IRRI, which are encouraged for identification of favorable gene for agronomic traits. The introduction of rice varieties was complied with MTA of IRRI and China’s legislation, and all the experiments were conducted in accordance with the regulations of CAAS, and did not involve any endangered or protected species.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12864-017-4221-5) contains supplementary material, which is available to authorized users.
Contributor Information
Jian Zhang, Email: 0603zhj@163.com.
Kai Chen, Email: kaiserchenkai@163.com.
Yunlong Pang, Email: pangyunlong8804@126.com.
Shahzad Amir Naveed, Email: shahzad.nibge@gmail.com.
Xiuqin Zhao, Email: zhaoxiuqin@caas.cn.
Xiaoqian Wang, Email: xqwang1988@126.com.
Yun Wang, Email: 18810522880@163.com.
Michael Dingkuhn, Email: michael.dingkuhn@cirad.fr.
Julie Pasuquin, Email: j.pasuquin@irri.org.
Zhikang Li, Email: zhkli1953@126.com.
Jianlong Xu, Email: xujlcaas@126.com.
References
- 1.Nagajyoti PC, Lee KD, Sreekanth TVM. Heavy metals, occurrence and toxicity for plants: a review. Environ Chem Lett. 2010;8(3):199–216. doi: 10.1007/s10311-010-0297-8. [DOI] [Google Scholar]
- 2.De Dorlodot S, Lutts S, Bertin P: Effects of ferrous iron toxicity on the growth and mineral composition of an Interspecific Rice. J Plant Nutr 2005, 28(1):1-20.
- 3.Becker M, Asch F. Iron toxicity in rice—conditions and management concepts. J Plant Nutr Soil Sci. 2005;168(4):558–573. doi: 10.1002/jpln.200520504. [DOI] [Google Scholar]
- 4.Gu H-H, Zhan S-S, Wang S-Z, Tang Y-T, Chaney R, Fang X-H, Cai X-D, Qiu R-L. Silicon-mediated amelioration of zinc toxicity in rice (Oryza Sativa L.) seedlings. Plant Soil. 2012;350(1–2):193–204. doi: 10.1007/s11104-011-0894-8. [DOI] [Google Scholar]
- 5.Vromman D, Lutts S, Lefèvre I, Somer L, De Vreese O, Šlejkovec Z, Quinet M: Effects of simultaneous arsenic and iron toxicities on rice (Oryza Sativa L.) development, yield-related parameters and as and Fe accumulation in relation to as speciation in the grains. Plant Soil 2013, 371(1–2):199-217.
- 6.Borkert CM, Cox FR, Tucker M. Zinc and copper toxicity in peanut, soybean, rice, and corn in soil mixtures. Commun Soil Sci Plant Anal. 1998;29(19–20):2991–3005. doi: 10.1080/00103629809370171. [DOI] [Google Scholar]
- 7.Song A, Li P, Li Z, Fan F, Nikolic M, Liang Y. The alleviation of zinc toxicity by silicon is related to zinc transport and antioxidative reactions in rice. Plant Soil. 2011;344(1–2):319–333. doi: 10.1007/s11104-011-0749-3. [DOI] [Google Scholar]
- 8.Dufey I, Hakizimana P, Draye X, Lutts S, Bertin P. QTL mapping for biomass and physiological parameters linked to resistance mechanisms to ferrous iron toxicity in rice. Euphytica. 2009;167(2):143–160. doi: 10.1007/s10681-008-9870-7. [DOI] [Google Scholar]
- 9.Dong Y, Ogawa T, Lin D, Koh HJ, Kamiunten H, Matsuo M, Cheng S. Molecular mapping of quantitative trait loci for zinc toxicity tolerance in rice seedling (Oryza Sativa L.) Field Crop Res. 2006;95(2–3):420–425. doi: 10.1016/j.fcr.2005.03.005. [DOI] [Google Scholar]
- 10.Manara A. Plant responses to heavy metal toxicity. In: Furini A, editor. Plants and heavy metals. Dordrecht: Springer Netherlands; 2012. pp. 27–53. [Google Scholar]
- 11.Dufey I, Hiel MP, Hakizimana P, Draye X, Lutts S, Koné B, Dramé KN, Konaté KA, Sie M, Bertin P. Multienvironment quantitative trait loci mapping and consistency across environments of resistance mechanisms to ferrous iron toxicity in rice. Crop Sci. 2012;52(2):539–550. doi: 10.2135/cropsci2009.09.0544. [DOI] [Google Scholar]
- 12.Chandel G, Banerjee S, Verulkar SB: Expression profiling of metal homeostasis related candidate genes in rice (Oryza spp.) using semi quantitative RT-PCR analysis. Rice Genetics Newsletter 2010.
- 13.Liu H, Soomro A, Zhu Y, Qiu X, Chen K, Zheng T, Yang L, Xing D, Xu J. QTL underlying iron and zinc toxicity tolerances at seedling stage revealed by two sets of reciprocal introgression populations of rice ( Oryza Sativa L.) Crop J. 2016;4(4):280–289. doi: 10.1016/j.cj.2016.05.007. [DOI] [Google Scholar]
- 14.Wu P, Hu B, Liao CY, Zhu JM, Wu YR, Senadhira D, Paterson AH. Characterization of tissue tolerance to iron by molecular markers in different lines of rice. Plant Soil. 1998;203(2):217–226. doi: 10.1023/A:1004321218387. [DOI] [Google Scholar]
- 15.Wan J-l, H-q Z, Wan J-m, Ikehashi H. Detection and analysis of QTLs for ferrous iron toxicity tolerance in rice, Oryza Sativa L. Euphytica. 2003;131(2):201–206. doi: 10.1023/A:1023915710103. [DOI] [Google Scholar]
- 16.Zhang J, Aijaz AS, Chai L, Cui Y, Wang X, Zheng T, Jianlong XU, Zhikang LI. Mapping of QTL for iron and zinc toxicity tolerance at seedling stage using a set of reciprocal introgression lines of Rice. Acta Agron Sin. 2013;39(10):1754. doi: 10.3724/SP.J.1006.2013.01754. [DOI] [Google Scholar]
- 17.Dufey I, Draye X, Lutts S, Lorieux M, Martinez CP, Bertin P. Novel QTLs in an interspecific backcross Oryza Sativa × Oryza Glaberrima for resistance to iron toxicity in rice. Euphytica. 2015;204(3):609–625. doi: 10.1007/s10681-014-1342-7. [DOI] [Google Scholar]
- 18.Dufey I, Mathieu A-S, Draye X, Lutts S, Bertin P. Construction of an integrated map through comparative studies allows the identification of candidate regions for resistance to ferrous iron toxicity in rice. Euphytica. 2015;203(1):59–69. doi: 10.1007/s10681-014-1255-5. [DOI] [Google Scholar]
- 19.Holland JB. Genetic architecture of complex traits in plants. Curr Opin Plant Biol. 2007;10(2):156–161. doi: 10.1016/j.pbi.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Ingvarsson PK, Street NR. Association genetics of complex traits in plants. New Phytol. 2011;189(4):909–922. doi: 10.1111/j.1469-8137.2010.03593.x. [DOI] [PubMed] [Google Scholar]
- 21.Matthus E, Wu LB, Ueda Y, Holler S, Becker M, Frei M. Loci, genes, and mechanisms associated with tolerance to ferrous iron toxicity in rice (Oryza Sativa L.) Theor Appl Genet. 2015;128(10):2085–2098. doi: 10.1007/s00122-015-2569-y. [DOI] [PubMed] [Google Scholar]
- 22.Qiu X, Pang Y, Yuan Z, Xing D, Xu J, Michael D, Li Z, Ye G. Genome-wide association study of grain appearance and milling quality in a worldwide collection ofIndicaRice Germplasm. PLoS One. 2015;10(12):e0145577. doi: 10.1371/journal.pone.0145577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshida S, Fomo DA, Cock J, Gomez KA: Routine procedure for growing rice plants in culture solution; 1976.
- 24.Mccouch SR, Wright MH, Tung CW, Maron LG, Mcnally KL, Fitzgerald M, Singh N, Declerck G, Agostoperez F, Korniliev P. Open access resources for genome-wide association mapping in rice. Nat Commun. 2016;7:10532. doi: 10.1038/ncomms10532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z. GAPIT: genome association and prediction integrated tool. Bioinformatics. 2012;28(18):2397. doi: 10.1093/bioinformatics/bts444. [DOI] [PubMed] [Google Scholar]
- 26.Project IRGS The map-based sequence of the rice genome. Nature. 2005;436(7052):793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 27.Blum A. Drought resistance - is it really a complex trait? Funct Plant Biol. 2011;38(10):753–757. doi: 10.1071/FP11101. [DOI] [PubMed] [Google Scholar]
- 28.Tiwari R, Mamrutha HM. Precision Phenotyping for mapping of traits for Abiotic stress tolerance in crops. In: Salar RK, Gahlawat SK, Siwach P, Duhan JS, editors. Biotechnology: prospects and applications. New Delhi: Springer India; 2013. pp. 79–85. [Google Scholar]
- 29.Lafitte R: Genetic variation in performance under reproductive-stage water deficit in a doubled haploid rice population in upland fields. 2000.
- 30.Yue B, Xue W, Xiong L, Yu X, Luo L, Cui K, Jin D, Xing Y, Zhang Q. Genetic basis of drought resistance at reproductive stage in Rice: separation of drought tolerance from drought avoidance. Genetics. 2006;172(2):1213. doi: 10.1534/genetics.105.045062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu JL, Lafitte HR, Gao YM, Fu BY, Torres R, Li ZK. QTLs for drought escape and tolerance identified in a set of random introgression lines of rice. Theor. Appl. Genet. 2005;111(8):1642–1650. doi: 10.1007/s00122-005-0099-8. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Zhang Q, Zheng T, Cui Y, Zhang W, Xu J, Li Z. Drought-tolerance QTLs commonly detected in two sets of reciprocal introgression lines in rice. Crop Pasture Sci. 2014;65(2):171. doi: 10.1071/CP13344. [DOI] [Google Scholar]
- 33.Fukuda A, Shiratsuchi H, Fukushima A, Yamaguchi H, Mochida H, Terao T, Ogiwara H. Detection of chromosomal regions affecting iron concentration in Rice shoots subjected to excess ferrous iron using chromosomal segment substitution lines betweenJaponica and Indica. Plant Prot. Sci. 2012;15(3):183–191. doi: 10.1626/pps.15.183. [DOI] [Google Scholar]
- 34.Bughio N, Yamaguchi H, Nishizawa NK, Nakanishi H, Mori S. Cloning an iron-regulated metal transporter from rice. J Exp Bot. 2002;53(374):1677–1682. doi: 10.1093/jxb/erf004. [DOI] [PubMed] [Google Scholar]
- 35.Sichul L, An GH. Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant Cell Environ. 2009;32(4):408–416. doi: 10.1111/j.1365-3040.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- 36.Shimizu A, Guerta CQ, Gregorio GB, Kawasaki S, Ikehashi H. QTLs for nutritional contents of rice seedlings ( Oryza Sativa L.) in solution cultures and its implication to tolerance to iron-toxicity. Plant Soil. 2005;275(1):57–66. doi: 10.1007/s11104-004-4683-5. [DOI] [Google Scholar]
- 37.Leon TBD, Linscombe S, Subudhi PK. Molecular dissection of seedling salinity tolerance in Rice (Oryza Sativa L.) using a high-density GBS-based SNP linkage map. Rice. 2016;9(1):52. doi: 10.1186/s12284-016-0125-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chini A, Grant JJ, Seki M, Shinozaki K, Loake GJ. Drought tolerance established by enhanced expression of the CC-NBS-LRR gene, ADR1, requires salicylic acid, EDS1 and ABI1. Plant J Cell Mol Biol. 2004;38(5):810–822. doi: 10.1111/j.1365-313X.2004.02086.x. [DOI] [PubMed] [Google Scholar]
- 39.Belkhadir Y, Subramaniam R, Dangl JL. Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr Opin Plant Biol. 2004;7(4):391. doi: 10.1016/j.pbi.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP. F-box proteins in Rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and Abiotic stress. Plant Physiol. 2007;143(4):1467–1483. doi: 10.1104/pp.106.091900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ki-Hong J, Hyun-Jung G, Hoi-Khoanh G, Nalini CAK, Quynh-Nga N, Heebak C, Zhang T, Wang W, Jin-Hyun K, Hong-Kyu C. Genome-wide identification and analysis ofJaponicaandIndicacultivar-preferred transcripts in rice using 983 Affymetrix array data. Rice. 2013;6(1):19. doi: 10.1186/1939-8433-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang WC, Chinghuei K. Enhanced peroxidase activity in rice leaves in response to excess iron, copper and zinc. Plant Sci. 2000;158(2):71–76. doi: 10.1016/S0168-9452(00)00307-1. [DOI] [PubMed] [Google Scholar]
- 43.Wu LB, Shhadi MY, Gregorio G, Matthus E, Becker M, Frei M. Genetic and physiological analysis of tolerance to acute iron toxicity in rice. Rice. 2014;7(1):8. doi: 10.1186/s12284-014-0008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou GA, Chang RZ, Qiu LJ. Overexpression of soybean ubiquitin-conjugating enzyme gene GmUBC2 confers enhanced drought and salt tolerance through modulating abiotic stress-responsive gene expression in Arabidopsis. Plant Mol Biol. 2010;72(4):357. doi: 10.1007/s11103-009-9575-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang S-D, Ling L-Z, Yi T-S. Evolution and divergence of SBP-box genes in land plants. BMC Genomics. 2015;16(1):1–10. doi: 10.1186/1471-2164-16-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atwell S, Huang YS, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT. Genome-wide association study of 107 phenotypes in Arabidopsis Thaliana inbred lines. Nature. 2010;465(7298):627. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruengphayak S, Ruanjaichon V, Saensuk C, Phromphan S, Tragoonrung S, Kongkachuichai R, Vanavichit A. Forward screening for seedling tolerance to Fe toxicity reveals a polymorphic mutation in ferric chelate reductase in rice. Rice. 2015;8(1):3. doi: 10.1186/s12284-014-0036-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Satoh-Nagasawa N, Mori M, Nakazawa N, Kawamoto T, Nagato Y, Sakurai K, Takahashi H, Watanabe A, Akagi H. Mutations in rice (Oryza Sativa) heavy metal ATPase 2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant Cell Physiol. 2012;53(1):213–224. doi: 10.1093/pcp/pcr166. [DOI] [PubMed] [Google Scholar]
- 49.Wang N, Wang ZP, Liang XL, Weng JF, Lv XL, Zhang DG, Yang J, Yong HJ, Li MS, Li FH. Identification of loci contributing to maize drought tolerance in a genome-wide association study. Euphytica. 2016;210(2):1–15. doi: 10.1007/s10681-016-1688-0. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCA 3D plot of the first three principal components (PC) in 222 accessions (a) and 211 accessions (b); Table S1. List of indica accessions used in the present study. (DOCX 201 kb)
ANOVA results of the measured traits under Fe toxicity for 211 indica accessions. (DOCX 25 kb)
ANOVA results for the measured traits under zinc toxicity for 211 indica accessions. (DOCX 26 kb)
Correlations between all measured traits under control (upper triangular) and stress (lower triangular) conditions in Fe (a) and Zn (b) experiments. (DOCX 275 kb)
Significant SNPs associated with measured traits in Fe experiment. (XLSX 34 kb)
Significant SNPs associated with measured traits in Zn experiment. (XLSX 40 kb)
Data Availability Statement
The data sets supporting the results of this article are included within the article and its additional files.


