Abstract
Background
Termination of translation in eukaryotes is controlled by two interacting polypeptide chain release factors, eRFl and eRF3. eRFl recognizes nonsense codons UAA, UAG and UGA, while eRF3 stimulates polypeptide release from the ribosome in a GTP- and eRFl – dependent manner. Recent studies has shown that proteins interacting with these release factors can modulate the efficiency of nonsense codon readthrough.
Results
We have isolated a nonessential yeast gene, which causes suppression of nonsense mutations, being in a multicopy state. This gene encodes a protein designated Itt1p, possessing a zinc finger domain characteristic of the TRIAD proteins of higher eukaryotes. Overexpression of Itt1p decreases the efficiency of translation termination, resulting in the readthrough of all three types of nonsense codons. Itt1p interacts in vitro with both eRFl and eRF3. Overexpression of eRFl, but not of eRF3, abolishes the nonsense suppressor effect of overexpressed Itt1p.
Conclusions
The data obtained demonstrate that Itt1p can modulate the efficiency of translation termination in yeast. This protein possesses a zinc finger domain characteristic of the TRIAD proteins of higher eukaryotes, and this is a first observation of such protein being involved in translation.
Background
The final step of protein biosynthesis represents the termination codon-dependent release of a nascent completed peptide chain from the ribosome. In eukaryotes, this process is controlled by two protein factors: eRFl, recognizing all three types of nonsense codons, and eRF3, which stimulates polypeptide release in a GTP- and eRFl- dependent manner [1-3]. In the yeast Saccharomyces cerevisiae, the eRFl and eRF3 release factors are encoded by the SUP45 and SUP35 genes, respectively, and are often designated as the Sup45 and Sup35 proteins [4]. Partial inactivation of these release factors by mutations results in enhanced nonsense codon readthrough, which can be revealed in yeast by suppression of nonsense mutations, while deletions of the corresponding genes are lethal. It was shown for vertebrates and yeast that eRF3 and eRFl interact with each other to form a heterodimeric complex both in vivo and in vitro[2,4-6]. Yeast eRF3 has a complex structure and is composed of the amino-terminal region and carboxy-terminal (C) domain of 253 and 432 amino acids, respectively [7-9]. The conserved C domain of Sup35p is responsible for its function in translation termination and is essential for cell viability, while the N-terminal region is neither conserved, nor essential. This region may be further subdivided into the middle (M) domain of unknown function and N-terminal (N) domain of 123 amino acids, which is responsible for the prion properties of eRF3.
In human cells, eRFl, being in excess, enhances the efficiency of translation termination, which is consistent with its function in translation [10]. However, in yeast only simultaneous overexpression of both release factors is required for the antisuppressor effect [4], suggesting that a complex of these factors is active in vivo in translation termination.
Premature termination of translation is not the only consequence of the occurrence of nonsense mutations: they can also enhance decay rate of the corresponding mRNA (Nonsense Mediated Decay; NMD). This phenomenon has been observed in both prokaryotic and eukaryotic cells. Several factors involved in NMD have been identified in yeast S. cerevisiae. Among them, the UPF1, UPF2, and UPF3 genes are characterized best [for review, see [11]]. Mutations in these genes selectively stabilize mRNAs containing early nonsense codons without affecting the decay rate of most wild-type mRNAs. These mutations manifest themselves as nonsense suppressors and initially it was concluded that the suppression was solely due to the increase of abundance of nonsense-containing mRNAs and corresponding readthrough proteins. However, later it was demonstrated that it is not so and nonsense suppression was due to the impaired ability of Upf1 protein to enhance translation termination at nonsense codons. The upf1 mutations were identified that suppress nonsense mutations, but do not stabilize nonsense codon-containing mRNAs [12,13]. The involvement of the Upf1 protein in translation termination was confirmed by the finding that it physically interacts with both eRF1 and eRF3 [14]. Recent data indicate that Upf2p and Upf3p are also involved in the control of translation termination serving as activators of Upf1p function [15]. Upf1p contains a cysteine- and histidine-rich region near its amino terminus and all the motifs characteristic of the superfamily group I helicases [12,13]. Another member of this protein superfamily, Mtt1p, whose overexpression enhances the level of nonsense codon readthrough, was recently identified [16]. This protein also interacts with the translation termination factors, but in contrast to Upf1p is not involved in the control of NMD and exerts an opposite effect on translation termination.
Here we present a study directed for identification of additional proteins involved in translation termination in yeast. A screen for multicopy nonsense suppressors revealed a novel protein that inhibits translation termination by binding to polypeptide chain release factors. Interestingly, this protein belongs to a recently described class of TRIAD zinc finger proteins, many of which are presumed to be involved in transcription.
Results
Isolation and characterization of the ITT1 gene
S. cerevisiae strain 2G-DM8, carrying the non-suppressible ura3-52 mutation and nonsense mutations his7-1 (UAA), trp1-289 (UAG) and lys2-187 (UGA) was transformed with the yeast genomic library. The transformants were then replica plated on the media selective for the plasmid marker URA3 and lacking histidine, tryptophane or lysine. Fifty transformants able to grow on either one of these media were selected. Only one of them grew on all media. The plasmid-less Ura- colonies appearing after streaking this transformant on YPD medium, became His-, Trp- and Lys-, which confirmed that suppressor phenotype of the transformant depended on the presence of plasmid.
Plasmid DNA recovered from this transformant carried a genomic DNA insert of approximately 5 kb. The sequence responsible for multicopy suppression was delimited to a XbaI-HindIII region of 2.7 kb. Sequencing of this fragment showed that it contains the open reading frame (ORF) YML068w (GenBank # CAA86252.1), which encodes a polypeptide of 464 amino acids with estimated molecular mass of 54 kDa. No function was ascribed to this ORF, which we designated as ITT1 (Inhibitor of Translation Termination). The codon adaptation index (CAI) of the ITT1 gene (Yeast Protein Database) is 0.127, which suggests that this gene is expressed at low levels. The deduced Itt1 protein possesses a zinc finger domain. This domain starts from residue 180 and belongs to a recently described TRIAD family [17]. It includes three double zinc fingers: a RING C3HC4 element followed by two elements of C6HC and C7HC structure. A search for Itt1p homologues revealed 8 proteins in Caenorhabditis elegans and 8 proteins in man with similarity ranging from 20 to 35% (Figure 1). S. cerevisiae has one more TRIAD protein, YKR017c, but its similarity to Itt1p is lower than that of some TRIAD proteins from man and C elegans. Although the TRIAD proteins are not well characterized, conservation of the TRIAD zinc fingers suggests that they may have important functions (see Discussion).
Figure 1.
A comparison of ammo acid sequences of the Itt1 protein of S. cerevisiae and its best homologues from Schizosaccharomyces pombe (GenBank CAB65614.1), man (androgen receptor activator ARA54, NP_004281.1) and C. elegans (AAB52683.1). Sequences were aligned by introducing gaps (...). Conservative amino acids are highlighted in black (cysteines and histidines) and gray and shown as "consensus". The putative nuclear import signal is underlined. The similarities between the second and third double zinc finger elements are shown by lines. In the third element, the residues presumably involved in zinc binding are numbered. In the earlier assignment [17], these are 1,2, &,$ , 3, 4, 5, 6, with $ corresponding to histidine of some other TRIAD sequences.
To characterize the Itt1p function, the diploid strain H8 was disrupted for ITT1. The obtained strain was sporulated, and tetrads were dissected. The 2+:2- segregation for the LEU2 disruption marker observed for 39 tetrads analyzed demonstrated that the ITT1 gene is not essential for viability. Curiously, despite this gene being inessential, our attempts to disrupt it in a haploid strain were unsuccessful.
To quantify the suppressor effect of overexpressed Itt1p, the 1A-H8-B2 psi- strain, carrying the itt1::LEU2 disruption, was transformed with either multicopy YEplac112-ITT1 or centromeric YCplac22-ITT1 plasmids. Northern blot analysis revealed that amount of ITT1 mRNA in the transformant with multicopy plasmid increased approximately 10-fold compared to the transformant with centromeric one (data not shown). Overexpression of Itt1p increased the readthrough levels of all types of stop codons, which confirms omnipotence of the ITT1 multicopy suppression (Figure 2). The levels of nonsense suppression did not noticeably depend on the presence of wild-type ITT1 gene. Similar data were obtained for the strain with enhanced level of nonsense suppression due to deletion of UPF1 (data not shown).
Figure 2.
The levels of nonsense suppression related to Itt1p overexpression. Suppression efficiency was determined in the strain 1A-H8-B2 (itt1::LEU2) and its derivative, 1A-H8-B2-R (itt1::LEU2 SUP35-C). The strains carried either one of the plasmids YCplac22-ITT1 (centromeric ITT1), YEplac112-ITT1 (multicopy ITT1) or pITT1-SUP45 (multicopy plasmid with both ITT1 and SUP45), in combinations with pUKC815, pUKC817, pUKC818 or pUKC819. The levels of readthrough of UAA (a), UAG (b) or UGA (c) nonsense codons were determined as described in Materials and Methods. Values are the mean of three independent assays ± SD. The expression status of ITT1 and other relevant genes is shown below the graph: Single, ITT1 expressed from centromeric plasmid; Multi, expression of ITT1 or SUP45 from multicopy plasmids; C, chromosomal SUP35-C deletion allele; WT, chromosomal SUP35 or SUP45 wild-type alleles.
Itt1p interacts with eRF1 and eRF3
The inhibition of translation termination by excess Itt1p could be due to decreased levels of release factors. However, neither extra copies, nor deletion of the ITT1 gene affected the abundance of eRF1 and eRF3 (data not shown). Itt1p could also inhibit termination by binding to the eRF1 and eRF3 release factors. To examine the interaction of Itt1p with eRF1 and eRF3, we studied the ability of immobilized Itt1p to bind purified eRF1 and eRF3. These proteins were individually expressed in E. coli as fusions with either 6-histidine or GST tags. Also, we used eRF3 expressed in yeast as GST-eRF3 fusion, since it was observed earlier that eRF3 N-terminal part may be folded incorrectly in bacteria [[18] and our unpublished data]. The purified GST-Itt1p associated with the glutathione agarose beads was incubated with the purified bacterially-expressed His6-eRF3, His6-eRF3N2 (eRF3 amino acids 1 – 153), eRF3C (amino acids 254 – 685) and eRF1, as well as with eRF3 isolated from yeast. Then unbound proteins were removed by washing, and GST-Itt1p with associated proteins were eluted and analyzed by Western blotting using polyclonal antibodies against eRF3 and eRF1. Itt1p specifically bound eRF1, eRF3 and eRF3N2, but did not bind the eRF3C fragment and GST protein (Figure 3). The interaction of Itt1p with eRF1 and eRF3 was also studied using yeast lysates as a source of these proteins. Immobilized GST-Itt1p precipitated eRF3 and eRF1 from lysates of cells with the multicopy SUP35 and SUP45 plasmids, but not from wild-type lysates (data not shown). This probably indicates that interaction between the studied proteins is relatively weak.
Figure 3.
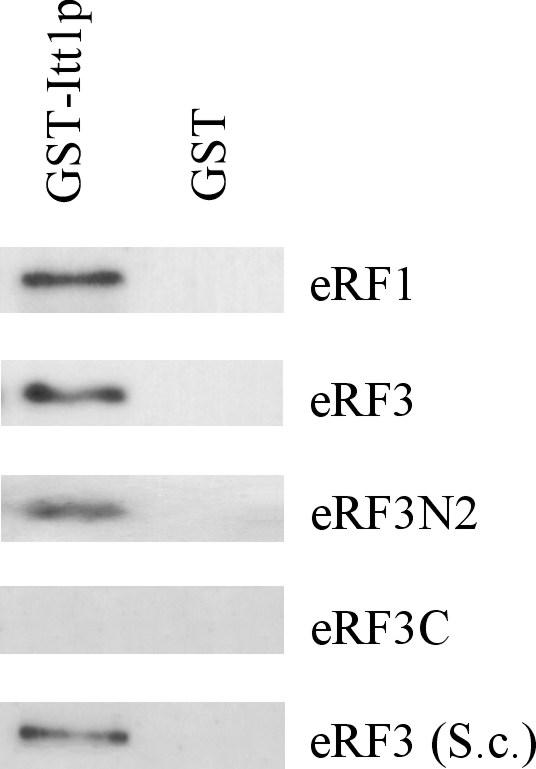
Itt1p interacts with both eRF3 and eRF1. All proteins were isolated from E. coli, except eRF3 (S.c.), isolated from S. cerevisiae. The indicated proteins were incubated with GST-Itt1p or GST proteins immobilized on glutathione-Sepharose 4B. Following washing, bound proteins were eluted and analyzed by Western blotting with polyclonal antibodies against eRF3 and eRF1.
The suppressor effect of excess Itt1p depends on both eRF1 and eRF3 release factors
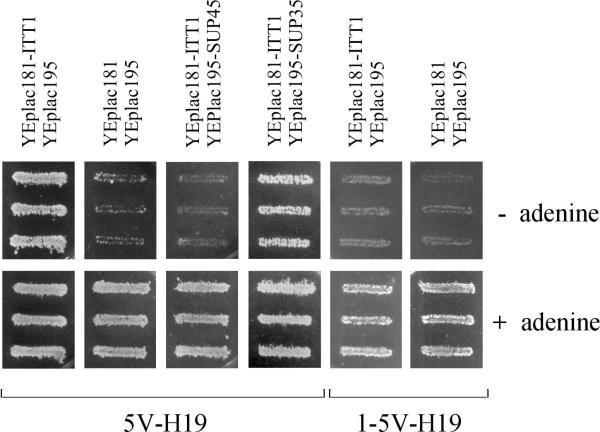
To test the role of eRF1 in the ITT1 suppressor effect, the multicopy plasmids with ITT1 and SUP45 were simultaneously introduced into the strain 5V-H19. These transformants did not express the suppressor phenotype (Figure 4). In contrast to eRF1, the overexpression of eRF3 did not abolish the suppressor effect of the Itt1p overexpression. It is noteworthy that overexpression of eRF1 alone does not cause antisuppressor effect [4] and overproduction of eRF3 does not suppress the ade2-1 UAA mutation in the 5V-H19 strain (data not shown).
Figure 4.
Overexpression of eRF1 and N-terminal truncation of eRF3 inhibits nonsense suppressor phenotype of transformants with multicopy ITT1 plasmid. The strain 5V-H19 harboring pairs of plasmids YEplac181-ITTl/YEplacl95, YEplac181/YEplac195, YEplacl81- ITTl/YEplacl95-SUP45 or YEplacl81-ITTl/YEplacl95-SUP35 and the strain 1-5V-H19 (SUP35-C) with YEplac181- ITT1/YEplac19 5 or YEplac181/YEplac195 were grown on medium selective for plasmids, patched on SC-Ade and SC+Ade plates and incubated for three days. The growth of three independently obtained transformants of each strain is shown.
These data suggest that eRF1 is a primary target for the Itt1p inhibition, while the Itt1p interaction with eRF3 may not play an important role in the suppressor effect. To check the latter, we examined the effect of excess Itt1p in the strain 1-5V-H19, which encodes only the eRF3 C-terminal domain unable to interact with Itt1p. This strain and 5V-H19 were transformed with the multicopy ITT1 plasmid and the suppressor effect was scored by studying growth of transformants on adenine omission medium. The suppression of ade2-1 UAA mutation was observed in both strains, although the efficiency of suppression was much lower in transformants of 1-5V-H19 (Figure 4). Thus, the interaction of Itt1p with eRF3 could play a role in the Itt1p suppressor effect.
The influence of eRF1 and eRF3 on the suppressor effect of elevated Itt1p levels revealed in the strains 5V-H19 and 1-5V-H19 in a plate assay was further quantified in the strains 1A-H8-B2 and 1A-H8-B2-R containing the wild-type SUP35 gene and its 5'-deletion SUP35-C allele, respectively (Figure 2). The absence of the N domain of eRF3 reduced the readthrough of nonsense codons caused by multicopy ITT1 approximately 2–3-fold. Examination of transformants with the pITTI-SUP45 plasmid, which carried both ITT1 and SUP45, revealed that the overexpression of eRF1 completely abolished suppression caused by overexpression of the Itt1 protein.
Discussion
This paper describes a novel protein, Itt1p, involved in the control of translation termination in yeast. Two lines of evidence support this conclusion: (i) overexpression of Itt1p enhances the readthrough of UAA, UAG and UGA nonsense codons; (ii) Itt1p interacts with both eRF1 and eRF3 polypeptide chain release factors. It is noteworthy that in contrast to deletions of SUP45 and SUP35 the knockout of ITT1 is not lethal and does not noticeably influence the nonsense codon readthrough. This suggests that Itt1p is not essential for the release of completed polypeptide chains from the ribosome.
The suppressor effect of Itt1p was observed at increased Itt1p to eRF1 ratio, but did not occur when this ratio was normal, including the case when both proteins were overexpressed. This suggests that eRF1 experiences quantitative, rather than qualitative alteration. The simplest explanation of these and other data is that the binding of Itt1p to eRF1 makes it inactive in translation termination. It is important that Itt1p is likely to be expressed at lower level than eRF1. According to published estimates [19], the difference in CAI of eRF1 (0.334) and Itt1p (0.127) could mean that eRF1 is expressed at about 3-fold higher level than Itt1p. The role of eRF3 in the suppressor effect of Itt1p is less clear. The suppressor effect of excess Itt1p was not affected by overexpression of eRF3, but was reduced in the presence of N-terminally truncated eRF3, which does not interact with Itt1p. To explain this, it is possible to suggest that the interaction of Itt1p with eRF3 is weaker than with eRF1, but it strengthens the binding of Itt1p to eRF1/eRF3 complex, thus enhancing the suppressor effect of Itt1p. However, the effect of the N-terminally truncated eRF3 can also be explained by observation that it promotes translation termination better than complete protein [8]. It may be difficult to distinguish these two mechanisms and we consider it likely that both take place.
It is not clear whether the inhibitory effect of Itt1p represents its main function, or whether it is a consequence of recruiting eRFs for some function different from the translation termination. The second opportunity looks more appealing, and it is supported by some structural features of Itt1p. Itt1p belongs to a unique family of zinc finger proteins called TRIAD. It contains three double zinc finger elements, one of which belongs to a RING class [17]. The similarity of Itt1p with its homologues from other eukaryotes is not high (20–30%), but it spans the whole length of Itt1p with the cysteines and histidines of zinc finger elements being highly conserved. This suggests a functional similarity of Itt1p to at least some TRIAD proteins.
The alignment of Itt1p with its closest homologues (Figure 1) and other TRIAD proteins (not shown) reveals notable similarity of the second and third double zinc finger elements, manifested in similar spacing of cysteines and some conserved residues. This is in contrast to the earlier assignment of the third finger as belonging to a RING class C3HC4[17]. One cause of this difference is that the histidine of the proposed RING signature is poorly conserved (marked $ in Figure 1). On the other hand, highly conserved histidine and cysteine residues (marked 7 and 8) were disregarded previously [17]. Intriguingly, in our version the third element contains an odd number of conserved residues, nine. Either one of the residues is unimportant for binding zinc (marked &), or there could be alternative configurations for zinc binding by this element.
Many of RING finger-containing proteins bind ubiquitin-conjugating enzymes and are the substrates for E2-dependent ubiquitination [20]. It was proposed that this mechanism can be used to target the RING-containing protein or associated proteins for degradation in a regulated manner. If so, the excess Itt1p could cause suppression by accelerating degradation of eRF3 and eRFl. However, this is not the case, since the overexpression of Itt1p did not affect the levels of eRF3 and eRF1. It is also known that RING finger proteins can be transcriptional factors [21] and nuclear localization was predicted for many of the TRIAD proteins [17]. However, only few of these proteins were functionally characterized. Two of them are human androgen receptor activator ARA54 (Figure 1) and rat protein kinase C-associated protein [22,23]. For both proteins it may be suggested that they function in cytoplasm and nucleus and play a role in transcription. Itt1p may also be involved in transcription regulation since two-hybrid analysis has shown that it interacts with the Snf11p transcription factor [24]. This, together with the fact that Itt1p contains a putative nuclear import signal (Figure 1), allows to speculate that Itt1p may perform coupling of translation termination with transcription of certain genes.
Itt1p is not the only protein that could link translation termination with other cellular processes in yeast. At present, two such proteins are known. The first is Upf1p, which is involved in the control of NMD pathway and stimulates translation termination probably by binding to eRF3 and eRF1 release factors [13,14]. Strikingly, this protein and its partners, Upt2p and Upf3p, are also required to control the total accumulation of large number of mRNAs in addition to their role in RNA surveillance, though mechanisms of such control are unknown [25]. The second is Mtt1p, a homologue of Upf1p, which interacts with both release factors, but is not involved in the NMD control and inhibits translation termination [16]. This protein has 5'→ 3' DNA-dependent helicase activity and is thought to be involved in chromosome replication [26-28]. However, the question is still open, whether such proteins can mediate the interdependence of these cellular processes.
Conclusions
The data presented in this work show that the increased expression of the ITT1 (YML068w) gene reduces efficiency of translation termination. The Itt1 protein can bind to the translation termination factors eRF1 and eRF3 and we propose that the resulting complex(es) are incompetent for termination. eRF1 appears to be a primary target for the inhibition by Itt1p, since the suppressor effect of the excess Itt1p is reverted by overexpression of eRF1, but not of eRF3. Itt1p possesses a zinc finger domain characteristic of the TRIAD proteins of higher eukaryotes, and this is a first observation of such protein being involved in translation. However, the role of this protein is probably not restricted by translation. Itt1p interacts with the transcriptional factor Snf11 and therefore could function in transcription, similarly to some of its homologues from other species.
Materials and Methods
Genetic methods
We used standard organic (YEPD) and synthetic complete (SC) media for yeast [29] and LB medium for bacteria [30]. Appropriate amounts of amino acids, bases, and antibiotics were added when necessary. The 5'-fluoroorotic acid (5FOA) medium was prepared according to [31]. The final concentration of 5FOA was 400 μg/ml. All solid media contained 2.0% agar. Yeast cells were grown at 30°C, and bacteria at 37°C. Standard yeast genetic procedures for mating, sporulation, and tetrad analysis were used [29]. DNA transformation of yeast and Escherichia coli cells was performed as described previously [32,33].
Strains and plasmids
The yeast strains 2G-DM8 (MATα ade2-144,717 pheA10 his7-1 lys2-l87 trp1-1 ura3-52) and 5V-H19 (MATa ade2-1 SUQ5 can1-100 leu2-3,112 ura3-52) were described in [8]. The strain 1-5V-H19 was obtained by replacing the wild-type SUP35 gene of 5V-H19 with the SUP35-C deletion allele, which encodes amino acids 254-685 of eRF3 [9]. The diploid H8 strain (MA Tα/MA Ta trp1-289/ trp1-289 leu2-3,112/ leu2-3,112 ura3-52/ ura3-52 his3-Δ1/ his3-Δ1) was described in [34]. Construction of other strains is described below. All strains were [psi].-
DNA manipulations were carried out by standard protocols [30]. The DH5 α E. coli strain [supE44 Δlac U169 (φ 80 lacZΔ M15) hsdR17recA1endA1gyrA96 thi-1 relA1] was used for plasmid construction [30]. To create yeast genomic library, chromosomal DNA of the 5V-H19 strain was partially digested with Sau3A and fractionated on agarose gel. DNA fragments ranging from 6 to 12 kb were isolated. The ends of chromosomal DNA fragments were partially filled in with Klenow enzyme and ligated to partially filled in SalI site of the YEplac195 plasmid [35]. The ligation products were used for E. coli transformation.
The plasmids containing ITT1, SUP45 and SUP35 were constructed as follows. The 2.7 kb XbaI-HindIII genomic fragment carrying the ITT1 gene was cloned into the same sites of pBluescript II KS(+) (Stratagene, USA), and the resulting plasmid was designated as pITT1. The ITT1 SacI-HindIII fragment of this plasmid was inserted into the multicopy vectors YEplac195, YEplacl8l, YEplac112, with URA3, LEU2 and TRP1 selectable markers, respectively, and into the TRP1-carrying centromeric vector YCplac22 [35]. The 2.5 kb XhoI-SalISUP45-csarymg fragment of pEMBLyex4-SUP45 [4] was cloned into the SalI site of YEplacl95 to yield YEplac195-SUP45. The 2.7 kb SacI-SalIITT1 fragment of the plasmid pITTl was inserted into the same sites of YEplac195-SUP45 to obtain the pITTl-SUP45 plasmid. The 3.6 kb XhoI-XbaI SUP35-carrying fragment of pEMBLyex4-SUP35 [8] was inserted into the SalIand XbaI sites of YEplac195 which resulted in the plasmid YEplac195-SUP35.
The construction of the hybrid genes encoding N-terminal fusions of GST (glutathione S-transferase) to eRF1, eRF3 and eRF3C was described earlier [18,36]. To construct the pGST-ITT1 plasmid encoding GST-Itt1p fusion protein, the 1.7 kb BamHI-HindIII fragment carrying the entire ITT1 gene (BamHI site was engineered at the 5' end of the ITT1 ORF) was cloned into the same sites of pGEX-2TH, which was obtained from the pGEX-2T plasmid (Pharmacia, Sweden) by EcoRI to HindIII replacement. To obtain plasmids expressing His6-eRF3N2 and His6-eRF3, the SUP35 coding sequence from engineered BglII site at position-12 before the start codon to BalI site at codon 154 or XbaI site in the 3' non-coding region, which was filled-in by Klenow, was cloned into BamHI and HincII sites of pQE-10 (Qiagen) in-frame and downstream of the 6-histidine tag sequence. Both proteins have N-terminal extension MRGSHHHHHHTDLAT. All fusion proteins were expressed in the E. coli strain TG1 {supE HsdΔ5 thiΔ (lac-proAB) F' [traD36 proAB+lacIqlacZ M15]}.
The ITT1 gene was disrupted as follows. The 1.0 kb EcoRI-XbaI fragment internal to the ITT1 ORF was replaced with the 2.0 kb LEU2-carrymg SacI-XbaI fragment of the plasmid pJJ283 [37]. The EcoRI and SacI ends of these fragments were filled in with Klenow enzyme prior to ligation. The obtained plasmid was cleaved with BclI and HindIII and the obtained 3.6 kb fragment carrying the itt1::LEU2 disruption allele was used to transform the strain H8. The Ura+ transformant was designated as H8-B2 and confirmed to be heterozygous for the ITT1 disruption by Southern blot analysis (data not shown). The strain 1A-H8-B2 carrying the itt1::LEU2 allele was obtained as a meiotic segregant of the diploid H8 heterozygous for the ITT1 disruption allele.
The strain 1A-H8-B2-R was obtained from 1A-H8-B2 by replacing the wild-type SUP35 gene with the SUP35-C deletion allele using the integration/excision method [38]. The pFL44ScΔ-SUP35C integrative plasmid (lacking the ClaI fragment with 2 μm replication origin) bearing a SUP35-C allele [8] was linearized by SalI site internal to the SUP35 ORF and integrated into the SUP35 gene of the strain 1A-H8-B2 by selecting of transformants on uracil omission medium. The Ura+ integrants expressing both eRF3 and eRF3C were selected by Western blotting and placed onto the 5-FOA containing medium to select for the alternative URA3-SUP35 or URA3-SUP35-C excision. The obtained Ura- clones of one integrant were screened by Western blotting to select a clone expressing only the eRF3C protein (data not shown).
β-Galactosidase assays of yeast strains transformed with the pUKC815/817/181/819 series vectors
The URA3-carrying plasmid pUKC815 encodes a PGKl-lacZ gene fusion, while the pUKC817, pUKC818 and pUKC819 plasmids are identical to pUKC815 except that one of the three termination codons, UAA, UAG and UGA, respectively, is present in-frame at the junction of the PGK1 and lacZ genes [39]. Suppression of the in-frame premature stop codons will result in β-galactosidase activity and the levels of β-galactosidase activity can therefore be used to quantify the readthrough of nonsense codons. The nonsense suppression levels were determined as ratio of β-galactosidase activities in the cells transformed with plasmids pUKC817, pUKC818 and pUKC819 to that of the transformant with pUKC815. Individual transformants were grown selectively in SC supplemented with the required amino acids and bases to mid-exponential phase at 30°C.
Preparation of yeast cell lysates
Yeast cultures were grown to OD600 of 1.5, harvested, washed in water and lysed by vortexing with glass beads in buffer A (25 mM Tris-HCl [pH 7.4], 100 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol) containing 1 mM phenylmethylsulfonylfluoride to limit proteolytic degradation. Cell debris was removed by centrifugation at 15,000 g for 10 min.
Purification of eRF1, eRF3 and eRF3 fragments and assay for interaction with immobilized GST-Itt1p
The GST-Itt1p, GST-eRF1, GST-eRF3C fusion proteins expressed in E. coli and GST-eRF3 expressed in yeast were isolated by affinity chromatography on glutathione-Sepharose 4B (Pharmacia). The GST extension from GST-eRF1 and GST-eRF3 was removed with thrombin (Sigma), and from GST-eRF3C with factor Xa (Promega), as described [40]. The His6-eRF3 and His6-eRF3N2 proteins expressed in E. coli were isolated using TALON resin (Clontech) according to manufacturer's instructions. The glutathione-Sepharose 4B resin with immobilized GST-Itt1p was incubated with yeast lysates or with the purified eRFl, eRF3, eRF3N2 and eRF3C proteins for 2 h at 4°C and then washed with a 40-fold resin volume of buffer A. Bound proteins were eluted with 2% SDS and analyzed by Western blotting.
Protein gel electrophoresis and Western blot analysis
Protein samples were separated on an SDS polyacrylamide gel as described [41] and electrophoretically transferred to nitrocellulose sheets [42]. Western blots were probed with polyclonal rabbit anti-eRF3, anti-eRF3C or anti-eRF1 (a gift of K. M. Jones, University of Kent) antibody. Bound antibodies were detected with the Amersham ECL system as instructed by manufacturer.
Acknowledgments
Acknowledgements
We are grateful to S. Peltz for the plasmid pKOM and for M. Agaphonov for helpful discussion. The work was supported by grants from Howard Hughes Medical Institutes, INTAS, Russian Foundation for Basic Research (M.D.T-A.) and The Wellcome Trust (V.V.K.).
Contributor Information
Valery N Urakov, Email: molgen@cardio.ru.
Igor A Valouev, Email: molgen@cardio.ru.
Eugeny I Lewitin, Email: elewitin@hotmail.com.
Sergey V Paushkin, Email: paushkin@hhmi.apenn.edu.
Vyacheslav S Kosorukov, Email: molgen@cardio.ru.
Vitaly V Kushnirov, Email: vita@cardio.ru.
Vladimir N Smirnov, Email: yakusheva@cardio.ru.
Michael D Ter-Avanesyan, Email: ter@cardio.ru.
References
- Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman L, Haenni AL, Celis JE, Philippe M, Justesen J, Kisselev LL. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov SG, Kisselev LL, Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M, Kisselev L. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 1996;2:334–341. [PMC free article] [PubMed] [Google Scholar]
- Stansfield I, Jnes KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, Tuite MF. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L, Simonsen JL, Merkulova TI, Litvinov DY, Martensen PM, Rechinsky VO, Camonis JH, Kisselev LL, Justesen J. Functional expression of eukaryotic polypeptide chain release factors 1 and 3 by means of baculovirus/insect cells and complex formation between the factors. Eur J Biochem. 1998;256:36–44. doi: 10.1046/j.1432-1327.1998.2560036.x. [DOI] [PubMed] [Google Scholar]
- Ito K, Ebihara K, Nakamura Y. The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. RNA. 1998;4:958–972. doi: 10.1017/S1355838298971874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov VV, Ter-Avanesyan MD, Telckov MV, Surguchov AP, Smimov VN, Inge SG-Vechtomov. Nucleotide sequence of the SUP2(SUP35) gene of Saccharomyces cerevisiae. Gene. 1988;66:45–54. doi: 10.1016/0378-1119(88)90223-5. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Kushnirov VV, Dagkesamanskaya AR, Didichenko SA, Chernoff YO, Inge-Vechtomov SG, Smirnov VN. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol. 1993;7:683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN. The SUP35 omnipotent suppressor is involved in the maintenance of the non-Mendelian determinant [PSI+] in the yeast Saccharomyces cerevisiae. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff X, Philippe M, Jean-Jean O. Overexpression of human release factor 1 alone has an antisuppressor effect in human cells. Mol Cell Biol. 1997;17:3164–3172. doi: 10.1128/mcb.17.6.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A, Peltz SW. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW. Genetic and biochemical characterization of the mutation in the ATPase and helicase regions of Upf1 protein. Mol Cell Biol. 1996;16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf1 protein complex, but not mRNA turnover. Mol Cell Biol. 1996;16:5491–5506. doi: 10.1128/mcb.16.10.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maderazo AB, He F, Mangus DA, Jacobson A. Upf1p control of nonsense mRNA translation is regulated by NMD2p and Upf3p. Mol Cell Biol. 2000;20:4591–5603. doi: 10.1128/MCB.20.13.4591-4603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Majlesi N, Banerjee T, Peltz SW. Mtt1 is a Upf1-like helicase that interacts with the translation termination factors and whose overexpression can modulate termination efficiency. RNA. 2000;6:730–743. doi: 10.1017/S1355838200992392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Reijden BA, Erpelinck-Verschueren CA, Lowenberg B, Jansen JH. TRIADs: a new class of proteins with a novel cysteine-rich signature. Protein Sci. 1999;8:1557–1561. doi: 10.1110/ps.8.7.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Interaction between yeast Sup45p (eRF1) and Sup35p (eRF3) polypeptide chain release factors: implication for prion-dependent regulation. Mol Cell Biol. 1997;17:2798–2805. doi: 10.1128/mcb.17.5.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher B, Latter GI, Monardo P, McLaughlin CS, Garrels JI. A sampling of the yeast proteome. Mol Cell Biol. 1999;19:7357–7368. doi: 10.1128/mcb.19.11.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin AJ, Borden KL, Boddy MN, Freemont PS. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208–214. doi: 10.1016/0968-0004(96)10036-0. [DOI] [PubMed] [Google Scholar]
- Kang H-Y, Yeh S, Fujimoto N, Chang C. Cloning and characterization of human prostate coactivator ARA54, a novel protein that associates with the androgen receptor. J Biol Chem. 1999;274:8570–8576. doi: 10.1074/jbc.274.13.8570. [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Tokunaga C, Nakagawa N, Tanizawa K, Kuroda S, U Kikkawa. Transcriptional activity of RBCK1 protein (RBCC protein interacting with PKC 1): requirement of RING-finger and B-Box motifs and regulation by protein kinases. Biochem Biophys Res Commun. 1998;247:392–396. doi: 10.1006/bbrc.1998.8795. [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Lelivelt MJ, Culbertson MR. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol Cell Biol. 1999;19:6710–6719. doi: 10.1128/mcb.19.10.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas EE, Chen PH, Leszyk J, Biswas SB. Biochemical and genetic characterization of a replication protein A dependent DNA helicase from the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;206:850–856. doi: 10.1006/bbrc.1995.1121. [DOI] [PubMed] [Google Scholar]
- Biswas EE, Fricke WM, Chen PH, Biswas SB. Yeast DNA helicase A: cloning expression, purification, and enzymatic characterization. Biochemistry. 1997;36:13277–13284. doi: 10.1021/bi971292s. [DOI] [PubMed] [Google Scholar]
- Biswas EE, Chen PH, Biswas SB. Purification and characterization of DNA polymerase-associated replication protein A-dependent yeast DNA helicase A. Biochemistry. 1997;36:13270–13276. doi: 10.1021/bi9712910. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor Press; 1986.
- Sambrook J, Fritsch EE, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edCold Spring Harbor Laboratory, Cold Spring Harbor Press; 1989.
- McCusker JH, Davis RW. The use of proline as a nitrogen source causes hypersensitivity to, and allows more economical use of 5FOA in Saccharomyces cerevisiae. Yeast. 1991;7:607–608. doi: 10.1002/yea.320070608. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Techniques for transformation of Escherichia coli. DNA Cloning: A Practical Approach (Edited by Glover DM) Oxford, England, IRL Press; 1985. pp. 109–135.
- Dagkesamanskaya AR, Ter-Avanesyan MD. Interaction of the yeast omnipotent suppressor SUP1(SUP45) and SUP2(SUP35) with non-Mendelian factors. Genetics. 1991;128:513–520. doi: 10.1093/genetics/128.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter MD-Avanesyan. Propagation of the yeast prion-like [PSI+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3133. [PMC free article] [PubMed] [Google Scholar]
- Jones JS, Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990;6:363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor Press; 1990.
- Stansfield , Akhmaloka , Tuite MF. A mutant allele of the SUP45 (SAL4) gene of Saccharomyces cerevisiae shows temperature-dependent allosuppressor and omnipotent suppressor phenotypes. Curr Genet. 1995;27:417–426. doi: 10.1007/BF00311210. [DOI] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriphage T4. Nature. 1970;227:680–689. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]