Abstract
Background
Biological interpretation of DNA microarray data may differ depending on underlying assumptions and statistical tests of bioinformatics tools used. We used Gene Set Enrichment Analysis (GSEA) and Ingenuity Pathway Analysis (IPA) to analyze previously generated DNA microarray data from human monocytes stimulated with N. meningitidis and IL-10 (“the model system”), and with meningococcal sepsis plasma before and after immunodepletion of IL-10 (“the patient plasma system”). The objectives were to compare if the two bioinformatics methods resulted in similar biological interpretation of the datasets, and to identify whether GSEA provided additional insight compared with IPA about the monocyte host response to meningococcal activation.
Results
In both experimental models, GSEA and IPA identified genes associated with pro-inflammatory innate immune activation, including TNF-signaling, Toll-like receptor signaling, JAK-STAT-signaling, and type I and type II interferon signaling. GSEA identified genes regulated by the presence of IL-10 with similar gene sets in both the model system and the patient plasma system. In the model system, GSEA and IPA in sum identified 170 genes associated with oxidative phosphorylation/mitochondrial function to be down-regulated in monocytes stimulated with meningococci. In the patient plasma system, GSEA and IPA in sum identified 122 genes associated with oxidative phosphorylation/mitochondrial dysfunction to be down-regulated by meningococcal sepsis plasma depleted for IL-10. Using IPA, we identified IL-10 to up-regulate 18 genes associated with oxidative phosphorylation/mitochondrial function that were down-regulated by N. meningitidis.
Conclusions
Biological processes associated with the gene expression changes in the model system of meningococcal sepsis were comparable with the results found in the patient plasma system. By combining GSEA with IPA, we discovered an inhibitory effect of N. meningitidis on genes associated with mitochondrial function and oxidative phosphorylation, and that IL-10 partially reverses this strong inhibitory effect, thereby identifying, to our knowledge, yet another group of genes where IL-10 regulates the effect of LPS. We suggest that relying on a single bioinformatics tool together with an arbitrarily chosen filtering criteria for data analysis may result in overlooking relevant biological processes and signaling pathways associated with genes differentially expressed between compared experimental conditions.
Electronic supplementary material
The online version of this article (10.1186/s12865-017-0229-5) contains supplementary material, which is available to authorized users.
Keywords: Gene expression, mRNA, Bioinformatics, Meningococcal sepsis, N. Meningitidis, Interleukin-10, Ingenuity pathway analysis, Gene set enrichment analysis
Background
Sepsis, defined as life-threatening organ dysfunction caused by a dysregulated host response to infection [1], remains a serious cause of mortality in intensive care units worldwide [2, 3]. The past decade has seen a large number of studies aiming to investigate the pathophysiology of sepsis through systems biology approaches, with genome-wide expression profiling being a widely used method [4–6]. This approach has in part been motivated by the failure of a large number of clinical trials directed towards the specific inhibition of endotoxin and cytokines [7, 8], and the need to increase our understanding of the complexity of the sepsis syndrome [9, 10].
Meningococcal sepsis is an overwhelming form of the sepsis syndrome, with previous studies identifying mortality within 12–24 h in previously healthy children and adults [11]. The intense and rapidly evolving pro-inflammatory response resulting from high levels of meningococci and lipopolysaccharides (LPS, endotoxin) in blood [12–16] is suggested to be the main determinant of septic shock with multi-organ failure, coagulopathy and mortality. Our research group has over the past two decades used a primary cell model consisting of elutriated and cryopreserved human monocytes to elucidate cellular mechanisms activated by meningococcal sepsis plasma [14, 17–20]. We have viewed human monocytes as a relevant cellular model for several reasons, including their pro-coagulant activity in meningococcal sepsis [21, 22], and their role as phagocytic cells in various tissues after differentiation to macrophages [23].
Our group has recently investigated the gene expression profile induced by N. meningitidis in human monocytes. The first study demonstrated that N. meningitidis and purified LPS can differentially regulate the expression of over 4600 genes [18]. Follow-up studies have investigated the differential expression of genes induced by plasma from patients with meningococcal sepsis, with a special focus on the biological significance of the anti-inflammatory cytokine IL-10. The highest concentrations of IL-10 are found in non-survivors of meningococcal septic shock [16, 17, 24]. We have studied the effect of IL-10 in two previous studies, one stimulating monocytes with wild-type N. meningitidis [25] and recombinant IL-10, and the other using plasma samples from patients with severe meningococcal sepsis or septic shock [26]. In both studies we showed that IL-10 regulates a group of genes that are induced by N. meningitidis, and that these genes are associated with a broad range of functional categories.
Over a decade ago, the main approach to gene expression analysis was to focus on genes representing the largest difference between two experimental conditions, namely those at the top and bottom of a list. However, as described by Subramanian et al. [27], this approach has had a number of limitations. First, an extremely large list may make selection of relevant genes daunting. Secondly, individual genes may not meet the threshold for statistical significance once correction for multiple testing is performed. Finally, focusing on individual genes may overlook important effects suggested by a set of genes that are coordinately expressed, but where the fold change (FC) level does not exceed the cut-off point. It is in response to these challenges that gene set enrichment analysis (GSEA), as initially described by Mootha et al. [28] and later developed by Subramanian et al. [27], was established.
Our previous gene expression studies have used the commercially available tool, Ingenuity Pathway Analysis (IPA) for data analysis [29]. The IPA software analyzes the gene expression profile against molecular relationships in the Ingenuity Knowledge Base, which is a repository of functional annotations, biological interactions and modelled relationships between proteins, genes, cells, tissues, drugs and diseases reported from primary literature sources, including peer-reviewed journal articles and review articles, and other databases (e.g., KEGG, EntrezGene, the Gene Ontology Project). IPA has become among the most widely used tools for gene expression analysis. Similar to GSEA, IPA is a form of enrichment analysis, which is an analytic strategy aiming to identify whether a group of genes is significantly associated with a particular biological process or signaling pathway [30]. GSEA and IPA differ with respect to the statistical approach taken to identifying the significantly enriched groups of genes [27, 31].
Over time, the increasing number of bioinformatics tools have made it easier for investigators to make biological interpretation of genes differentially expressed between different experimental conditions. However, the increasing number of tools have also made comparisons between gene expression studies, such as those for sepsis [5], more difficult, since biological interpretation may differ depending on the methods’ underlying assumptions and statistical tests. One approach to overcome these challenges is to expose the dataset to two different data analytical methods, and consider whether the methods yield similar biological interpretation. In this study we used IPA and GSEA to analyze previously generated DNA microarray data from studies of human monocyte host response to N. meningitidis, IL-10 and meningococcal sepsis plasma [25, 26]. The objectives were to compare whether these two methods resulted in similar biological interpretation of the datasets; and identify whether GSEA provided additional insight about the human monocyte host response to meningococcal activation.
Methods
Definitions of the compared experimental conditions and models
Gene expression data from two different, previously published models of transcriptomic changes induced in human monocytes were used for this study (Table 1). The data and protocols of both studies are compliant with the minimum information about a microarray experiment (MIAME) guidelines [32]. In the first model [25], gene expression changes were induced in human monocytes stimulated with the N. meningitidis reference strain 44/76 and recombinant IL-10. The control group was unstimulated human monocytes. This model is hereafter denoted the “the model system”. In the second model [26] gene expression changes were induced in human monocytes stimulated with plasma from patients with severe meningococcal sepsis or septic shock containing high (≥130 endotoxin units (EU)/mL) levels of LPS, before and after immunodepletion of native IL-10. Plasma samples with IL-10 are denoted “patient plasma with IL-10”. Plasma samples after IL-10 immunodepletion are denoted “IL-10 immunodepleted patient plasma”. The control group was human monocytes stimulated with plasma from non-shock patients with meningococcal meningitis or mild meningococcemia, containing low (≤1.4 EU/mL) levels of LPS and IL-10. These plasma samples are as a group denoted “low LPS plasma”. The clinical course, microbiological data and cytokine measurements of each patient have previously been reported [26]. This model is hereafter denoted the “ The patient plasma system ”. In both models the monocytes were stimulated for three hours. This study compared results between three experimental conditions in each model system. In the model system the following conditions were compared: monocytes stimulated with N. meningitidis versus unstimulated monocytes (Nm vs ctr ), monocytes stimulated with N. meningitidis in combination with IL-10 versus unstimulated monocytes (Nm + IL-10 vs ctr), and monocytes stimulated with N. meningitidis and IL-10 versus monocytes stimulated with N. meningitidis only (Nm + IL-10 vs Nm ). In the patient plasma system the following experimental conditions were compared: IL-10 immunodepleted plasma vs low LPS plasma, patient plasma with IL-10 vs low LPS plasma, and IL-10 immunodepleted plasma vs patient plasma with IL-10.
Table 1.
Experimental conditions from which DNA microarray data was generated
| In vitro experimental model | Experimental conditions | |||
|---|---|---|---|---|
| Model system | Unstimulated (“Ctr”) | 106/mL N. meningitidis (“Nm”) | 106/mL N. meningitidis + 25 ng/mL IL-10 (“Nm + IL-10”) | 25 ng/mL IL-10 (“IL-10”)a |
| Patient plasma system | Plasma from patients with meningococcal meningitis or mild meningococcemia (“low LPS plasma”) | Plasma from patients with severe meningococcal sepsis or septic shock, depleted for IL-10 (“IL-10 immunodepleted plasma”) | Plasma from patients with severe meningococcal sepsis or septic shock (“patient plasma with IL-10”) | Plasma from patients with meningococcal meningitis or mild meningococcemia, depleted for IL-10 (“low LPS plasma immunodepleted for IL-10”)a |
aData from these experimental conditions were not used in the present study
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) [27, 28] was performed using GSEA version 2.0.14 [33]. Data from previously generated gene expression profiles induced in the model system [25] and in the patient plasma system [26] were imported into the GSEA software. Normalized and background corrected log2-transformed signal intensities after using the Robust Multichip Analysis (RMA) algorithm implemented in Partek Genomic Suit software were used. For this study, we analyzed the expression profiles against the hallmark gene sets available from the Molecular Signatures Database [34]. The hallmark gene sets are a priori defined genes that have been identified by computational methods to be coordinately expressed in various biological states and processes. GSEA was run according to default parameters: probes for the same gene were collapsed into a single gene symbol (identified by its HUGO gene symbol), permutation number was set to 1000, and permutation type was set to “gene sets.” The conventional cut-off value for statistical significance used in GSEA is a false discovery rate (FDR) of 25%. In order to reduce the likelihood of false positive results, this study used FDR 5% as cut-off value for enriched gene sets.
Ingenuity pathway analysis
Gene lists (Excel files) containing gene identifiers (probe set IDs), and corresponding P-values were uploaded to Ingenuity Pathway Analysis (IPA, Ingenuity Systems, www.ingenuity.com). A cut-off at FDR <5% was set to identify significantly differentially expressed genes. Several tools in IPA were used to analyze the gene expression data. The canonical pathway tool was used to identify the top canonical pathways associated with the genes differentially expressed between compared conditions. Biological functions associated with the differentially expressed genes were identified by mapping each gene to its corresponding function in the Ingenuity Knowledge Base. The right-tailed Fisher’s exact test was performed in IPA to calculate a P-value determining the probability that each biological function assigned to the data set was due to chance alone. The P-value was corrected for multiple comparisons using the Benjamini-Hochberg method for correcting the FDR [35]. Permission was granted by QIAGEN Silicon Valley to use copyrighted figures generated from Ingenuity Pathways Analysis in this publication. Figures produced from IPA are available under an open-access CC-BY license for purposes of publication.
Canonical pathway analysis in IPA
There are two main groups of canonical pathways in IPA – metabolic and signaling – and these are hierarchically grouped according to a number of sub-categories (Panel 1). In order to identify those pathways most relevant to the experimental condition and disease of study, disease- and cell-specific pathways not deemed relevant to the experimental conditions under study were excluded from the analysis. Canonical pathways significantly enriched by the differentially expressed genes in the datasets were identified with the right-tailed Fisher’s Exact Test, which calculates a P-value determining the probability that the canonical pathway is associated with the data set due to random chance alone. The P-values were corrected for multiple testing using the Benjamini-Hochberg method for correcting the false discovery rate [31].
Statistical analysis
The GSEA methodology calculates three statistically important values. The first is the enrichment score, which reflects the degree to which an a priori defined list of genes S is overrepresented in a ranked list of genes L. In this study, the a priori defined list of genes S are the hallmark gene sets described above, and the list L are genes ranked according to log2-transformed signal intensities from each experimental condition studied. The enrichment score is calculated by walking down the list L, increasing a running-sum statistic when a gene in S is encountered, and decreasing it when encountering a gene not in S. The magnitude of the increment depends on the correlation of the genes with the phenotype. The enrichment score is the maximum deviation from zero encountered in the random walk, and corresponds to a weighted Kolmogorov–Smirnov-like statistic, as described more extensively in the supplementary material by Subramanian et al. [27]. The second is the nominal P value which is calculated to estimate the significance of the enrichment score. The nominal P value is then normalized to account for the size of the gene set, and then adjusted for multiple testing by calculating the false discovery rate (FDR), which is the third important statistical value. The FDR estimates the probability that a given enrichment score represents a false positive finding. The threshold for statistical significance was set to FDR 5%. The differentially expressed genes for IPA were identified using two-way ANOVA in Partek Genomic Suite, as previously described [25, 26]. In the present study, P-values were corrected using the Benjamini-Hochberg method [35] for controlling the FDR, and a FDR of <5% was set as requirement for inclusion in the analysis. No FC-value was set as cut-off.
Results
GSEA and IPA associated gene expression induced in the model system with similar sets of signaling pathways
We first examined the gene expression changes induced in the model system, beginning with GSEA. The enriched gene sets used in GSEA represent genes that previous genome-wide expression studies have demonstrated to be coordinately expressed in different experimental conditions and biological states [34]. In Nm vs ctr, we identified 15 hallmark gene sets to be significantly enriched (FDR <5%) (Table 2). Twelve of these 15 gene sets were significantly enriched in Nm + IL-10 vs ctr (Table 3), only differing with respect to the gene sets epithelial mesenchymal transition, coagulation, and estrogen response late which no longer were significantly expressed.
Table 2.
Gene sets enriched in human monocytes stimulated with N. meningitidis vs control
| Name of hallmark gene set | Descriptiona | FDR | Number of genes in the gene set | Founder gene setsb |
|---|---|---|---|---|
| Interferon alpha response | Genes up-regulated in response to alpha interferon proteins | <0.001 | 97 | 82 |
| Interferon gamma response | Genes up-regulated in response to IFNG | <0.001 | 200 | 82 |
| TNF signaling via NfkB | Genes regulated by NF-kB in response to TNF | <0.001 | 200 | 120 |
| Inflammatory response | Genes defining inflammatory response | <0.001 | 200 | 120 |
| Allograft rejection | Genes up-regulated during transplant rejection | <0.001 | 200 | 190 |
| IL6 JAK-STAT3 signalling | Genes up-regulated by IL6 [via STAT3, e.g., during acute phase response] | <0.001 | 87 | 24 |
| IL2 STAT5 signaling | Genes up-regulated by STAT5 in response to IL2 stimulation | <0.001 | 200 | 13 |
| KRAS signaling down | Genes down-regulated by KRAS activation | <0.001 | 200 | 16 |
| KRAS signaling up | Genes up-regulated by KRAS activation | <0.001 | 200 | 14 |
| Apoptosis | Genes mediating programmed cell death (apoptosis) by activation of caspases | <0.001 | 161 | 80 |
| Complement | Genes encoding components of the complement system, which is part of the innate immune system | 0.001 | 200 | 71 |
| Epithelial mesenchymal transition | Genes defining epithelial-mesenchymal transition, as in wound healing, fibrosis and metastasis | 0.001 | 200 | 106 |
| Coagulation | Genes encoding components of blood coagulation system; also up-regulated in platelets | 0.015 | 138 | 71 |
| Estrogen response late | Genes defining late response to estrogen | 0.007 | 200 | 59 |
| Hypoxia | Genes up-regulated in response to low oxygen levels (hypoxia) | 0.015 | 200 | 87 |
aDescription of the gene sets were obtained by the Molecular Signatures Database
bFounder gene sets constitute the original experiments associating the genes with specified functions or biological processes
Table 3.
Gene sets enriched in human monocytes stimulated with N. meningitidis and IL-10 vs control
| Name of hallmark gene set | Descriptiona | FDR | Number of genes in the gene set | Founder gene setsb |
|---|---|---|---|---|
| Interferon gamma response | Genes up-regulated in response to IFNG | <0.001 | 200 | 82 |
| Interferon alpha response | Genes up-regulated in response to alpha interferon proteins. | <0.001 | 97 | 82 |
| TNF signaling via NfkB | Genes regulated by NF-kB in response to TNF | <0.001 | 200 | 120 |
| Inflammatory response | Genes defining inflammatory response | <0.001 | 200 | 120 |
| IL2 STAT5 signaling | Genes up-regulated by STAT5 in response to IL2 stimulation. | <0.001 | 200 | 13 |
| IL6 JAK-STAT3 signaling | Genes up-regulated by IL6 [via STAT3, e.g., during acute phase response] | <0.001 | 87 | 24 |
| Allograft rejection | Genes up-regulated during transplant rejection | <0.001 | 200 | 190 |
| Complement | Genes encoding components of the complement system, which is part of the innate immune system | <0.001 | 200 | 71 |
| KRAS signaling up | Genes up-regulated by KRAS activation | 0.003 | 200 | 14 |
| KRAS signaling down | Genes down-regulated by KRAS activation | 0.007 | 200 | 16 |
| Apoptosis | Genes mediating programmed cell death (apoptosis) by activation of caspases | 0.007 | 161 | 80 |
| Hypoxia | Genes up-regulated in response to low oxygen levels (hypoxia) | 0.039 | 200 | 87 |
aDescription of the gene sets were obtained from the Molecular Signatures Database
bFounder gene sets constitute the original experiments associating the genes with specified functions or biological processes
The significantly enriched gene sets in each experimental condition were then compared against significantly enriched canonical pathways in IPA. In Nm vs ctr (Fig. 1), both GSEA and IPA identified differentially expressed genes to be associated with apoptosis (denoted “Death Receptor Signaling” by IPA), hypoxia (in IPA denoted “Hypoxia in the Cardiovascular System”), interferon signaling and IL-6 signaling. In Nm + IL-10 vs ctr (Fig. 2), both GSEA and IPA identified apoptosis, interferon signaling, and JAK-STAT signaling to be affected. A greater number of canonical pathways were significantly enriched in monocytes stimulated with N. meningitidis together with IL-10 than with N. meningitidis alone, indicating the biological effect induced by the presence of IL-10.
Fig. 1.
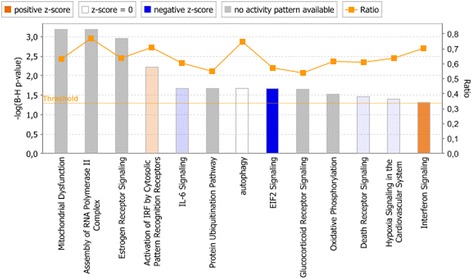
Canonical pathways significantly enriched in human monocytes when comparing Nm vs ctr. Significantly enriched canonical pathways were identified with a right-tailed Fisher’s Exact Test that calculates a P-value determining the probability that each canonical pathway associated with the dataset was due to chance alone. The P-values were corrected for multiple testing using the Benjamini-Hochberg method for correcting the FDR. The z-score indicates predicted activation state of the canonical pathway. Blue color or lighter shades of blue indicate a negative z-score and down-regulation of the pathway, and orange color or lighter shades of orange indicate a positive z-score and up-regulation of the pathway. Ratio denotes the number of significantly expressed genes compared with the total number of genes associated with the canonical pathway
Fig. 2.
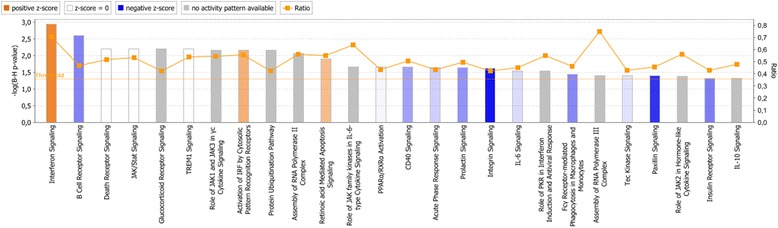
Canonical pathways significantly enriched in human monocytes when comparing Nm + IL-10 vs ctr. Significantly enriched canonical pathways were identified with a right-tailed Fisher’s Exact Test that calculates a P-value determining the probability that each canonical pathway associated with the dataset was due to chance alone. The P-values were corrected for multiple testing using the Benjamini-Hochberg method for correcting the FDR. The z-score indicates predicted activation state of the canonical pathway. Blue color or lighter shades of blue indicate a negative z-score and down-regulation of the pathway, and orange color or lighter shades of orange indicate a positive z-score and up-regulation of the pathway. Ratio denotes the number of significantly expressed genes compared with the total number of genes associated with the canonical pathway
GSEA and IPA associated gene expression induced in the patient plasma system with similar sets of signaling pathways
We next used GSEA and IPA to examine the gene expression profiles induced in the patient plasma system. The GSEA findings (Tables 4 and 5) identified enrichment of biologically relevant gene sets similar to those in the model system, including those related to inflammatory response, interferon response, JAK-STAT signaling, TNF signaling, IL-6 signaling and IL-2 signaling, complement, apoptosis and hypoxia. These signaling pathway and biological processes were identified to be regulated in both experimental conditions (patient plasma with IL-10 vs low LPS plasma, and IL-10 immunodepleted plasma vs low LPS plasma ).
Table 4.
Gene sets enriched in human monocytes stimulated with IL-10 immunodepleted plasma vs low LPS plasma
| Name of hallmark gene set | Descriptiona | FDR | Number of genes in the gene set | Founder gene setsb |
|---|---|---|---|---|
| TNF signaling via NfkB | Genes regulated by NF-kB in response to TNF | <0.001 | 200 | 120 |
| Inflammatory response | Genes defining inflammatory response. | <0.001 | 200 | 120 |
| Interferon gamma response | Genes up-regulated in response to IFNG | <0.001 | 200 | 82 |
| Interferon alpha response | Genes up-regulated in response to alpha interferon proteins. | <0.001 | 97 | 82 |
| IL2 STAT5 signaling | Genes up-regulated by STAT5 in response to IL2 stimulation. | <0.001 | 200 | 13 |
| IL6 JAK-STAT3 signaling | Genes up-regulated by IL6 [via STAT3, e.g., during acute phase response. | <0.001 | 87 | 24 |
| KRAS signaling up | Genes up-regulated by KRAS activation. | 0.001 | 200 | 14 |
| Epithelial mesenchymal transition | Genes defining epithelial-mesenchymal transition, as in wound healing, fibrosis and metastasis | <0.001 | 200 | |
| Allograft rejection | Genes up-regulated during transplant rejection. | <0.001 | 200 | 190 |
| MYC targets_v2 | A subgroup of genes regulated by MYC | 0.004 | 58 | 6 |
| Complement | Genes encoding components of the complement system, which is part of the innate immune system. | 0.004 | 200 | 71 |
| Coagulation | Genes encoding components of blood coagulation system | 0.005 | 138 | 71 |
| Apoptosis | Genes mediating programmed cell death (apoptosis) by activation of caspases. | 0.016 | 161 | 80 |
| Estrogen response early | Genes defining early response to estrogen | 0.018 | 200 | |
| KRAS signaling down | Genes down-regulated by KRAS activation. | 0.029 | 200 | 16 |
| Hypoxia | Genes up-regulated in response to low oxygen levels (hypoxia). | 0.031 | 200 | 87 |
| UV response up | Genes up-regulated in response to ultraviolet (UV) radiation | 0.044 | 58 | 16 |
aDescription of the gene sets were obtained from the Molecular Signatures Database
bFounder gene sets constitute the original experiments associating the genes with specified functions or biological processes
Table 5.
Gene sets enriched in human monocytes stimulated with patient plasma with IL-10 vs low LPS plasma
| Name of hallmark gene set | Descriptiona | FDR | Number of genes in the gene set | Founder gene setsb |
|---|---|---|---|---|
| TNF signaling via NfkB | Genes regulated by NF-kB in response to TNF | <0.001 | 200 | 120 |
| Interferon gamma response | Genes up-regulated in response to IFNG | <0.001 | 200 | 82 |
| Inflammatory response | Genes defining inflammatory response | <0.001 | 200 | 120 |
| Interferon alpha response | Genes up-regulated in response to alpha interferon proteins | <0.001 | 97 | 82 |
| IL6 JAK-STAT3 signaling | Genes up-regulated by IL6 [via STAT3, e.g., during acute phase response] | <0.001 | 87 | 24 |
| IL2 STAT5 signaling | Genes up-regulated by STAT5 in response to IL2 stimulation | <0.001 | 200 | 13 |
| MYC targets_v2 | A subgroup of genes regulated by MYC | <0.001 | 58 | 6 |
| Epithelial mesenchymal transition | Genes defining epithelial-mesenchymal transition, as in wound healing, fibrosis and metastasis | <0.001 | ||
| Complement | Genes encoding components of the complement system | <0.001 | 200 | 71 |
| Allograft rejection | Genes up-regulated during transplant rejection | <0.001 | 200 | 190 |
| KRAS signaling up | Genes up-regulated by KRAS activation | 0.001 | 200 | 14 |
| Estrogen response early | Genes defining early response to estrogen | 0.001 | ||
| Coagulation | Genes encoding components of blood coagulation system | 0.001 | 138 | 71 |
| UV response up | Genes up-regulated in response to ultraviolet (UV) radiation | 0.005 | 158 | 16 |
| Hypoxia | Genes up-regulated in response to low oxygen levels (hypoxia) | 0.008 | 200 | 87 |
| Apoptosis | Genes mediating programmed cell death (apoptosis) by activation of caspases | 0.013 | 161 | 80 |
| Unfolded protein response | Genes up-regulated during unfolded protein response, a cellular stress response related to the endoplasmic reticulum | 0.036 | 113 | 22 |
aDescription of the gene sets were obtained from the Molecular Signatures Database
bFounder gene sets constitute the original experiments associating the genes with specified functions or biological processes
We then compared GSEA data with the canonical pathways identified to be significantly enriched by IPA. IL-10 immunodepleted plasma induced a significant enrichment of genes associated with apoptosis signaling, TNF-signaling, Toll-like receptor signaling, and interferon response (in IPA identified as “Activation of IRF by Cytosolic Pattern Recognition Receptors”), similar to what was identified by GSEA (Fig. 3). In patient plasma with IL-10, IPA only identified the significant enrichment of three canonical pathways, suggesting that the presence of IL-10 had a strong regulatory effect on induction of genes associated with canonical pathways (Fig. 4). However, when we loosened the statistical stringency by setting an uncorrected P value of 0.05 as the threshold for significant enrichment of canonical pathways, IPA identified patient plasma with IL-10 to significantly enrich a larger number of canonical pathways, including IL-6 signaling, JAK/STAT signaling, apoptosis and Toll-like receptor signaling (Additional file 1), as also identified by GSEA.
Fig. 3.
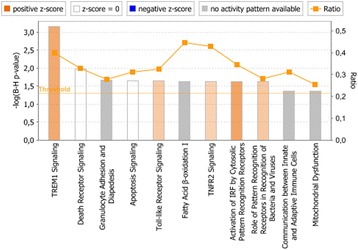
Canonical pathways significantly enriched in human monocytes stimulated with IL-10 immunodepleted plasma vs low LPS plasma. Significantly enriched canonical pathways were identified with a right-tailed Fisher’s Exact Test that calculates a P-value determining the probability that each canonical pathway associated with the dataset was due to chance alone. The P-values were corrected for multiple testing using the Benjamini-Hochberg method for correcting the FDR. The z-score indicates predicted activation state of the canonical pathway. Blue color or lighter shades of blue indicate a negative z-score and down-regulation of the pathway, and orange color or lighter shades of orange indicate a positive z-score and up-regulation of the pathway. Ratio denotes the number of significantly expressed genes compared with the total number of genes associated with the canonical pathway
Fig. 4.
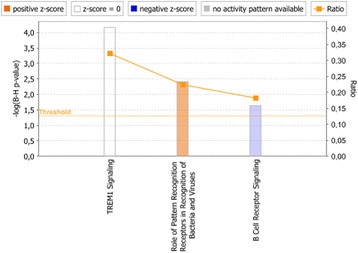
Canonical pathways significantly enriched in human monocytes stimulated with patient plasma with IL-10 vs low LPS plasma. Significantly enriched canonical pathways were identified with a right-tailed Fisher’s Exact Test, that calculates a P-value determining the probability that each canonical pathway associated to the dataset was due to chance alone. The P-values were corrected for multiple testing using the Benjamini-Hochberg method for correcting the FDR, and a P-value of <0.05 was set as threshold for statistical significance. The z-score indicates predicted activation state of the canonical pathway. Blue color or lighter shades of blue indicate a negative z-score and down-regulation of the pathway, and orange color or lighter shades of orange indicate a positive z-score and up-regulation of the pathway. Ratio denotes the number of significantly expressed genes compared with the total number of genes associated with the canonical pathway
Gene set enrichment analysis identified genes differentially induced by IL-10 to be associated with similar gene sets in both the model system and the patient plasma system
We next used GSEA to investigate the effect of IL-10 on meningococci-induced gene expression in the model system and in the patient plasma system. In the model system, when comparing Nm + IL-10 vs Nm , the differentially expressed genes were associated with the significant enrichment of 10 gene sets (Table 6, detailed reports in Additional files 2, 3, 4, 5), with oxidative phosphorylation being the gene set with the highest enrichment score. This indicates that genes associated with oxidative phosphorylation have greater expression levels in monocytes stimulated by N. meningitidis and IL-10, than by N. meningitidis alone. Comparison of the corresponding experimental conditions in the patient plasma system - patient plasma with IL-10 vs IL-10 immunodepleted plasma - identified oxidative phosphorylation to be the most significantly enriched gene set, suggesting that genes associated with oxidative phosphorylation have greater expression in monocytes stimulated with meningococcal LPS together with IL-10 compared with meningococcal LPS alone. In addition, seven of the eight significant gene sets in the model system were identified also in the patient plasma system.
Table 6.
Overview and comparison of significantly enriched gene sets in the model system and the patient plasma systema
| Gene sets enriched in Nm + IL-10 vs Nm | Gene sets enriched in patient plasma with IL-10 vs IL-10 immunodepleted plasma | Gene sets enriched in Nm vs Nm + IL-10 | Gene sets enriched in IL-10 immunodepleted plasma vs patient plasma with with IL-10 |
|---|---|---|---|
| Oxidative phosphorylation | Oxidative phosphorylation | TNF signaling | Interferon alpha response |
| MYC Targets_v1 | Peroxisome | KRAS signaling DN | TNF signaling |
| DNA repair | MYC Targets_v1 | Epithelial-mesenchymal transition | UV response |
| Adipogenesis | Adipogenesis | UV response | Interferon gamma response |
| Peroxisome | MYC targets_v2 | Inflammatory response | Allograft rejection |
| MYC targets_v2 | P53 Pathway | Kras signaling up | Kras signaling up |
| P53 Pathway | Reactive oxygen species | Apical surface | Hedgehog signaling |
| Protein secretion | DNA repair | Cholesterol homeostasis | Hallmark angiogenesis |
| Fatty Acid Metabolism | Allograft rejection | Mitotic spindle | |
| Glycolysis | Interferon alpha response | ||
| Bile acid metabolism | Coagulation |
We next compared gene sets enriched in Nm vs Nm + IL-10. As expected, GSEA identified the enrichment of gene sets associated with the innate immune response, in addition to several other gene sets with probably limited biological relevance to meningococcal infection (such as epithelial-mesenchymal transition, UV response, apical surface, allograft rejection, cholesterol homeostasis). Four of these gene sets (TNF signaling, Interferon alpha response, UV response, Kras signaling up) were significantly enriched when comparing the corresponding experimental conditions in the patient plasma system – IL-10 immunodepleted vs patient plasma with IL-10. In comparison, IPA enriched Nm + IL-10 vs Nm (Fig. 5) for biological processes associated with inflammatory signaling (increased activity of TREM1 signaling, and decreased activity of HMGB1 signaling) and cellular integrity and adhesion (including epithelial adherens junction signaling, tight junction signaling, integrin signaling, and granulocyte adhesion and diapedesis). Analysis of IL-10 immunodepleted vs patient plasma with IL-10 did not significantly enrich any biological processes or signaling pathways after Benjamini-Hochberg correction for the false discovery rate (results not shown).
Fig. 5.
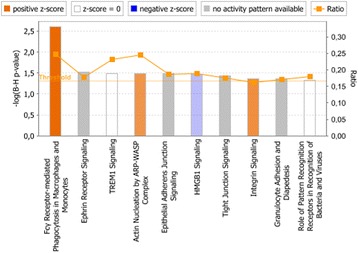
Canonical pathways significantly enriched in human monocytes stimulated with Nm + IL-10 vs Nmabc. aSignificantly enriched canonical pathways were identified with a right-tailed Fisher’s Exact Test, that calculates a P-value determining the probability that each canonical pathway associated to the dataset was due to chance alone. The P-values were corrected for multiple testing using the Benjamini-Hochberg method for correcting the FDR. bThe z-score indicates predicted activation state of the canonical pathway. Blue color or lighter shades of blue indicate a negative z-score and down-regulation of the pathway, and orange color or lighter shades of orange indicate a positive z-score and up-regulation of the pathway. cRatio denotes the number of significantly expressed genes compared with the total number of genes associated with the canonical pathway
Overall, GSEA analysis suggested that the biological processes associated with the gene expression changes in the model system of meningococcal sepsis were comparable with the results found in the patient plasma system, but similar biological processes were not enriched in IPA.
GSEA and IPA identified significant down-regulation of genes associated with mitochondrial function and oxidative phosphorylation in human monocytes stimulated by meningococcal LPS, and partial up-regulation of this response by IL-10
In the model system, oxidative phosphorylation was identified by GSEA to be enriched in unstimulated monocytes vs monocytes stimulated with N. meningitidis (Additional file 6). In the patient plasma system, oxidative phosphorylation was the most enriched gene set in low LPS plasma vs IL-10 immunodepleted plasma (Additional file 7). These two findings suggest that the presence of meningococcal LPS is associated with down-regulation of genes related to oxidative phosphorylation in monocytes.
We therefore examined the association between meningococcal activation of monocytes, and down-regulation of these genes in more detail. In IL-10 immunodepleted plasma vs low LPS plasma, GSEA identified 104 genes to be down-regulated (Additional file 8), while IPA identified 36 genes to be down-regulated (Additional file 9). Eighteen of these genes were identified by both bioinformatics tools. In sum, GSEA and IPA identified 122 genes associated with mitochondrial function/oxidative phosporylation to be down-regulated after depleting meningococcal sepsis plasma for IL-10. When we compared Nm vs ctr, GSEA identified 130 genes to be down-regulated (Additional file 10), and IPA identified 89 genes to be down-regulated (Additional file 11). Here, 49 genes were identified by both tools. Considering the model system and the patient plasma system together, GSEA and IPA identified 170 genes associated with oxidative phosphorylation and mitochondrial function to be down-regulated in monocytes stimulated by N. meningitidis and meningococcal LPS.
Oxidative phosphorylation was also the most enriched gene set when comparing Nm + IL-10 vs Nm, and patient plasma with IL-10 vs IL-10 immunodepleted plasma (Table 6), suggesting that the presence of IL-10 up-regulated genes associated with oxidative phosphorylation. To visualize the effect of IL-10 on the Nm-induced down-regulation, we generated the pathway labeled “mitochondrial dysfunction” in IPA, and overlaid P-values and FC values calculated from comparing gene expression levels in Nm + IL-10 vs Nm (Fig. 6a). We also overlaid this pathway with P-values and FC values generated from comparing Nm vs ctr (Fig. 6b) and N m + IL-10 vs ctr (Fig. 6c). Using IPA, we identified 18 genes to be up-regulated when comparing Nm + IL-10 vs Nm (Table 7), suggesting that the presence of IL-10 partially reversed the down-regulation of genes induced by N. meningitidis. We also overlaid P-values and FC values generated from comparing experimental conditions in the patient plasma system (Fig. 7). Comparing patient plasma with IL-10 vs IL-10 immunodepleted plasma (Fig. 7a) did not identify the presence of IL-10 to significantly up-regulate genes that were down-regulated in IL-10 immunodepleted plasma. However, a greater down-regulation of genes was associated with IL-10 immunodepleted plasma vs low LPS plasma (Fig. 7b) than patient plasma with IL-10 vs low LPS plasma (Fig. 7c). This indicates that the presence of IL-10 in meningococcal sepsis plasma increase the expression of genes associated with mitochondrial function/oxidative phosphorylation.
Fig. 6.
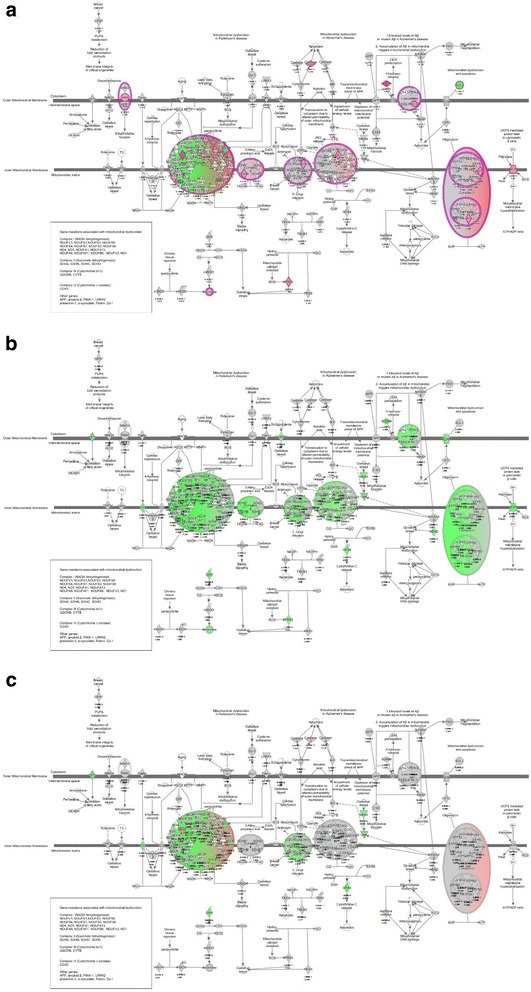
Mitochondrial dysfunction in IPA in human monocytes. a Expression levels when comparing Nm + IL-10 vs Nm. b Expression levels when comparing Nm vs ctr. c Expression levels when comparing Nm + IL-10 vs ctr
Table 7.
Genes associated with mitochondrial function identified to be regulated by IL-10 in monocytes stimulated by N. meningitidis
| Symbol | Entrez gene name | Location | Type(s) | Fold change1 | ||
|---|---|---|---|---|---|---|
| Nm vs ctr | Nm + IL-10 vs ctr | Nm + IL-10 vs Nm | ||||
| ACO2 | aconitase 2, mitochondrial | Cytoplasm | Enzyme | −1.66 | −1.33 | 1.25 |
| ATP5A1 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle | Cytoplasm | Transporter | −1.83 | −1.34 | 1.37 |
| ATP5H | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit d | Cytoplasm | Enzyme | −1.50 | −1.18 | 1.27 |
| BCL2 | B-cell CLL/lymphoma 2 | Cytoplasm | Transporter | NC | NC | −1.46 |
| CASP3 | caspase 3, apoptosis-related cysteine peptidase | Cytoplasm | Peptidase | NC | 1.86 | 1.79 |
| COX7C | cytochrome c oxidase subunit VIIc | Cytoplasm | Enzyme | −1.66 | NC | 1.45 |
| CYB5A | cytochrome b5 type A (microsomal) | Cytoplasm | Enzyme | −1.69 | NC | 1.48 |
| FURIN | furin (paired basic amino acid cleaving enzyme) | Cytoplasm | Peptidase | −2.47 | 1.42 | 3.51 |
| NCSTN | nicastrin | Plasma Membrane | Peptidase | −1.41 | 1.17 | 1.21 |
| NDUFA6 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14 kDa | Cytoplasm | Enzyme | −2.00 | −1.53 | 1.31 |
| NDUFA9 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39 kDa | Cytoplasm | Enzyme | 1.28 | 3.10 | 2.43 |
| NDUFA10 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42 kDa | Cytoplasm | Transporter | −1.38 | −1.14 | 1.22 |
| NDUFAB1 | NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1, 8 kDa | Cytoplasm | Enzyme | −2.26 | −1.56 | 1.45 |
| NDUFB5 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16 kDa | Cytoplasm | Enzyme | −1.48 | NC | 1.37 |
| NDUFS2 | NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49 kDa (NADH-coenzyme Q reductase) | Cytoplasm | Enzyme | −2.04 | NC | 1.92 |
| NDUFS6 | NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13 kDa (NADH-coenzyme Q reductase) | Cytoplasm | Enzyme | −1.74 | −1.42 | 1.22 |
| NDUFV2 | NADH dehydrogenase (ubiquinone) flavoprotein 2, 24 kDa | Cytoplasm | Enzyme | NC | −1.61 | −1.44 |
| PRDX3 | peroxiredoxin 3 | Cytoplasm | Enzyme | −1.63 | NC | 1.47 |
| SDHB | succinate dehydrogenase complex, subunit B, iron sulfur (Ip) | Cytoplasm | Enzyme | −1.91 | −1.41 | 1.35 |
| TRAK1 | trafficking protein, kinesin binding 1 | Nucleus | Other | −1.29 | NC | 1.25 |
| UCP2 | uncoupling protein 2 (mitochondrial, proton carrier) | Cytoplasm | Transporter | −2.07 | −1.49 | 1.39 |
| UQCR10 | ubiquinol-cytochrome c reductase, complex III subunit X | Cytoplasm | Enzyme | −1.75 | −1.38 | 1.26 |
| VPS9D1 | VPS9 domain containing 1 | Other | Transporter | NC | NC | 1.86 |
1Significantly expressed genes identified with two-way ANOVA, P < 0.05 after correction for multiple testing using the Benjamini-Hochberg method for correcting the false discovery rate [36]. NC denotes P-value >0.05
Fig. 7.
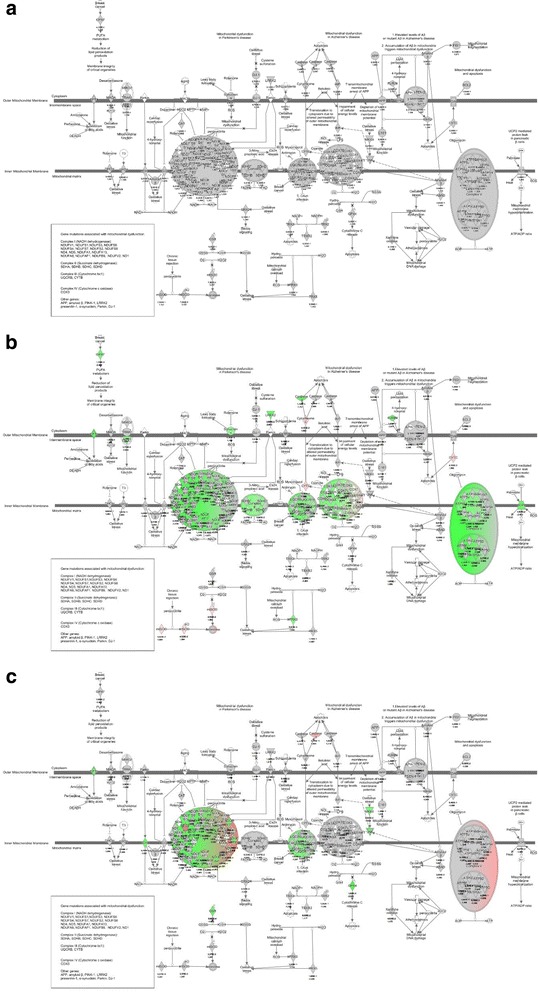
Mitochondrial dysfunction in IPA before and after immunodepletion of IL-10. a Expression levels when comparing patient plasma with IL-10 vs IL-10 immunodepleted plasma. b Expression levels when comparing patient plasma with IL-10 vs low LPS plasma. c Expression levels when comparing IL-10 immunodepleted plasma vs low LPS plasma
Discussion
The emergence of a large number of bioinformatics tools over the past decade have made it easier for researchers to interpret the vast amount of transcriptomic data generated from DNA microarrays, RNA sequencing and other gene expression methods. However, the diverse range of tools have also created a number of challenges. First, differences in assumptions underlying the methods used have made it more difficult to compare studies that have used genome-wide expression profiling to study similar pathophysiological phenomena, such as sepsis [5]. Secondly, depending on the methods used, the same gene expression dataset may result in different interpretations, thereby requiring close attention to the underlying statistical assumptions that have affected data interpretation.
The first aim of this study was to compare the biological interpretation resulting from two different bioinformatics tools – GSEA and IPA. Specifically, we investigated the pro-inflammatory response induced in two different model systems of meningococcal activation of human monocytes. Overall, both GSEA and IPA associated the differentially regulated genes to similar biological processes and functional groups, identifying that N. meningitidis and meningococcal sepsis plasma depleted for IL-10, respectively, induced genes associated with pro-inflammatory innate immune activation, including TNF-signaling, Toll-like receptor signaling, JAK-STAT-signaling, type I and type II interferon, as well as apoptosis and mitochondrial dysfunction. Some enriched gene sets in GSEA, such as KRAS signaling, allograft rejection, epithelial mesenchymal transition, and estrogen late response did not immediately appear relevant to the experimental condition and disease under study, and were, except for estrogen late response, not detected by IPA. In addition, when we focused specifically on genes differentially induced by the presence of IL-10, GSEA and IPA differed in the enrichment of biological processes and signaling pathways.
Overall, the rapid induction of pro-inflammatory genes by sepsis plasma is consistent with comparable previous studies of sepsis-induced gene expression at an early stage of the sepsis syndrome [36–40]. It should be noted that all these studies investigated gene expression in whole-blood or leukocyte isolates collected within 24 h of admission, which differs from our dataset which is generated by inducing gene expression in human monocytes elutriated from heparinized whole blood from healthy blood donors combined with sepsis plasma samples [26]. These studies have therefore the advantage of reporting data generated from leukocytes isolated directly from patients during the clinical course of sepsis. An advantage of our model is that the onset of pathogenic activation of the monocytes as well as the bacterial load is known. In most clinical sepsis studies, the exact onset of the disease is unknown. Furthermore, the initial blood samples are drawn from various time-points during the first 24 h of admission, and the bacterial load in plasma or the microbial molecules driving the innate immune reaction often remain unknown [36–40].
A few differences exist between GSEA and IPA. The first and foremost is that GSEA does not implement a filtering criteria prior to analysis, and therefore consider the expression of all the genes in the dataset. The main strength of GSEA is to identify the extent to which an a priori defined list of genes, which previous experiments have associated with biological processes or signaling pathways, is coordinately expressed in the dataset. This differs from the conventional approach to gene expression analysis in IPA, which normally implies setting a pre-defined cut-off based on a corrected or uncorrected P-value, and FC values, such as 1.5 and 2.0. A risk with this approach is that the investigator may unintentionally exclude relevant genes, and risks the loss of biologically relevant information which the bioinformatics tools are unable to detect [41]. In comparison, the underlying statistics of GSEA enables the detection of biological functions associated with coordinated, but subtle changes in gene expression levels.
In our case, the previous interpretation of gene expression profiling of human monocyte response to meningococcal infection, generated both in a model system [25] and in a patient plasma system [26], overlooked the large number of significantly regulated genes associated with mitochondrial dysfunction and oxidative phosphorylation. This was primarily due to the implemented filtering criteria consisting of a cut-off for statistical significance (P < 0.01) and FC (1.5). However, in the present study, using a filter of FDR 5% only, the IPA analysis suggested that N. meningitidis induces a strong down-regulation of a large number of genes associated with oxidative phosphorylation, consistent with what was identified by GSEA.
Another challenge to directly comparing GSEA with IPA is that the gene sets in GSEA and canonical pathways in IPA do not directly correspond to each other. The gene sets in GSEA are curated based on gene expression patterns identified in previously conducted experimental conditions. In many cases, the gene sets correspond to a broadly defined biological process (such as “inflammatory response”, “complement” or “coagulation”), or are named after the response to a particular stimulus (such as interferon-gamma or TNF). In comparison, the canonical pathways in IPA delineate more detailed biochemical steps known to result from specific exposures or receptor-binding, or describe mechanistic interactions occurring in specific diseases (refer to Panel 1). Furthermore, IPA does not limit itself to interactions observed in gene expression studies, which is the case for the hallmark gene sets from the Molecular Signatures Database [34] used for the present study. Another advantage is that IPA is also more frequently updated.
We used these two methods in a complementary manner. The Gene Set Enrichment Analysis, using the hallmark gene sets or other gene sets from the Molecular Signatures Database [34] when relevant, produced an unbiased, initial analysis of the extent to which there was coordinated expression of genes related to defined biological functions. Verification of these results was conducted in IPA, which enabled a more detailed examination of whether the differential regulation of genes was associated with mechanistic pathways, as well as locating the position and direction of change of specific genes in pathways.
The second aim was to investigate whether the biological processes associated with the gene expression changes in our model system of meningococcal sepsis was comparable to the results found in our patient plasma system. GSEA suggests that the simple model system, consisting of human monocytes stimulated in a normal pooled plasma with wild-type N. meningitidis and IL-10, induces transcriptomic changes associated with similar functional groups observed by the more complex plasma from patients with meningococcal sepsis. This finding has two biological interpretations. First, our simple human monocyte model system appears robust for the study of cellular mechanisms and biological processes associated with the host response to N. meningitidis. It might thus be applicable to other infectious agents. Second, the gene expression changes in the model system, where the transcriptomic response is initiated by the treatment of monocytes with N. meningitidis and IL-10, are reproduced in the patient plasma system before and after IL-10 immunodepletion. It can therefore be suggested that the dominant inducer of gene expression in meningococcal sepsis plasma is N. meningitidis, and that the dominant modulator of the Nm-induced gene expression pattern is IL-10.
Finally, this study wanted to identify whether GSEA and IPA together could generate additional insight about the human monocyte host response to meningococcal infection. The IPA analysis, supported by GSEA, suggested that N. meningitidis is a strong down-regulator of genes associated with oxidative phosphorylation and mitochondrial function. The reduced expression of genes associated with mitochondrial function and oxidative phosphorylation has also been identified in white blood cells isolated from sepsis patients [42, 43], and in large animal sepsis models [40]. Tang et al. have reported the inhibited expression of genes associated with mitochondrial dysfunction in a study isolating neutrophils from sepsis patients [42]. However, their study reported that gene expression was even lower in the non-septic controls. Wong et al. used IPA and identified that genes up-regulated in neutrophils from sepsis patients enriched pathways related to mitochondrial dysfunction [40]. Two more recent studies have more specifically studied the differential regulation of genes associated with oxidative phosphorylation in sepsis patients [44, 45]. One study by Weiss et al. [45] determined the differential expression of 296 nuclear-encoded mitochondrial genes in whole blood collected within 24 h of admission from pediatric patients with septic shock compared with non-septic controls. They identified that 118 of 296 genes were differentially regulated (48 up-regulated and 70 down-regulated) in septic shock patients. Between survivors and non-survivors of septic shock, the genes cytochrome C oxidase subunit VIIb and NADH dehydrogenase flavoprotein were differentially regulated. They also identified that a sub-group of patients with higher rate of multiple-organ failure and higher mortality rate had a greater repression of nuclear-encoded mitochondrial genes, compared with two other sub-groups. Another smaller study, reported in a letter to the editor by Raman et al. [44] describes gene expression in whole-blood in previously healthy children with meningococcal septicemia at 0, 4, 8, 12, 24, and 48 h from time of admission. They identified an immediate reduction of gene expression associated with oxidative phosphorylation processes, and it continued to decrease over the 48 h, leading the authors to suggest that mitochondrial dysfunction contributed to multi-organ failure. However, the letter does not report the clinical outcomes of the patients investigated. The findings from our and previous gene expression studies are consistent with a long held view that mitochondrial dysfunction and altered oxidative phosphorylation have implications in sepsis pathophysiology [46, 47]—a theme that has recently received renewed attention [44, 48, 49]. In the context of meningococcal sepsis, the intense down-regulation of genes associated with oxidative phosphorylation indicates that N. meningitidis as part of a pro-inflammatory response may induce dysfunction of oxidative phosphorylation and deranged energy metabolism [50, 51]—which has also been referred to as “cytopathic hypoxia” [52]—which contributes to rapidly evolving multiple organ failure. We identified furthermore that IL-10 partially reverses the down-regulation of genes associated with oxidative phosphorylation. To our knowledge, this has not been reported in previous studies of the effect of IL-10 on gene expression induced by LPS [53–56]. We have in our previous studies shown that IL-10, in response to the meningococci, both up- and down-regulates genes associated with a broad range of cellular functions [25, 26]. The increased expression of genes associated with oxidative phosphorylation induced by IL-10 indicates that activation of IL-10-signaling may improve cellular respiration, and that this may part of IL-10’s well-known attenuation of the harmful effects of meningococcal activation.
Overall, these findings should motivate functional validation by research groups specialized in the study of mitochondrial function and the oxidative phosphorylation pathway in order to generate stronger evidence about the effect of meningococci on oxidative phosphorylation in human monocytes and possibly other immune cells, and the regulatory effect of IL-10.
We propose that our study adds three additional insights to existing studies on gene expression changes related to oxidative phosphorylation in sepsis. First, we have identified that a specific pathogen – N. meningitidis - is able to induce down-regulation of a large number of genes associated with mitochondrial function and oxidative phosphorylation in monocytes within 3 h. Second, meningococcal sepsis plasma immunodepleted for IL-10 induces down-regulation to a similar extent. Third, the presence of IL-10 appears to partially reverse this down-regulation, adding new knowledge to the biological effects of IL-10 in meningococcal disease and possibly in other infections activating the IL-10 signaling system.
Conclusion
We have investigated two previously generated microarray datasets of meningococcal activation of human monocytes with two different bioinformatics tools – GSEA and IPA. We have shown that relying on a single bioinformatics tool together with arbitrarily chosen filtering criteria for data analysis may result in overlooking relevant biological processes and signaling pathways associated with genes differentially expressed between compared experimental conditions. By combining GSEA with IPA, we discovered an inhibitory effect exerted by N. meningitidis on genes associated with mitochondrial function and oxidative phosphorylation, and that IL-10 partially reverses this strong inhibitory effect, thereby identifying, to our knowledge, yet another area where IL-10 regulates the effect of LPS.
Additional files
Canonical - Fisher p0.05 - Plasma w. IL-10 vs low LPS plasma. (TIFF 6585 kb)
GSEA report - Nm + IL-10 vs Nm. (XLS 33 kb)
GSEA report - Enriched in patient plasma w. IL-10 vs IL-10 immunodepleted. (XLS 31 kb)
GSEA report - Nm vs Nm + IL-10.txt. (XLS 2 kb)
GSEA report - Enriched in IL-10 immunodepleted vs patient plasma w. IL-10.tx. (XLS 30 kb)
GSEA report - Enriched in ctr vs Nm.txt. (XLS 32 kb)
GSEA report - Enriched in low LPS plasma vs IL-10 immunodepleted. (XLS 29 kb)
GSEA core enrichment - Oxphos - IL-10 immunodepleted vs low LPS plasma. (XLS 61 kb)
Mitochondrial dysfunction - IL-10 immunodepleted vs low LPS plasma. (XLS 16 kb)
GSEA core enrichment- Oxphos - Ctr vs Nm. (XLS 62 kb)
Mitochondrial dysfunction - Nm vs ctr - FDR5. (XLS 46 kb)
Acknowledgements
Not applicable.
Funding
This research was funded by the Research Council of Norway through the Medical Student Research Programme.
Availability of data and materials
The datasets supporting the conclusions of this article are available in the United States National Center for Biotechnology Information Gene Expression Omnibus (GEO) repository under accession number GSE82316 at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE82166.
Abbreviations
- GSEA
Gene set enrichment analysis
- IL-10
Interleukin-10
- IPA
Ingenuity Pathway Analysis
- Nm
N. meningitidis
- TNF
Tumor Necrosis Factor
Authors’ contributions
JPB, RØ and PB supervised the project. UG used GSEA and IPA to analyze gene expression data sets from experiments conducted for previously published studies. UG lead the work on drafting the manuscript, including tables and figures, while JPB, OKO, BB, PK, PB and RØ commented multiple iterations until completion of the final manuscript, which all authors approved.
Ethics approval and consent to participate
This study used DNA microarray data generated from the use of patient plasma samples (n = 6), previously collected after informed consent from parents, relatives, or patients, and used in accordance with ethics approval from the Regional Medical Ethics Committee of Health Region I in Norway (Biobank material access number 948; Studies of meningococcal disease; Oslo University Hospital, Oslo, Norway). The clinical and microbiological data on each patient has previously been reported [26]. Human monocytes elutriated from heparinized whole blood collected from consenting, healthy donors was also used for the experiments that generated the data for this study, and were used in accordance with ethics approval from the Regional Medical Ethics Committee of Health Region I in Norway (Biobank material access number 908; Human monocytes and lymphocytes; Oslo University Hospital, Oslo, Norway).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12865-017-0229-5) contains supplementary material, which is available to authorized users.
Contributor Information
Unni Gopinathan, Phone: +47 48294074, Email: unni.gnathan@gmail.com.
Reidun Øvstebø, Email: reidun.ovstebo@medisin.uio.no.
Berit Sletbakk Brusletto, Email: berit.brusletto@medisin.uio.no.
Ole Kristoffer Olstad, Email: o.k.olstad@medisin.uio.no.
Peter Kierulf, Email: peter.kierulf@medisin.uio.no.
Petter Brandtzaeg, Email: petter.brandtzag@medisin.uio.no.
Jens Petter Berg, Email: j.p.berg@medisin.uio.no.
References
- 1.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA. 2016;315(8):801–10. [DOI] [PMC free article] [PubMed]
- 2.Kaukonen K-M, Bailey M, Suzuki S, Pilcher D, Bellomo R. Mortality related to severe sepsis and septic shock among critically ill patients in Australia and New Zealand, 2000-2012. JAMA. 2014;311(13):1308–1316. doi: 10.1001/jama.2014.2637. [DOI] [PubMed] [Google Scholar]
- 3.Stevenson EK, Rubenstein AR, Radin GT, Wiener RS, Walkey AJ. Two decades of mortality trends among patients with severe sepsis: a comparative meta-analysis. Crit Care Med. 2014;42(3):625–631. doi: 10.1097/CCM.0000000000000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong HR. Genome-wide expression profiling in pediatric septic shock. Pediatr Res. 2013;73(4 Pt 2):564–569. doi: 10.1038/pr.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang BM, Huang SJ, McLean AS. Genome-wide transcription profiling of human sepsis: a systematic review. Crit Care Lond Engl. 2010;14(6):R237. doi: 10.1186/cc9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russell JA. Gene expression in human sepsis: what have we learned? Crit Care Lond Engl. 2011;15(1):121. doi: 10.1186/cc9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marshall JC. Why have clinical trials in sepsis failed? Trends Mol Med. 2014;20(4):195–203. doi: 10.1016/j.molmed.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Angus DC. The search for effective therapy for sepsis: back to the drawing board? JAMA. 2011;306(23):2614–2615. doi: 10.1001/jama.2011.1853. [DOI] [PubMed] [Google Scholar]
- 9.Liu X, Ren H, Peng D. Sepsis biomarkers: an omics perspective. Front Med. 2014;8(1):58–67. doi: 10.1007/s11684-014-0318-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skibsted S, Bhasin MK, Aird WC, Shapiro NI. Bench-to-bedside review: future novel diagnostics for sepsis - a systems biology approach. Crit Care Lond Engl. 2013;17(5):231. doi: 10.1186/cc12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Greeff SC, de Melker HE, Schouls LM, Spanjaard L, van Deuren M. Pre-admission clinical course of meningococcal disease and opportunities for the earlier start of appropriate intervention: a prospective epidemiological study on 752 patients in the Netherlands, 2003-2005. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc. Clin Microbiol. 2008;27(10):985–992. doi: 10.1007/s10096-008-0535-1. [DOI] [PubMed] [Google Scholar]
- 12.Hackett SJ, Guiver M, Marsh J, Sills JA, Thomson APJ, Kaczmarski EB, et al. Meningococcal bacterial DNA load at presentation correlates with disease severity. Arch Dis Child. 2002;86(1):44–46. doi: 10.1136/adc.86.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Øvstebø R, Brandtzaeg P, Brusletto B, Haug KBF, Lande K, Høiby EA, et al. Use of robotized DNA isolation and real-time PCR to quantify and identify close correlation between levels of Neisseria meningitidis DNA and lipopolysaccharides in plasma and cerebrospinal fluid from patients with systemic meningococcal disease. J Clin Microbiol. 2004;42(7):2980–2987. doi: 10.1128/JCM.42.7.2980-2987.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bjerre A, Brusletto B, Øvstebø R, Joø GB, Kierulf P, Brandtzaeg P. Identification of meningococcal LPS as a major monocyte activator in IL-10 depleted shock plasmas and CSF by blocking the CD14-TLR4 receptor complex. J Endotoxin Res. 2003;9(3):155–163. doi: 10.1177/09680519030090030301. [DOI] [PubMed] [Google Scholar]
- 15.Brandtzaeg P, Kierulf P, Gaustad P, Skulberg A, Bruun JN, Halvorsen S, et al. Plasma endotoxin as a predictor of multiple organ failure and death in systemic meningococcal disease. J Infect Dis. 1989;159(2):195–204. doi: 10.1093/infdis/159.2.195. [DOI] [PubMed] [Google Scholar]
- 16.Brandtzaeg P, Bjerre A, Øvstebø R, Brusletto B, Joø GB, Kierulf P. Neisseria meningitidis lipopolysaccharides in human pathology. J Endotoxin Res. 2001;7(6):401–420. [PubMed] [Google Scholar]
- 17.Brandtzaeg P, Osnes L, Ovstebø R, Joø GB, Westvik AB, Kierulf P. Net inflammatory capacity of human septic shock plasma evaluated by a monocyte-based target cell assay: identification of interleukin-10 as a major functional deactivator of human monocytes. J Exp Med. 1996;184(1):51–60. doi: 10.1084/jem.184.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ovstebø R, Olstad OK, Brusletto B, Møller AS, Aase A, Haug KBF, et al. Identification of genes particularly sensitive to lipopolysaccharide (LPS) in human monocytes induced by wild-type versus LPS-deficient Neisseria meningitidis strains. Infect Immun. 2008;76(6):2685–2695. doi: 10.1128/IAI.01625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Møller A-SW, Øvstebø R, Westvik A-B, Joø GB, Haug K-BF, Kierulf P. Effects of bacterial cell wall components (PAMPs) on the expression of monocyte chemoattractant protein-1 (MCP-1), macrophage inflammatory protein-1alpha (MIP-1alpha) and the chemokine receptor CCR2 by purified human blood monocytes. J Endotoxin Res. 2003;9(6):349–360. doi: 10.1177/09680519030090060801. [DOI] [PubMed] [Google Scholar]
- 20.Henriksson CE, Klingenberg O, Ovstebø R, Joø G-B, Westvik A-B, Kierulf P. Discrepancy between tissue factor activity and tissue factor expression in endotoxin-induced monocytes is associated with apoptosis and necrosis. Thromb Haemost. 2005;94(6):1236–1244. doi: 10.1160/TH05-07-0463. [DOI] [PubMed] [Google Scholar]
- 21.Osterud B, Flaegstad T. Increased tissue thromboplastin activity in monocytes of patients with meningococcal infection: related to an unfavourable prognosis. Thromb Haemost. 1983;49(1):5–7. [PubMed] [Google Scholar]
- 22.Osterud B, Bjorklid E. Tissue factor in blood cells and endothelial cells. Front Biosci Elite Ed. 2012;4:289–299. doi: 10.2741/e376. [DOI] [PubMed] [Google Scholar]
- 23.Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat Rev Immunol. 2011;11(11):762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehmann AK, Halstensen A, Sørnes S, Røkke O, Waage A. High levels of interleukin 10 in serum are associated with fatality in meningococcal disease. Infect Immun. 1995;63(6):2109–2112. doi: 10.1128/iai.63.6.2109-2112.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gopinathan U, Ovstebø R, Olstad OK, Brusletto B, Dalsbotten Aass HC, Kierulf P, et al. Global effect of interleukin-10 on the transcriptional profile induced by Neisseria meningitidis in human monocytes. Infect Immun. 2012;80(11):4046–4054. doi: 10.1128/IAI.00386-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gopinathan U, Brusletto BS, Olstad OK, Kierulf P, Berg JP, Brandtzaeg P, et al. IL-10 immunodepletion from meningococcal sepsis plasma induces extensive changes in gene expression and cytokine release in stimulated human monocytes. Innate Immun. 2015;21(4):429–449. doi: 10.1177/1753425914547743. [DOI] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 29.Krämer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinforma Oxf Engl. 2014;30(4):523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ingenuity Systems. Calculating and Interpreting the p-values for Functions, Pathways and Lists in IPA [Internet]. 2016 [cited 2016 Feb 1]. Available from: https://www.ingenuity.com/wp-content/themes/ingenuity-qiagen/pdf/ipa/functions-pathways-pval-whitepaper.pdf.
- 32.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29(4):365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 33.Broad Institute. Gene Set Enrichment Analysis [Internet]. 2015 [cited 2015 Nov 1]. Available from: http://www.broadinstitute.org/gsea/downloads.jsp.
- 34.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinforma Oxf Engl. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B Methodol. 1995;57(1):289–300. [Google Scholar]
- 36.Shanley TP, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Doctor A, et al. Genome-level longitudinal expression of signaling pathways and gene networks in pediatric septic shock. Mol Med Camb Mass. 2007;13(9–10):495–508. doi: 10.2119/2007-00065.Shanley. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cvijanovich N, Shanley TP, Lin R, Allen GL, Thomas NJ, Checchia P, et al. Validating the genomic signature of pediatric septic shock. Physiol Genomics. 2008;34(1):127–134. doi: 10.1152/physiolgenomics.00025.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cazalis M-A, Lepape A, Venet F, Frager F, Mougin B, Vallin H, et al. Early and dynamic changes in gene expression in septic shock patients: a genome-wide approach. Intensive Care Med Exp. 2014;2(1):20. doi: 10.1186/s40635-014-0020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maslove DM, Tang BM, McLean AS. Identification of sepsis subtypes in critically ill adults using gene expression profiling. Crit Care Lond Engl. 2012;16(5):R183. doi: 10.1186/cc11667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong HR, Freishtat RJ, Monaco M, Odoms K, Shanley TP. Leukocyte subset-derived genomewide expression profiles in pediatric septic shock. Pediatr Crit Care Med J Soc Crit Care Med World Fed Pediatr Intensive Crit Care Soc. 2010;11(3):349–355. doi: 10.1097/PCC.0b013e3181c519b4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dalman MR, Deeter A, Nimishakavi G, Duan Z-H. Fold change and p-value cutoffs significantly alter microarray interpretations. BMC Bioinformatics. 2012;13(Suppl 2):S11. doi: 10.1186/1471-2105-13-S2-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang BMP, McLean AS, Dawes IW, Huang SJ, Lin RCY. The use of gene-expression profiling to identify candidate genes in human sepsis. Am J Respir Crit Care Med. 2007;176(7):676–684. doi: 10.1164/rccm.200612-1819OC. [DOI] [PubMed] [Google Scholar]
- 43.Weiss SL, Selak MA, Tuluc F, Perales Villarroel J, Nadkarni VM, Deutschman CS, et al. Mitochondrial dysfunction in peripheral blood mononuclear cells in pediatric septic shock. Pediatr Crit Care Med J Soc Crit Care Med World Fed Pediatr Intensive Crit Care Soc. 2015;16(1):e4–12. doi: 10.1097/PCC.0000000000000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raman S, Klein N, Kwan A, Hubank M, Rahman S, Rashid A, et al. Oxidative phosphorylation gene expression falls at onset and throughout the development of meningococcal sepsis-induced multi-organ failure in children. Intensive Care Med. 2015;41(8):1489–1490. doi: 10.1007/s00134-015-3817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss SL, Cvijanovich NZ, Allen GL, Thomas NJ, Freishtat RJ, Anas N, et al. Differential expression of the nuclear-encoded mitochondrial transcriptome in pediatric septic shock. Crit Care Lond Engl. 2014;18(6):623. doi: 10.1186/s13054-014-0623-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brealey D, Brand M, Hargreaves I, Heales S, Land J, Smolenski R, et al. Association between mitochondrial dysfunction and severity and outcome of septic shock. Lancet Lond Engl. 2002;360(9328):219–23. [DOI] [PubMed]
- 47.Singer M. Mitochondrial function in sepsis: acute phase versus multiple organ failure. Crit Care Med. 2007;35(9 Suppl):S441–8. [DOI] [PubMed]
- 48.Lee I, Hüttemann M. Energy crisis: the role of oxidative phosphorylation in acute inflammation and sepsis. Biochim Biophys Acta. 2014;1842(9):1579–86. [DOI] [PMC free article] [PubMed]
- 49.Lorente L, Martín MM, López-Gallardo E, Blanquer J, Solé-Violán J, Labarta L, et al. Decrease of oxidative phosphorylation system function in severe septic patients. J Crit Care. 2015. [DOI] [PubMed]
- 50.Arulkumaran N, Deutschman CS, Pinsky MR, Zuckerbraun B, Schumacker PT, Gomez H, et al. MITOCHONDRIAL FUNCTION IN SEPSIS. Shock Augusta Ga. 2016;45(3):271–81. [DOI] [PMC free article] [PubMed]
- 51.Fink MP. Cytopathic hypoxia and sepsis: is mitochondrial dysfunction pathophysiologically important or just an epiphenomenon. Pediatr Crit Care Med J Soc Crit Care Med World Fed Pediatr Intensive Crit Care Soc. 2015;16(1):89–91. [DOI] [PubMed]
- 52.Fink MP. Cytopathic hypoxia. Is oxygen use impaired in sepsis as a result of an acquired intrinsic derangement in cellular respiration? Crit Care Clin. 2002;18(1):165–75. [DOI] [PubMed]
- 53.Williams L, Jarai G, Smith A, Finan P. IL-10 expression profiling in human monocytes. J Leukoc Biol. 2002;72(4):800–9. [PubMed] [Google Scholar]
- 54.Jung M, Sabat R, Krätzschmar J, Seidel H, Wolk K, Schönbein C, et al. Expression profiling of IL-10-regulated genes in human monocytes and peripheral blood mononuclear cells from psoriatic patients during IL-10 therapy. Eur J Immunol. 2004;34(2):481–93. [DOI] [PubMed]
- 55.Lang R, Patel D, Morris JJ, Rutschman RL, Murray PJ. Shaping gene expression in activated and resting primary macrophages by IL-10. J Immunol Baltim Md 1950. 2002;169(5):2253–63. [DOI] [PubMed]
- 56.Antoniv TT, Park-Min K-H, Ivashkiv LB. Kinetics of IL-10-induced gene expression in human macrophages. Immunobiology. 2005;210(2–4):87–95. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Canonical - Fisher p0.05 - Plasma w. IL-10 vs low LPS plasma. (TIFF 6585 kb)
GSEA report - Nm + IL-10 vs Nm. (XLS 33 kb)
GSEA report - Enriched in patient plasma w. IL-10 vs IL-10 immunodepleted. (XLS 31 kb)
GSEA report - Nm vs Nm + IL-10.txt. (XLS 2 kb)
GSEA report - Enriched in IL-10 immunodepleted vs patient plasma w. IL-10.tx. (XLS 30 kb)
GSEA report - Enriched in ctr vs Nm.txt. (XLS 32 kb)
GSEA report - Enriched in low LPS plasma vs IL-10 immunodepleted. (XLS 29 kb)
GSEA core enrichment - Oxphos - IL-10 immunodepleted vs low LPS plasma. (XLS 61 kb)
Mitochondrial dysfunction - IL-10 immunodepleted vs low LPS plasma. (XLS 16 kb)
GSEA core enrichment- Oxphos - Ctr vs Nm. (XLS 62 kb)
Mitochondrial dysfunction - Nm vs ctr - FDR5. (XLS 46 kb)
Data Availability Statement
The datasets supporting the conclusions of this article are available in the United States National Center for Biotechnology Information Gene Expression Omnibus (GEO) repository under accession number GSE82316 at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE82166.


