Abstract
While the beta-cells of the endocrine pancreas are defined as cells with high levels of insulin production and tight stimulus-secretion coupling, the existence of functional heterogeneity among them has been known for decades. Recent advances in molecular technologies, in particular single cell profiling on both the protein and messenger RNA level, have uncovered that beta-cells exist in several antigenically and molecularly definable states. Using antibodies to cell surface markers or multi-dimensional clustering of beta-cells using more than 20 protein markers by mass cytometry, four distinct groups of beta-cells could be differentiated. However, whether these states represent permanent cell lineages or are readily interconvertible from one group to another remains to be determined. Nevertheless, future analysis of the pathogenesis of type 1 and type 2 diabetes will certainly benefit from a growing appreciation of beta-cell heterogeneity. Here, we aim to summarize concisely the recent advances in the field and their possible impact on our understanding of beta-cell physiology and pathophysiology.
Keywords: Single-cell transcriptomics, beta-cell, beta-cell heterogeneity, mass cytometry
1. Introduction
We have known for decades that not all beta-cells are equal, but the analytical tools to identify inherent variation in beta-cell function and health have been limited. Through technological advances, it is now possible to interrogate single cells at the DNA, mRNA, protein, and functional levels. Recent discoveries show that human beta-cells exist in multiple subtypes and that islet cell types can interconvert in T2D. These findings raise the question whether the relative abundance of functionally distinct subpopulations vary in the glucose intolerant state or in diabetes, whether the different subtypes have unique antigenic properties in the context of type 1 diabetes, and to what extent and at which rate the different beta-cell subpopulations are interconvertible. These questions are not academic, since their answers will provide insights into the pathophysiology of Type 1 and Type 2 diabetes, and will likely be relevant in our search for novel targets for therapeutic intervention.
By definition, all adult beta-cells express high levels of insulin, which is released in response to appropriate stimuli to help maintain blood glucose homeostasis. The primary secretagogue for beta-cells is glucose, and the set-point and response curve for glucose are determined by the expression of the low affinity glucokinase enzyme instead of hexokinase in these cells. However, in vivo multiple other factors contribute to control the net insulin secretion of beta-cells, among them amino acids, hormones, fatty acids, and neuronal input. While for convenience’ sake it is often assumed that all beta-cells respond to these inputs equally, evidence for functional heterogeneity among beta-cells was in fact already reported in the 1980s by Salomon and Meda, who were able to study the response of rat beta-cells to glucose on the single cell level using the reverse hemolytic plaque assay [1]. In quick succession, multiple studies confirmed and extended these findings [2, 3, 4, 5], showing for instance that beta-cells with high rates of insulin protein synthesis were preferential glucose responders [2]. Until recently, neither the physiological significance of this in vitro phenomenon nor the molecular mechanisms driving it were known. This review summarizes some of the recent technological advances and exciting results that have begun to elucidate these issues.
2. Functional heterogeneity among rodent and human beta-cells – novel approaches and insights
Advanced in-situ Ca2+ imaging was recently brought to bear on the question of beta-cell heterogeneity by Rutter’s group in London. First, they employed in-situ Ca2+ imaging approaches, together with large scale mapping of cellular connectivity, to characterize the secretory behavior of human beta cells [6]. When stimulated by high glucose alone, human beta-cells exhibited only moderate cooperativity; however, in the presence of the incretin GLP-1, connectivity was established among sub-networks of beta-cells. Importantly, these beta- to beta-cell connections were inhibited by the addition of high concentrations of free fatty acids, simulating lipotoxicity. Importantly, the beta-cell response to GLP-1 was inversely correlated with body mass index, suggesting that beta-cell connectivity might play a role in the pathogenesis of type 2 diabetes. While these studies demonstrated altered beta-cell behavior given changing metabolic conditions, they did not directly address beta-cell heterogeneity.
A critical question regarding the functional coupling of beta-cells is whether the beta-cell networks consists of equivalent beta-cells which all have the same impact on the timing of the Ca2+ oscillation, or if a hierarchy exists between certain ‘pacemaker’ and ‘follower’ beta-cells, and thus true functional heterogeneity. Rutter and colleagues recently addressed this issue using elegant opotogenetic methods to determine that islet cells contain a small minority (less than 10% of beta-cells) that when silenced disrupt beta-cell networks, while calcium dynamic and insulin secretion where not affected when ‘follower’ beta-cells were silenced [7]. They concluded that the minority ‘hub’ cells establish long-range connectivity to control and synchronize the remaining beta-cells. On the molecular level, hub cells appeared to exhibit a less mature phenotype and have higher mitochondrial membrane potential. At present, the molecular properties of hub versus follower beta-cells have not been determine on either the transcript or protein level, but novel single cell technologies (see below) will hopefully soon be able to answer these important outstanding questions.
A different approach to islets cell heterogeneity was taken by Lickert’s group in Munich, who had been studying the planar cell polarity pathway in mouse islets in various transgenic mouse models. Planar cell polarity is the process that results in the collective directed orientation of cells within an epithelial plane, such as the defined orientation of hair cells in the inner air. In seminal work by Grapin-Botton and colleagues, it had been shown that the planar cell polarity (PCP) pathway is critical during embryonic development for the differentiation of endocrine cells from polarized progenitors [8]. To track the activity of the planar cell polarity pathway in islet cells throughout life, Lickert and colleagues derived a gene replacement allele at the Flattop (Fltp) locus, replacing a normal copy with the cDNA encoding bacterial β-galactosidase plus the Venus fluorescent protein [9]. Using this tool, they followed Venus protein expression as a surrogate for Fltp promoter activation, and found that the percentage of Venus-positive beta-cells increased during development, topping out at 80% of beta-cells in adult mice. The other endocrine cell types were also Fltp-Venus positive to varying degrees, suggesting that all endocrine cells display some heterogeneity, at least as this marker is concerned. It should be noted, however, that by design the Fltp-Venus allele produced a null mutation in the Fltp gene; thus, all data were obtained in heterozygous mice. Despite this caveat, the authors found that the minority of beta-cells that had not activated the Fltp-Venus allele had higher rates of beta-cell proliferation in very young, pre-weaning mice.
In an effort to identify the molecular underpinnings of the differences between Venus-positive and -negative cells, the authors performed a transcriptome analysis of sorted beta-cell populations. The global transcriptome analysis was consistent with Fltp-Venus positive cells exhibiting a more mature phenotype. Importantly, when enriched Fltp-Venus positive and negative beta-cells were re-aggregated and assayed in vitro for their insulin secretory behavior, the Fltp-Venus-positive pseudoislets secreted more insulin in response to glucose than their counterparts. In addition, at least in certain settings, the Fltp-Venus negative beta-cells displayed higher proliferation rates than those that were Fltp-Venus positive, such as during pregnancy, though not in non-pregnant adult mice. Differences in gene expression profiles of proliferating and quiescent beta-cells where previously reported by Dor and colleagues, who found that beta-cell function genes are relatively reduced in expression in replicating beta-cells [10].
A different approach for the study of beta-cell heterogeneity, this time in human islets, was taken by Grompe’s group, who screened for cell surface antibodies that could subdivide beta-cells in FACS analysis [11]. Using this approach, they discovered that human beta-cells elaborate different levels of the cell surface proteins ST8SIA1 and CD9, and can therefore be sorted into four sub-populations. Importantly, the relative abundance of the various subtype depended on metabolic state, with different frequencies being observed in type 2 diabetes. Thus, it is possible that the diabetic state causes a redistribution of the beta-cell subtypes, or even that altered subtype patterns contribute to the etiology of type 2 diabetes. It is important to note that at present it is unknown if these beta-cell subtypes represent stable or semi-stable cell types or more rapidly interchangeable states. This key question in the field of human beta-cell heterogenetity will have to be addressed using advanced lineage tracing. In the case of the mouse, it is clear, however, that Fltp-negative cells can convert to Fltp-positive cells over time [9].
The studies described above all have in common that they begin with a specific functional property of the beta-cell – i.e. calcium currents – or the expression levels of preselected makers – i.e. Flattop, CD9 or ST8SIA1. Thus, it is likely that unbiased, genome-wide approaches will uncover beta-cell heterogeneity to unprecedented detail and granularity. Fortunately, recent advances in single cell biology, in particular single cell transcriptomics, have already begun to uncover important features of beta cell heterogeneity.
3. Beta-cell heterogeneity: Insights from single-cell technologies
Multiple cutting-edge technologies have recently been employed to explore beta-cell heterogenetic. In one approach, Wang and colleagues employed mass cytometry, short for ‘single cell time-of-flight mass spectrometry’ also known as ‘CyTOF’, to determine the abundance of 24 proteins in single cells from human islets simultaneously [12]. Analyzing human islets from multiple donors, ranging in age from three month to 70 years, the authors confirmed that the replication rates of human islet cells decrease with age, with maximal rates of Ki67-positive of 4% for beta- and 7% for alpha cells in infants. Strikingly, they noted that the replication rate of alpha-cells is nearly twice that of beta- and delta-cells throughout life. Because the percentage of alpha-cells in the endocrine pancreas does not change appreciably with age, these findings suggest that either human alpha-cells have a higher rate of cell death than the other islet cell types, or that excess alpha-cells might serve as a source of ‘reserve’ cells for trans-differentiation into beta-cells. This is not as far-fetched as it may seem, given the strong evidence from rodent models that alpha-cells can be made to express beta-cell genes under certain experimental conditions [13, 14], and the fact that human alpha-cells retain many beta-cell specific genes in a ‘bivalent’ epigenetic state [15, 16]. Regarding beta-cell heterogeneity in itself, Wang and colleagues found that beta-cells displayed clear ‘substructure’ based on expression of the 24 proteins analyzed, with three to four subtypes detectable depending on the donor (see Figure 1 [12]). The great advantage of mass cytometry is that it does not suffer from the spectral overlap of the fluorophores employed for FACS, and can in principle detect up to 100 different heavy metal isotopes. Thus, with the development of many more heavy-metal conjugated antibodies, the technology promises to provide even more resolution in the future. Importantly, the platform has recently been adopted to tissue sections – termed ‘imaging mass cytometry’, with sub-cellular resolution and enabling in-depth phenotyping of cell-cell interactions in pancreatic tissue sections [17, 18]. In particular, the application of imaging mass cytometry to pancreatic samples from type 1 diabetics promises to map the interplay between the immune system and the endocrine pancreas in unprecedented detail.
Figure 1.
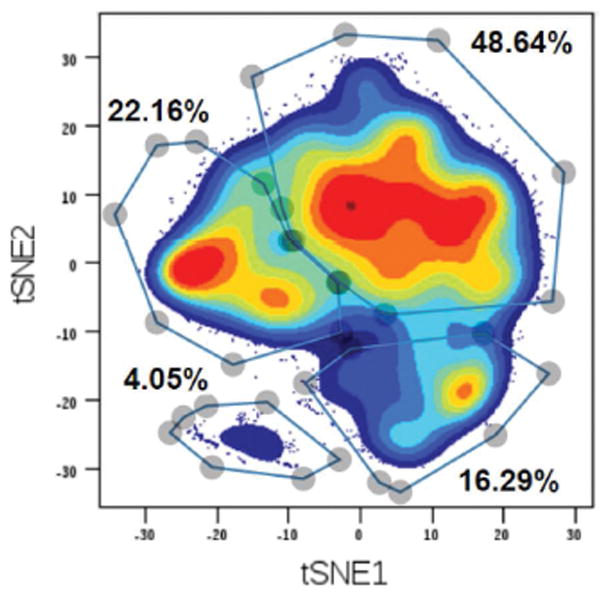
Mass cytometry defines four human beta-cells states or subtypes. Single cell suspensions were prepared from human islets and subjected to mass cytometry (CyTOF) with 24 heavy isotope-labeled antibodies. After gating for only C-peptide positive beta-cells, beta cells were next analyzed by the t-distributed stochastic neighbor embedding (t-SNE) algorithm for dimensionality reduction to visualized beta-cell subtypes. Colors indicate density of the cells, and subgroups were manually gated based on natural occurring groups. From Wang et al., 2016. Cell Metabolism (Reference 12), with permission.
Transcriptome analysis has undergone dramatic advances over the past five years, so that multiple methodologies now exist to determine mRNA levels on the single cell level near genome-wide. Because the initial capture of mRNA molecules and their conversion to cDNA is of limited efficacy, very low abundance transcripts cannot yet be determined with precision. Because these rare transcripts will – by chance – be captured in some cells but not in others, this can lead to an artefactual overestimation of heterogeneity among cells in a given population. It is expected that the efficacy of reverse transcription and initial amplification will be improved in the near future to minimize the impact of this issue. In addition, by computationally combining rare transcripts into gene sets, groups of genes can be assessed more accurately. Nevertheless, depending on methodology, up to 5,000 high and medium abundance mRNAs can be captured routinely even today, allowing single cell analysis with exceptional detail. There is an obvious trade-off between the depth of sequencing per cell and the number of cells analyzed in each single-cell transcriptome study, which will depend on the specific question to be answered in each experiment.
Over the past two years, several groups have employed single-cell RNA sequencing (scRNAseq) technologies to the analyses of mouse and human islets. Li and colleagues [19] were the first to publish single cell RNA sequencing, but only on 70 human islet cells. Thus, while they were able to determine the expression of known marker genes such as insulin and pre-pro-glucagon to determine major cell types, the number of cells analyzed was too limited to characterize cell subtypes. This study was quickly followed by single-cell transcriptomic publications with larger numbers of islet cells from control and T2D deceased organ donors [20–23]. Xin and colleagues found about 250 genes to be differentially expressed in T2D compared to control islets, with the differentially expressed genes active in different endocrine cell types, not just in beta-cells. Heterogeneity among beta-cells was not analyzed in this report. In contrast, Muraro and colleagues [21] did identify a small set of genes that were differentially expressed among beta-cells, SRXN1, SQSTM1, and the three ferritin subunits FTH1P3, FTH1, and FTL, which all are thought to function in the ER and oxidative stress response a [24, 25].
Two groups elected to sequence large numbers of cells at relatively low sequence depth. Baron and colleagues sequenced a large number of islet cells but at low coverage, and reported transcriptome variability among beta cells, in particular for genes associated with beta-cell maturation such as Urocortin 3 (UCND) and the ER stress response (DDIT3, HERPUD1, and HSPA5; [20]. Among ductal cells, Baron and colleagues found two expression profiles, which they related to the morphologically distinct cell types identified previously [26]. Thus, ductal cells that form the terminal duct are distinct from those that connect to the acinus (centro-acinar cells). Baron and colleagues used immunostaining to relate their transcriptome-driven subtypes to the spatial separation among the ductal and centro-acinar cells. Subpopulations among alpha-, beta- and acinar cells were reported by Segerstolpe and colleagues [22]. In particular, among alpha cells they found multiple proliferating cells from different donors, which were identified by high transcript levels for cell cycle-associated genes such as Ki67 or CENPF. Interestingly, steady-state mRNA levels for pre-pro-glucagon were not altered in the replicating cells, indicating either that proliferation does not require a whole-sale deactivation of alpha-cell maturation markers, or that the half-life of the pre-pro-glucagon mRNA is so long as to mask any transient decrease in its promoter activity. Using expression levels of five marker genes (RBP4, FFAR4/GPR120, ID1, ID2 and ID3) the authors subdivided beta-cells into five clusters, again supporting at least some level of gene expression heterogeneity among beta-cells.
We generated single cell transcriptomes of high sequencing depths from multiple organ donors, including children, adults, T1D and T2D [27]. The high sequencing depth allowed us to discovered many islet cells that display divergent expression patterns within each canonical endocrine cell type. Importantly, we found that insulin transcript levels can vary by more than 100-fold in human beta-cells, an effect that would be difficult to capture using immunostaining methods (See Figure 2). In a rare replicating alpha-cells, we found that multiple genes of the hedgehog signaling pathway were activated, demonstrating how single cell transcriptomics can implicate novel regulatory pathways in islet cell biology. Perhaps most strikingly, we found that in alpha- and beta-cells from type 2 diabetics, the gene expression profiles of juvenile endocrine cells were re-acquired, suggesting a partial dedifferentiation process.
Figure 2.
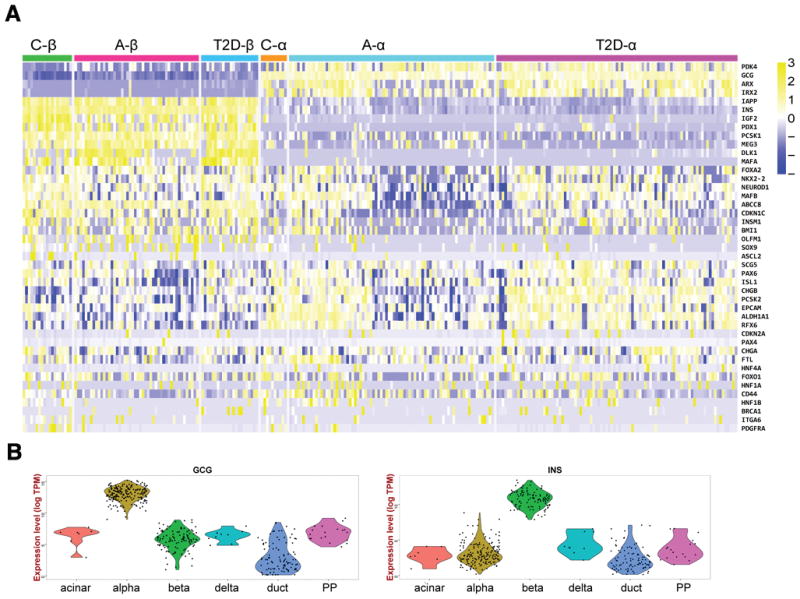
Single-cell RNA-seq of human islet cells identifies pancreatic cell types, gene signatures, and large expression level variation of hormone genes. Single-cell expression data from multiple human deceased organ donors. A: Heatmap showing hierarchical clustering of alpha- and beta-cells from different donor types. C, Child, A, Adult, T2D, type 2 diabetic. Note the the major cell types cluster together regardless of type of organ donor. Adult alpha-cells, but not alpha cells from T2D organ donors clearly display two subtypes based on transcript levels of multiple genes. B: Violin plots confirm that annotated alpha- and beta-cells have high expression of the signature hormone gene, but also establish that glucagon and insulin transcript levels can vary widely among alpha- and beta-cells, respectively. From Wang et al., Diabetes, 2016 (Reference 27). Copyright American Diabetes Association.
A topic of much discussion is the issue of islet cell plasticity, or the degree and frequency with which one adult endocrine cell type can convert into another. Data from mouse models, for instance with forced overexpression of key transcriptional regulators, have clearly demonstrated that islet cells can be forced to change fate [28]. One would imagine that single cell transcriptomics could rapidly determine to which extent transitory cells exist in human islets. Unfortunately, single cell transcriptomic studies are hampered by certain technical limitations, in particular the fact that the methods to capture single cells are not perfectly reliable, meaning that occasionally two cells are captured simultaneously. Therefore, the publications discussed above all employed stringent measures to exclude potential doublets, for instance by filtering out wells that express transcripts typically seen in alpha- and beta-cells. By definition, this filter could be removing data from a true doublet, but also from a cell that is half way through its trans-differentiation process from alpha- to beta-cell. Therefore, by applying stringent criteria, bi-hormonal cells, if they do exist, will be excluded from analysis. Novel tools will need to be brought to bear in order to address this critical question in islet biology.
4. Multihormonal islet cells
Among all the excitement about novel technologies, it should not be forgotten that biologists have for decades analyzed single islet cells in situ by immunohistochemical and immunofluorescent means. Of course, this type of analysis is limited by the number of proteins that can be assayed simultaneously (about four, as the channels available in most confocal microscropes) and issues of sensitivity, background, autofluorescence, and dynamic range of the assay. For instance, endocrine cells positive form more than one of the canonical islet hormones have been observed multiple times. In mice, it appears that double hormone positive are present during fetal life but eliminated by birth [29]. In the human pancreas, double hormone positive cells appear to be more persistent, and their frequency has been reported to increase in type 2 diabetes, and suggested as a new mechanism of beta cell failure via de-differentiation [30]. This notion found further support by the recent finding by Dahan and colleagues that a fraction of beta- and delta-cells activate the gastrin promoter in type 2 diabetes [31]. In addition, islet cell fate is often dependent on the continued presence of even a single key transcription factor, as seen in mutation that lead to loss of the precise beta-cell phenotype by co-expression of other hormones or even cell fate conversion [32–35]. Thus, evidence is accumulating from mouse and human studies that islet cells retain at least a minimal degree of plasticity even in adulthood. At present, it appears that only a small subset of islet cells changes fate under conditions of metabolic stress (transcription factor null mutations playing only a very limited role in human disease). A critical question thus arises: is islet cell plasticity inherent to only a small number of cells, perhaps retained from a fetal precursor state, or can all islet cells interconvert given the appropriate stimulus? Only by answering this key question can we hope to harness islet cell plasticity to therapeutic gain.
As introduced briefly above with the example of imaging mass cytometry, medium and high throughput methodologies are begging to be applied with spatial resolution. Exciting recent advances include “spatial genomics” [36] and “spatial proteomics” [37, 38]. In addition to in situ mass spectrometry, the multiplexing of single molecule RNA fluorescence in situ hybridization promising to allow for the simultaneous detection and quantification of dozens or even hundreds of mRNA species in tissue sections [39, 40]. Undoubtedly, these novel methodologies will uncover important new features of islet cell biology, including endocrine cell heterogeneity and plasticity. On the other hand, single cell genomics will be advanced further through the application of single cell ATACseq for the mapping of open chromatin states, and single cell DNA methylation analysis to islet cells.
5. Summary and Conclusions
The past five years have brought us great progress in our understanding of beta-cell heterogeneity on both the functional and molecular levels, and give us a glimpse of what the future will bring, when single cell proteomic, transcriptomic, and epigenomic technologies are applied to large numbers of islet cells from multiple organ donors with different pathologies. It appears highly likely that we will be able to obtain a detailed molecular understanding of very rare human islet cells, such as those undergoing transdifferentiation into another hormone phenotype, those undergoing ER stress or apoptosis, and those that are actively proliferating, and especially to what degree different beta-cell subtypes contribute to the etiology of diabetes. One could envision, for instance, that the molecular analysis of replicating human beta-cells will reveal unique vulnerabilities that prevent productive expansion of insulin-producing cells under conditions of high metabolic demand. If these vulnerabilities, such as increased susceptibility to oxidative stress, could be targeted, an effective increase in beta-cell mass might become achievable for the treatment of type 2 diabetes. While we cannot see into the future and predict what we will find when we study the molecular heterogeneity of the human endocrine pancreas in ever increasing detail, one thing is certain: we are entering an exciting new phase of discovery in islet biology.
Acknowledgments
Funding Information
Related work in our labs is supported through NIH grants UC4DK104119 (to K.H.K., D.A., and B.G.) and UC4DK112217 (to K.H.K.), the BIRAX Regenerative Medicine Initiative (14BX14NHBG to B.G.) and Israel Science Foundation - Juvenile Diabetes Research Foundation Joint Program in Type 1 Diabetes Research (1506/12, to B.G.).
Callouts
“Single cell technologies are revolutionizing our understanding of islet cell biology”
“Novel proteomic data indicate the existence of at least four subtypes or states of human beta-cells”
“Optogenetic studies have established the existence of pacemaker beta-cells in the mouse islet”
Footnotes
The authors have no conflicts of interest to declare.
References
- 1.Salomon D, Meda P. Heterogeneity and contact-dependent regulation of hormone secretion by individual B cells. Experimental cell research. 1986;162:507–520. doi: 10.1016/0014-4827(86)90354-x. [DOI] [PubMed] [Google Scholar]
- 2.Bosco D, Meda P. Actively synthesizing beta-cells secrete preferentially after glucose stimulation. Endocrinology. 1991;129:3157–3166. doi: 10.1210/endo-129-6-3157. [DOI] [PubMed] [Google Scholar]
- 3.Pipeleers D, Kiekens R, Ling Z, Wilikens A, Schuit F. Physiologic relevance of heterogeneity in the pancreatic beta-cell population. Diabetologia. 1994;37(Suppl 2):S57–64. doi: 10.1007/BF00400827. [DOI] [PubMed] [Google Scholar]
- 4.Van Schravendijk CF, Kiekens R, Pipeleers DG. Pancreatic beta cell heterogeneity in glucose-induced insulin secretion. The Journal of biological chemistry. 1992;267:21344–21348. [PubMed] [Google Scholar]
- 5.Pipeleers DG. Heterogeneity in pancreatic beta-cell population. Diabetes. 1992;41:777–781. doi: 10.2337/diab.41.7.777. [DOI] [PubMed] [Google Scholar]
- 6.Hodson DJ, Mitchell RK, Bellomo EA, et al. Lipotoxicity disrupts incretin-regulated human β cell connectivity. The Journal of clinical investigation. 2013;123:4182–4194. doi: 10.1172/JCI68459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston NR, Mitchell RK, Haythorne E, et al. Beta Cell Hubs Dictate Pancreatic Islet Responses to Glucose. Cell metabolism. 2016;24:389–401. doi: 10.1016/j.cmet.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortijo C, Gouzi M, Tissir F, Grapin-Botton A. Planar cell polarity controls pancreatic beta cell differentiation and glucose homeostasis. Cell Rep. 2012;2:1593–1606. doi: 10.1016/j.celrep.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bader E, Migliorini A, Gegg M, et al. Identification of proliferative and mature β-cells in the islets of Langerhans. Nature. 2016;535:430–434. doi: 10.1038/nature18624. [DOI] [PubMed] [Google Scholar]
- 10.Klochendler A, Caspi I, Corem N, et al. The Genetic Program of Pancreatic beta-Cell Replication In Vivo. Diabetes. 2016;65:2081–2093. doi: 10.2337/db16-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorrell C, Schug J, Canaday PS, et al. Human islets contain four distinct subtypes of β cells. Nature communications. 2016;7:11756. doi: 10.1038/ncomms11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang YJ, Golson ML, Schug J, et al. Single-Cell Mass Cytometry Analysis of the Human Endocrine Pancreas. Cell Metabolism. 2016;24:616–626. doi: 10.1016/j.cmet.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chera S, Baronnier D, Ghila L, et al. Diabetes recovery by age-dependent conversion of pancreatic δ-cells into insulin producers. Nature. 2014;514:503–507. doi: 10.1038/nature13633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorel F, Népote V, Avril I, et al. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 2010;464:1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bramswig NC, Everett LJ, Schug J, et al. Epigenomic plasticity enables human pancreatic alpha to beta cell reprogramming. J Clin Invest. 2013;123:1275–1284. doi: 10.1172/JCI66514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bramswig NC, Kaestner KH. Transcriptional and epigenetic regulation in human islets. Diabetologia. 2014;57:451–454. doi: 10.1007/s00125-013-3150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giesen C, Wang HA, Schapiro D, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11:417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 18.Schuffler PJ, Schapiro D, Giesen C, Wang HA, Bodenmiller B, Buhmann JM. Automatic single cell segmentation on highly multiplexed tissue images. Cytometry A. 2015;87:936–942. doi: 10.1002/cyto.a.22702. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Klughammer J, Farlik M, et al. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO reports. 2016;17:178–187. doi: 10.15252/embr.201540946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baron M, Veres A, Wolock SL, et al. A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure. Cell Systems. 2016:1–35. doi: 10.1016/j.cels.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muraro MJ, Dharmadhikari G, Grün D, et al. A Single-Cell Transcriptome Atlas of the Human Pancreas. Cell Systems. 2016;3:385–394. e383. doi: 10.1016/j.cels.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segerstolpe Å, Palasantza A, Eliasson P. Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes. Cell metabolism. 2016 doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xin Y, Kim J, Okamoto H, Ni M, Wei Y, Adler C. RNA Sequencing of Single Human Islet Cells Reveals Type 2 Diabetes Genes. Cell metabolism. 2016 doi: 10.1016/j.cmet.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 24.Orino K, Lehman L, Tsuji Y, Ayaki H, Torti SV, Torti FM. Ferritin and the response to oxidative stress. The Biochemical journal. 2001;357:241–247. doi: 10.1042/0264-6021:3570241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y, Duan S, Zhou Y, et al. Sulfiredoxin-1 attenuates oxidative stress via Nrf2/ARE pathway and 2-Cys Prdxs after oxygen-glucose deprivation in astrocytes. J Mol Neurosci. 2015;55:941–950. doi: 10.1007/s12031-014-0449-6. [DOI] [PubMed] [Google Scholar]
- 26.Rovira M, Scott SG, Liss AS, Jensen J, Thayer SP, Leach SD. Isolation and characterization of centroacinar/terminal ductal progenitor cells in adult mouse pancreas. Proc Natl Acad Sci U S A. 2010;107:75–80. doi: 10.1073/pnas.0912589107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang YJ, Schug J, Won K-J, et al. Single-Cell Transcriptomics of the Human Endocrine Pancreas. Diabetes. 2016;65:3028–3038. doi: 10.2337/db16-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang YP, Thorel F, Boyer DF, Herrera PL, Wright CV. Context-specific alpha- to-beta-cell reprogramming by forced Pdx1 expression. Genes Dev. 2011;25:1680–1685. doi: 10.1101/gad.16875711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrera PL. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development. 2000;127:2317–2322. doi: 10.1242/dev.127.11.2317. [DOI] [PubMed] [Google Scholar]
- 30.Talchai C, Xuan S, Lin HV, Sussel L, Accili D. Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell. 2012;150:1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dahan T, Ziv O, Horwitz E, et al. Pancreatic beta-Cells Express the Fetal Islet Hormone Gastrin in Rodent and Human Diabetes. Diabetes. 2017;66:426–436. doi: 10.2337/db16-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ediger BN, Lim HW, Juliana C, et al. LIM domain-binding 1 maintains the terminally differentiated state of pancreatic beta cells. J Clin Invest. 2017;127:215–229. doi: 10.1172/JCI88016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gutierrez GD, Bender AS, Cirulli V, et al. Pancreatic beta cell identity requires continual repression of non-beta cell programs. J Clin Invest. 2017;127:244–259. doi: 10.1172/JCI88017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swisa A, Avrahami D, Eden N, et al. PAX6 maintains beta cell identity by repressing genes of alternative islet cell types. J Clin Invest. 2017;127:230–243. doi: 10.1172/JCI88015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, McKenna LB, Bogue CW, Kaestner KH. The diabetes gene Hhex maintains delta-cell differentiation and islet function. Gene Dev. 2014;28:829–834. doi: 10.1101/gad.235499.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stahl PL, Salmen F, Vickovic S, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 37.Rizzo DG, Prentice BM, Moore JL, Norris JL, Caprioli RM. Enhanced Spatially Resolved Proteomics Using On-Tissue Hydrogel-Mediated Protein Digestion. Anal Chem. 2017;89:2948–2955. doi: 10.1021/acs.analchem.6b04395. [DOI] [PubMed] [Google Scholar]
- 38.Spraggins JM, Rizzo DG, Moore JL, Noto MJ, Skaar EP, Caprioli RM. Next-generation technologies for spatial proteomics: Integrating ultra-high speed MALDI-TOF and high mass resolution MALDI FTICR imaging mass spectrometry for protein analysis. Proteomics. 2016;16:1678–1689. doi: 10.1002/pmic.201600003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shah S, Lubeck E, Schwarzkopf M, et al. Single-molecule RNA detection at depth by hybridization chain reaction and tissue hydrogel embedding and clearing. Development. 2016;143:2862–2867. doi: 10.1242/dev.138560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah S, Lubeck E, Zhou W, Cai L. In Situ Transcription Profiling of Single Cells Reveals Spatial Organization of Cells in the Mouse Hippocampus. Neuron. 2016;92:342–357. doi: 10.1016/j.neuron.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


