Abstract
Objectives
Tamoxifen bioactivation to endoxifen is primarily mediated by CYP2D6; however, substantial variability remains unexplained. Our objective was to conduct a comprehensive assessment of the effect of genetic variation in tamoxifen-relevant enzymes and transporters on steady-state endoxifen concentrations.
Methods
Comprehensive genotyping of CYP enzymes and transporters was conducted using the iPLEX® ADME PGx Pro Panel in 302 tamoxifen treated breast cancer patients. Predicted activity phenotype for 19 enzymes and transporters were analyzed for univariate association with endoxifen concentration, and then adjusted for CYP2D6 and clinical covariates.
Results
In univariate analysis, higher activity of CYP2C8 (regression β=0.22, p=0.020) and CYP2C9 (β=0.20, p=0.04), lower body weight (β=−0.014, p<0.0001), and endoxifen measurement during winter (each β<−0.39, p=0.002), were associated with higher endoxifen concentration. After adjustment for CYP2D6 diplotype, weight and season, CYP2C9 remained significantly associated with higher concentration (p=0.02), but only increased the overall model R-squared by 1.3%.
Conclusions
Our results further support a minor contribution of CYP2C9 genetic variability on steady state endoxifen concentration. Integration of clinician and genetic variables into individualized tamoxifen dosing algorithms would marginally improve their accuracy, and potentially enhance tamoxifen treatment outcomes.
Keywords: tamoxifen, endoxifen, pharmacogenomics, CYP2C9, CYP2D6, season
Introduction
Tamoxifen, a selective estrogen receptor modulator, is used to prevent the recurrence of estrogen receptor positive (ER+) breast cancer in the adjuvant treatment of pre- and post-menopausal women, as well as for treatment of metastatic disease and ductal carcinoma in situ[1]. Tamoxifen is metabolized to several metabolites with greater anti-estrogenic potency including 4-hydroxy-tamoxifen and endoxifen[2]. Reaching therapeutic concentrations of these metabolites, particularly endoxifen, may be critical to the efficacy of tamoxifen treatment; patients above a therapeutic threshold have been reported to have lower recurrence rates than women with lower concentrations[3, 4].
The enzyme primarily responsible for metabolic conversion of tamoxifen to endoxifen is CYP2D6, but other enzymes and transporters are also active in this metabolic pathway[5]. Genetic variation in CYP2D6 has a profound effect on endoxifen serum concentrations as it is the main pathway for the conversion of the primary tamoxifen metabolite, N-desmethyltamoxifen to endoxifen, and substantially contributes to the formation of 4-OH tamoxifen from tamoxifen[3][6]. Patients with genotypes that confer CYP2D6 poor metabolizer (PM) phenotype have lower endoxifen concentrations than those with intermediate (IM), normal (NM, formerly referred to as extensive [EM]) or ultra-rapid (UM) metabolizer status[7].
Aside from CYP2D6, there are several enzymes and transporters known to play a role in the metabolism and/or distribution of tamoxifen and endoxifen. Prior studies have reported associations between tamoxifen and/or its metabolites for common functionally consequential single nucleotide polymorphisms (SNPs) in CYP2C9[4, 5, 8]and CYP2C19[4, 8, 9]. In addition to genetics, there are some patient specific factors such as height, weight, and the calendar date that may contribute to steady state endoxifen concentrations[10]. It may be possible to use genetic and clinical information to predict a patient’s endoxifen concentration prior to initiation, and use this information to guide personalized tamoxifen dose adjustment to achieve therapeutic endoxifen concentrations. Several groups, including ours, have piloted this approach by dose escalating patients with low-activity CYP2D6 genotypes and have made progress towards normalizing endoxifen concentrations without increasing treatment related toxicity[2, 11–14]. In addition, one study dose escalated patients solely based on baseline endoxifen[15]; however, these studies have been unable to achieve comparable endoxifen levels in CYP2D6 PM patients suggesting other genetic data and clinical variables need to be incorporated into a prediction algorithm.
Further research is needed to determine the impact of SNPs in enzymes and transporters on endoxifen concentrations in order to improve the accuracy of personalized tamoxifen dosing algorithms. Prior pharmacogenetic studies have used a candidate SNP approach with limited allelic coverage. Our objective was to use a comprehensive genotyping approach that includes the vast majority of functionally consequential SNPs in the genes relevant to endoxifen pharmacokinetics (CYP2C9, CYP2C19, CYP3A4, CYP3A5)[4, 5, 8, 9, 16–20] to discover genetic predictors of steady-state endoxifen concentration in patients receiving tamoxifen treatment. We also included additional enzymes and transporters that have insufficient available information to rule out their involvement in endoxifen concentrations, several of which have been previously associated with tamoxifen efficacy and/or toxicity[21–23].
Methods
Patient Cohort
Patients included in this secondary analysis were originally enrolled in a prospective clinical study of CYP2D6 genotype-guided tamoxifen dose escalation (UNC Lineberger Comprehensive Cancer Center 0801)[2]. Detailed inclusion and exclusion criteria were previously reported, but briefly: women 18 years of age and older and taking tamoxifen 20 mg for at least 4 months prior to study entry were eligible to enroll unless they had Eastern Cooperative Oncology Group (ECOG) performance status >2, impaired liver, bone marrow or kidney function, history of thromboembolic disease, prior liver or bone marrow transplant, received blood transfusions within 3 months of study entry, or were taking strong CYP2D6 inhibitors. Patients taking medications known to inhibit CYP2D6 including amiodarone, bupropion, cimetidine, haloperidol, indinavir, ritonavir, terbinafine, quinidine, duloxetine, paroxetine, or fluoxetine were excluded. Those taking weak inhibitors of CYP2D6 (venlafaxine, citalopram, sertraline, or escitalopram) were included. After an initial pilot study of 120 patients[2] the study was expanded to a total of 500 participants[24].
Sample and Data Collection
Patient demographic and treatment information including age, weight, menopausal status, self-reported race, duration of tamoxifen therapy, and concomitant medication were collected at study entry. Genomic DNA was isolated from the blood samples collected at entry into the clinical trial for germline genotyping. Baseline tamoxifen and metabolite concentrations were measured at baseline when all patients were taking tamoxifen 20 mg daily and had reached steady state. The following metabolites were measured in addition to tamoxifen: 4-OH-tamoxifen, N-desmethyl tamoxifen, and endoxifen. Concentrations were determined by liquid chromatography-mass spectrometry by collaborators at Indiana University using a previously described assay[2].
Genotyping and Activity Phenotype Prediction
At study enrollment, CYP2D6 status was determined for each participant using the Roche AmpliChip™ CYP450 Test (Indianapolis, IN). Genotype was categorized into diplotypes (i.e. NM/IM, NM/PM, IM/IM, IM/PM) and then phenotypes (UM, NM, IM, PM) as previously described[25]. Phenotypes UM and NM were combined due to small number of UM phenotypes and prior work in the cohort that confirmed differences in activity are negligible[25]. Residual DNA samples were genotyped for 36 additional genes, primarily CYP450 enzymes and drug transporters, on the iPLEX® ADME PGx Pro Panel by Agena Bioscience (San Diego, CA) for this secondary pharmacogenetic analysis. All genetic information underwent appropriate quality control. SNPs with genotyping call rate <95% and samples with a call rate of <90% across all genotyped SNPs were excluded from analysis. Of the 36 genes genotyped on the panel, the following 19 genes were chosen to be included in the analysis based on their potential contribution to tamoxifen metabolism or transport: CYP2C9, CYP3A4, CYP3A5, ABCB1, SLOCO1B1, SULT1A1, SULT1A2, UGT2B7, UGT2B15, UGT2B17, CYP1A1, CYP1A2, CYP2A6, CYP2B6, CYP2C19, CYP2C8, CYP2E1, ABCC2 and ABCG2. Each allele was assigned a predicted activity determination (low, normal, high) based on published data and each patient’s activity diplotype was translated into predicted activity phenotypes (poor, intermediate, normal or ultra-rapid) for analysis, as described in Supplemental Digital Content, Tables 1 and 2.
Statistical Analysis
Patients who were successfully genotyped and had baseline endoxifen concentration data measured on the modified assay used for the expansion cohort were included in the analysis. Steady state endoxifen concentrations underwent square root transformation to improve distributional normality. Predicted activity for each enzyme or transporter was analyzed assuming an additive effect with the number of levels (2, 3 or 4) corresponding to the number of phenotypes described in Supplemental Digital Content, Table 2. Clinical variables analyzed included age and weight as continuous variables, season of sample collection (January–March, April–June, July–September, October–December), CYP2D6 inhibitor status (none or weak CYP2D6 inhibitor), menopausal status (pre- or post-menopausal), and self-reported ethnicity (white, African-American, or other) as categorical variables. Associations between predicted activity phenotype or clinical variables and baseline steady state concentration for tamoxifen and its metabolites were analyzed using univariate linear regression. Following univariate analysis, genetic associations were adjusted for clinical covariates that were significant in the univariate setting. Multivariable models were constructed with each combination of CYP2C8, CYP2C9, and CYP2C19 activity phenotypes to assess the independent contribution of each gene. The most parsimonious model was selected based on Akaike Information Criterion (AIC), and R-squared values were estimated from the linear regression models. As a hypothesis generating secondary screen, the association of each predicted activity phenotype with tamoxifen, its other metabolites (4-OH-tamoxifen and N-desmethyl-tamoxifen) and their metabolic ratios was evaluated. An uncorrected p-value threshold of p<0.05 was considered statistically significant for all analyses, which were conducted in SAS v9.4 statistical software (Cary, NC).
Results
Patient and Demographic Data
After excluding subjects that were missing endoxifen concentration or genetic data, 302 patients were included in the final analysis (Figure 1). Patient demographic data including age, weight, race, date of sample collection, CYP2D6 diplotype, and concomitant use of a CYP2D6 inhibitor (i.e. citalopram, escitalopram, sertraline or venlafaxine) can be found in Table 1. The average age in this cohort was 52.8, 85% of the patients self-reported as Caucasian, and 45% were pre-menopausal. The CYP2D6 phenotype distribution was similar to previously reported in this cohort with 37% having NM phenotype (NM/NM diplotype), 4% having PM phenotype (PM/PM diplotype) and the remaining ~60% being IM phenotype that can be subdivided into several diplotypes as reported in Table 1[25].
Figure 1. CONSORT Diagram of Patient Matriculation from Clinical Trial to this Pharmacogenetic Analysis.
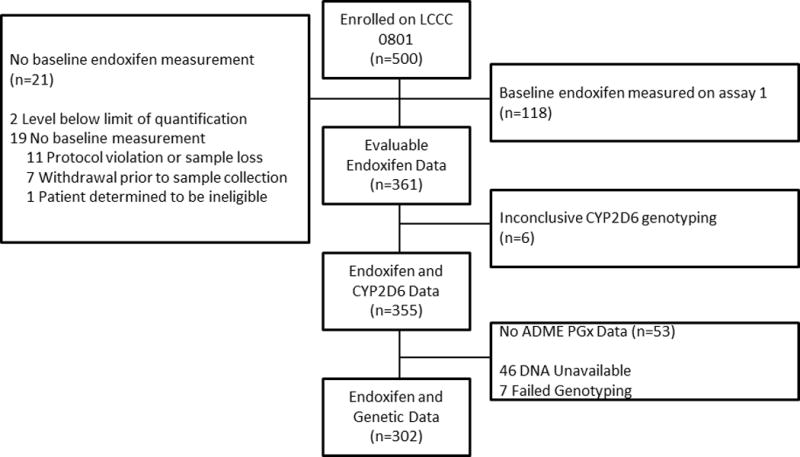
Of the 500 patients enrolled in the original clinical trial, 361 had evaluable endoxifen data, 355 had usable CYP2D6 genetic information, and 302 were successfully genotyped on the ADME PGx Pro Panel and were included in this analysis.
Table 1.
Patient Demographics
| Clinical Variable | Level | Median (range) or n (%) |
|---|---|---|
| Age | Years | 52.8 (24.8–95.1) |
| Weight | Kg | 70.8 (32.1–149.1) |
| Self-Reported Race | Caucasian | 253 (84.6%) |
| African American | 38 (12.7%) | |
| Other/Unknown | 8 (2.7%) | |
| Season of Baseline Sample Collection for Endoxifen Measurement | Winter (Jan–Mar) | 58 (19.4%) |
| Spring (April–June) | 67 (22.3%) | |
| Summer (July–Sept) | 61 (20.3%) | |
| Fall (Oct–Dec) | 114 (38.0%) | |
| Concomitant CYP2D6 Weak Inhibitor Use | Yes (total) citalopram escitalopram sertraline |
23 (7.7%) 12 (4.0%) 10 (3.3%) 1 (0.33%) |
| No | 277 (92.3%) | |
| Menopausal Status | Pre-menopausal | 134 (44.7%) |
| Post-menopausal | 166 (55.3%) | |
| Time on Tamoxifen Prior to Enrollment | Years | 0.8 (0.3–9.7) |
| CYP2D6 Diplotype as defined in[26] (UM Alleles Considered NM alleles) | NM/NM | 110 (37%) |
| NM/IM | 58 (19.5%) | |
| NM/PM | 79 (26.6%) | |
| IM/IM | 10 (3.4%) | |
| IM/PM | 27 (9.1%) | |
| PM/PM | 13 (4.4%) |
Univariate Associations of Clinical and Genetic Data
In the univariate analysis, an increase in steady-state endoxifen concentration was detected for patients with increased CYP2C9 phenotype activity (Linear Regression β coefficient=0.20, 95% Confidence Interval: 0.01–0.39, p=0.04) and CYP2C8 phenotype activity (β=0.22, 95% CI: 0.03–0.40, p=0.02, Table 2, Figure 2). None of the other genes analyzed were associated with endoxifen concentration in the univariate analysis, though there was a suggestive trend for SLCO1B1 (p=0.07, Table 2) and CYP2C19 (p=0.11, Table 2, Figure 2).
Table 2.
Mean Endoxifen Concentration by Metabolizer Status for All Genes Included in Analysis
| Mean Endoxifen (Std. Dev.) Stratified by Predicted Activity Phenotype in ng/mL | Univariate and Adjusted Association of Each Gene with Endoxifen Concentration | |||||
|---|---|---|---|---|---|---|
| Gene | Poor | Intermediate | Normal | Ultra-rapid | Unadjusted Association | Adjusted Association* |
| CYP1A1 | 3.19 (1.53) n=2 |
7.07 (4.66) n=52 |
8.05 (5.51) n=248 |
β = 0.20 (−0.06, 0.46) p=0.13 |
β = 0.18 (−0.05, 0.42) p=0.12 |
|
| CYP1A2 | 9.57 (6.18) n=6 |
6.15 (3.76) n=22 |
8.13 (4.48) n=22 |
7.93 (5.53) n=252 |
β = 0.04 (−0.11, 0.20) p=0.61 |
β =−0.02 (−0.16, 0.12) p=0.80 |
| CYP2A6 | 7.46 (9.71) n=5 |
7.65 (4.50) n=74 |
7.95 (5.57) n=220 |
B = 0.03 (−0.19, 0.25) p=0.79 |
β = 0.13 (−0.06, 0.33) p=0.18 |
|
| CYP3A4 | NA n=0 |
9.28 (4.99) n=14 |
7.78 (5.39) n=288 |
β =−0.33 (−0.83, 0.18) p=0.20 |
β =−0.35 (−0.80, 0.11) p=0.13 |
|
| CYP3A5 | 7.71 (5.29) n=240 |
8.35 (5.40) n=54 |
8.51 (7.70) n=8 |
β = 0.06 (−0.16, 0.28) p=0.58 |
β = 0.05 (−0.14, 0.24) p=0.61 |
|
| CYP2B6 | 8.20 (4.81) n=24 |
7.48 (5.53) n=127 |
8.11 (5.33) n=151 |
β = 0.06 (−0.11, 0.22) p=0.50 |
β = 0.01 (−0.14, 0.16) p=0.94 |
|
| CYP2C8 | 6.71 (5.04) n=13 |
6.73 (4.29) n=84 |
8.38 (5.72) n=205 |
β = 0.22 (0.03, 0.40) p=0.02 |
β = 0.13 (−0.03, 0.30) p=0.11 |
|
| CYP2C9 | 6.26 (4.82) n=9 |
7.18 (4.90) n=104 |
8.29 (5.61) n=189 |
β = 0.20 (0.01, 0.39) p=0.04 |
β = 0.20 (0.03, 0.37) p=0.02 |
|
| CYP2C19 | 7.95 (5.66) n=13 |
7.25 (4.72) n=91 |
7.64 (5.97) n=109 |
8.71 (5.16) n=89 |
β = 0.10 (−0.02, 0.22) p=0.11 |
β = 0.10 (−0.01, 0.21) p=0.07 |
| CYP2E1 | 8.27 (5.28) n=23 |
8.03 (5.40) n=84 |
7.72 (5.39) n=195 |
β =−0.05 (−0.22, 0.11) p=0.53 |
β =−0.10 (−0.26, 0.05) p=0.18 |
|
| ABCB1 | 8.93 (7.53) n=37 |
7.90 (5.15) n=205 |
7.01 (4.41) n=60 |
β =−0.13 (−0.31, 0.06) p=0.19 |
β =−0.01 (−0.18, 0.16) p=0.92 |
|
| ABCC2 | 9.30 (5.34) n=10 |
7.52 (5.42) n=69 |
8.29 (4.99) n=89 |
7.62 (5.61) n=134 |
β =−0.03 (−0.15, 0.09) p=0.63 |
β =−0.02 (−0.13, 0.08) p=0.65 |
| ABCG2 | 12.82 (10.85) n=3 |
7.30 (4.28) n=50 |
7.90 (5.48) n=249 |
β = 0.01 (−0.25, 0.26) p=0.96 |
β =−0.08 (−0.31, 0.15) p=0.51 |
|
| SLCO1B1 | 6.97 (4.21) n=62 |
7.76 (5.64) n=153 |
8.63 (5.57) n=87 |
β = 0.14 (−0.01, 0.29) p=0.07 |
β = 0.09 (−0.04, 0.22) p=0.18 |
|
| SULT1A1 | 7.81 (6.59) n=45 |
8.35 (5.56) n=102 |
7.56 (4.51) n=111 |
7.45 (5.65) n=44 |
β =−0.04 (−0.16, 0.07) p=0.49 |
β =−0.00 (−0.11, 0.10) p=0.94 |
| SULT1A2 | 7.72 (5.70) n=74 |
7.86 (5.31) n=144 |
7.94 (5.23) n=84 |
β = 0.01 (−0.14, 0.16) p=0.88 |
β = 0.06 (−0.07, 0.19) p=0.40 |
|
| UGT2B7 | 8.38 (5.36) n=63 |
7.97 (5.49) n=166 |
7.11 (5.09) n=73 |
β =−0.12 (−0.28, 0.03) p=0.12 |
β =−0.11 (−0.25, 0.03) p=0.12 |
|
| UGT2B15 | 7.75 (4.48) n=75 |
7.88 (5.61) n=149 |
8.07 (5.82) n=71 |
β =−0.00 (−0.15, 0.15) p=0.98 |
β = 0.04 (−0.09, 0.18) p=0.52 |
|
| UGT2B17 | 6.60 (5.48) n=35 |
8.27 (5.45) n=138 |
7.83 (5.25) n=122 |
β = 0.08 (−0.08, 0.24) p=0.30 |
β = 0.06 (−0.08, 0.20) p=0.41 |
|
P-values were adjusted for season, weight, and CYP2D6 diplotype
Gray boxes denote phenotype groups that did not exist for that gene (See Supplementary Table 2)
Figure 2. Endoxifen Concentration by CYP2C8, CYP2C9, and CYP2C19 Activity.
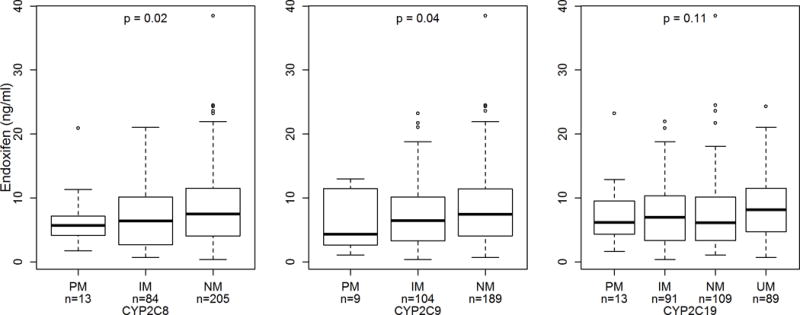
Box and whisker plot (circles represent outliers) of steady-state endoxifen concentration stratified by predicted activity phenotype for CYP2C8 (left), CYP2C9 (middle), and CYP2C19 (right). Endoxifen concentration increased as predicted phenotypic activity increased for each gene (p=0.02, p=0.04, p=0.11, respectively).
A significant association was found with higher weight associated with lower endoxifen concentration (β=−0.014, 95% CI: −0.020 – −0.008, p<0.0001, Figure 3). We also found that patients whose samples were collected in the fall (β=−0.55, 95% CI: −0.84 – −0.26, p=0.0002), summer (β=−0.55, 95% CI: −0.88 – −0.23, p=0.0009) or spring (β=−0.39, 95% CI: −0.71 – −0.07, p=0.02) had significantly lower endoxifen concentration compared with samples collected in the winter (Figure 3). No evidence of an association was found with other clinical variables: age, race, menopausal status, or concomitant administration of weak CYP2D6 inhibitors (Table 3).
Figure 3. Association of Endoxifen Concentration with Clinical Variables.
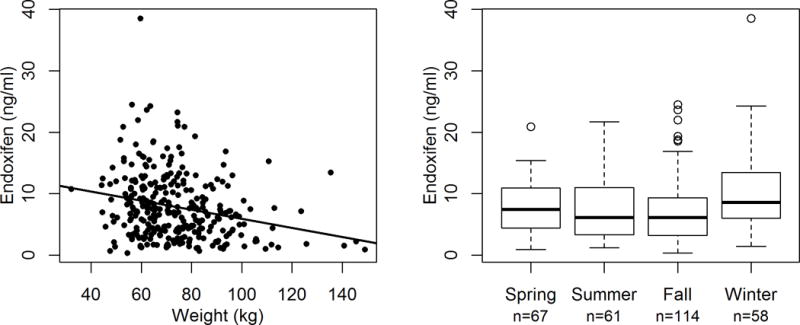
Steady-state endoxifen concentration was associated with body weight and the season of sample collection. Figure 3 (left): Increased patient body weight (kg) was associated with lower endoxifen concentrations (β=−0.014, p<0.0001). Figure 3 (right): Endoxifen concentrations were lower in samples collected during fall (β=−0.55, p=0.0002) summer (β=−0.55, p=0.0009) and spring (β=−0.39, p=0.02) compared with those collected during winter.
Table 3.
Univariate Association of Clinical Variables with Endoxifen Concentration
| Clinical Variable | B Coefficient (95% Confidence Interval) | P-value | |
|---|---|---|---|
| Weight (kg) | −0.014 (−0.020, −0.008) | < 0.0001 | |
| Age (10 years) | 0.05 (−0.05,0.14) | 0.31 | |
| Self-reported Race (vs. white) | African American | −0.13 (−0.45,0.18) | 0.41 |
| Other | 0.15 (−0.50,0.81) | 0.65 | |
| Season of Sample Collection (vs. winter) | Fall | −0.55 (−0.84, −0.26) | 0.0002 |
| Summer | −0.55 (−0.88, −0.23) | 0.0009 | |
| Spring | −0.39 (−0.71, −0.07) | 0.02 | |
| Post-Menopausal (vs. pre) | 0.14 (−0.08,0.35) | 0.21 | |
| Concomitant CYP2D6 Weak Inhibitor Use | 0.33 (−0.07,0.73) | 0.11 | |
Adjusted Associations for Genetic Variables
After adjustment for clinical variables with significant univariate associations (CYP2D6 diplotype, weight and season) the association for CYP2C9 maintained significance (p=0.02) while the association for CYP2C8 was no longer significant (p=0.11). Interestingly, the association for CYP2C19 was nearly significant after adjustment (p=0.07) whereas the trend for SLCO1B1 disappeared (p=0.18, Table 2).
The trends seen with CYP2C8, CYP2C9, and CYP2C19 are likely due to known linkage disequilibrium between the high activity CYP2C19*17 and the wild-type CYP2C8 and CYP2C9 (D’>0.9)[26]. Therefore, we constructed multivariable models including different combinations of the three genes adjusted for CYP2D6, weight, and season (Supplemental Digital Content, Table 3). Comparing the goodness of fit statistics (AIC), the optimal model included only CYP2C9 (p=0.002, AIC 728.2378, Supplemental Digital Content, Table 3). CYP2C9 maintained significance when included in a model with CYP2C19 (p=0.045) and was borderline significant in a model with CYP2C19 and CYP2C8 (p=0.07), but neither of these other genes maintained significance when included in a model with CYP2C9 (all p>0.05), suggesting that CYP2C9 drives the identified associations for all three genes. In the final multivariable model, the addition of CYP2D6 diplotype explained 15.3% of the variability in steady-state endoxifen concentration while CYP2C9 (1.3%), weight (5.3%), and season (3.8%) explained relatively little of the residual variability (Table 4).
Table 4.
Final multivariable model of genetic and clinical variables associated with steady-state endoxifen concentration.
| Clinical Variable | B Coefficient (95% Confidence Interval) | P-value | r2 Contribution to Final Model | |
|---|---|---|---|---|
| CYP2C9 Activity Phenotype | .20 (0.03, 0.37) | .025 | 1.3 | |
| Weight (kg) | −.013 (−.02, −.01) | <0.0001 | 5.3 | |
| Season of Sample Collection (vs. winter) | Fall | −.48 (−.74, −.22) | 0.0004 | 3.8 |
| Summer | −.49 (−.79, −.19) | 0.0017 | ||
| Spring | −.41 (−.7, −.19) | 0.0064 | ||
| CYP2D6 Diplotype as defined in [25] (vs. NM/NM, UM Alleles Considered NM alleles) | NM/IM | −.35 (−.61, −.08) | .0155 | 15.3 |
| NM/PM | −.49 (−.73, −.26) | <0.0001 | ||
| IM/IM | −.87 (−1.4, −.34) | 0.0015 | ||
| IM/PM | −.96 (−1.3, −.61) | <0.0001 | ||
| PM/PM | −1.32 (−1.79, −.85) | <0.0001 | ||
Secondary Screen of Genetic and Clinical Predictors of Tamoxifen, Metabolites, and Metabolite Ratios
Screening of other metabolites and metabolite ratios for hypothesis generation led to identification of associations with weight and season and two nominal associations with genetic factors. Similar to the association seen with endoxifen, greater weight was associated with lower concentrations of OH-tamoxifen (β=−0.0038, 95% CI: −0.0062 – −0.0015, p=0.0021, Supplemental Digital Content, Table 4) and N-desmethyltamoxifen (β=−0.025, 95% CI: −0.044 – −0.0048, p=0.017). In addition, OH-tamoxifen concentrations were lower in the fall (β=−0.21, 95% CI: −0.32 – −0.96, p=0.0004, Supplemental Digital Content, Table 4), and summer (β=−0.13, 95% CI: −0.26 – −0.0038, p=0.049) and N-desmethyltamoxifen concentrations were lower in the fall (β=−1.053, 95% CI: −1.99 – −0.12, p=0.031,) as compared to winter, similar to the associations detected for endoxifen. Screening predicted activity phenotype, higher SULT1A1 activity was nominally associated with greater endoxifen/4-OH-tamoxifen ratio (β =0.54, 95% CI: 0.04–1.04, p=0.03) and increased CYP2C8 activity was associated with a higher ratio of endoxifen to N-desmethyltamoxifen (β=0.01, 95% CI: 0.00, 0.02, p=0.04, Supplemental Digital Content, Table 5).
Discussion
The effect of genetic variability in CYPs and transporters, aside from CYP2D6, and clinical variables on steady-state endoxifen concentrations has not been thoroughly explored. CYP2D6 is thought to account for around 50% of the metabolism of tamoxifen to endoxifen[7], leaving a large portion of the metabolism unexplained. In this study, dozens of functionally relevant SNPs were genotyped to predict patient’s phenotypic activity of nineteen CYPs and transporters, in order to determine whether this activity was associated with variability in endoxifen concentrations. Our findings further support previous analyses suggesting that genetic variation in CYP2C9, weight, and the season of sample collection contribute to this residual variability. Although there were intriguing trends for CYP2C8 and CYP2C19, multivariable models suggest these may be spurious associations caused by linkage disequilibrium with CYP2C9.
Our findings support previous work indicating a contribution of CYP2C9 to endoxifen production. Human liver microsomes homozygous for low-activity CYP2C9 genotypes have lower rates of 4-OH-tamoxifen formation than wild-type microsomes[18]. This in vitro finding has been confirmed in clinical cohorts of tamoxifen treated patients. Lower steady-state endoxifen[5] or 4-OH-tamoxifen[4] concentrations have been detected in patients carrying common, low-activity CYP2C9 polymorphisms (CYP2C9*2 and *3). Recently, Powers et al. utilized a more comprehensive CYP2C9 activity phenotyping approach by integrating uncommon low-activity variants (i.e. *4 and *11) and concomitant inhibitor treatment (i.e. fluoxetine and omeprazole), confirming that patients with diminished CYP2C9 activity have lower endoxifen concentrations[17]. Our results are in line with these previous findings, though we did not have information on CYP2C9 inhibitor co-administration. Other studies that did not detect associations between CYP2C9 phenotype and concentrations of endoxifen or upstream metabolites were likely underpowered due to the rarity of low-activity CYP2C9 variants in patients of Asian descent[7, 8]. Other clinical analyses have reported associations between common functionally consequential polymorphisms in CYP2C19 (i.e. low-activity *2 and *3 and high-activity *17) and concentrations of tamoxifen and its metabolites[8, 9]. Our multivariable model suggests that CYP2C19 itself has a negligible, if any, contribution to endoxifen concentration and these associations can likely be attributable to linkage disequilibrium between the high activity CYP2C19*17 and the wild-type CYP2C8 and CYP2C9[26].
This study, similar to other reports examining BMI, found that higher body weight was associated with lower endoxifen[3, 4]. Body weight was utilized instead of body mass index because height information was unavailable. We also detected a significant association with season of sample collection, in which samples collected during the winter had higher concentrations of endoxifen than those collected in other seasons. Slightly higher concentrations of tamoxifen metabolites, but not tamoxifen itself, were also identified in the winter, suggesting this may be a metabolic phenomenon. Previously, two groups have reported the opposite effect, with samples collected in the spring and summer having greater endoxifen concentration than those collected in the winter[10, 27]. The mechanism for this seasonal effect is unclear as the original hypothesis, that seasonal sun-associated vitamin D induced CYP3A4 activity, was not confirmed by analyses of vitamin D and CYP3A activity in these publications[27]. The reason for the discrepancy in the direction of effect between our study and these prior publications is also not clear, but may have to do with geographical differences in sun exposure, as our study was conducted in North Carolina, USA. Also differences in the prevalence of vitamin D supplementation between cohorts could contribute, however we did not collect this information. One final possible explanation is increased tamoxifen adherence during winter months due to increased tolerability of hot flashes. Contrary to that hypothesis, tamoxifen concentrations were not different between the seasons. Lastly, we did not detect an association with CYP2D6 inhibitor co-administration, which is known to be a major determinant of endoxifen concentration[10, 28]. This is unsurprising, as patients taking strong or moderate CYP2D6 inhibitors were excluded from this study and relatively few patients (n=23) were taking weak inhibitors, which likely have a marginal effect on enzyme activity.
Several limitations of this analysis should be considered. The primary limitation is the lack of confirmation from prospective clinical trials that endoxifen serum concentration is associated with tamoxifen treatment efficacy, or perhaps toxicity, in breast cancer patients[29, 30]. Additionally, despite our attempt to perform a comprehensive assessment of the known, functionally consequential polymorphisms within genes putatively related to endoxifen concentration, it is likely that our panel did not include some relevant genes and some variants within the genes analyzed, particularly less common variants that have not yet been discovered. The panel utilized was as comprehensive as possible considering current knowledge in the field. The predicted functionality for polymorphisms was extrapolated from known data, but could have been incorrectly categorized. Further research efforts are needed, likely in well-controlled in vitro experiments, to estimate metabolic activity of individual alleles, perhaps accounting for substrate-dependent effects.
Our analysis explained approximately a quarter of the variability in endoxifen concentration, which is lower than previously reported for CYP2D6 genotype[7]. One possible explanation is that our samples were collected irrespective of the timing of the last dose. Additionally, endoxifen concentration was measured at baseline, assuming patients were adherent to tamoxifen treatment, as self-reported. Tamoxifen adherence is highest within the first year of therapy, around 77–88%, and decreases over time[31]; our patient cohort was taking tamoxifen on average 0.8 years prior to study entry, suggesting adherence was relatively high. Lastly, we did not perform multiple comparisons correction, increasing the possibility of false positive findings; however, the agreement of our findings with prior literature somewhat alleviates this concern.
Conclusion
In this retrospective secondary analysis of data collected from a prospectively enrolled cohort, genetic variation that confers diminished CYP2C9 activity was associated with decreased steady-state endoxifen concentrations. In adjusted analyses, CYP2C9 activity seemed to be independently contributory while CYP2C8 and CYP2C19 were not associated with endoxifen concentration, likely due to known linkage disequilibrium. These results suggest that individualized treatment approaches should consider several genetic and clinical factors in addition to CYP2D6 status to improve treatment efficacy. Further research is needed to integrate these genetic variables into an endoxifen prediction algorithm to guide personalized tamoxifen dosing. A combined analysis of existing datasets could be conducted to develop a more comprehensive and accurate algorithm. However, for these algorithms to be clinically useful endoxifen concentration needs to be definitively associated with tamoxifen treatment outcomes, ideally therapeutic efficacy.
Supplementary Material
Acknowledgments
Supported by Susan G. Komen Career Catalyst Award KG100355, Grant No. CA58223 from the National Cancer Institute Specialized Programs of Research Excellence, North Carolina University Cancer Research Fund, University of North Carolina at Chapel Hill Investments for the Future Grant No. 6231, Laboratory Corporation of America, Roche Diagnostics, American Society of Clinical Oncology Foundation and Breast Cancer Research Foundation, National Institute of General Medical Sciences Pharmacogenomics Research Network Award (U-01GM061373) to the Consortium on Breast Cancer Pharmacogenomics.
Footnotes
This work was presented in part at the 2015 San Antonio Breast Cancer Symposium and 2017 American Society of Clinical Pharmacology and Therapeutics Annual Meeting
Conflict of Interest
None declared
Supplemental Digital Content is available at Pharmacogenetics and Genomics’ website.
References
- 1.Early Breast Cancer Trialists’ Collaborative G. Dowsett M, Forbes JF, Bradley R, Ingle J, Aihara T, et al. Aromatase inhibitors versus tamoxifen in early breast cancer: patient-level meta-analysis of the randomised trials. Lancet (London, England) 2015;386(10001):1341–52. doi: 10.1016/S0140-6736(15)61074-1. [DOI] [PubMed] [Google Scholar]
- 2.Irvin WJ, Jr, Walko CM, Weck KE, Ibrahim JG, Chiu WK, Dees EC, et al. Genotype-guided tamoxifen dosing increases active metabolite exposure in women with reduced CYP2D6 metabolism: a multicenter study. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29(24):3232–9. doi: 10.1200/JCO.2010.31.4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madlensky L, Natarajan L, Tchu S, Pu M, Mortimer J, Flatt SW, et al. Tamoxifen Metabolite Concentrations, CYP2D6 Genotype, and Breast Cancer Outcomes. Clinical pharmacology and therapeutics. 2011;89(5):718–25. doi: 10.1038/clpt.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saladores P, Murdter T, Eccles D, Chowbay B, Zgheib NK, Winter S, et al. Tamoxifen metabolism predicts drug concentrations and outcome in premenopausal patients with early breast cancer. The pharmacogenomics journal. 2015;15(1):84–94. doi: 10.1038/tpj.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murdter TE, Schroth W, Bacchus-Gerybadze L, Winter S, Heinkele G, Simon W, et al. Activity levels of tamoxifen metabolites at the estrogen receptor and the impact of genetic polymorphisms of phase I and II enzymes on their concentration levels in plasma. Clinical pharmacology and therapeutics. 2011;89(5):708–17. doi: 10.1038/clpt.2011.27. [DOI] [PubMed] [Google Scholar]
- 6.Dehal SS, Kupfer D. CYP2D6 catalyzes tamoxifen 4-hydroxylation in human liver. Cancer Res. 1997;57(16):3402–6. [PubMed] [Google Scholar]
- 7.Jin Y, Desta Z, Stearns V, Ward B, Ho H, Lee KH, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97(1):30–9. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 8.Lim JS, Sutiman N, Muerdter TE, Singh O, Cheung YB, Ng RC, et al. Association of CYP2C19*2 and Associated Haplotypes with Lower Norendoxifen Levels in Tamoxifen-treated Asian Breast Cancer Patients. Br J Clin Pharmacol. 2016 doi: 10.1111/bcp.12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gjerde J, Geisler J, Lundgren S, Ekse D, Varhaug JE, Mellgren G, et al. Associations between tamoxifen, estrogens, and FSH serum levels during steady state tamoxifen treatment of postmenopausal women with breast cancer. BMC cancer. 2010;10 doi: 10.1186/1471-2407-10-313. 313-2407-10-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teft WA, Gong IY, Dingle B, Potvin K, Younus J, Vandenberg TA, et al. CYP3A4 and seasonal variation in vitamin D status in addition to CYP2D6 contribute to therapeutic endoxifen level during tamoxifen therapy. Breast cancer research and treatment. 2013;139(1):95–105. doi: 10.1007/s10549-013-2511-4. [DOI] [PubMed] [Google Scholar]
- 11.Dezentje VO, Opdam FL, Gelderblom H, Hartigh den J, Van der Straaten T, Vree R, et al. CYP2D6 genotype- and endoxifen-guided tamoxifen dose escalation increases endoxifen serum concentrations without increasing side effects. Breast cancer research and treatment. 2015;153(3):583–90. doi: 10.1007/s10549-015-3562-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barginear MF, Jaremko M, Peter I, Yu C, Kasai Y, Kemeny M, et al. Increasing tamoxifen dose in breast cancer patients based on CYP2D6 genotypes and endoxifen levels: effect on active metabolite isomers and the antiestrogenic activity score. Clinical pharmacology and therapeutics. 2011;90(4):605–11. doi: 10.1038/clpt.2011.153. [DOI] [PubMed] [Google Scholar]
- 13.Kiyotani K, Mushiroda T, Imamura CK, Tanigawara Y, Hosono N, Kubo M, et al. Dose-adjustment study of tamoxifen based on CYP2D6 genotypes in Japanese breast cancer patients. Breast cancer research and treatment. 2012;131(1):137–45. doi: 10.1007/s10549-011-1777-7. [DOI] [PubMed] [Google Scholar]
- 14.Martinez de Duenas E, Ochoa Aranda E, Blancas Lopez-Barajas I, Ferrer Magdalena T, Bandres Moya F, Chicharro Garcia LM, et al. Adjusting the dose of tamoxifen in patients with early breast cancer and CYP2D6 poor metabolizer phenotype. Breast (Edinburgh, Scotland) 2014;23(4):400–6. doi: 10.1016/j.breast.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Fox P, Balleine RL, Lee C, Gao B, Balakrishnar B, Menzies AM, et al. Dose Escalation of Tamoxifen in Patients with Low Endoxifen Level: Evidence for Therapeutic Drug Monitoring-The TADE Study. Clin Cancer Res. 2016;22(13):3164–71. doi: 10.1158/1078-0432.CCR-15-1470. [DOI] [PubMed] [Google Scholar]
- 16.Crewe HK, Ellis SW, Lennard MS, Tucker GT. Variable contribution of cytochromes P450 2D6, 2C9 and 3A4 to the 4-hydroxylation of tamoxifen by human liver microsomes. Biochem Pharmacol. 1997;53(2):171–8. doi: 10.1016/s0006-2952(96)00650-8. [DOI] [PubMed] [Google Scholar]
- 17.Powers JL, Buys SS, Fletcher D, Melis R, Johnson-Davis K, Lyon E, et al. Multi-gene and Drug Interaction Approach for Tamoxifen Metabolite Patterns Reveals Possible Involvement of CYP2C9, CYP2C19 and ABCB1. J Clin Pharmacol. 2016 doi: 10.1002/jcph.771. [DOI] [PubMed] [Google Scholar]
- 18.Coller JK, Krebsfaenger N, Klein K, Endrizzi K, Wolbold R, Lang T, et al. The influence of CYP2B6, CYP2C9 and CYP2D6 genotypes on the formation of the potent antioestrogen Z-4-hydroxy-tamoxifen in human liver. Br J Clin Pharmacol. 2002;54(2):157–67. doi: 10.1046/j.1365-2125.2002.01614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beelen K, Opdam M, Severson TM, Koornstra RH, Vincent AD, Hauptmann M, et al. CYP2C19 2 predicts substantial tamoxifen benefit in postmenopausal breast cancer patients randomized between adjuvant tamoxifen and no systemic treatment. Breast Cancer Res Treat. 2013;139(3):649–55. doi: 10.1007/s10549-013-2568-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desta Z, Ward BA, Soukhova NV, Flockhart DA. Comprehensive Evaluation of Tamoxifen Sequential Biotransformation by the Human Cytochrome P450 System in Vitro: Prominent Roles for CYP3A and CYP2D6. Journal of Pharmacology and Experimental Therapeutics. 2004;310(3):1062–75. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 21.Kiyotani K, Mushiroda T, Imamura CK, Hosono N, Tsunoda T, Kubo M, et al. Significant Effect of Polymorphisms in CYP2D6 and ABCC2 on Clinical Outcomes of Adjuvant Tamoxifen Therapy for Breast Cancer Patients. Journal of Clinical Oncology. 2010;28(8):1287–93. doi: 10.1200/JCO.2009.25.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greer AK, Dates CR, Starlard-Davenport A, Edavana VK, Bratton SM, Dhakal IB, et al. A potential role for human UDP-glucuronosyltransferase 1A4 promoter single nucleotide polymorphisms in the pharmacogenomics of tamoxifen and its derivatives. Drug Metab Dispos. 2014;42(9):1392–400. doi: 10.1124/dmd.114.058016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez-Santander A, Gaibar M, Novillo A, Romero-Lorca A, Rubio M, Chicharro LM, et al. Relationship between genotypes Sult1a2 and Cyp2d6 and tamoxifen metabolism in breast cancer patients. PLoS One. 2013;8(7):e70183. doi: 10.1371/journal.pone.0070183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hertz DL, Deal A, Ibrahim JG, Walko CM, Weck KE, Anderson S, et al. Tamoxifen Dose Escalation in Patients With Diminished CYP2D6 Activity Normalizes Endoxifen Concentrations Without Increasing Toxicity. Oncologist. 2016;21(7):795–803. doi: 10.1634/theoncologist.2015-0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hertz DL, Snavely AC, McLeod HL, Walko CM, Ibrahim JG, Anderson S, et al. In Vivo Assessment of the Metabolic Activity of CYP2D6 Diplotypes and Alleles. British journal of clinical pharmacology. 2015 doi: 10.1111/bcp.12665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pedersen RS, Brasch-Andersen C, Sim SC, Bergmann TK, Halling J, Petersen MS, et al. Linkage disequilibrium between the CYP2C19*17 allele and wildtype CYP2C8 and CYP2C9 alleles: identification of CYP2C haplotypes in healthy Nordic populations. Eur J Clin Pharmacol. 2010;66(12):1199–205. doi: 10.1007/s00228-010-0864-8. [DOI] [PubMed] [Google Scholar]
- 27.Antunes MV, Timm TA, de Oliveira V, Staudt DE, Raymundo S, Gossling G, et al. Influence of CYP2D6 and CYP3A4 Phenotypes, Drug Interactions, and Vitamin D Status on Tamoxifen Biotransformation. Ther Drug Monit. 2015;37(6):733–44. doi: 10.1097/FTD.0000000000000212. [DOI] [PubMed] [Google Scholar]
- 28.Borges S, Desta Z, Li L, Skaar TC, Ward BA, Nguyen A, et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: Implication for optimization of breast cancer treatment. Clinical pharmacology and therapeutics. 2006;80(1):61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Lintermans A, Van Asten K, Jongen L, Blomme C, Lambrechts D, Van Calster B, et al. Prospective study evaluating the effect of impaired tamoxifen metabolisation on efficacy in breast cancer patients receiving tamoxifen in the neo-adjuvant or metastatic setting. ASCO Meeting Abstracts. 2016;34(15_suppl):523. [Google Scholar]
- 30.Stearns V, O’Neill AM, Schneider BP, Flockhart DA, Skaar TC, Liu MC, et al. A phase II prospective trial correlating progression-free survival (PFS) with CYP2D6 activity in patients with metastatic breast cancer treated with tamoxifen: ECOG-ACRIN E3108. ASCO Meeting Abstracts. 2016;34(15_suppl):546. [Google Scholar]
- 31.Gotay C, Dunn J. Adherence to long-term adjuvant hormonal therapy for breast cancer. Expert review of pharmacoeconomics & outcomes research. 2011;11(6):709–15. doi: 10.1586/erp.11.80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


