Abstract
Pathologists rely on microscopy to diagnose disease states in tissues and organs. They utilize both high-resolution, high-magnification images to interpret the staining and morphology of individual cells, as well as low-magnification overviews to give context and location to these cells. Intravital imaging is a powerful technique for studying cells and tissues in their native, live environment and can yield sub-cellular resolution images similar to those used by pathologists. However, technical limitations prevent the straightforward acquisition of low-magnification images during intravital imaging, and they are hence not typically captured. The serial acquisition, mosaicking, and stitching together of many high-resolution, high-magnification fields of view is a technique that overcomes these limitations in fixed and ex vivo tissues. The technique however, has not to date been widely applied to intravital imaging as movements caused by the living animal induce image distortions that are difficult to compensate for computationally. To address this, we have developed techniques for the stabilization of numerous tissues, including extremely compliant tissues, that have traditionally been extremely difficult to image. We present a novel combination of these stabilization techniques with mosaicked and stitched intravital imaging, resulting in a process we call Large-Volume High-Resolution Intravital Imaging (LVHR-IVI). The techniques we present are validated and make large volume intravital imaging accessible to any lab with a multiphoton microscope.
Keywords: Mosaic, Stitching, Intravital Imaging, Multi-scale imaging
1. Introduction
In histopathologic analysis of fixed, stained tissues, low magnification images inform about the tissue wide structure and architecture, and are used to identify regions of the tissue for further analysis at high magnification. The high magnification analysis then reveals the identity of individual cells (based upon their staining and cellular and nuclear morphology), and the spatial relationships between them. While this switch between low and high magnification occurs repeatedly throughout histopathologic analysis, the low magnification perspective is most often not obtained in live tissue imaging, leading to a loss of tissue wide context.
The major reason for this underutilization of low magnification views arises from multiphoton microscopy’s reliance upon high-magnification, high-numerical-aperture objective lenses for efficient and bright signal generation. Low magnification lenses typically do not generate enough multiphoton signal to create high quality images. An alternative approach that we have adopted from the field of digital pathology [1] is to acquire many high-magnification, high-resolution images in a mosaic pattern and stitch them together to produce a low-magnification image, a process we call Large-Volume, High-Resolution (LVHR) imaging.
Here we describe a method for combining LVHR imaging with intravital imaging to create Large-Volume High-Resolution Intravital Imaging or LVHR-IVI - obtaining large area, high-resolution imaging in living animals which combines the advantages of histopathologic analysis with the live dynamics of intravital imaging. The method is composed of both a surgical technique for exposing and stabilizing the tissue of interest, and for the acquisition with multiphoton microscopy of multiple high-magnification tiles that are stitched together to form a large-area, low-magnification image. We also provide, as examples for the reader, designs for tissue stabilization tools, an example Fiji [2] macro, based upon a published algorithm [3], for automating the stitching of images over time and depth, and an example excel spreadsheet which can assist in generating and importing the numerous xy coordinate locations needed to control the microscope translation stage.
The novelty of this method lies in its combination of mosaicked-stitched imaging with intravital microscopy of live tissues. Here we provide a detailed description of the method and several examples of its utilization in a variety of tissues. To our knowledge, images utilizing this technique have not been previously reported in the published literature. Although we focus in this method on the imaging of compliant tissues, the procedure can be easily adapted to many other organs susceptible to physiologic motion–induced artifacts. These approaches do not require customized hardware or software, and are compatible with nearly all automated xy-stage equipped multiphoton imaging systems that are accessible to nearly all laboratories.
2. Materials and methods
2.1. Experimental design
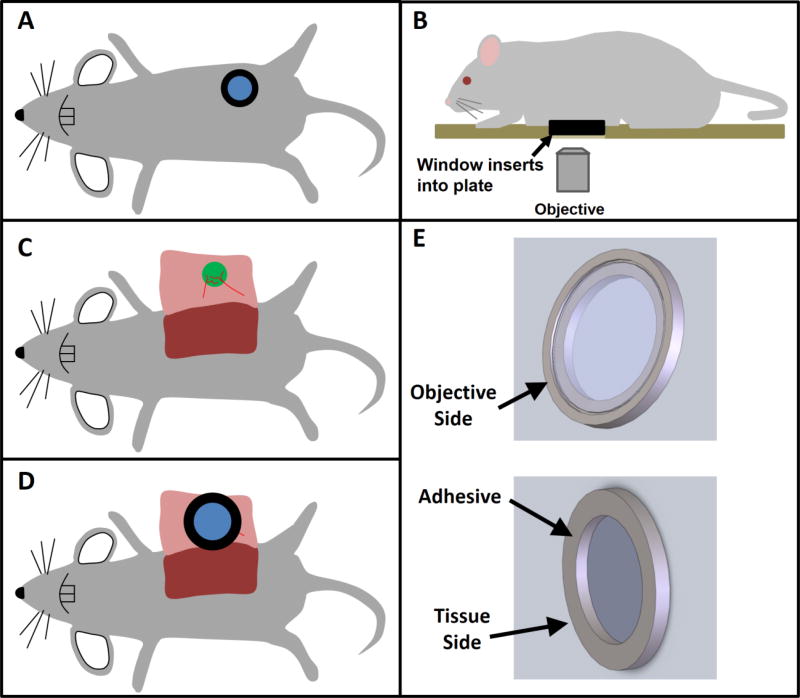
Key to the success of LVHR-IVI in living animals is tissue stabilization. In our prior work we developed several techniques for stabilized intravital imaging using skin flap surgeries [4, 5]. We have also developed surgical protocols for imaging mammary tumors [6] and lung tissues [7] using imaging windows (Figure 1A). Our experience with these window surgeries has shown us that, when implanted correctly, tissues are well stabilized relative to the windows and any movement from the heartbeat or intercostal muscle contractions are transmitted to the entire window and displace the window and the tissue together as one. Thus, complete immobilization of the tissue relative to the objective lens can be achieved by immobilizing the window frame. This is accomplished by capturing the window in a tightly toleranced recess that is bored into a custom-made xy stage plate (Figure 1B & Supplemental Figure 1).
Figure 1. Overview of surgical protocol.
A) With implantable imaging windows such as the mammary, abdominal, or even lung imaging windows, the window itself provides a stable frame which keeps the tissue hydrated and immobilized relative to the glass. However, involuntary movements such as heart beat and respiration still cause movement of the tissue and window together. B) Inserting the window frame into a tight-fitting stage plate which is fixed relative to the objective lens completely immobilizes both the window frame and tissue. C) With skin flap surgeries, the tissue of interest (e.g. tumor, mammary fat pad, lymph node, salivary gland, etc.) is exposed by surgical incision through the skin. D) The conditions for stable imaging that are possible when using imaging windows can be recreated by attaching a cover-glass inset into a shallow window frame over the tissue of interest. Adhesive placed under the window frame, away from the tissue under study, immobilizes the tissue relative to the frame. The window frame can then be inserted as in B into a tight-fitting stage plate. E) 3D computer aided designs of the shallow window frame and cover-glass described in D.
Using this concept as a basis for the design, we developed a method which is applicable to many different tissues, which includes compliant tissues such as the mammary fat-pad, lymph nodes, and liver which are particularly challenging as they are extremely compliant and easily transmit motion and vibrations from the animal’s involuntary movements.
The method begins with the surgical exposure of the tissue of interest. Figure 1C demonstrates this using the mammary fat-pad or inguinal lymph node as an example location. Once surgically exposed, adhesive is applied to the underside of a shallow window frame holding a circular cover-glass (Supplemental Figure 2). A small droplet of PBS is placed in the center of the cover-glass and the window is positioned so that the tissue of interest is centered under the cover-glass (Figure 1D & 1E). Finally, as in Figure 1B, the animal is placed on the custom xy stage plate with the window inserted into the recess. Placing the adhesive on the bottom of the window frame ensures that the imaged tissue is kept clear and unaffected. The adhesive additionally creates a seal surrounding the tissue of interest and keeps it moist, preventing dehydration. As was shown by Ritsma et al. [8], the use of adhesive on the rim of the window does not impact the integrity and viability of the tissue within the imaging area.
2.2. Microscope setup
Many commercially available microscopes utilize multiple objective lenses, however, we have found that a single low-magnification, high-numerical aperture, long-working distance objective lens (25× 1.05NA) is sufficient for nearly all imaging tasks. As discussed in Entenberg et al. [9] this lens provides: the working distance needed for intravital imaging deep into tissues; rapid switching through a wide range of magnifications by utilizing electronic zoom; nearly the same resolution (~0.25 µm lateral and 1.3 µm axial) as high magnification water immersion lenses; and high light throughput across the entire visible and near infrared spectrum. While advantageous, this method does not however rely on utilization of this objective lens and can be performed with many different lenses. All of the images presented in this manuscript are single optical slices acquired with this objective lens.
Similarly, while we use a custom designed multiphoton microscope [10], this procedure can be performed on any commercially available multiphoton or confocal microscope so long as it is equipped with an automated xy stage that is capable of recording a list of preset positions to acquire sequentially.
Finally, many commercially available microscopes come with either the ability to automatically program their automated xy stage for mosaicked acquisition, or the ability to import collections of xy coordinates. For the latter, we have provided an excel spreadsheet (Supplemental Materials, Mosaic Creator) which, with minor modification, can be used to generate the list of coordinates for automated import. The easiest way to accomplish this is to use the stage controller software to export a list of manually selected coordinates. Using this file, determine the format for the output text file. This format typically consists of numbers for x, y, and z coordinates and some surrounding text. In the Mosaic Creator file, the text within columns F, G, and H can be altered to match the output format for the stage controller. If all coordinates are located on a single output line, columns F, G, and H can be copied directly into a text file and uploaded into the stage controller after updating the imaging parameters described in section 2.5 below. If each coordinate is to be placed on a separate line, then column L can be copied to a text file.
2.3. Materials
Fluorescent dyes such as Tetramethylrhodamine isothiocyanate–Dextran (T1287, Sigma-Aldrich), Evan’s Blue (E2129, Sigma-Aldrich), or Texas Red Dextran (D1830, Thermo Fisher Scientific)
Phosphate Buffered Saline (14190136, ThermoFisher Scientific Inc.)
Isoflurane (029405, Henry-Schein)
Anesthesia unit (100 Series Vaporizer, SurgiVet)
Charcoal Filters (F-Air CS, Braintree Scientific, Inc.)
Tubing (6516T14, McMaster Carr)
Oxygen supply (TechAir)
PBS (14190136, ThermoFisher Scientific Inc.)
Transfer pipette (13-711-37, Fisher Scientific Inc.)
Puralube Vet Ointment (039886, Butler Schein Animal Health)
Petroleum Jelly (29704, Fisher Scientific)
Cyanoacrylate (1647358, Henkel Adhesives)
Transfer Pipette (13-711-37, Fisher Scientific Inc.)
Stiff rubber (1310N14, McMaster Carr)
12mm #1.5 Cover-glass (72230-01, Electron Microscopy Sciences)
Pulse-oximeter (MouseSTAT, Kent Scientific)
Low magnification, high-resolution objective lens (XLPLN25XWMP2, Olympus)
Custom x-y stage plate with bored recess (Supplemental Figure 1)
Custom shallow imaging window (Supplemental Figure 2)
2.4. Protocol
2.4.1. General ethics statement
All experiments dealing with live animals must be conducted in accordance with all relevant animal use and care guidelines and regulations. The procedures used in this study were carried out in accordance with the National Institutes of Health regulations concerning the care and use of experimental animals and with the approval of the Einstein College of Medicine Animal Care and Use Committee.
2.4.2. Mouse setup
Prior to starting the surgery, use cyanoacrylate to affix a cover-glass to the shallow window frame as shown in Supplemental Figure 3A. This should be done at least 1 hour before using the window to allow the adhesive to set (Supplemental Figure 3B).
Before beginning the surgery, prepare the microscope for imaging by turning on the imaging lasers and all electronics.
Turn on the anesthesia unit with a flow of 100% oxygen at a rate of 1.5–2 liters per minute mixed with 4–5% (vol/vol) isoflurane delivered to an anesthesia chamber. Place the mouse in the chamber. Take precautions to avoid inhalation of isoflurane during the procedure by using a well-ventilated room and a scavenging system. Activated-charcoal filters can be used to capture exhaust from the anesthesia chamber or from the ventilator. Runoff gas from the nose cone anesthesia unit can be removed using a suction hose connected to a ventilated house vacuum line.
Place the anesthetized mouse on the lab bench with a nose cone providing 100% oxygen mixed with 1.5–2% (vol/vol) isoflurane. The mouse’s body temperature should be maintained at physiologic temperatures with an overhead heat lamp or a warming pad. Assure that the mouse is propery anesthetized by performing a toe-pinch test to determine the absence of the pain reflex before the start of the surgical procedure. If the animal withdraws in response to pain, increase the dosage of isoflurane by 0.5%, wait for 1–2 min and repeat the toe-pinch test. Cover the animal’s eyes with ophthalmic ointment to prevent drying since mice are unable to close their eyes under the influence of anesthesia.
Next, construct and insert an i.v. catheter following the procedure published by Harney et al. [11]. Avoid introducing bubbles into the catheter line which can lead to air emboli and be fatal to the mouse. Administer 50–100 µL of sterile PBS via tail vein catheter to provide hydration during the surgical procedure. To prevent volume overload, be careful to not inject more than 200 µL. After the surgery, and during imaging, additional doses of ~50 µL per hour should be injected to maintain proper hydration of the animal.
2.4.3. Surgical procedure
To begin the surgical procedure, expose the tissue of interest through an appropriate sterile surgical technique (Supplemental Figure 3C). Several existing publications describe surgical protocols to expose various tissues including mammary tumors, [4, 11, 12] and fat-pads [13], salivary glands [14], liver [15], lymph nodes [16, 17] and kidney [18]. These tissues fall into the category of compliant (e.g. mammary fat-pad, lymph node, salivary gland, etc.), which more readily transmit vibrations due to their jelly-like consistency, or rigid (mammary tumor, kidney, etc.) which are easier to stabilize due to their increased stiffness and inflexible shape. If the tissue is compliant, additional stabilization will be required. This is accomplished with a rigid backing formed by a small piece of stiff rubber, cut to just larger than the exposed tissue. Take the rubber, apply a thin film of cyanoacrylate to one face, and affix it to the back side of the tissue, as shown in Supplemental Figure 3D.
Next, take the shallow imaging window and apply a small film of cyanoacrylate to the window frame in the location indicated in Figure 1E and Supplemental Figure 3E. Using a transfer pipette, apply a small droplet of PBS (~10–20 µL) to the center of the cover-glass on the side that will face the tissue (see Supplemental Figure 3F). Make sure that the PBS droplet on the cover-glass does not come into contact with the cyanoacrylate on the rim of the window as this will cause the cyanoacrylate to polymerize and set prematurely. The droplet should be ~2–3 mm in diameter. If made larger, the droplet may not cling to the cover-glass when transferred to the tissue or may contact the adhesive causing premature setting. Invert and center the window over the tissue of interest and gently press the window frame to the tissue to adhere (Supplemental Figure 3G). Make sure that the area of interest is kept in the center of the window and the surface of the tissue is relatively dry before attempting to adhere the window.
2.4.4. Mouse positioning on stage
Apply a thin layer of petroleum jelly to the bored recess on the underside of the imaging stage plate. The petroleum jelly will form a hydrophobic barrier between the shallow imaging window and the stage plate preventing wicking of water between the two via capillary action. Snap the stage plate onto xy stage. Transfer mouse and anesthesia unit to the xy stage.
Invert the mouse and set the shallow window into the bored recess (Supplemental Figure 3H). Be careful not to dislodge the window or the tail vein catheter at this time. Place environmental enclosure around the animal and turn on the heater. The tail vein catheter may be kept outside of the box for easy access. Using a transfer pipette, place a large drop of water between the objective and the cover-glass and visualize tissue by eye through the ocular. Verify that the vasculature of the tissue is perfused by observing flowing erythrocytes.
Crucial to the success of long term intravital imaging is the maintenance of the animal’s proper physiology. As mice lose their ability to regulate body temperature under the influence of anesthesia, it is important to heat the animal with either a warming pad, or a chamber flooded with temperature-regulated forced air. Monitor the vitals of the mouse using a pulse-oximeter and adjust the isoflurane level to maintain a surgical plane of anesthesia.
2.5. Mosaic image acquisition
Position the xy stage at the upper-left-most corner of the area to be imaged and zero the xy stage controller. Using the Mosaic Creator Excel spreadsheet (Supplemental Materials), enter the size of the microscope’s field of view, the desired number of x-tiles, y-tiles and field overlap. Upload the positions generated into the xy stage controlling software. Visit a sampling of the positions to see if the laser power and detector gains are set correctly for the tissue. Acquire images from all of the mosaic positions and save them to disk.
2.6. Data Analysis
There are multiple software packages that can be used for analyzing the acquired 3D and 4D data including the commercially available (and somewhat expensive) packages Imaris (Bitplane Inc.), Volocity (PerkinElmer Inc.), and Amira (FEI Inc.), and the freely available open source Fiji (NIH). While the choice of which software to use is based partly on personal preference and familiarity with a particular package, as well as availability within one’s home institution, each does offer some advantage over the others. An excellent review of these and other software packages, along with highlights of some of their advantages and limitations is presented by Walter et al. [19]. One advantage to Fiji is the free availability of many plugins to accomplish common tasks and the capability of writing custom macros to automate repetitive tasks. Included in this protocol is one such macro (Supplemental Materials, Moasic Merger Macro.ijm) that can be used to stitch the individually acquired tiles together and create a 3D or 4D data set known as a Hyperstack for analysis.
Once the images have been saved, use the Mosaic Merger Macro to load the images into Fiji and stitch them together. Make sure to set the correct number of x-tiles and y-tiles in the macro. The macro may need to be altered to accommodate the image file naming scheme for your microscope. Once the mosaic has been loaded and stitched together, the ROI Manager in Fiji can be used to toggle on and off an overlay showing the outline of each of the original acquired fields of view.
If the mosaic is acceptable, image acquisition can be continued and mosaic-z stacks or mosaic-z stack-time lapses can be captured and saved. After all of the images have been acquired and saved to disk, the 3D or 4D mosaic can be stitched together as a hyperstack in Fiji. Examine the mosaic for events and locations of interest for further study. One can proceed to perform further time-lapse imaging on just those specific regions to investigate events with a high temporal resolution.
2.7. Limitations
There are several limitations to the techniques presented in this method. The first and foremost is that the acquisition of large volumes of data takes a much longer time than acquisition of individual fields of view. For typical multiphoton microscopes with acquisition speeds of 1 frame per second (fps), a 10×10 mosaic takes ~100 seconds to capture. This is the time required for each z-slice. In typical applications, tissues are imaged to a depth of 100 µm with a step size of 5 µm between each z-slice (a sampling size sufficient to capture 15–20 µm diameter cells at greater than Nyquist conditions).
If time-lapse imaging is required in order to capture the cellular dynamics in a field, the total acquisition time for a single time point becomes on the order of 30 minutes. Balancing the conflicting need for resolution, large area, large depth, and speed will depend heavily upon the type of biological process that is being studied. 30 minutes per volume may be sufficient to capture slow events such as cellular division or collective migration of tumor cells, whereas extremely rapid events such as the visualization of calcium signaling or circulating immune cells may be addressed by capturing only one low-magnification high-resolution image for the tissue at the beginning of the imaging session and using the single mosaicked image to inform which individual fields of view warrant further, high-speed, analysis. Processes such as tumor or immune cell motility in tissue fall in between and are best addressed by limiting the number of z slices to a maximum of 2 or 3. Of course, microscope systems with a much higher frame rate [20, 21] (e.g. video rate systems running at ~30 fps) can dramatically reduce this acquisition time from 30 minutes to 1 minute per time point.
Another limitation is due to the large size of the data sets that result from LVHR-IVI. These data sets are memory intensive (occupying upwards of several gigabytes) and can present a challenge to efficiently and quickly stitch and to store. These two issues are not of great concern as the constant reduction in cost of hard disk space and increase in speed of processors will greatly ameliorate these issues with time. There is also currently a large push to address these issues in the field of digital pathology [22, 23].
Finally, closely related to the issue of size, these data sets present challenges when it comes to analysis and are somewhat time consuming to analyze fully given the volume of the data. Efficient image analysis and data mining will require the development of automated machine learning/vision algorithms to properly address. We have already begun investigating these avenues [24], though new approaches will be advantageous as well.
2.8. Troubleshooting
There are multiple issues that can arise during this method. First, the smell of anesthetic may be noted during setup or imaging. This is most likely a result of a leak in the anesthesia tubing or an issue with the scavenging system. It is important to correct these issues with the anesthesia equipment as they can lead to the mouse being inappropriately anesthetized or to personnel being inadvertently exposed to anesthesia.
Second, during the mouse set up, the window can accidently become unglued from the tissue flap either from insufficient cyanoacrylate or prematurely cyanoacrylate polymerization due to excess liquid. This can be addressed by removing the window, appropriately drying the tissue, and applying glue to the rim of a clean window. Since the adhesive is placed far outside of the imagable area this should not affect the viability of the imaged tissue,
Third, after the mouse is placed on the stage and imaging is attempted, excess movement from breathing maybe noted. This can be addressed by ensuring that the tissue connecting the imaged area and the body of the animal is not pulled taut and/or by applying gentle pressure (as described in the protocol by Masedunskas et al. [14]) to the top surface of the tissue (or, in the case of compliant tissues, the rigid rubber backing that was glued to the tissue). It is important not to apply too much pressure as blood vessels can easily become compressed and blood flow stopped. This is easily verified visually.
Finally, after the images have been stitched, it might be noted that the stitched image is doubled or is numbered incorrectly. This is likely due to either the x-y offset or the number of x and y tiles being incorrectly entered into the macro. This can be manually adjusted in the macro and the image reprocessed.
3. Theory
3.1. Development of the protocol
Traditionally, intravital imaging has been considered to be a practice that requires specialized equipment [4, 25] and significant skill, leading it to be regarded as a “technically difficult” process [26]. However, recent commercialization of the microscope instrumentation along with the development of numerous protocols [4, 11, 14, 27] and surgical techniques [8, 28] have simplified the process to the point where it has become accessible to an ever increasing number of labs [29]. In many applications of intravital imaging, just a few fields of view (FOVs), randomly positioned within the tissue, are recorded. Since these FOVs represent a small minority of the total imageable tissue, the overall architecture and context of the tissue is lost. This concept is illustrated in Figure 2A with a photograph commonly used in the image analysis literature.
Figure 2. Multi-scale imaging provides detail and context by acquiring many high-magnification images sequentially and stitching them together to form a low magnification overview image.
A) The acquisition of just a few individual fields of view does not provide enough context to understand fully the imaged subject. B) Information about the subject is improved by increasing the number of acquisitions, however, without maintaining the spatial relationship between the individual images, the overall context remains obscured. C) Acquisition and arrangement of the images in a specific order (yellow numbers) maintains the spatial relationship of the images. D) Final stitching of the image reveals the overall context while preserving the original resolution of the underlying images.
In the past, we have successfully compensated for some of this loss of information by greatly increasing the number of FOVs acquired and applying mathematical algorithms designed to extract complex or hidden relationships between imaged parameters [24]. In that work, we developed new insights into the mechanisms of metastasis by utilizing a support vector machine (SVM) algorithm (a nonlinear, multiparametric classification algorithm suitable for the analysis of systems with arbitrary distributions and/or non-linear parameters) to analyze hundreds of fields of view and dissect the microenvironmental conditions that are responsible for invasive tumor phenotypes [24].
Though the large volume of data comprising the complete acquisition of all FOVs does give much more information about the underlying subject, a coherent picture is still lacking (Figure 2B). What is missing in these studies is the reconstruction of the spatial relationship between the acquired fields. By acquiring the fields in a specific order and mosaicking them together (Figure 2C) the spatial relationships of the images can be maintained and the overall architecture of the tissue reconstructed (Figure 2D). This information may then be used to inform about the spatial ecology (identity, quantity and location) of the constituent components (cells, stroma, matrix, etc.) [30–32] or inform about which areas are most suitable for further analysis (e.g. time-lapse imaging).
While not common in the field of intravital imaging, mosaicking is a technique that has been used in multiple areas of research, such as biology, oceanography, and X-ray microscopy [33–37]. It has even been combined with multiphoton microscopy, though only to image large areas of fixed and mechanically sectioned tissues [33]. Only one study in the published literature [13], our investigation into the mechanisms of early tumor cell dissemination from the mammary fat pad, has ever mentioned having utilized mosaic intravital imaging, albeit without reporting either the method or any mosaicked images.
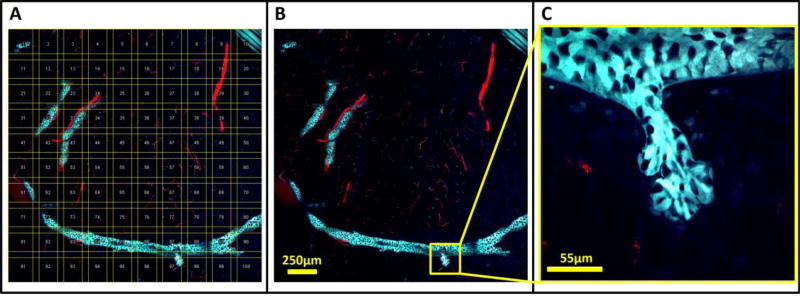
This study relied heavily on LVHR-IVI for imaging the untransformed mammary fat-pad [13]. Given the very sparse structure of the mammary ductal tree, LVHR-IVI proved crucial to the successful identification of regions for further high-resolution time-lapse imaging. Since this tissue is extremely compliant, however, intercostal muscle contractions produced significant motion artifacts which were only overcome with the application of the techniques described in this method (shallow window and custom xy-stage plate). With the tissue stabilized, large areas were imaged with high-resolution by the serial acquisition of individual high-magnification high-resolution tiles which were then mosaicked and stitched together. Figure 3A shows an example of acquired fields of view (yellow boxes) arranged in order along with the sequence of their acquisition (yellow numbers). To compensate for non-uniform field illumination, the positions of the individual images are overlapped somewhat (10–20%). Depending upon the degree of non-uniformity in field illumination, dark bands may still be visible on the stitched images. Though not utilized in this work, optimized stitching algorithms may be used to further reduce these effects [38].
Figure 3. Ordered acquisition and stitching of intravital images produces multi-scale, low-magnification, high-resolution images that provide histologic context to tissues while preserving single-cell resolution.
A) A 10×10 mosaic of an untransformed fat pad in a 10 week old MMTV-Her2-CFP mouse whose mammary epithelia were transgenically labeled with CFP (cyan) and vasculature labeled with fluorescently labeled dextran (red) injected iv. Yellow boxes show the outline of each acquired field of view and numbers indicate their order of acquisition. Fields of view are overlapped to compensate for non-uniform flatness of field illumination. B) Stitched images show the overall structure of the tissue. The formation of the ductal tree by the epithelial cells (cyan) is clearly visible as are the blood vessels that support them. C) A single lobule consisting of several acini can be observed budding off of the ductal tree.
These mosaics thus provided a low-magnification view similar to those utilized by pathologists during initial histopathological assessment of tissues (Figure 3B). This low-magnification overview is crucial for understanding the biological context of the imaged cells. For example, Figure 3C shows a zoomed in image of the indicated area in panel B. In panel C the structure could easily be mistaken for a microinvasion of tumor cells into stroma. However, the low-magnification view in panel B reveals this structure to be a normal branching duct. The risk of this potential misidentification is high since malignant foci frequently overlap with benign structures. The low-magnification view provides essential information necessary to distinguish benign from malignant processes, and to identify regions of interest for further high-resolution time-lapse imaging.
4. Results
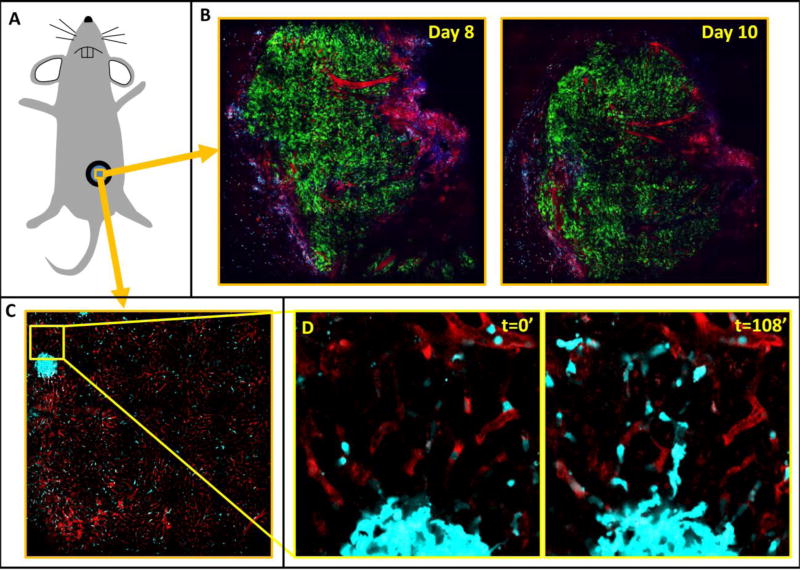
The method presented here enables the combination of large-volume high resolution imaging with intravital microscopy. As such, it consists of two parts. The first part is a method for the acquisition of low-magnification, high-resolution intravital images built up by acquiring many individual, high-magnification tiles in a sequential manner and stitching them together. A prerequisite for the success of this method is the stabilization of the tissue of interest. Any of the methods that we and others have developed for the stabilization of primary mammary tumors [4, 5] and other tissues [9, 14, 39] may be employed with success. This includes the use of mammary [6] and abdominal imaging windows [40] (Figure 4A) which provide the additional benefit of being able to return to the tissue day after day. This is demonstrated by two large mosaics of a mammary tumor xenograft imaged through a mammary imaging window taken 8 days and 10 days post implantation (Figure 4B). Growth of the tumor over the intervening time has changed the overall morphology of the tissue, however a similar blood vessel structure can still be recognized.
Figure 4. Mammary and abdominal imaging windows provide sufficient tissue stabilization in order to perform mosaicking.
A) Cartoon representing a mouse with an imaging window surgically implanted. B) 10×10 mosaics covering 2.8 mm of Dendra2 labeled MBA-231 orthotopic xenograft tumor implanted in the 4th mammary fat-pad of a Rag2−/− mouse and imaged through a mammary imaging window on days 8 and 10 post-surgical implantation. Green = GFP labeled tumor cells, Red = TMR labeled vasculature, Blue = SHG. C) 10×10 mosaic covering 1.4 mm of healthy liver tissue taken through an abdominal imaging window a C57/BL6 mouse whose myeloid cells were transgenically labeled with CFP [48]. Despite the low magnification, the high-resolution of the underlying fields of view is maintained. A collection of macrophages forming a granuloma-like structure can be seen. Cyan = CFP labeled macrophages, Red = TMR labeled vasculature. D) Stills from a subregion of the time-lapse mosaic shown in panel C showing single macrophages migrating out of the granuloma-like structure. FOV = 170 µm.
Despite the large number of frame acquisitions, microscopes with a frame acquisition rate of just 1 fps can still capture time-lapsed, z-slice mosaics with a great enough repetition rate to capture single cell dynamics. This can be seen in Figure 4C using an abdominal imaging window to view the liver. In this 10×10 mosaic covering 1.4×1.4 mm of liver tissue, a cluster of macrophages (cyan) forming a granuloma-like structure can be seen. The vasculature has been labeled with high molecular weight (155kD) tetramethyl-rhodamine (TMR) which does not leak out over time. Time lapse imaging of this tissue (Supplemental Movie 1) shows the dynamics of the macrophages. A subregion of the mosaic, focused on the collection of macrophages forming the granuloma-like structure (Figure 4D and Supplemental Movie 1 inset) shows the motility of the single macrophages as they migrate out of the cluster.
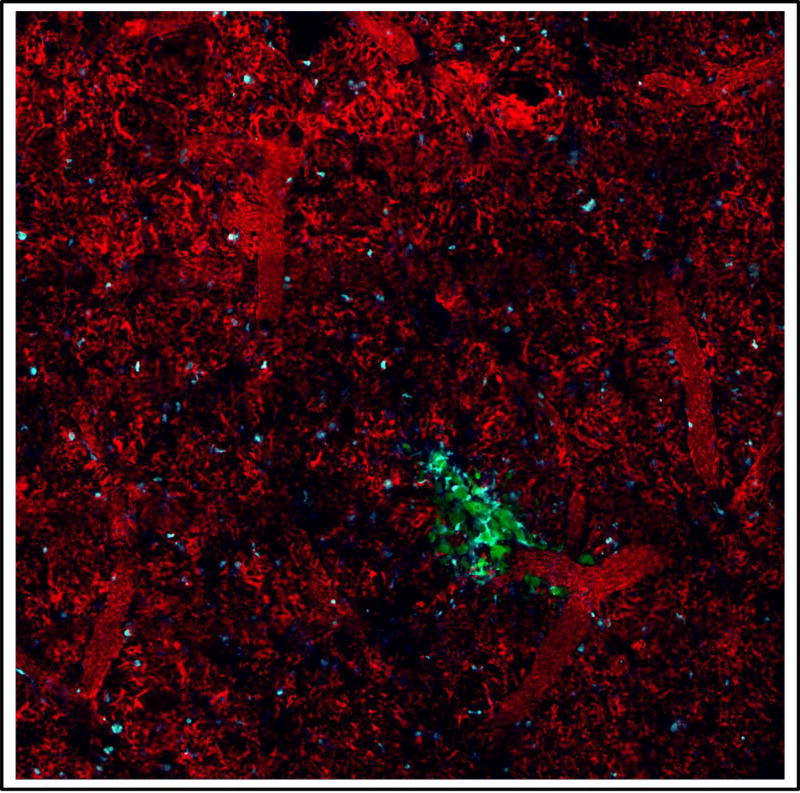
We have also performed the mosaicking protocol with our published vacuum stabilized lung imaging window [7, 9]. The image stability of this technique is even sufficient to enable the use of our published blood averaging technique which averages together all of the time points for the blood (red) channel into a single image and then replicates this image as the background for each frame of the movie containing the other channels. This produces a clear view of the boundaries of the vasculature of the lung despite the numerous interruptions in vascular signal caused by the passage of unlabeled erythrocytes and leukocytes [7, 9] (Figure 5).
Figure 5. Low-magnification, high-resolution, 6×6 mosaic of 1.2×1.2 mm of lung tissue containing a micro-metastatic lesion imaged through a vacuum-stabilized lung imaging window.
The high mechanical stability provided by the vacuum stabilized imaging window allows LVHR-IVI of E0771 tumor cells in the lungs of a C57/BL6 mouse whose myeloid cells were transgenically labeled with CFP [48]. Green = GFP tumor cells, Cyan = CFP labeled macrophages and monocytes, Red = TMR labeled vasculature
We and other labs have been successful in developing techniques that expose and stabilize tissues for intravital imaging over long periods of time. This includes mammary tumors [41] and various other organs [14, 16, 17, 40], some of which have traditionally been unimageable due to their inherent motion [13, 39, 42]. Still, there has been no generalizable technique for the immobilization of living tissues, especially as these tissues become more compliant. As such, the second part of the method presented here offers a generalizable procedure applicable to the stabilization of a range of tissue types and stiffness.
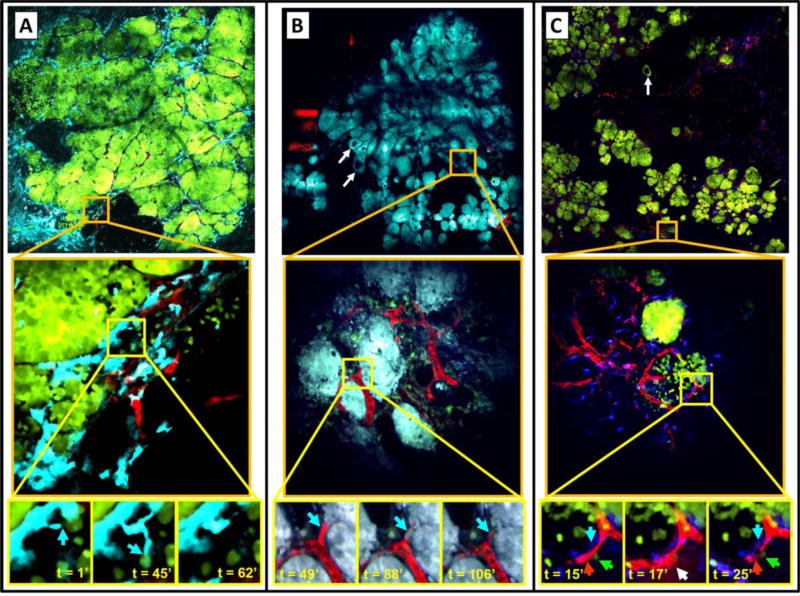
We have used this procedure to image all stages of progression of transgenic mammary tumors. This includes the often imaged late-stage carcinoma (Figure 6A), as well as the more challenging earlier stages including tissues with mixtures of late- and early-stage carcinoma (Figure 6B), tissues with mixtures of early-stage and ductal-hyperplasia (Figure 6C), and even the untransformed fat-pad (Figure 3). In each of these cases, the low-magnification, high-resolution images generated by the method are able to be examined at single-cell resolution to identify regions of interest for further study. For instance, we have been able to identify regions of macrophage infiltration (Figure 6A, middle) in the late-stage carcinoma, and close examination of high-resolution time-lapse images of this region reveal tumor cells acquiring an invasive phenotype (Figure 6A, blue arrows in bottom panels). In Figure 6B, areas of tumor bed vascularization can be identified (Figure 6B, middle) and the vasculature monitored to observe hematogenous dissemination of tumor cells (Figure 6B, blue arrows in bottom panels). Finally, in Figure 6C, we have been able to use the low-magnification, high-resolution images of early carcinoma, a tissue traditionally unimageable intravitally due to its extreme compliance and instability, to identify regions with distinctly different growth patterns (Figure 6C, middle). High-resolution time-lapse imaging of these regions has captured the function of the micro-anatomical tripartite structure known as the Tumor Microenvironment of Metastasis (TMEM) (Figure 6C, white arrow in t=17’ panel). TMEM consist of a tumor cell and perivascular macrophage in direct contact with an endothelial cell (blood vessel) and have been previously shown to act as sites of both cancer cell intravasation and transient vascular permeability [43, 44].
Figure 6. Mosaic time lapse and z-series intravital imaging in transgenic mice allows acquisition of very large volumes of tumor heterogeneity that can be zoomed to follow single cell phenotypes in real time.
A–C) Top panels: A single z plane showing large area imaging of a mammary tumor within a transgenic PyMT mouse expressing fluorescent proteins. Middle panels: One individual tile (indicated by orange squares) from which the mosaic is composed. Bottom panels: Time lapse imaging of the sub-region (indicated by yellow squares).
A) Top panel: 6×6 mosaic covering 1.6×1.6 mm of a late-stage carcinoma. Cyan = CFP expressing macrophages, Red = Fluorescent dextran labeled vasculature, Green = GFP expressing tumor cells. Middle panel: Peritumoral region of local inflammation indicated by CFP positive myeloid cells located at the tumor-stroma interface. FOV = 340×340 µm. Bottom panel: Stills from a time-lapse movie showing macrophages migrating to interact with tumor cells (blue arrows). FOV=60×60 µm.
B) Top panel: 6×6 mosaic covering 1.6×1.6 mm of a few ducts undergoing hyperplastic changes (white arrows) and carcinoma showing areas of late (top) and early (bottom) stage. Cyan = CFP expressing tumor cells, Red = Fluorescent dextran labeled vasculature, Green = GFP expressing macrophages. Middle panel: Region showing sheets of cancer cells with intersecting vasculature. FOV = 340×340 µm. Bottom panel: Stills from a time-lapse of an intravasated tumor cell (Blue arrow). FOV=60×60 µm.
C) Top panel: 10×10 mosaic covering 4×4 mm of an early carcinoma. Green = Dendra expressing tumor cells, Red = Fluorescent dextran labeled vasculature, Blue = CFP expressing macrophages. Middle panel: Image shows two groups of cancer cells with distinctly different growth patterns. FOV = 512×512 µm. Bottom panel: TMEM (Blue arrow=macrophage, Green arrow=tumor cell, Red arrow=endothelial cell in t=15’ panel) induced transient vascular leakage (white arrow in t=17’ panel). FOV = 133×133 µm. TMEM remains after leakage has ceased at t=25’.
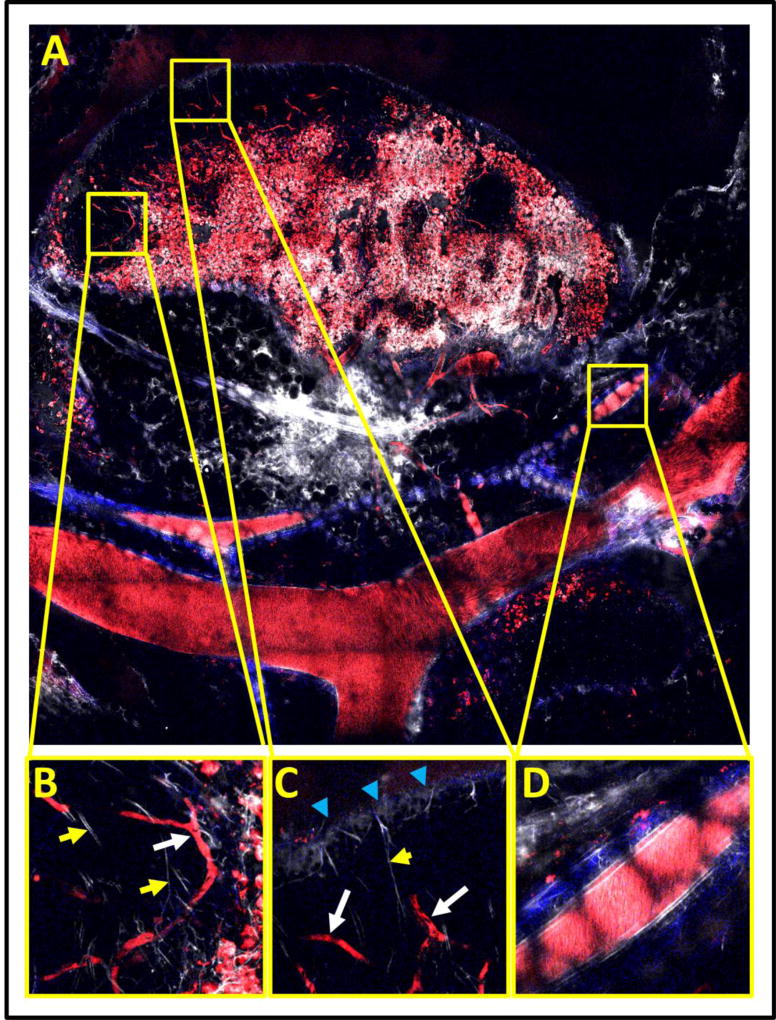
In addition, we have been able to employ both parts of the method presented here to image the intact lymph node (Figure 7). The low-magnification, high-resolution image (Figure 7A) is an extremely useful guide for studies of the lymph node since its large extent (~3.5 mm) makes it very difficult to characterize the specific areas contained in any single field alone. This is demonstrated by the individual fields of view presented in Figures 7B–D. From any one of these fields of view alone, it is nearly impossible to determine if the imaged field is internal or external to the node or the identity of the structures. The context provided by the low magnification view clearly identifies panels B and C as sub-capsular sinuses with slow flowing capillaries (as evidenced by shadows of erythrocytes, white arrows) and fine lymphatic capillaries (yellow arrows). Particularly difficult to identify without the low magnification overview is the lymph node capsule shown in panel C (blue arrows). Panel D demonstrates a fast flowing blood vessel traveling external to and alongside the node. The honeycomb shadow pattern is indicative of an overlaying layer of fat cells.
Figure 7. Large scale mosaics reveal the structure and distribution of vessels and cells within compliant tissues such at the lymph nodes.
A) 10×10 mosaic covering a 4×4 mm area of lymph node, blood vessels and lymphatics. B) and C) High resolution subfields showing subcapsular sinuses within the lymph node. Shadows of slow flowing erythrocytes (white arrows) and fine lymphatic capillaries (yellow arrows) can be observed within these zones. FOV = 300 µm. D) A high resolution subfield showing a fast flowing blood vessel external to the lymph node. FOV = 300 µm. All panels: Red = Texas Red labeled vessels and macrophages, White = Evan’s Blue labeled lymph. Blue = SHG..
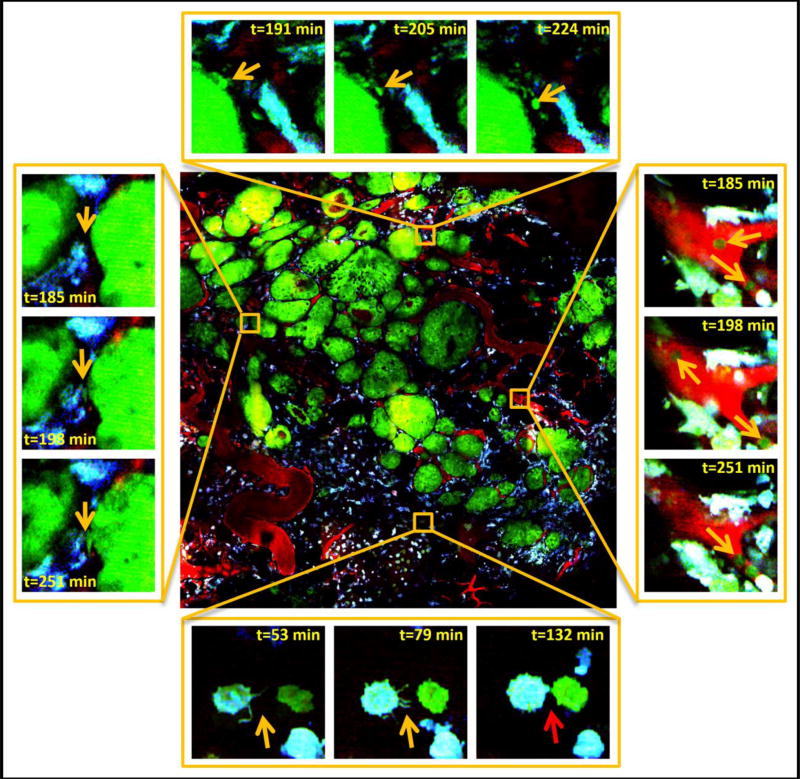
Finally, the time-lapsed LVHR-IVI allows the simultaneous capture and evaluation of the dynamics of the entire tissue. These time-lapsed multiscale images can allow a much more complete investigation of cellular dynamics than is possible with ordinary intravital imaging. Figure 8 shows time-lapse LVHR-IVI of an entire tumor that is a mixture of early and late carcinoma. Careful examination of the images at high resolution reveals many processes occurring simultaneously including: single cell migration (Figure 8, top); circulating tumor cells (Figure 8 right); tumor cell-macrophage interactions (Figure 8, bottom); and tumor cell invasion into stroma (Figure 8, left).
Figure 8. Large-volume high-resolution intravital imaging captures time-lapse movies of large volumes of tumor tissue at single cell resolution and in real time.
Center is a stitched mosaic of 36 (6×6) high-resolution (0.24 µm pixel size) image tiles taken of a transgenic PyMT tumor. Stills from simultaneously acquired sub-regions reveal single cell dynamics with subcellular resolution. Top: A chain of single tumor cells (orange arrows) can be observed separating from the main tumor and invading the stroma. Right: Individual tumor cells (orange arrows) can be seen in the vasculature. Left: A tumor cell protrusion (orange arrows) can be seen as the cell starts separating from the main tumor and moving toward macrophages. Bottom: As an example of the subcellular resolution of the imaging, an isolated macrophage can be observed extending filopodia (orange arrows) towards an isolated tumor cell. As the two cells make contact, the filopodia disappear (red arrow). Green = GFP tumor cells, Cyan = CFP macrophages, Red = fluorescently labeled blood serum. Field of View (FOV) of center image = 1.6×1.6 mm. FOV of surrounding images = 66µm.
5. Discussion
Intravital imaging (IVI) allows for a unique look at living tissue where the dynamics of individual cells can be directly observed and recorded in health and disease. This is accomplished through a combination of multiphoton microscopy’s ability to perform non-destructive optical sectioning deep into thick tissues, with the ability to label the structure and function of cells and whole tissues using fluorescent dyes and genetically encoded fluorescent proteins. This combination produces images of the live tissue similar to those generated by mechanical sectioning and staining of fixed tissues.
While the acquisition of LVHR images of organs or tissues is straightforward when motion of the sample is not present, intravital imaging involves imaging a living animal which imparts to the tissue many motion artifacts arising from breathing, heartbeat, and tissue dehydration. Thus, the limitation to successful combination of LVHR imaging with intravital imaging lies not in the hardware or instrumentation, but in the ability to stabilize the living tissues sufficiently. While this is true for ordinary high-resolution microscopy, where motion artifacts on the order of microns can distort and destroy an image, it is even more crucial for mosaic imaging where stability requirements are exceedingly stringent given the extended scale of the imaging.
The live multiscale view generated by these techniques allows for a better understanding of cellular dynamics in the context of the overall tissue architecture, guides the selection of regions for further high-resolution imaging and provides data on large volumes of tissue that can be analyzed using machine learning algorithms [24].
To make this combination of LVHR imaging and IVI feasible, we have recently developed new approaches for the stabilization of tissues [5, 9, 13, 45, 46], that enable both long time-lapse and LVHR-IVI, even in compliant tissues such as lymphatics [45, 46], lymph nodes [45], lung [9], liver, and the mammary fat-pad [13].
By combining these techniques with our previously published mammary imaging window [6], these studies can be extended over multiple imaging sessions spanning days to weeks. This enables the capture of events spanning orders of magnitude in space and time and directly addresses the restrictions of inadequate resolution and limited imaging periods that have precluded “the direct detection of slow processes, such as collective invasion with structural ECM degradation and remodeling that develop over days” [47].
6. Conclusions
In summary, the method presented here is a generalizable technique for the stabilization and large volume intravital imaging of living tissues. It is compatible with many types of tissues where cells and other structures of interest have been labeled using either genetically encoded fluorescent proteins or injectable fluorescent dyes. This method requires no specialized equipment and can be accomplished with any multiphoton microscope equipped with an automated xy stage and controlling software.
Supplementary Material
XY stage insert design schematics.
Shallow window design schematics.
A) Preparation of the shallow imaging window. A thin layer of adhesive is placed on the inset rim of the window frame (red line) and the cover glass seated in the recess. B) Waiting time for adhesive to completely set. A setting time of one hour ensures that the cover glass cannot become dislodged and forms a water tight seal. C) Surgical exposure of the tissue of interest. Previously published protocols can be used to expose the tissue of interest. D) Stabilization of compliant tissue. The application of adhesive to a rigid piece of rubber allows it to be adhered to the back side of the tissue of interest. E) Application of adhesive to the frame of the shallow imaging window. Applying adhesive only to the rim of the imaging window (red line) will stabilize the tissue within the window, while avoiding exposure of the tissue of interest to the adhesive. F) Application of PBS to the cover glass. Before applying the window to the tissue, a 2–3 mm droplet of PBS is suspended from the center of the cover glass. When sealed to the tissue, a hydration chamber is formed which prevents dehydration of the tissue. G) Mouse with window adhered. Once window is adhered over the tissue of interest, the mouse may be positioned on the microscope. H) Mouse positioned on microscope stage. To begin imaging, the mouse is inverted and the window placed into the recess in the imaging stage plate.
Frames are 4.7 minutes apart.
A method for the combination of mosaicked-stitched imaging with intravital microscopy of live tissues is presented.
This is a generalizable method for stabilization and large-volume high-resolution intravital imaging (LVHR-IVI) of living tissues.
LVHR-IVI is applicable to many different tissues including extremely compliant ones.
Enables the capture of dynamic events spanning orders of magnitude in space and time.
Acknowledgments
This technology was developed in the Gruss-Lipper Biophotonics Center and the Integrated Imaging Program at Einstein College of Medicine. We acknowledge the support of these Centers in this work: Einstein’s Integrated Imaging Program, Montefiore’s Ruth L. Kirschstein T32 Training Grant of Surgeons for the Study of the Tumor Microenvironment (CA200561), and NIH CA100324, and CA216248. We thank Mike Rottenkolber and Anthony Leggiadro of the Einstein Machine Shop for their skilled craftsmanship and design insight.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
DE, JC and MO defined the biological and imaging goals for the technique, DE conceived of the technical solution and designed the window and stage plate. DE & MS imaged the fat-pad, DE & YW imaged the liver, DE & SV imaged the lung, DE & JP imaged the different stages of carcinoma, JP imaged the lymph node and mammary xenograft, DE, JP, MO, JC, JAG wrote the paper.
Competing financial interests
None
References
- 1.Farahani N, Parwani A, Pantanowitz L. Whole slide imaging in pathology: advantages, limitations, and emerging perspectives. Pathol. Lab Med. Int. 2015;7:23–33. [Google Scholar]
- 2.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25(11):1463–5. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Entenberg D, Kedrin D, Wyckoff J, Sahai E, Condeelis J, Segall JE. Imaging tumor cell movement in vivo. Curr Protoc Cell Biol. 2013:Unit19 7. doi: 10.1002/0471143030.cb1907s58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams JK, Entenberg D, Wang Y, Avivar-Valderas A, Padgen M, Clark A, Aguirre-Ghiso JA, Castracane J, Condeelis JS. Validation of a device for the active manipulation of the tumor microenvironment during intravital imaging. Intravital. 2016;5(2):e1182271. doi: 10.1080/21659087.2016.1182271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kedrin D, Gligorijevic B, Wyckoff J, Verkhusha VV, Condeelis J, Segall JE, van Rheenen J. Intravital imaging of metastatic behavior through a mammary imaging window. Nat Methods. 2008;5(12):1019–21. doi: 10.1038/nmeth.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez-Tirado C, Kitamura T, Kato Y, Pollard JW, Condeelis JS, Entenberg D. Long-term High-Resolution Intravital Microscopy in the Lung with a Vacuum Stabilized Imaging Window. J Vis Exp. 2016;(116) doi: 10.3791/54603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ritsma L, Steller EJ, Beerling E, Loomans CJ, Zomer A, Gerlach C, Vrisekoop N, Seinstra D, van Gurp L, Schafer R, Raats DA, de Graaff A, Schumacher TN, de Koning EJ, Rinkes IH, Kranenburg O, van Rheenen J. Intravital microscopy through an abdominal imaging window reveals a pre-micrometastasis stage during liver metastasis. Science translational medicine. 2012;4(158):158ra145. doi: 10.1126/scitranslmed.3004394. [DOI] [PubMed] [Google Scholar]
- 9.Entenberg D, Rodriguez-Tirado C, Kato Y, Kitamura T, Pollard JW, Condeelis J. In vivo subcellular resolution optical imaging in the lung reveals early metastatic proliferation and motility. Intravital. 2015;4(3):1–11. doi: 10.1080/21659087.2015.1086613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Entenberg D, Wyckoff J, Gligorijevic B, Roussos ET, Verkhusha VV, Pollard JW, Condeelis J. Setup and use of a two-laser multiphoton microscope for multichannel intravital fluorescence imaging. Nat Protoc. 2011;6(10):1500–20. doi: 10.1038/nprot.2011.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harney AS, Wang Y, Condeelis JS, Entenberg D. Extended Time-lapse Intravital Imaging of Real-time Multicellular Dynamics in the Tumor Microenvironment. J Vis Exp. 2016;(112):e54042. doi: 10.3791/54042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyckoff J, Gligorijevic B, Entenberg D, Segall J, Condeelis J. High-resolution multiphoton imaging of tumors in vivo. Cold Spring Harbor Protocols. 2011;2011(10):1167–84. doi: 10.1101/pdb.top065904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harper KL, Sosa MS, Entenberg D, Hosseini H, Cheung JF, Nobre R, Avivar-Valderas A, Nagi C, Girnius N, Davis RJ, Farias EF, Condeelis J, Klein CA, Aguirre-Ghiso JA. Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature. 2016;540:589–612. doi: 10.1038/nature20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masedunskas A, Porat-Shliom N, Tora M, Milberg O, Weigert R. Intravital microscopy for imaging subcellular structures in live mice expressing fluorescent proteins. J Vis Exp. 2013;(79) doi: 10.3791/50558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dasari S, Weber P, Makhloufi C, Lopez E, Forestier CL. Intravital Microscopy Imaging of the Liver following Leishmania Infection: An Assessment of Hepatic Hemodynamics. J Vis Exp. 2015;(101):e52303. doi: 10.3791/52303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liou HL, Myers JT, Barkauskas DS, Huang AY. Intravital imaging of the mouse popliteal lymph node. J Vis Exp. 2012;(60) doi: 10.3791/3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sellers SL, Payne GW. Intravital microscopy of the inguinal lymph node. J Vis Exp. 2011;(50) doi: 10.3791/2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunn KW, Sutton TA, Sandoval RM. Live-animal imaging of renal function by multiphoton microscopy. Curr Protoc Cytom Chapter. 2007;12:Unit12 9. doi: 10.1002/0471142956.cy1209s41. [DOI] [PubMed] [Google Scholar]
- 19.Walter T, Shattuck DW, Baldock R, Bastin ME, Carpenter AE, Duce S, Ellenberg J, Fraser A, Hamilton N, Pieper S, Ragan MA, Schneider JE, Tomancak P, Heriche JK. Visualization of image data from cells to organisms. Nat Methods. 2010;7(3 Suppl):S26–41. doi: 10.1038/nmeth.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Entenberg D, Roorda RD, Toledo-Crow R. Non-linear microscope for imaging of the neural systems in live drosophila. OSA BioMed. 2004 [Google Scholar]
- 21.Entenberg D, Aranda I, Li Y, Toledo-Crow R, Schaer D, Li Y. Multimodal microscopy of immune cells and melanoma for longitudinal studies. Proc of SPIE. 2006;6081:62–73. [Google Scholar]
- 22.Deroulers C, Ameisen D, Badoual M, Gerin C, Granier A, Lartaud M. Analyzing huge pathology images with open source software. Diagn Pathol. 2013;8:92. doi: 10.1186/1746-1596-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clunie DA, Dennison DK, Cram D, Persons KR, Bronkalla MD, Primo HR. Technical Challenges of Enterprise Imaging: HIMSS-SIIM Collaborative White Paper. J Digit Imaging. 2016;29(5):583–614. doi: 10.1007/s10278-016-9899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gligorijevic B, Bergman A, Condeelis J. Multiparametric classification links tumor microenvironments with tumor cell phenotype. PLoS Biol. 2014;12(11):e1001995. doi: 10.1371/journal.pbio.1001995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zinselmeyer BH, Lynch JN, Zhang X, Aoshi T, Miller MJ. Video-rate two-photon imaging of mouse footpad - a promising model for studying leukocyte recruitment dynamics during inflammation. Inflamm Res. 2008;57(3):93–6. doi: 10.1007/s00011-007-7195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cahalan MD, Parker I, Wei SH, Miller MJ. Real-time imaging of lymphocytes in vivo. Curr Opin Immunol. 2003;15(4):372–7. doi: 10.1016/s0952-7915(03)00079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wyckoff J, Gligorijevic B, Entenberg D, Segall J, Condeelis J. Live Cell Imaging: A Laboratory Manual. Cold Spring Harbor Laboratory Press; 2010. High-Resolution multiphoton imaging of tumors in vivo; pp. 441–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorand RD, Barkauskas DS, Evans TA, Petrosiute A, Huang AY. Comparison of intravital thinned skull and cranial window approaches to study CNS immunobiology in the mouse cortex. Intravital. 2014;3(2) doi: 10.4161/intv.29728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weigert R, Porat-Shliom N, Amornphimoltham P. Imaging cell biology in live animals: ready for prime time. J Cell Biol. 2013;201(7):969–79. doi: 10.1083/jcb.201212130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heindl A, Nawaz S, Yuan Y. Mapping spatial heterogeneity in the tumor microenvironment: a new era for digital pathology. Lab Invest. 2015;95(4):377–84. doi: 10.1038/labinvest.2014.155. [DOI] [PubMed] [Google Scholar]
- 31.Kothari S, Phan JH, Stokes TH, Wang MD. Pathology imaging informatics for quantitative analysis of whole-slide images. Journal of the American Medical Informatics Association. 2013;20(6):1099–1108. doi: 10.1136/amiajnl-2012-001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lloyd MC, Rejniak KA, Brown JS, Gatenby RA, Minor ES, Bui MM. Pathology to enhance precision medicine in oncology: lessons from landscape ecology. Adv Anat Pathol. 2015;22(4):267–72. doi: 10.1097/PAP.0000000000000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price DL, Chow SK, Maclean NA, Hakozaki H, Peltier S, Martone ME, Ellisman MH. High-resolution large-scale mosaic imaging using multiphoton microscopy to characterize transgenic mouse models of human neurological disorders. Neuroinformatics. 2006;4(1):65–80. doi: 10.1385/NI:4:1:65. [DOI] [PubMed] [Google Scholar]
- 34.De Zanet S, Rudolph T, Richa R, Tappeiner C, Sznitman R. Retinal slit lamp video mosaicking. Int J Comput Assist Radiol Surg. 2016;11(6):1035–41. doi: 10.1007/s11548-016-1377-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seshamani S, Lau W, Hager G. Real-time endoscopic mosaicking. Med Image Comput Comput Assist Interv. 2006;9(Pt 1):355–63. doi: 10.1007/11866565_44. [DOI] [PubMed] [Google Scholar]
- 36.Kwasnitschka T, Koser K, Sticklus J, Rothenbeck M, Weiss T, Wenzlaff E, Schoening T, Triebe L, Steinfuhrer A, Devey C, Greinert J. DeepSurveyCam--A Deep Ocean Optical Mapping System. Sensors (Basel) 2016;16(2):164. doi: 10.3390/s16020164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loo BW, Jr, Meyer-Ilse W, Rothman SS. Automatic image acquisition, calibration and montage assembly for biological X-ray microscopy. J Microsc. 2000;197(Pt 2):185–201. doi: 10.1046/j.1365-2818.2000.00644.x. [DOI] [PubMed] [Google Scholar]
- 38.Legesse FB, Chernavskaia O, Heuke S, Bocklitz T, Meyer T, Popp J, Heintzmann R. Seamless stitching of tile scan microscope images. J Microsc. 2015;258(3):223–32. doi: 10.1111/jmi.12236. [DOI] [PubMed] [Google Scholar]
- 39.Vinegoni C, Lee S, Gorbatov R, Weissleder R. Motion compensation using a suctioning stabilizer for intravital microscopy. Intravital. 2012;1(2):115–121. doi: 10.4161/intv.23017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ritsma L, Steller EJ, Ellenbroek SI, Kranenburg O, Borel Rinkes IH, van Rheenen J. Surgical implantation of an abdominal imaging window for intravital microscopy. Nat Protoc. 2013;8(3):583–94. doi: 10.1038/nprot.2013.026. [DOI] [PubMed] [Google Scholar]
- 41.Nakasone ES, Askautrud HA, Kees T, Park JH, Plaks V, Ewald AJ, Fein M, Rasch MG, Tan YX, Qiu J, Park J, Sinha P, Bissell MJ, Frengen E, Werb Z, Egeblad M. Imaging tumor-stroma interactions during chemotherapy reveals contributions of the microenvironment to resistance. Cancer Cell. 2012;21(4):488–503. doi: 10.1016/j.ccr.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Looney MR, Thornton EE, Sen D, Lamm WJ, Glenny RW, Krummel MF. Stabilized imaging of immune surveillance in the mouse lung. Nat Methods. 2011;8(1):91–6. doi: 10.1038/nmeth.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harney AS, Arwert EN, Entenberg D, Wang Y, Guo P, Qian BZ, Oktay MH, Pollard JW, Jones JG, Condeelis JS. Real-Time Imaging Reveals Local, Transient Vascular Permeability, and Tumor Cell Intravasation Stimulated by TIE2hi Macrophage-Derived VEGFA. Cancer Discov. 2015;5(9):932–43. doi: 10.1158/2159-8290.CD-15-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karagiannis GS, Pastoriza JM, Wang Y, Harney AS, Entenberg D, Pignatelli J, Sharma VP, Xue EA, Cheng E, D'Alfonso TM, Jones JG, Anampa J, Rohan TE, Sparano JA, Condeelis JS, Oktay MH. Neoadjuvant chemotherapy induces breast cancer metastasis through a TMEM-mediated mechanism. Science translational medicine. 2017;9(397) doi: 10.1126/scitranslmed.aan0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Das S, Sarrou E, Podgrabinska S, Cassella M, Mungamuri SK, Feirt N, Gordon R, Nagi CS, Wang Y, Entenberg D, Condeelis J, Skobe M. Tumor cell entry into the lymph node is controlled by CCL1 chemokine expressed by lymph node lymphatic sinuses. J Exp Med. 2013;210(8):1509–28. doi: 10.1084/jem.20111627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zolla V, Nizamutdinova IT, Scharf B, Clement CC, Maejima D, Akl T, Nagai T, Luciani P, Leroux JC, Halin C, Stukes S, Tiwari S, Casadevall A, Jacobs WR, Jr, Entenberg D, Zawieja DC, Condeelis J, Fooksman DR, Gashev AA, Santambrogio L. Aging-related anatomical and biochemical changes in lymphatic collectors impair lymph transport, fluid homeostasis, and pathogen clearance. Aging Cell. 2015;14(4):582–94. doi: 10.1111/acel.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedl P, Locker J, Sahai E, Segall JE. Classifying collective cancer cell invasion. Nat Cell Biol. 2012;14(8):777–83. doi: 10.1038/ncb2548. [DOI] [PubMed] [Google Scholar]
- 48.Ovchinnikov DA, van Zuylen WJ, DeBats CE, Alexander KA, Kellie S, Hume DA. Expression of Gal4-dependent transgenes in cells of the mononuclear phagocyte system labeled with enhanced cyan fluorescent protein using Csf1r-Gal4VP16/UAS-ECFP double-transgenic mice. J Leukoc Biol. 2008;83(2):430–3. doi: 10.1189/jlb.0807585. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
XY stage insert design schematics.
Shallow window design schematics.
A) Preparation of the shallow imaging window. A thin layer of adhesive is placed on the inset rim of the window frame (red line) and the cover glass seated in the recess. B) Waiting time for adhesive to completely set. A setting time of one hour ensures that the cover glass cannot become dislodged and forms a water tight seal. C) Surgical exposure of the tissue of interest. Previously published protocols can be used to expose the tissue of interest. D) Stabilization of compliant tissue. The application of adhesive to a rigid piece of rubber allows it to be adhered to the back side of the tissue of interest. E) Application of adhesive to the frame of the shallow imaging window. Applying adhesive only to the rim of the imaging window (red line) will stabilize the tissue within the window, while avoiding exposure of the tissue of interest to the adhesive. F) Application of PBS to the cover glass. Before applying the window to the tissue, a 2–3 mm droplet of PBS is suspended from the center of the cover glass. When sealed to the tissue, a hydration chamber is formed which prevents dehydration of the tissue. G) Mouse with window adhered. Once window is adhered over the tissue of interest, the mouse may be positioned on the microscope. H) Mouse positioned on microscope stage. To begin imaging, the mouse is inverted and the window placed into the recess in the imaging stage plate.
Frames are 4.7 minutes apart.