Abstract
Microorganisms are in constant competition for growth niches and environmental resources. In Gram-negative bacteria, contact-dependent growth inhibition (CDI) systems link the fate of one cell with its immediate neighbor through touch-dependent, receptor-mediated toxin delivery. Though discovered for their ability to confer a competitive growth advantage, CDI systems also play significant roles in inter-sibling cooperation, promoting both auto-aggregation and biofilm formation. In this review, we detail the mechanisms of CDI toxin delivery and consider how toxin exchange between isogenic sibling cells could regulate gene expression.
INTRODUCTION
Within the polymicrobial communities that predominate in nature, bacteria adapt their physiology in response to the myriad of metabolites and signaling molecules in their environment. This dynamic interplay reflects the continuous adjustment to fluctuating nutrient/chemical landscapes in the environment and within animal and plant hosts. For example, some enteric pathogens establish transient syntrophic relationships with members of the microbiota, using the sugars released by saccharolytic bacteria to fuel growth and colonization (1). Invaders also compete with resident bacteria whose metabolic requirements overlap with their own. For example, beneficial commensal bacteria appear to suppress pathogen colonization by competing for preferred carbohydrates sources (2, 3). As metabolic demands evolve, so does the demeanor of established microbial interactions. Such changes can be abrupt when antibiotics or host immune responses impinge on the community. Previously antagonistic associations may turn collegial or vice versa, sometimes leading to co-infections (4–6).
Bacterial contact-dependent growth inhibition (CDI) is a common mechanism used by some Gram-negative bacteria to either initiate hostilities or forge cooperative interactions. CDI was discovered in Escherichia coli isolate EC93, which rapidly kills laboratory strains of E. coli K-12 during co-culture (7, 8). E. coli EC93 uses its CdiB and CdiA two-partner secretion proteins to deliver an ionophoric toxin into other E. coli cells (9). CdiA is an extended filamentous protein that is exported to the cell surface by the outer-membrane localized CdiB transporter. Upon binding its receptor on a neighboring cell, CdiA delivers its C-terminal toxin domain (CdiA-CT) into the target (Fig. 1). E. coli EC93 cells protect themselves from toxins delivered from neighboring siblings with CdiI immunity proteins. Thus, CDI imparts a competitive growth advantage over strains that lack immunity. However, CDI systems also promote cooperation between sibling cells, facilitating social behaviors that underlie biofilm formation and pathogenesis. Here, we review the molecular mechanism of CDI mediated competition and cooperation.
Figure 1. CDI-mediated toxin delivery.
CdiA is exported to the cell surface following Sec-dependent secretion across the inner membrane. CdiA binds to outer-membrane protein (OMP) receptors on target bacteria using a centrally located domain. Following receptor recognition, the C-terminal toxin domain is delivered into the periplasm where it presumably interacts with specific integral membrane proteins (IMP) to enter the lipid bilayer (for pore-forming toxins) or cross the membrane to enter the cytosol. Target bacteria are usually inhibited through dissipation of the proton-motive force or degradation of nucleic acids. Toxin domains delivered into sibling cells are neutralized by the direct binding of cognate CdiI immunity proteins. The outer-membrane (OM), peptidoglyan (PG) and cytoplasmic membrane (CM) are indicated.
DISTRIBUTION AND ORGANIZATION OF cdi LOCI
Since its discovery in E. coli EC93, cdi loci have been identified in a wide variety of Gram-negative proteobacteria (10, 11). The systems are invariably encoded within genomic/pathogenicity islands or plasmids (12); therefore not all strains of a given species necessarily carry cdi genes. Further, CdiA-CT and CdiI sequences are extraordinarily polymorphic between bacteria, such that strains often deploy different toxin-immunity pairs (10, 11, 13). CDI systems are typically organized as cdiBAI gene clusters, though most loci in the Burkholderiales are arranged in an alternative cdiAIB order (Fig. 2). In all instances, the toxic 3´-coding sequence of cdiA is closely linked to the downstream immunity gene, with the cdiI initiation codon often overlapping the cdiA termination codon. Consistent with their presence on mobile genetic elements, cdiA-CT/cdiI sequences appear to be shared between bacteria through horizontal gene transfer. Moreover, related toxin-immunity modules are often associated with bacteriocins and type VI and type VII secretion systems (10, 11, 14–19), indicating that the toxins are deployed by several competition systems. Further evidence of horizontal exchange is manifest by the tandem arrays of 'orphaned' cdiA-CT/cdiI gene pairs often found downstream of cdiBAI gene clusters (Fig. 2) (15, 17, 20). Insertion sequence elements, integrases and transposases are usually interspersed amongst the orphan toxin-immunity gene pairs. The function of orphan gene clusters remains largely unexplored. Because a number of orphan toxin-immunity pairs retain growth inhibition and immunity functions, they may expand the cell's toxic repertoire through recombination with the upstream cdiA gene (17, 20). Alternatively, orphan pairs may represent ancestral toxins that were displaced during the integration of new incoming cdiA-CT/cdiI sequences.
Figure 2. Variation in cdi gene clusters.
The cdi loci of selected bacterial strains are presented. Where available, ordered locus designations are provided below each cdiA gene. Secretion and accessory genes (cdiB, bcpO and hlyC) are depicted in yellow, and predicted immunity genes are shown in green. Genes encoding transposases, integrases and other proteins associated with mobile genetic elements are presented in light cyan. Orphan cdiA-CT/cdiI gene pairs are indicated by number (e.g. o1, o2, etc.) corresponding to their position downstream of the main cdiBAI cluster. The downward pointing carats indicate the position of sequences encoding VENN (or ELYN) peptide motifs that demarcate the variable CdiA-CT toxin region. Asterisks indicate that the orphan cdiA-CT sequence harbors mutations that are predicted to inactivate the encoded toxin domain.
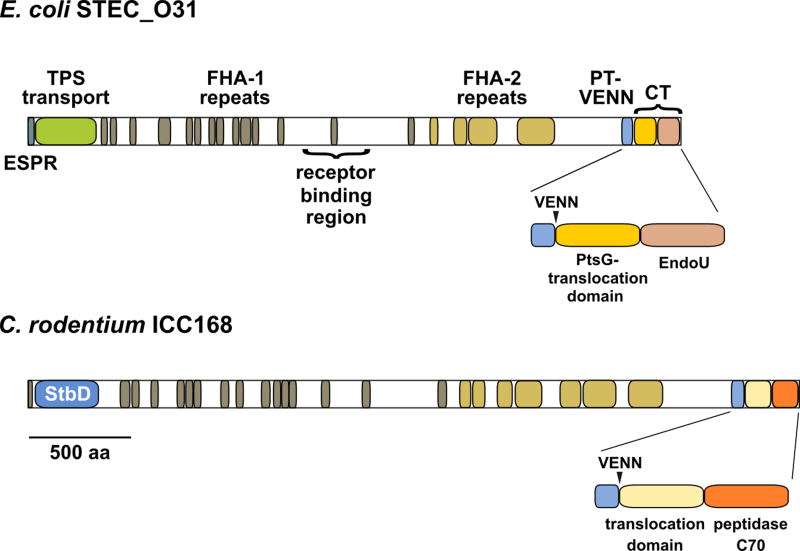
Though cdiB, cdiA and cdiI constitute the minimal core of a CDI system, some loci contain additional genes that are important for function. A subset of Burkholderia systems encode a small lipoprotein (BcpO) between cdiI and cdiB (Fig. 2). BcpO plays a significant role in CdiA secretion and toxin delivery for these systems (21). Other loci encode a putative lysyl acyltransferase related to the hemolysin activator HlyC (Fig. 2) (22, 23). This latter association suggests that some CdiA effectors may be subject to post-translational modification with fatty acyl chains. We also note that Citrobacter rodentium ICC168 encodes an unusual CdiA annotated as a fimbrial adhesin (Fig. 2). This protein contains the characteristic hemagglutinin-peptide repeats and pretoxin-VENN domain of CdiA, but lacks the N-terminal TPS transport domain required for export by CdiB (Fig. 3). Instead, the Citrobacter effector contains an N-terminal domain that is closely related to the StbD fimbrial protein from Salmonella species (Fig. 3). Collectively, these observations suggest that the fimbrial chaperone and usher proteins encoded immediately upstream mediate secretion and presentation of this unique CdiA effector (Fig. 2).
Figure 3. CdiA protein domain architecture.
Predicted domain structures of CdiA effectors from E. coli STEC_O31 (NCBI identifier: WP_001385946.1) and C. rodentium ICC168 (WP_012907078.1). The N-terminal extended signal peptide region (ESPR) and TPS transport domain (Pfam: PF05860) are required for CdiA secretion across cytoplasmic and outer membranes, respectively. FHA-1 peptide repeats likely form a right-handed β-helix, and FHA-2 repeats are also predicted to form a predominately β-structure. The pretoxin-VENN domain (PF04829) demarcates the variable C-terminal (CT) region. The CdiA-CT region is often composed of two variable domains. The N-terminal domain mediates translocation across the target-cell cytoplasmic membrane, and the extreme C-terminal domain is responsible for toxicity. The toxin domain of CdiASTECO31 is a predicted bacterial EndoU RNase (PF14436), and the toxin domain of CdiAICC168 is a predicted cysteine protease (PF12385).
TOXIN DELIVERY AND ACTIVATION
Outer membrane receptors
CdiA binds to specific cell-surface receptors on target bacteria (Fig. 1). The molecular basis of CdiA-receptor interactions is best understood for CDI systems deployed by E. coli strains EC93 and 536. The CdiAEC93 effector binds to BamA – a key subunit of the essential outer membrane β-barrel assembly machine (BAM) complex (24, 25). BamA is found in all Gram-negative bacteria, and its sequence is highly conserved amongst the Enterobacteriaceae. However, E. coli EC93 cells are unable to inhibit the growth of closely related enterobacteria because the surface exposed loops of BamA vary considerably between species (8). Expression of E. coli bamA in Salmonella Typhimurium, Citrobacter freundii and Enterobacter aerogenes sensitizes these bacteria to CdiAEC93-mediated growth inhibition. Similarly, exchange of E. coli bamA with alleles from E. cloacae or S. Typhimurium protects cells from growth inhibition and abrogates CdiAEC93-dependent cell-cell adhesion (8, 25). CdiAEC536 recognizes heterotrimeric complexes of OmpF and OmpC (26). OmpC and OmpF are highly expressed outer membrane osmoporins that are subject to intense selective pressure from bacteriophages and adaptive immune systems (27–29). Consequently, the surface exposed residues of OmpC vary between E. coli isolates and this antigenic variation protects many potential target strains from CdiAEC536 activity (26). Intriguingly, group A colicins appear to use OmpF as a conduit to translocate nuclease domains across the outer membrane (30, 31). It is presently unclear whether CdiAEC536 also uses OmpC/OmpF for toxin translocation. However, because CdiA proteins exploit a variety of cell-surface receptors, including lipopolysaccharide ((32) and unpublished data), it appears that porins are not an obligate part of the CDI toxin translocation pathway.
Recent work has localized the receptor-binding domain within the CdiA filament. Alignment of the closely related CdiAEC93 and CdiAEC536 proteins shows divergence in a central region between the FHA-1 and FHA-2 peptide repeats (Fig. 3), suggesting that this region could be responsible for differential receptor binding activity (26). This conclusion is somewhat unexpected, because prevailing models predict that the C-terminal toxin domain forms the distal end of the CdiA filament. However, truncated CdiAEC93 lacking ~1,000 C-terminal residues retains BamA-binding activity (25), demonstrating that the C-terminus is not required to bind target cells. Further, CdiAEC93 fragments containing the central region bind directly to purified BamA, and receptor specificity is altered when this region is exchanged between CdiA proteins (33). These findings underscore the modular nature of CdiA, but also raise important questions about effector structure and topology on the inhibitor cell surface.
Toxin translocation into the cytoplasm
Most characterized CDI toxins are nucleases, which must be translocated into the target-cell cytoplasm to degrade substrates. Crystallography and comparative sequence analyses indicate that the variable CdiA-CT region is often composed of two domains with distinct functions during CDI (22, 34, 35). The extreme C-terminal domain is responsible for toxicity, whereas the N-terminal domain governs transport from the periplasm into the target-cell cytoplasm (Fig. 3). Genetic evidence suggests that the N-terminal domain interacts directly with membrane protein receptors to mediate this translocation (Fig. 1) (36). Integral membrane proteins implicated in CDI include several ABC-transporters (GltJ/GltK, RbsC and MetI), the D-glucose transporter PtsG, the ATP-dependent zinc metallopeptidase FtsH, and a predicted inner-membrane protein of unknown function (YciB) (36). How these disparate membrane proteins mediate toxin translocation is not clear. One possibility is that N-terminal CdiA-CT domains use membrane proteins as receptors to bring tethered nuclease domains into close proximity with the lipid bilayer. Because toxin transport requires the proton-motive force (37), this electrochemical gradient could energize translocation. Additionally, the proton gradient lowers the periplasmic pH, which could induce toxin unfolding for insertion directly into the lipid bilayer. This mechanism is similar to that proposed for the import of colicin nuclease domains, which adopt a molten globule-like state upon interaction with membrane lipids (38–40). This destabilization of tertiary structure suggests that the toxins must unfold during transport across the cytoplasmic membrane.
Toxin activation
In some instances, CDI toxins are inherently inactive, with activation occurring only upon entry into target cells. The CdiA-CTEC536 toxin deployed by uropathogenic E. coli 536 is a latent anticodon nuclease that is only active when bound to the cysteine biosynthetic enzyme CysK (41). Purified CdiA-CTEC536 does not cleave tRNA in vitro unless the reaction is supplemented with CysK, and ∆cysK mutants are completely resistant to CdiA-CTEC536 toxin (41, 42). Intriguingly, CdiA-CTEC536 inserts its C-terminal GYGI peptide into the active-site cleft of CysK, mimicking the highly conserved 'cysteine synthase' complex, in which CysE uses its C-terminal GDGI motif to bind the CysK active site (43, 44). CysK significantly increases toxin thermostability and also promotes binding to tRNA substrates (44, 45). Because CDI toxins must presumably unfold to enter target cells, the interaction with CysK may have evolved to compensate for the toxin's intrinsic instability. In this model, the binding partner ensures that the toxin refolds efficiently after delivery into the cytoplasm. More recently, we have discovered that diverse CdiA-CT toxins from E. coli strains EC869, NC101 and 96.154 interact functionally with translation factors EF-Tu and EF-Ts (46). CdiA-CTEC869 specifically cleaves within the aminoacyl acceptor stem of tRNAGln and tRNAAsn molecules and binds to EF-Tu with high affinity. Remarkably, the toxin only cleaves substrate in the context of tRNA•EF-Tu•GTP ternary complexes (46), suggesting that the interaction with EF-Tu is required for nuclease activity. Moreover, EF-Ts is required for toxin activity in vivo, though guanine nucleotide exchange activity per se is not required for the nuclease reaction. Instead, it appear that EF-Ts promotes the formation of tRNA•EF-Tu•GTP complexes for rapid cleavage by CdiA-CTEC869. Finally, the trimeric integral membrane protein AcrB may play a role in the activation of membrane-pore toxins. E. coli ∆acrB mutants are resistant to the CdiA-CTEC93 pore-forming toxin during CDI (24), but also grow normally when the toxin is produced intra-cellularly (unpublished data). Thus, AcrB is required for intoxication regardless of whether the toxin is delivered from a neighboring cell or expressed internally. These observations raise the possibility that AcrB anchors CdiA-CTEC93 in the membrane or perhaps activates the toxin to allow ion flow.
THE YIN AND YANG OF BACTERIAL COMPETITION AND COOPERATION
Cell-cell adhesion and cooperative behavior
The fitness advantage of E. coli EC93 over laboratory strains led to the competition model of CDI function. However, the adhesive properties of CdiA also promote collaboration between sibling cells (Fig. 4). Expression of cdi is associated with auto-aggregation and biofilm formation in several species including Erwinia chysanthemii (47), Xylella fastidiosa (48), Neisseria meningitidis (49), Xanthomonas axonopodis (50), B. thailandensis (21), E. coli (25), and Pseudomonas aeruginosa (51). Biofilm formation is a prime example of cooperative behavior, in which individual bacteria collaborate to build multi-cellular structures. Such communities are beneficial because they protect inhabitants from predation and antimicrobial compounds. Moreover, this collective behavior often contributes to virulence, perhaps accounting for the large number of bacterial pathogens that carry CDI systems. For example, HecA/CdiA of Erwinia (Dickeya) chrysanthemii promotes adherence to leaf epidermal cells and allows planktonic bacteria to join pre-established cell aggregates (Fig. 4) (47). Similarly, CdiA homologs from N. meningitidis (HrpA) (52), Moraxella catarrhalis (MhaB) (53) and X. fastidiosa (HxfA) (54, 55) are proposed to mediate initial adhesion to host cells. For X. fastidiosa, HxfA/CdiA contributes to the colonization of insect vectors, thereby promoting subsequent transmission into plant hosts. In some instances, CDI may play a more active role in pathogenesis. N. meningitidis hrpA/cdiA mutants are less fit inside human cells and are also defective for endosomal escape into the cytoplasm (56).
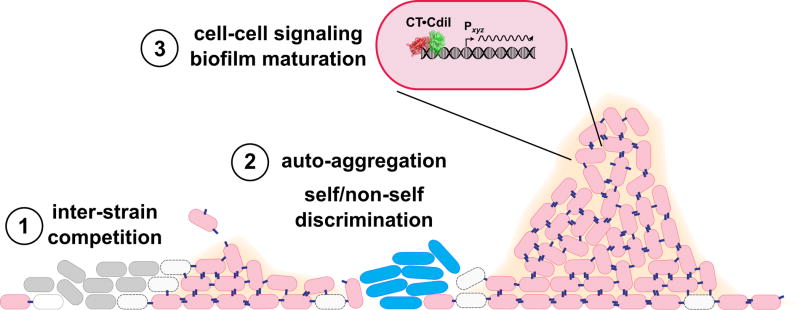
Figure 4. Cellular interactions mediated by CDI.
CDI systems mediate antagonistic and cooperative interactions in polymicrobial communities. During initial surface colonization, CDI mediates competition between strains of the same species (1). CDI+ cells (pink) use CdiA effectors (blue filaments) to inhibit CDI− strains (gray cells) of the same species. Inhibited CDI− bacteria are depicted as empty cells with dashed envelopes. CdiA adhesin activity promote auto-aggregation and the development of single-species microcolonies (2). Interactions between CdiA and its receptor also contribute to self/nonself discrimination, because unrelated species usually lack the specific receptor required for CDI-dependent cell adhesion (2). Finally, CDI appears to mediate cell-cell signaling to promote pillar formation in mature biofilms (3). Signaling entails changes in biofilm gene expression, suggesting that neutralized CdiA-CT•CdiI complexes could influence transcription as depicted in the expanded CDI+ cell.
The role of CDI in cooperative biofilm formation has been examined most extensively in B. thailandensis E264. The B. thailandensis cdi locus is critical for wild-type biofilm formation, and the number and size of pillar structures are altered in cdiA mutants (Fig. 4) (57). Further, B. thailandensis cells that constitutively express cdi genes produce abnormally dense and flat biofilms that lack discrete pillar structures. This latter finding demonstrates that cdi expression must be precisely regulated for cooperation. Transcription of the B. thailandensis cdi locus is induced by quorum-sensing homoserine lactones (58), and there is evidence of complex spatiotemporal regulation (21). Thus, cdi genes are preferentially expressed at high cell densities when cell-cell contacts are maximized. Of course, CDI-mediated cooperation is only possible between cells that express the appropriate cognate CdiI immunity protein. Non-isogenic bacteria are excluded from the biofilm community through toxin delivery (59). However, when non-isogenic cells are provided with the appropriate immunity gene, the unrelated populations are able to congregate and form mixed pillars. These studies demonstrate that self/non-self discrimination in bacteria can be controlled by specific toxin-immunity protein binding interactions.
Contact-dependent cell-cell signaling
Remarkably, CdiA-mediated cell-cell adhesion is not sufficient to promote full biofilm formation in B. thailandensis, and the exchange of catalytically active toxin domains is also required (57). This observation suggests that delivered toxin domains have an unanticipated signaling function when delivered into immune sibling cells. Recent work from Cotter and colleagues has revealed that toxin delivery is associated with changes in gene expression, including the upregulation of a putative transcriptional regulator involved in biofilm formation (60). The molecular pathway linking toxin delivery to transcriptional regulation is not yet known, but there are several possible mechanisms. CdiA-CTE264 has Mn2+-dependent DNase activity (21, 61), and therefore its intrinsic DNA-binding activity could contribute to signaling. Consistent with this model, we have found that the neutralized CdiA-CT•CdiIE264 complex binds to DNA (unpublished data), suggesting that the toxin-immunity protein complex could act as a transcription factor to regulate gene expression (Fig. 4). Alternatively, the complex may exhibit residual (and sub-lethal) 'nickase' activity, thereby modulating DNA supercoiling and indirectly influencing transcription. Whether contact-dependent cell signaling extends to other types of toxins remains an open question. We note that many CDI toxins cleave tRNA (34, 61, 62), and that tRNA fragments regulate protein synthesis and contribute to stress responses in vertebrates (63). Angiogenin represents an intriguing example of RNase-mediated response regulation. Angiogenin is a potent angiogenesis factor and a paralog of RNase A/RNase 1. Circulating angiogenin is endocytosed and trafficked to the nucleolus, where it cleaves a promoter-associated RNA to de-repress transcription of ribosomal RNA (64–66). The resulting increase in ribosome production is critical to support cell growth and proliferation. In addition, cytoplasmic angiogenin is activated to cleave the anticodons of mature tRNA during stress (67, 68). The resulting tRNA fragments inhibit protein synthesis, promote stress granule assembly and enhance cell survival (69, 70). Though these regulatory mechanisms have only been described in vertebrates, tRNA fragments accumulate in Streptomyces coelicolor during aerial hyphae formation and diminish later in development as spores are produced (71). These latter findings suggest that tRNA fragments have regulatory activities in prokaryotes and raise the possibility that CDI delivered RNase toxins could also participate in signaling. Determining how CDI toxin exchange regulates gene expression represents an outstanding and exciting challenge for the field.
Acknowledgments
Research in the Low and Hayes laboratories is supported by grants from the National Science Foundation (MCB1545720) and the National Institutes of Health (GM117373 and GM117930). A.M.J. is supported in part by the Ralph M. Parsons Fellowship.
References
- 1.Ng KM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502(7469):96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamada N, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336(6086):1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS One. 2013;8(1):e53957. doi: 10.1371/journal.pone.0053957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mashburn LM, Jett AM, Akins DR, Whiteley M. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol. 2005;187(2):554–566. doi: 10.1128/JB.187.2.554-566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen AT, Jones JW, Ruge MA, Kane MA, Oglesby-Sherrouse AG. Iron Depletion Enhances Production of Antimicrobials by Pseudomonas aeruginosa . J Bacteriol. 2015;197(14):2265–2275. doi: 10.1128/JB.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wakeman CA, et al. The innate immune protein calprotectin promotes Pseudomonas aeruginosa and Staphylococcus aureus interaction. Nat Commun. 2016;7:11951. doi: 10.1038/ncomms11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aoki SK, et al. Contact-dependent inhibition of growth in Escherichia coli. Science. 2005;309(5738):1245–1248. doi: 10.1126/science.1115109. [DOI] [PubMed] [Google Scholar]
- 8.Ruhe ZC, Wallace AB, Low DA, Hayes CS. Receptor polymorphism restricts contact-dependent growth inhibition to members of the same species. MBio. 2013;4(4) doi: 10.1128/mBio.00480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aoki SK, Webb JS, Braaten BA, Low DA. Contact-dependent growth inhibition causes reversible metabolic downregulation in Escherichia coli . J Bacteriol. 2009;191(6):1777–1786. doi: 10.1128/JB.01437-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aoki SK, et al. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature. 2010;468(7322):439–442. doi: 10.1038/nature09490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct. 2012;7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruhe ZC, et al. CDI Systems Are Stably Maintained by a Cell-Contact Mediated Surveillance Mechanism. PLoS Genet. 2016;12(6):e1006145. doi: 10.1371/journal.pgen.1006145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morse RP, et al. Diversification of beta-Augmentation Interactions between CDI Toxin/Immunity Proteins. J Mol Biol. 2015;427(23):3766–3784. doi: 10.1016/j.jmb.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang D, Iyer LM, Aravind L. A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res. 2011;39(11):4532–4552. doi: 10.1093/nar/gkr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poole SJ, et al. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet. 2011;7(8):e1002217. doi: 10.1371/journal.pgen.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holberger LE, Garza-Sanchez F, Lamoureux J, Low DA, Hayes CS. A novel family of toxin/antitoxin proteins in Bacillus species. FEBS Lett. 2012;586(2):132–136. doi: 10.1016/j.febslet.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koskiniemi S, et al. Selection of orphan Rhs toxin expression in evolved Salmonella enterica serovar Typhimurium. PLoS Genet. 2014;10(3):e1004255. doi: 10.1371/journal.pgen.1004255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghequire MG, De Mot R. Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol Rev. 2014;38(4):523–568. doi: 10.1111/1574-6976.12079. [DOI] [PubMed] [Google Scholar]
- 19.Walker D, Lancaster L, James R, Kleanthous C. Identification of the catalytic motif of the microbial ribosome inactivating cytotoxin colicin E3. Protein Sci. 2004;13(6):1603–1611. doi: 10.1110/ps.04658504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arenas J, Schipper K, van Ulsen P, van der Ende A, Tommassen J. Domain exchange at the 3´ end of the gene encoding the fratricide meningococcal two-partner secretion protein A. BMC Genomics. 2013;14:622. doi: 10.1186/1471-2164-14-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson MS, Garcia EC, Cotter PA. The Burkholderia bcpAIOB genes define unique classes of two-partner secretion and contact dependent growth inhibition systems. PLoS Genet. 2012;8(8):e1002877. doi: 10.1371/journal.pgen.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willett JL, Ruhe ZC, Goulding CW, Low DA, Hayes CS. Contact-Dependent Growth Inhibition (CDI) and CdiB/CdiA Two-Partner Secretion Proteins. J Mol Biol. 2015;427(23):3754–3765. doi: 10.1016/j.jmb.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogier JC, Duvic B, Lanois A, Givaudan A, Gaudriault S. A New Member of the Growing Family of Contact-Dependent Growth Inhibition Systems in Xenorhabdus doucetiae. PLoS One. 2016;11(12):e0167443. doi: 10.1371/journal.pone.0167443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aoki SK, et al. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol Microbiol. 2008;70(2):323–340. doi: 10.1111/j.1365-2958.2008.06404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruhe ZC, et al. CdiA promotes receptor-independent intercellular adhesion. Mol Microbiol. 2015;98(1):175–192. doi: 10.1111/mmi.13114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beck CM, et al. CdiA Effectors from Uropathogenic Escherichia coli Use Heterotrimeric Osmoporins as Receptors to Recognize Target Bacteria. PLoS Pathog. 2016;12(10):e1005925. doi: 10.1371/journal.ppat.1005925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snyder JA, et al. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect Immun. 2004;72(11):6373–6381. doi: 10.1128/IAI.72.11.6373-6381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen SL, et al. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli : a comparative genomics approach. Proc Natl Acad Sci U S A. 2006;103(15):5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen L, Bollback JP, Dimmic M, Hubisz M, Nielsen R. Genes under positive selection in Escherichia coli. Genome Res. 2007;17(9):1336–1343. doi: 10.1101/gr.6254707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Housden NG, et al. Intrinsically disordered protein threads through the bacterial outer-membrane porin OmpF. Science. 2013;340(6140):1570–1574. doi: 10.1126/science.1237864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Housden NG, Kleanthous C. Colicin translocation across the Escherichia coli outer membrane. Biochem Soc Trans. 2012;40(6):1475–1479. doi: 10.1042/BST20120255. [DOI] [PubMed] [Google Scholar]
- 32.Koskiniemi S, et al. Genetic analysis of the CDI pathway from Burkholderia pseudomallei 1026b. PLoS One. 2015;10(3):e0120265. doi: 10.1371/journal.pone.0120265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruhe ZC, et al. CdiA effectors use modular receptor-binding domains to recognize target bacteria. mBio. 2017;8(2):e00290–17. doi: 10.1128/mBio.00290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morse RP, et al. Structural basis of toxicity and immunity in contact-dependent growth inhibition (CDI) systems. Proc Natl Acad Sci U S A. 2012;109(52):21480–21485. doi: 10.1073/pnas.1216238110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beck CM, et al. CdiA from Enterobacter cloacae delivers a toxic ribosomal RNase into target bacteria. Structure. 2014;22(5):707–718. doi: 10.1016/j.str.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willett JL, Gucinski GC, Fatherree JP, Low DA, Hayes CS. Contact-dependent growth inhibition toxins exploit multiple independent cell-entry pathways. Proc Natl Acad Sci U S A. 2015;112(36):11341–11346. doi: 10.1073/pnas.1512124112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruhe ZC, Nguyen JY, Beck CM, Low DA, Hayes CS. The proton-motive force is required for translocation of CDI toxins across the inner membrane of target bacteria. Mol Microbiol. 2014;94(2):466–481. doi: 10.1111/mmi.12779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mosbahi K, et al. The cytotoxic domain of colicin E9 is a channel-forming endonuclease. Nat Struct Biol. 2002;9(6):476–484. doi: 10.1038/nsb797. [DOI] [PubMed] [Google Scholar]
- 39.Mosbahi K, et al. Destabilization of the colicin E9 endonuclease domain by interaction with negatively charged phospholipids: implications for colicin translocation into bacteria. J Biol Chem. 2004;279(21):22145–22151. doi: 10.1074/jbc.M400402200. [DOI] [PubMed] [Google Scholar]
- 40.Mosbahi K, Walker D, James R, Moore GR, Kleanthous C. Global structural rearrangement of the cell penetrating ribonuclease colicin E3 on interaction with phospholipid membranes. Protein Sci. 2006;15(3):620–627. doi: 10.1110/ps.051890306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diner EJ, Beck CM, Webb JS, Low DA, Hayes CS. Identification of a target cell permissive factor required for contact-dependent growth inhibition (CDI) Genes Dev. 2012;26(5):515–525. doi: 10.1101/gad.182345.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beck CM, Diner EJ, Kim JJ, Low DA, Hayes CS. The F pilus mediates a novel pathway of CDI toxin import. Mol Microbiol. 2014;93(2):276–290. doi: 10.1111/mmi.12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campanini B, et al. Moonlighting O-acetylserine sulfhydrylase: New functions for an old protein. Biochim Biophys Acta. 2015;1854(9):1184–1193. doi: 10.1016/j.bbapap.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson PM, et al. Unraveling the essential role of CysK in CDI toxin activation. Proc Natl Acad Sci U S A. 2016;113(35):9792–9797. doi: 10.1073/pnas.1607112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaundal S, Uttam M, Thakur KG. Dual Role of a Biosynthetic Enzyme, CysK, in Contact Dependent Growth Inhibition in Bacteria. PLoS One. 2016;11(7):e0159844. doi: 10.1371/journal.pone.0159844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones AM, Garza-Sanchez F, So J, Hayes CS, Low DA. Activation of contact-dependent antibacterial tRNase toxins by translation elongation factors. Proc Natl Acad Sci U S A. 2017;114(10):E1951–E1957. doi: 10.1073/pnas.1619273114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rojas CM, Ham JH, Deng WL, Doyle JJ, Collmer A. HecA, a member of a class of adhesins produced by diverse pathogenic bacteria, contributes to the attachment, aggregation, epidermal cell killing, and virulence phenotypes of Erwinia chrysanthemi EC16 on Nicotiana clevelandii seedlings. Proc Natl Acad Sci U S A. 2002;99(20):13142–13147. doi: 10.1073/pnas.202358699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guilhabert MR, Kirkpatrick BC. Identification of Xylella fastidiosa antivirulence genes: hemagglutinin adhesins contribute X. fastidiosa biofilm maturation and colonization and attenuate virulence. Mol Plant Microbe Interact. 2005;18(8):856–868. doi: 10.1094/MPMI-18-0856. [DOI] [PubMed] [Google Scholar]
- 49.Neil RB, Apicella MA. Role of HrpA in biofilm formation of Neisseria meningitidis and regulation of the hrpBAS transcripts. Infect Immun. 2009;77(6):2285–2293. doi: 10.1128/IAI.01502-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gottig N, Garavaglia BS, Garofalo CG, Orellano EG, Ottado J. A filamentous hemagglutinin-like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PLoS One. 2009;4(2):e4358. doi: 10.1371/journal.pone.0004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mercy C, Ize B, Salcedo SP, de Bentzmann S, Bigot S. Functional Characterization of Pseudomonas Contact Dependent Growth Inhibition (CDI) Systems. PLoS One. 2016;11(1):e0147435. doi: 10.1371/journal.pone.0147435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmitt C, et al. A functional two-partner secretion system contributes to adhesion of Neisseria meningitidis to epithelial cells. J Bacteriol. 2007;189(22):7968–7976. doi: 10.1128/JB.00851-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balder R, Hassel J, Lipski S, Lafontaine ER. Moraxella catarrhalis strain O35E expresses two filamentous hemagglutinin-like proteins that mediate adherence to human epithelial cells. Infect Immun. 2007;75(6):2765–2775. doi: 10.1128/IAI.00079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Killiny N, Almeida RP. Factors affecting the initial adhesion and retention of the plant pathogen Xylella fastidiosa in the foregut of an insect vector. Appl Environ Microbiol. 2014;80(1):420–426. doi: 10.1128/AEM.03156-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Killiny N, Almeida RP. Xylella fastidiosa afimbrial adhesins mediate cell transmission to plants by leafhopper vectors. Appl Environ Microbiol. 2009;75(2):521–528. doi: 10.1128/AEM.01921-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tala A, et al. The HrpB-HrpA two-partner secretion system is essential for intracellular survival of Neisseria meningitidis. Cell Microbiol. 2008;10(12):2461–2482. doi: 10.1111/j.1462-5822.2008.01222.x. [DOI] [PubMed] [Google Scholar]
- 57.Garcia EC, Anderson MS, Hagar JA, Cotter PA. Burkholderia BcpA mediates biofilm formation independently of interbacterial contact-dependent growth inhibition. Mol Microbiol. 2013;89(6):1213–1225. doi: 10.1111/mmi.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Majerczyk C, et al. Global analysis of the Burkholderia thailandensis quorum sensing-controlled regulon. J Bacteriol. 2014;196(7):1412–1424. doi: 10.1128/JB.01405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anderson MS, Garcia EC, Cotter PA. Kind discrimination and competitive exclusion mediated by contact-dependent growth inhibition systems shape biofilm community structure. PLoS Pathog. 2014;10(4):e1004076. doi: 10.1371/journal.ppat.1004076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garcia EC, Perault AI, Marlatt SA, Cotter PA. Interbacterial signaling via Burkholderia contact-dependent growth inhibition system proteins. Proc Natl Acad Sci U S A. 2016;113(29):8296–8301. doi: 10.1073/pnas.1606323113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nikolakakis K, et al. The toxin/immunity network of Burkholderia pseudomallei contact-dependent growth inhibition (CDI) systems. Mol Microbiol. 2012;84(3):516–529. doi: 10.1111/j.1365-2958.2012.08039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson PM, et al. Functional Diversity of Cytotoxic tRNase/Immunity Protein Complexes from Burkholderia pseudomallei. J Biol Chem. 2016;291(37):19387–19400. doi: 10.1074/jbc.M116.736074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Diebel KW, Zhou K, Clarke AB, Bemis LT. Beyond the Ribosome: Extra-translational Functions of tRNA Fragments. Biomark Insights. 2016;11(Suppl 1):1–8. doi: 10.4137/BMI.S35904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu ZP, Tsuji T, Riordan JF, Hu GF. The nuclear function of angiogenin in endothelial cells is related to rRNA production. Biochem Biophys Res Commun. 2002;294(2):287–292. doi: 10.1016/S0006-291X(02)00479-5. [DOI] [PubMed] [Google Scholar]
- 65.Xu ZP, Tsuji T, Riordan JF, Hu GF. Identification and characterization of an angiogenin-binding DNA sequence that stimulates luciferase reporter gene expression. Biochemistry. 2003;42(1):121–128. doi: 10.1021/bi020465x. [DOI] [PubMed] [Google Scholar]
- 66.Hoang TT, Raines RT. Molecular basis for the autonomous promotion of cell proliferation by angiogenin. Nucleic Acids Res. 2017;45(2):818–831. doi: 10.1093/nar/gkw1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fu H, et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583(2):437–442. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 68.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185(1):35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Emara MM, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285(14):10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43(4):613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haiser HJ, Karginov FV, Hannon GJ, Elliot MA. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res. 2008;36(3):732–741. doi: 10.1093/nar/gkm1096. [DOI] [PMC free article] [PubMed] [Google Scholar]