Abstract
Obesity during maturation can affect the growing skeleton directly and indirectly, although these effects and the mechanisms behind them are not fully understood. Our objective was to determine how a high-fat diet with or without metformin treatment affects skeletal development. We also sought to characterize changes that occur in white adipose tissue, circulating metabolites, lipids, and gut microbiota. A diet-induced obesity C57BL/6J mouse model was used to test the effects of obesity and metformin on bone using bone histomorphometry and microcomputed tomography. Bone marrow adipose tissue was quantified with osmium tetroxide microcomputed tomography and histology. Dual-energy x-ray absorptiometry was used to analyze body composition. Hematoxylin and eosin staining was used to assess changes in white adipose depots, mass spectrometry was used for circulating lipids and protein metabolite analysis, and ribosomal RNA sequencing was used for gut microbiome analysis. Mice fed a high fat-diet since wean displayed increased medullary areas and decreased osteoblast numbers in the long bones; this phenotype was partially normalized by metformin. Marrow and inguinal adipose expansion was also noted in obese mice, and this was partially normalized by metformin. A drug-by-diet interaction was noted for circulating lipid molecules, protein metabolites, and gut microbiome taxonomical units. Obesity was not detrimental to trabecular bone in growing mice, but bone marrow medullary expansion was observed, likely resulting from inhibition of osteoblastogenesis, and this was partially reversed by metformin treatment.
In this work, a comprehensive analysis was performed on mice in response to high-fat diet and metformin in terms of bone, adipose, circulating metabolites and lipids, and gut microbiome phenotypes.
Obesity is a common and mounting global disease with complex and integrated consequences, including systemic inflammation, insulin resistance, and nonalcoholic fatty liver disease. Obese adults have higher bone mineral density (BMD) and greater bone strength in the tibia (1), lumbar spine (2), femoral neck, distal radius (2), and lower extremities (3). However, fracture risk is also increased in obese individuals with either prediabetes or normal glucose tolerance, even when adjusted for age, physical activity, and the use of drugs known to increase the risk of fractures (4); this highlights the complicated relationship between bone condition and obesity. Thus, it is crucial that the bone health of these patients be carefully considered, especially in relation to diabetes treatments. Thus, a more comprehensive analysis of the interactions among obesity, antidiabetic drugs, and bone phenotypes would benefit the field and is undertaken here.
Metformin is the most widely prescribed treatment of type 2 diabetes mellitus worldwide and is also used for obese, prediabetic patients. A more complete understanding of the skeletal response to metformin is confounded by the fact that the impact of dietary excess on skeletal turnover is itself not well understood. Metformin improves glucose tolerance, insulin sensitivity, and peripheral glucose uptake, and decreases insulin-induced suppression of fatty acid oxidation, low-density lipoprotein cholesterol, and triglyceride levels (5). Despite minimal weight loss effects, metformin improves immune function and decreases inflammation by reducing C-reactive proteins and proinflammatory cytokines such as tumor necrosis factor-α (5). Metformin also affects white adipose tissue (WAT) and suppresses abnormal extracellular matrix remodeling in adipose tissue in mice with diet-induced obesity (DIO) (6); however, the actions of metformin on the skeleton are not clear (7, 8). Results from human studies are conflicting and show either a neutral or beneficial effect on the skeleton, in contrast to thiazolidinediones, which have unfavorable effects on bone (7). We hypothesized that metformin has different effects in lean vs obese individuals and tested this here using a well-established DIO mouse model that shares many characteristics with human obesity (9). We aimed to characterize the full spectrum of effects of metformin in both lean and obese mice, including effects on skeletal phenotype, bone marrow adipose tissue (MAT), WAT, systemic lipid and protein metabolites, and gut microbiome composition.
Methods
Animal care, euthanization, and blood collection
Male and female C57BL/6J mice (stock# 000664, The Jackson Laboratory, Bar Harbor, ME) were purchased and bred at the Maine Medical Center Research Institute animal facility or the Dana-Farber Cancer Institute animal facility and used for experiments. Male mice were randomly assigned to each group and were used for all experiments. Mice had ad libitum access to food and water. Mice were euthanized by CO2 inhalation or exsanguination. For exsanguination, mice were anesthetized using 100 mg/kg ketamine and 15 mg/kg xylazine (Sigma-Aldrich, St. Louis, MO) and blood (2 mL) was collected into a sterile microcentrifuge tube from mice after surgical removal of their eye. Blood was allowed to clot for 45 minutes at room temperature, and centrifuged for 15 minutes at 12,000 relative centrifugal force to separate serum (top, used for lipidomic and metabolomics profiling) from the cellular component (bottom). Experiments were performed in accordance with approved protocols from the appropriate Institutional Animal Care and Use Committee (Dana-Farber Cancer Institute, Boston, MA, or Maine Medical Center Research Institute, Scarborough, ME). No adverse effects were observed in our studies.
Diet and drug treatments
From the age of wean, mice were fed a high-fat diet (HFD) (60% kcal from fat) (D12492, Research Diets, Inc., New Brunswick, NJ) or chow control diet (RMH 3000 Prolab® Isopro® LabDiets, St. Louis, MO, or Tekland 2018, Envigo, Madison WI) from weening (∼3 weeks) for 15 weeks. For the following 6 weeks, mice were also treated with or without 300 mg/kg metformin in drinking water, as described previously (10, 11). Metformin HCl 1000 mg tablets, (Drug NDC: 683820030, PHR1084-500MG, Sigma-Aldrich), were crushed and added to mouse drinking water (2 g/L) for an equivalent dose of 300 mg/kg per day, based on estimates that mice drink 1.5 mL/10 g body weight per day.
Body composition
Dual-energy x-ray absorptiometry for whole-body composition exclusive of the head were performed on mice at 16, 20, and 22 weeks at age using the PIXImus (GE Lunar, Fairfield, CT) as described previously (12). The PIXImus was calibrated daily with a phantom provided by the manufacturer.
Bone histomorphometry
Femur and tibia were dissected from mice, fixed in 10% formalin for 48 hours, transferred to phosphate-buffered saline, dehydrated, embedded in methyl methacrylate, sectioned into 4-μm slices, and stained with von Kossa as described previously (13). Consecutive sections were stained by tartrate-resistant acid phosphatase and toluidine blue to quantitate structure and cellular parameters by bone histomorphometry. Bone histomorphometry was performed using the Osteomeasure system (Osteo-Metrics, Decatur, GA), and the results were analyzed as described and expressed according to the standardized nomenclature (14).
Microcomputed tomography
A high-resolution desktop microtomographic imaging system (µCT40, Scanco Medical AG, Brüttisellen, Switzerland) was used to assess trabecular bone microarchitecture and cortical bone morphology, in the proximal metaphysis and middiaphysis of the tibia. This machine was also used to assess trabecular bone microarchitecture and cortical bone morphology in the distal metaphysis and middiaphysis of the femur, respectively. Scans of the entire tibia and femur were acquired using a 10-µm3 isotropic voxel size, 70-kVp peak x-ray tube potential, 114-µA x-ray intensity, and 200-ms integration time, and were subjected to Gaussian filtration and segmentation. Image acquisition and analysis protocols adhered to guidelines for microcomputed tomography (µCT) assessment of rodent bone microstructure (15). Trabecular bone was identified by manually contouring the endocortical region and using a threshold of 338 mgHA/cm3 to segment bone from soft tissue. In the proximal tibia, trabecular bone architecture was analyzed in a 1000-µm long region using 100 transverse 10-µm slices that began 100 µm inferior to the growth plate and extended distally. In the femur, trabecular bone architecture was analyzed in a region that began 200 µm superior to the distal femoral growth plate and extended proximally 1500 µm, using 150 transverse 10-µm slices. The segmented images were analyzed with the standard Scanco trabecular bone morphology script to measure trabecular bone volume fraction (%), trabecular BMD (mgHA/cm3), trabecular thickness (mm), trabecular number (mm−1), trabecular separation (mm), and connectivity density (1/mm3). Cortical bone morphology was analyzed in a 500-µm (50 transverse slices) long region that began 2 mm superior to the distal tibiofibular junction and extended distally (for the tibia) or at the femoral middiaphysis (for the femur). Cortical bone was segmented using a threshold of 700 mgHA/cm3 and the standard Scanco script for cortical bone morphology was used to measure total cross-sectional area (mm2), cortical bone area (mm2), medullary area (mm2), bone area fraction (cortical bone area/total cross-sectional area, %), cortical tissue mineral density (mgHA/cm3), cortical thickness (mm), cortical porosity (%), periosteal perimeter (mm), endocortical perimeter (mm), and the maximum, minimum, and polar moments of inertia (mm4).
OsO4 µCT
Quantification and visualization of marrow adipose tissue was performed as we previously described (16). Briefly, long bones were dissected free of soft tissues and fixed in 10% neutral buffered formalin (Fisher Scientific, Pittsburgh, PA) overnight at 4°C with gentle agitation. MAT was quantified via osmium tetroxide (OsO4) staining followed by µCT as described (16). Following µCT scanning for trabecular and cortical structure, the tibiae were demineralized in 4.1% EDTA, pH 7.4, for 14 days. These were then washed with cold tap water and then placed into 2-mL cryovials with a four-way divider (four tibiae per vial). In a fume hood, equal volumes of 5% potassium dichromate and 2% OsO4 (Electron Microscopy Sciences, Hatfield, PA, catalog no. 19152) (1% final OsO4 concentration) were mixed and added to each cryovial. Then cryovials were capped and the tibiae were allowed to incubate for 48 hours at room temperature. The tibiae were washed in cool tap water for 2 hours to remove unbound OsO4 and then transferred to fresh tubes containing phosphate-buffered saline. OsO4-stained tibiae were imaged using a µCT scanner (µCT-35 or µCT-40) (Scanco Medical). Scans were acquired of the whole tibia using a 10-µm3 isotropic voxel size, 55-kVp peak x-ray tube potential, 145-µA x-ray intensity, and 300- to 500-ms integration time, and were subjected to Gaussian filtration and segmentation. The medullary cavity for each tibia was manually contoured in the same regions where proximal medullary volume and medullary volumes were measured in the mineralized µCT scans. OsO4 staining in the distal regions was segmented using thresholds equivalent to 1584 mgHA/cm3 and volumetric analysis was run to measure MAT volume (mm3). MAT volumes were normalized by the medullary volumes from the mineralized µCT analysis and are reported as the distal MAT volume fractions (%).
Immunohistochemistry
For hematoxylin and eosin staining, tissues were fixed in 10% formalin for 3 days, paraffin embedded, and processed for staining. Bones were either decalcified until soft in EDTA or plastic embedded and sectioned without decalcification. Images were taken using a Nikon Eclipse 80i microscope. The ImageJ length tool and cell count macro was used to determine adipocyte number and average diameter of all adipocytes in one field of view (image taken from 20× objective). Four fields of view per animal were taken and averaged together to obtain an average adipocyte diameter (d) for each animal. Adipocyte area was approximated by using the equation area = π*(d/2)2.
Serum lipidomic analysis
Lipidomic analysis was performed on serum from mouse eye bleeds using direct infusion and sequential precursor ion fragmentation methods using our water-free dichloromethane/methanol lipid isolation approach (17). Briefly, 10 μL of mouse serum was combined with water, methanol, and dichloromethane. After vortexing, a dichloromethane/methanol/sample monophase was obtained and allowed to stand for 30 minutes at room temperature. The monophase was broken into a two-phase system following addition of dichloromethane and methanol by centrifugation for 10 minutes at 300 relative centrifugal force in a Heraeus Instruments (Hanau, Germany) Megafuge 2R. The lower dichloromethane organic phase was removed to a new glass tube and dried under nitrogen and a partial vacuum using a Visiprep manifold (Supelco, Bellefonte, PA). Lipid extracts were dissolved in methanol/dichloromethane (50:50, v/v, containing 5 mM ammonium acetate) for mass spectrometry (MS) analysis. MS was performed by direct infusion in 50% dichloromethane, 50% methanol, and 5 mM ammonium acetate onto a Sciex 5600 TripleTOF mass spectrometer. Lipids were analyzed using a bias-free global lipid profiling acquisition technique, termed MS/MSALL, using sequential precursor ion fragmentation. Duplicate MS runs were conducted in two ion modes: positive (e.g., for triacylglycerols, sphingomyelins, cholesteryl esters) and negative (e.g., for glycerophospholipids). Minimum signal-to-noise and minimum percent intensities for MS and MS/MS were: triacylglycerols (MS 5/1 and MS/MS, 10/1, 2%); sphingomyelins (SMs; MS 5/1 and MS/MS, 5/1, 2%); diacylglycerols (MS 5/1 and MS/MS, 2/1, 2%); and glycerophospholipids (MS 5/2 and MS/MS, 10/1, 2%). Otherwise, MS/MSALL parameters were as previously published (17, 18). For sample comparisons, a t test was applied using MarkerView software (Sciex, Framingham, MA).
Serum metabolite analysis
Metabolites were extracted from mouse blood in ice-cold methanol and endogenous metabolite profiles were obtained using two liquid chromatography-tandem mass spectrometry methods as described (19). Metabolomic profiling was done at the Beth Israel Mass Spectrometry Core Facility (Beth Israel Deaconess Medical Center, Boston, MA). Data were acquired using a 5500 QTRAP triple quadrupole mass spectrometer (AB/Sciex) coupled to a Prominence UFLC system (Shimadzu, Kyoto, Japan) via selected reaction monitoring of 289 endogenous water soluble metabolites for steady-state analyses of samples. All values that were nondetectable were assigned the lowest detectable value (3000). Excel software was used for analysis, statistically significant metabolites were identified if they met the requirements of a fold change cutoff over 1.5, and P < 0.05 using a two-tailed homoscedastic t test with n = 3.
Microbiome sequencing analysis
At age 23 weeks, five to seven pellets of feces per mouse were taken from mice immediately upon excretion, frozen at −80°C, and shipped on dry ice to Second Genome Inc. (San Francisco, CA) for processing and analysis.
Sample preparation
Nucleic acid isolation was performed with the MoBio PowerMag® Microbiome kit (Qiagen, Carlsbad, CA) according to the manufacturer’s guidelines and optimized for high-throughput processing. All samples were quantified via the Qubit® Quant-iT dsDNA High Sensitivity Kit (Invitrogen, Carlsbad, CA) to ensure that they met minimum concentration and mass of DNA.
Library preparation
To enrich the sample for bacterial 16S V4 rDNA region, DNA was amplified using fusion primers designed against the surrounding conserved regions that are tailed with sequences to incorporate Illumina adapters and indexing barcodes. Each sample was polymerase chain reaction (PCR)–amplified with two differently barcoded V4 fusion primers. Samples that met the post-PCR quantification minimum were advanced for pooling and sequencing. For each sample, amplified products were concentrated using a solid-phase reversible immobilization method for the purification of PCR products and quantified by quantitative PCR.
Profiling methods
A pool containing 16S V4 enriched, amplified, barcoded samples were loaded into a MiSeq® reagent cartridge, and then onto the instrument along with the flow cell. After cluster formation on the MiSeq instrument, the amplicons were sequenced for 250 cycles with custom primers designed for paired-end sequencing. Samples are processed in a Good Laboratory Practices–compliant service laboratory running Quality Management Systems for sample and data tracking. The laboratory implements detailed standard operating procedures, equipment and process validation, training, audits and document control measures. Quality control and quality assurance metrics are maintained for all sample handling, processing and storage procedures.
Ordination, clustering, and classification methods
Principal coordinate analysis was used for two-dimensional ordination plotting to help visualize complex relationships between samples. Principal coordinate analysis uses the sample-to-sample dissimilarity values to position the points relative to each other by maximizing the linear correlation between the dissimilarity values and the plot distances. To create dendrograms, the samples from the distance matrix are clustered hierarchically using the ward method.
Statistics
All data are expressed as the mean ± standard deviation (SD), unless otherwise noted. Ordinary two-way analysis of variance (ANOVA) tests were used to determine diet, drug, and diet by drug significance, using P < 0.05 as the cutoff for significance, with Tukey multiple comparison post hoc testing unless otherwise noted. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05. Prism 6.02 software was used to compute all statistical calculations unless otherwise noted.
Results
Metformin has minimal effects on body mass, fat mass, or lean mass in DIO
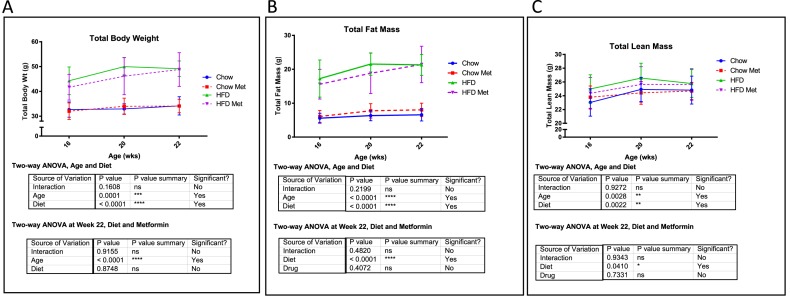
Mice were fed an HFD or chow for 15 weeks and treated in the last 6 weeks with or without metformin. This diet regimen has been shown previously (20), and herein confirmed to induce obesity and metabolic dysfunction (Fig. 1A–2B, Supplemental Fig. 1 (149.4KB, pptx) ). Metformin (300 mg/kg per day) effectively normalized blood glucose levels as early as day 10 of treatment (Supplemental Fig. 1 (149.4KB, pptx) ). HFD significantly increased total mass and fat mass by dual-energy x-ray absorptiometry at 16, 20, and 22 weeks vs chow, and metformin did not rescue this (Fig. 1A–1B). No substantial differences in total lean mass were observed based on diet or drug (Fig. 1C). Overall, metformin had little effect on the diet-induced obesity body composition phenotype (body mass, fat mass, or lean mass).
Figure 1.
Effects of metformin (met) on mouse weight and body composition. (A) PIXImus data for total body weight at 16, 20, and 22 weeks. Significant increases in weight were observed with HFD and age by two-way ANOVA. Two-way ANOVA showed no effect of metformin on total weight in HFD or chow mice at each time point. (B) Total fat mass assessed by PIXImus at 16, 20, and 22 weeks. (C) Total lean mass assessed by PIXImus at 16, 20, and 22 weeks. Data represent mean ± SD analyzed with two-way ANOVA with Tukey multiple comparison testing, n ≥ 8 mice for all groups. ns, not significant.
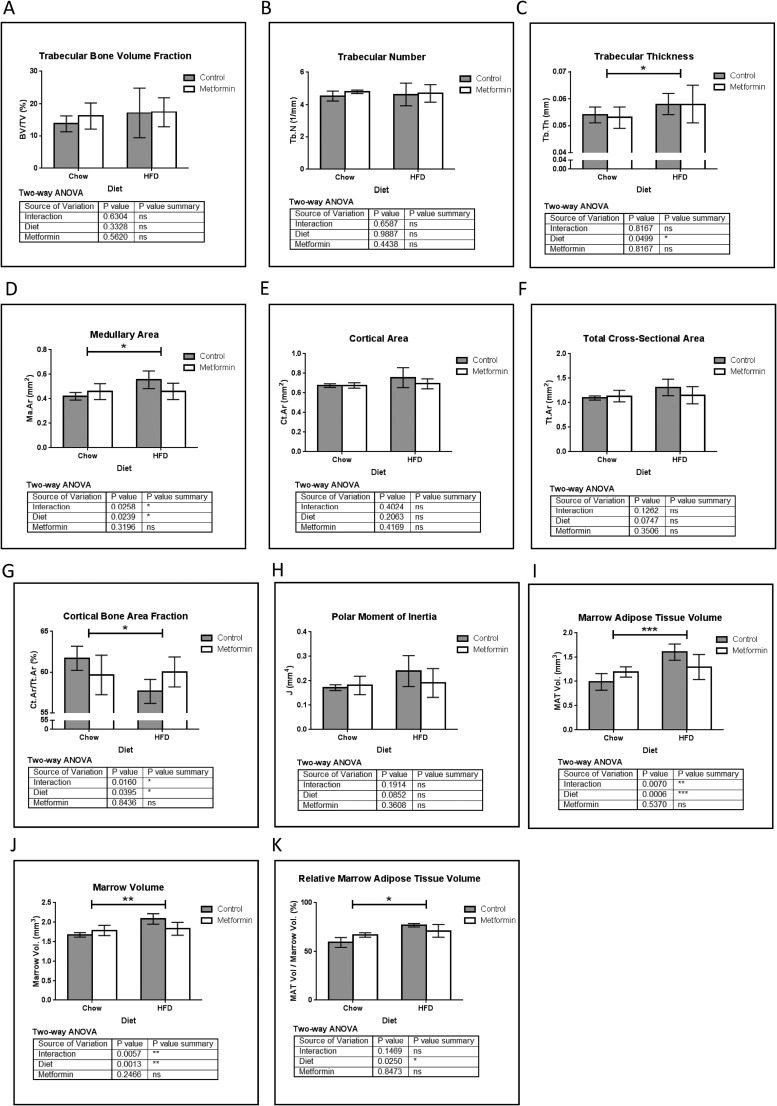
Figure 2.
μCT analysis of tibia. (A) Trabecular bone volume fraction (BV/TV). (B) Trabecular number (Tb.N). (C) Trabecular thickness (Tb.Th). (D) Medullary area (Ma.Ar) was significantly increased by HFD, and a substantial interaction effect was observed whereby metformin tended to normalize this result. (E) Cortical area (Ct.Ar) and (F) total cross-sectional area (Tt.Ar). (G) Cortical bone area fraction decreased in HFD; metformin tended to normalize this bone phenotype with a substantial interaction effect observed. (H) Changes in cortical bone result in a trend of a net increase in the polar moment of inertia (J) in mice on HFD, which is negated upon treatment with metformin. Using OsO4 μCT analysis of tibial MAT, both (I) marrow adipose tissue volume and (J) marrow volume were increased by HFD vs chow and showed a substantial interaction effect between metformin and HFD. (K) Relative MAT volume was significantly increased by HFD, and this was partially reversed with metformin. Data represent mean ± SD analyzed with two-way ANOVA, n = 5 mice for all groups.
Metformin inhibits the obesity-induced skeletal adaptive changes in the tibia
We then analyzed the effects of metformin on tibial bone parameters using μCT, OsO4 μCT, and static histomorphometry. Overall, we observed HFD feeding in growing animals had no effect on trabecular bone volume fraction (Fig. 2A) or trabecular number (Fig. 2B), but did significantly increase trabecular thickness (Fig. 2C). HFD significantly increased medullary area (Fig. 2D), which was partially rescued by metformin, as evidenced by a substantial interaction coefficient between metformin and HFD. In contrast to the femur, no substantial findings were observed in HFD tibia for cortical area, total cross-sectional area, periosteal perimeter, or endocortical perimeter (Fig. 2E–2F and data not shown). However, changes in tibial cortical bone led to a substantial decrease in cortical bone area fraction in HFD mice (Fig. 2G), which was partially rescued by metformin. These changes in bone mass resulted in an increased polar moment of inertia (Fig. 2H), as well as maximum and minimum moments of inertia (data not shown) in HFD mice. HFD also significantly increased total marrow adipose tissue volume, (Fig. 2I) and even when normalized to marrow volume (which was also increased by HFD, Fig. 2J), HFD increased MAT, as seen in the relative marrow adipose tissue volume (Fig. 2K). These phenotypes were again partially rescued with metformin (Fig. 2I–2K). Overall, the substantial interaction effects observed in medullary area, cortical bone area fraction, marrow volume, and MAT volume demonstrate that metformin may correct the tibial phenotype of HFD.
To elucidate the mechanisms behind the μCT data, we performed static bone histomorphometry. We observed that HFD decreased osteoblasts per total area (Supplemental Fig. 2A (149.4KB, pptx) ), but had no effect on number of osteoblasts per bone perimeter, osteoblast surface per bone perimeter (Supplemental Fig. 2B (149.4KB, pptx) –2C (149.4KB, pptx) ) or osteoid surface per bone surface (Supplemental Fig. 2D (149.4KB, pptx) ). HFD also increased MAT, reinforcing OsO4 μCT data (Supplemental Fig. 2E (149.4KB, pptx) –2F (149.4KB, pptx) ). Metformin tended to normalize all of these parameters altered by HFD (Supplemental Fig. 2 (149.4KB, pptx) ). Osteoclasts and osteoclast activity, assessed as number of osteoclasts per bone perimeter (Supplemental Fig. 2G (149.4KB, pptx) ), osteoclast surface per bone perimeter (Supplemental Fig. 2H (149.4KB, pptx) ), and eroded surface per bone surface (Supplemental Fig. 2I (149.4KB, pptx) ) tended to be increased with HFD, which appeared to be normalized with met treatment, but none of these findings was significantly different. In sum, metformin decreases MAT induced by HFD and may activate osteoblasts that are inhibited by HFD. Interestingly, for some parameters, substantial interaction effects demonstrate a restorative action of metformin on bones at the macroscopic level that is not seen in chow-fed mice.
Metformin inhibits the obesity-induced skeletal adaptive phenotype in the femur
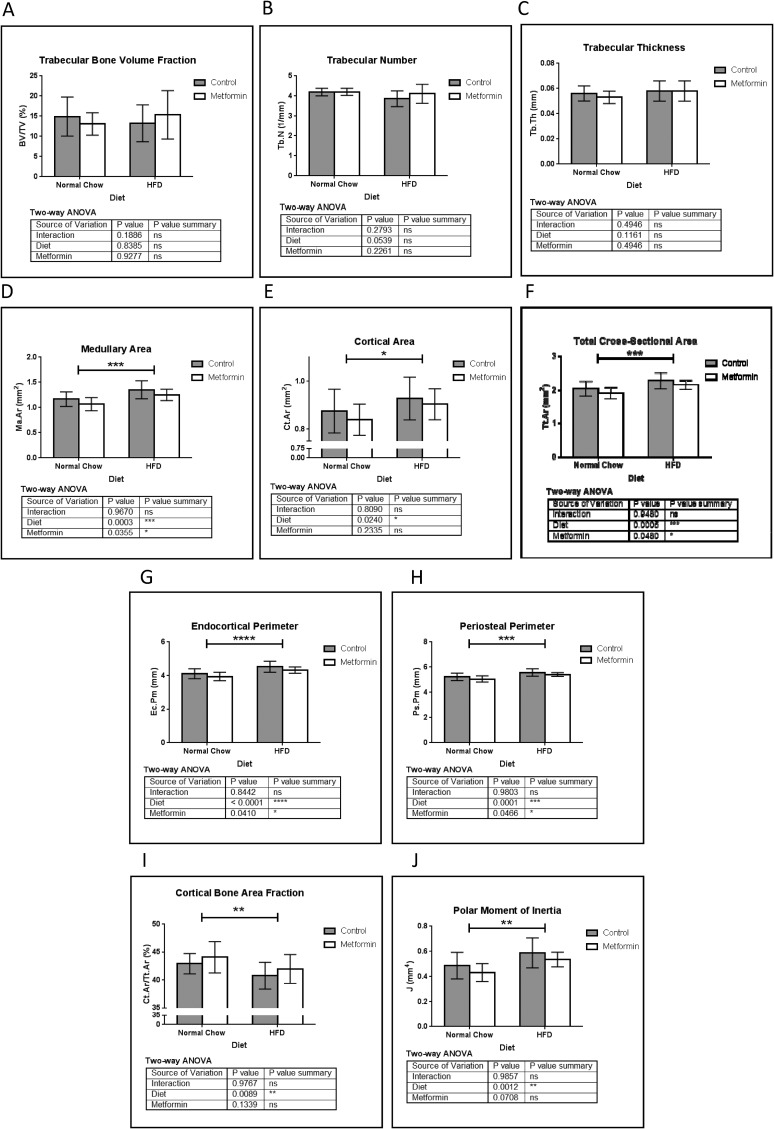
To further strengthen our data, mouse femurs were also analyzed using μCT and static bone histomorphometry. Similar to tibia data, no net changes in bone volume fraction (Fig. 3A) or trabecular number (Fig. 3B) were observed with HFD or metformin, and opposed to tibial data, no substantial trabecular thickness differences were observed (Fig. 3C). Interestingly, cortical bone demonstrated the same general response to HFD and metformin in the femur and tibia. HFD increased femoral medullary area (Fig. 3D), cortical area (Fig. 3E), total cross-sectional area (Fig. 3F), endocortical perimeter (Fig. 3G), and periosteal perimeter (Fig. 3H), which were all partially rescued with metformin. As a result, cortical bone area fraction was decreased by HFD, which also was partially rescued by metformin (Fig. 3I). In HFD, redistribution of bone mass from the endocortical to the periosteal region resulted in substantial increases in polar (Fig. 3J), maximum, and minimum moments of inertia (data not shown), which were partly normalized by metformin. Overall, the HFD femoral bone cortical phenotype was partially corrected through metformin treatment.
Figure 3.
μCT analysis of femur. Similar to tibia, (A) femoral bone volume fraction (BV/TV) and (B) trabecular number (Tb.N) showed no significant differences between all groups. No changes were observed in (C) Tb.N . Cortical bone showed a DIO response similar to the tibia. (D) Femur medullary area (Ma.Ar) was increased with HFD, and this was normalized by metformin. (E) Cortical area (Ct.Ar) and (F) total cross-sectional area (Tt.Ar) were significantly increased with HFD. Similarly, endocortical perimeter (Ec.Pm) (G) and periosteal perimeter (Ps.Pm) (H) were significantly increased by HFD and decreased by metformin. (I) Ct.Ar fraction was significantly decreased by HFD, and this was normalized partially by metformin treatment. These changes resulted in a significant increase in polar moment of inertia by HFD (J), which was partially restored to normal by metformin. Data represent mean ± SD analyzed with two-way ANOVA, n = 10 mice for all groups.
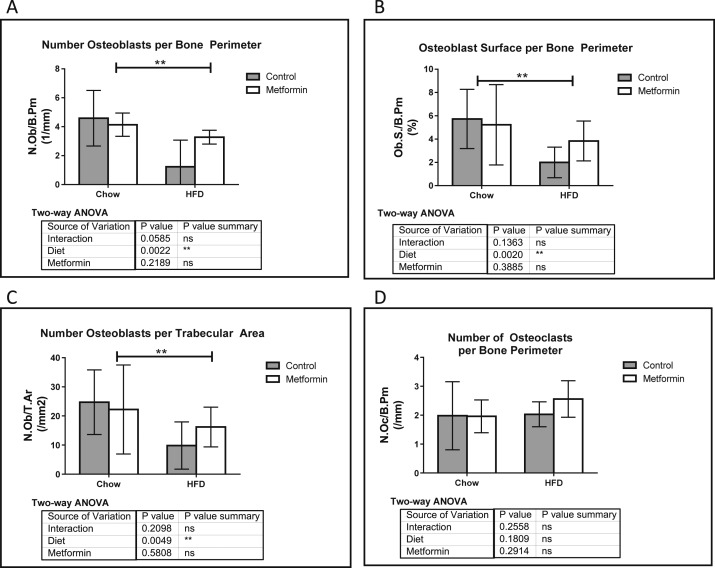
Similar to tibia static histomorphometry, osteoblast numbers were decreased in HFD vs chow and partially rescued with metformin in femora (Fig. 4A–C). Femoral osteoclast parameter changes were undetectable, suggesting that the increased medullary area was mostly driven by a decrease in osteoblast number and activity induced by HFD rather than osteoclast activation (Fig. 4D). In sum, these data suggest that metformin acts in an adversarial role to HFD in the cortical compartments of the femur.
Figure 4.
Femur static histomorphometry demonstrates metformin’s ability to normalize HFD phenotype. (A) Number of osteoblasts per bone perimeter (N.Ob/B.Pm), (B) osteoblast surface per bone perimeter (Ob.S./B.Pm), and (C) number of osteoblasts per trabecular area (N.Ob/T.Ar) were all decreased with HFD, and rescued with metformin. (D) Number of osteoclasts per bone perimeter (N.Oc/B.Pm), as well as all other osteoclast parameters (not shown) did not change in response to the diet or drug treatment. Data represent mean ± SD. Statistical analysis performed with two-way ANOVA using n ≥ 9 for all groups.
Adipose tissue phenotype is affected by obesity and metformin
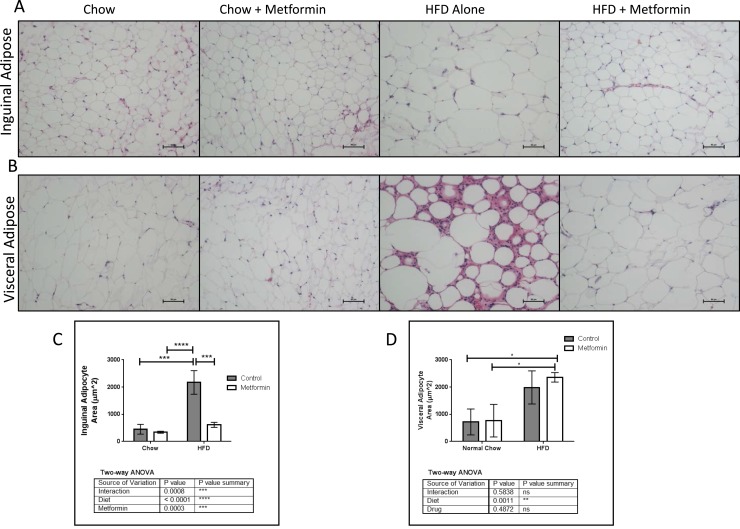
To address systemic effects of metformin on WAT, we measured adipocyte size in visceral adipose tissue (VAT) and inguinal adipose using hematoxylin and eosin staining. HFD significantly increased inguinal adipocyte size, and metformin normalized this phenotype by significantly reducing adipocyte size (Fig. 5A and 5C). HFD also significantly increased VAT adipocyte cell size, but metformin did not rescue this response (Fig. 5B and 5D). However, there was less inflammatory cell infiltration in HFD adipose depots when these mice were treated with metformin, suggesting that metformin’s anti-inflammatory properties specifically affected VAT (Fig. 5B). In contrast, metformin had no observable effects in mice on chow (Fig. 5A–5B).
Figure 5.
Normalizing effects of metformin on white adipose tissue in HFD. Representative hematoxylin and eosin sections of (A) inguinal and (B) visceral adipose depots. Scale bar = 50 μm. (C) Metformin significantly decreased inguinal adipocyte size in HFD mice. (D) HFD mice with or without metformin treatment have hypertrophic adipocytes vs chow mice, which was not rescued with metformin treatment. Data represent mean ± SD, n = 3, analyzed with two-way ANOVA and Tukey post hoc testing.
Circulating metabolite profiles are affected by obesity and metformin
To better understand the effects of metformin on circulating lipid and metabolite levels, we measured blood serum lipid and metabolite profiles immediately before euthanizing mice using gas chromatography and MS. Metabolite analysis revealed substantial differences between all groups (Tables 1–4). We compared HFD vs chow (Table 1), HFD + metformin vs chow + metformin (Table 2), HFD + metformin vs HFD (Table 3), and chow + metformin vs chow (Table 4). Metabolites were found to be dictated by diet: 10 upregulated and 15 downregulated metabolites were identified in HFD vs chow groups. Interestingly, we observed that the chow + metformin vs chow alone induced the fewest number of significantly changed metabolites (6 upregulated and 3 downregulated), whereas HFD + metformin vs chow + metformin induced the greatest number of altered metabolites (36 upregulated and 12 downregulated). Some of the metabolites were decreased in HFD vs chow, regardless of metformin treatment (e.g., shikimate, maleic acid, fumarate, malate), whereas other metabolites were only decreased in HFD vs chow when metformin was not present (e.g., 2-isopropylmalic acid, hydroxyproline, 3-methylphenylacetic acid). In both chow and HFD, metformin had the effect of decreasing spermidine and increasing pyrophosphate. In sum, metabolomic analysis revealed more examples of the “drug-by-diet interaction” effect in which metformin induces different effects on mice based on their diet, and provided data of type and number of metabolite class molecules altered by HFD and metformin.
Table 1.
Serum Metabolites Significantly Different for HFD vs Chow (N = 3)
| Sample ID | HFD | HFD | HFD | Chow | Chow | Chow | Fold-change | P |
|---|---|---|---|---|---|---|---|---|
| Shikimate | 1,089,275 | 1,687,554 | 1,774,052 | 24,346,440 | 36,264,443 | 37,236,094 | 0.04651 | 0.0017 |
| 3-Methylphenylacetic acid | 50,374.47 | 12,528.38 | 37,123.99 | 261,828.1 | 374,982.6 | 540,307.4 | 0.08498 | 0.0117 |
| 2-Aminooctanoic acid | 3,705,619 | 3,336,997 | 3,106,194 | 6,186,280 | 11,259,663 | 8,351,355 | 0.39341 | 0.02436 |
| N-acetyl-glutamine | 348,255.6 | 1,368,072 | 741,499.4 | 1,862,150 | 2,034,418 | 1,530,678 | 0.45287 | 0.04058 |
| Maleic acid | 1,894,292 | 1,763,624 | 2,659,257 | 3,979,273 | 4,135,781 | 4,259,294 | 0.51051 | 0.00226 |
| Fumarate | 1,886,339 | 1,688,418 | 2,866,039 | 4,116,645 | 3,979,963 | 4,342,941 | 0.51777 | 0.0062 |
| 2-Keto-isovalerate | 1,876,718 | 1,848,292 | 2,531,636 | 3,828,446 | 3,989,420 | 4,239,557 | 0.5189 | 0.00158 |
| Malate | 13,036,534 | 13,338,714 | 20,444,932 | 25,481,291 | 27,355,471 | 28,282,788 | 0.57718 | 0.01106 |
| 2-Isopropylmalic acid | 435,794.8 | 544,200.1 | 501,269.3 | 716,197.6 | 850,253.7 | 806,456 | 0.62424 | 0.00417 |
| Hydroxyproline | 1,129,533 | 971,637.6 | 1,400,073 | 1,754,945 | 2,126,389 | 1,698,862 | 0.62744 | 0.01947 |
| Serine | 10,864,799 | 9,120,770 | 9,096,639 | 6,100,208 | 6,902,703 | 6,321,955 | 1.50491 | 0.00678 |
| 4-Aminobutyrate | 19,742.1 | 18,701.03 | 14,974.13 | 11,219.59 | 11,486.34 | 12,302.38 | 1.52585 | 0.01442 |
| Purine | 444,057.3 | 496,184.5 | 421,698 | 254,463.2 | 323,655.7 | 313,140 | 1.52811 | 0.00703 |
| Uridine | 774,870.4 | 1,007,281 | 1,010,560 | 534,301.5 | 624,173.9 | 588,797.5 | 1.59833 | 0.01332 |
| Methylcysteine | 2,116,275 | 2,522,708 | 2,473,738 | 1,321,345 | 1,693,198 | 1,307,515 | 1.64568 | 0.00665 |
| Deoxyuridine | 166,050.2 | 190,394.9 | 184,623.3 | 113,833.8 | 120,865.1 | 891,59.99 | 1.67069 | 0.00392 |
| Aminoimidazole carboxamide ribonucleotide | 11,724.92 | 9084.187 | 12,305.94 | 4272.649 | 6163.236 | 5877.462 | 2.02994 | 0.00829 |
| Kynurenic acid | 303,590.8 | 406,552.9 | 254,063.9 | 144,964.3 | 155,164.5 | 159,147.8 | 2.09941 | 0.02028 |
| Ornithine | 11,732,045 | 11,815,352 | 7,731,882 | 3,400,980 | 6,215,043 | 5,270,134 | 2.10123 | 0.0259 |
| Dimethylglycine | 6,246,739 | 9,722,711 | 6,381,083 | 3,568,381 | 3,546,751 | 3,498,425 | 2.10585 | 0.02628 |
| Homocysteine | 67,440.13 | 58,050.68 | 51,315.81 | 27,962.43 | 20,051.53 | 34,720.26 | 2.13704 | 0.00766 |
| Nicotinamide | 21,645,145 | 16,344,710 | 14,481,236 | 1,928,119 | 8,393,518 | 8,048,776 | 2.85628 | 0.01934 |
| 3-S-methylthiopropionate | 332,935.7 | 323,819.6 | 400,830.5 | 68,402.46 | 111,655.7 | 113,215.4 | 3.60614 | 0.00085 |
| N1-methyl-2_4-pyridone-3_5-carboximide | 21,978.47 | 23,191.8 | 27,399.03 | 3000 | 5991.789 | 4821.317 | 5.25366 | 0.00046 |
| Pantothenate | 126,600.8 | 111,466.3 | 71,988.17 | 11,066.49 | 15,090.77 | 19,685.47 | 6.76346 | 0.00589 |
Table 2.
Serum Metabolites Significantly Different for HFD + Met vs Chow + Met (N = 3)
| Sample ID | HFD + Met | HFD + Met | HFD + Met | Chow + Met | Chow + Met | Chow + Met | Fold-change | P |
|---|---|---|---|---|---|---|---|---|
| Shikimate | 605,719.963 | 790,075.7 | 1,132,579 | 2E + 07 | 15,714,190 | 3E + 07 | 0.0376 | 0.00581 |
| p-Hydroxybenzoate | 419,791.166 | 460,338.3 | 459,449 | 1E + 06 | 35,29,902 | 3,859,770 | 0.1535 | 0.03574 |
| Guanine | 9556.31493 | 3000 | 6317.29 | 54,031 | 23,618.46 | 35,415.2 | 0.1669 | 0.02562 |
| Guanosine | 3000 | 3000 | 3000 | 5988 | 7753.123 | 4533.46 | 0.4925 | 0.02935 |
| Spermine | 5606.12565 | 13,090.07 | 13,003.8 | 21,017 | 19,736.19 | 20,876 | 0.5144 | 0.01655 |
| Biotin | 70,864.04 | 112,564.1 | 92,945.3 | 2E + 05 | 192,707.6 | 171,522 | 0.5161 | 0.00349 |
| 5-Methyl-THF | 53,306.9446 | 75,387.88 | 62,464.5 | 1E + 05 | 157,550 | 94,422.2 | 0.5246 | 0.04362 |
| Aconitate | 3,247,657.85 | 2,689,442 | 3,482,053 | 7E + 06 | 4,232,556 | 5,119,315 | 0.5937 | 0.03798 |
| Maleic acid | 2,794,039.31 | 2,597,995 | 3,135,051 | 4E + 06 | 4,415,480 | 4,403,549 | 0.6518 | 0.00076 |
| Fumarate | 2,942,438.32 | 2,454,856 | 3,261,639 | 5E + 06 | 4,320,662 | 4,454,487 | 0.6518 | 0.00307 |
| Citraconic acid | 3,772,059.64 | 2,8134,38 | 4,113,861 | 6E + 06 | 4,858,139 | 5,805,660 | 0.6531 | 0.0184 |
| 2-Keto-isovalerate | 2,730,426.57 | 2,563,682 | 3,071,544 | 4E + 06 | 4,106,779 | 43,77,090 | 0.6676 | 0.00154 |
| Malate | 19018590.1 | 16,745,201 | 2.2E + 07 | 3E + 07 | 28,011,756 | 2.9E + 07 | 0.6724 | 0.00355 |
| d-gluconate | 670,774.588 | 784,246.6 | 644,759 | 4E + 05 | 584,131.7 | 437,898 | 1.5058 | 0.0362 |
| Glycerate | 1,309,515.05 | 1,557,792 | 1,348,045 | 8E + 05 | 991,361 | 967,198 | 1.5138 | 0.00679 |
| Aminoadipic acid | 710,505.938 | 665,427.6 | 633,801 | 5E + 05 | 512,968.3 | 342,082 | 1.518 | 0.01494 |
| 1-Methyl-histidine | 22,870,625.4 | 29,057,640 | 2.8E + 07 | 1E + 07 | 20,544,071 | 1.6E + 07 | 1.5692 | 0.02118 |
| Indole-3-carboxylic acid | 778,561.738 | 766,996.2 | 652,174 | 5E + 05 | 533,004.3 | 316,826 | 1.5714 | 0.03504 |
| Sarcosine | 2,998,972.43 | 3,498,907 | 3,753,705 | 2E + 06 | 2,637,517 | 2,128,288 | 1.5908 | 0.02323 |
| d-glucono-delta-lactone-6-phosphate | 15,412.1783 | 16,734.72 | 21,961 | 11,282 | 12,955.61 | 8665.03 | 1.6445 | 0.03999 |
| Serine | 11,445,636.7 | 12,470,874 | 1.3E + 07 | 6E + 06 | 8,087,514 | 8,308,067 | 1.665 | 0.00591 |
| Tyrosine | 7,462,204.81 | 9,349,107 | 9,367,042 | 3E + 06 | 5,801,518 | 6,571,261 | 1.6716 | 0.04066 |
| Purine | 454,234.356 | 412,670.6 | 441,460 | 2E + 05 | 302,269.5 | 283,971 | 1.7653 | 0.01694 |
| Deoxyuridine | 181,350.726 | 204,080.1 | 223,233 | 1E + 05 | 119,641 | 107,828 | 1.7757 | 0.00214 |
| N-carbamoyl-l-aspartate-nega | 309,050.244 | 356,079 | 458,034 | 2E + 05 | 189,559.2 | 246,257 | 1.785 | 0.02588 |
| Cytidine | 5,040,124.51 | 4,642,244 | 5,145,783 | 2E + 06 | 3,907,352 | 2,507,073 | 1.7931 | 0.02489 |
| N1-methyl-2_4-pyridone-3_5-carboximide | 11,275.692 | 16,421 | 12,607.9 | 7985 | 7983.588 | 6337.27 | 1.8069 | 0.02147 |
| Adenine | 1,866,588.36 | 2,415,479 | 2,173,534 | 9E + 05 | 1,276,336 | 1,372,603 | 1.8447 | 0.01201 |
| Cytosine | 527,972.157 | 595,812 | 641,231 | 3E + 05 | 409,074.6 | 274,988 | 1.8591 | 0.00878 |
| Methylcysteine | 2,101,963.8 | 2,662,049 | 2,577,650 | 9E + 05 | 1,440,804 | 1,612,409 | 1.8745 | 0.01622 |
| Methionine | 12,467,259.6 | 12,521,314 | 1E + 07 | 4E + 06 | 8,487,193 | 5,832,900 | 1.8852 | 0.01768 |
| N-acetyl-glucosamine-1-phosphate | 171,904.099 | 211,826.7 | 144,505 | 86,279 | 121,083.4 | 62,691.1 | 1.956 | 0.02922 |
| Lysine | 8,866,233.09 | 8,419,670 | 7,657,754 | 3E + 06 | 5,504,385 | 3,755,480 | 1.9716 | 0.00524 |
| Ornithine | 6,705,806.78 | 10,903,272 | 1E + 07 | 3E + 06 | 5,335,247 | 5,395,010 | 1.988 | 0.03583 |
| Kynurenic acid | 241,377.738 | 310,478.2 | 232,505 | 2E + 05 | 137,071 | 94,389.1 | 2.0176 | 0.01289 |
| Valine | 7,014,835.72 | 7,397,898 | 6,332,323 | 2E + 06 | 44,35,899 | 3,404,522 | 2.0636 | 0.00754 |
| Threonine | 15,489,462 | 17,160,731 | 1.7E + 07 | 7E + 06 | 9,283,102 | 7,746,320 | 2.0742 | 0.00063 |
| sn-Glycerol-3-phosphate | 3,531,076.72 | 2,745,981 | 2,147,569 | 1E + 06 | 2,020,982 | 1,026,274 | 2.0899 | 0.04925 |
| Dephosphocoenzyme A | 8017.57731 | 4848.703 | 6414.78 | 3000 | 3000 | 3000 | 2.1423 | 0.02001 |
| 3-S-methylthiopropionate | 195852.635 | 268,270.3 | 267,664 | 1E + 05 | 86,598.02 | 123,599 | 2.1621 | 0.00876 |
| Thiamine-phosphate | 279,054.966 | 273,780.9 | 192,910 | 70,555 | 124,249 | 131,393 | 2.2862 | 0.01448 |
| CMP | 221,807.553 | 175,820.4 | 180,646 | 68,817 | 113,825.3 | 69,540.6 | 2.2931 | 0.00645 |
| Uracil | 5,199,782.25 | 5,508,057 | 4,118,841 | 9E + 05 | 2,546,003 | 2,384,553 | 2.559 | 0.01154 |
| GMP | 6,103,903.27 | 3,896,003 | 3,719,601 | 2E + 06 | 2,076,211 | 1,258,296 | 2.7239 | 0.02264 |
| Guanosine 5-diphosphate,3-diphosphate | 8006.50112 | 16,605.46 | 13,358.2 | 3000 | 5877.247 | 4816.68 | 2.7728 | 0.03764 |
| UMP | 545,588.157 | 498,907.7 | 511,887 | 2E + 05 | 213,678.6 | 86,489.6 | 2.8421 | 0.00273 |
| AMP | 3,2101,229.2 | 30,582,462 | 2.8E + 07 | 1E + 07 | 11,490,037 | 5,333,311 | 2.9069 | 0.00256 |
| Deoxycytidine triphosphate | 70,539.6893 | 87,640.02 | 63,068.4 | 13907 | 7587.078 | 43302.6 | 3.4145 | 0.01677 |
| IMP | 3,974,063.04 | 3,214,610 | 3,454,655 | 1E + 06 | 1,051,939 | 344,493 | 3.7765 | 0.00254 |
Table 3.
Serum Metabolites Significantly Different for HFD + Met vs HFD (N = 3)
| Sample ID | HFD + Met | HFD + Met | HFD + Met | HFD | HFD | HFD | Fold-change | P |
|---|---|---|---|---|---|---|---|---|
| 4-Pyridoxic acid | 3000 | 3000 | 3000 | 195,637 | 91,172.59 | 77,957.75 | 0.02467 | 0.03334 |
| p-Hydroxybenzoate | 419,791.2 | 46,0338.3 | 459,448.6 | 2,348,553 | 3,806,022 | 1,953,398 | 0.16522 | 0.01608 |
| Pantothenate | 71,093.75 | 3000 | 5143.925 | 126,600.8 | 111,466.3 | 71,988.17 | 0.25556 | 0.04968 |
| Hydroxyphenylpyruvate | 3000 | 16,395.87 | 3000 | 26,619.25 | 25,889.42 | 31,300.46 | 0.26722 | 0.01279 |
| Spermidine | 186,016.7 | 202,543.1 | 126,621.9 | 631,357.3 | 608,266.1 | 516,912.6 | 0.29329 | 0.00059 |
| Guanine | 9,556.315 | 3000 | 6317.29 | 15,171.26 | 23,440.06 | 15,135.71 | 0.35116 | 0.02555 |
| N1-methyl-2_4-pyridone-3_5-carboximide | 11,275.69 | 16,421 | 12,607.9 | 21,978.47 | 23,191.8 | 27,399.03 | 0.55539 | 0.00881 |
| Uridine | 764,327.4 | 559,337 | 528,960 | 774,870.4 | 1,007,281 | 1,010,560 | 0.66338 | 0.04343 |
| Threonine | 15,489,462 | 17,160,731 | 1,6842,064 | 9,955,130 | 11,772,739 | 10,654,448 | 1.52837 | 0.0015 |
| Allantoin | 10,120,555 | 9,403,367 | 8,723,334 | 5,809,824 | 6,276,761 | 5,675,186 | 1.59034 | 0.00139 |
| Lysine | 8,866,233 | 8,419,670 | 7,657,754 | 5,086,581 | 5,636,138 | 4,093,039 | 1.68359 | 0.00415 |
| Arginine | 2,9376,288 | 30,684,375 | 25,458,372 | 12,168,850 | 15,009,274 | 20,806,835 | 1.7822 | 0.01383 |
| Thiamine-phosphate | 279,055 | 273,780.9 | 192,909.7 | 134,290.1 | 133,617.1 | 141,202.1 | 1.82285 | 0.016 |
| Hydroxyproline | 2,409,287 | 2,351,086 | 2,040,864 | 1,129,533 | 971,637.6 | 1,400,073 | 1.94252 | 0.00291 |
| Putrescine | 30,665.33 | 34,054.71 | 51,655.73 | 16,354.23 | 20,759.58 | 21,909.11 | 1.9717 | 0.04669 |
| Dephosphocoenzyme A | 8017.577 | 4848.703 | 6414.775 | 3000 | 3000 | 3000 | 2.14234 | 0.02001 |
| 2-Aminooctanoic acid | 9,545,403 | 6,850,644 | 6,343,098 | 3,705,619 | 3,336,997 | 3,106,194 | 2.24057 | 0.01415 |
| Inosine 5′-diphosphate | 16,109.84 | 11,763.31 | 7412.48 | 4808.097 | 2672.499 | 3205.988 | 3.30186 | 0.03405 |
| AMP | 32,101,229 | 30,582,462 | 27,657,966 | 149,164.4 | 68,649.81 | 22,688,635 | 3.94394 | 0.04228 |
| UMP | 545,588.2 | 498,907.7 | 511,887.1 | 11,332.91 | 3000 | 339,231.4 | 4.40198 | 0.0229 |
| GMP | 6,103,903 | 3,896,003 | 3,719,601 | 136,798.7 | 6412.838 | 2,904,594 | 4.50144 | 0.04314 |
| IMP | 3,974,063 | 3,214,610 | 3,454,655 | 15,843.59 | 4809.649 | 2,096,385 | 5.02746 | 0.01769 |
| 2'-Deoxyguanosine 5′-diphosphate | 123,920.6 | 43,271.81 | 99,391.99 | 4827.277 | 3000 | 32,071.06 | 6.68159 | 0.04216 |
| Pyrophosphate | 205,145.3 | 149,011.4 | 59,543.77 | 22,271.93 | 3776.937 | 28,480.04 | 7.58681 | 0.04974 |
| 2,3-Dihydroxybenzoic acid | 495,845 | 698,138.4 | 655,879.4 | 3000 | 101,327.2 | 3000 | 17.2357 | 0.00114 |
Table 4.
Serum Metabolites Significantly Different for Chow + Met vs Chow (N = 3)
| Sample ID | Chow + Met | Chow + Met | Chow + Met | Chow | Chow | Chow | Fold-change | P |
|---|---|---|---|---|---|---|---|---|
| Spermidine | 117,505 | 313,967 | 396,913 | 759,230 | 602,290 | 649,046 | 0.41202 | 0.01429 |
| 3-Phosphoglycerate | 13,459.1 | 24,102.2 | 27,214.6 | 40,630.7 | 43,537.6 | 49,656.9 | 0.48403 | 0.00961 |
| S-adenosyl-l-homocysteine | 121,800 | 226,595 | 208,520 | 318,811 | 455,565 | 339,844 | 0.49983 | 0.02541 |
| Homocysteic acid | 14,080.5 | 24,518.1 | 14,460.5 | 29,196.1 | 30,297.2 | 44,274.1 | 0.51133 | 0.0465 |
| Aminoimidazole carboxamide ribonucleotide | 3000 | 3000 | 3000 | 4272.65 | 6163.24 | 5877.46 | 0.5517 | 0.01434 |
| d-glucosamine-1-phosphate | 3000 | 3754.83 | 3000 | 4812.82 | 4809.59 | 5321.56 | 0.65276 | 0.0047 |
| Guanosine | 5988.01 | 7753.12 | 4533.46 | 3000 | 3000 | 3440.89 | 1.93568 | 0.03537 |
| Spermine | 21,017.3 | 19,736.2 | 20,876 | 9540.61 | 12,384.6 | 4746.84 | 2.31064 | 0.00677 |
| Pyrophosphate | 55,963.7 | 66,967.6 | 95,109.8 | 20,345.6 | 28,787.1 | 42,259.1 | 2.38578 | 0.03363 |
Lipidomic profiles are affected by obesity and metformin
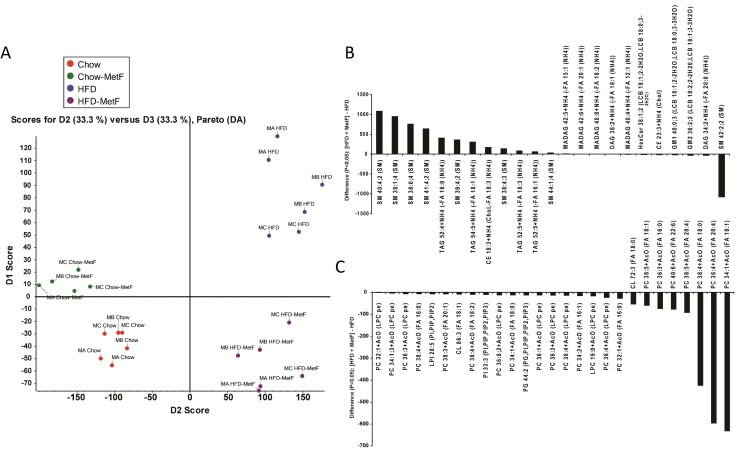
Multivariate statistical analysis was used to identify the alteration of global serum metabolites associated with obese mice as compared with healthy controls and differential responses to drug treatment. Lipid profiling showed the largest numbers of distinct lipid species changing significantly (P < 0.05) in HFD vs chow diets (Supplemental Table 1 (16.7KB, docx) ). This overall trend is reflected by principal component analysis (e.g., for chow vs HFD treatments; Fig. 6A). Interestingly, principal component analysis suggested that metformin had a more pronounced effect on global serum lipid composition for HFD mice compared with mice on a chow diet (Fig. 6A).
Figure 6.
SWATH (MS/MSALL)-based lipid profiling of mouse serum. (A) PCA of chow and HFD with and without metformin (MetF). Most changed (B) positive ion and (C) negative ion lipids characteristic of MetF effects on HFD-fed mice. Bars indicate lipid differences with P ≤ 0.05, n = 3 mice for all groups, labeled as MA, MB, and MC (mouse A, B, and C, respectively). CE, cholesteryl ester; CL, cardiolipin; DAG, diacylglycerol; GM1, GM2, gangliosides M1 and M2; HexCer, monohexosylceramide; LPC, lysophosphatidylcholine; MAG, monoalkyldiacylglycerol; PCA, principal component analysis; PG, phosphatidylglycerol; PI, phosphatidylinositol; TAG, triacylglycerol.
Although all classes of lipids comprised differentially regulated distinct molecular species, the classes exhibiting the most molecular species with the greatest changes were SMs and phosphatidylcholines (PCs) (Fig. 6B and 6C, respectively). SM molecular species were the most dramatically changed by metformin in chow-fed mice, supporting the idea that metformin’s effects are mediated, at least in part, by changes in SM metabolism. The strong depression of multiple PCs and other glycerophospholipid molecular species (e.g., LPI28:5, PI33:3, PC34:1, PC36:4) is consistent with previously published results that indicate metformin reduces lysophosphatidylcholine production in human hepatocytes (21). However, our results suggest that, although SM42:2:2 is significantly decreased, other SM molecular species appeared increased in metformin HFD-fed mouse serum (Fig. 6B). This discrepancy may reflect a shift in the clearance of SMs in metformin HFD-fed mice. Overall, circulating markers of systemic health and metabolism were dependent on both diet and drug, suggesting that metformin may differentially affect health, biomarkers, lipid utilization and production, and cellular metabolism in obese compared with lean individuals.
Gut microbiome is affected by obesity and metformin
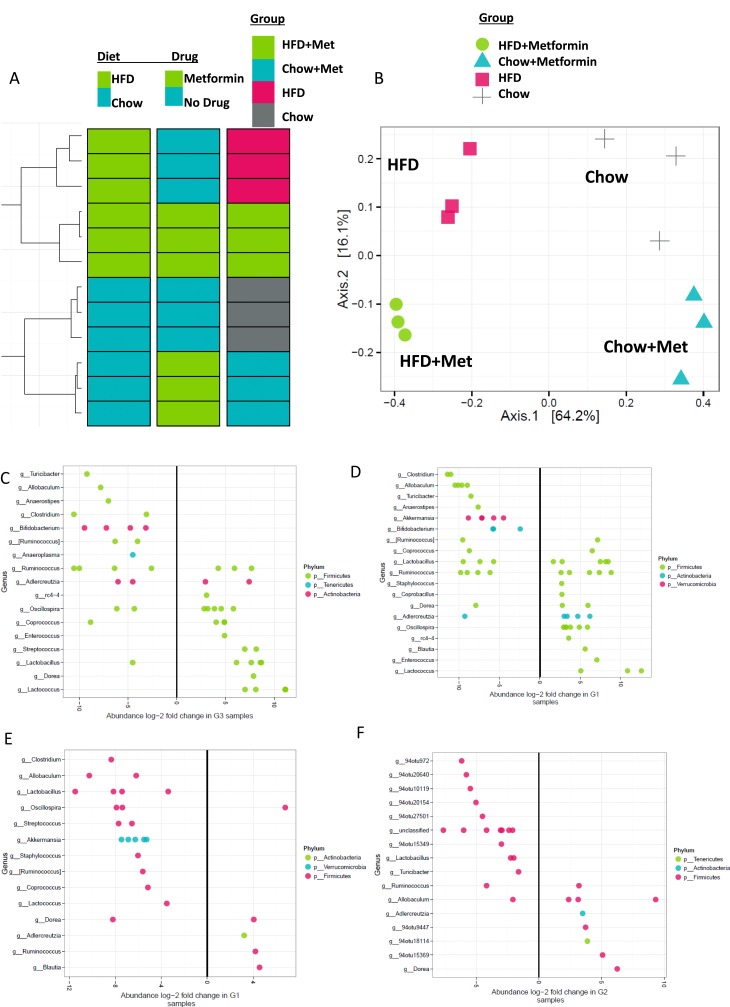
Last, we sought to identify differences in the gut microbiome of these groups by analyzing their 16s rRNA sequences. Operational taxonomic units (OTUs, similar to a species and sometimes referred to as phylotypes) analyzed in fecal samples from mice were used to assess diversity and populations within groups using high-throughput sequencing and bioinformatics tools. Hierarchical clustering analysis (Fig. 7A) and principal coordinate analysis (Fig. 7B) revealed the largest differences in microbiome were due to diet, and metformin further subcategorized the groups. In mice not treated with metformin, there were 170 differentially abundant OTUs in HFD vs chow mice (Fig. 7C). Within mice on metformin, 160 operational taxonomic units (OTUs) were differentially abundant, including members of a number of different genera from Firmicutes, Actinobacteria, and Verrucomicrobia, in HFD vs chow mice (Figure 7D). Interestingly, 72 OTUs, including taxa from Lactococcus, Coprococcus, Ruminococcus, Staphylococcus, Akkermansia, Streptococcus, Oscillospira, Lactobacillus, Allobaculum, Clostridium, and Dorea, were found to be differentially abundant in HFD mice treated with metformin vs HFD mice alone (Fig. 7E). Metformin did not induce such a large shift in chow mice: only 27 OTUs significantly changed in chow-fed mice treated with metformin vs untreated (Fig. 7F). Of the important features that could be identified down to the genus level, OTUs from Lactobacillus and Turicibacter were reduced in abundance in chow-metformin–treated mice, and OTUs from genera Dorea and Adlercreutzia increased in abundance in chow-metformin vs untreated mice. Overall, the data demonstrate that gut microbiota respond to both diet and drug, and that microbiota responses to metformin are dependent on mouse diet.
Figure 7.
Cluster analysis of gut microbiome from 16S rRNA sequencing. 16S sequencing was performed on feces-derived DNA. (A) Hierarchical clustering revealed diet and drug effects to be driven mostly by diet, and secondarily by metformin. (B) Principal component analysis of microbial compositions demonstrated that diet is the overwhelming driver of each sample’s microbial community composition. Within each diet, metformin (met) treatment influences the microbiota composition. Abundance long-twofold changes for OTUs in (C) HFD vs chow, (D) HFD + met vs HFD + chow, (E) HFD + met vs HFD, and (F) chow + met vs chow mice. n = 3 mice.
Discussion
In this study, we sought to determine the direct and indirect effects of DIO and metformin on peak skeletal acquisition. Our data revealed the complex nature of the interactions between obesity, bone, bone marrow adipose tissue, and the gut microbiome. Interestingly, we found that skeletal compartments, including cortical bone and bone marrow adipose tissue, as well as WAT, are altered in young obese mice; this is partially ameliorated by metformin. In DIO, endocortical bone expansion was observed in the femur; this was in part reversed by metformin. On the other hand, trabecular bone volumes in both femur and tibia were not altered by an HFD or by metformin, except in the tibia where HFD increased trabecular thickness. Possibly longer treatment with metformin would induce greater effects on the trabecular skeleton. Interestingly, other groups have found HFD decreases bone mass and increases osteoclast numbers in C57BL/6J mice, but these studies used older mice, shorter term diets, or both (22–24), suggesting that HFD initiation age and duration affect osteoclast response, likely from the high energy requirements of growing mice or increase in mouse weight and hence bone loading in longer term HFD feeding.
There has been extensive research into the effects of obesity on bone health, with most findings suggesting that obesity increases mechanical loading on bones, resulting in increased bone volumes (25). Interestingly, obesity has also been shown to correlate with increased risk of fracture (4), and increased visceral adipose specifically is inversely associated with bone mechanical properties in adult males (26). Our data suggest that obesity in growing mice leads to cortical bone expansion and increased endocortical resorption, resulting in a net increase in the long bones’ moments of inertia, and thus greater rigidity, in response to increased body weight. This typically translates to stronger bone, but could also produce an increased fracture risk when the bone is loaded in unconventional directions. More studies into the mechanical strength of bones in multiple directions will determine how the cortical expansion we observed translates to real life consequences, and will also elucidate if the opposing action of metformin on this process is beneficial or deleterious. Three-point bending or compression mechanical testing should be performed in future studies to help correlate μCT and histomorphometric data to functional outcomes in the long bones and vertebrae respectively.
It is challenging to ascertain the exact mechanisms behind the bone phenotypes observed here because abnormal glucose tolerance, systemic or local inflammation, increased oxidative stress and excess accumulation of advanced glycation end products in bone extracellular matrix can all affect bone remodeling and inhibit osteoblast proliferation and differentiation (27, 28). Moreover, there is disagreement in the field about the effects of hypercholesteremia and lipotoxicity on bone health, but most of this work is from adults and not maturing animals, as investigated here. Some findings show that hypercholesterolemic mice also present an osteoporotic phenotype, and statins, the first-line drug for hypercholesterolemia, increase BMD and in some studies reduce fracture risk in humans (29, 30). Yet other studies found no long-term effects of cholesterol, low-density lipoprotein, or high-density lipoprotein levels on BMD, after correcting for confounding variables (31, 32). What is becoming clear through numerous studies, however, is that the net negative effect of obesity on the skeleton appears to be driven by a myriad of interconnected pathologies (22) and local as well as systemic (humoral and neuronal) signaling factors such as leptin (33) and chemerin (34).
This report demonstrates a reduction in MAT in DIO after treatment with metformin. Similar to published studies (20), we observed that a long-term HFD in growing mice induced greater MAT volume, which occurred because of both an increase in adipocyte number and size. Interestingly, MAT is also elevated in obese humans and is inversely associated with volumetric BMD (25), perhaps because of pro-osteoclastic molecules such as RANKL, which has recently been identified as a marrow adipocyte-derived factor (35). As Yue et al. recently described, HFD promotes bone marrow adipogenesis through enhanced leptin receptor signaling in bone marrow stromal cells (23), so it is possible that this cell population is being targeted and modulated by DIO to shift their lineage allocation from osteoblasts to adipocytes, and that metformin might correct this. Indeed, in vitro studies confirm that metformin can induce Runx2 and AMPK, which hasten calvarial osteoblast differentiation (36). If lineage commitment is altered in these mice, lineage tracing of osterix + cells using an inducible fluorescent marker would be useful to determine from when and where the osteoblasts and bone marrow adipocytes were derived. Changes in progenitor cells could also be identified in future studies by performing flow cytometry for mesenchymal stem/progenitor cells to determine changes in their frequency. It is also possible that endocortical osteoblasts underwent apoptosis in DIO mice and that this was rescued via metformin treatment. Similarly, the driver behind the decrease in MAT observed with metformin treatment may be lipolysis of mature adipocytes rather than cell fate change; this has been previously reported with metformin treatment (37). Future work will be aimed at identifying other mechanisms besides cell fate switching that may be operative by which HFD inhibits osteoblast number or function, such as lipotoxicity, which could be reversed by metformin.
Our study also demonstrated that metformin reduces inguinal WAT volume and visceral WAT crowning in DIO but not in chow-fed mice. These data recapitulate findings from prior studies that found metformin ameliorated HFD-induced liver inflammatory responses and hepatic steatosis, and decreased white adipocyte diameter (38–40). Both in vivo and in vitro studies suggest that metformin decreases gene expression for adipogenic pathways and increases browning gene expression, suggesting that metformin may induce beiging in WAT (8, 41). Our study also provides data regarding changes in the gut flora from diet or metformin. Metformin may alter the microbiome through increasing intestinal glucose uptake and lactate production, changing glucagonlike peptide-1 concentrations, and altering the bile acid pool within the intestine, thereby potentially altering the microbiome (42). Future work may aim at determining how specific gut microbiomes contribute to the skeletal or systemic effects of metformin and/or DIO. It would also be beneficial in future studies to determine the effects of HFD and metformin on muscle mass and intermuscular adipose tissue, as these were not examined here.
There are a few limitations to our study. First, although it is clear that there are both direct and indirect effects from metformin on the skeleton in DIO, the precise mechanisms responsible for the changes in adipogenesis and osteoclastogenesis have not been fully delineated. To determine how metformin affects bone marrow adipocytes, future research could test direct treatment of metformin on mature and maturing adipocytes. Gene expression analysis could determine if metformin induces autophagy, lipolysis, apoptosis, dedifferentiation, or cell death in bone marrow adipocytes. If no direct effects on adipocytes or preadipocytes are observed, this is evidence that metformin’s effects depend on systemic signaling. In addition, analyzing white and bone marrow adipocyte bioenergetics (oxidative phosphorylation and glycolysis) with and without metformin would determine changes in metabolic pathways and substrate utilization with metformin treatment, such as increases in lipid utilization, induced by metformin that would explain changes in adipocyte or lipid droplet size.
Second, it is often difficult to uncouple obesity, diet, and metabolism in DIO in mice and humans. Also, the type of dietary fat, rather than total intake, is a more accurate determinant of fat’s contribution to human health, and in mice, HFD may better translate to the Western or “cafeteria” diet rather than an actual high-fat/low carbohydrate diet in humans (43). Therefore, exploring other models of diabetes without obesity (e.g., the streptozotocin model), HFD without obesity [e.g., the FVB obesity-resistant mouse (44)], obesity without diabetes [e.g., the Brd2 knockout (45)], and diabetes and obesity [leptin- or leptin receptor-deficient mice (9)], will help to disentangle effects of metabolism, diet content, and obesity on bone health.
Herein we characterized the effects of obesity and metformin on bone, MAT, protein and lipid metabolites, and gut microbiome bacteria. In DIO in growing mice, metformin improves osteoblastogenesis and suppresses bone resorption in the cortical region and reduces marrow adiposity. Overall, these data provide the most complete characterization of phenotype changes in mice in response to metformin and/or DIO. Importantly, we find that metformin tends to normalize DIO bone and MAT phenotypes, suggesting that it be considered for use not only in patients with diabetes, but perhaps even for obese patients with high risk of fracture.
Acknowledgments
We thank Michael Armanini for assistance with µCT, Dr. Frank Ko for assistance with osmium tetroxide staining, Dr. John Asara, Beth Israel Deaconess Medical Center, for performing metabolomic profiling, and Dr. Michael Erard (Maine Medical Center Research Institute) for careful criticism and editing of the written manuscript.
Financial Support: This work was supported by funding from the Center for Skeletal Research Core [National Institutes of Health (NIH) Grant P30 AR066261], the National Institute of General Medical Sciences (NIGMS)/NIH Grant P30 GM106391, the Histopathology Core of NIH Grants NIGMS P30 GM103392 (R. Friesel, principal investigator) and P30GM106391 (D. Wojchowski, principal investigator), startup funds from the Maine Medical Center Research Institute, the NIH/National Institute of Diabetes and Digestive and Kidney Diseases (Grant R24 DK092759-01), and the American Cancer Society (Research Grant IRG-16-191-33).
Author Contributions: S.B., M.M., Y.K., and M.R.R. performed experiments. A.S., D.H., S.M., A.M.R., H.F., and C.F. assisted with experiments. D.B. and M.B. performed and analyzed μCT data. K.N. and R.B. performed and analyzed histomorphometry data. C.V. performed the lipidomic screen and data analysis. M.M., C.J.R., and M.R.R. performed experimental design, manuscript preparation, and editing. All authors provided feedback on the manuscript and provided final approval. M.R.R. accepts final responsibility for the integrity of the data analysis.
Acknowledgments
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- ANOVA
- analysis of variance
- BMD
- bone mineral density
- DIO
- diet-induced obesity
- HFD
- high-fat diet
- MAT
- marrow adipose tissue
- MS
- mass spectrometry
- OsO4
- osmium tetroxide
- OTU
- operational taxonomic unit
- PC
- phosphatidylcholine
- PCR
- polymerase chain reaction
- SD
- standard deviation
- SM
- sphingomyelin
- VAT
- visceral adipose tissue
- WAT
- white adipose tissue
- μCT
- microcomputed tomography.
References
- 1.Evans AL, Paggiosi MA, Eastell R, Walsh JS. Bone density, microstructure and strength in obese and normal weight men and women in younger and older adulthood. J Bone Miner Res. 2015;30(5):920–928. [DOI] [PubMed] [Google Scholar]
- 2.Sornay-Rendu E, Boutroy S, Vilayphiou N, Claustrat B, Chapurlat RD. In obese postmenopausal women, bone microarchitecture and strength are not commensurate to greater body weight: the Os des Femmes de Lyon (OFELY) study. J Bone Miner Res. 2013;28(7):1679–1687. [DOI] [PubMed] [Google Scholar]
- 3.Morin S, Tsang JF, Leslie WD. Weight and body mass index predict bone mineral density and fractures in women aged 40 to 59 years. Osteoporos Int. 2009;20(3):363–370. [DOI] [PubMed] [Google Scholar]
- 4.Neglia C, Argentiero A, Chitano G, Agnello N, Ciccarese R, Vigilanza A, Pantile V, Argentiero D, Quarta R, Rivezzi M, Di Tanna GL, Di Somma C, Migliore A, Iolascon G, Gimigliano F, Distante A, Piscitelli P. Diabetes and obesity as independent risk factors for osteoporosis: updated results from the ROIS/EMEROS Registry in a population of five thousand post-menopausal women living in a region characterized by heavy environmental pressure. Int J Environ Res Public Health. 2016;13(11):E1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pernicova I, Korbonits M. Metformin--mode of action and clinical implications for diabetes and cancer. Nat Rev Endocrinol. 2014;10(3):143–156. [DOI] [PubMed] [Google Scholar]
- 6.Luo T, Nocon A, Fry J, Sherban A, Rui X, Jiang B, Xu XJ, Han J, Yan Y, Yang Q, Li Q, Zang M. AMPK activation by metformin suppresses abnormal extracellular matrix remodeling in adipose tissue and ameliorates insulin resistance in obesity. Diabetes. 2016;65(8):2295–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandran M. Diabetes drug effects on the skeleton. Calcif Tissue Int. 2017;100(2):133–149. [DOI] [PubMed] [Google Scholar]
- 8.Molinuevo MS, Schurman L, McCarthy AD, Cortizo AM, Tolosa MJ, Gangoiti MV, Arnol V, Sedlinsky C. Effect of metformin on bone marrow progenitor cell differentiation: in vivo and in vitro studies. J Bone Miner Res. 2010;25(2):211–221. [DOI] [PubMed] [Google Scholar]
- 9.Lutz TA, Woods SC. Overview of animal models of obesity. Curr Protoc Pharmacol. 2012;58:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao Y, Xia R, Wang L, Wang YQ, Gao FB. Multimodality imaging assessments of response to metformin therapy for breast cancer in nude mice. Chin Med J (Engl). 2013;126(19):3717–3722. [PubMed] [Google Scholar]
- 11.Chandel NS, Avizonis D, Reczek CR, Weinberg SE, Menz S, Neuhaus R, Christian S, Haegebarth A, Algire C, Pollak M. Are metformin doses used in murine cancer models clinically relevant? Cell Metab. 2016;23(4):569–570. [DOI] [PubMed] [Google Scholar]
- 12.Delahunty KM, Shultz KL, Gronowicz GA, Koczon-Jaremko B, Adamo ML, Horton LG, Lorenzo J, Donahue LR, Ackert-Bicknell C, Kream BE, Beamer WG, Rosen CJ. Congenic mice provide in vivo evidence for a genetic locus that modulates serum insulin-like growth factor-I and bone acquisition. Endocrinology. 2006;147(8):3915–3923. [DOI] [PubMed] [Google Scholar]
- 13.Swami A, Reagan MR, Basto P, Mishima Y, Kamaly N, Glavey S, Zhang S, Moschetta M, Seevaratnam D, Zhang Y, Liu J, Memarzadeh M, Wu J, Manier S, Shi J, Bertrand N, Lu ZN, Nagano K, Baron R, Sacco A, Roccaro AM, Farokhzad OC, Ghobrial IM. Engineered nanomedicine for myeloma and bone microenvironment targeting. Proc Natl Acad Sci USA. 2014;111(28):10287–10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dempster DW, Compston JE, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, Ott SM, Recker RR, Parfitt AM. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 2013;28(1):2–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Müller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res. 2010;25(7):1468–1486. [DOI] [PubMed] [Google Scholar]
- 16.Scheller EL, Troiano N, Vanhoutan JN, Bouxsein MA, Fretz JA, Xi Y, Nelson T, Katz G, Berry R, Church CD, Doucette CR, Rodeheffer MS, Macdougald OA, Rosen CJ, Horowitz MC. Use of osmium tetroxide staining with microcomputerized tomography to visualize and quantify bone marrow adipose tissue in vivo. Methods Enzymol. 2014;537:123–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liaw L, Prudovsky I, Koza RA, Anunciado-Koza RV, Siviski ME, Lindner V, Friesel RE, Rosen CJ, Baker PRS, Simons B, Vary CPH. Lipid profiling of in vitro cell models of adipogenic differentiation: relationships with mouse adipose tissues. J Cell Biochem. 2016;117(9):2182–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simons B, Kauhanen D, Sylvänne T, Tarasov K, Duchoslav E, Ekroos K. Shotgun lipidomics by sequential precursor ion fragmentation on a hybrid quadrupole time-of-flight mass spectrometer. Metabolites. 2012;2(1):195–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo B, Groenke K, Takors R, Wandrey C, Oldiges M. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J Chromatogr A. 2007;1147(2):153–164. [DOI] [PubMed] [Google Scholar]
- 20.Doucette CR, Horowitz MC, Berry R, MacDougald OA, Anunciado-Koza R, Koza RA, Rosen CJ. A high fat diet increases bone marrow adipose tissue (MAT) but does not alter trabecular or cortical bone mass in C57BL/6J mice. J Cell Physiol. 2015;230(9):2032–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wanninger J, Neumeier M, Weigert J, Liebisch G, Weiss TS, Schäffler A, Aslanidis C, Schmitz G, Schölmerich J, Buechler C. Metformin reduces cellular lysophosphatidylcholine and thereby may lower apolipoprotein B secretion in primary human hepatocytes. Biochim Biophys Acta. 2008;1781(6-7):321–325. [DOI] [PubMed] [Google Scholar]
- 22.Chen X, Wang C, Zhang K, Xie Y, Ji X, Huang H, Yu X. Reduced femoral bone mass in both diet-induced and genetic hyperlipidemia mice. Bone. 2016;93:104–112. [DOI] [PubMed] [Google Scholar]
- 23.Yue R, Zhou BO, Shimada IS, Zhao Z, Morrison SJ. Leptin receptor promotes adipogenesis and reduces osteogenesis by regulating mesenchymal stromal cells in adult bone marrow. Cell Stem Cell. 2016;18(6):782–796. [DOI] [PubMed] [Google Scholar]
- 24.Shu L, Beier E, Sheu T, Zhang H, Zuscik MJ, Puzas EJ, Boyce BF, Mooney RA, Xing L. High-fat diet causes bone loss in young mice by promoting osteoclastogenesis through alteration of the bone marrow environment. Calcif Tissue Int. 2015;96(4):313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu EW, Greenblatt L, Eajazi A, Torriani M, Bredella MA. Marrow adipose tissue composition in adults with morbid obesity. Bone. 2017;97:38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bredella MA, Lin E, Gerweck AV, Landa MG, Thomas BJ, Torriani M, Bouxsein ML, Miller KK. Determinants of bone microarchitecture and mechanical properties in obese men. J Clin Endocrinol Metab. 2012;97(11):4115–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu R, Peng F, Liu H, Wang Y, Li L, Wang G, Song J, Shao Z. Clinical significance of osteoblast precursors and osteoclast precursors in earlier diagnosis and monitoring of myeloma bone disease. Ann Hematol. 2016;95(7):1099–1106. [DOI] [PubMed] [Google Scholar]
- 28.Fernández JM, Molinuevo MS, Sedlinsky C, Schurman L, Cortizo AM, McCarthy AD. Strontium ranelate prevents the deleterious action of advanced glycation endproducts on osteoblastic cells via calcium channel activation. Eur J Pharmacol. 2013;706(1-3):41–47. [DOI] [PubMed] [Google Scholar]
- 29.Lupattelli G, Scarponi AM, Vaudo G, Siepi D, Roscini AR, Gemelli F, Pirro M, Latini RA, Sinzinger H, Marchesi S, Mannarino E. Simvastatin increases bone mineral density in hypercholesterolemic postmenopausal women. Metabolism. 2004;53(6):744–748. [DOI] [PubMed] [Google Scholar]
- 30.Chan KA, Andrade SE, Boles M, Buist DS, Chase GA, Donahue JG, Goodman MJ, Gurwitz JH, LaCroix AZ, Platt R. Inhibitors of hydroxymethylglutaryl-coenzyme A reductase and risk of fracture among older women. Lancet. 2000;355(9222):2185–2188. [DOI] [PubMed] [Google Scholar]
- 31.Samelson EJ, Cupples LA, Hannan MT, Wilson PWF, Williams SA, Vaccarino V, Zhang Y, Kiel DP. Long-term effects of serum cholesterol on bone mineral density in women and men: the Framingham Osteoporosis Study. Bone. 2004;34(3):557–561. [DOI] [PubMed] [Google Scholar]
- 32.Solomon DH, Avorn J, Canning CF, Wang PS. Lipid levels and bone mineral density. Am J Med. 2005;118(12):1414. [DOI] [PubMed] [Google Scholar]
- 33.Ducy P, Amling M, Takeda S, Priemel M, Schilling AF, Beil FT, Shen J, Vinson C, Rueger JM, Karsenty G. Leptin inhibits bone formation through a hypothalamic relay: a central control of bone mass. Cell. 2000;100(2):197–207. [DOI] [PubMed] [Google Scholar]
- 34.Ramos-Junior ES, Leite GA, Carmo-Silva CC, Taira TM, Neves KB, Colón DF, da Silva LA, Salvador SL, Tostes RC, Cunha FQ, Fukada SY. Adipokine chemerin bridges metabolic dyslipidemia and alveolar bone loss in mice. J Bone Miner Res. 2017;32(5):974–984. [DOI] [PubMed] [Google Scholar]
- 35.Fan Y, Hanai JI, Le PT, Bi R, Maridas D, DeMambro V, Figueroa CA, Kir S, Zhou X, Mannstadt M, Baron R, Bronson RT, Horowitz MC, Wu JY, Bilezikian JP, Dempster DW, Rosen CJ, Lanske B. Parathyroid hormone directs bone marrow mesenchymal cell fate. Cell Metab. 2017;25(3):661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jang WG, Kim EJ, Lee K-N, Son H-J, Koh J-T. AMP-activated protein kinase (AMPK) positively regulates osteoblast differentiation via induction of Dlx5-dependent Runx2 expression in MC3T3E1 cells. Biochem Biophys Res Commun. 2011;404(4):1004–1009. [DOI] [PubMed] [Google Scholar]
- 37.Lettieri Barbato D, Tatulli G, Aquilano K, Ciriolo MR. FoxO1 controls lysosomal acid lipase in adipocytes: implication of lipophagy during nutrient restriction and metformin treatment. Cell Death Dis. 2013;4(10):e861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woo S-L, Xu H, Li H, Zhao Y, Hu X, Zhao J, Guo X, Guo T, Botchlett R, Qi T, Pei Y, Zheng J, Xu Y, An X, Chen L, Chen L, Li Q, Xiao X, Huo Y, Wu C. Metformin ameliorates hepatic steatosis and inflammation without altering adipose phenotype in diet-induced obesity. PLoS One. 2014;9(3):e91111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim EK, Lee SH, Jhun JY, Byun JK, Jeong JH, Lee S-Y, Kim JK, Choi JY, Cho M-L. Metformin prevents fatty liver and improves balance of white/brown adipose in an obesity mouse model by inducing FGF21. Mediators Inflamm. 2016;2016:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Souza-Mello V, Gregório BM, Cardoso-de-Lemos FS, de Carvalho L, Aguila MB, Mandarim-de-Lacerda CA. Comparative effects of telmisartan, sitagliptin and metformin alone or in combination on obesity, insulin resistance, and liver and pancreas remodelling in C57BL/6 mice fed on a very high-fat diet. Clin Sci (Lond). 2010;119(6):239–250. [DOI] [PubMed] [Google Scholar]
- 41.Zhao J, Yue W, Zhu MJ, Sreejayan N, Du M. AMP-activated protein kinase (AMPK) cross-talks with canonical Wnt signaling via phosphorylation of beta-catenin at Ser 552. Biochem Biophys Res Commun. 2010;395(1):146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCreight LJ, Bailey CJ, Pearson ER. Metformin and the gastrointestinal tract. Diabetologia. 2016;59(3):426–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Risérus U, Willett WC, Hu FB. Dietary fats and prevention of type 2 diabetes. Prog Lipid Res. 2009;48(1):44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu CC, Qing K, Chen Y. Diet-induced changes in stearoyl-CoA desaturase 1 expression in obesity-prone and -resistant mice. Obes Res. 2004;12(8):1264–1270. [DOI] [PubMed] [Google Scholar]
- 45.Wang F, Liu H, Blanton WP, Belkina A, Lebrasseur NK, Denis GV. Brd2 disruption in mice causes severe obesity without type 2 diabetes. Biochem J. 2009;425(1):71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]