Abstract
The GATOR1/SEACIT complex consisting of Iml1-Npr2-Npr3 inhibits Target of Rapamycin Complex 1 (TORC1) in response to amino acid insufficiency. In glucose medium, Saccharomyces cerevisiae mutants lacking the function of this complex grow poorly in the absence of amino acid supplementation, despite hallmarks of increased TORC1 signaling. Such mutants perceive they are amino acid-replete and thus repress metabolic activities that are important for achieving this state. We find that npr2Δ mutants have defective mitochondrial TCA cycle activity and retrograde response. Supplementation of glutamine, and especially aspartate, which are nitrogen-containing forms of TCA cycle intermediates, rescue growth of npr2Δ mutants. These amino acids are then consumed in biosynthetic pathways that require nitrogen to support proliferative metabolism. Our findings reveal that negative regulators of TORC1 such as GATOR1/SEACIT regulate the cataplerotic synthesis of these amino acids from the TCA cycle in tune with the amino acid and nitrogen status of cells.
INTRODUCTION
The budding yeast S. cerevisiae regulates energy homeostasis and metabolism through the conserved Target of Rapamycin Complex 1 (TORC1)1. Genes encoding the kinase component of TORC1 (TOR1, TOR2) were first cloned in yeast, as mutants are resistant to the small molecule immunosuppressant rapamycin2. Subsequently, work by many groups has shown that TORC1 is responsive to amino acids and nitrogen sources to regulate an extensive network of energy-consuming processes important for cell growth. Upon amino acid or nitrogen starvation, decreased TORC1 activity triggers autophagy and other metabolic adaptions important for cell survival1, 3, 4. Major targets of TORC1 in yeast include Sch9p, an AGC family kinase and functional analog of mammalian S6K5, and Gln3p, a transcriptional activator that promotes the expression of nitrogen catabolite repression (NCR)-sensitive genes, required for the assimilation of poor nitrogen sources such as proline or urea4. Low TORC1 activity during nitrogen starvation results in the dephosphorylation of Gln3p, which enables its nuclear translocation and activation6.
The conserved, vacuole-associated Rag GTPases Gtr1p and Gtr2p form a heterodimer to activate TORC1 in response to nutrient signals7, 8. A variety of regulators of TORC1 have been shown to function by modulating the activity of these small GTPases, through their action as GEFs (Guanine nucleotide Exchange Factor) or GAPs (GTPase Activating Protein). A conserved, vacuole-associated complex consisting of three proteins Iml1p, Npr2p, and Npr3p (named SEACIT in yeast, or GATOR1 in mammals)9, 10, has been identified as a GAP for the Rag GTPases11, 12, thereby enabling the complex to function as a major negative regulator of TORC1 in response to amino acid insufficiency.
Several components of GATOR1 (Npr2p and Npr3p) were first identified as “nitrogen permease regulators” in yeast important for growth in poor nitrogen sources13–15. Loss of GATOR1 leads to insensitivity to amino acid starvation11, 12. Without Npr2p, yeast cells bypass autophagy and continue to grow in minimal medium containing lactate as the carbon source9, 16. Under such conditions that demand mitochondrial oxidative metabolism, npr2Δ mutants synthesize and consume glutamine for the synthesis of nitrogen-containing metabolites, revealing that activation of TORC1 promotes the synthesis and utilization of glutamine to support proliferative metabolism17. However, in minimal glucose medium, it has been observed that npr2Δ mutants grow poorly in the presence of the most commonly used nitrogen source, ammonium15. The slow growth of these mutants is unexpected and seemingly inconsistent with its reported function as a negative regulator of TORC1. Due to the phenomenon of glucose repression and a dramatic alteration of yeast physiology in the absence of glucose, it is conceivable that mutants lacking GATOR1 function may differentially affect metabolism and proliferation in a manner dependent on the carbon source18.
The importance of glutamine for the growth and proliferation of a variety of cell types has long been known19. However, the extent through which TORC1 influences other metabolic pathways beyond glutamine metabolism in the regulation of cell growth and homeostasis remain incompletely understood. In this study, we have systematically dissected why npr2Δ mutants grow poorly in the most commonly utilized synthetic defined glucose medium. This has enabled us to decipher how TORC1 more broadly directs cellular metabolism under conditions that favor a highly glycolytic metabolism, and point to a fundamental metabolic function for the GATOR1/SEACIT complex in balancing anaplerotic and cataplerotic reactions of the mitochondrial TCA cycle.
RESULTS
npr2Δ mutants grow poorly in minimal glucose (SD) medium
We previously determined that the GATOR1 complex consisting of Iml1p-Npr2p-Npr3p is required for prototrophic yeast cells to induce autophagy upon switch from rich to minimal medium with lactate as a carbon source, conditions that require mitochondrial respiration9. In minimal lactate medium, not only do iml1Δ, npr2Δ, and npr3Δ mutants bypass autophagy, but they also grow faster than wild type (WT) cells, consistent with the role of this complex as a negative regulator of TORC116, 17.
However, in the more commonly used minimal glucose medium (SD), iml1Δ, npr2Δ, and npr3Δ mutants each exhibited notably slower growth than WT (Fig. 1a, Supplementary Results, Supplementary Fig. 1a). Moreover, a GTP-locked mutant of Gtr1p (Q65L) that exhibits constitutively activated TORC120, also grew poorly, similar to the npr2Δ mutant (Supplementary Fig. 1b). Deletion of GTR1 alleviated the slow growth phenotype of npr2Δ mutants, consistent with the finding that Npr2p functions specifically through Gtr1p (Supplementary Fig. 1b)12. The slow growth phenotype observed was specifically due to the absence of Npr2p as reintroduction of a construct expressing NPR2-flag completely rescued the growth of npr2Δ mutants (Supplementary Fig. 1c).
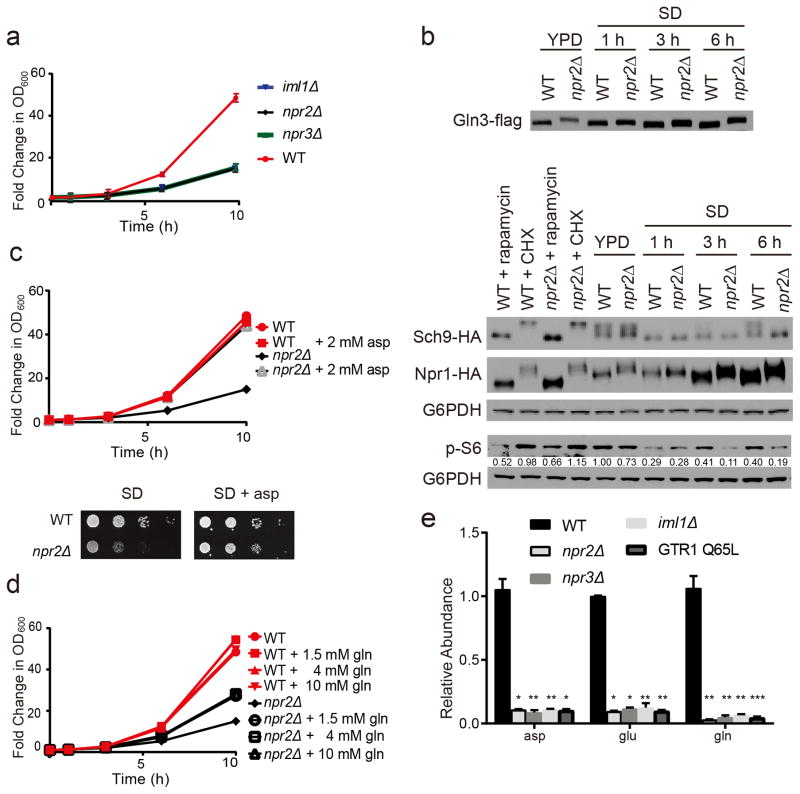
Figure 1. The defective growth of npr2Δ mutants can be rescued by aspartate or glutamine.
(a) Growth of wild type (WT), iml1Δ, npr2Δ, and npr3Δ strains in SD medium. Data were mean +/- s.d. from one set of triplicates, 3 independent experiments were performed.
(b) Western blots depicting Gln3p, Sch9p, Npr1p and phospho-S6 proteoforms in WT and npr2Δ cells switched from YPD to SD medium. The Sch9 protein is cleaved by NTCB treatment to observe the mobility shift of the C-terminal peptide. 200 ng/mL rapamycin or 25 μg/mL cycloheximide was added as controls for inhibition or stimulation of TORC1 signaling. G6PDH serves as the loading control. p-S6 amounts were quantified against G6PDH by ImageJ and normalized against WT in YPD. Full gels are shown in Supplementary Fig. 5a.
(c) Growth of WT or npr2Δ cells in SD with or without supplementation of aspartate (2 mM) in liquid culture (upper panel) or agar plate (lower panel). Data were mean ± s.d. from one set of triplicates, 3 independent experiments were performed.
(d) Growth of WT or npr2Δ cells with or without supplementation of indicated amounts of glutamine. Data were mean ± s.d. from one set of triplicates, 3 independent experiments were performed.
(e) Relative abundance of intracellular glutamine, glutamate and aspartate in WT, npr2Δ, npr3Δ, iml1Δ and GTR1 Q65L cells quantitated by LC-MS/MS. Yeast cells were collected after switch from YPD to SD for 6 hours. Data were mean ± s.d. from one set of 4 repeats, 2 independent experiments were performed. *p < 0.05, **p < 0.01, ***p < 0.001 by two-tailed Student’s t-test.
Despite their slow growth phenotype, npr2Δ mutants exhibited increased TORC1 signaling as assessed by the phosphorylation status of the responsive substrates Gln3p and Npr1p (Fig. 1b, Supplementary Fig. 1d)21. Supplementation of low concentrations of rapamycin could rescue the growth of npr2Δ, npr3Δ, and iml1Δ mutants, consistent with slow growth due to hyperactive TORC1 (Supplementary Fig. 1e). However, Sch9p, a reported ortholog of mammalian S6K and major substrate of TORC15, became less phosphorylated in npr2Δ mutants over time in SD medium (Fig. 1b, Supplementary Fig. 1d). Nonetheless, multiple lines of evidence indicate that despite having hallmarks of increased TORC1 signaling, npr2Δ mutants still exhibited defective growth in minimal glucose medium.
Why would a cell with hyperactive TORC1 signaling grow more slowly? SD medium and the majority of defined yeast media contain ammonium (NH4+) as the nitrogen source, requiring prototrophic yeast strains to synthesize all twenty amino acids using ammonium. We surmised that npr2Δ mutants in glucose might perceive themselves to be in an amino acid and nitrogen-replete state, regardless of the nutrient environment. Consistent with this idea, the poor growth of npr2Δ mutants in SD medium was rescued upon supplementation of a standard amino acid mixture (SD + CSM) (Supplementary Fig. 1f). As such, we interpret these results to suggest that npr2Δ mutants inappropriately repress metabolic activities, such as the synthesis of amino acids, because they perceive themselves to already be amino acid-replete. Therefore, growth deficits may emerge especially under conditions that require the synthesis of amino acids (e.g., minimal SD medium).
We next tried to rescue the growth of npr2Δ mutants by supplementation of single amino acids. Strikingly, the sole addition of modest concentrations of aspartate (2 mM) could completely rescue the growth of npr2Δ cells (Fig. 1c). Glutamine could partially rescue growth of npr2Δ cells, but very high concentrations did not have an additional benefit (Fig. 1d). Asparagine also fully rescued the growth of npr2Δ mutants while methionine had a partial effect. Other amino acids had no observable effect on growth (Supplementary Fig. 1g). Aspartate was also able to fully rescue growth of other GATOR1 mutants (iml1Δ and npr3Δ) (Supplementary Fig. 1e). Interestingly, the branched-chain amino acids, leucine and isoleucine, modestly inhibited the growth of WT but not npr2Δ cells. These rescue experiments suggest that npr2Δ mutants might exhibit defects in the synthesis of particular amino acids, especially aspartate, and to a lesser extent, glutamine.
To further understand the metabolic basis of the slow growth of npr2Δ mutants in minimal SD medium without amino acids, we measured the relative abundance of intracellular metabolites in npr2Δ cells compared to WT. Focusing first on amino acids, glutamine, glutamate and aspartate amounts were extremely low in npr2Δ, as well as npr3Δ, iml1Δ, and GTR1 Q65L cells compared to WT (Fig. 1e), consistent with the observation that glutamine and aspartate could rescue their growth. These results suggest that the GATOR1 complex might play a role in the regulation of glutamine and aspartate synthesis, which led us to further investigate pathways required for their production. Under all conditions tested thus far, we observed the phenotypes of iml1Δ, npr2Δ, and npr3Δ mutants to be identical (Fig. 1a,e, Supplementary Fig. 1a,e). In subsequent experiments, we focused on npr2Δ mutants to assess the consequences of loss of GATOR1 function.
npr2Δ mutants have defective mitochondrial functions
Glutamine can be synthesized from the TCA cycle intermediate α-ketoglutarate by the sequential actions of glutamate dehydrogenase (Gdh1p/Gdh3p) and glutamine synthetase (Gln1p). Aspartate can be synthesized from the TCA cycle intermediate oxaloacetate, by the action of the aspartate aminotransferases (Aat1p/Aat2p). Since both glutamine and aspartate can be derived from TCA cycle metabolites, we investigated whether mitochondrial functions might be regulated by Npr2p and thus the GATOR1 complex. Upon measuring dissolved oxygen amounts from exponential growth to the diauxic shift, we observed that npr2Δ and other GATOR1 mutants, as well as the GTR1 Q65L mutant, consumed much less oxygen compared to WT during growth in SD medium (Fig. 2a, Supplementary Fig. 2a), despite having comparable amounts of mitochondria as assessed by mtDNA content (Supplementary Fig. 2b). Even though WT cells grew faster initially, WT and npr2Δ cells consumed glucose at a similar rate from the growth medium (Fig. 2a, Supplementary Fig. 2c). Therefore, npr2Δ cells consume less oxygen per glucose under these conditions. Notably, aspartate supplementation did not restore the reduced respiratory activity of npr2Δ cells (Supplementary Fig. 2a), despite its ability to fully rescue their growth.
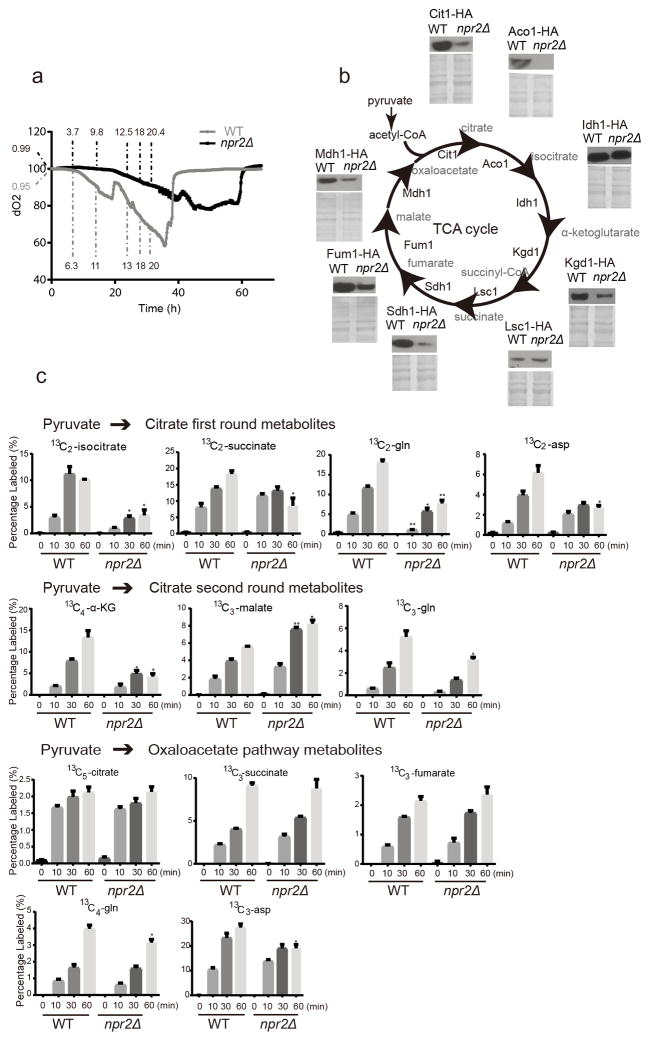
Figure 2. npr2Δ mutant cells have defective mitochondrial function.
(a) Dissolved oxygen levels as a function of growth following inoculation into SD medium. WT and npr2Δ strains were grown in parallel bioreactor vessels, and dissolved oxygen amounts were monitored continuously. The OD600 at corresponding time points were labeled for WT (grey) and npr2Δ (black). Representative data were selected from 3 independent experiments.
(b) Western blots indicating amounts of HA-tagged TCA cycle enzymes: Aco1p (aconitase), Cit1p (citrate synthase), Idh1p (isocitrate dehydrogenase), Kgd1p (α-ketoglutarate dehydrogenase), Lsc1p (succinyl-CoA ligase), Sdh1p (succinate dehydrogenase), Fum1p (fumarase) and Mdh1p (mitochondrial malate dehydrogenase) in WT and npr2Δ cells grown in SD for 6 hours. Total protein level (Coomassie blue stain) is shown as loading control. Full gels are shown in Supplementary Fig. 5b.
(c) Relative abundance of labeled TCA cycle metabolites from glucose-13C6 tracing experiments. Labeled species are shown as percentage of all isotopomers of the respective metabolite. Glucose-13C6 is converted into pyruvate. Pyruvate can be converted into acetyl-CoA, and all first round TCA cycle metabolites are M+2 labeled. The final product M+2 oxaloacetate in combination with M+2 labeled acetyl-CoA can give rise to second round metabolites. Pyruvate can also be converted into M+3 oxaloacetate and the TCA products are labeled as listed in Supplementary Table 2. Data were mean ± s.d. from 3 independent experiments. *p < 0.05, **p < 0.01, ***p<0.001, by two-tailed Student’s t-test.
We then examined TCA cycle enzyme levels and found that npr2Δ cells express significantly reduced amounts of most TCA cycle enzymes, including Aco1p, Cit1p, Kgd1p, Sdh1p, Fum1p and Mdh1p (Fig. 2b). We also measured the corresponding TCA cycle metabolites. However, when compared at similar growth stages, there was no significant difference in amounts of most TCA cycle metabolites between WT and npr2Δ cells, except for α-ketoglutarate (Supplementary Fig. 2d).
However, bulk measurements of metabolite pools do not inform about the rates of synthesis or consumption. We then turned to a glucose-13C6 labeling experiment to assess rates of TCA cycle metabolite production. WT and npr2Δ cells consumed similar amounts of glucose over the time period examined. After entering the cell, glucose-13C6 is converted into labeled pyruvate. There are two ways for the labeled pyruvate to enter the TCA cycle. It can be converted either to acetyl-CoA with loss of one carbon as CO2, or carboxylated to oxaloacetate with a gain of one carbon. The 13C-labeling patterns of metabolites from each pathway were assessed as described previously22. For the pyruvate to citrate pathway, npr2Δ cells produced significantly less early TCA cycle metabolites (isocitrate and α-ketoglutarate), glutamine and aspartate, throughout a period of 1 h (Fig. 2c). For the pyruvate to oxaloacetate pathway, npr2Δ cells also produced less glutamine and aspartate from this route (Fig. 2c), despite comparable labeling of most TCA intermediates. These data suggest that npr2Δ mutants are compromised in their ability to synthesize glutamine and aspartate from TCA cycle intermediates. The addition of cell-permeable α-ketoglutarate (dimethyl 2-oxoglutarate) was unable to rescue growth of these mutants, despite largely restoring α-ketoglutarate to levels comparable to WT (Supplementary Fig. 2e).
We next observed that npr2Δ mutants exhibited hallmarks of oxidative metabolism characteristic of differentiated or non-proliferating cells. The amount of the glutamate synthase enzyme Glt1p was markedly reduced in npr2Δ cells, consistent with the idea that these mutants inappropriately perceive themselves to be glutamate or nitrogen-replete23 (Supplementary Fig. 2f). Upon examining the enzymes directly involved in aspartate synthesis, pyruvate carboxylase enzyme (Pyc1/Pyc2) amounts were comparable to WT, but amounts of the aspartate aminotransferase enzyme Aat2p were higher in npr2Δ cells (Supplementary Fig. 2g). The expression profile of AAT2 is correlated with mitochondrial biogenesis, as opposed to growth, across the yeast metabolic cycle (Supplementary Fig. 2h)24, suggesting this particular enzyme is typically induced to promote oxaloacetate synthesis to support mitochondrial activities. In addition, we observed that the pyruvate dehydrogenase complex (Pda1p) was significantly less phosphorylated in npr2Δ cells, suggesting they prefer the routing of pyruvate into acetyl-CoA for oxidation in the mitochondria (Supplementary Fig. 2g). Consistent with this idea, npr2Δ cells also expressed higher amounts of the respiratory mitochondrial pyruvate carrier subunit Mpc3p (Supplementary Fig. 2g)25. Taken together, these data all support the idea that mutants lacking GATOR1 function perceive themselves to be amino acid- and nitrogen-replete, and are wired in a state that favors utilization of the mitochondria for oxidation of glucose-derived pyruvate, instead of aspartate and glutamine synthesis.
Furthermore, npr2Δ cells also exhibited a modest reduction in the NAD+/NADH ratio, consistent with reduced TCA cycle and respiratory activity (Supplementary Fig. 3a). However, supplementation of pyruvate and other attempts to restore the NAD+/NADH ratio were not successful in rescuing the growth defect of npr2Δ cells. Notably, npr2Δ cells secreted substantial amounts of acetate, suggesting a deficiency in acetate utilization for mitochondrial functions (Supplementary Fig. 2c). Therefore, not only do npr2Δ mutants appear wired for mitochondrial oxidative metabolism, they also have defects in the actual utilization of the TCA cycle for mitochondrial respiration.
npr2Δ mutants cannot activate the retrograde pathway
We next sought to understand the basis of the inefficient TCA cycle utilization in npr2Δ mutants. The expression of several TCA cycle enzymes that were decreased in npr2Δ mutants is regulated by the mitochondrial retrograde response transcription factors Rtg1p/Rtg3p. Mitochondrial dysfunction causes Rtg1p/Rtg3p to translocate into the nucleus and activate genes that are important for adaptation to respiratory deficiency26. These target genes include several enzymes of the TCA cycle, such as CIT1 and ACO1. This is because even in the absence of respiration, the first enzymes of the TCA cycle are still required for the synthesis of the non-essential amino acid glutamate, which is derived from the TCA cycle intermediate α-ketoglutarate. Intriguingly, the retrograde response is also regulated by TORC1, as rapamycin induces nuclear translocation of Rtg1p/Rtg3p27. Due to low amounts of glutamate and glutamine, reduced amounts of TCA cycle enzymes, and dysregulation of TORC1 in npr2Δ cells, we suspected that the retrograde response might be defective in such mutants. Consistent with this possibility, rtg1Δ mutants have reduced citrate synthase, aconitase and isocitrate dehydrogenase protein and enzyme activities and behave as glutamate and aspartate auxotrophs28.
Indeed in SD medium, WT cells exhibited nuclear localization of Rtg1-GFP, but the transcription factor remained in the cytosol in npr2Δ mutants (Fig. 3a). Thus, under minimal glucose conditions, cells normally activate the retrograde response for the synthesis of glutamate and glutamine, from the nitrogen source ammonium. In contrast, npr2Δ cells appeared tricked into thinking they are aspartate, glutamine, and nitrogen-replete, and do not turn on the retrograde response. As expected, the addition of either the electron transport chain complex III inhibitor antimycin or the TORC1 inhibitor rapamycin was sufficient to cause Rtg1-GFP to enter the nucleus in both WT and npr2Δ cells (Fig. 3a). In contrast, glutamine as well as aspartate addition was sufficient to cause export of Rtg1-GFP out of the nucleus in WT cells (Fig. 3a). We observed a significant decrease in downstream targets of Rtg1p/Rtg3p, such as CIT1, CIT2, at both the mRNA and protein level in npr2Δ cells (Fig. 3b, Supplementary Fig. 3b). Taken together, these phenotypes are all indicative of a defective retrograde response in npr2Δ cells, and reveal they do not appropriately utilize the mitochondria to boost the synthesis of glutamate, glutamine, and aspartate.
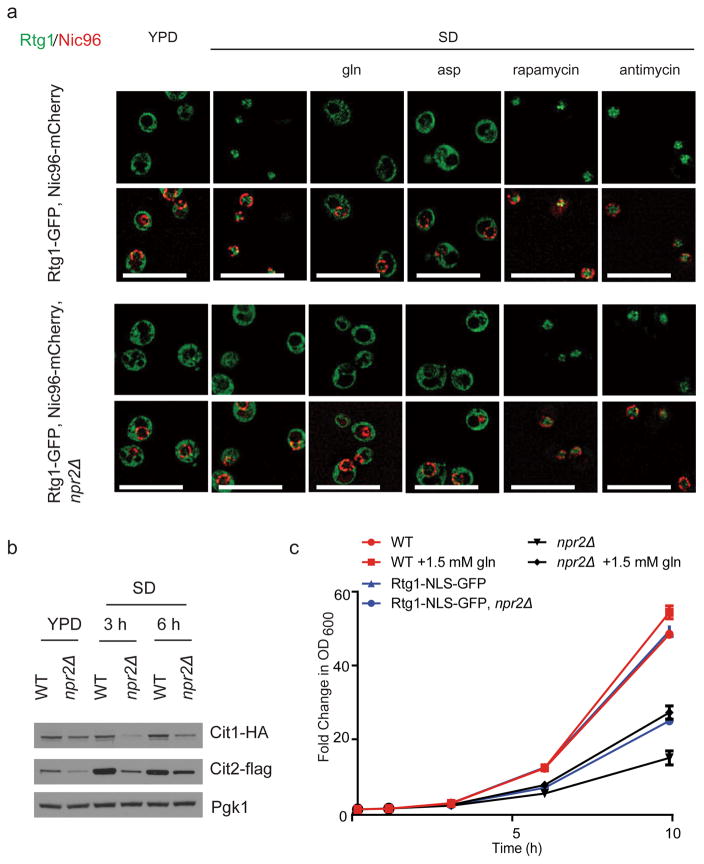
Figure 3. npr2Δ mutants exhibit a defective mitochondria-to-nucleus retrograde (RTG) response.
(a) WT or npr2Δ cells expressing Rtg1-GFP, Nic96-mCherry (nuclear pore complex marker) were transferred from YPD to SD for 6 hours and treated with mock, 50 μM antimycin, 50 nM rapamycin, 2 mM glutamine or 2 mM aspartate (pH 5.1) for 30 min prior to imaging. Antimycin and rapamycin treatment activated the retrograde response by in both strains, while glutamine or aspartate addition inactivated the pathway in WT cells. Scale bar 5 μm.
(b) Western blot depicting amounts of retrograde response targets Cit1p and Cit2p in WT and npr2Δ cells switched from YPD to SD for indicated times. Note that npr2Δ mutants have consistently lower amounts of these enzymes in SD medium. Full gels are shown in Supplementary Fig. 5c.
(c) Forced nuclear localization of Rtg1p can partially rescue the growth of npr2Δ mutants. Growth of WT or npr2Δ cells expressing a version of Rtg1-GFP containing a strong nuclear localization signal (NLS) (Rtg1-NLS-GFP). Two duplicates at each time point were measured, in two independent experiments.
We then asked whether restoration of the retrograde response might improve the growth of npr2Δ mutants in SD medium. We constructed a strain expressing a version of Rtg1-GFP containing a strong SV40 large T antigen nuclear localization signal (NLS) SPKKKRKV29 from its endogenous locus. Imaging of these cells in rich YPD medium confirmed that Rtg1-NLS-GFP was constitutively present in the nucleus under conditions that would normally trigger its exclusion (Supplementary Fig. 3c). In SD medium, the forced nuclear localization of Rtg1p was able to rescue the growth of npr2Δ cells to a similar extent as supplementation of glutamine (Fig. 3c). Taken together, these findings confirm that the inability to turn on the retrograde response is partially responsible for the poor growth of npr2Δ cells in minimal glucose medium in the absence of amino acid supplementation.
npr2Δ mutants exhibit compromised nucleotide production
Beyond their functions as amino acids, glutamine and aspartate play additional roles in cellular metabolism and biosynthesis. Glutamine is a nitrogen donor in both de novo purine and pyrimidine synthesis, while aspartate donates nitrogen for purine synthesis and its carbons for pyrimidine synthesis. We observed that npr2Δ, npr3Δ and iml1Δ cells contained reduced nucleotide amounts compared to WT (Fig. 4a). Aspartate supplementation rescued the intracellular levels of nucleotide metabolites in npr2Δ cells, while glutamine or retrograde pathway activation by Rtg1-NLS-GFP did not, consistent with the ability of aspartate but not glutamine to fully rescue cell growth (Fig. 4b, Supplementary Fig. 4a). The α-ketoglutarate, aspartate, glutamine, and glutamate levels were also rescued to a significant extent by aspartate in npr2Δ cells (Fig. 4b). The overall differences between the metabolomes of WT and npr2Δ cells are diminished by aspartate supplementation, further indicating the importance of GATOR1 to the regulation of aspartate synthesis in minimal medium (Supplementary Fig. 4b).
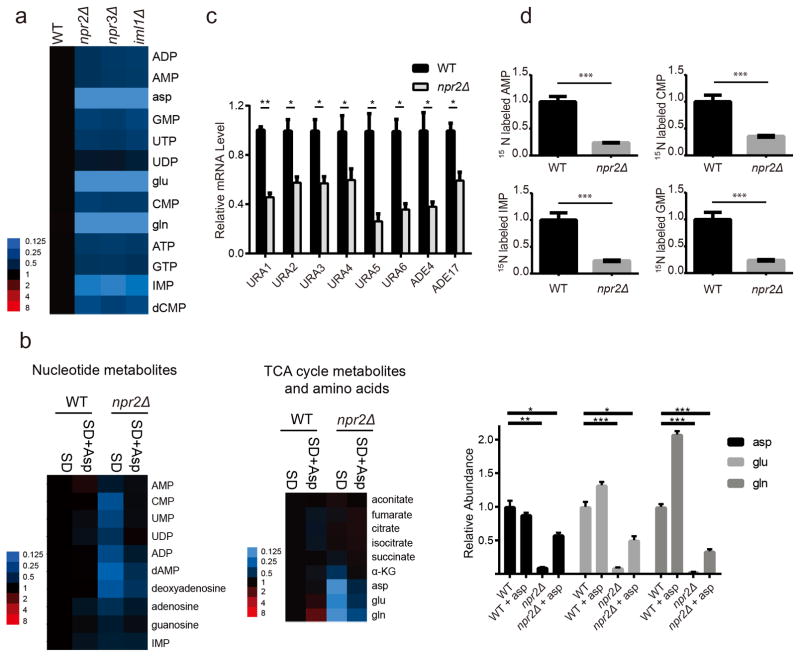
Figure 4. npr2Δ cells produce less nucleotide metabolites.
(a) Heat map depicting abundance of nucleotide metabolites in WT, npr2Δ, npr3Δ and iml1Δ cells. Cells were grown in YPD and then transferred into SD for 6 h prior to extraction. Intracellular nucleotide metabolites levels were measured with targeted LC-MS/MS methods. Data were collected from 3 independent experiments.
(b) Heat map of relative abundance of nucleotide metabolites (left) or TCA cycle metabolites, glutamine, glutamate and aspartate (middle) following aspartate supplementation, normalized against WT. Cells were switched into SD for 6 h as in (a), but with or without addition of 2 mM aspartate. Metabolites were extracted and analyzed by LC-MS/MS. Aspartate, glutamate and glutamine levels are also shown in bar graph format (right). Data were mean ± s.d. from 3 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 by two-tailed Student’s t-test.
(c) mRNA levels of pyrimidine synthesis pathway enzymes (URA1, URA2, URA3, URA4, URA5, URA6) and purine synthesis pathway (ADE4, ADE17) from WT and npr2Δ cells collected as shown in (a). Data were mean ± s.d. from 2 independent experiments with duplicates for each genotype. *p < 0.05, **p < 0.01, by two-tailed Student’s t-test.
(d) Relative abundance of labeled nucleotide metabolites from 15N-ammonium tracing experiments. Cells were grown in YPD and then transferred into SD for 3 h, and then switched into SD-N plus 5 g/L ammonium-15N2 sulfate for 1 h. Data were mean ± s.d. from 3 independent experiments. ***p<0.001, by two-tailed Student’s t-test.
Analysis of mRNA amounts further showed that npr2Δ cells exhibited reduced mRNA expression of pyrimidine biosynthetic enzymes, including URA1, URA2, URA3, URA4, URA5, URA6, as well as two purine biosynthetic enzymes, ADE4 (5-phosphoribosyl-1-pyrophosphate amidotransferase), the rate-determining enzyme, and ADE17 (5-aminoimidazole-4-carboxamide ribonucleotide transformylase) (Fig. 4c)30. We then confirmed using ammonium-15N tracing that npr2Δ cells produce less nucleotide metabolites (Fig. 4d). Therefore, they are compromised in their ability to utilize ammonium for the synthesis of the nitrogenous bases present in these nucleotides. Reduced de novo synthesis of nucleotides is consistent with the reduced synthesis of aspartate and glutamine in npr2Δ cells.
To assess whether the decrease in nucleotide metabolism is the basis of the growth defect of npr2Δ mutants, we tested whether supplementation of nucleosides or nitrogenous bases could rescue growth in SD medium. None of the nucleoside or nitrogenous base combinations could improve the growth of npr2Δ mutants (Supplementary Fig. 4c,d). Therefore, the deficiencies in nucleotide synthesis are not the primary cause of the poor growth. It is likely that additional functions for aspartate and glutamine are important for growth, such as their roles in protein synthesis and other downstream cellular functions. For example, aspartate is also required for asparagine and methionine synthesis, which could explain the partial rescue of growth by these additional amino acids (Supplementary Fig. 1d). Nonetheless, these findings support a role for TORC1 in promoting the synthesis of glutamine, and especially aspartate, from the TCA cycle for purposes of nucleotide synthesis. Loss of the GATOR1 complex deprives cells of the ability to properly regulate the synthesis of these amino acids from the TCA cycle, and therefore nucleotide synthesis is also compromised.
DISCUSSION
The GATOR1 complex has emerged as a conserved negative regulator of TORC1 that functions in response to amino acid insufficiency. Here we have shown that in high glucose, npr2Δ mutants lacking GATOR1 function grow slowly unless they are supplemented with amino acids, despite having hallmarks of increased TORC1 signaling. We reasoned that npr2Δ mutants perceive themselves to be in an amino acid- and nitrogen-replete state, and therefore repress metabolic activities that are required to achieve this state. Through analysis and rescue of the slow growth phenotype of npr2Δ mutants, we deduced how TORC1 alters cellular metabolism to achieve the amino acid- and nitrogen-replete state. Notably, despite the many alterations in transcription, protein levels, and phosphorylation status of metabolic enzymes caused by loss of Npr2p, the sole addition of aspartate was sufficient to completely rescue the growth of npr2Δ mutant cells. Our findings therefore reveal that a fundamental metabolic function of TORC1 is to promote the synthesis and utilization of the amino acids aspartate and glutamine, which are derived from cataplerotic reactions of the mitochondrial TCA cycle (Fig. 5).
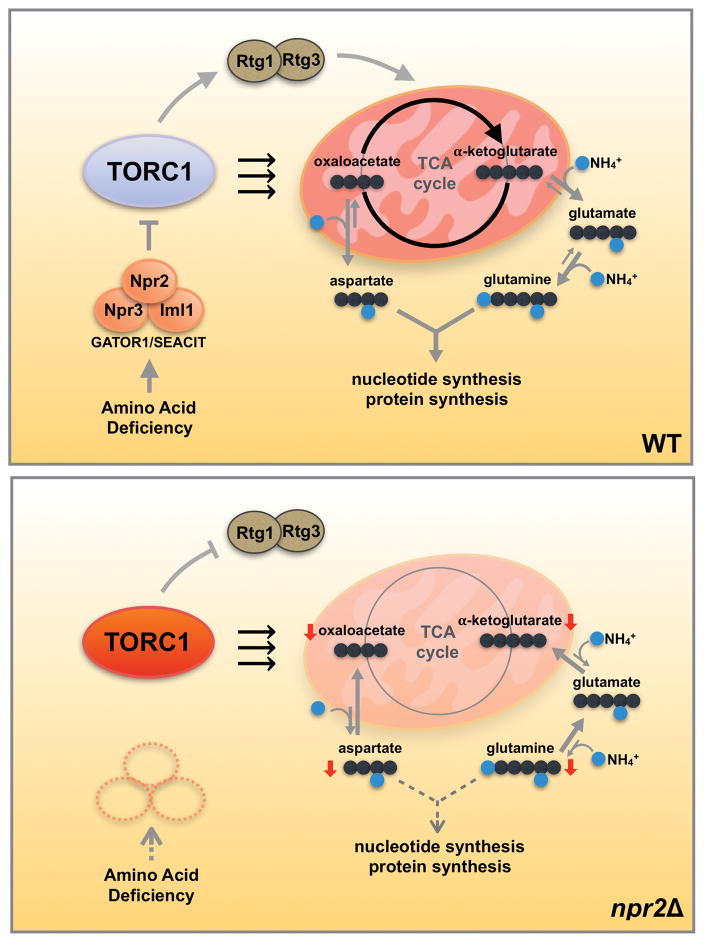
Figure 5. The GATOR1/SEACIT complex regulates cataplerotic reactions of the mitochondrial TCA cycle in tune with the amino acid and nitrogen status of cells.
In glucose medium, yeast cells preferentially perform glycolysis and reduce mitochondrial TCA cycle activity. When starved for amino acids (SD medium), WT cells are able to sense the lack of amino acids and regulate TORC1 activity through Npr2p and the GATOR1/SEACIT complex. As a result, WT cells boost TCA cycle activity as well as the retrograde response pathway for the production of nitrogen-containing glutamine and aspartate from α-ketoglutarate and oxaloacetate (cataplerotic reactions). Glutamine and aspartate are then consumed for nucleotide and protein synthesis, which have a high nitrogen demand. In npr2Δ mutants, cells inappropriately perceive themselves to be amino acid-replete. Constitutively active TORC1 inhibits the replenishment of α-ketoglutarate and oxaloacetate (anaplerotic reactions) due to dysregulation of the retrograde response pathway, and thus utilization of the mitochondria for synthesis of aspartate and glutamine is reduced due to the perception that cells are already amino acid-replete. Therefore, the GATOR1/SEACIT complex regulates TORC1 to balance cataplerotic and anaplerotic reactions of the TCA cycle in tune with the amino acid and nitrogen status of cells. Since glutamine and particularly aspartate rescue the consequences of loss of GATOR1/SEACIT, high TORC1 activity signifies that cells have the appropriate capacity for biosynthetic demands requiring these amino acids.
Why might this represent a sensible function for TORC1? Since its original discovery in budding yeast in 1991, the regulation of TORC1 signaling has been closely linked to the quality of nitrogen sources21, 27, 31. While yeasts have the capability to utilize a variety of different nitrogenous compounds to synthesize all essential nitrogen-containing metabolites, they exhibit a hierarchical preference with glutamine as the most favorable nitrogen source32. When given less preferred nitrogen sources, yeast cells activate Gln3p, the transcriptional activator of nitrogen catabolite repressed (NCR) genes, to enable their assimilation to glutamine33. This is largely achieved through inhibition of TORC1-mediated phosphorylation of Gln3p, which promotes its nuclear localization34. In addition to Gln3p, yeast cells are also dependent on the Rtg1p/Rtg3p transcription factors for the synthesis of glutamine. Since both rapamycin and mitochondrial dysfunction lead to nuclear translocation of Rtg1p/Rtg3p and induction of the mitochondrial retrograde response, the cellular response to either amino acid starvation or respiratory deficiency involves induction of a gene expression program geared towards the synthesis of glutamate and glutamine. Under such conditions, cells decrease the consumption of glutamine and instead focus on its preservation. Consistent with this idea, we previously observed that induction of autophagy leads cells to accumulate glutamine17. In contrast, during growth, glutamine is consumed rapidly due to its requirement as a nitrogen donor for the biosynthesis of nitrogen-containing metabolites such as nucleotides17.
Our findings reveal that the GATOR1 complex enables TORC1 to function in response to nitrogen quality and the availability of the preferred nitrogen source glutamine. Although these pathways are clearly responsive to glutamine, how glutamine is sensed in cells to elicit the proper regulation of these transcription factors remains an open question35–38. However, glutamine might not be sensed directly, as yeast cells can exhibit increased TORC1 signaling and proliferative metabolism despite substantially lower absolute amounts of intracellular glutamine as a result of its rapid consumption for biosynthesis during growth17. Moreover, we observed a discrepancy between the phosphorylation status of Sch9p, the most commonly used readout of TORC1 in yeast, and other TORC1 activity markers. In high glucose conditions lacking free amino acids, npr2Δ cells have hyperphosphorylated Gln3p and Npr1p, yet exhibit reduced Sch9p and Rps6p phosphorylation. However, the fact that the npr2Δ mutant exhibits a rapamycin-reversible, slow growth phenotype just as a GTP-locked GTR1 Q65L mutant suggests they in fact have hyperactive TORC1 signaling. Thus, Sch9p and S6 phosphorylation status may be more correlated with growth rate than TORC1 activity. Possible reasons for this discrepancy could be due to the influence of both TORC1 and TORC2 on phospho-S639. Moreover, the AGC kinase Ypk3p, rather than Sch9p, might be the true yeast ortholog of S6K, and the activity of Sch9p or Rps6p phosphatases could also be affected in GATOR1 mutants39, 40. Nonetheless, our findings suggest caution when correlating the phosphorylation status of TORC1 substrates to the amino acid or growth status of cells.
Despite the importance of glutamine, aspartate may in fact be a more critical and demanding output of TORC1. Our findings show that TORC1 utilizes mitochondrial TCA cycle activity for aspartate synthesis rather than for ATP, as the growth of npr2Δ cells with low TCA cycle activity can be completely rescued by adding modest concentrations of aspartate (Fig. 1d). Aspartate, like glutamine, is also involved in protein, purine and pyrimidine synthesis. However, the supplementation of various combinations of nucleosides and nucleobases was unable to rescue the growth of npr2Δ cells, suggesting that the effects of aspartate and glutamine extend beyond just nucleotide synthesis. It is notable that aspartate synthesis from oxaloacetate requires glutamate as the nitrogen donor, as opposed to ammonium. Glutamate synthesis from α-ketoglutarate consumes NAD(P)H, while aspartate synthesis would be impaired by high NADH due to the reversibility of malate dehydrogenase and depletion of oxaloacetate to malate (Fig. 2c). Thus, aspartate synthesis would be dependent on glutamate availability as well as a more proper NAD+/NADH ratio, perhaps explaining why npr2Δ cells with mitochondrial dysfunction might be more dependent on aspartate than glutamine.
Several recent studies proposed a major function of mitochondrial respiration in mammalian cells is to support aspartate synthesis41–43. Our findings in yeast are consistent with aspartate as a key biosynthetic output of the mitochondria. However, we propose that the defect in aspartate synthesis in npr2Δ cells is not just due to a redox issue, but stems from a collective set of impairments due to dysregulated TORC1 signaling. TORC1 exerts its effects via a variety of transcriptional, translational, and post-translational mechanisms to rewire cellular metabolism in a manner to promote aspartate synthesis, in addition to glutamine. Furthermore, in mammalian cells mTORC1 was reported to stimulate both purine and pyrimidine synthesis44–46. Here we show that the ability of mTORC1 to promote nucleotide synthesis is likely a consequence of its activation of glutamine and aspartate synthesis. Moreover, the metabolic functions of TORC1 extend beyond just nucleotide synthesis, but to other important metabolites important for proliferative growth, such as NAD+ and glutathione, with their common link being dependency on these nitrogenous amino acids17. It follows that activation of aspartate and glutamine synthesis can impact numerous additional downstream processes, such as protein translation and ribosome biogenesis, which are dependent on increased nucleotide synthesis for rRNA transcription.
Therefore, taken together our findings point to a specific metabolic function for inhibitors of TORC1 signaling such as GATOR1 – that is to regulate key cataplerotic reactions of the mitochondrial TCA cycle in tune with the amino acid and nitrogen status of cells (Fig. 5). It is not a coincidence that aspartate and glutamine are essentially nitrogenous forms of the TCA cycle intermediates oxaloacetate and α-ketoglutarate. While TORC1 activation promotes such cataplerotic reactions and their subsequent use for biosynthesis to support proliferative metabolism, TORC1 inhibition promotes anaplerotic reactions that aim to restore TCA cycle intermediates to ensure that the TCA cycle can be utilized for ATP synthesis under starvation or stress conditions. Consistent with this idea, the majority of transcriptional targets of the retrograde response, which is activated upon inhibition of TORC1 signaling, promote anaplerotic reactions that function to replenish TCA cycle intermediates26, 27. Properly balancing these disparate TCA cycle functions is likely to be intrinsically important throughout the life of a cell, and loss of this capability might be especially detrimental under nutritional or metabolic stress. Lastly, the findings reported here might offer an additional explanation for the metabolic basis of aerobic glycolysis and the Warburg effect that is often observed in rapidly proliferating cell types: if the mitochondrial TCA cycle intermediates α-ketoglutarate and oxaloacetate must be consumed for the synthesis of glutamine and aspartate to support biosynthetic demands, then cells have no choice but to become less reliant on the TCA cycle and respiration for ATP generation.
ONLINE METHODS
Yeast strains, gene deletion and tagging
The prototrophic CEN.PK strain background was used in all experiments unless specified. Strains are listed in Supplementary Table 1. Gene deletions or tagging was performed using a PCR-based strategy, by gene replacement using KanR/NatR/HygroR cassettes via homologous recombination47. Rich medium YPD contained 1% yeast extract, 2% peptone and 2% glucose, amino acid starved SD medium contains 6.7 g/L yeast nitrogen base + ammonium sulfate without amino acids (DIFCO), plus 2% glucose. Unless specified, cells were grown in YPD overnight and then switched to YPD and SD. Cell growth curves were typically initiated at OD600 ~ 0.1.
DNA/RNA purifications and RT-qPCR
RNA was isolated with the MasterPure Yeast RNA kit (Epicentre). Reverse transcription was performed on 1 μg of purified RNA, using SuperScript II reverse transcriptase (Invitrogen). Quantitative PCR was performed using SYBR Green (Bio-Rad), primers, and template cDNA. Transcript abundance was normalized to glucose-6-phosphate dehydrogenase (G6PD). For mitochondrial DNA content measurement, DNA was extracted following Hoffman’s protocol, then digested with RNase48. COX2 gene (COX2 F: GTGATGAAGTTATTTCACCAGC, COX2 R: ATTCAACAGTTTCACCACTATC) was measured as mtDNA content, ACT1 (ACT1 F: CCCAGGTATTGCCGAAAGAATGC, ACT1 R: GGAAGATGGAGCCAAAGCGG) was measured for genomic DNA content.
Cell collection, protein extraction and detection
Equal numbers of cells were collected from respective cultures and stored in −80°C. Cells were lysed in 8% trichloroacetic acid (TCA) solution by bead-beating with glass beads, following the TCA precipitation method. Protein concentrations from extracts were measured using bicinchoninic acid assay (BCA assay, Thermo Scientific). Equal amounts of samples were resolved on 4–12% bis-tris gels. Coomassie blue–stained gels were used as loading controls. The following primary antibodies were used: monoclonal FLAG M2 (F1804, 1:3000, Sigma-Aldrich), HA (12CA5, 1:3000, Roche), GFP (11814460001, 1:3000, Roche), and pS6 (2211, 1:1000, Cell Signaling). The pS6 antibody recognizes phosphorylation of Ser232, Ser233 in yeast Rps6p40. Secondary antibodies are horseradish peroxidase–conjugated secondary antibodies (1706516, 1706515, both 1:10000, anti-mouse and anti-rabbit, Bio-Rad). Standard enhanced chemiluminescence (Thermo Scientific) was used for Western blot development.
Detection of phosphorylated Sch9 by NTCB cleavage
Sch9 phosphorylation was detected using minor adjustments to previously published methods5. Collected samples were quenched in 10% trichloroacetic acid at 4°C, and rapidly harvested by centrifugation. The pellets were stored in −80°C and lysed in 50 mM Tris (pH 7.5), 5 mM EDTA, 6 M urea, 1% SDS, 1 mM phenylmethylsulfonyl fluoride (PMSF), and protein phosphatase inhibitors (50 mM sodium fluoride, 2 mM sodium orthovanadate) with bead-beating. The lysates were heated at 65°C then centrifuged. The protein lysates were equalized according to protein concentration, then cleaved by treatment with 2 mM 2-nitro-5-thiocyanatobenzoic acid (NTCB) in 0.1 M CHES (pH 10.5) at room temperature overnight. Further analysis was done by 8% SDS–polyacrylamide gel electrophoresis and immunoblot with the HA antibody.
Cells for metabolite extractions
Cells were grown in YPD overnight, then diluted in YPD (OD600 = 0.2) and grown to OD600 ~1.0. The cells were either collected for metabolite extractions or transferred to SD and then collected for metabolite extractions at specific time points. Collected cells were cold quenched in a −40°C methanol bath, spun down at 4°C, extracted with HPLC grade 75% ethanol and heated for 3 minutes at 85°C as described previously49. The supernatants with metabolites were collected and dried down.
Metabolite analysis by LC-MS/MS
Extracted metabolites were measured using targeted LC-MS/MS methods described previously17. The relative amounts were either compared directly among genotypes or normalized against that of WT at the first time point.
15N-Ammonium sulfate labeling and tracer analysis
Cells were grown in YPD to OD600 ~ 1.0 and switched to SD at OD600 = 0.2 and grown to OD600 ~ 0.5. Then cells were collected for metabolite extraction or switched to SD where all the ammonium sulfate was 15N-labeled ammonium sulfate [(15NH4)2SO4] (Cambridge Isotope Laboratories Inc). 15N-labeled metabolites were extracted at indicated times and detected by LC-MS/MS.
13C-Glucose labeling and tracer analysis
Cells were grown as above and switched to SD where all the glucose was 13C6-glucose (Sigma-Aldrich). 13C-labeled metabolites were extracted at indicated times and detected by LC-MS/MS, with the targeted parent and daughter ions specific to the 13C forms of the metabolites (Supplementary Table 2). Percent labeled was calculated by dividing the specific labeled metabolite (e.g., M+2) against the total metabolite (unlabeled plus all detected labeled isotopomers).
Glucose and NAD+/NADH measurements
Medium was collected from different time points and spun to collect the supernatant. The glucose concentration of the medium was measured with Glucose(GO) Assay Kit (Sigma-Aldrich) at A570, the exact values were calculated according to standard curve. NAD+/NADH ratios were measured as described previously50.
Dissolved oxygen level measurement
Dissolved oxygen levels during batch growth was measured with a Multifors Fermentor (Infors HT). 10 ODs of cells were inoculated into 500 mL of SD in dual fermentor vessels running side by side with the same medium. IRIS v5 software was used to record the pH and dissolved oxygen concentration.
Live cell imaging
Cells were grown in YPD to log phase then transferred to SD and subjected to the indicated treatments before imaging using a DeltaVision RT imaging system with a 100× oil-immersion objective. Images were deconvoluted then processed by ImageJ.
Hierarchical clustering analysis and heat maps
The normalized abundances of metabolites were log2-transformed, centered about the mean, and clustered by Spearman rank correlation with the software Cluster 3.0. The data were visualized as heat maps with the software Java TreeView (created by Alok).
Statistical analysis
All data represent mean values ± s.d. collected from at least two independent biological experiments performed in technical triplicates. P values were calculated by two-tailed Student’s t test by GraphPad Prism 7 and are indicated in the figure legends with *p < 0.05, **p < 0.01, ***p < 0.001.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (R01GM094314), CPRIT (RP140655), and the Welch Foundation (I-1797). We thank Yu-San Yang for assistance with illustrations.
Footnotes
Author contributions
J.C., L.S., B.M.S., and B.P.T. conceived and designed the study. J.C., B.M.S., and L.S. conducted the experiments. L.S. first observed the ability of aspartate to rescue growth of GATOR1/SEACIT mutants. J.C. and B.P.T. wrote the manuscript.
Competing financial interests
The authors declare no competing interests.
Data availability
No datasets or accession numbers.
References
- 1.Loewith R, Hall MN. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011;189:1177–1201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science (New York, NY ) 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 3.Diaz-Troya S, Perez-Perez ME, Florencio FJ, Crespo JL. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy. 2008;4:851–865. doi: 10.4161/auto.6555. [DOI] [PubMed] [Google Scholar]
- 4.Courchesne WE, Magasanik B. Regulation of nitrogen assimilation in Saccharomyces cerevisiae: roles of the URE2 and GLN3 genes. Journal of bacteriology. 1988;170:708–713. doi: 10.1128/jb.170.2.708-713.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urban J, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Molecular cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 6.Crespo JL, Powers T, Fowler B, Hall MN. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6784–6789. doi: 10.1073/pnas.102687599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nature cell biology. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu X, Tu BP. Selective regulation of autophagy by the Iml1-Npr2-Npr3 complex in the absence of nitrogen starvation. Molecular biology of the cell. 2011;22:4124–4133. doi: 10.1091/mbc.E11-06-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dokudovskaya S, et al. A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Molecular & cellular proteomics: MCP. 2011;10:M110.006478. doi: 10.1074/mcp.M110.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bar-Peled L, et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science (New York, NY ) 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panchaud N, Peli-Gulli MP, De Virgilio C. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Science signaling. 2013;6:ra42. doi: 10.1126/scisignal.2004112. [DOI] [PubMed] [Google Scholar]
- 13.Rousselet G, Simon M, Ripoche P, Buhler JM. A second nitrogen permease regulator in Saccharomyces cerevisiae. FEBS letters. 1995;359:215–219. doi: 10.1016/0014-5793(95)00038-b. [DOI] [PubMed] [Google Scholar]
- 14.Spielewoy N, et al. Npr2, yeast homolog of the human tumor suppressor NPRL2, is a target of Grr1 required for adaptation to growth on diverse nitrogen sources. Eukaryotic cell. 2010;9:592–601. doi: 10.1128/EC.00192-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neklesa TK, Davis RW. A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 complex. PLoS genetics. 2009;5:e1000515. doi: 10.1371/journal.pgen.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sutter BM, Wu X, Laxman S, Tu BP. Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell. 2013;154:403–415. doi: 10.1016/j.cell.2013.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laxman S, Sutter BM, Shi L, Tu BP. Npr2 inhibits TORC1 to prevent inappropriate utilization of glutamine for biosynthesis of nitrogen-containing metabolites. Science signaling. 2014;7:ra120. doi: 10.1126/scisignal.2005948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kayikci O, Nielsen J. Glucose repression in Saccharomyces cerevisiae. FEMS yeast research. 2015;15 doi: 10.1093/femsyr/fov068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. The Journal of clinical investigation. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakashima N, Noguchi E, Nishimoto T. Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics. 1999;152:853–867. doi: 10.1093/genetics/152.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt A, Beck T, Koller A, Kunz J, Hall MN. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. The EMBO journal. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buescher JM, et al. A roadmap for interpreting (13)C metabolite labeling patterns from cells. Current opinion in biotechnology. 2015;34:189–201. doi: 10.1016/j.copbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valenzuela L, Ballario P, Aranda C, Filetici P, Gonzalez A. Regulation of expression of GLT1, the gene encoding glutamate synthase in Saccharomyces cerevisiae. Journal of bacteriology. 1998;180:3533–3540. doi: 10.1128/jb.180.14.3533-3540.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuang Z, et al. High-temporal-resolution view of transcription and chromatin states across distinct metabolic states in budding yeast. Nature structural & molecular biology. 2014;21:854–863. doi: 10.1038/nsmb.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bender T, Pena G, Martinou JC. Regulation of mitochondrial pyruvate uptake by alternative pyruvate carrier complexes. The EMBO journal. 2015;34:911–924. doi: 10.15252/embj.201490197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Molecular cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 27.Komeili A, Wedaman KP, O’Shea EK, Powers T. Mechanism of metabolic control. Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. The Journal of cell biology. 2000;151:863–878. doi: 10.1083/jcb.151.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Small WC, et al. Enzymatic and metabolic studies on retrograde regulation mutants of yeast. Biochemistry. 1995;34:5569–5576. doi: 10.1021/bi00016a031. [DOI] [PubMed] [Google Scholar]
- 29.Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 30.Smolina VS, Bekker ML. Properties of 5-phosphoryl-1-pyrophosphate amidotransferase from the yeast Saccharomyces cerevisiae wild type and mutant with altered purine biosynthesis regulation. Biokhimiia (Moscow, Russia) 1982;47:162–167. [PubMed] [Google Scholar]
- 31.MacGurn JA, Hsu PC, Smolka MB, Emr SD. TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell. 2011;147:1104–1117. doi: 10.1016/j.cell.2011.09.054. [DOI] [PubMed] [Google Scholar]
- 32.Godard P, et al. Effect of 21 different nitrogen sources on global gene expression in the yeast Saccharomyces cerevisiae. Molecular and cellular biology. 2007;27:3065–3086. doi: 10.1128/MCB.01084-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rai R, et al. Nuclear Gln3 Import Is Regulated by Nitrogen Catabolite Repression Whereas Export Is Specifically Regulated by Glutamine. Genetics. 2015;201:989–1016. doi: 10.1534/genetics.115.177725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 35.Duran RV, et al. Glutaminolysis activates Rag-mTORC1 signaling. Molecular cell. 2012;47:349–358. doi: 10.1016/j.molcel.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 36.Nicklin P, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jewell JL, et al. Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science (New York, NY ) 2015;347:194–198. doi: 10.1126/science.1259472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stracka D, Jozefczuk S, Rudroff F, Sauer U, Hall MN. Nitrogen source activates TOR (target of rapamycin) complex 1 via glutamine and independently of Gtr/Rag proteins. The Journal of biological chemistry. 2014;289:25010–25020. doi: 10.1074/jbc.M114.574335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yerlikaya S, et al. TORC1 and TORC2 work together to regulate ribosomal protein S6 phosphorylation in Saccharomyces cerevisiae. Molecular biology of the cell. 2016;27:397–409. doi: 10.1091/mbc.E15-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzalez A, et al. TORC1 promotes phosphorylation of ribosomal protein S6 via the AGC kinase Ypk3 in Saccharomyces cerevisiae. PloS one. 2015;10:e0120250. doi: 10.1371/journal.pone.0120250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sullivan LB, et al. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015;162:552–563. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Birsoy K, et al. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cardaci S, et al. Pyruvate carboxylation enables growth of SDH-deficient cells by supporting aspartate biosynthesis. Nature cell biology. 2015;17:1317–1326. doi: 10.1038/ncb3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ben-Sahra I, Hoxhaj G, Ricoult SJ, Asara JM, Manning BD. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science (New York, NY ) 2016;351:728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science (New York, NY ) 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robitaille AM, et al. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science (New York, NY ) 2013;339:1320–1323. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- 47.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast (Chichester, England) 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 48.Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 49.Castrillo JI, Hayes A, Mohammed S, Gaskell SJ, Oliver SG. An optimized protocol for metabolome analysis in yeast using direct infusion electrospray mass spectrometry. Phytochemistry. 2003;62:929–937. doi: 10.1016/s0031-9422(02)00713-6. [DOI] [PubMed] [Google Scholar]
- 50.Kern SE, Price-Whelan A, Newman DK. Extraction and measurement of NAD(P)(+) and NAD(P)H. Methods in molecular biology (Clifton, NJ ) 2014;1149:311–323. doi: 10.1007/978-1-4939-0473-0_26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.