Abstract
The mammalian gut microbiota has been linked to host developmental, immunologic, and metabolic outcomes. This collection of trillions of microbes inhabits the gut and produces a myriad of metabolites, which are measurable in host circulation and contribute to the pathogenesis of human diseases. The link between endogenous metabolite availability and chromatin regulation is a well-established and active area of investigation, however, whether microbial metabolites can elicit similar effects is less understood. In this review, we focus on seminal and recent research that establishes chromatin regulatory roles for both endogenous and microbial metabolites. We also highlight key physiologic and disease settings where microbial metabolite-host chromatin interactions have been established and/or may be pertinent.
Introduction
The static information contained in the eukaryotic genome is made remarkably more dynamic and complex via its association with other nuclear proteins and nucleic acids. In eukaryotes, genomic DNA is organized and compacted into what is known as chromatin, which consists of nucleic acids, histone proteins, and other chromatin-associated proteins. Adding to this complexity, these proteins and nucleic acids undergo chemical modification, and small and non-coding RNAs can also exert regulatory effects on the genome (reviewed in (1–3)). This collection of factors, that exists independently of the DNA sequence itself, comprises the epigenome and exerts regulatory control over processes such as transcription and DNA replication and repair.
These epigenetic factors allow eukaryotes to sense and respond to environmental cues. Intermediary metabolites, such as acetyl coenzyme A (acetyl-CoA) and alpha-ketoglutarate (α-KG; also known as 2-oxoglutarate, 2-OG) are key messengers to the epigenome of stimuli and stressors. In addition to intermediates of macronutrient metabolism, micronutrients and minerals (e.g., B-vitamins and iron) also regulate the activity of chromatin-modulating enzymes. While the interplay between chromatin and endogenous metabolites has been a subject of intense investigation for nearly two decades, recent findings have revealed the mammalian gut microbiota as a major producer of metabolites, many of which have been demonstrated to play regulatory roles on host physiology (reviewed in (4)). Analogous to endogenous metabolites, gut microbial metabolites may signal to host chromatin to alter host genetic responses to environmental signals.
The mammalian gut microbiota consists of trillions of bacteria that inhabit the mammalian gut. Fungi and viruses are also detectable within the host gut microbiome, however data regarding regulation of host physiology by these organisms are scarce, and there is no evidence, to our knowledge, of commensal fungi (5,6) and viruses (7) directly impacting host epigenetic programming. Thus, this review will primarily focus on gut bacteria. The gut microbiota has been demonstrated to play both protective and contributory roles in the setting of human disease. Several associations have been made between the gut microbiota and host metabolic disease, including obesity and adipose tissue inflammation (8–13), metabolic syndrome and type 2 diabetes mellitus (T2DM) (14–23), cardiovascular disease (24–27), and non-alcoholic fatty liver disease (NAFLD) (28–30). The gut microbiota has also been linked to autoimmune, inflammatory, and allergic disease, including type 1 diabetes mellitus (T1DM) (31,32), inflammatory bowel disease (33–36), allergy and asthma (37–39). In the setting of colon cancer, the gut microbiota has been associated with both therapeutic effects (40,41) and promotion of disease (42). Thus, the gut microbiota exerts effects on a variety of host organ systems and either contributes to or provides protection against metabolic, immunologic, inflammatory, and oncologic disease.
Host diet and environment also affect the functional capacity and composition of the gut microbiota. Both over-nutrition (9,10,12) and malnutrition (43–46) affect the gut microbiome. In humans, an altered diet can affect gut microbial community composition and function within as little as one day after the altered diet reaches the distal gut (47). Common dietary additives such as non-caloric artificial sweeteners and emulsifiers have also been linked with gut dysbiosis (alteration in the gut microbiota associated with pathogenesis) and glucose intolerance and inflammatory bowel disease, respectively (16,34). Additionally, there are known differences in microbiota composition, gene-richness, and metabolite production associated with agrarian or plant-based diets vs. “Westernized” diets (47–51). There are also lasting consequences of obesity and a “Western” lifestyle on the microbiome, both in the setting of post-diet weight gain (52) and multigenerational consumption of a diet low in microbial accessible carbohydrates (MACs), which result in a progressive loss of diversity that cannot be replenished by reintroduction of dietary MACs (53). In addition to dietary factors, altered gut anatomy in the setting of gastric bypass surgery influences the gut microbiota and its function (54,55). Finally, the early life environment, including in-utero, has been shown to impact the gut microbiota and metabolic outcomes (32,56–58).
Thus, the interactions between mammalian hosts and their gut microbiota are myriad and complex, affected by environmental factors such as diet and lifestyle, and are dynamic throughout the lifespan. While many associations have been made between host phenotypes and microbial community composition and metabolite production, the molecular mechanisms underlying these phenotypes remain largely unexplored. Understanding how the microbiota communicate environmental cues to the host epigenome provides an additional avenue for mechanistic exploration. Here, we review seminal and recent literature focused on regulation of chromatin states by endogenous and gut microbial metabolite availability. Additionally, we highlight clinically relevant settings in which these interactions occur.
Chromatin Dynamics
Genomic DNA in eukaryotes is packaged into chromatin, a highly structured nucleo-protein complex that compresses DNA by a factor of approximately 1,000 to 10,000-fold in interphase and mitotic chromosomes, respectively (59). The fundamental unit of chromatin is the nucleosome core particle, which is comprised of an octamer of core histone proteins (2 copies each of histone H2A, H2B, H3, and H4) wrapped by 146bp of genomic DNA (60). Nucleosome core particles are separated from each other by linker DNA that varies in length from ~10–80bp. Roughly 75–90% of genomic DNA interacts with nucleosomes, which are spaced, on average, every 200 bp. Nucleosome positioning is dictated, in part, by the underlying DNA sequence. The numerous salt bridges, hydrogen bonds, and ionic interactions that occur between histone residues and the DNA phosphate backbone induce significant bending of the DNA as it wraps around the histone core 1.65 times, and sequence-dependent DNA flexibility pays a key role in this process (60). About 50% of nucleosome positioning in yeast can be explained by the intrinsic DNA sequence, wherein nucleosome occupancy can be predicted based on nucleosome affinity for a particular sequence of genomic DNA (61). Packaging of DNA into nucleosomes generally inhibits binding of other non-histone proteins with DNA-binding motifs. In support of this, accessibility of DNA bound to a nucleosome core particle has been reported to be 73% that of bare DNA (60). However, nucleosomes can also recruit specific chromatin “readers”, including bromodomain and chromo- and tudor-domain containing proteins, which recognize and bind to acetylated and methylated histone residues, respectively (62,63).
Histones are small, highly basic proteins with flexible N-terminal tails that are subject to a variety of covalent post-translational modifications (PTMs). Although histone methylation, acetylation, and phosphorylation are the most commonly studied modifications, a growing list of other chemical modifications has been identified on histones, including various acylations (propionylation (64), butyrylation (64), 2-hydroxyisobutyrylation (65), malonylation (66), succinylation (66), and crotonylation (67,68)), sumoylation (69), O-linked N-acetylglucosamine (70–72), ubiquitination (70,73), formylation (74), glutathionylation (75), and ADP-ribosylation (76) (reviewed in (77)). These modifications and their functional significance, if known, have been extensively catalogued (78). Briefly, histone PTMs affect chromatin structure (i.e. open vs. closed) and/or recruit other factors that recognize specific histone PTM states, ultimately impacting processes involving interaction with genomic DNA. It is noteworthy that linker DNA can also be bound by the linker histone H1, and that selective incorporation of histone H1 is generally associated with closed chromatin. Finally, there exists extensive crosstalk between histone PTMs, which are thought to collectively comprise a “histone code,” a complex and combinatorial cipher that integrates environmental cues to regulate transcription, replication, and repair processes (3).
The activity of most histone-modifying enzymes is dependent upon sufficient levels of intermediary metabolites, vitamins, and minerals. These small molecules act as necessary co-substrates, activators, or inhibitors, thereby coupling cellular metabolic state to chromatin regulation. Subsequent sections will focus on regulation of chromatin by endogenous and gut microbial metabolites. Given the focus on gut microbiota-mediated regulation of host chromatin here, we cannot exhaustively cover chromatin regulation by endogenous metabolites, but direct readers to high yield reviews on those subjects.
Regulation of Chromatin Modification by Endogenous Metabolite Availability
Histone Acetylation and Central Carbon Metabolism
Histone acetylation was first identified in 1963 (79), and was demonstrated to de-repress histone-mediated inhibition of RNA synthesis in 1964, at which point it was hypothesized not only to regulate transcription, but also to be reversible (80). More than 50 years later, histone acetylation remains an active area of investigation, particularly in relation to cellular metabolism and in the setting of cancer pathogenesis and treatment. Histone acetylation has been reported on lysine, arginine, threonine, and serine residues, and occurs on canonical histone proteins H2A, H2B, H3, and H4 as well as the variant histones H2A.Z and H3.3 and the linker histone H1 (reviewed in (77,78)).
Acetylation of lysine residues neutralizes the positive charge on the lysine epsilon amino group, which decreases the ionic interaction between histones and the negatively charged phosphate backbone of DNA. This results in a relative opening of chromatin, and histone acetylation is broadly associated with activate transcription. Acetylated lysine residues are “read” or recognized by bromodomain-containing proteins, which in addition to a bromodomain, often contain a variety of other functional domains that allow for HAT activity, DNA binding, histone methyltransferase activity, helicase activity, ATPase activity and more (reviewed in (81)).
Histone acetylation most commonly occurs on lysine residues and is catalyzed by histone acetyltransferases (HATs; also known as KATs, lysine acetyltransferases). HATs comprise a superfamily of enzymes that is divided into 5 sub-families, based on sequence and structural similarities. These families include Hat1, Gcn5/PCAF, MYST, p300/CBP, and Rtt109, all of which possess a conserved acetyl-CoA binding region, but their N- and C-terminal regions play roles in substrate binding (reviewed in (82)). All 5 sub-families transfer an acetyl moiety from the thioester of acetyl-CoA to a lysine epsilon amino group on histones, generating a molecule of CoA, but each has a unique catalytic mechanism (reviewed in (83) and (82)). HATs also display rather promiscuous substrate specificity, and the molecular mechanisms underlying this process are not well understood.
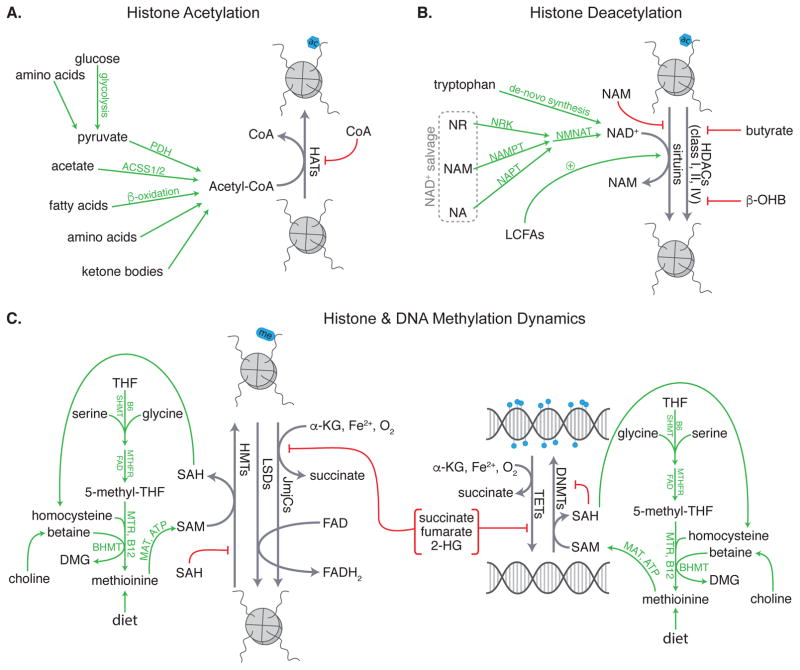
In addition to its role in histone acetylation, acetyl-CoA lies at the center of both catabolic and anabolic metabolism. Acetyl-CoA is produced by a number of processes including (1) oxidation of glucose to pyruvate and oxidative decarboxylation of pyruvate to acetyl-CoA via glycolysis and the pyruvate dehydrogenase complex, (2) activation of acetate to acetyl-CoA via mitochondrial and cytosolic acetyl-CoA synthetase 1 and 2, respectively (ACSS1 and 2), (3) β-oxidation of free fatty acids, and (4) breakdown of amino acids and ketone bodies (Figure 1A). In a fed state, acetyl-CoA production is mostly associated with breakdown of glucose, whereas in a fasted state, the majority of acetyl-CoA comes from fatty acid β-oxidation. Depending on cellular state, acetyl-CoA is either fed into the TCA cycle to support ATP production or is shuttled into biosynthetic pathways, including lipid, cholesterol, and amino acid production.
Figure 1.
Endogenous metabolites regulate (A) histone acetylation, (B) histone deacetylation, and (C) histone and DNA methylation and demethylation. PDH = pyruvate dehydrogenase, ACSS1/2 = acetyl-CoA synthetase 1/2, HAT = histone acetyltransferase, NR = nicotinamide riboside, NA = nicotinic acid, NAM = nicotinamide, NMNAT = nicotinamide/nicotinic acid mononucleotide adenylyltransferase, LCFAs = long-chain fatty acids, NAD+ = nicotinamide adenine dinucleotide, HDACs = histone deacetylases, β-OHB = β-hydroxybutyrate, SHMT = serine hydroxymethyltransferase, MTHFR = methylenetetrahydrofolate reductase, 5-methyl-THF = 5-methyl- tetrahydrofolate, THF = tetrahydrofolate, MTR = methionine synthase, DMG = dimethylglycine, MAT = S-adenosylmethionine synthase, SAH = S-adenosyl homocysteine, SAM = S-adenosylmethionine, HMTs = histone methyltransferases, LSDs = lysine specific demethylases, JmjCs = Jumonji C family histone demethylases, α-KG = α-ketoglutarate, TETs = Ten Eleven Translocation methylcytosine dioxygenases, DNMTs = DNA methyltransferases
Given that acetyl-CoA is the acetyl donor for all HATs, availability of this metabolic intermediate also affects HAT activity and functions as a cellular rheostat linking metabolic and chromatin states. Indeed, oscillations in acetyl-CoA availability during the yeast metabolic cycle coincide with enrichment of histone acetylation at key growth genes (84). Mammalian cells have a similar dependence on acetyl-CoA availability for histone acetylation. In contrast to single-celled eukaryotes, which rely upon acetate conversion to acetyl-CoA via ACS2 for histone acetylation (85), metazoans rely on glucose as a primary carbon source. Thus, in mammalian cells, nucleocytosolic acetyl-CoA is produced, in part, from glucose-derived citrate that is shuttled out of mitochondria and converted to acetyl-CoA via ATP-citrate lyase (ACL). Wellen et al. demonstrated that ACL is necessary for glucose-dependent histone acetylation in mammalian cells and that supplementation with acetate can partially rescue histone H3 acetylation in siACL-treated mammalian cells (86). Thus, histone acetylation in both single-celled and metazoan eukaryotes is mediated by acetyl-CoA availability, and although ACL plays a primary role in histone acetylation in mammalian cells, AceCS1-mediated (also known as ACSS2) production of acetyl-CoA may contribute as well. Indeed, both ACL and AceCS1 have been demonstrated to be important for histone acetylation in the setting of mammalian tumorigenesis (87,88).
Interestingly, the cytosolic isoform of acetyl-CoA synthetase in yeast and mammalian cells (ACS2 and AceCS1, respectively) is localized to both the cytosol and nucleus (86,89). ACL is also localized to both the cytosol and nucleus (86). Since both acetate and citrate are small enough to diffuse through nuclear pores (90), this suggests that acetyl-CoA can be produced in both the cytosol and nucleus, possibly supporting localized production for histone acetylation. A number of metabolic enzymes have been reported to “moonlight” in the nucleus, including pyruvate kinase (PK) and the pyruvate dehydrogenase complex (PDC), which can translocate to the nucleus and facilitate local acetyl-CoA production for histone acetylation (91). Further, PK and PDC form a complex with the HAT p300 to promote histone acetylation and activation of transcription (92). This phenomenon of “moonlighting” metabolic enzymes in the nucleus is not limited to those involved in histone acetylation. A number of other mitochondrial enzymes whose products regulate histone-modifying enzymes have also been reported to translocate to the nucleus under various conditions (reviewed in (93)). Therefore, local production of key regulatory metabolites may represent a key mode of communication of metabolic status to the nucleus.
Histone Deacetylation and Intermediary Metabolites
Histone acetylation is a balance between the activity of HATs and histone deacetylases (HDACs). Mammalian HDACs are grouped into four classes, based on their homology to yeast deacetylases, enzymatic activities, and subcellular localization. Class I, IIa-b, and IV HDACs are Zn2+-dependent, and are inhibited by small molecules that chelate the active-site Zn2+, including trichostatin A (TSA) and the pharmacologic HDAC inhibitor (HDACi) Vorinostat (suberoylanilide hydroxamic acid, SAHA). While Class I HDACs are localized to the nucleus, Class IIa are nucleocytosolic, Class IIb are primarily cytosolic, and Class IV are cytoplasmic (reviewed in (94)). The Class III HDACs, also known as sirtuins, are a family of enzymes that carry out NAD+-dependent lysine deacetylation, generating nicotinamide and 2’-O-acetyl-ADP-ribose (reviewed in (95) and (96)). Although sirtuins have a conserved NAD+-binding site and catalytic domain, they diverge based on their subcellular location, tissue-specific expression patterns, and substrate specificity (95). There are 7 mammalian sirtuins, of which Sirt1 and Sirt6 are nuclear, Sirt2 is nucleocytosolic, and Sirt7 is localized to the nucleolus.
Regulation of histone deacetylation by metabolic intermediates is shown in Figure 1B. Sirtuins require NAD+ as a co-substrate for histone deacetylation. NAD+ can be synthesized de-novo from tryptophan via the kynurenine pathway, or salvaged from niacin (vitamin B3), which is comprised of nicotinamide (NAM) and nicotinic acid (NA), or nicotinamide riboside (NR). NAD+ production, salvage, and biology is thoroughly reviewed in (97). NAD+ functions as a redox cofactor in multiple oxidative pathways, including glycolysis, the TCA cycle, oxidative phosphorylation, and β-oxidation of fatty acids. Therefore, sirtuins can sense cellular metabolic status via NAD+ availability.
Sirtuin histone deacetylase activity was first linked to nutrient availability in 2000. Imai et al. demonstrated that yeast and mouse Sirt2 (H4 K16 deacetylase), which was already associated with silenced chromatin, required NAD+ for histone deacetylase activity (98). Sirt1 (H3 K9 and K14 deacetylase) activity is also affected under conditions where NAD+ salvage is impaired, including aging and diet- and age-induced T2DM. Gomes et al. have identified a PGC1α/β-independent pathway that links the age-related decline in NAD+ availability to decreased Sirt1 activity, which ultimately stabilizes Hif1α and decreases expression of mitochondrial encoded OXPHOS genes (99). Caloric excess associated with high fat feeding can overwhelm mammalian metabolism. Indeed, mice fed a high fat diet display impaired NAD+ biosynthesis and develop T2DM. This diet-induced T2DM is rescued by supplementation with an NAD+ precursor via a mechanism that is partially mediated by Sirt1 (100).
Given that Sirt6 (H3 K9 and K56 deacetylase) has a higher affinity for NAD+ and can bind it in the absence of a an acetylated substrate, it is less likely that Sirt6 is regulated by availability of this metabolite (101). Interestingly, Sirt6 has very low in-vitro histone deacetylase activity but is activated up to 35-fold by select long-chain fatty acids (LCFAs), including myristic, oleic, and linoleic acid (95). Sirt6 plays a role in genome stability and glucose homeostasis, and loss of Sirt6 function plays a role in the metabolic switch that occurs in human tumors (102,103).
Unlike sirtuins, the Zn2+-dependent HDACs do not require NAD+ as a co-substrate. However, these HDACs are still regulated by small molecule metabolites. The discovery that butyrate inhibits HDACs in Friend erythroleukemic cells was one of the first examples of metabolite-mediated regulation of epigenetic machinery (104,105). The structural analog and ketone body, β-hydroxybutyrate (β-OHB, also known as 3-hydroxybutyrate), also inhibits these HDACs. During fasting, the liver switches to oxidation of fatty acids rather than glucose. When the acetyl-CoA produced by fatty acid oxidation exceeds the oxidative capacity of the liver, it is channeled into synthesis of ketone bodies, two of which are organic acids: acetoacetate and β-OHB. Interestingly, the ratio of acetoacetate to β-OHB produced is dependent upon the NAD+/NADH ratio, which is low during fatty acid oxidation, thus favoring production of β-OHB. β-OHB inhibits Class I and IIa HDACs with a median IC50 in the range of 2–5 mM (106). Human serum levels are typically ~100 μM, but levels can increase up to 6 mM following a 2–3 day fast or strenuous exercise, up to 25 mM in uncontrolled diabetes mellitus, and up to 2 mM with a ketogenic diet that is almost devoid of carbohydrates (107–109). Thus, physiological fluctuations in serum ketone bodies are sufficient to exert effects on HDAC activity. Though not a primary focus here, the functional effects of ketone body inhibition of HDACs are reviewed in (110).
Histone Methylation and One-Carbon Metabolism
Lysine and/or arginine methylation occurs on all core histone proteins, variant histones H3.3, H2A.Z, and macroH2A; and the linker histone H1 (histone methylation is reviewed in (62)). Lysine residues can be monomethylated (me1), dimethylated (me2), or trimethylated (me3), whereas arginine residues can be either monomethylated or dimethylated, and arginine dimethylation can occur either asymmetrically or symmetrically. Both the site and degree of methylation determine function. For example, methylation of H3K4, H3K36, and H3K79 is associated with active transcription, whereas methylation of H3K9, H3K27, and H4K20 are associated with repressed transcription (78). In contrast to histone acetylation, methylation of histones does not alter the positive charge of lysine and arginine residues, and thus does not have a significant impact on chromatin structure. Rather, histone methylation serves as a docking site for other factors that contain methyl-binding domains, including PhD fingers, WD40 repeats, CW domains, PWWP domains, Ankyrin repeats, and members of the Royal superfamily (includes chromodomains and Tudor domains).
As with acetylation, histone methylation is dynamic and informed by intermediate metabolites (Figure 1C). Histone methylation is carried out by histone methyltransferases (HMTs) and is removed by demethylases. All HMTs utilize the methyl donor, S-adenosylmethionine (SAM), an intermediary metabolite in the one-carbon (1C) cycle. SAM availability is maintained by the folate and 1C cycles, which not only support histone and DNA methylation, but also provide intermediates for nucleic acid synthesis, amino acid homeostasis, and redox homeostasis. Thus, SAM availability connects several metabolic processes to histone methylation.
One-carbon metabolism is reviewed in (111). Although plants and select bacteria and yeast can synthesize folate and methionine, humans cannot and must obtain both via diet. Notably, while a number of dietary nutrients can serve as sources of 1C units, choline, serine, and glycine are the most important sources (111). The folate and 1C cycles converge at re-methylation of homocysteine to form methionine via a vitamin B12-dependent reaction catalyzed by methionine synthase (MTR). Homocysteine can also be re-methylated to methionine via betaine-homocysteine methyltransferase (BHMT), which uses betaine as a methyl donor. The formation of SAM from methionine is then catalyzed by methionine adenosyltransferase (MAT) in an ATP-dependent reaction that is conserved across all branches of life (112,113). Thus, vitamin and nutrient availability affects SAM levels, which may regulate the activity of HMTs.
Indeed, restriction of either methionine or folate in the growth medium of methionine-dependent Δmet or folate-dependent Δfol yeast strains, respectively, results in decreased H3K4me2 and H4K4me3 (114). Similar evidence is present in higher eukaryotes as well. In mouse embryonic stem cells (mESCs), threonine catabolism provides a significant fraction of cellular glycine, which feeds into production of SAM. Restriction of threonine in the growth medium of mESCs and knockdown of Tdh (threonine dehydrogenase) results in decreased SAM availability and H3K4me3, which slowed growth and increased differentiation (115). Methionine restriction in Hct116 cells results in decreased H3K4me3 in select regions of the genome, which corresponded with decreased expression of enzymes involved in 1C metabolism, suggesting direct feedback on 1C metabolism to maintain homeostasis (116). Further, methionine restriction in mice results in decreased plasma methionine and liver SAM levels and a concomitant decrease in liver H3K4me3, demonstrating that dietary restriction of 1C sources affects histone methylation in-vivo (116).
Histone Demethylation and TCA cycle intermediates
There are two families of histone demethylases, both of which are regulated by intermediates of the TCA cycle: (1) the FAD-dependent LSD (lysine specific demethylase) family of enzymes, and (2) the alpha-ketoglutarate- (α-KG), iron-, and oxygen-dependent JmjC (Jumonji C) family of enzymes (Figure 1C). These enzymes are reviewed in detail in (117). FAD is produced in the cytoplasm and mitochondria from vitamin B2 (riboflavin), and is involved in the TCA cycle, oxidative phosphorylation, and fatty acid β-oxidation. Loss of the H3K4 and H3K9 demethylase LSD1 in cultured adipocytes, either by small molecule inhibition or by knockdown, leads to increased expression of genes involved in energy expenditure via increased H3K4me3 at promoters (118). α-KG is a TCA cycle intermediate that is produced in mitochondria by IDH2/3 (isocitrate dehydrogenase 2 and 3) and in the cytosol by IDH1. While the role of α-KG levels in regulation of JmjC family histone demethylases has not yet been shown, both oxygen and iron availability mediate the activity of these enzymes. Hypoxia has been reported to increase histone methylation and modulate downstream gene expression in a number of settings and cell types via inhibition of JmjC demethylases (119–121), and pharmacologic iron chelation in mouse myoblast cells results in increased histone methylation at JmjC target sites (122). Notably, although the direct role of α-KG as a regulator of JmjC family members remains to be elucidated, its structural analog 2-hydroxyglutarate (2-HG), which is produced by cancer-associated IDH1 and IDH2 mutants, is a competitive inhibitor of these enzymes, resulting in aberrant histone methylation that contributes to the cancer phenotype (123,124). The role of 2-HG as an oncometabolite is reviewed in (125). A subset of cancers also display mutations in FDH (fumarate dehydrogenase) and SDH (succinate dehydrogenase), which result in accumulation of fumarate and succinate, respectively, both of which inhibit JmjC demethylases (126,127).
While not a focus of this review, it is also worth noting that DNA methyltransferases (DNMTs) and the Ten Eleven Translocation (TET) methylcytosine dioxygenases, which oxidize 5-methylcytosines to promote DNA demethylation, have similar metabolite dependencies as histone methyltransferases and demethylases (Figure 1C). Dietary restriction of 1C sources results in altered DNA methylation (DNAm) patterns in mice and humans (128,129), and the cancer-associated FDH and SDH mutations induce DNA hypermethylation via inhibition of α-KG-dependent TET enzymes (126,130,131).
Regulation of chromatin by gut microbial metabolites
Given the many roles endogenous metabolites play in chromatin regulation, the question arises whether gut microbial metabolites can exert similar effects. The interaction between microbial metabolites and histone acetylation, histone methylation, and DNA methylation is depicted in Figures 2–4, respectively. Mammalian hosts (and all other coelomates) have coevolved with a collection of trillions of microorganisms that inhabit the gut, known as the gut microbiota (132–134). The gut microbiome, which collectively contains roughly 9.9 million bacterial genes, profoundly expands the genetic capacity and diversity of the human host by a factor of 500-fold (135,136). This collection of symbiotic microorganisms, which is shaped by complex trophic and competitive interactions, provides the host with considerable metabolic flexibility. Complex dietary substrates, including polysaccharides and resistant starches (known as microbial accessible carbohydrates or MACs), are not metabolized by host enzymes in the upper gastrointestinal tract and pass to the distal gut where they are subject to microbial fermentative reactions that produce a wide variety of metabolites, including short chain fatty acids (SCFAs). Acetate (C2), priopionate (C3), and butyrate (C4) comprise ≥95% of SCFAs produced by the gut microbiota, but lactate (C3), valerate (C5), capropate (C6), and branched SCFAs (bSCFAs) such as isobutyrate (C4) and isovalerate (C5) are also produced in lesser quantities (137). The gut bacteria also degrade proteins and amino acids, producing branched-chain fatty acids, amines, volatile sulfur compounds, and phenolic compounds (137). The gut microbiota and its metabolites have been associated with a number of host developmental, immunologic, and metabolic outcomes, including proper nervous and immune system development (138–140), obesity (8,9,13,141), diabetes mellitus (14,15,32) (142), cardiovascular disease (24), non-alcoholic liver disease (28), inflammatory bowel disease (33,34), and colorectal cancer (40–42).
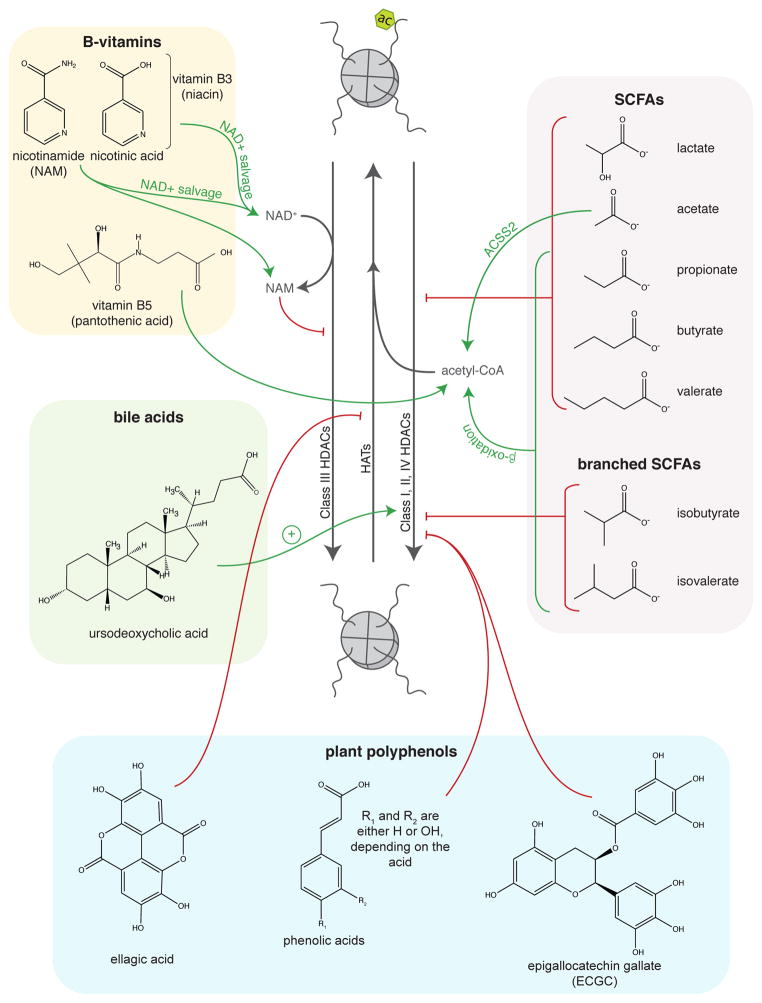
Figure 2.
Regulation of histone acetylation by gut microbial metabolites.
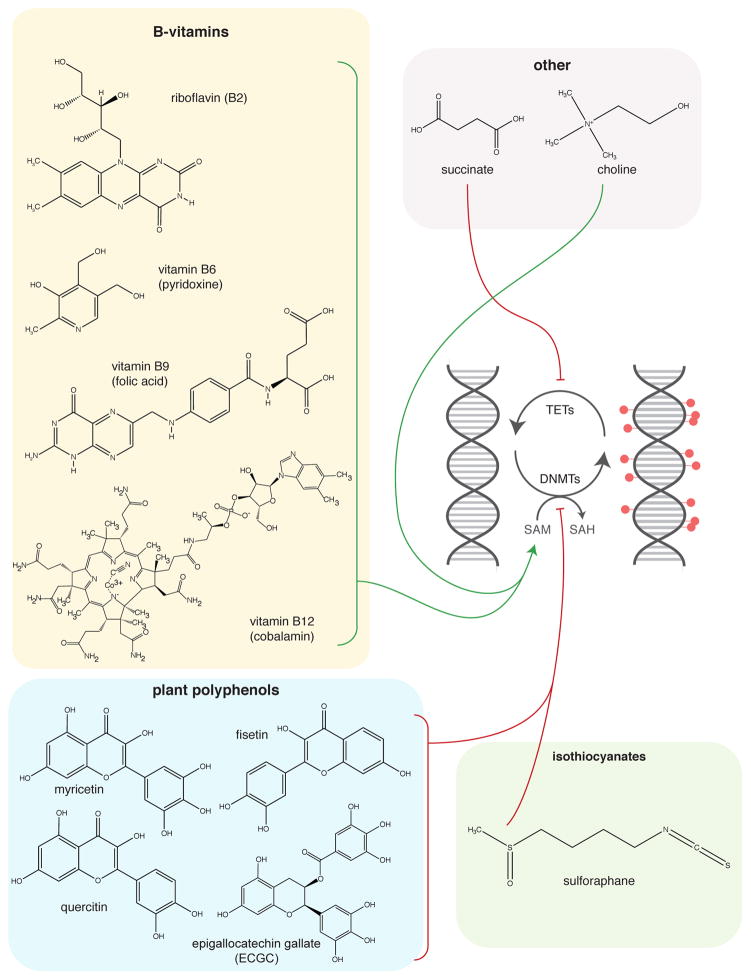
Figure 4.
Regulation of DNA methylation by gut microbial metabolites. PUFAs = polyunsaturated fatty acids
Here we discuss evidence for gut microbial regulation of the host epigenetic machinery via de-novo synthesis, co-metabolism of dietary and host intermediates, direct competition with the host for dietary nutrients, and conversion of dietary substrates to bioactive compounds.
Organic Acids
While direct modification of chromatin states by gut microbial metabolites in vivo has not yet been revealed for the vast majority of microbially produced small molecules, there is direct evidence that SCFAs impact host tissue histone modification (Figure 2) (40,143). In a mouse model of colorectal cancer (CRC), colonization with a minimal core microbiome plus the butyrate producer B. fibrisolvens and high fiber feeding resulted in increased butyrate production by the microbiota and accumulation of butyrate in Warburg-like cancer cells, leading to HDAC inhibition and increased histone H3 acetylation (40,41). These mice also had a lower tumor burden and altered expression of genes involved in apoptosis and proliferation in tumors relative to controls (40). Butyrate and histone acetylation levels were also higher in human CRC tumor samples vs. nearby normal tissue, suggesting that this diet-microbiota-host chromatin interactions occurs in human tissue as well (40). We recently demonstrated that global host histone methylation and acetylation are driven by the gut microbiota in a diet-dependent manner in multiple tissues, including those outside the alimentary tract (143). Further, the microbiota-driven chromatin effects were partially mimicked by supplementation of germ-free mice with a mixture of the three most abundance SCFAs (acetate, propionate, and butyrate) (143).
Treatment of HT-29 human CRC cells with propionate, butyrate, and valerate induces histone H4 acetylation, causes cell cycle arrest, and induces expression of markers of cellular differentiation (144). Notably, although acetate and caproate were tested in this study as well, there were no appreciable effects of these organic acids (144). However, acetate can induce histone acetylation in Hct116 cells (86). Waldecker et al. show that C3–C5 SCFAs and C4–C5 bSCFAs inhibit HDACs in HT-29 cells, but that acetate has no HDACi activity, even at concentrations as high as 20 mM (145). Lactate (C3), another fermentation product, also displays HDACi activity (146). Finally, the gut microbiota produce succinate, which is a known inhibitor of both the JmjC family of histone demethylases and the TET family enzymes that facilitate DNA demethylation (126,130).
B-vitamins
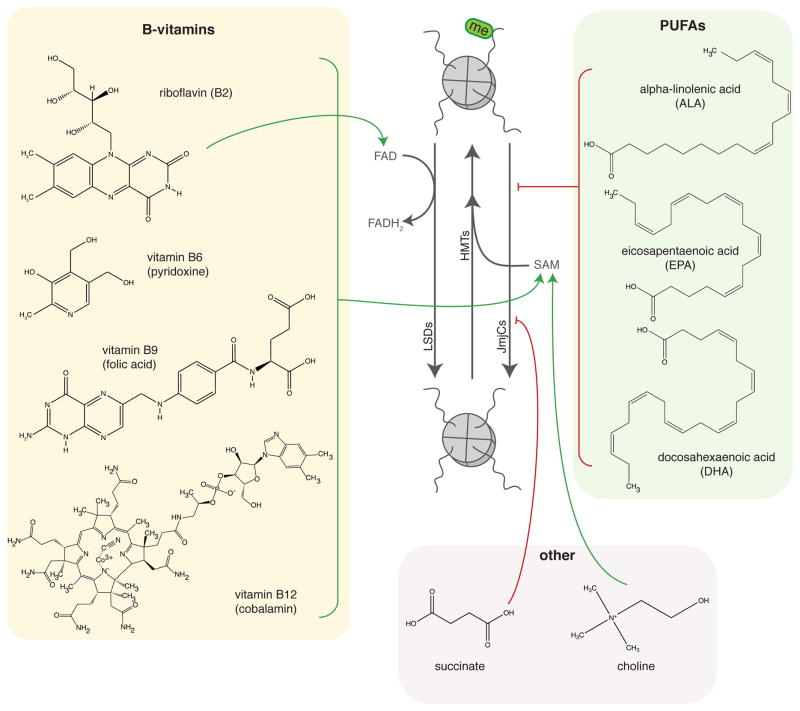
The gut bacteria can synthesize de novo a number of B-vitamins, including riboflavin (B2), niacin (B3), pantothenic acid (B5), pyridoxine (B6), folate (B9), and cobalamin (B12) (reviewed in (147)). B-vitamins play a role in histone acetylation (Figure 2), histone methylation (Figure 3), and DNA methylation (Figure 4). Vitamins B6, B9, and B12 are all important components and cofactors of the folate and 1C cycles, and promote availability of the universal methyl donor SAM for HMTs and DNMTs. More specifically, vitamins B6 and B12 are both necessary cofactors in 1C metabolism (Figure 1C). Animals cannot synthesize folate, and thus depend on exogenous sources such as dietary intake. Another potential source, however, is the colonic microbiota, which can synthesize equal or more folate than typical adult dietary intake (148,149). Further, microbially produced folate is readily absorbed across the gut in adult human subjects (149). Vitamin B2 is a precursor for FAD, a necessary cofactor for methylenetetrahydrofolate reductase (MTHFR) in the folate cycle.
Figure 3.
Regulation of histone methylation by gut microbial metabolites.
Vitamins B3 and B5 affect histone acetylation. Vitamin B3 (niacin) is comprised of nicotinic acid (NA) and nicotinamide (NAM), both of which contribute to the NAD+ pool via the NAD+ salvage pathway via distinct enzymes (150). Increasing the NAD+ pool may impact the activity of the NAD+-dependent sirtuin family of histone deacetylases. It is also worth noting that NAM is a product inhibitor of sirtuins (151). VItamin B5 is a precursor for coenzyme A (CoA), and is phosphorylated by pantothenate kinase during the first step of CoA synthesis. CoA is necessary for the production of the HAT substrate acetyl-CoA. Although it is unclear how bacterial production of vitamin B5 affects availability in host circulation, RNAi-mediated silencing of pantothenate kinase in Drosophila S2 cells reduces CoA levels and acetylation of histone H3, histone H4, and tubulin (152). This decrease in protein acetylation is associated with impaired DNA damage repair and survival that is partially rescued by supplementation with pantethine or HDACi treatment (152). Thus, gut microbial production of B-vitamins may promote histone and DNA methylation by supporting SAM availability and may impact histone acetylation via regulation of sirtuins and HATs, though direct regulation by the gut microbiota of these processes remains to be demonstrated.
Conversion of Dietary Substrates to Chromatin-Modulatory Compounds
In addition to degradation of complex plant polysaccharides, resistant starches, and proteins, the gut microbiota also chemically transforms plant polyphenol and isothiocyanate metabolites into bioactive compounds. Plant polyphenols are found in various fruits, vegetables, nuts, and teas and undergo extensive transformations by the gut microbiota prior to entering enterohepatic circulation and undergoing further metabolism by the host (153,154). Although these compounds have limited bioavailability and are found at low levels in plasma, in-vitro and cell culture experiments reveal that these compounds affect HATs, HDACs, DNMTs, and even non-coding RNAs. It is also worth noting that a number of polyphenol metabolites are aromatic organic acids, which may account for their weak HDACi activity. Epigallocatechin (ECGC), phenolic acids, stilbenes, and coumarins exert weak HDACi activity (145,155,156). Additionally, the stilbene resveratrol has been demonstrated to target the class III HDAC Sirt1, the HAT p300, and non-coding RNA expression (156–158). Unlike other polyphenol metabolites, ellagic acid does not affect HDAC activity, but rather has been shown to decrease HAT activity in TNF-stimulated THP-1 cells (159). The polyphenol metabolites ECGC and flavonoids (catechin and quercetin) and the isothiocyanate sulforaphane have been demonstrated to inhibit DNMTs (160–162).
Host-microbe Cometabolites
In addition to the myriad of metabolites produced by the gut microbiota, there is also considerable cometabolism of circulating host and microbial metabolites. For example, bile acids are synthesized from cholesterol in the liver, excreted into the gut following a meal, and about 95% are reabsorbed across the gut to circulate back to the liver in a process called enterohepatic circulation. These primary bile acids are metabolized by the gut microbiota in the ileum and colon to generate secondary bile acids, which function as potent signaling molecules and regulate a number of metabolic processes, including synthesis of primary bile acids (reviewed in (163)). Of note, the gut microbiota influences both the bile acid pool size and composition (163). Ursodeoxycholic acid, which is a primary bile acid in mice but a secondary bile acid in humans, induces expression of HDAC6 and subsequent hypoacetylation of histones H2A, H3, and H4 in cultured cells (164). However, it is unclear how ursodeoxycholic acid affects the expression of HDAC6, and more investigation is required to better understand how bile acids may affect histone modification.
Microbial metabolites are also metabolized by the host. Trimethylamine (TMA) is a microbial product of dietary choline metabolism (165). There is also evidence that TMA-producing gut microbes compete with the host for dietary choline availability (26). TMA enters host circulation and is metabolized to trimethylamine-N-oxide (TMAO) in the liver by flavin-dependent monooxygenase 3 (FMO3) (166). TMAO has recently been associated with both non-alcoholic fatty liver disease (167) and cardiovascular disease (24). Therefore, gut microbial competition with the host for dietary choline may impact SAM availability and ultimately histone and DNA methylation. Whether this competition contributes to a chromatin-mediated mechanism underlying these diseases remains unclear.
The gut microbiota also participates in metabolism of dietary PUFAs (poly-unsaturated fatty acids). Gut bacteria isolated from humans are able to metabolize PUFAs such as linoleic acid and alpha-linolenic acid into conjugated linoleic acids and conjugated linolenic acids (168). The conjugated PUFAs are detectable in cecal contents of mice supplemented with dietary PUFAs, but not in plasma, suggesting that the effects of these compounds is likely confined to the intestine (168). Given that the gut microbiota can metabolize PUFAs, it is also possible that microbes compete with the host for these compounds. Linoleic acid is a known activator of the nuclear HDAC Sirt6 in-vitro (95), therefore, it is possible that bacterial co-metabolism of this PUFA may affect its availability to Sirt6 in mammalian hosts.
Interplay Between Environment, Microbiota, and Chromatin in Disease
The mammalian gut microbiota has emerged as a key mediator of host metabolism and health. A major route of chemical communication between this vast group of microorganisms and the host is via microbial metabolism of dietary nutrients and production of metabolites. Microbial metabolites have been demonstrated to play roles as signaling molecules and metabolic substrates (15,18,19,52,163,169), a topic which has been thoroughly reviewed (4,163,170). There is evidence that exposure to antibiotics (171), cold ambient temperatures (172), natural seasonal variation in food intake and ambient temperature during hibernation (173–175), natural seasonal variation in food sources (176), geographical and cultural differences in dietary habits or dietary scarcity (48–50,177,178), hormonal cues (31,58), alteration in dietary macronutrient composition (10,47,53,179), food additives (16,34), and more can affect gut microbial communities.
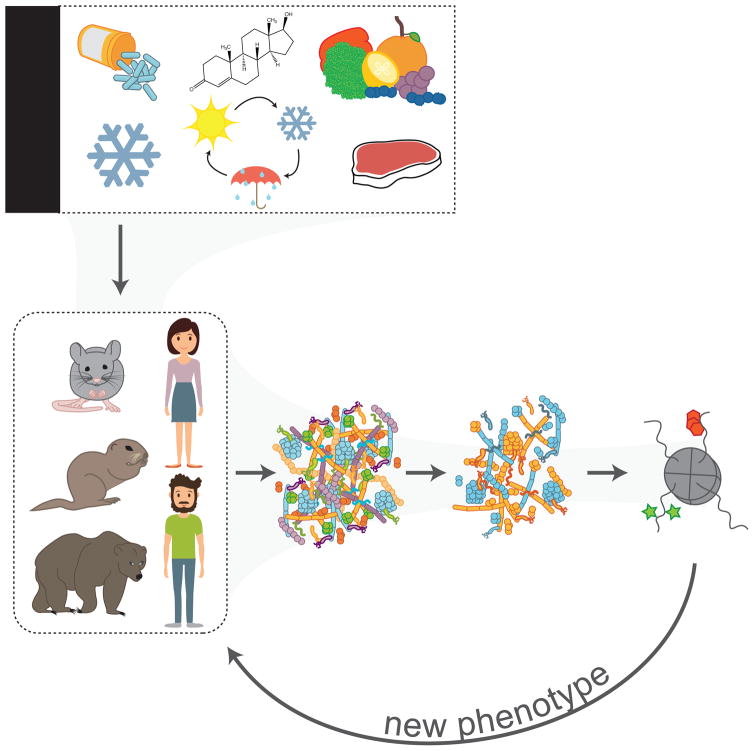
This prompts a new framework for investigation of gut microbiota-host interactions, wherein communication between the host and its microbiota occurs not only through ligand:receptor interactions, but also through modification of chromatin via regulation of histone- and DNA-modifying enzymes by small molecule metabolites. Within this model (Figure 5), mammalian hosts encounter any number of environmental exposures throughout life. These exposures are sensed by the gut microbiota, which are in direct contact with the environment given their residence in the alimentary tract. Microbial community composition and activity are altered in response to changes in environment, resulting in altered microbial metabolite profiles. Metabolites are then absorbed across the host gut and serve as effector molecules at the level of host chromatin, where they elicit changes in gene expression programs, ultimately leading to a new developmental and/or metabolic phenotype. Below, we highlight known relationships between gut microbial metabolites and host chromatin in the setting of host health and disease.
Figure 5.
A model wherein the mammalian gut microbiota mediates communication between environmental exposures and host epigenetic programming of downstream phenotypes. Environmental exposures known to affect mammalian microbiota include: antibiotics, cold exposure, hormones, natural seasonal cycles in ambient temperature and food sources, and dietary composition, excess, and scarcity. The composition of the mammalian gut microbiota, which resides in the host alimentary tract and is in direct contact with the environment, is altered in response to environmental factors. Alteration of microbial community composition leads to differences in microbial metabolite production, and ultimately altered chemical signaling to host chromatin. Modifications to host chromatin then drive new transcriptional programs to produce an adapted phenotype. The proposed cycle may repeat in response to continued exposure to the same environmental factor or in response to a new exposure.
Innate and Adaptive Immunity
Gut microbial metabolites and microbial components signal to the host immune system, regulating both proper immune system development and function. This includes production of SCFAs, aryl hydrocarbon receptor ligands, polyamine metabolites, and activation of host pattern recognition receptors via microbial components such as lipopolysaccharide, flagellin, peptidoglycan, and formyl peptides. This topic is vast and readers are directed to a recent comprehensive review (140) for specific information about microbiota-host immunity interactions. Here, we specifically highlight instances in the literature that associate the gut microbiota with epigenetic control of host immunity.
Blood-brain barrier (BBB) permeability is increased in GF mice relative to specific pathogen free (SPF) control mice (180). GF mice have decreased expression of the tight junction proteins occludin and claudin-5, but colonization of GF mice with SPF fecal microbiota increased expression of these tight junction proteins and decreased BBB permeability (180). Since SCFAs are known to enhance barrier integrity in the intestinal epithelium via proper assembly of tight junctions, Braniste et al. investigated whether the SCFA butyrate could induce similar effects at the BBB, and found increased histone acetylation in brain lysates from GF mice either treated with sodium butyrate (an HDACi) or mono-colonized with the butyrate producer C.tyrobuticum (180). Priming of NK cells in the innate immune system has also been shown to be partially affected by differential histone H3K4me3 enrichment at transcriptional start sites of microbiota-mediated inflammatory response genes, and treatment of conventionally raised (ConvR) mice with antibiotics erased H3K4me3 at these genes (181). Differences in chromatin accessibility at enhancers in GF mice relative to their ConvR counterparts have also been identified in purified intraepithelial lymphocytes (IEL), revealing transcription factor circuits that are differentially regulated in GF and ConvR mice (182). Enhancers in IEL populations also stratify by their history of microbial exposure, where IEL enhancers from GF mice clustered separately from those in ConvR mice, which were colonized at birth, and from IELs in conventionalized mice (ConvD), which were colonized at 3 weeks of age (182). Collectively, these studies suggest that the gut microbiota signals to innate and adaptive host defense systems via mechanisms that are at least partly tuned by chromatin responses.
SCFAs in Cancer and Diet-Induced Obesity
Gut microbial butyrate is protective in the setting of colon cancer via its action as an HDACi (40). Conversely, butyrate has also been associated with microbiota-dependent promotion of cancer in a genetic mouse model of CRC (APCMin/+, MSH2−/−) (42). This genetic mouse model of CRC has a loss of the tumor suppressor gene APC (Adenomatous Polyposis Coli) and the DNA mismatch repair gene MSH2 (MutS Homolog 2), suggesting that the protective effects of microbial butyrate in the setting of CRC are partly mediated through regulation of these genes or their downstream effects.
We have also recently demonstrated that the gut microbiota alters global histone methylation and acetylation in a diet-dependent manner (143). Mass spectrometry analysis of >50 unique histone PTM states revealed that the gut microbiota drives changes in global histone acetylation and methylation in multiple tissues, including colon, liver, and white adipose tissue (143). Feeding a western-type diet, which is lower in fermentable substrates, attenuated microbiota-driven changes in global histone acetylation and methylation (143). Further, supplementation of GF mice with SCFAs was sufficient to partially recapitulate the colonization-associated chromatin signature. Changes in chromatin state were also associated with altered expression of genes involved in processes such as glucose homeostasis, lipid metabolism, and immunity (143). These results support a role for bacterial SCFAs and other metabolites as key mediators of host phenotype via signaling to host chromatin.
Early Life Environments
There are also interesting parallels between the gut microbiota and host chromatin during early life. Environmental exposures that occur early in life, particularly during key developmental windows for the host and its gut microbiota, can have lasting impacts on host chromatin and microbial communities. For example, both maternal and paternal nutrition and early life adversity have been shown to alter DNA methylation in humans and mice (128,129,183–185). Early host life is also a critical developmental time for the mammalian gut microbiota, which matures over the course of the first several years of life in humans (57,184,186–188).
Maternal prenatal stress, BMI, and weight gain during pregnancy affect offspring microbial community composition and activity (189,190). Recent evidence also suggests that the infant gut microbiome may be established in a stepwise manner that begins in utero, challenging the concept of a sterile in utero environment (191). There is also evidence that the microbial community composition in human meconium is indicative of maternal diabetes status (192), and that high fat feeding during pregnancy alters the offspring metabolome and microbiome in primates (193,194). Further, maternal high fat feeding in primates induces acetylation of histone H3K14 (195). Although there is no evidence to-date that definitively links the maternal gut microbiota to epigenetic programming in offspring, these studies collectively suggest a possible role for gut microbial mediation of host epigenetic states in utero. Notably, Sonnenburg et al. demonstrate that changes in microbial community composition in response to a low-MAC diet over several generations results in progressive loss of diversity that is not recoverable simply by switching to a higher-MAC diet (53). This study provides evidence for transgenerational inheritance of the gut microbiota that is influenced by diet. Although the early life environment was not explicitly examined in this study, it was an implicit part of the study design. While it remains unclear how early life factors and microbiota-host chromatin communication may be involved in this setting, it suggests the possibility that diet-microbiota interactions may program host chromatin responses across generations.
Antibiotics represent an extremely prevalent early-life exposure that can affect both host and microbial community development and metabolic outcomes. Extrapolations from 2010 data comprised of information on >70% of prescriptions in the United States (US) suggest that an average child in the US has received roughly 3, 10, and 17 courses of antibiotics by the age of 2, 10, and 20 years of life (187). For decades, administration of sub-therapeutic doses of antibiotics has been used to promote growth and fattening in livestock (196,197). Importantly, if antimicrobials are given later in the animal’s life the effects on growth are less than if the exposure occurred in early life, and a study of germ-free chickens showed loss of the growth-promoting effects of antibiotics in the absence of a gut microbiota (187,198,199). Similar effects have been reported in murine models, where exposure to subtherapeutic doses of antibiotics either from birth or early in life altered gut microbial community composition and increased host adiposity and susceptibility to metabolic disease (56,200,201). Further, these effects were synergistic when coupled with high fat feeding, and the metabolic impacts on the host persisted even after microbial communities have recovered, suggesting metabolic reprogramming occurs in response to early life exposure to antibiotics (56,200). While there is currently no evidence of epigenetic reprogramming in the setting of early life exposure to antibiotics, it is possible that gut microbiota-mediated reprogramming of the host epigenome may mediate some of the persistent metabolic effects of this transient exposure.
Total Parenteral Nutrition-Induced Gut Dysfunction and Extreme Stress
Total parenteral nutrition (TPN) is a therapeutic intervention that provides nutrients intravenously, allowing for complete bowel rest in patients that cannot be fed enterally (202). Although TPN is critical for the survival of these patients, it is not without sequelae. Enteral deprivation adversely affects the gastrointestinal tract in a number of ways, including the disruption of the epithelial and mucosal immune system; the induction of an inflammatory response to the decrease of lymphocytes (202,203) and an increase in mucosal proinflammatory cytokines (e.g. TNF-α) (204); reduced epithelial barrier function, which in many cases results in sepsis (205); hepatitis due to oxidative stress (206) and hepatobiliary complications (ex: steatosis, cholestasis, and fibrosis) (207); and dysbiosis resulting from changes in the gut microbiota community structure and a loss of microbial diversity. This alteration of the microbiome may play a central role in the deleterious outcomes resulting from TPN. Previous studies have shown that the lack of enteral intake favors phyla such as Proteobacteria and Bacteroidetes, and contributes to a loss of Firmicutes (208). Firmicutes rely heavily on dietary carbohydrates which are absent during prolonged fasting or TPN, whereas Proteobacteria can metabolize a broader range of substrates, including the amino acid leucine, which permeates to the lumen during TPN feeding and causes dysbiosis (209). Akkermansia muciniphila, a species in the phylum Verrucomicrobia, also increases markedly in TPN-fed mice (210). Like Proteobateria, A. muciniphila demonstrates resistance to fasting by relying on host mucin as a carbon source. The consequences of these alterations in intestinal microbiome communities have been linked to hepatic disease via activation of toll-like receptor 4 (TLR4)-induced inflammation (211). Furthermore, the loss of epithelial barrier function increases the susceptibility of the host to the transport of microbial products (212). To our knowledge, however, no studies to-date have examined the link between TPN-induced changes in gut microbial communities and their significance to the epigenetic architecture of the host.
TPN provides an interesting and clinically relevant setting for investigation of how bypassing the GI tract for nutrient assimilation may affect microbiota-host chromatin interactions. However, the animal adaptation known as hibernation represents a natural, recurring state of chronic enteral nutrient deprivation that occurs over the lifetime of an animal. Hibernation shares a number of similarities with TPN, the foremost being the exclusion of the gastrointestinal tract from nutrient acquisition, yet the lack of enteral nutrient sources is a normal part of a hibernator’s annual feeding cycle, and these animals are resistant to maladies associated with prolonged fasting. Hibernation is a circannual suppression of metabolism that facilitates survival during periods in which food resources in the environment are reduced, and maintenance of constant, high body temperature and metabolism are energetically expensive due to low ambient temperatures. In the case of small hibernating mammals such as thirteen-lined ground squirrels (Ictidomys tridecimlineatus), basal metabolic rates fall to less than 5% of summer active rates as body temperatures approach near-freezing temperatures in a state called torpor (213). Bouts of torpor (which last 1–3 weeks) are interspersed with periodic interbout arousals (IBAs) during which metabolic rate rises and body temperatures briefly return to 37°C for less than 24 hours (213), however complete fasting is maintained throughout the hibernation season.
The absence of enteral nutrients causes shifts in microbial community composition, some of which are shared between animals on TPN and hibernators. Firmicutes, which specialize in the processing of plant polysaccharides are reduced in both models, whereas Protobacteria and Verrucomicrobia that can process host-derived mucin in conditions of limiting substrates tend to increase (173,214,215). The natural shifts in microbial communities observed in hibernators may also be driven by the marked changes in body temperature, but the effects of body temperature independent of changes in feeding behavior and metabolic depression on microbial community dynamics remain poorly defined. During torpor when body temperatures are lower than 10ºC, microbes have limited metabolic and proliferative activity while IBAs provide brief periods of rewarming and microbial proliferation, particularly for those species capable of degrading and utilizing host-derived compounds (216). These large shifts in temperature during torpor-arousal cycles do not appear to cause extinction of summer-active populations. Microbial communities associated with enteral feeding remain present in lower abundance, and are available for reseeding as dietary intake shifts the circannual cycle. In hibernating thirteen-lined ground squirrels, the concentrations of SCFAs are 25% of that found in the summer active period (213). Additionally, the relative molar amounts of acetate, propionate, and butyrate change, likely due to differences in substrate availability over the winter hibernation season and relative microbial taxa abundance during different metabolic states (137).
The key distinguishing feature between TPN and hibernation is that the association between hibernating animals and the gut microbiome represents a unique coevolution that has adapted to the extreme dietary and thermal shifts that occur during their annual cycle. For example, the increase in gut permeability that occurs during hibernation is not associated with an increase in the inflammatory response (217,218), reductions in tight junction proteins like occludin, or increased enterocyte apoptosis (see (215) for review). Investigating the effects of parenteral nutrition in conventional laboratory animal models using approaches such as germ-free mice have many advantages and are an essential investigative tool, but their limitations include the exposure to stressors to which the animal has not evolved that distance these experimental systems from the animal’s natural physiology. Hibernators, which represent the most extreme example of fasting in mammals, provide a unique opportunity to investigate the implications of naturally occurring changes in gut microbial populations and metabolite production on chromatin states. Additionally, hibernating mammals exhibit a robust resistance to other extreme stressors that have profound deleterious effects to most mammals, including hemorrhagic shock and ischemia-reperfusion injury (219,220), radiation (221,222), and thermal stress (213) that may be linked to chromatin and the microbiome.
Conclusions
The mammalian gut is a bioreactor that contains a vast population of microbes. These complex communities supply the host with ~10% of the calories harvested from the diet and produce a multitude of metabolites whose roles in signaling and epigenetic regulation are multifaceted and only beginning to be understood. There is a paucity of data that directly link microbial metabolites to host chromatin regulation relative to the rapidly growing body of literature regarding gut microbial mediation of human health and disease. This is partly due to the highly complex nature of gut microbiota-host interactions. Host genetics affect gut microbial community composition and functional capacity (223,224), however environmental factors appear to exert stronger effects (18). The massive genetic and environmental diversity within the human population poses a significant challenge for the study of gut microbial metabolite-host chromatin interactions in human cohorts, and study of these interactions in gnotobiotic mouse models is relatively resource intensive. Nonetheless, the fact that environmental factors have been shown to outweigh host genetics is a harbinger for the role of gut microbial communication of environmental changes to host chromatin, particularly given that chromatin allows for dynamic responses to external stimuli by an otherwise static genome. Given the ever-increasing prevalence of metabolic disease worldwide, understanding how microbial metabolites signal to host chromatin in this setting offers new opportunities to develop preventive and treatment measures for environment- and lifestyle-associated disorders.
Acknowledgments
We apologize for any omission of relevant publications due to space limitations. K.A.K. is supported by NIH F30 DK108494. R.S.D is supported by NIH R03 AG052390-01. J.M.D is supported by NIH GM059785. H.V.C. is supported by NSF IOS1558044. The authors have read and understood the Translational Research policy on declaration of interests and declare that we have no competing interests to disclose. All authors have read the Translational Research authorship agreement and approve of this manuscript.
Abbreviations
- α-KG
alpha-ketoglutarate
- acetyl-CoA
acetyl-coenzyme A
- BBB
blood brain barrier
- bSCFA
branched short chain fatty acid
- CoA
coenzyme A
- ConvD
conventionalized
- ConvR
conventionally raised
- CRC
colorectal cancer
- DNMT
DNA methyltransferase
- GF
germ-free
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HDACi
histone deacetylase inhibitor
- HMT
histone methyltransferase
- IBA
interbout arousal
- LC-MS/MS
liquid chromatography coupled to tandem mass spectrometry
- MACs
microbial accessible carbohydrates
- NAFLD
non-alcoholic fatty liver disease
- PTMs
post-translational modifications
- SCFA
short chain fatty acid
- T2DM
type 2 diabetes mellitus
- TPN
total parenteral nutrition
References
- 1.Peschansky VJ, Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. 2014 Jan 1;9(1):3–12. doi: 10.4161/epi.27473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nature Reviews Genetics. 2013 Mar 1;14(3):204–20. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 3.Jenuwein T, Allis CD. Translating the histone code. Science American Association for the Advancement of Science. 2001 Aug 10;293(5532):1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 4.Sonnenburg JL, Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016 Jul 7;535(7610):56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. 2014 Jun 1;14(6):405–16. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sokol H, Leducq V, Aschard H, Pham H-P, Jegou S, Landman C, et al. Fungal microbiota dysbiosis in IBD. Gut. 2017 Jun;66(6):1039–48. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogilvie LA, Jones BV. The human gut virome: a multifaceted majority. Front Microbiol. 2015 Jan 1;6:918–8. doi: 10.3389/fmicb.2015.00918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004 Nov 2;101(44):15718–23. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006 Dec 21;444(7122):1027–131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 10.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013 Sep 6;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005 Aug 2;102(31):11070–5. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009 Jan 22;457(7228):480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani PD, Bäckhed F. Crosstalk between Gut Microbiota and Dietary Lipids Aggravates WAT Inflammation through TLR Signaling. Cell Metabolism. 2015 Oct 6;22(4):658–68. doi: 10.1016/j.cmet.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012 Oct 4;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 15.Perry RJ, Peng L, Barry NA, Cline GW, Zhang D, Cardone RL, et al. Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome. Nature. 2016 Jun 9;534(7606):213–7. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suez J, Korem T, Zeevi D, Zilberman-Schapira G, Thaiss CA, Maza O, et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. 2014 Oct 9;514(7521):181–6. doi: 10.1038/nature13793. [DOI] [PubMed] [Google Scholar]
- 17.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science American Association for the Advancement of Science. 2010 Apr 9;328(5975):228–31. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ussar S, Griffin NW, Bezy O, Fujisaka S, Vienberg S, Softic S, et al. Interactions between Gut Microbiota, Host Genetics and Diet Modulate the Predisposition to Obesity and Metabolic Syndrome. Cell Metabolism. 2015 Sep 1;22(3):516–30. doi: 10.1016/j.cmet.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014 Jan 16;156(1–2):84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 20.De Vadder F, Kovatcheva-Datchary P, Zitoun C, Duchampt A, Bäckhed F, Mithieux G. Microbiota-Produced Succinate Improves Glucose Homeostasis via Intestinal Gluconeogenesis. Cell Metabolism Elsevier Inc. 2016 Jul 12;24(1):151–7. doi: 10.1016/j.cmet.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 21.Grasset E, Puel A, Charpentier J, Collet X, Christensen JE, Tercé F, et al. A Specific Gut Microbiota Dysbiosis of Type 2 Diabetic Mice Induces GLP-1 Resistance through an Enteric NO-Dependent and Gut-Brain Axis Mechanism. Cell Metabolism Elsevier Inc. 2017 May 2;25(5):1075–5. doi: 10.1016/j.cmet.2017.04.013. [DOI] [PubMed] [Google Scholar]
- 22.Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature Nature Publishing Group. 2013 May 28;498(7452):99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 23.Singh V, Chassaing B, Zhang L, San Yeoh B, Xiao X, Kumar M, et al. Microbiota-Dependent Hepatic Lipogenesis Mediated by Stearoyl CoA Desaturase 1 (SCD1) Promotes Metabolic Syndrome in TLR5-Deficient Mice. Cell Metabolism. 2015 Oct 27;22(6):983–96. doi: 10.1016/j.cmet.2015.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, DuGar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011 Apr 7;472(7341):57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gregory JC, Buffa JA, Org E, Wang Z, Levison BS, Zhu W, et al. Transmission of atherosclerosis susceptibility with gut microbial transplantation. J Biol Chem. 2015 Feb 27;290(9):5647–60. doi: 10.1074/jbc.M114.618249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romano KA, Vivas EI, Amador-Noguez D, Rey FE. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. MBio. 2015;6(2):e02481. doi: 10.1128/mBio.02481-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karlsson FH, kFFA, Nookaew I, Tremaroli V, Fagerberg BOR, Petranovic D, et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun Nature Publishing Group. 1AD;3:1–8. doi: 10.1038/ncomms2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature Nature Publishing Group. 2012 Jan 31;482(7384):179–85. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metabolism Elsevier Inc. 2017 May 2;25(5):1054–5. doi: 10.1016/j.cmet.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bashiardes S, Shapiro H, Rozin S, Shibolet O, Elinav E. Non-alcoholic fatty liver and the gut microbiota. Molecular Metabolism. 2016 Sep 1;5(9):782–94. doi: 10.1016/j.molmet.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013 Aug 22;39(2):400–12. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livanos AE, Greiner TU, Vangay P, Pathmasiri W, Stewart D, McRitchie S, et al. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat Microbiol. 2016 Aug 22;1(11):16140. doi: 10.1038/nmicrobiol.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chu H, Khosravi A, Kusumawardhani IP, Kwon AHK, Vasconcelos AC, Cunha LD, et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science. 2016 May 27;352(6289):1116–20. doi: 10.1126/science.aad9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chassaing B, Koren O, Goodrich JK, Poole AC, Srinivasan S, Ley RE, et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 2015 Mar 5;519(7541):92–6. doi: 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walters WA, Xu Z, Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett Federation of European Biochemical Societies. 2014 Nov 17;588(22):4223–33. doi: 10.1016/j.febslet.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ananthakrishnan AN, Luo C, Yajnik V, Khalili H, Garber JJ, Stevens BW, et al. Gut Microbiome Function Predicts Response to Anti- integrin Biologic Therapy in Inflammatory Bowel Diseases. Cell Host and Microbe Elsevier Inc. 2017 May 10;21(5):603–3. doi: 10.1016/j.chom.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu Di, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009 Oct 29;461(7268):1282–6. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO reports. 2012 May 1;13(5):440–7. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujimura KE, Lynch SV. Microbiota in Allergy and Asthma and the Emerging Relationship with the Gut Microbiome. Cell Host and Microbe Elsevier Inc. 2015 May 13;17(5):592–602. doi: 10.1016/j.chom.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donohoe DR, Holley D, Collins LB, Montgomery SA, Whitmore AC, Hillhouse A, et al. A Gnotobiotic Mouse Model Demonstrates That Dietary Fiber Protects against Colorectal Tumorigenesis in a Microbiota- and Butyrate-Dependent Manner. Cancer Discov. 2014 Dec 1;4(12):1387–97. doi: 10.1158/2159-8290.CD-14-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metabolism. 2011 May 4;13(5):517–26. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, Rubino S, et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell. 2014 Jul 17;158(2):288–99. doi: 10.1016/j.cell.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 43.Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013 Feb 1;339(6119):548–54. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014 Jun 19;510(7505):417–21. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanton LV, Charbonneau MR, Salih T, Barratt MJ, Venkatesh S, Ilkaveya O, et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science. 2016 Feb 18;351(6275):aad3311–1. doi: 10.1126/science.aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reyes A, Blanton LV, Cao S, Zhao G, Manary M, Trehan I, et al. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. PNAS. 2015 Sep 8; doi: 10.1073/pnas.1514285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature [Internet] 2013 Dec 11;505(7484):559–63. doi: 10.1038/nature12820. Available from: http://www.nature.com/doifinder/10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu GD, Compher C, Chen EZ, Smith SA, Shah RD, Bittinger K, et al. Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut. 2016 Jan;65(1):63–72. doi: 10.1136/gutjnl-2014-308209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lozupone CA, Stombaugh J, Gonzalez A, Ackermann G, Wendel D, Vázquez-Baeza Y, et al. Meta-analyses of studies of the human microbiota. Genome Res. 2013 Oct;23(10):1704–14. doi: 10.1101/gr.151803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012 May 9;:1–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013 Aug 29;500(7464):585–8. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 52.Thaiss CA, Itav S, Rothschild D, Meijer M, Levy M, Moresi C, et al. Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature Publishing Group. 2016 Nov 24;540(7634):544–51. doi: 10.1038/nature20796. [DOI] [PubMed] [Google Scholar]
- 53.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature Nature Publishing Group. 2016 Jan 14;529(7585):212–5. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liou AP, Paziuk M, Luevano J-M, Machineni S, Turnbaugh PJ, Kaplan LM. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med. 2013 Mar 27;5(178):178ra41–1. doi: 10.1126/scitranslmed.3005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tremaroli V, Karlsson F, Werling M, Ståhlman M, Kovatcheva-Datchary P, Olbers T, et al. Roux-en-Y Gastric Bypass and Vertical Banded Gastroplasty Induce Long-Term Changes on the Human Gut Microbiome Contributing to Fat Mass Regulation. Cell Metabolism. 2015 Aug 4;22(2):228–38. doi: 10.1016/j.cmet.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the Intestinal Microbiota during a Critical Developmental Window Has Lasting Metabolic Consequences. Cell Elsevier Inc. 2014 Aug 14;158(4):705–21. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016 Jun 15;8(343):343ra82–2. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Bäckhed HK, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012 Aug 3;150(3):470–80. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alberts B, Johnson A, Lewis J, Raff M, Roberts K. Molecular Biology of the Cell. 4. New York: Garland Science; 2002. Chromosomal DNA and its Packaging in the Chromatin Fiber. [Google Scholar]
- 60.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2. A resolution. Nature. 1997 Sep 18;389(6648):251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 61.Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, Moore IK, et al. A genomic code for nucleosome positioning. Nature. 2006 Aug 17;442(7104):772–8. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nature Reviews Genetics. 2012 May;13(5):343–57. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Etchegaray J-P, Mostoslavsky R. Interplay between Metabolism and Epigenetics: A Nuclear Adaptation to Environmental Changes. Molecular Cell. 2016 Jun 2;62(5):695–711. doi: 10.1016/j.molcel.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Molecular & Cellular Proteomics. 2007 May;6(5):812–9. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Unoki M, Masuda A, Dohmae N, Arita K, Yoshimatsu M, Iwai Y, et al. Lysyl 5-hydroxylation, a novel histone modification, by Jumonji domain containing 6 (JMJD6) Journal of Biological Chemistry. 2013 Mar 1;288(9):6053–62. doi: 10.1074/jbc.M112.433284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xie Z, Dai J, Dai L, Tan M, Cheng Z, Wu Y, et al. Lysine succinylation and lysine malonylation in histones. Molecular & Cellular Proteomics. 2012 May 1;11(5):100–7. doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011 Sep 16;146(6):1016–28. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sabari BR, Tang Z, Huang H, Yong-Gonzalez V, Molina H, Kong HE, et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Molecular Cell. 2015 Apr 16;58(2):203–15. doi: 10.1016/j.molcel.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci USA. 2003 Nov 11;100(23):13225–30. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fujiki R, Hashiba W, Sekine H, Yokoyama A, Chikanishi T, Ito S, et al. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011 Nov 27;480(7378):557–60. doi: 10.1038/nature10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang S, Roche K, Nasheuer H-P, Lowndes NF. Modification of histones by sugar β-N-acetylglucosamine (GlcNAc) occurs on multiple residues, including histone H3 serine 10, and is cell cycle-regulated. J Biol Chem. 2011 Oct 28;286(43):37483–95. doi: 10.1074/jbc.M111.284885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sakabe K, Wang Z, Hart GW. β-N-acetylglucosamine (O-GlcNAc) is part of the histone code. 2010:19915–20. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gatti M, Pinato S, Maspero E, Soffientini P, Polo S, Penengo L. A novel ubiquitin mark at the N-terminal tail of histone H2As targeted by RNF168 ubiquitin ligase. cc. 2012 Jul 1;11(13):2538–44. doi: 10.4161/cc.20919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang T, Zhou X, Taghizadeh K, Dong M, Dedon PC. N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proc Natl Acad Sci USA. 2007 Jan 2;104(1):60–5. doi: 10.1073/pnas.0606775103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.García-Giménez JL, Olaso G, Hake SB, Bönisch C, Wiedemann SM, Markovic J, et al. Histone h3 glutathionylation in proliferating mammalian cells destabilizes nucleosomal structure. Antioxid Redox Signal. 2013 Oct 20;19(12):1305–20. doi: 10.1089/ars.2012.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hottiger MO. ADP-ribosylation of histones by ARTD1: an additional module of the histone code? FEBS Lett. 2011 Jun 6;585(11):1595–9. doi: 10.1016/j.febslet.2011.03.031. [DOI] [PubMed] [Google Scholar]
- 77.Arnaudo AM, Garcia BA. Proteomic characterization of novel histone post-translational modifications. Epigenetics Chromatin BioMed Central Ltd. 2013;6(1):24. doi: 10.1186/1756-8935-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao Y, Garcia BA. Comprehensive Catalog of Currently Documented Histone Modifications. Cold Spring Harb Perspect Biol. 2015;7(9) doi: 10.1101/cshperspect.a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Phillips DM. The presence of acetyl groups of histones. Biochem J Portland Press Ltd. 1963 May;87(2):258–63. doi: 10.1042/bj0870258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and Methylation of Histones and Their Possible Role in the Regulation of RNA Synthesis. Proc Natl Acad Sci USA National Academy of Sciences. 1964 May;51(5):786–94. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fujisawa T, Filippakopoulos P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nature Publishing Group. 2017 Apr;18(4):246–62. doi: 10.1038/nrm.2016.143. [DOI] [PubMed] [Google Scholar]
- 82.Marmorstein R, Zhou M-M. Cold Spring Harb Perspect Biol [Internet] 7. Vol. 6. Cold Spring Harbor Lab; 2014. Jul 1, Writers and readers of histone acetylation: structure, mechanism, and inhibition; pp. a018762–2. Available from: http://cshperspectives.cshlp.org/lookup/doi/10.1101/cshperspect.a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fan J, Krautkramer KA, Feldman JL, Denu JM. Metabolic regulation of histone post-translational modifications. ACS Chem Biol. 2015 Jan 16;10(1):95–108. doi: 10.1021/cb500846u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Molecular Cell. 2011 May 20;42(4):426–37. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic Acetyl-Coenzyme A Synthetase Is Required for Histone Acetylation and Global Transcription. Molecular Cell. 2006 Jan 1;23(2):207–17. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 86.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-Citrate Lyase Links Cellular Metabolism to Histone Acetylation. Science American Association for the Advancement of Science. 2009 May 21;324(5930):1076–80. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee JV, Carrer A, Shah S, Snyder NW, Wei S, Venneti S, et al. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metabolism. 2014 Aug 5;20(2):306–19. doi: 10.1016/j.cmet.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Comerford SA, Huang Z, Du X, Wang Y, Cai L, Witkiewicz AK, et al. Acetate dependence of tumors. Cell. 2014 Dec 18;159(7):1591–602. doi: 10.1016/j.cell.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Molecular Cell. 2006 Jul 21;23(2):207–17. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 90.Paine PL, Moore LC, Horowitz SB. Nuclear envelope permeability. Nature. 1975 Mar 13;254(5496):109–14. doi: 10.1038/254109a0. [DOI] [PubMed] [Google Scholar]
- 91.Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, Haromy A, et al. A Nuclear Pyruvate Dehydrogenase Complex Is Important for the Generation of Acetyl-CoA and Histone Acetylation. Cell Elsevier Inc. 2014 Jul 3;158(1):84–97. doi: 10.1016/j.cell.2014.04.046. [DOI] [PubMed] [Google Scholar]
- 92.Matsuda S, Adachi J, Ihara M, Tanuma N, Shima H, Kakizuka A, et al. Nuclear pyruvate kinase M2 complex serves as a transcriptional coactivator of arylhydrocarbon receptor. Nucleic Acids Research. 2016 Jan 29;44(2):636–47. doi: 10.1093/nar/gkv967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Boukouris AE, Zervopoulos SD, Michelakis ED. Metabolic Enzymes Moonlighting in the Nucleus: Metabolic Regulation of Gene Transcription. Trends in Biochemical Sciences. 2016 Aug;41(8):712–30. doi: 10.1016/j.tibs.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 94.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014 Jan 2;124(1):30–9. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem American Society for Biochemistry and Molecular Biology. 2013 Oct 25;288(43):31350–6. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012 Apr;13(4):225–38. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Houtkooper RH, Cantó C, Wanders RJ, Auwerx J. The Secret Life of NAD +: An Old Metabolite Controlling New Metabolic Signaling Pathways. Endocrine Reviews. 2010 Apr;31(2):194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403(6771):795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 99.Gomes AP, Price NL, Ling AJY, Moslehi JJ, Montgomery MK, Rajman L, et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013 Dec 19;155(7):1624–38. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yoshino J, Mills KF, Yoon MJ, Imai S-I. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metabolism. 2011 Oct 5;14(4):528–36. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pan PW, Feldman JL, Devries MK, Dong A, Edwards AM, Denu JM. Structure and biochemical functions of SIRT6. J Biol Chem American Society for Biochemistry and Molecular Biology. 2011 Apr 22;286(16):14575–87. doi: 10.1074/jbc.M111.218990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kugel S, Feldman JL, Klein MA, Silberman DM, Sebastian C, Mermel C, et al. Identification of and Molecular Basis for SIRT6 Loss- of-Function Point Mutations in Cancer. CellReports The Authors. 2015 Oct 20;13(3):479–88. doi: 10.1016/j.celrep.2015.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kugel S, Sebastian C, Fitamant J, Ross KN, Saha SK, Jain E, et al. SIRT6 Suppresses Pancreatic Cancer through Control of Lin28b. Cell. 2016 Jun 2;165(6):1401–15. doi: 10.1016/j.cell.2016.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Riggs MG, Whittaker RG, Neumann JR, Ingram VM. n-Butyrate causes histone modification in HeLa and Friend erythroleukaemia cells. Nature. 1977 Aug 4;268(5619):462–4. doi: 10.1038/268462a0. [DOI] [PubMed] [Google Scholar]
- 105.Candido EP, Reeves R, Davie JR. Sodium butyrate inhibits histone deacetylation in cultured cells. Cell. 1978 May;14(1):105–13. doi: 10.1016/0092-8674(78)90305-7. [DOI] [PubMed] [Google Scholar]
- 106.Shimazu T, Hirschey MD, Newman J, He W, Shirakawa K, Le Moan N, et al. Suppression of oxidative stress by β-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science American Association for the Advancement of Science. 2013 Jan 11;339(6116):211–4. doi: 10.1126/science.1227166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim DY, Rho JM. Current Opinion in Clinical Nutrition & Metabolic Care. 2008. The ketogenic diet and epilepsy. [DOI] [PubMed] [Google Scholar]
- 108.Robinson AM, Williamson DH. Physiological roles of ketone bodies as substrates and signals in mammalian tissues. Physiological Reviews. 1980 Jan;60(1):143–87. doi: 10.1152/physrev.1980.60.1.143. [DOI] [PubMed] [Google Scholar]
- 109.Cahill GF, Jr, Herrera MG, Morgan AP, Soeldner JS, Steinke J, Levy PL, et al. Hormone-fuel interrelationships during fasting. J Clin Invest. 1966 Nov 1;45(11):1751–69. doi: 10.1172/JCI105481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Newman JC, Verdin E. Ketone bodies as signaling metabolites. Trends in Endocrinology & Metabolism. 2014 Jan 1;25(1):42–52. doi: 10.1016/j.tem.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ducker GS, Rabinowitz JD. One-Carbon Metabolism in Health and Disease. Cell Metabolism. 2016 Sep 15; doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Thomas D, Surdin-Kerjan Y. The synthesis of the two S-adenosyl-methionine synthetases is differently regulated in Saccharomyces cerevisiae. Molecular and General Genetics MGG. 1991;226–226(1–2):224–32. doi: 10.1007/BF00273607. [DOI] [PubMed] [Google Scholar]
- 113.Sakata SF, Shelly LL, Ruppert S, Schutz G, Chou JY. Cloning and expression of murine S-adenosylmethionine synthetase. Journal of Biological Chemistry. 1993 Jul 5;268(19):13978–86. [PubMed] [Google Scholar]
- 114.Sadhu MJ, Guan Q, Li F, Sales-Lee J, Iavarone AT, Hammond MC, et al. Nutritional control of epigenetic processes in yeast and human cells. Genetics. 2013 Nov;195(3):831–44. doi: 10.1534/genetics.113.153981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013 Jan 11;339(6116):222–6. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mentch SJ, Mehrmohamadi M, Huang L, Liu X, Gupta D, Mattocks D, et al. Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metabolism. 2015 Sep 22; doi: 10.1016/j.cmet.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007 Mar 7;8(4):307–18. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 118.Hino S, Sakamoto A, Nagaoka K, Anan K, Wang Y, Mimasu S, et al. FAD-dependent lysine-specific demethylase-1 regulates cellular energy expenditure. Nat Commun. 2012 Mar 27;3:758–8. doi: 10.1038/ncomms1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhou X, Sun H, Chen H, Zavadil J, Kluz T, Arita A, et al. Hypoxia Induces Trimethylated H3 Lysine 4 by Inhibition of JARID1A Demethylase. Cancer Res. 2010 May 12;70(10):4214–21. doi: 10.1158/0008-5472.CAN-09-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tausendschön M, Dehne N, Brüne B. Hypoxia causes epigenetic gene regulation in macrophages by attenuating Jumonji histone demethylase activity. Cytokine. 2011 Feb;53(2):256–62. doi: 10.1016/j.cyto.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 121.Park S-J, Kim J-G, Son TG, Yi JM, Kim ND, Yang K, et al. Biochemical and Biophysical Research Communications. 4. Vol. 434. Elsevier Inc; 2013. May 17, The histone demethylase JMJD1A regulates adrenomedullin-mediated cell proliferation in hepatocellular carcinoma under hypoxia; pp. 722–7. [DOI] [PubMed] [Google Scholar]
- 122.Rensvold JW, Krautkramer KA, Dowell JA, Denu JM, Pagliarini DJ. Iron Deprivation Induces Transcriptional Regulation of Mitochondrial Biogenesis. J Biol Chem. 2016 Sep 30;291(40):20827–37. doi: 10.1074/jbc.M116.727701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. Journal of Experimental Medicine. 2010 Feb 15;207(2):339–44. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature Nature Publishing Group. 2012 Mar 12;483(7390):474–8. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ward PS, Thompson CB. Metabolic Reprogramming: A Cancer Hallmark Even Warburg Did Not Anticipate. Cancer Cell. 2012 Mar;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, et al. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes & Development. 2012 Jun 15;26(12):1326–38. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Smith EH, Janknecht R, Maher LJ. Succinate inhibition of alpha-ketoglutarate-dependent enzymes in a yeast model of paraganglioma. Human Molecular Genetics. 2007 Dec 15;16(24):3136–48. doi: 10.1093/hmg/ddm275. [DOI] [PubMed] [Google Scholar]
- 128.Dominguez-Salas P, Moore SE, Baker MS, Bergen AW, Cox SE, Dyer RA, et al. Maternal nutrition at conception modulates DNA methylation of human metastable epialleles. Nat Commun. 2014 Jan 1;5:3746–6. doi: 10.1038/ncomms4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Radford EJ, Ito M, Shi H, Corish JA, Yamazawa K, Isganaitis E, et al. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science. 2014 Aug 15;345(6198):1255903. doi: 10.1126/science.1255903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Letouzé E, Martinelli C, Loriot C, Burnichon N, Abermil N, Ottolenghi C, et al. SDH Mutations Establish a Hypermethylator Phenotype in Paraganglioma. Cancer Cell. 2013 Jun 10;23(6):739–52. doi: 10.1016/j.ccr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 131.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012 Mar 22;483(7390):479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, et al. Animals in a bacterial world, a new imperative for the life sciences. PNAS National Acad Sciences. 2013 Feb 26;110(9):3229–36. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Micro. 2008 Oct;6(10):776–88. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008 Jun 20;320(5883):1647–51. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kaoutari El A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Micro. 2013 Jul;11(7):497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 136.Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014 Aug 1;32(8):834–41. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 137.Besten den G, van Eunen K, Groen AK, Venema K, Reijngoud D-J, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013 Sep 1;54(9):2325–40. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Goyal MS, Venkatesh S, Milbrandt J, Gordon JI, Raichle ME. Feeding the brain and nurturing the mind: Linking nutrition and the gut microbiota to brain development. PNAS. 2015 Nov 17;112(46):14105–12. doi: 10.1073/pnas.1511465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013 Aug 2;341(6145):569–73. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nature Publishing Group Nature Publishing Group. 2016 Jun 1;16(6):341–52. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Samuel BS, Shaito A, Motoike T, Rey FE, Bäckhed F, Manchester JK, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. PNAS. 2008 Oct 28;105(43):16767–72. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mariño E, Richards JL, McLeod KH, Stanley D, Yap YA, Knight J, et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol. 2017 May;18(5):552–62. doi: 10.1038/ni.3713. [DOI] [PubMed] [Google Scholar]
- 143.Krautkramer KA, Kreznar JH, Romano KA, Vivas EI, Barrett-Wilt GA, Rabaglia ME, et al. Diet-Microbiota Interactions Mediate Global Epigenetic Programming in Multiple Host Tissues. Molecular Cell. 2016 Dec 1;64(5):982–92. doi: 10.1016/j.molcel.2016.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hinnebusch BF, Meng S, Wu JT, Archer SY, Hodin RA. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr. 2002 May;132(5):1012–7. doi: 10.1093/jn/132.5.1012. [DOI] [PubMed] [Google Scholar]
- 145.Waldecker M, Kautenburger T, Daumann H, Busch C, Schrenk D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J Nutr Biochem. 2008 Sep;19(9):587–93. doi: 10.1016/j.jnutbio.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 146.Lukovac S, Belzer C, Pellis L, Keijser BJ, de Vos WM, Montijn RC, et al. Differential Modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of Host Peripheral Lipid Metabolism and Histone Acetylation in Mouse Gut Organoids. MBio. 2014 Jul 1;5(4):e01438-14–e01438–14. doi: 10.1128/mBio.01438-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Current Opinion in Biotechnology. 2013 Apr;24(2):160–8. doi: 10.1016/j.copbio.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 148.Kim TH, Yang J, Darling PB, O'Connor DL. A large pool of available folate exists in the large intestine of human infants and piglets. J Nutr American Society for Nutrition. 2004 Jun;134(6):1389–94. doi: 10.1093/jn/134.6.1389. [DOI] [PubMed] [Google Scholar]
- 149.Aufreiter S, Gregory JF, Pfeiffer CM, Fazili Z, Kim Y-I, Marcon N, et al. Folate is absorbed across the colon of adults: evidence from cecal infusion of (13)C-labeled [6S]-5-formyltetrahydrofolic acid. Am J Clin Nutr. 2009 Jul 1;90(1):116–23. doi: 10.3945/ajcn.2008.27345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cantó C, Menzies KJ, Auwerx J. NAD(+) Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell Metabolism. 2015 Jul 7;22(1):31–53. doi: 10.1016/j.cmet.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Feldman JL, Dittenhafer-Reed KE, Denu JM. Sirtuin catalysis and regulation. J Biol Chem. 2012 Dec 14;287(51):42419–27. doi: 10.1074/jbc.R112.378877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Siudeja K, Srinivasan B, Xu L, Rana A, de Jong J, Nollen EAA, et al. Impaired Coenzyme A metabolism affects histone and tubulin acetylation in Drosophila and human cell models of pantothenate kinase associated neurodegeneration. EMBO Mol Med. 2011 Dec 1;3(12):755–66. doi: 10.1002/emmm.201100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.van Duynhoven J, Vaughan EE, Jacobs DM, Kemperman RA, van Velzen EJJ, Gross G, et al. Metabolic fate of polyphenols in the human superorganism. Proc Natl Acad Sci USA. 2011 Mar 15;108( Suppl 1):4531–8. doi: 10.1073/pnas.1000098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Silberberg M, Morand C, Mathevon T, Besson C, Manach C, Scalbert A, et al. The bioavailability of polyphenols is highly governed by the capacity of the intestine and of the liver to secrete conjugated metabolites. Eur J Nutr. 2006 Mar;45(2):88–96. doi: 10.1007/s00394-005-0568-5. [DOI] [PubMed] [Google Scholar]
- 155.Nandakumar V, Vaid M, Katiyar SK. (-)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis. 2011 Mar 29;32(4):537–44. doi: 10.1093/carcin/bgq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Vahid F, Zand H, Nosrat-Mirshekarlou E, Najafi R, Hekmatdoost A. The role dietary of bioactive compounds on the regulation of histone acetylases and deacetylases: A review. Gene Elsevier BV. 2015 May 10;562(1):8–15. doi: 10.1016/j.gene.2015.02.045. [DOI] [PubMed] [Google Scholar]
- 157.Bonkowski MS, Sinclair DA. Slowing ageing by design: the rise of NAD(+) and sirtuin-activating compounds. Nature Publishing Group. 2016 Nov;17(11):679–90. doi: 10.1038/nrm.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Shakibaei M, Buhrmann C, Mobasheri A. Resveratrol-mediated SIRT-1 Interactions with p300 Modulate Receptor Activator of NF-kappa B Ligand (RANKL) Activation of NF-kappa B Signaling and Inhibit Osteoclastogenesis in Bone-derived Cells. Journal of Biological Chemistry American Society for Biochemistry and Molecular Biology. 2011;286(13):11492–505. doi: 10.1074/jbc.M110.198713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kiss AK, Granica S, Stolarczyk M, Melzig MF. Epigenetic modulation of mechanisms involved in inflammation: Influence of selected polyphenolic substances on histone acetylation state. Food Chemistry Elsevier Ltd. 2012 Apr 1;131(3):1015–20. [Google Scholar]
- 160.Rajavelu A, Tulyasheva Z, Jaiswal R, Jeltsch A, Kuhnert N. The inhibition of the mammalian DNA methyltransferase 3a (Dnmt3a) by dietary black tea and coffee polyphenols. BMC Biochem. 2011;12(1):16. doi: 10.1186/1471-2091-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lee WJ, Shim J-Y, Zhu BT. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol Pharmacol. 2005 Oct;68(4):1018–30. doi: 10.1124/mol.104.008367. [DOI] [PubMed] [Google Scholar]
- 162.Wong CP, Hsu A, Buchanan A, Palomera-Sanchez Z, Beaver LM, Houseman EA, et al. Effects of Sulforaphane and 3,3′-Diindolylmethane on Genome-Wide Promoter Methylation in Normal Prostate Epithelial Cells and Prostate Cancer Cells. In: Futscher BW, editor. PLoS ONE. 1. Vol. 9. 2014. Jan 22, pp. e86787–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wahlström A, Sayin SI, Marschall H-U, Bäckhed F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metabolism Elsevier Inc. 2016 Jul 12;24(1):41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 164.Akare S, Jean-Louis S, Chen W, Wood DJ, Powell AA, Martinez JD. Ursodeoxycholic acid modulates histone acetylation and induces differentiation and senescence. Int J Cancer. 2006;119(12):2958–69. doi: 10.1002/ijc.22231. [DOI] [PubMed] [Google Scholar]
- 165.Craciun S, Balskus EP. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. PNAS. 2012 Dec 26;109(52):21307–12. doi: 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Krueger SK, Williams DE. Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol Ther. 2005 Jun;106(3):357–87. doi: 10.1016/j.pharmthera.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dumas M-E, Barton RH, Toye A, Cloarec O, Blancher C, Rothwell A, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci USA. 2006 Aug 15;103(33):12511–6. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Druart C, Neyrinck AM, Vlaeminck B, Fievez V, Cani PD, Delzenne NM. Role of the lower and upper intestine in the production and absorption of gut microbiota-derived PUFA metabolites. PLoS ONE. 2014;9(1):e87560. doi: 10.1371/journal.pone.0087560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nøhr MK, Pedersen MH, Gille A, Egerod KL, Engelstoft MS, Husted AS, et al. GPR41/FFAR3 and GPR43/FFAR2 as cosensors for short-chain fatty acids in enteroendocrine cells vs FFAR3 in enteric neurons and FFAR2 in enteric leukocytes. Endocrinology. 2013 Oct;154(10):3552–64. doi: 10.1210/en.2013-1142. [DOI] [PubMed] [Google Scholar]
- 170.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. 2012 Jun 8;336(6086):1262–7. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 171.Blaser MJ. Antibiotic use and its consequences for the normal microbiome. Science. 2016 Apr 29;352(6285):544–5. doi: 10.1126/science.aad9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ziętak M, Kovatcheva-Datchary P, Markiewicz LH, Ståhlman M, Kozak LP, Bäckhed F. Altered Microbiota Contributes to Reduced Diet-Induced Obesity upon Cold Exposure. Cell Metabolism The Author(s) 2016 Jun 14;23(6):1216–23. doi: 10.1016/j.cmet.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Carey HV, Walters WA, Knight R. Seasonal restructuring of the ground squirrel gut microbiota over the annual hibernation cycle. AJP: Regulatory, Integrative and Comparative Physiology. 2013 Jan 1;304(1):R33–R42. doi: 10.1152/ajpregu.00387.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Stevenson TJ, Duddleston KN, Buck CL. Effects of Season and Host Physiological State on the Diversity, Density, and Activity of the Arctic Ground Squirrel Cecal Microbiota. Applied and Environmental Microbiology. 2014 Aug 21;80(18):5611–22. doi: 10.1128/AEM.01537-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sommer F, Ståhlman M, Ilkayeva O, Arnemo JM, Kindberg J, Josefsson J, et al. The Gut Microbiota Modulates Energy Metabolism in the Hibernating Brown Bear Ursus arctos. CellReports Elsevier Ltd. 2016 Feb 23;14(7):1655–61. doi: 10.1016/j.celrep.2016.01.026. [DOI] [PubMed] [Google Scholar]
- 176.Maurice CF, Knowles SC, Ladau J, Pollard KS, Fenton A, Pedersen AB, et al. Marked seasonal variation in the wild mouse gut microbiota. Nature Publishing Group. 2015 May 29;9(11):2423–34. doi: 10.1038/ismej.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hehemann J-H, Kelly AG, Pudlo NA, Martens EC, Boraston AB. Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. PNAS National Acad Sciences. 2012 Nov 27;109(48):19786–91. doi: 10.1073/pnas.1211002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Subramanian S, Blanton LV, Frese SA, Charbonneau M, Mills DA, Gordon JI. Cultivating Healthy Growth and Nutrition through the Gut Microbiota. Cell Elsevier Inc. 2015 Mar 26;161(1):36–48. doi: 10.1016/j.cell.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The Effect of Diet on the Human Gut Microbiome: A Metagenomic Analysis in Humanized Gnotobiotic Mice. Sci Transl Med American Association for the Advancement of Science. 2009;1(6):6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Braniste V, Al-Asmakh M, Kowal C, Anuar F, Abbaspour A, Tóth M, et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med. 2014 Nov 19;6(263):263ra158-8. doi: 10.1126/scitranslmed.3009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012 Jul 27;37(1):171–86. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 182.Semenkovich NP, Planer JD, Ahern PP, Griffin NW, Lin CY, Gordon JI. Impact of the gut microbiota on enhancer accessibility in gut intraepithelial lymphocytes. PNAS. 2016 Dec 20;113(51):14805–10. doi: 10.1073/pnas.1617793113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. PNAS. 2008 Nov 4;105(44):17046–9. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Majnik AV, Lane RH. The relationship between early-life environment, the epigenome and the microbiota. Epigenomics. 2015 Oct;7(7):1173–84. doi: 10.2217/epi.15.74. [DOI] [PubMed] [Google Scholar]
- 185.Crews D, Gillette R, Scarpino SV, Manikkam M, Savenkova MI, Skinner MK. Epigenetic transgenerational inheritance of altered stress responses. Proc Natl Acad Sci USA. 2012;109(23):9143–8. doi: 10.1073/pnas.1118514109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yassour M, Vatanen T, Siljander H, Hämäläinen A-M, Härkönen T, Ryhänen SJ, et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med. 2016 Jun 15;8(343):343ra81–1. doi: 10.1126/scitranslmed.aad0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Cox LM, Blaser MJ. Antibiotics in early life and obesity. Nat Rev Endocrinol. 2015 Mar 1;11(3):182–90. doi: 10.1038/nrendo.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zeissig S, Blumberg RS. Life at the beginning: perturbation of the microbiota by antibiotics in early life and its role in health and disease. Nat Immunol. 2014 Apr 1;15(4):307–10. doi: 10.1038/ni.2847. [DOI] [PubMed] [Google Scholar]
- 189.Collado MC, Isolauri E, Laitinen K, Salminen S. Effect of mother“s weight on infant”s microbiota acquisition, composition, and activity during early infancy: a prospective follow-up study initiated in early pregnancy. Am J Clin Nutr. 2010 Oct 20;92(5):1023–30. doi: 10.3945/ajcn.2010.29877. [DOI] [PubMed] [Google Scholar]
- 190.Zijlmans MAC, Korpela K, Riksen-Walraven JM, de Vos WM, de Weerth C. Maternal prenatal stress is associated with the infant intestinal microbiota. Psychoneuroendocrinology. 2015 Mar 1;53:233–45. doi: 10.1016/j.psyneuen.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 191.Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. 2016 Mar 22;6:23129. doi: 10.1038/srep23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hu J, Nomura Y, Bashir A, Fernandez-Hernandez H, Itzkowitz S, Pei Z, et al. Diversified Microbiota of Meconium Is Affected by Maternal Diabetes Status. In: Tse H, editor. PLoS ONE. 11. Vol. 8. 2013. Nov 6, pp. e78257–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ma J, Prince AL, Bader D, Hu M, Ganu R, Baquero K, et al. High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat Commun. 2014 Jan 1;5:3889–9. doi: 10.1038/ncomms4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Cox J, Williams S, Grove K, Lane RH, Aagaard-Tillery KM. A maternal high-fat diet is accompanied by alterations in the fetal primate metabolome. Am J Obstet Gynecol. 2009 Sep 1;201(3):281–9. doi: 10.1016/j.ajog.2009.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Aagaard-Tillery KM, Grove K, Bishop J, Ke X, Fu Q, McKnight R, et al. Developmental origins of disease and determinants of chromatin structure: maternal diet modifies the primate fetal epigenome. Journal of Molecular Endocrinology. 2008 Aug 1;41(2):91–102. doi: 10.1677/JME-08-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Butaye P, Devriese LA, Haesebrouck F. Antimicrobial Growth Promoters Used in Animal Feed: Effects of Less Well Known Antibiotics on Gram-Positive Bacteria. Clinical Microbiology Reviews. 2003 Apr 1;16(2):175–88. doi: 10.1128/CMR.16.2.175-188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Dibner JJ, Richards JD. Antibiotic growth promoters in agriculture: History and mode of action. Poult Sci Oxford University Press. 2005 Apr;84(4):634–43. doi: 10.1093/ps/84.4.634. [DOI] [PubMed] [Google Scholar]
- 198.Gaskins HR, Collier CT, Anderson DB. Antibiotics as growth promotants: Mode of action. Anim Biotechnol. 2002;13(1):29–42. doi: 10.1081/ABIO-120005768. [DOI] [PubMed] [Google Scholar]
- 199.Coates ME, Fuller R, Harrison GF, Lev M, Suffolk SF. British Journal of Nutrition. 1. Vol. 17. Cambridge University Press; 1963. Feb 1, A comparision of the growth of chicks in the Gustafsson germ-free apparatus and in a conventional environment, with and without dietary supplements of penicillin; pp. 141–50. [DOI] [PubMed] [Google Scholar]
- 200.Mahana D, Trent CM, Kurtz ZD, Bokulich NA, Battaglia T, Chung J, et al. Antibiotic perturbation of the murine gut microbiome enhances the adiposity, insulin resistance, and liver disease associated with high-fat diet. Genome Medicine. Genome Medicine. 2016 Apr 25;:1–20. doi: 10.1186/s13073-016-0297-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. 2012 Aug 22;488(7413):621–6. doi: 10.1038/nature11400. Available from: http://www.nature.com/doifinder/10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Pierre JF. Gastrointestinal immune and microbiome changes during parenteral nutrition. Am J Physiol Gastrointest Liver Physiol. 2017 Mar 1;312(3):G246–56. doi: 10.1152/ajpgi.00321.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Suzuki K, Kawamoto S, Maruya M, Fagarasan S. Adv Immunol. Vol. 107. Elsevier; 2010. Jan 1, GALT-Chapter 6:Organization and Dynamics Leading to IgA Synthesis; pp. 153–85. [DOI] [PubMed] [Google Scholar]
- 204.Feng Y, Teitelbaum DH. Tumour necrosis factor--induced loss of intestinal barrier function requires TNFR1 and TNFR2 signalling in a mouse model of total parenteral nutrition. J Physiol (Lond) 2013 Aug 1;591(15):3709–23. doi: 10.1113/jphysiol.2013.253518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kudsk KA, Croce MA, Fabian TC, Minard G, Tolley EA, Poret HA, et al. Ann Surg. 5. Vol. 215. Lippincott, Williams, and Wilkins; 1992. May, Enteral versus parenteral feeding. Effects on septic morbidity after blunt and penetrating abdominal trauma; pp. 503–11. discussion511–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Tsai J-J, Kuo H-C, Lee K-F, Tsai T-H. Proteomic analysis of plasma from rats following total parenteral nutrition-induced liver injury. Proteomics. 2015 Nov;15(22):3865–74. doi: 10.1002/pmic.201500128. [DOI] [PubMed] [Google Scholar]
- 207.Koelfat KVK, Schaap FG, Hodin CMJM, Visschers RGJ, Svavarsson BI, Lenicek M, et al. Parenteral nutrition dysregulates bile salt homeostasis in a rat model of parenteral nutrition-associated liver disease. Clin Nutr. 2016 Sep 24; doi: 10.1016/j.clnu.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 208.Ralls MW, Miyasaka E, Teitelbaum DH. Intestinal microbial diversity and perioperative complications. JPEN J Parenter Enteral Nutr. 2014 Jan 1;38(3):392–9. doi: 10.1177/0148607113486482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ralls MW, Demehri FR, Feng Y, Raskind S, Ruan C, Schintlmeister A, et al. Bacterial nutrient foraging in a mouse model of enteral nutrient deprivation: insight into the gut origin of sepsis. Am J Physiol Gastrointest Liver Physiol. 2016 Oct 1;311(4):G734–43. doi: 10.1152/ajpgi.00088.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Miyasaka EA, Feng Y, Poroyko V, Falkowski NR, Erb-Downward J, Gillilland MG, et al. Total parenteral nutrition-associated lamina propria inflammation in mice is mediated by a MyD88-dependent mechanism. J Immunol. 2013 Jun 15;190(12):6607–15. doi: 10.4049/jimmunol.1201746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Harris JK, Kasmi El KC, Anderson AL, Devereaux MW, Fillon SA, Robertson CE, et al. Specific microbiome changes in a mouse model of parenteral nutrition associated liver injury and intestinal inflammation. PLoS ONE. 2014;9(10):e110396–6. doi: 10.1371/journal.pone.0110396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Demehri FR, Barrett M, Teitelbaum DH. Changes to the Intestinal Microbiome With Parenteral Nutrition: Review of a Murine Model and Potential Clinical Implications. Nutr Clin Pract. 2015 Dec;30(6):798–806. doi: 10.1177/0884533615609904. [DOI] [PubMed] [Google Scholar]
- 213.Carey HV, Andrews MT, Martin SL. Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiological Reviews. 2003 Oct;83(4):1153–81. doi: 10.1152/physrev.00008.2003. [DOI] [PubMed] [Google Scholar]
- 214.Dill-McFarland KA, Neil KL, Zeng A, Sprenger RJ, Kurtz CC, Suen G, et al. Hibernation alters the diversity and composition of mucosa-associated bacteria while enhancing antimicrobial defence in the gut of 13-lined ground squirrels. Mol Ecol. 2014 Sep 3;23(18):4658–69. doi: 10.1111/mec.12884. [DOI] [PubMed] [Google Scholar]
- 215.Carey HV, Assadi-Porter FM. The Hibernator Microbiome: Host-Bacterial Interactions in an Extreme Nutritional Symbiosis. Annual Review of Nutrition. 2017;37 doi: 10.1146/annurev-nutr-071816-064740. [DOI] [PubMed] [Google Scholar]
- 216.Carey HV, Duddleston KN. Animal-microbial symbioses in changing environments. J Therm Biol. 2014 Aug;44:78–84. doi: 10.1016/j.jtherbio.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Carey HV. Seasonal changes in mucosal structure and function in ground squirrel intestine. Am J Physiol. 1990 Aug;259(2 Pt 2):R385–92. doi: 10.1152/ajpregu.1990.259.2.R385. [DOI] [PubMed] [Google Scholar]
- 218.Kurtz CC, Carey HV. Seasonal changes in the intestinal immune system of hibernating ground squirrels. Dev Comp Immunol. 2007;31(4):415–28. doi: 10.1016/j.dci.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 219.Lindell SL, Klahn SL, Piazza TM, Mangino MJ, Torrealba JR, Southard JH, et al. Natural resistance to liver cold ischemia-reperfusion injury associated with the hibernation phenotype. AJP: Gastrointestinal and Liver Physiology. 2005 Mar;288(3):G473–80. doi: 10.1152/ajpgi.00223.2004. [DOI] [PubMed] [Google Scholar]
- 220.Otis JP, Pike AC, Torrealba JR, Carey HV. Hibernation reduces cellular damage caused by warm hepatic ischemia-reperfusion in ground squirrels. Journal of Comparative Physiology B. 2017 May;187(4):639–48. doi: 10.1007/s00360-017-1056-y. [DOI] [PubMed] [Google Scholar]
- 221.Griko YV, Rask JC, Raychev R. Advantage of Animal Models with Metabolic Flexibility for Space Research Beyond Low Earth Orbit. Astrobiology. 2017 Jan 1;1(1) [Google Scholar]
- 222.Cerri M, Tinganelli W, Negrini M, Helm A, Scifoni E, Tommasino F, et al. Life Sciences in Space Research. C. Vol. 11. Elsevier Ltd; 2016. Nov 1, Hibernation for space travel: Impact on radioprotection; pp. 1–9. [DOI] [PubMed] [Google Scholar]
- 223.Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, et al. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host and Microbe. 2016 May 11;19(5):731–43. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. PNAS. 2010 Nov 2;107(44):18933–8. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]