Abstract
The duality of group II introns, capable of carrying out both self-splicing and retromobility reactions, is hypothesized to have played a profound role in the evolution of eukaryotes. These introns likely provided the framework for the emergence of eukaryotic retroelements, spliceosomal introns and other key components of the spliceosome. Group II introns are found in all three domains of life and are therefore considered to be exceptionally successful mobile genetic elements. Initially identified in organellar genomes, group II introns are found in bacteria, chloroplasts and mitochondria of plants and fungi, but not in nuclear genomes. Although there is no doubt that prokaryotic and organellar group II introns are evolutionary related, there are remarkable differences in survival strategies between them. Furthermore, an evolutionary relationship of group II introns to eukaryotic retroelements, including telomeres, and spliceosomes is unmistakable.
Keywords: ribozyme, retrotransposon, spliceosome, telomerase
Group II introns are ribozymes and mobile genetic elements
Mobile group II introns are ribonucleoproteins (RNPs), which consist of a catalytic RNA (ribozyme) and an intron-encoded protein (IEP) containing reverse transcriptase (RT) activity (Figure 1) [1, 2]. The RNP is formed prior to IEP-assisted intron splicing from pre-mRNA and the IEP then plays an instrumental role in the mobility of that intron [1, 3–5].
Figure 1. Structural organization of a mobile group II intron.
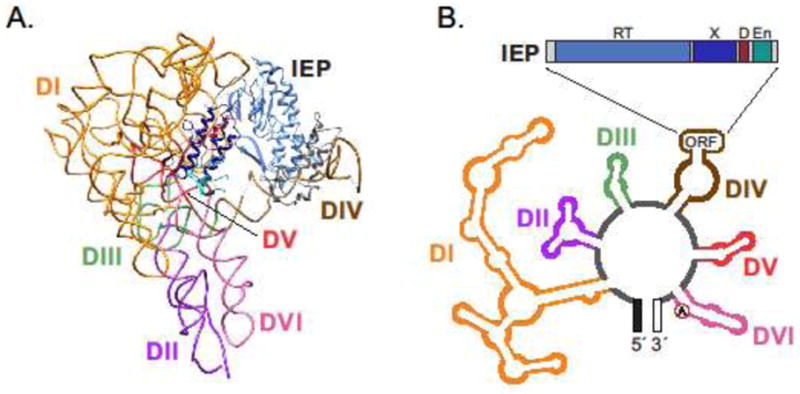
A. Molecular structure of the bacterial group II intron RNP complex. The catalytic RNA (DI–DVI) is complexed with the IEP [2].
B. Group II intron RNA secondary structure (bottom) and IEP domain organization (top) [4]. Six conserved structural domains are indicated (DI–DVI) in RNA structure with IEP ORF looped out of DIV (not to scale). The domains of both the RNA and IEP are color-coded as in A. Conserved domains shown in IEP are reverse transcriptase (RT), domain associated with RNA binding and splicing activity (maturase, X), DNA binding domain (D), and endonuclease domain (En).
Since the RNA folding is key to ribozyme activity, it is not surprising that group II intron RNAs possess a common secondary and tertiary structure, with minor variation, despite very little conservation of their primary sequences. The group II intron RNA secondary structure consists of six double-helical domains, DI to DVI, with DI, DV and DVI playing essential roles in splicing (Figure 1B) [1, 3–5]. DI is the largest domain and forms a structural scaffold for the molecule and also contains the exon recognition elements (exon binding sites, EBSs) [2, 6–8]. DV is the most highly conserved domain and contains the so-called “catalytic triad” (usually AGC, but often CGC), which binds catalytically important Mg2+ ions and, in combination with parts of DI, forms the active site. DVI contains a bulged adenosine, the nucleophile involved in initiation of the splicing reaction that results in intron lariat formation [2, 6–11].
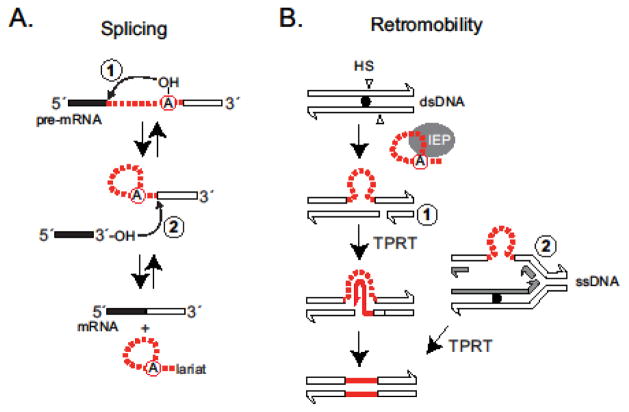
Group II intron splicing involves two successive transesterification reactions that produce ligated exons and release the intron (Figure 2A). Although group II introns are catalytic RNA molecules that are capable of self-splicing in vitro, the IEP is required to achieve and stabilize the catalytically active RNA structure in vivo. IEPs are multifunctional proteins containing conserved RT domains as well as a domain X associated with RNA binding and maturase activity involved in splicing. There are also DNA binding and DNA endonuclease domains involved in mobility (Figure 1) [1–4, 8].
Figure 2. Group II intron splicing and retromobility.
A. RNA splicing of group II introns. Splicing includes two transesterification reactions (1 and 2). In the first reaction, the 2′-OH of the branch-point adenosine of the intron (red circle) acts as nucleophile to attack the 5′ splice site. During the second reaction, the newly released 3′-OH of the upstream exon (black rectangle) attacks the 3′ splice site, resulting in ligated exons and an intron lariat. Both reactions are reversible.
B. Group II introns utilize a target DNA-primed reverse transcription (TPRT) mechanism for their mobility. Retrohoming of group II introns usually occurs into a specific dsDNA homing site (HS, black dot) with high efficiency. A primer for TPRT initiation is often provided by the endonuclease of the IEP, which cleaves the bottom strand of the target site (indicated as 1). Alternatively, during group II intron integration into ssDNA, an Okazaki fragment (indicated as 2) serves as primer for reverse transcription.
In order to mobilize, the two components of the intron RNP, the RNA core and the IEP, collaborate in recognition and invasion of intronless DNA alleles (retrohoming) or ectopic sequences (retrotransposition). Just like eukaryotic non-LTR retrotransposons, mobile group II introns utilize a target DNA-primed reverse transcription (TPRT) mechanism for their mobility [1, 3, 4]. During TPRT, in both retrohoming and retrotransposition, intron RNA reverse splices into DNA followed by reverse transcription of the intron. The integration occurs into dsDNA for most cases of retrohoming, and the primer for cDNA synthesis is provided by a nick introduced by the IEP’s endonuclease activity, leading to high-efficiency intron inheritance events (Figure 2B). In a low-frequency retrotransposition pathway, the intron recognizes degenerate sites and targets predominantly ssDNA, often at replication forks. Here, Okazaki fragments usually serve as primers for reverse transcription.
Diversity of group II introns
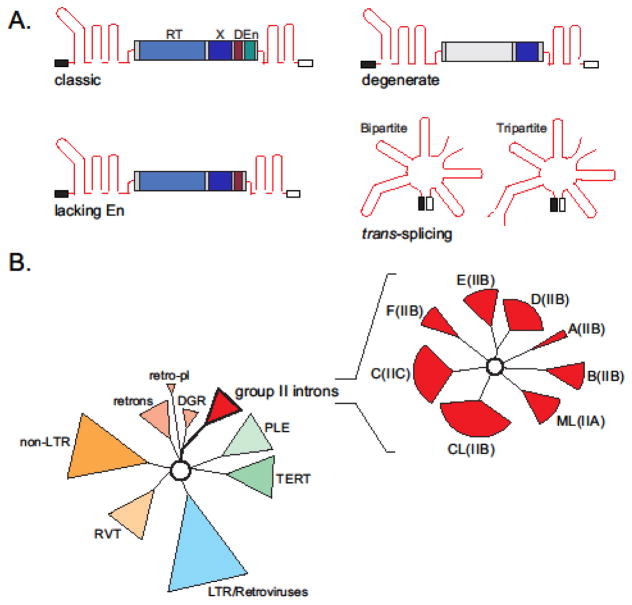
Group II introns come in diverse forms, which deviate from the classic description above (Figure 3A) [5]. For example, some group II introns lack the endonuclease domain in their IEPs and retrohome into ssDNA [12, 13]. Others have been split into two or more pieces that are located at distant loci in the genome [14–16]. There are additional examples of group II introns that carry homing endonucleases rather than reverse transcriptase coding sequences [17, 18]. There are also introns nested inside other introns, called twintrons, and introns with degenerate ribozyme structures incapable of self-splicing [5, 19, 20].
Figure 3. Diversity of group II introns and their relationship to other prokaryotic and eukaryotic RT-containing elements.
A. Variations in group II intron structural organization. Introns lacking the En domain, degenerate introns lacking some or all RT activity but maintaining the maturase function (e.g. chloroplast gene trnK and its IEP, MatK), and trans-splicing introns are shown [4, 5, 15, 16].
B. Evolutionary connections between RT-containing elements (right) (based on [26, 28] with modifications). Group II introns are related to all other RT-containing elements found in prokaryotes and eukaryotes (left) (based on [21] with modifications). LTR, long terminal repeat retrotransposons; DGR, diversity-generating retroelements; retro-pl, retro-plasmids; RVT, reverse transcriptase-related cellular genes; TERT, telomerase RT; PLE, Penelope-like element retrotransposons. Eight major lineages of the group II introns based on their RT domain are shown as red sectors (A–F, CL, and ML) (right). Intron subclasses based on RNA structure characteristics are indicated in parenthesis (IIA–IIC). Although the number of known group II introns is steadily growing, the evolutionary relationship between major lineages of introns remain unresolved [1, 5, 26, 28].
Classification of group II introns is based on either RNA or IEP components (Figure 3B). According to phylogenetic analysis of RT amino acid sequences, all prokaryotic and eukaryotic RT-containing elements are related to each other [21]. Moreover, group II introns seem to be closely related to non-LTR retrotransposons and telomerase (Figure 3B) [2, 5, 22–24]. Group II introns are represented by eight major lineages: A–F, chloroplast-like (CL), and mitochondrial-like (ML), as well as a few less prominent varieties [25–27]. Although, as noted above, the RNA fold is highly conserved, there are three major variations in group II intron RNA structures which were identified as three distinct subgroups: IIA (lineage ML), IIB (lineages A, B, D–F, and CL), and IIC (lineage C) [1, 20, 25–27]. These two classification systems allow one to categorize all known group II introns regardless of differences in their organization.
Bacterial group II introns are scarce
Bacterial group II introns are highly variable in abundance and diversity even between closely related species and strains [20, 25, 26, 28]. Most bacterial genomes harbor only a few introns [25, 26, 29], which represent a minuscule fraction of the genome when compared to other bacterial mobile elements [30]. This paucity of group II introns likely reflects host-cell defense mechanisms against intron proliferation [31]. The majority of known bacterial group II introns are far from the ‘classic’ full-length mobile forms and are represented by inactive, truncated vestiges often associated with sequences non-essential to the host, including conjugative plasmids and other mobile elements [32, 33], and in non-coding regions such as downstream of transcription terminator motifs or other inverted repeats [34, 35]. It should be noted that there are no algorithms to identify novel group II introns if they lack an IEP [36], and therefore the diversity and distribution of bacterial group II introns and their remnants might be underestimated [37].
Studies from diverse bacterial species show that group II introns undergo waves of proliferation, albeit rarely, and these bursts are in response to stressors, such as starvation [38–40]. These proliferative bursts are followed by rapid elimination of the resulting copies even without saturation of target sites, indicating a selective constraint on proliferation, likely due to the deleterious nature of the intron. Similar cycles of gain and loss are common among other prokaryotic mobile genetic elements as well [41]. Nevertheless, purifying selection against group II intron proliferation is somewhat unexpected, as the group II intron has the ability to splice precisely from the RNA transcript, which should, at least in theory, limit the negative effect of the insertion on bacterial fitness. However, splicing efficiency is known to vary dramatically among group II introns and under different conditions [42–46]. Other deleterious effects of intron proliferation may include non-specific insertions via retrotransposition, increased genomic instability due to recombination between newly emerged intron copies, and an increased replication cost for highly streamlined bacterial genomes. Thus, bacteria harboring new intron copies are likely rapidly removed from the population due to intron burden to the host.
One way to circumvent elimination from the population would be to keep the number of copies in check by targeting loci with low transcription rates, e.g. regions downstream of transcription terminators [34, 35]. Invading other mobile genetic elements is another successful evasion strategy, which is common among bacterial group II introns [32, 33]. By inserting into intergenic regions or other mobile genetic elements, bacterial group II introns can result in genome plasticity. For example, a specific lineage of group IIC introns is often found inside integrons [35, 47]. Integrons are genetic elements able to acquire and rearrange open reading frames (ORFs) into gene cassette units playing important roles in the evolution of multiresistance plasmids and horizontal gene transfer (for review see [48]). A major unanswered question is how genes are recruited into cassettes. It was proposed that group II introns provide RT activity during genesis of integron gene cassettes [35, 47].
Although data are scarce, bacterial group II introns may have post-transcriptional regulatory potential. One of the few introns within essential genes is the Avi.groEL intron from Azotobacter vinelandii, which is located within the termination codon of the housekeeping gene groEL encoding heat-shock protein Hsp60. Surprisingly, splicing of the Avi.groEL intron in vitro is activated only at elevated temperatures, suggesting a potential role in heat-shock regulation [42, 49]. Finally, some group II intron vestiges are stably maintained in the genomes of nitrogen-fixing Sinorhizobium species, hinting at functional importance [37]. A remnant of the RmInt1 intron, FRE652, is located in intergenic sequence that may be involved in establishing the symbiotic relationship with the plant host. Retention of FRE652 over evolutionary time led to the hypothesis that FRE652 evolved into a cis-regulatory element [37].
Organellar group II introns are abundant
Mitochondrial (mtDNA) genomes of fungi, protists and a few primitive metazoans as well as chloroplast genomes (cpDNA) often contain a plethora of group II introns. They are particularly prevalent in the mtDNA of higher plants [5, 50]. Both cpDNA and mtDNA are highly dynamic and exhibit substantial variation in content and organization even among closely related species. There is a myriad of processes driving the divergence, including ongoing intracellular transfer of genes to the nucleus, gene duplication and loss, pervasive rearrangements, and recurrent horizontal gene transfer. Group II introns are actively involved in shaping their host genomes via mobility and horizontal transfer. Indeed, waves of group II intron proliferation were reported for some green algae [51, 52]. A remarkable example of intron propagation was found in cpDNA of the freshwater euglenid Euglena gracilis with almost all of its protein-coding genes interrupted [53]. Additionally, at least two bursts of intron gain in cpDNA were described in another euglenid Monomorphina aenigmatica [54]. Moreover, horizontal transfer was suggested based on cpDNA intron similarity between diatoms and green algae [55] as well as between cryptophytes and euglenids [56].
Organellar group II introns often exhibit the features of “domestication”, whereby they have adapted to their host. They reside almost exclusively in housekeeping genes, lacking one or another key component necessary for self-splicing and mobility. Often, only one of the group II introns within an organellar genome retains a protein-coding sequence. For example, in land plant cpDNA, often only the trnK intron within a tRNA gene contains an ORF that encodes a protein, MatK, with RT and maturase activities [57, 58]. In mtDNA, only the fourth intron of the nad1 gene has an ORF, which encodes a similar protein, MatR [59, 60]. It has been proposed that MatK in plastids and MatR in mitochondria promote splicing of multiple organellar introns in trans [58, 60]. In contrast, IEPs from bacterial group II introns usually serve only their cognate intron.
Both MatK and MatR, with their RT and maturase activities, are somewhat conserved between plants species. MatK and MatR work in combination with nuclear RNA processing factors and represent “a model for an early proto-spliceosomal activity” [58, 60], possibly providing insight into spliceosome evolution in early eukaryotes [50]. These organellar “proto-spliceosomes” are quite complex, facilitating splicing of different introns, and involving multiple proteins with diverse properties. Among the protein factors identified so far are nuclear-encoded maturases (nMATs), RNA chaperones, RNA stabilizing proteins, and RNA helicases [50, 61]. Recently, a multimolecular RNP supercomplex was characterized for trans-splicing group II introns from cpDNA of the green alga Chlamydomonas reinhardtii, revealing striking similarity to the nuclear spliceosome, such as dynamic association of trans-acting RNAs with a set of protein splicing factors [62].
The matK ORF as well as trnK/matK datasets are widely used as markers in plant phylogeny reconstructions due to their rapid evolution, in contrast to other cpDNA regions [63]. More specifically, studying organellar group II introns and their splicing machinery might help us to resolve mysteries of spliceosomal intron emergence and the origin of the spliceosomal proteins. Keeping in mind that one of the key spliceosomal proteins, Prp8, displays homologies to group II intron IEPs and likely evolved from an ancestral maturase [2, 7, 23, 50, 64], understanding how organellar maturases act on multiple targets may help to answer the question of how maturase evolved into a core spliceosomal factor.
Breaking bad and giving rise to a spliceosomal catalytic core
Despite the putative ancestral relationship between group II introns and spliceosomal introns, no group II introns have been found in eukaryotic nuclei. However, virtually all extant eukaryotic nuclear genomes carry spliceosomal introns, which, unlike self-splicing group II introns, rely on a multitude of trans-acting factors that comprise a spliceosome. Interestingly, when introduced into one of several yeast nuclear genes, the bacterial and mitochondrial group II introns prevent their transcript from translation, even when successfully spliced, and it remains unclear why [65–67]. The group II intron-spliceosome transition therefore remains mysterious and is speculated upon below.
The spliceosome has been described as one of the most complex macromolecular machines known [68, 69]. It is a highly dynamic RNP complex that comprises five small nuclear RNAs (snRNAs) and a multitude of proteins forming a tight interacting network of molecules [64]. In the past few years, cryo-EM has led to profound structural insights of a number of spliceosomes at different stages of splicing [70–73]. Although the origin and evolution of spliceosomal introns and spliceosomes can be a polarizing question [74, 75], structural and functional comparison between the spliceosomal and group II intron complexes make their close relationship irrefutable. It is widely assumed that mobile group II introns from a bacterial progenitor of mitochondria invaded the chromosomes of the emerging eukaryote and rapidly proliferated to multiple genomic sites [24, 76].
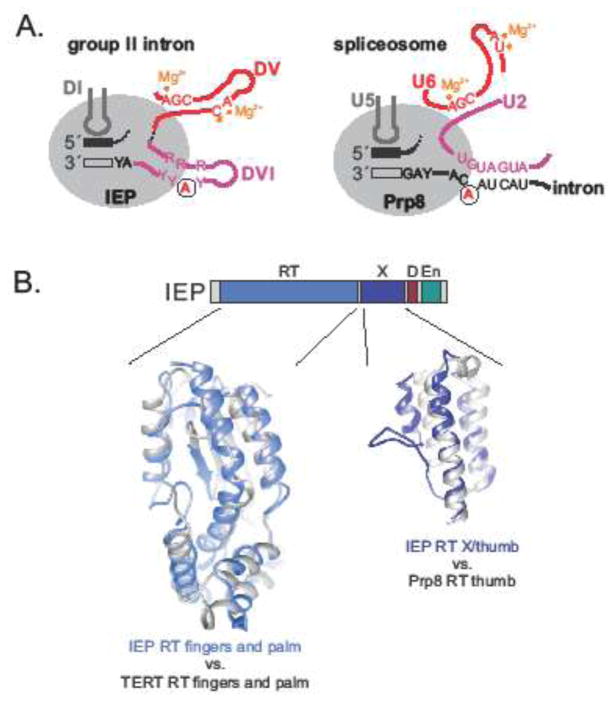
There are many similarities between group II and spliceosomal introns in terms of splicing mechanism and structure that are hard to explain by convergent evolution alone. First, both group II and spliceosomal introns undergo two identical transesterification reactions resulting in a lariat form of the intron and ligated exons (Figure 2A). Second, structural and functional parallels between group II intron RNAs and spliceosomal snRNAs are striking [2, 4, 5, 7, 77]. The most frequently invoked feature is the branch-site motif with the bulged A involved in the very first step of splicing. Whereas the bulged A is located within a helix of DVI in group II introns, a similar structure with the bulged A is formed by the pairing of the U2 snRNA to the spliceosomal intron’s branch point sequence (Figure 4A). Furthermore, in group II introns the catalytic triad in DV corresponds to a triad in the U6 snRNA, both binding catalytic Mg2, and interactions between DI in group II introns and intron binding sequences resemble the U5 snRNA base-pairing to 5′ and 3′ exons (Figure 4A) (recently reviewed in [4]). Finally, a recent cryo-EM structure of a bacterial group II intron RNP complex demonstrated a striking structure-function similarity in active site organization between the group II intron RNP that includes the IEP and the yeast spliceosome [2, 23]. Importantly, not only the RNA component but also specific domains of the IEP and Prp8 form highly similar structures within their respective active sites, making the case for an evolutionary connection between group II introns and the spliceosome even stronger. A pertinent example is the buttressing of group II intron DI or U5 snRNA interactions with their respective exons by Prp8 (Figure 4A). Strengthening the case, the crystal structures of the maturase domain from two different group II introns were shown to be similar in architecture to Prp8, and, interestingly, they also share similarity with the flaviviral RNA-dependent RNA polymerases [7].
Figure 4. Similarities between group II intron RNP and spliceosome active site.
A. Interactions in group II intron RNP active site and in activated spliceosome (recently reviewed in [4]). DI, domain I; DV, domain V; DVI, domain VI; IEP, intron-encoded protein; Y corresponds to C or U; R corresponds to A or G. Only some of the spliceosome components are shown for clarity. The 5′ exon is shown as a black bar and the 3′ exon is shown as an open bar, the bulged adenosine is in a circle, and residues involved in Mg2+ coordination are labeled with orange dots. Molecules showing homology in structure and function are in the same color.
B. Comparison of tertiary structure of group II IEP, Prp8 and telomerase. Group II intron IEP fingers and palm (PDB code: 5G2X) and telomerase RT (PDB code: 3DU6) domains are on the left, and IEP X/thumb domain and Prp8 (PDB code: 3JB9) are on the right. Corresponding structures are superimposed [2].
When and why have spliceosomal introns and the spliceosome emerged? The question of an intron-poor versus an intron-rich eukaryal ancestor is not a trivial one. While multicellular eukaryotes such as plants and animals are generally intron-rich, most unicellular eukaryotes are intron-poor. Thus, it may be that introns accumulated in the course of evolution of eukaryotes. However, multiple studies suggest the opposite, namely that an intron-rich ancestor existed and that intron loss occurred in many eukaryal lineages [78, 79]. Regardless, the transition from group II introns to the spliceosome and spliceosomal intron accumulation would have occurred prior to the last eukaryotic common ancestor.
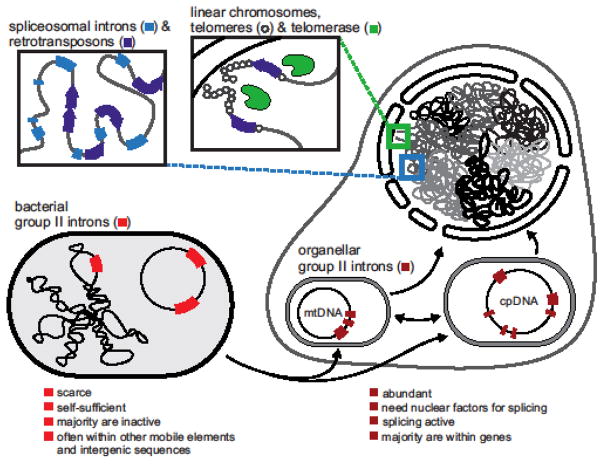
It has been proposed that rapid proliferation of group II introns transferred from bacterial progenitors of the mitochondria into the chromosome of a proto-eukaryotic organism caused genome instability (Key Figure, Figure 5). The emergence of the spliceosome, and likely the nucleus, was necessary to curb the deleterious effects of this proliferation [24, 76]. Under this scenario, first, group II introns acted as selfish mobile elements spreading across chromosomes. Next, some of the newly emerged intron copies were fixed and subsequently fragmented into the spliceosomal introns and evolved into the snRNAs and Prp8 [64], the active molecules of the spliceosome itself.
Key Figure, Figure 5. Mobile group II introns as ancestral eukaryotic elements.
Group II introns are related to spliceosomes, telomerase and other eukaryotic RT-containing elements [4, 21, 24, 81, 84]. Bacterial (bright red) and organellar (dark red) group II introns are thought to have been involved in key evolutionary transitions (black arrows) which led to the emergence of eukaryotic nuclear genome organization and function such as spliceosomal introns (light blue), linear chromosomes with telomeres plus telomerase (green), and retrotransposons (dark blue arrows). Contrasting characteristics of bacterial (bright red) and organellar group II introns (dark red) are listed below. Under the endosymbiotic scenario of the origin of the eukaryote cell, it is assumed that group II introns invaded the nuclear genome from either a bacterial progenitor of mitochondria or a proto-mitochondrion [24, 76]. An extensive proliferation of mobile group II introns to multiple genomic sites gave rise to the spliceosomal introns (light blue) and drove the evolution of a general splicing machinery to curb the deleterious effects of group II intron invasion [24, 76]. Additionally, a switch from circular to linear chromosomes could have occurred in response to the genome instability due to recombination between intron copies within a circular genome [24, 76, 80]. Linear chromosomes require maintenance of their telomeres (circles), performed either by telomerase (green) or, in some cases, by retrotransposons (dark blue arrows) [81–83].
Chromosome linearization could also have been triggered by group II intron proliferation and emerged as a defense against deletions caused by recombination between intron copies within a circular genome [24, 76, 80]. Linear chromosomes require maintenance of their telomeres, performed either by telomerase or, in some cases by retrotransposons [81–83]. The fruit-fly Drosophila, which lacks telomerase, has long head-to-tail arrays of retrotransposons that maintain chromosome ends [82]. Furthermore, Penelope-like elements (PLEs) lacking endonuclease domains are located at telomeres in organisms as diverse as rotifers and plants and have been shown to be closely related to telomerases [21, 81]. Remarkably, RT activity of telomerase, PLEs and non-LTR retroelements likely share a common ancestor with group II introns (Figure 3B and 4B) [2, 21, 24, 81, 84].
Thus, several characteristic features of eukaryotic nuclear genome organization and function such as exon-intron architecture of the gene, some components of the spliceosome, linear chromosomes, telomerase and retrotransposons are evolutionarily linked, directly or indirectly, to mobile group II introns. These self-splicing introns and their associated proteins must, therefore, be considered important elements in the evolution of the ancestral eukaryotic cell. Not only are group II introns pivotal elements in the prokaryotic-eukaryotic transition but they have also been exploited for biotechnological application in two different ways (reviewed in [85]). First, given that the introns can target DNA in site-specific fashion, their target-site specificity can be reprogrammed to recognize virtually any DNA sequence. These so-called “targetrons” have been used for site-specific genome modification in a variety of bacteria with intractable genetic systems. Second, thermostable group II intron RTs are being used for applications ranging from qRT-PCR through RNA-seq to sequence highly structured and modified RNAs [86].
Concluding remarks and further perspectives
Group II introns are unique retroelements possessing ribozyme activity. In bacteria, group II introns seem to be scarce but active mobile retroelements. One of the questions for the future is how such robust retrotransposons are silenced in a way that is consistent with cellular well-being, while being capable of bursts of movement when triggered by environmental stresses (See Outstanding Questions). In organellar genomes, on the other hand, they are often domesticated, lack mobility and are tightly regulated, again by mechanisms that remain to be determined. While group II introns are versatile retroelements, they are also thought to have given rise to a multitude of diverse eukaryotic sequences and functions. Among them are RNA and protein components of the spliceosome. Intriguing ancestral relationships also exist among group II introns, non-LTR retrotransposons and telomerase. Although there has been great progress in expanding our understanding of the inner workings of group II introns, non-LTR retrotransposons, spliceosomal introns and telomerases during the last decade, many more mysteries remain as to their origins and precise evolutionary relationships. Meanwhile, application of group II introns in biotechnology is proceeding apace.
OUTSTANDING QUESTIONS BOX.
Do bacterial group II introns act exclusively as mobile elements or do they have roles in bacterial genome function?
Is there a potential function of group II introns in organelles and how is their splicing regulated?
How did group II intron proliferation trigger emergence of the spliceosome?
How did group II intron maturase evolve into core spliceosomal factor Prp8?
What is the evolutionary relationship between group II introns, retrotransposons and telomerases?
TRENDS BOX.
Mobile group II introns are both ribozymes and retroelements. They are believed to be the progenitors of spliceosomal introns and some components of the spliceosome and are also closely related to eukaryotic retroelements.
Whereas prokaryotic group II introns act predominantly as mobile elements, organellar group II introns exhibit the features of domestication. Studying both provides clues into their evolutionary dynamics.
Group II intron ribonucleoproteins also bear similarities to retrotransposons, telomerases and viral polymerases.
Acknowledgments
The authors acknowledge Rebecca McCarthy, Xiaolong (Eren) Dong, Cathleen Green, Danielle Kelley, Carol Lyn Piazza, Dorie Smith and Justin Waldern for help with or comments on the manuscript. This work was supported by the National Institutes of Health (NIH) grants GM39422 and GM44844 to M.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lambowitz AM, Zimmerly S. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol. 2011;3:a003616. doi: 10.1101/cshperspect.a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qu G, et al. Structure of a group II intron in complex with its reverse transcriptase. Nat Struct Mol Biol. 2016;23:549–559. doi: 10.1038/nsmb.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toro N, et al. Bacterial group II introns: not just splicing. FEMS Microbiol Rev. 2007;31:342–358. doi: 10.1111/j.1574-6976.2007.00068.x. [DOI] [PubMed] [Google Scholar]
- 4.Lambowitz AM, Belfort M. Mobile DNA III. Microbiol Spectr./ASM Press; 2015. Mobile bacterial group II introns at the crux of eukaryotic evolution; pp. 1209–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zimmerly S, Semper C. Evolution of group II introns. Mob DNA. 2015;6:7. doi: 10.1186/s13100-015-0037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Somarowthu S, et al. Visualizing the ai5gamma group IIB intron. Nucleic Acids Res. 2014;42:1947–1958. doi: 10.1093/nar/gkt1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao C, Pyle AM. Crystal structures of a group II intron maturase reveal a missing link in spliceosome evolution. Nat Struct Mol Biol. 2016;23:558–565. doi: 10.1038/nsmb.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao C, Pyle AM. Structural insights into the mechanism of group II Intron splicing. Trends Biochem Sci. 2017 doi: 10.1016/j.tibs.2017.03.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toor N, et al. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marcia M, Pyle AM. Visualizing group II intron catalysis through the stages of splicing. Cell. 2012;151:497–507. doi: 10.1016/j.cell.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters JK, Toor N. Group II intron lariat: Structural insights into the spliceosome. RNA Biol. 2015;12:913–917. doi: 10.1080/15476286.2015.1066956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toro N, Martinez-Abarca F. Comprehensive phylogenetic analysis of bacterial group II intron-encoded ORFs lacking the DNA endonuclease domain reveals new varieties. PLoS One. 2013;8:e55102. doi: 10.1371/journal.pone.0055102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nisa-Martinez R, et al. Host factors influencing the retrohoming pathway of group II intron RmInt1, which has an intron-encoded protein naturally devoid of endonuclease activity. PLoS One. 2016;11:e0162275. doi: 10.1371/journal.pone.0162275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knoop V, et al. A tripartite group II intron in mitochondria of an angiosperm plant. Mol Gen Genet. 1997;255:269–276. doi: 10.1007/s004380050497. [DOI] [PubMed] [Google Scholar]
- 15.Bonen L. Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion. 2008;8:26–34. doi: 10.1016/j.mito.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 16.Kamikawa R, et al. Group II intron-mediated trans-splicing in the gene-rich mitochondrial genome of an enigmatic eukaryote, Diphylleia rotans. Genome Biol Evol. 2016;8:458–466. doi: 10.1093/gbe/evw011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toor N, Zimmerly S. Identification of a family of group II introns encoding LAGLIDADG ORFs typical of group I introns. RNA. 2002;8:1373–1377. doi: 10.1017/s1355838202023087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullineux ST, et al. A group II intron encodes a functional LAGLIDADG homing endonuclease and self-splices under moderate temperature and ionic conditions. RNA. 2010;16:1818–1831. doi: 10.1261/rna.2184010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai L, Zimmerly S. ORF-less and reverse-transcriptase-encoding group II introns in archaebacteria, with a pattern of homing into related group II intron ORFs. RNA. 2003;9:14–19. doi: 10.1261/rna.2126203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Candales MA, et al. Database for bacterial group II introns. Nucleic Acids Res. 2012;40(Database issue):D187–D190. doi: 10.1093/nar/gkr1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gladyshev EA, Arkhipova IR. A widespread class of reverse transcriptase-related cellular genes. Proc Natl Acad Sci U S A. 2011;108:20311–20316. doi: 10.1073/pnas.1100266108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koonin EV, et al. Origins and evolution of viruses of eukaryotes: The ultimate modularity. Virology. 2015;479–480:2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agrawal RK, et al. Forks in the tracks: Group II introns, spliceosomes, telomeres and beyond. RNA Biol. 2016:1–5. doi: 10.1080/15476286.2016.1244595. [DOI] [PMC free article] [PubMed]
- 24.Koonin EV. Viruses and mobile elements as drivers of evolutionary transitions. Philos Trans R Soc Lond B Biol Sci. 2016;371:20150442. doi: 10.1098/rstb.2015.0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon DM, et al. A broadscale phylogenetic analysis of group II intron RNAs and intron-encoded reverse transcriptases. Mol Biol Evol. 2009;26:2795–2808. doi: 10.1093/molbev/msp193. [DOI] [PubMed] [Google Scholar]
- 26.Toro N, Nisa-Martinez R. Comprehensive phylogenetic analysis of bacterial reverse transcriptases. PLoS One. 2014;9:e114083. doi: 10.1371/journal.pone.0114083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimmerly S, Wu L. An unexplored diversity of reverse transcriptases in bacteria. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0058-2014. MDNA3-0058-2014. [DOI] [PubMed] [Google Scholar]
- 28.Simon DM, Zimmerly S. A diversity of uncharacterized reverse transcriptases in bacteria. Nucleic Acids Res. 2008;36:7219–7229. doi: 10.1093/nar/gkn867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leclercq S, Cordaux R. Selection-driven extinction dynamics for group II introns in Enterobacteriales. PLoS One. 2012;7:e52268. doi: 10.1371/journal.pone.0052268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Y, et al. Dynamics of bacterial insertion sequences: can transposition bursts help the elements persist? BMC Evol Biol. 2015;15:288. doi: 10.1186/s12862-015-0560-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beauregard A, et al. The take and give between retrotransposable elements and their hosts. Annu Rev Genet. 2008;42:587–617. doi: 10.1146/annurev.genet.42.110807.091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein JR, Dunny GM. Bacterial group II introns and their association with mobile genetic elements. Front Biosci. 2002;7:d1843–d1856. doi: 10.2741/klein1. [DOI] [PubMed] [Google Scholar]
- 33.Staddon JH, et al. Conserved target for group II intron insertion in relaxase genes of conjugative elements of gram-positive bacteria. J Bacteriol. 2004;186:2393–2401. doi: 10.1128/JB.186.8.2393-2401.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robart AR, et al. Insertion of group II intron retroelements after intrinsic transcriptional terminators. Proc Natl Acad Sci U S A. 2007;104:6620–6625. doi: 10.1073/pnas.0700561104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quiroga C, et al. The S.ma.I2 class C group II intron inserts at integron attC sites. Microbiology. 2008;154:1341–1353. doi: 10.1099/mic.0.2007/016360-0. [DOI] [PubMed] [Google Scholar]
- 36.Abebe M, et al. A pipeline of programs for collecting and analyzing group II intron retroelement sequences from GenBank. Mob DNA. 2013;4:28. doi: 10.1186/1759-8753-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toro N, et al. Insights into the history of a bacterial group II intron remnant from the genomes of the nitrogen-fixing symbionts Sinorhizobium meliloti and Sinorhizobium medicae. Heredity (Edinb) 2014;113:306–315. doi: 10.1038/hdy.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coros CJ, et al. Global regulators orchestrate group II intron retromobility. Mol Cell. 2009;34:250–256. doi: 10.1016/j.molcel.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leclercq S, et al. Remarkable abundance and evolution of mobile group II introns in Wolbachia bacterial endosymbionts. Mol Biol Evol. 2011;28:685–697. doi: 10.1093/molbev/msq238. [DOI] [PubMed] [Google Scholar]
- 40.Belfort M. Mobile self-splicing introns and inteins as environmental sensors. Curr Opin Microbiol. 2017;38:51–58. doi: 10.1016/j.mib.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner A. Periodic extinctions of transposable elements in bacterial lineages: evidence from intragenomic variation in multiple genomes. Mol Biol Evol. 2006;23:723–733. doi: 10.1093/molbev/msj085. [DOI] [PubMed] [Google Scholar]
- 42.Ferat JL, et al. A group II intron has invaded the genus Azotobacter and is inserted within the termination codon of the essential groEL gene. Mol Microbiol. 2003;49:1407–1423. doi: 10.1046/j.1365-2958.2003.03649.x. [DOI] [PubMed] [Google Scholar]
- 43.Chee GJ, Takami H. Housekeeping recA gene interrupted by group II intron in the thermophilic Geobacillus kaustophilus. Gene. 2005;363:211–220. doi: 10.1016/j.gene.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 44.Chillon I, et al. Splicing of the Sinorhizobium meliloti RmInt1 group II intron provides evidence of retroelement behavior. Nucleic Acids Res. 2011;39:1095–1104. doi: 10.1093/nar/gkq847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinez-Rodriguez L, et al. Insights into the strategies used by related group II introns to adapt successfully for the colonisation of a bacterial genome. RNA Biol. 2014;11:1061–1071. doi: 10.4161/rna.32092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.LaRoche-Johnston F, et al. Recent horizontal transfer, functional adaptation and dissemination of a bacterial group II intron. BMC Evol Biol. 2016;16:223. doi: 10.1186/s12862-016-0789-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leon G, Roy PH. Potential role of group IIC-attC introns in integron cassette formation. J Bacteriol. 2009;191:6040–6051. doi: 10.1128/JB.00674-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gillings MR. Integrons: past, present, and future. Microbiol Mol Biol Rev. 2014;78:257–277. doi: 10.1128/MMBR.00056-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adamidi C, et al. A group II intron inserted into a bacterial heat-shock operon shows autocatalytic activity and unusual thermostability. Biochemistry. 2003;42:3409–3418. doi: 10.1021/bi027330b. [DOI] [PubMed] [Google Scholar]
- 50.Schmitz-Linneweber C, et al. Organellar maturases: A window into the evolution of the spliceosome. Biochim Biophys Acta. 2015;1847:798–808. doi: 10.1016/j.bbabio.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 51.Brouard JS, et al. Proliferation of group II introns in the chloroplast genome of the green alga Oedocladium carolinianum (Chlorophyceae) PeerJ. 2016;4:e2627. doi: 10.7717/peerj.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turmel M, et al. Mitochondrion-to-chloroplast DNA transfers and intragenomic proliferation of chloroplast group II introns in Gloeotilopsis green algae (Ulotrichales, Ulvophyceae) Genome Biol Evol. 2016;8:2789–2805. doi: 10.1093/gbe/evw190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hallick RB, et al. Complete sequence of Euglena gracilis chloroplast DNA. Nucleic Acids Res. 1993;21:3537–3544. doi: 10.1093/nar/21.15.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pombert JF, et al. Evidence for transitional stages in the evolution of euglenid group II introns and twintrons in the Monomorphina aenigmatica plastid genome. PLoS One. 2012;7:e53433. doi: 10.1371/journal.pone.0053433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brembu T, et al. The chloroplast genome of the diatom Seminavis robusta: new features introduced through multiple mechanisms of horizontal gene transfer. Mar Genomics. 2014;16:17–27. doi: 10.1016/j.margen.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 56.Khan H, Archibald JM. Lateral transfer of introns in the cryptophyte plastid genome. Nucleic Acids Res. 2008;36:3043–3053. doi: 10.1093/nar/gkn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hausner G, et al. Origin and evolution of the chloroplast trnK (matK) intron: a model for evolution of group II intron RNA structures. Mol Biol Evol. 2006;23:380–391. doi: 10.1093/molbev/msj047. [DOI] [PubMed] [Google Scholar]
- 58.Zoschke R, et al. An organellar maturase associates with multiple group II introns. Proc Natl Acad Sci U S A. 2010;107:3245–3250. doi: 10.1073/pnas.0909400107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo W, Mower JP. Evolution of plant mitochondrial intron-encoded maturases: frequent lineage-specific loss and recurrent intracellular transfer to the nucleus. J Mol Evol. 2013;77:43–54. doi: 10.1007/s00239-013-9579-7. [DOI] [PubMed] [Google Scholar]
- 60.Sultan LD, et al. The Reverse Transcriptase/RNA Maturase Protein MatR Is Required for the Splicing of Various Group II Introns in Brassicaceae Mitochondria. Plant Cell. 2016;28:2805–2829. doi: 10.1105/tpc.16.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brown GG, et al. Group II intron splicing factors in plant mitochondria. Front Plant Sci. 2014;5:35. doi: 10.3389/fpls.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reifschneider O, et al. A ribonucleoprotein supercomplex involved in trans-splicing of organelle group II introns. J Biol Chem. 2016;291:23330–23342. doi: 10.1074/jbc.M116.750570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kelchner SA. Group II introns as phylogenetic tools: structure, function, and evolutionary constraints. Am J Bot. 2002;89:1651–1669. doi: 10.3732/ajb.89.10.1651. [DOI] [PubMed] [Google Scholar]
- 64.Dlakic M, Mushegian A. Prp8, the pivotal protein of the spliceosomal catalytic center, evolved from a retroelement-encoded reverse transcriptase. RNA. 2011;17:799–808. doi: 10.1261/rna.2396011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chalamcharla VR, et al. Nuclear expression of a group II intron is consistent with spliceosomal intron ancestry. Genes Dev. 2010;24:827–836. doi: 10.1101/gad.1905010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zerbato M, et al. The brown algae Pl.LSU/2 group II intron-encoded protein has functional reverse transcriptase and maturase activities. PLoS One. 2013;8:e58263. doi: 10.1371/journal.pone.0058263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qu G, et al. RNA-RNA interactions and pre-mRNA mislocalization as drivers of group II intron loss from nuclear genomes. Proc Natl Acad Sci U S A. 2014;111:6612–6617. doi: 10.1073/pnas.1404276111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- 69.Wahl MC, et al. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 70.Yan C, et al. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015;349:1182–1191. doi: 10.1126/science.aac7629. [DOI] [PubMed] [Google Scholar]
- 71.Nguyen TH, et al. CryoEM structures of two spliceosomal complexes: starter and dessert at the spliceosome feast. Curr Opin Struct Biol. 2016;36:48–57. doi: 10.1016/j.sbi.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Galej WP, et al. Cryo-EM structure of the spliceosome immediately after branching. Nature. 2016;537:197–201. doi: 10.1038/nature19316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bertram K, et al. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature. 2017;542:318–323. doi: 10.1038/nature21079. [DOI] [PubMed] [Google Scholar]
- 74.Catania F, et al. Endogenous mechanisms for the origins of spliceosomal introns. J Hered. 2009;100:591–596. doi: 10.1093/jhered/esp062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Doolittle WF. The spliceosomal catalytic core arose in the RNA world... or did it? Genome Biol. 2013;14:141. doi: 10.1186/gb4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006;440:41–45. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- 77.Costa M, et al. Crystal structures of a group II intron lariat primed for reverse splicing. Science. 2016;354:aaf9258. doi: 10.1126/science.aaf9258. [DOI] [PubMed] [Google Scholar]
- 78.Csuros M, et al. Extremely intron-rich genes in the alveolate ancestors inferred with a flexible maximum-likelihood approach. Mol Biol Evol. 2008;25:903–911. doi: 10.1093/molbev/msn039. [DOI] [PubMed] [Google Scholar]
- 79.Csuros M, et al. A detailed history of intron-rich eukaryotic ancestors inferred from a global survey of 100 complete genomes. PLoS Comput Biol. 2011;7:e1002150. doi: 10.1371/journal.pcbi.1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de Lange T. A loopy view of telomere evolution. Front Genet. 2015;6:321. doi: 10.3389/fgene.2015.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gladyshev EA, Arkhipova IR. Telomere-associated, endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc Natl Acad Sci USA. 2007;104:9352–9357. doi: 10.1073/pnas.0702741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pardue ML, DeBaryshe PG. Retrotransposons that maintain chromosome ends. Proc Natl Acad Sci U S A. 2011;108:20317–20324. doi: 10.1073/pnas.1100278108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fulcher N, et al. If the cap fits, wear it: an overview of telomeric structures over evolution. Cell Mol Life Sci. 2014;71:847–865. doi: 10.1007/s00018-013-1469-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gillis AJ, et al. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 85.Enyeart PJ, et al. Biotechnological applications of mobile group II introns and their reverse transcriptases: gene targeting, RNA-seq, and non-coding RNA analysis. Mob DNA. 2014;5:2. doi: 10.1186/1759-8753-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zheng G, et al. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods. 2015;12:835–837. doi: 10.1038/nmeth.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]