Abstract
Well-studied and promising antimicrobial peptides (AMPs), with potent bactericidal activity, in vitro, have yet to have a significant impact in human medicine beyond topical applications. We previously showed that interactions of AMPs with concentrated human erythrocytes inhibit many of them, and suggested that screens and assays should be done in their presence to mimic host cell inhibition. Here, we use AMPs to characterize the activity of proteases that are associated with human erythrocytes. The representative AMPs, ARVA and indolicidin, are degraded significantly during incubation with dilute, washed erythrocytes and yield a variety of degradation products, suggesting significant exopeptidase activity. Comparison of these fragments with those obtained from incubation with serum shows that the proteolytic activity associated with cells yields unique products that are not explained by residual serum proteases. By separately testing the membrane and cytosolic fractions, we show that erythrocyte proteolytic activity is found only in the cytosol. Finally, we incubated a diverse cross-section of natural and synthetic linear AMPs with human erythrocyte cytosolic extracts and observed degradation of all of them. These results show that, in addition to cell binding, proteolysis can also contribute significantly to host cell inhibition of AMPs in vitro and possibly also in vivo.
Keywords: red blood cell, antibiotic, peptidase, proteolysis
Graphical Abstract
Introduction
Peptide drug candidates possess several characteristics that complicate their development as therapeutics. A non-exhaustive list includes issues of bioavailability (1), cytotoxicity (2), rapid metabolism via degradation (3) and systemic clearance (4). Despite these issues, a number of peptide drugs, including the HIV fusion inhibitor enfuvirtide (5) and the peptide hormones insulin and glucagon, used in the control of diabetes mellitus (6), play important roles in the clinical management and treatment of disease. Further, as of 2015, more than 100 peptide therapeutics were in clinical trials for wide spectrum of clinical conditions, a number that is expected to increase in the future (7). It has also been shown that cell-penetrating peptides (CPPs) and other peptide delivery vehicles may be used to deliver macromolecules such as oligonucleotide (8) and promising small molecules to their intracellular targets (9). Results like these combined with a vast available sequence space will continue to drive peptide-based biomedical research.
One of the more promising classes of peptide drugs are the antimicrobial peptides (AMPs), which are key effectors of innate immunity for organisms in many clades of the animal kingdom (10). Since their discovery several decades ago (11, 12), researchers have studied whether their potent antimicrobial and immunomodulatory effects could be harnessed and amplified for applications in clinical medicine. Despite the vast amount of research that has been conducted and the several thousand active sequences, both natural and synthetic, that have been discovered (13), progress thus far is not encouraging, as there are few FDA approved antimicrobial peptides (14). As opposed to traditional antibiotics that often target specific enzymes and biosynthetic processes, AMPs usually exert their antimicrobial effects through interactions with and destabilization of the bacterial cytoplasmic membrane (2). In favor of AMPs, this less specific mechanism often conveys broad-spectrum antimicrobial activity and is not as susceptible to the emergence of resistant phenotypes (15). Conversely, this lack of specificity can lead to off-target effects, including the potential to harm symbiotic microflora, cytotoxicity to eukaryotic cells, and loss of activity under physiological conditions, complicating their transition into clinical medicine (16).
Recently, we investigated a critical barrier to the clinical activity of antimicrobial peptides: interactions with host cells, modelled with concentrated human erythrocytes (17). While it is well established that eukaryotic cells may be permeabilized by certain AMPs, we sought to show that, for at least some AMPs, interactions with host cells can inhibit their activity against prokaryotes when both cell types are present, at least when AMPs encounter human cells prior to bacteria. By comparing the behavior of the L and D amino acid isomers of the synthetic, 12-residue peptide, ARVA (18), along with several other well-studied AMPs, we demonstrated that host cell interactions, including binding, have the potential to be major contributors to AMP activity loss in vivo. We concluded that the processes of screening for, and characterization of, potentially clinically active AMPs could be improved by consistently including concentrated host cells in these assays, creating an environment more representative of that encountered in vivo. Continuation of this work has revealed that even peptides that do not strongly associate with eukaryotic membranes may be subject to diminished antimicrobial efficacy in the presence of host cells. Here, we show that proteolysis can also be a significant factor in AMP activity loss in our suggested model system of human erythrocytes plus AMPs and bacteria. This activity cannot be attributed to serum proteases, which are the most common concern for AMPs and peptide drugs in general. Instead, we demonstrate that proteolytic degradation of peptides in our system was caused by unique enzymes directly associated with the cytosol of human erythrocytes.
While the presence of proteolytic activity associated with RBCs is well established, it has not been examined in the context of antimicrobial peptides, or peptide therapeutics in general. We show here that these proteases are able to degrade antimicrobial peptides with seemingly low sequence specificity, as evidenced by the variety of fragments produced. Further, we show definitively that this activity is directly associated with the erythrocytes themselves and is not a residual component of serum. The proteolytic activity is initially sequestered in the cytoplasm and thus may be innocuous to many potential peptide therapeutics, but given the propensity for AMPs to induce hemolysis, even at low levels, these proteases have the potential to interfere with activity in in vitro model systems and perhaps also in vivo. Finally, we show that a wide variety of AMP sequence variants, including synthetic and natural AMPs, are susceptible to degradation by this proteolytic activity, although the degradation rates vary substantially.
Materials and Methods
Peptides
All peptides used in this study were synthesized using solid-phase FMOC chemistry and purified to >95% by Bio-synthesis Inc. (Lewisville, TX). Peptides were dissolved in 0.025% acetic acid solution and concentrations were determined by absorbance at 280 nm, if possible. In the absence of tryptophan or tyrosine residues, concentrations were determined by measured weight of peptide and volume of solvent.
Red blood cell and serum preparation
Human red blood cells and serum were purchased from Interstate Bloodbank (Memphis, TN). Red blood cells were from O+ donors and were collected in a citrate phosphate dextrose (CPD) anticoagulant solution. Upon receipt, the cells were washed three times with sterile PBS and stored at 4° C at a density of 5×109 cells/mL until use. Before experiments were performed, cells were diluted to a working concentration of 1.33×108 cells/mL and washed three additional times, collecting and storing the supernatant with each wash. Serum was devoid of clotting factors (OTC) and was stored at −20° C until use. It was sterile filtered with a 0.22 micron vacuum filter to remove any particulates and precipitates before dilution to 2% of the initial concentration in PBS.
Preparation of red blood cell cytosolic extracts and membrane ghosts
Red blood cell ghosts were prepared per the method published by Steck and Kant (19). Approximately 1 mL of washed, packed RBCs were added to a 50-mL centrifuge tube and 40 mL of cold, 5 mM phosphate buffer (pH 8.0) was added to lyse the cells. The lysed cells were rocked while incubating on ice for 30 minutes and then centrifuged at 18,000xg to separate the membrane fraction from the cytosolic components. At this point, the supernatant was collected to serve as the cytosolic extract for analysis. The membrane fractions were subjected to two additional rounds of washing with cold, 5 mM phosphate buffer and centrifugation at 18,000xg. The membrane fractions were then resuspended in warm PBS (pH 8.0) and incubated with rocking for 45 minutes at 37° C to reseal the membranes. The resealed membranes were washed three times with room temperature PBS and centrifuged at 18,000xg following each wash. Both cytosolic and membrane fractions were stored at 4° C until use.
Peptide degradation experiments with cells
For each time point, a washed cell suspension was mixed with peptide to give final concentrations of 20 μM peptide and 1.0×108 cells/mL. All experiments contained 5 μM FMOC-aspartic acid as an internal HPLC standard. The mixtures were incubated at 37° C with agitation. At the appropriate time points, the mixtures were centrifuged at 1000xg to pellet the RBCs and the supernatant was removed for analysis by HPLC.
Peptide degradation with erythrocyte ghosts and cytosolic extracts
Membrane ghosts or cytosolic extracts were prepared as above. To match the cell-based experiments, peptide and cytosol or ghosts were mixed to give 20 μM peptide and a final concentration of cytosol/ghosts equivalent to 1.0×108 cells/mL. The mixtures were incubated at 37°C with agitation. At the appropriate time points, the mixtures were analyzed via HPLC.
Peptide degradation with serum
Serum was diluted to 2% in PBS to match the dilution of the cells used in other experiments. As with other experiments, the final peptide concentration was 20 μM. The mixtures were incubated at 37° C with agitation. At the appropriate time points, the mixtures were analyzed via HPLC.
HPLC analysis of peptide degradation
Analysis of peptides and degradation products was performed using reversed-phase chromatography. The stationary phase was a 100 mm × 4.6 mm C-18 column from Kromasil (Bohus, Sweden). The mobile phase was composed of a gradient of distilled water (+0.1% trifluoroacetic acid) and acetonitrile (+0.1% trifluoroacetic acid) with a flow rate of 1 mL/min. Where possible, peptide and peptide fragments were analyzed using tryptophan fluorescence (285ex/340em). In the absence of tryptophan residues, peptide was analyzed by absorbance at 220 nm.
Fragment collection and mass spectrometry
HPLC was used to collect potential peptide fragments eluted during analysis. Each potential fragment was subjected to MALDI-TOF mass spectrometry using a α-Cyano-4-hydroxycinnamic acid (CHCA) as a matrix. Masses observed in this analysis were compared to a list of all potential cleavage products. An error of less 0.5 Da was considered acceptable for peptide identification.
Results
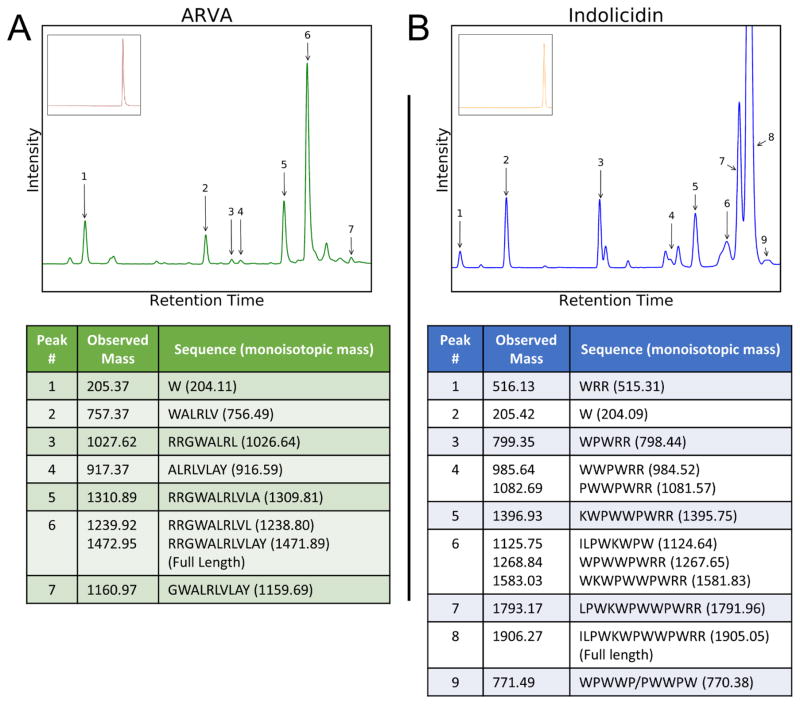
We previously reported that even a few minutes of preincubation of some antimicrobial peptides (AMPs) with human erythrocytes caused severe inhibition of bactericidal activity (17). We further showed for a synthetic 12-residue AMP called ARVA, host cell binding can account for the measured inhibition. However, we also observed the emergence of degraded peptide fragments upon extended incubation of ARVA and other AMPs with human red blood cells (RBCs). In the work presented here, we further pursue this phenomenon. We first incubated the well-studied peptides, ARVA (18) and indolicidin (20) (Table 1), with 1×108 red blood cells/mL (2% physiological) for four hours, removing the cells via centrifugation, and analyzing the supernates with reversed-phase HPLC. The resulting chromatographs are displayed in figure 1 and show several peaks in addition to the main peptide peak that we suspected were fragments of the full-length peptide. These potential fragments were isolated using HPLC and analyzed and identified by MALDI-TOF mass spectrometry. Components of these isolated peaks that matched theoretical peptide fragments are reported in figure 1. The results are highly supportive of proteolytic activity resulting from the incubation of peptide with RBCs. Additionally, the large number of fragments apparent for both peptides suggests either a lack of sequence specificity or the presence of multiple active proteases. Based on proteomics studies, both possibilities are realistic (21, 22). The fragments observed suggest at least the presence of both C-terminal and N-terminal exopeptidases. An interesting observation is that degradation appears to proceed more rapidly from the terminus that does not contain charged residues (C-terminal for ARVA, N-terminal for indolicidin). It is unclear whether this behavior is a result of the charged residues actively preventing cleavage at their respective termini or if fragments containing the charged residues are more likely to be observed due to better solubility. It is important to note that our method of monitoring degradation using HPLC with tryptophan fluorescence means that we can rule out artifact peaks because we can show that there are few, if any, Trp-containing peaks in blank samples or samples incubated with D-amino acid ARVA or indolicidin (Figure 1). However, it also means that some fragments will not be detected because they do not contain tryptophan. Also, the data in Figure 1 represent a single time point where some fragments may have already disappeared, while others may have yet to form. Importantly, the generation of a comprehensive catalog of all cleavage fragments was not the goal of this work, and the results and conclusions are not affected by missed fragments.
Table 1.
The sequences of the peptides used in this work.
| Peptide | Sequence |
|---|---|
| ARVA | RRGWALRLVLAY-NH2 |
| Indolicidin | ILPWKWPWWPWRR-NH2 |
| Cecropin A | KWKLFKKIEKVGQNIRDGIIKAGPAVAVVGQATQIAK-NH2 |
| Magainin 2 | GIGKFLHSAKKFGKAFVGEIMNS |
| VVRG | WVLVLRLGY-NH2 |
| MSI-78 | GIGKFLKKAKKFGKAFVKILKK-NH2 |
| MelP5 | GIGAVLKVLATGLPALISWIKAAQQL-NH2 |
| WLBU2 | RRWVRRVRRWVRRVVRVVRRWVRR |
Figure 1.
Peptide degradation by host cells. Antimicrobial peptides ARVA (A), and indolicidin (B) at 20 μM were incubated with 1×108 human red blood cells/mL for 4 hours at 37°C. The cells were centrifuged from solution and the supernatant was analyzed using RP HPLC. The peaks were detected by monitoring tryptophan fluorescence using excitation at 285 nm and emission at 340 nm. Peaks were collected and subjected to MALDI-TOF mass spectrometry to obtain identities by comparison of observed to expected fragments of the peptides. Peaks without numbers are degradation products that could not be definitively identified by MALDI-TOF. Insets: Control experiments were performed with variants of ARVA and indolicidin synthesized using all D-amino acids. The HPLC chromatographs reveal no peptide degradation or unexplained peaks.
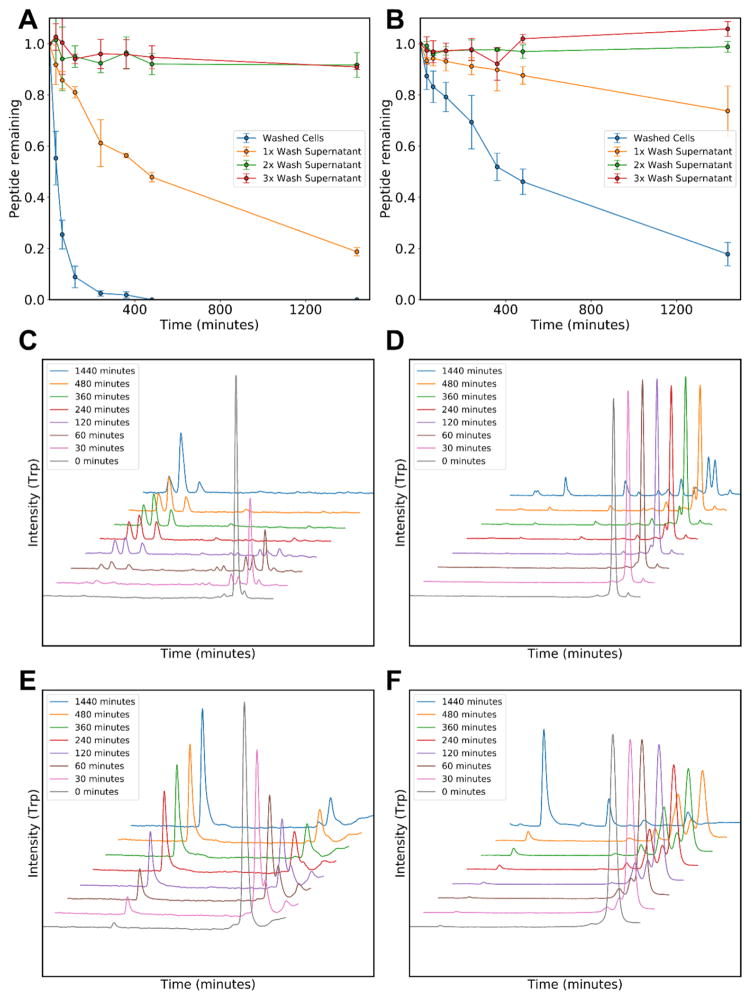
Having confirmed the presence of proteolytic activity in the RBC preparations, we sought to determine whether this activity could be attributed to the presence of residual serum proteases. In figures 2A and 2B, we incubated ARVA and indolicidin with the RBC wash supernates at each phase of RBC preparation (all preparations are washed at least three times). The data suggest that while there is some extracellular proteolytic activity before the cells are washed, this activity disappears in the second and third wash supernates where more than 90% of both peptides are recovered, even after 24 hours of incubation. Conversely, even after three washes, when either peptide is incubated with red blood cells (2% physiological), degradation occurs rapidly and does not decrease with the number of washes. Therefore, the observed proteolytic activity remains associated with the cells upon washing. We note that the rates of degradation are variable between the two peptides with intact ARVA being completely absent from solution after eight hours and some indolicidin being retained, even after 24 hours. While the number of fragments presented in figure 1 suggested a lack of sequence specificity, these data suggest that sequence may have an important role in the overall rate of degradation.
Figure 2.
Testing for residual serum protease activity. Antimicrobial peptides ARVA (A) and indolicidin (B) at 20 μM were incubated at 37°C with either the wash supernatants of 1×108 RBCs/mL or directly with RBCs after washing. The intact peptide remaining was assayed with HPLC after cells are centrifuged. C,E: ARVA HPLC traces, using tryptophan fluorescence detection, during incubation with RBCs (C) or with 2% serum (E). D,F: Indolicidin HPLC traces using tryptophan fluorescence detection during incubation with RBCs (D) or with 2% serum (F).
To further differentiate the pattern of serum protease degradation from the proteolytic activity that we observed in conjunction with RBCs, we performed parallel experiments with 2% human serum. Figures 2C and 2D show ARVA and indolicidin degradation with time in the presence of RBCs, detailing the numerous fragments that emerge and disappear throughout the 24-hour incubation. Nearly identical experiments were performed with human serum (Figs. 2E and 2F) and show a different pattern of peptide degradation. We collected the serum degradation products and identified them using MALDI-TOF mass spectrometry. Although, we are likely not capturing all cleavage products, the results in table 2 show that unique products are associated either with serum protease activity or with RBC-associated protease activity. For example, there is almost no overlap between serum and RBC degradation products of ARVA and only partial overlap between the degradation products of indolicidin (Table 2). A particular additional activity of note in serum is the removal of a single arginine residue from the N-terminus of ARVA or the C-terminus of indolicidin. These results show that the proteolytic activity observed in association with RBCs is distinct and unique from serum proteases.
Table 2.
Indolicidin and ARVA peptide fragments identified after incubation with human serum or RBCs. Termini that are still intact are marked by as asterisk. Blank cells indicate that a particular fragment was not observed in one set of experiments. These fragment lists are not meant to be comprehensive, but rather show a population of detectable species that are present at one time point during degradation.
| ARVA fragments observed | Indolicidin | ||
|---|---|---|---|
| Serum | RBC Cytosol | Serum | RBC Cytosol |
| RGWALRLVLAY* | *ILPWKWPWWPWR | ||
| *RRGWALRLVLA | LPWKWPWWPWR | ||
| *RRGWALRLVL | LPWKWPWWPWRR* | LPWKWPWWPWRR* | |
| WALRLVLAY* | WKWPWWPWRR* | WKWPWWPWRR* | |
| GWALRLVLAY* | WKWPWWPWR | ||
| *RRGWALRL | KWPWWPWRR* | KWPWWPWRR* | |
| RGWALRLVL | WPWWPWRR* | WPWWPWRR* | |
| WALRLVLAY* | *ILPWKWPW | *ILPWKWPW | |
| ALRLVLAY* | ALRLVLAY* | PWWPWRR* | |
| GWALRLVL | WPWWPWR | ||
| LRLVLAY* | WPWWP/PWWPW | ||
| *RRGWALR | WPWRR* | ||
| *RRGWAL *RGWALR |
WWPWRR* | WWPWRR* | |
| WALRLV | WWPWR | ||
| W | W | WRR* | |
| W | W | ||
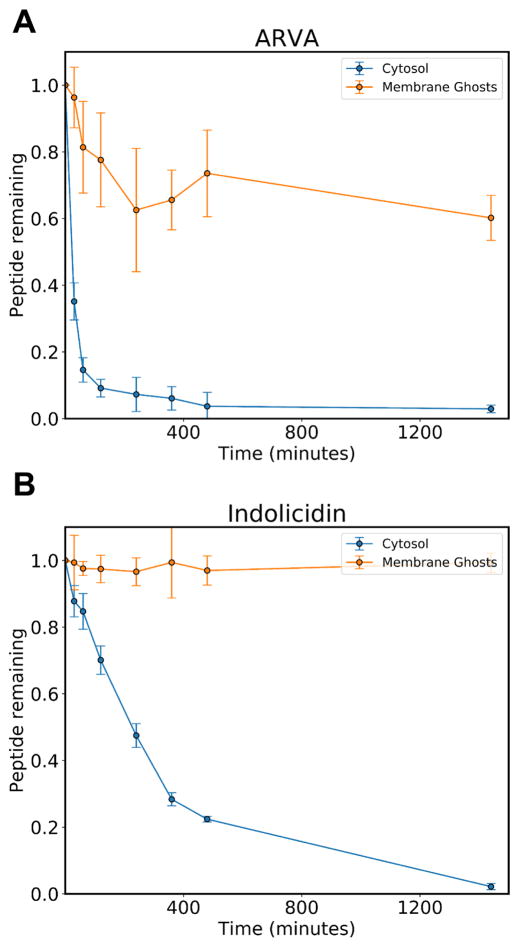
Having shown that peptide degradation is associated with erythrocytes, we sought to determine the subcellular localization of the proteolytic activity. Given that antimicrobial peptides have a high affinity for the membrane (2), it seemed possible that the proteolytic activity was cell surface associated, either transmembrane or surface-bound. However, because antimicrobial peptides can be lytic when acting on eukaryotic cells, it is possible that intracellular proteins were being released by peptide-dependent lysis. The latter effect would also occur due to background autolysis which occurs constantly at a low rate in RBC preparations. To separate the plasma membrane from the intracellular components, we used an RBC ghost preparation protocol (19) in which cells are lysed with a hypotonic buffer and high speed centrifugation is used to remove the membrane fragments from the suspension. This procedure enables separate collection of membranes and supernate containing intracellular proteins and other components. Repeated washes deplete any remaining intracellular components and a final resealing step (19) returns the membrane to a cell-like morphology. Once the cytosol and membrane solutions were collected, they were diluted to match the conditions of prior incubations with intact cells, equivalent to 1×108 cells/mL (2% physiological). We then performed time course experiments, monitoring the amount of peptide that was degraded over 24 hours at 37°C. The results are presented in figure 3 for ARVA (A) and indolicidin (B) and show convincingly that peptide degradation can be attributed to intracellular components. The membrane fractions have little, if any, detectable protease activity. Given this fact, it is not surprising that degradation is much more rapid in the presence of pure cytosolic extracts relative to intact cells, where only a fraction of cells would be expected to release their contents due to lysis. We note the complete lack of degradation of indolicidin upon incubation with ghost membrane, but the apparent, partial loss of ARVA. The absence of any cleavage fragments suggest that the loss of ARVA can be explained by strong membrane binding. In fact, the amount of peptide not recovered agrees very well with our previous measurements of RBC membrane binding (17). Indolicidin has a much lower affinity for eukaryotic membranes than ARVA (unpublished observation).
Figure 3.
Cellular localization of protease activity. The antimicrobial peptides ARVA (A) and indolicidin (B) at 20 μM were incubated with the cytosolic extract of human red blood cells or the isolated, washed membrane fractions. The fraction of intact peptide remaining was assayed by HPLC. The results show that proteolytic activity is present in the cytosolic fraction, but not the membrane fraction. The small peptide loss in experiments where ARVA is incubated with membrane fractions can be explained by strong membrane binding.
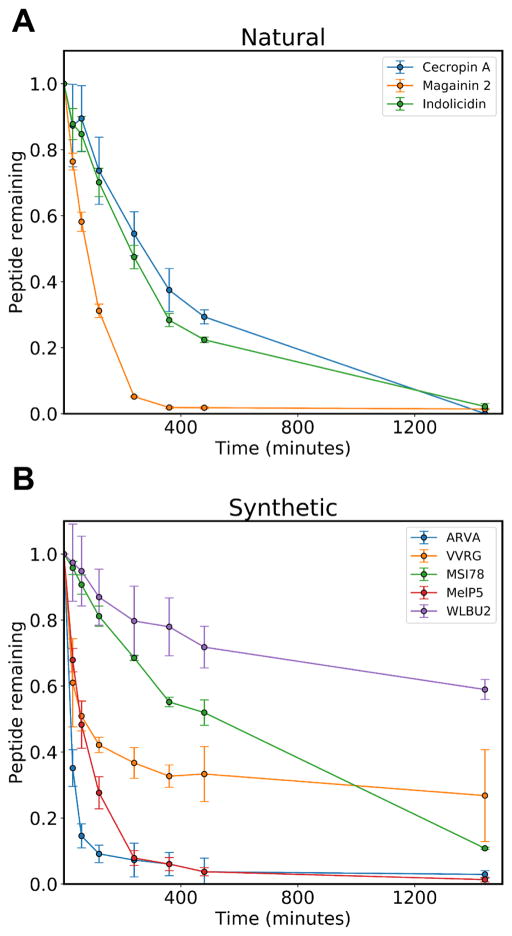
We expanded our study of AMP degradation by intracellular RBC proteases to a panel of natural (indolicidin, cecropin A, and magainin 2, Fig. 4A) and synthetically derived (ARVA, VVRG, MSI78, MelP5, and WLBU2, Fig. 4B) AMPs. These peptides represent a subset of those studied in our prior work on AMP-RBC interactions with a few omissions due to difficulty with chromatographic separation (17). Because these peptides cause different amounts of hemolysis, we standardized these experiments by incubating peptides in cytosolic extracts rather than with intact cells. The results indicate that all peptides studied were degraded significantly over the 24-hour observation period, however, the rate of degradation was highly variable between peptides. While the RBC concentration is 2% of expected physiological values, 100% of the cells were lysed for these experiments. Therefore, this experiment is equivalent to 2% hemolysis of physiologically concentrated RBCs, or 2% hemolysis in vivo. Even a reduction in hemolysis to 0.2%, which is low enough to be difficult to accurately measure, would nonetheless lead to substantial degradation for many of the peptides studied here.
Figure 4.
Peptide susceptibility to degradation. A panel of natural (A) and synthetic (B) antimicrobial peptides were incubated at 20 μM with the cytosolic extracts of 1×108 red blood cells/mL at 37°C. The fraction of intact peptide remaining was monitored by reverse phase HPLC.
A particularly interesting phenomenon in these experiments is the difference in stability between Magainin 2 and MSI-78. Magainin 2 is a naturally occurring AMP derived from the skin of the African clawed frog, Xenopus laevis (12), whereas MSI-78 is a synthetic peptide based on the magainin family and shares a substantial amount of sequence identity with Magainin 2 (23) (Table 1). The major difference between the two peptides is the substitution of five additional, cationic lysine residues throughout the peptide for polar, but uncharged residues in the parent sequence. These changes have a profound impact on the rates of degradation. The naturally occurring Magainin 2 is more than 50% degraded in under two hours while the synthetic variant does not reach this midpoint until longer than eight hours. Moreover, the natural peptide is completely degraded by the six hour time point whereas a small amount of synthetic peptide persists, even after twenty-four hours of incubation. An interesting note about MSI-78, is that in terms of success in clinical trials to treat human disease, this peptide has been the most successful. It has advanced to phase III trials for topical applications twice since its discovery, although it has yet to pass the final step of FDA approval (14). It is reasonable to hypothesize that a contributing factor to its high level of success is its relative resistance to proteolytic degradation.
Another peptide with a slow degradation profile is the synthetic AMP, WLBU2. This peptide was designed as an idealized amphipathic helix with robust hydrophobic (8 valines, 3 tryptophans) and polar faces (13 arginines) (24). Like MSI-78, this peptide carries a large positive charge, which based on available data, could potentially be a stability enhancing characteristic. Additionally, the bulky, beta-branched valine and aromatic tryptophan residues might both sterically obscure access to the peptide backbone. We initially became interested in this peptide due to reports that it retained activity in the presence of concentrated RBCs (24), something that we had not observed with other peptides. When tested in our model system, we did not observe full retention of activity as had been reported previously, however, we note that it did perform better than most of the other peptides studied (17). As with MSI-78, it seems likely that increased stability to proteases may be responsible for its above average performance.
Discussion
Biological stability and availability are primary concerns in the development of any potential therapeutic, but are especially challenging for peptide-based drugs (1). In the context of systemically administered peptides, serum proteases and serum protein binding are common concerns. In previous work with AMPs, we showed that peptide binding to host cells must also be considered when assessing the therapeutic potential of AMPs (17). We suggested that AMP screening, selection, and characterization assays should be modified to include the presence of host cells to create a more realistic model of the in vivo barriers to clinical activity. We further suggested that concentrated human erythrocytes are a convenient and meaningful mimic of host cells. Here, we expand on that work and show that erythrocyte intracellular proteases also negatively impact the efficacy of AMPs and must be considered in the design of peptides and in the development of experimental protocols.
The presence of proteolytic activity associated with the red blood cell has been known for some time (25, 26). Before the discovery of the proteasome and substantial evidence supporting its role as the major proteolytic machinery of the eukaryotic cell, a number of reports described the existence of an ATP-independent proteolytic activity in the cytosol of the human erythrocyte (27–30). It was speculated that an enzyme complex or pathway was responsible for the degradation of oxidized and damaged proteins, an idea that makes sense given the physiological role of the red blood cell and the lack of protein turnover after maturation (27, 29). The size of the complex reported in these early studies (670 – 700 kDa) (29) and a recent study on the activity of the 20S proteasomal subunit in mature erythrocytes (31) suggest that the proteasome is likely responsible for at least a portion of the observed proteolytic activity. In addition to the proteasome, proteomics-based studies have revealed the likely presence of several other proteolytic enzymes (21, 22); although to our knowledge, this has not been verified biochemically. The results of this work show that a variety of peptide sequences are susceptible to degradation by enzymatic activity sequestered within the RBC cytosol. The diversity of cleavage products suggests the activity of several unique proteases, or proteases with multiple activities and the pattern of cleavage suggests that exopeptidases are responsible for at least some of the degradation. Furthermore, the total sequestered RBC protease activity is similar to the total serum protease activity under physiological conditions.
The implications of this work for in vitro experiments are significant. The information presented here is critical for design of experiments in which RBCs are used as model systems. For example, human red blood cells are an important model cell type in many fields and are specifically used for the study of disease states like malaria, anemia and sickle cell disease (32, 33). The information provided here about the cytosolic proteases of RBCs should be especially important for the field of antimicrobial peptides, because many are susceptible to proteolysis and because RBCs have long been used as model systems to assess lytic and toxic activities of AMPs. As we show here, even fresh RBCs, used as a model system, have a significant rate of autolysis when stored in phosphate buffered saline and will contain released proteases unless thoroughly washed immediately prior to use. In addition, hemolysis induced by AMPs in vitro, which is common, will also release cytosolic proteases. While complete digestion of an AMP will eventually eliminate activity, protease-dependent artifacts could include either loss or gain of hemolytic or antibacterial activities due to the unpredictable behavior of the intermediate products.
The implications of RBC proteases for peptide drugs in vivo are less certain. With the exception of disease states like sickle cell disease, mature human erythrocytes do not display significant levels of endo- or exocytosis (34), or show significant amounts of hemolysis. Thus, release of cytosolic proteases is not normally expected. However, in acute bacteremia, or septicemia, conditions in which a systemically active AMP would be useful, the invading microorganism or the innate immune response could cause hemolysis, and the release of intracellular proteases (35, 36). There are also many other human disease states that cause abnormally high levels of hemolysis (37). The utility of a systemically administered antimicrobial peptide could be compromised in individuals with any of these conditions, for any peptide drug that is susceptible to RBC-associated proteases. For AMPs, perhaps the most likely source of hemolysis in vivo, is the peptide itself. In the in vitro experiments performed here, the concentration of RBCs was 1×108 RBCs/mL, 2% of human physiological concentration. It has been shown previously that both MIC values and hemolysis are directly dependent on cell density (38, 39). However, as cell counts are increased towards physiological concentrations, and fractional hemolysis decreases, the total pool of cytosolic proteases also increases. Thus, it remains possible that the amount of hemolysis induced by a therapeutic AMP at the MIC at physiological cell counts, while small, would be sufficient to cause release of an impactful quantity of intracellular proteases. Furthermore, some AMPs and other membrane active peptides can translocate spontaneously across lipid bilayers and access the cell cytosol without causing cell lysis (40, 41). Given the presence of concentrated proteolytic activity in the cytosol, translocation of susceptible peptides into RBCs will lead to their rapid degradation, even when no hemolysis has occurred.
Since the inception of peptides as therapeutics, there have been efforts to increase their stability in the biological context. To avoid or reduce proteolysis, researchers can engineer stability through amino acid composition changes, substitution of non-natural amino acids, termini modification, crosslinking or cyclization (42). An approach that can be especially effective is the substitution of individual L-amino acids with D-amino acids or those with side chains that do not occur naturally (43). A scanning approach that is sometimes applied is sequential deletion of one or a combination of residues to find a minimally active motif and minimize the opportunity for cleavage. A study by Nguyen and colleagues on the degradation of short, tryptophan-rich AMPs in serum examined, among other things, the effect of terminus modification on peptide stability (44). Interestingly, they discovered that while either N-terminal acetylation or C-terminal amidation were minimally effective alone, together, they acted synergistically and made a modest contribution to increased stability (44). A more successful approach employed in that study and in others is end-to-end cyclization, which eliminates potential sites of cleavage for exopeptidases and may alter the availability of internal sites (44, 45). Disulfide crosslinking also protects peptides from degradation (46). For example, we recently reported serum degradation experiments on a disulfide-crosslinked Ebola virus peptide with two conserved cysteine residues and observed nearly complete protection of the crosslinked core peptide after 24 hours (47). Similarly, one of the largest naturally-occurring AMP families, the defensins, are characterized by their multiple conserved cysteine residues and compact, disulfide crosslinked structures (48) and are probably protease resistant.
Given the amount of research already available on the topic of serum protease susceptibility, it is almost certain that a viable strategy also exists for protecting L-amino acid peptides from the RBC-associated proteases described herein. However, neither the determinants of proteolytic susceptibility nor the determinants of antimicrobial activity are completely understood. Thus, modifications must be tested individually and design changes must be made by trial and error, or by screening. The work reported here indicates that L-amino acid AMPs or variants will need to be tested for stability against serum proteases, as usual, but also against the cytosolic proteases of erythrocytes, since most AMPs cause at least some low level hemolysis (17). Ultimately, all D-amino acid AMPs may be a preferable approach as they will give complete protease resistance, while having comparable or identical bactericidal activity to L-amino acid variants of the same peptide (17). Developing a more complete understanding of antimicrobial peptide activity in vivo and of the barriers between the lab bench and clinic is of vital importance for the advancement of translational AMP research. At present, it may be more important than ever as the spread of antibiotic resistant pathogens accelerates and seriously threatens human health both inside and outside of the hospital (49). Recent research (17, 38), in which antimicrobial peptides are studied in the context of their interactions with both eukaryotic cells and prokaryotic cells, may suggest a critical paradigm shift that will bring antimicrobial peptides to the clinic. Taken together, our previous report (17) on erythrocyte-dependent AMP inhibition, and this work on erythrocyte intracellular proteases, reveal the dual effects of cell binding and proteolysis on AMP activity in the presence of erythrocytes. Both effects are significant and are expected to be important, in vitro and in vivo. These results provide further support for our previously stated (17) conclusion that host-cell dependent impediments to clinical utility should be an integral part of the entire AMP design, discovery and characterization process. Screens and assays used to identify and characterize AMPs should include both host and target cells from the beginning of the discovery and development pipeline. This will allow researchers to more rapidly identify peptide drug candidates that have true clinical potential, and that are less likely to have critical in vivo impediments to activity, including those related to host cell binding and host cell-dependent proteolysis.
Highlights.
The cytosol of human erythrocytes contains broad exopeptidase activity
The proteolytic activity in the cytosol of erythrocytes is unique from serum enzymes
L-amino acid antimicrobial peptides are highly susceptible to degradation in vitro
Erythrocyte proteolysis must be accounted for in the design of experiments
Acknowledgments
This work is funded by the National Institutes of Health (R21AI119104).
Abbreviations
- RBC
red blood cell
- AMP
antimicrobial peptide
Footnotes
Author contributions. C. Starr designed and conducted the experiments. C. Starr and W. Wimley analyzed data and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Morishita M, Peppas NA. Is the oral route possible for peptide and protein drug delivery? Drug Discov Today. 2006;11:905–910. doi: 10.1016/j.drudis.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 3.Mentlein R. Dipeptidyl-peptidase IV (CD26)-role in the inactivation of regulatory peptides. Regul Pept. 1999;85:9–24. doi: 10.1016/s0167-0115(99)00089-0. [DOI] [PubMed] [Google Scholar]
- 4.Leibach FH, Ganapathy V. Peptide transporters in the intestine and the kidney. Annu Rev Nutr. 1996;16:99–119. doi: 10.1146/annurev.nu.16.070196.000531. [DOI] [PubMed] [Google Scholar]
- 5.Lalezari JP, Henry K, O’Hearn M, Montaner JS, Piliero PJ, Trottier B, Walmsley S, Cohen C, Kuritzkes DR, Eron JJ, Chung J, DeMasi R, Donatacci L, Drobnes C, Delehanty J, Salgo M. Enfuvirtide, an HIV-1 Fusion Inhibitor, for Drug-Resistant HIV Infection in North and South America. New Engl J Med. 2003;348:2175–2185. doi: 10.1056/NEJMoa035026. [DOI] [PubMed] [Google Scholar]
- 6.Cramer J. A systematic review of adherence, treatment satisfaction and costs, in fixed-dose combination regimens in type 2 diabetes. Diabetes Care. 2004;27:1218–1224. doi: 10.1185/03007995.2011.570745. [DOI] [PubMed] [Google Scholar]
- 7.Fosgerau K, Hoffmann T. Peptide therapeutics: Current status and future directions. Drug Discov Today. 2015;20:122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Langel U, Gestin M, Kurrikoff K. Recent in vivo advances in cell-penetrating peptide-assisted drug delivery. Expert Opin Drug Deliv. 2016;13:373–387. doi: 10.1517/17425247.2016.1125879. [DOI] [PubMed] [Google Scholar]
- 9.Kauffman WB, Fuselier T, He J, Wimley WC. Mechanism Matters: A Taxonomy of Cell Penetrating Peptides. Trends Biochem Sci. 2015;40:749–764. doi: 10.1016/j.tibs.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 11.Steiner H, Hultmark D, Engström A, Bennich H, Boman HG. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature. 1981;292:246–248. doi: 10.1038/292246a0. [DOI] [PubMed] [Google Scholar]
- 12.Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci. 1987;84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44:1087–1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox JL. Antimicrobial peptides stage a comeback. Nat Biotechnol. 2013;31:379–382. doi: 10.1038/nbt.2572. [DOI] [PubMed] [Google Scholar]
- 15.Wimley WC. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem Biol. 2010;5:905–917. doi: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marr AK, Gooderham WJ, Hancock RE. Antibacterial peptides for therapeutic use: obstacles and realistic outlook. Curr Opin Pharmacol. 2006;6:468–472. doi: 10.1016/j.coph.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Starr CG, He J, Wimley WC. Host Cell Interactions Are a Significant Barrier to the Clinical Utility of Peptide Antibiotics. ACS Chem Biol. 2016;11:3391–3399. doi: 10.1021/acschembio.6b00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rathinakumar R, Walkenhorst WF, Wimley WC. Broad-spectrum antimicrobial peptides by rational combinatorial design and high-throughput screening: The importance of interfacial activity. J Am Chem Soc. 2009;131:7609–7617. doi: 10.1021/ja8093247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steck TL, Kant JA. Preparation of impermeable ghosts and inside-out vesicles from human erythrocyte membranes. Methods Enzymol. 1974;31:172–180. doi: 10.1016/0076-6879(74)31019-1. [DOI] [PubMed] [Google Scholar]
- 20.Selsted ME, Novotny MJ, Morris WL, Tang YQ, Smith W, Cullor JS. Indolicidin, a novel bactericidal tridecapeptide amide from neutrophils. J Biol Chem. 1992;267:4292–4295. [PubMed] [Google Scholar]
- 21.Roux-Dalvai F, Gonzalez de Peredo A, Simo C, Guerrier L, Bouyssie D, Zanella A, Citterio A, Burlet-Schiltz O, Boschetti E, Righetti PG, Monsarrat B. Extensive Analysis of the Cytoplasmic Proteome of Human Erythrocytes Using the Peptide Ligand Library Technology and Advanced Mass Spectrometry. Mol Cell Proteomics. 2008;7:2254–2269. doi: 10.1074/mcp.M800037-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Pasini EME, Kirkegaard M, Mortensen P, Lutz HU, Thomas AW, Mann M. In-depth analysis of the membrane and cytosolic proteome of red blood cells. Blood. 2006;108:791–801. doi: 10.1182/blood-2005-11-007799. [DOI] [PubMed] [Google Scholar]
- 23.Hallock KJ, Lee DK, Ramamoorthy A. MSI-78, an Analogue of the Magainin Antimicrobial Peptides, Disrupts Lipid Bilayer Structure via Positive Curvature Strain. Biophys J. 2003;84:3052–3060. doi: 10.1016/S0006-3495(03)70031-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deslouches B, Islam K, Craigo JK, Paranjape SM, Montelaro RC, Mietzner TA. Activity of the de novo engineered antimicrobial peptide WLBU2 against Pseudomonas aeruginosa in human serum and whole blood: Implications for systemic applications. Antimicrob Agents Chemother. 2005;49:3208–3216. doi: 10.1128/AAC.49.8.3208-3216.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarone G, Hamasaki N, Fukuda M, Marchesi VT. Proteolytic degradation of human erythrocyte band 3 by membrane-associated protease activity. J Membr Biol. 1979;48:1–12. doi: 10.1007/BF01869253. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto K, Marchesi VT. Purification and characterization of acid proteinase from human erythrocyte membranes. Biochim Biophys Acta. 1984;790:208–218. doi: 10.1016/0167-4838(84)90024-4. [DOI] [PubMed] [Google Scholar]
- 27.Fagan JM, Waxman L, Goldberg AL. Red blood cells contain a pathway for the degradation of oxidant-damaged hemoglobin that does not require ATP or ubiquitin. J Biol Chem. 1986;261:5705–5713. [PubMed] [Google Scholar]
- 28.Davies KJ, Goldberg AL. Proteins damaged by oxygen radicals are rapidly degraded in extracts of red blood cells. J Biol Chem. 1987;262:8227–8234. [PubMed] [Google Scholar]
- 29.Cells B, Pacifici RE, Salo DC, Davies KJA. Macroxyproteinase (M.O.P): A 670 kDa proteinase complex that degrades oxidatively denatured proteins in red blood cells. Free Radic Biol Med. 1989;7:521–536. doi: 10.1016/0891-5849(89)90028-2. [DOI] [PubMed] [Google Scholar]
- 30.Fagan JM, Waxman L. The ATP-independent pathway in red blood cells that degrades oxidant-damaged hemoglobin. J Biol Chem. 1992;267:23015–23022. [PubMed] [Google Scholar]
- 31.Neelam S, Kakhniashvili DG, Wilkens S, Levene SD, Goodman SR. Functional 20S proteasomes in mature human red blood cells. Exp Biol Med. 2011;236:580–591. doi: 10.1258/ebm.2011.010394. [DOI] [PubMed] [Google Scholar]
- 32.Cowman AF, Crabb BS. Invasion of red blood cells by malaria parasites. Cell. 2006;124:755–766. doi: 10.1016/j.cell.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Kato GJ, Gladwin MT, Steinberg MH. Deconstructing sickle cell disease: Reappraisal of the role of hemolysis in the development of clinical subphenotypes. Blood Rev. 2007;21:37–47. doi: 10.1016/j.blre.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knowles DW, Tilley L, Mohandas N, Chasis JA. Erythrocyte membrane vesiculation: Model for the molecular mechanism of protein sorting. Proc Natl Acad Sci. 1997;94:12969–12974. doi: 10.1073/pnas.94.24.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huycke MM, Spiegel CA, Gilmore MS. Bacteremia caused by hemolytic, high-level gentamicin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 1991;35:1626–1634. doi: 10.1128/aac.35.8.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McArthur HL, Dalal BI, Kollmannsberger C. Intravascular Hemolysis As a Complication of Clostridium Perfringens Sepsis. J Clin Oncol. 2006;24:2387–2388. doi: 10.1200/JCO.2005.03.4009. [DOI] [PubMed] [Google Scholar]
- 37.Dhaliwal G, Cornett PA, Tierney LM. Hemolytic anemia. Am Fam Physician. 2004;69:2599–2606. [PubMed] [Google Scholar]
- 38.Savini F, Luca V, Bocedi A, Massoud R, Park Y, Mangoni ML, Stella L. Cell-density dependence of host-defense peptide activity and selectivity in the presence of host cells. ACS Chem Biol. 2017;12:52–56. doi: 10.1021/acschembio.6b00910. [DOI] [PubMed] [Google Scholar]
- 39.Roversi D, Luca V, Aureli S, Park Y, Mangoni ML, Stella L. How many antimicrobial peptide molecules kill a bacterium? The case of PMAP-23. ACS Chem Biol. 2014;2014:2003–2007. doi: 10.1021/cb500426r. [DOI] [PubMed] [Google Scholar]
- 40.Marks JR, Placone J, Hristova K, Wimley WC. Spontaneous membrane-translocating peptides by orthogonal high-throughput screening. J Am Chem Soc. 2011;133:8995–9004. doi: 10.1021/ja2017416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fuselier T, Wimley WC. Spontaneous Membrane Translocating Peptides: The Role of Leucine-Arginine Consensus Motifs. Biophys J. 2017;113:835–846. doi: 10.1016/j.bpj.2017.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Werle M, Bernkop-Schnurch A. Strategies to improve plasma half life time of peptide and protein drugs. Amino Acids. 2006;30:351–367. doi: 10.1007/s00726-005-0289-3. [DOI] [PubMed] [Google Scholar]
- 43.Vlieghe P, Lisowski V, Martinez J, Khrestchatisky M. Synthetic therapeutic peptides: science and market. Drug Discov Today. 2010;15:40–56. doi: 10.1016/j.drudis.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen LT, Chau JK, Perry NA, de Boer L, Zaat SAJ, Vogel HJ. Serum stabilities of short tryptophan- and arginine-rich antimicrobial peptide analogs. PLoS One. 2010;5:1–8. doi: 10.1371/journal.pone.0012684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Molhoek EM, Van Dijk A, Veldhuizen EJA, Haagsman HP, Bikker FJ. Improved proteolytic stability of chicken cathelicidin-2 derived peptides by d-amino acid substitutions and cyclization. Peptides. 2011;32:875–880. doi: 10.1016/j.peptides.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 46.Fazio M, Oliveira V, Bulet P, Miranda M, Daffre S, Miranda A. Structure-Activity Relationship Studies of Gomesin: Importance of the Disulfide Bridges for Conformation, Bioactivities, and Serum Stability. Biopolymers. 2006;84:205–218. doi: 10.1002/bip.20396. [DOI] [PubMed] [Google Scholar]
- 47.He J, Melnik LI, Komin A, Wiedman G, Fuselier T, Morris CF, Starr CG, Searson PC, Gallaher WR, Hristova K, Garry RF, Wimley WC. Ebola Virus Delta Peptide is a Viroporin. J Virol. 2017 doi: 10.1128/JVI.00438-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 49.Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]