Abstract
Introduction
While lung cancer is generally thought to be environmentally provoked, anecdotal familial clustering has been reported suggesting there may be genetic susceptibility factors. We systematically tested whether germline mutations in eight candidate genes may be risk factors for lung adenocarcinoma.
Methods
We studied lung adenocarcinoma cases for whom germline sequence data had been generated as part of The Cancer Genome Atlas (TCGA) project, but that had not been previously analyzed. We selected eight genes, ATM, BRCA2, CHEK2, EGFR, PARK2, TERT, TP53, and YAP1, based on prior anecdotal association with lung cancer or genome wide association studies.
Results
Among 555 lung adenocarcinoma cases, we detected 14 pathogenic mutations in five genes; they occurred at a frequency of 2.5% and represented an odds ratio of 66 (95 confidence interval, 33 to 125, P<0.0001, chi-square test). The mutations fell most commonly in ATM (50%), followed by TP53, BRCA2, EGFR and PARK2. The majority (86%) of these variants had been reported in other familial cancer syndromes. Another 12 cases (2%) carried ultra-rare variants that were predicted to be deleterious by three protein prediction programs; these most frequently involved ATM and BRCA2.
Conclusions
A subset of lung adenocarcinoma patients, at least 2.5% to 4.5%, carries germline variants that have been linked to cancer risk in Mendelian syndromes. The genes fall most frequently in DNA repair pathways. Our data indicate that lung adenocarcinoma, similar to other solid tumors, contains a subset of patients with inherited susceptibility.
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide; it accounts for approximately 1.5 million deaths annually and cigarette smoke is the major culprit in 85–90% of these cases1. Several pieces of evidence point to inherited factors potentiating the carcinogenic effects of cigarette smoke. Rare familial forms of lung cancer have been described; they show a pattern consistent with autosomal dominant inheritance. The EGFR T790M mutation, PARK2 loss-of-function mutations, and, in Asian populations, a single gain-of-function variant, YAP1 R331W, have been associated with this phenotype2–4. In genome wide association studies, common BRCA2 and CHEK2 variants have been found to have strong association with lung cancer risk5. For lung adenocarcinoma, there is also a well-documented association with polymorphisms in the telomerase reverse transcriptase gene, TERT6. Moreover, for two well-recognized familial cancer syndromes, Li-Fraumeni syndrome and Ataxia Telengiectasia, lung cancer has been reported in smokers as well as never smokers7–9.
Methods
We sought to systematically examine whether the lung adenocarcinoma diagnosis enriches for individuals with germline pathogenic mutations in genes linked anecdotally to lung cancer risk, and, if present, to determine their prevalence. We focused on the lung adenocarcinoma histology since it has been reported to have familial clustering2, and it includes the largest group of never smokers (approximately 15%). Importantly, although the somatic landscape of lung adenocarcinoma has been studied and reported, the germline exome data have not10. We focused on eight candidate genes that had been associated with lung cancer cases in rare Mendelian syndromes or in genome wide association studies: ATM, BRCA2, CHEK2, EGFR, PARK2, TERT, TP53, and YAP1. After approval from the database of Genotypes and Phenotypes (dbGaP) data access committee, we designed an algorithm to download non-tumor sequence data that were curated as part of The Cancer Genome Atlas (TCGA) lung adenocarcinoma project.
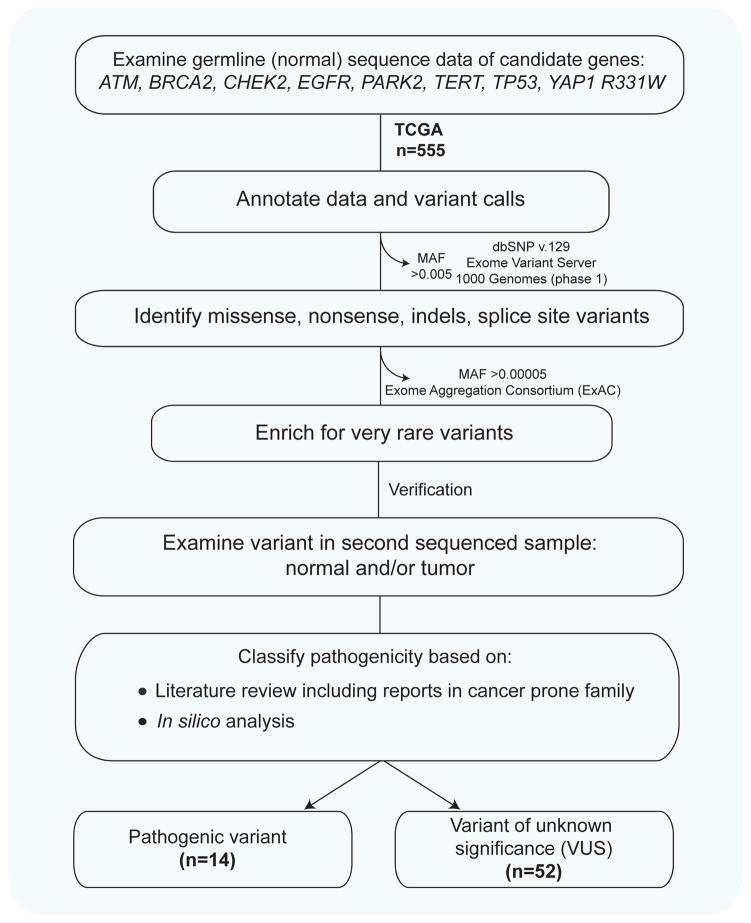
The filtering strategy is summarized in Figure 1 and the detailed methods are included in the supplementary materials. Briefly, sequence data for the eight candidate genes we selected, from normal/non-tumor tissue, were filtered for rare potentially disease risk alleles. We excluded common variants defined as greater than 0.005 minor allele frequency in each of dbSNP v.129, the Exome Variant Server and the 1000 Genomes project. In a second filtering step in order to enrich for ultra-rare variants, we filtered variants with frequencies greater than 0.00005 in the Exome Aggregation Consortium (ExAC) database (Figure 1). To exclude sequencing errors, we examined each of these remaining variants manually in an independently sequenced second normal and/or tumor sample and each of the variants had a minimum coverage depth of 20x. These validated rare variants were then studied in a literature search and were considered pathogenic if: 1. They had been previously reported in familial cancer syndromes, or 2. They were frameshift/nonsense mutations in known tumor suppressor genes. The remaining alterations were classified as variants of unknown significance (VUS), and their effect on protein function was examined in silico using three prediction programs.
Figure 1. Approach to identifying germline cancer-predisposing mutations in sequenced lung adenocarcinoma cases.
The flow chart delineates a priori designed strategy for identifying rare pathogenic variants and the numbers bolded indicate the number of cases examined and filtered. MAF, refers to minor allele frequency.
Results
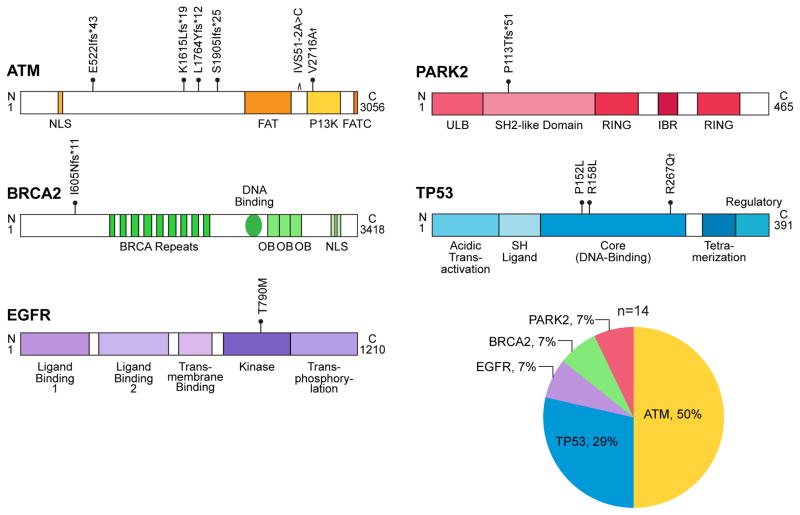
At the time of analysis, there were 555 unique lung adenocarcinoma cases which had non-tumor DNA sequenced. The characteristics for 497 of these subjects were publicly available and are summarized in Table 1. The mean age was younger than the United States lung adenocarcinoma population (65.3 vs.70.0 years) and there was a female predominance (53% vs. 49%). There were no pathogenic variants in CHEK2 or TERT and we found no cases with the YAP1 R331W variant. However, 14 cases (2.5%) carried 12 unique pathogenic variants in five other genes: ATM (n=7), followed by TP53 (n=4), BRCA2 (n=1), EGFR (n=1) and PARK2 (n=1) (Figure 2 and Table 2). Seven mutations altered the reading frame or disrupted splicing, and all five missense mutations fell in highly conserved domains. All 11 ATM and TP53 missense mutations had been previously reported in ataxia telengiectasia or Li-Fraumeni syndrome, respectively (Table 2). The BRCA2 frameshift mutation had been reported in an ovarian cancer patient11, and EGFR T790M has been documented in familial lung adenocarcionma2, 12. PARK2 variants had been identified in familial lung cancer, and the frameshift we identified similarly predicted loss-of-function because of a premature stop codon4, 13. The prevalence of these variants in 60,706 individuals in the ExAC database was 0.0004%, indicating that the lung adenocarcinoma phenotype enriches for the rare heterozygous alleles we identified by an odds ratio of 66 (95% confidence interval 33 to 125, P<0.0001, chi-square test with Yates’ correction, two-sided).
Table 1.
TCGA lung adenocarcinoma cohort characteristics
| n=497 (%)* | |
|---|---|
| Age at diagnosis | |
| Mean | 65.3 (range 33–88) |
| 31–40 | 4 (1%) |
| 41–50 | 31 (6%) |
| 51–60 | 117 (24%) |
| 61–70 | 166 (33%) |
| 71–80 | 136 (27%) |
| 81–90 | 24 (5%) |
| Unknown | 19 (4%) |
| Gender | |
| Male | 234 (47%) |
| Female | 263 (53%) |
| Race | |
| White | 367 (74%) |
| African-American | 46 (9%) |
| Hispanic | 7 (1%) |
| Asian | 8 (2%) |
| American Indian | 1 (0%) |
| Unknown | 68 (14%) |
| Smoking status | |
| Former | 292 (59%) |
| Current | 119 (24%) |
| Never | 72 (14%) |
| Unknown | 14 (3%) |
| Stage** | |
| I | 264 (53%) |
| II | 123 (25%) |
| III | 83 (17%) |
| IV | 26 (5%) |
| Unknown | 1 (<1%) |
| Prior malignancy | |
| No | 407 (82%) |
| Yes | 90 (18%) |
Data were available for 497 of 555 individuals for whom sequence data were analyzed.
Lung cancer stage was determined based on American Joint Commission on Cancer (AJCC) criteria, 5th to 7th editions (1991–2013).
Figure 2. Germline variants in lung adenocarcinoma cases.
Schema of ATM, BRCA2, EGFR, PARK2, and TP53 proteins with the 12 unique pathogenic mutations marked at their relative amino acid position with the protein domains schematized. These occurred at a frequency of 2.5% in the studied cohort. † Refers to the two mutations identified in two separate individuals, ATM V2716A and TP53 R287Q. The relative frequencies of the mutations by gene are shown in the adjacent pie chart.
Table 2.
Clinical characteristics of lung adenocarcinoma cases with germline pathogenic variants in cancer predisposing genes (n=14)
| Age | M/F | Mutation | Germline DNA source | Validation DNA source(s) | Smoking status | Pack-years | Prior Malignancy | Prior association with disease |
|---|---|---|---|---|---|---|---|---|
| 41 | M |
TP53 R267Q |
Blood | Tumor and Duplicate† | Current | 38 | LFS* | |
| 49 | M |
BRCA2 I605Nfs*11 |
Blood | Tumor | Former | na | BCC | Yang JAMA 2011 |
| 52 | M |
ATM E522Ifs*43 |
Blood | Tumor | Current | 36 | A-T* | |
| 60 | F |
EGFR T790M |
Blood | Tumor | Former | 40 | Haber Nat Gen 2005 | |
| 63 | F |
TP53 R267Q |
Normal tissue | Duplicate† | Never | 0 | LFS* | |
| 65 | F |
ATM L1764Yfs*12 |
Normal tissue | Tumor | Former | 10 | A-T* | |
| 67 | M |
ATM V2716A |
Blood | Tumor | Current | 67 | A-T* | |
| 68 | F |
ATM K1615Lfs*19 |
Normal tissue | Tumor | Former | 50 | Unknown | A-T* |
| 70 | M |
ATM IVS51-2A>C |
Blood | Tumor | Former | 43 | A-T Teraoka AJHG 1999 |
|
| 70 | F |
PARK2 P113Tfs*51 |
Blood | Tumor | Former | 20 | Xiong AJHG 2015 | |
| 73 | F |
TP53 P152L |
Blood | Tumor | Former | na | Thymus | LFS* |
| 74 | F |
ATM V2716A |
Normal tissue | Tumor | Never | 0 | A-T* | |
| na | na |
ATM S1905Ifs*25 |
Normal tissue | Tumor | na | na | na | A-T* |
| na | na |
TP53 R158L |
Normal tissue | Tumor | na | na | na | LFS* |
Refers to mutations in ataxia telengiectasia (A-T) and Li-Fraumeni syndrome (LFS) patients reported in respective disease specific databases http://chromium.lovd.nl/LOVD2 and http://p53.iarc.fr/ as outlined in the Methods.
Previously reported somatic mutations;
Duplicate samples refer to an independently sequenced normal tissue that was analyzed for confirmation of the mutation status.
Abbreviations: A-T, ataxia-telengiectasia; BCC, basal cell carcinoma; LFS, Li-Fraumeni syndrome; na, not available
We examined the clinical characteristics of mutation carriers. Twelve of the 14 cases had published clinical data and they had a mean age of 62.7 years at diagnosis, slightly younger than the 65.3 mean for the cohort. Two cases were 41 and 49 years, a finding suggestive of a genetic predisposition since only 7% of the entire TCGA cohort was under 50 years at diagnosis. Eight of the 12 cases were female (67%), and the two never smokers were female. Three cases had a history of prior malignancy (25%) including a TP53 mutation carrier who had a prior thymus cancer, a Li-Fraumeni-associated cancer. The stage at presentation for these mutation carriers reflected the distribution of stage for the entire TCGA cohort (Table 1) and all the patients with available data reported being white. The non-synonymous mutation burden in tumors was available for the six cases (for whom data had been available and published) and it matched what has been previously reported for the TCGA10. In the two never smokers, with ATM and TP53 mutations, it was 22 and 71, respectively, and the four smokers with ATM and TP53 mutations, had a mutation burden of 220 ± 19 s.e.m., also comparable to the known mutation burden in lung adenocarcinoma14. Family history data were not collected for this cohort, as for the remainder of the TCGA. In addition to this subset of cases with pathogenic mutations, we identified 52 other cases with ultra-rare or previously unreported variants (9.4%). These variants fell predominantly in BRCA2 (n=25, 48%) and ATM (15, n=29%) (Supplementary Table 1). Twelve of these 52 mutations (23%), or 2% of this entire lung adenocarcinoma cohort, were predicted to be deleterious by three protein prediction programs, PolyPhen, PROVEAN and SIFT (Supplemental Table 1).
Discussion
Lung cancer is considered environmentally-driven and smokers have at least a 20-fold increased risk1. While the rates of lung cancer have decreased in the United States as a result of successful prevention efforts, there has been no change in the number of cases in never smokers and individuals with relatively low cigarette smoke exposure history1. We found 2.5% of unselected lung adenocarcinoma patients carried pathogenic germline mutation in a cancer predisposing gene. Given that lung adenocarcinoma has an annual incidence of 500,000 worldwide, this is a sizable and clinically relevant subset. The 2.5% prevalence we report may under-estimate the overall frequency of germline mutations of major effect size on lung adenocarcinoma risk because our analysis relied on extremely stringent criteria for pathogenicity. If we also account for the ultra-rare variants that were consistently predicted to be deleterious to protein function (Supplementary Table 1), the prevalence of germline mutant cases could total 4.5%. Our data therefore add lung adenocarcinoma to the list of adult-onset cancers that includes a definable subset with germline mutations, similar to prostate and ovarian cancer15, 16. Further analysis of other large lung adenocarcinoma exome datasets will be important for estimating the overall prevalence of germline mutations in cancer predisposing genes in this cancer.
There are potential clinical implications of the findings we report. First, lung adenocarcinoma patients should be queried about a family history of all cancers, not just lung cancer, as their families may show clustering of Li-Fraumeni cancers (e..g sarcoma, leukemia, breast) or ATM-associated cancers. Second, as the use of next generation panel testing continues to expand in clinical practice, variants that are suspicious for germline status (e.g. near 50% of reads) in the genes we identified should prompt considering a referral for genetic counseling and dedicated evaluation for germline testing. The most common mutant genes we identified involve DNA repair pathways with ATM, TP53 and BRCA2 being most common. This observation is consistent with a carcinogenesis mechanism where cigarette smoke-induced genotoxic damage, or other environmental insults, are highly mutagenic in the background of defective DNA repair17. ATM polymorphisms have been associated with severe radiation-induced pneumonitis, illustrating another example where the presence of a germline mutation may also inform the risk of specific clinical complications for an individual patient18. Importantly, germline defects in DNA repair genes cause sensitivity to poly-ADP ribose polymerase (PARP) inhibitors in advanced solid tumors such as prostate, ovarian, pancreatic and breast cancer19–21. The data we report here suggest a similar therapy approach may be effective in a small, genetically definable subset of lung adenocarcinoma patients who carry germline mutations in DNA repair pathways.
Supplementary Material
Acknowledgments
We thank Corina Antonescu from the Johns Hopkins Computational Biology Core for help with the data analysis.
Funding: This work was supported by NIH grants CA160433 and HL119476, the Commonwealth Foundation and the Maryland Cigarette Restitution Fund (to M.A.) and NSF ABI-1356078 (to L.F.).
Footnotes
Disclosures: The authors declare no relevant conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alberg AJ, Brock MV, Ford JG, et al. Epidemiology of lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143:e1S–29S. doi: 10.1378/chest.12-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell DW, Gore I, Okimoto RA, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet. 2005;37:1315–1316. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- 3.Chen HY, Yu SL, Ho BC, et al. R331W Missense Mutation of Oncogene YAP1 Is a Germline Risk Allele for Lung Adenocarcinoma With Medical Actionability. J Clin Oncol. 2015;33:2303–2310. doi: 10.1200/JCO.2014.59.3590. [DOI] [PubMed] [Google Scholar]
- 4.Xiong D, Wang Y, Kupert E, et al. A recurrent mutation in PARK2 is associated with familial lung cancer. American journal of human genetics. 2015;96:301–308. doi: 10.1016/j.ajhg.2014.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, McKay JD, Rafnar T, et al. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat Genet. 2014;46:736–741. doi: 10.1038/ng.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landi MT, Chatterjee N, Yu K, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swift M, Morrell D, Massey RB, et al. Incidence of cancer in 161 families affected by ataxia-telangiectasia. N Engl J Med. 1991;325:1831–1836. doi: 10.1056/NEJM199112263252602. [DOI] [PubMed] [Google Scholar]
- 8.Hwang SJ, Cheng LS, Lozano G, et al. Lung cancer risk in germline p53 mutation carriers: association between an inherited cancer predisposition, cigarette smoking, and cancer risk. Human genetics. 2003;113:238–243. doi: 10.1007/s00439-003-0968-7. [DOI] [PubMed] [Google Scholar]
- 9.Lo YL, Hsiao CF, Jou YS, et al. ATM polymorphisms and risk of lung cancer among never smokers. Lung Cancer. 2010;69:148–154. doi: 10.1016/j.lungcan.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang D, Khan S, Sun Y, et al. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA. 2011;306:1557–1565. doi: 10.1001/jama.2011.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gazdar A, Robinson L, Oliver D, et al. Hereditary lung cancer syndrome targets never smokers with germline EGFR gene T790M mutations. J Thorac Oncol. 2014;9:456–463. doi: 10.1097/JTO.0000000000000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veeriah S, Taylor BS, Meng S, et al. Somatic mutations of the Parkinson’s disease-associated gene PARK2 in glioblastoma and other human malignancies. Nature genetics. 2010;42:77–82. doi: 10.1038/ng.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swanton C, Govindan R. Clinical Implications of Genomic Discoveries in Lung Cancer. N Engl J Med. 2016;374:1864–1873. doi: 10.1056/NEJMra1504688. [DOI] [PubMed] [Google Scholar]
- 15.Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N Engl J Med. 2016;375:443–453. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanchi KL, Johnson KJ, Lu C, et al. Integrated analysis of germline and somatic variants in ovarian cancer. Nature communications. 2014;5:3156. doi: 10.1038/ncomms4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alder JK, Guo N, Kembou F, et al. Telomere length is a determinant of emphysema susceptibility. Am J Respir Crit Care Med. 2011;184:904–912. doi: 10.1164/rccm.201103-0520OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Yang M, Bi N, et al. ATM polymorphisms are associated with risk of radiation-induced pneumonitis. Int J Radiat Oncol Biol Phys. 2010;77:1360–1368. doi: 10.1016/j.ijrobp.2009.07.1675. [DOI] [PubMed] [Google Scholar]
- 19.Mateo J, Carreira S, Sandhu S, et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol. 2015;33:244–250. doi: 10.1200/JCO.2014.56.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.