Abstract
Kruppel-associated box zinc finger proteins (KRAB-ZFPs) make up the largest family of transcription factors in humans. These proteins emerged in the last common ancestor of coelacanth and tetrapods and have expanded and diversified in the mammalian lineage. Although their mechanism of transcriptional repression has been well studied for over a decade, the DNA binding activities and the biological function of these proteins has been largely unexplored. Recent large scale ChIP-seq studies and loss-of-function experiments have revealed that KRAB-ZFPs play a major role in the recognition and transcriptional silencing of transposable elements (TEs), consistent with an “arms race model” of KRAB-ZFP evolution against invading TEs. However, this model is insufficient to explain the evolution of many KRAB-ZFPs that appear to domesticate TEs for novel host functions. We highlight some of the mammalian regulatory innovations driven by specific KRAB-ZFPs, including genomic imprinting, meiotic recombination hotspot choice, and placental growth.
Keywords: KRAB-zinc finger proteins (KRAB-ZFPs), Transposable elements (TEs), Mammalian evolution
Molecular architecture of KRAB-ZFPs
KRAB-ZFPs are the largest subgroup of the tandem C2H2-type zinc finger protein (ZFP) family, which is the largest transcription factor family in mouse and human genomes. KRAB-ZFPs are characterized by two domain structures: the N-terminal KRAB domain (see Glossary) and a tandem array of C2H2 zinc fingers at the C-terminus. The KRAB domain mediates the recruitment of TRIM28 (tripartite motif-containing protein 28, also known as KAP1, Tif1β or KRIP-1 [1]), which functions as a scaffold for other repressive histone-modifying and binding factors, including the histone H3K9me3 methyltransferase, Set Domain Bifurcated 1 (SETDB1) [2], the histone deacetylase containing NuRD complex [3, 4], and heterochromatin protein 1 (HP1) [5, 6] that catalyze heterochromatin formation and transcriptional repression (Figure 1A). If initiated during early embryogenesis, KRAB/TRIM28-induced gene silencing also promotes heritable epigenetic silencing by inducing DNA methylation that can be maintained even upon release from TRIM28 [7, 8]. Moreover, KRAB-TRIM28-induced heterochromatic silencing can spread in certain contexts along the chromosome over large genomic distances, which is likely facilitated by the H3K9me3 reader protein HP1 that can further propagate H3K9me3 via the independent recruitment of SETDB1 [9, 10].
Figure 1.
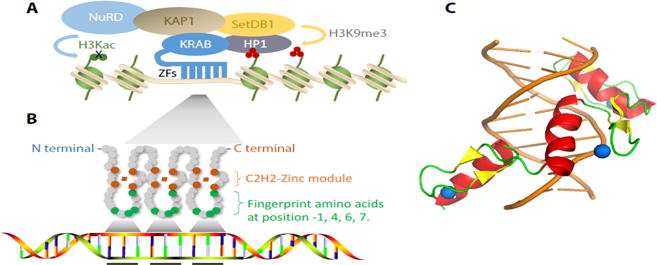
(A) Model of the KRAB-ZFPs repression complex with 6 zinc fingers (the average KRAB-ZFP in humans has more than 10 fingers). KRAB-ZFPs bind DNA via tandem C2H2 zinc finger (ZF) domains, and interact with TRIM28 to form a stable complex via the KRAB domain. TRIM28 then recruits other chromatin-related corepressors: SETDB1, a histone H3K9me3 methyltransferase; HP1, a histone H3K9me3 (red) reader of heterochromatin-induced silencing; NuRD complex, a major protein complex for deacetylation of multiple lysines on histone tails (green). (B) Linear model of a three-fingered zinc finger array’s interactions with DNA. For each C2H2 zinc finger domain, two cysteines and two histidines interact with a single zinc atom to stabilize the finger (orange), while the fingerprint amino acid residues at the -1, 4, 6 and 7 positions primarily make base specific contacts. (C) Structural model of a three-fingered ZFP interacting with its target sequence. Each zinc finger domain has two beta-sheets (yellow) and one alpha helix (red). The alpha-helix can bind in the major groove of DNA (orange) to make sequence-specific contacts to DNA bases. The loop region between two beta-sheets and C-terminal of the alpha helix wrap the zinc atom (blue) to form the stable invariant region.
The KRAB domain contains approximately 75 amino acids, consisting of KRAB-A and KRAB-B type modules. KRAB-A is the primary repressive module that interacts with TRIM28, while KRAB-B can enhance the binding affinity of KRAB-A to TRIM28 [11]. However, many KRAB-ZFPs such as HZF12 [12], ZFP120 [13], and ZK1 [14], are still effective transcriptional repressors while lacking the KRAB-B type module. In rare cases there have been reports of KRAB domains functioning as transcriptional activators, like Nizp1 [15], ZFP480 [16], and ZBRK1 [17], but the vast majority of KRAB domain containing proteins are potent repressors, which has led to the use of the KRAB domain in engineered repressors, including Cas9-KRAB fusions [18].
Unlike the two KRAB modules that are encoded by separate exons, the tandem C2H2 zinc fingers are always encoded by a single exon, usually the last exon in the KRAB-ZFP gene structure [19, 20]. The number of zinc fingers per KRAB-ZFP also varies; in humans, KRAB-ZFPs contain between 3 and 40 zinc fingers with an average of 12 [20]. Each zinc finger is stabilized by binding to a zinc ion, which is characteristically contacted by two cysteine and two histidine residues. The classic C2H2 zinc finger encompasses 28 amino acids, which can interact with 3 consecutive nucleotides of the primary DNA strand and one nucleotide of the secondary strand within the adjacent tri-nucleotide (Figure 1B) [21]. Structural studies have shown that amino acids at positions -1, 2, 3 and 6 of the C2H2 α-helix are responsible for the direct interactions with specific DNA target sites, thus determining the binding specificity of the zinc fingers [22–24]. These amino acid positions are thus referred to as the zinc “fingerprint” amino acids (Figure 1A–C). A number of computational prediction tools have been developed to predict the specific DNA target sequences of zinc finger arrays that are based on several parameters: 1) the assumption of the collinearity of ZNF arrays, thus targeting DNA triplets without gaps, 2) the concept of zinc fingerprints controlling target specificity, and 3) experimental data of single zinc fingers within small zinc finger arrays bound to short DNA stretches [25–29]. However, growing evidence supports that not all the C2H2 domains within a zinc finger array bind DNA. Some of the zinc fingers may be engaged in the interactions with RNA or proteins [25, 30, 31]. In addition, amino acids adjacent to fingerprint amino acids within zinc fingers can influence the binding specificity, and neighboring zinc fingers can influence a zinc finger’s DNA interactions [24]. Moreover, some epigenetic modifications such as DNA methylation can also influence zinc finger binding activities [32]. In sum, these can partially explain why zinc finger binding motifs identified by ChIP-seq (Box 1) are usually shorter than predicted and why predicted binding motifs often contain stretches of inaccuracies [25, 31, 33]. The lack of complete modularity of the zinc finger domain and the imprecision of predicting zinc finger array binding sites has provided the major limitation to the potential wider use of designer zinc fingers in genome editing applications.
Text Box 1. ~2/3 of human KRAB-ZFPs bind to TEs genome-wide.
ChIP-seq has been proven to be a reliable strategy to map the genome-wide binding sites and determine the DNA binding motifs of tandem zinc finger genes. Recent studies have used large scale ChIP-seq screens to map the binding sites of nearly all the human KRAB-ZFPs, as well as TRIM28 in a number of different human tissues [31, 33]. For these specialized ChIP-seq studies, cDNAs for predicted KRAB-ZFPs are cloned and over-expressed in cell lines, and cross-linked chromatin is immunoprecipitated with antibodies to the epitope tag. Libraries from bound regions are sequenced and mapped onto the human or mouse genome to determine regions of local enrichment. Special care is required to map reads to repeat elements, because in many cases reads may not map uniquely. In addition to identifying target motifs, these studies revealed that the majority of KRAB-ZFPs bind to TEs, including LTR, L1, SINE, and SVA families, as well as simple repeats and other variable number tandem repeats, including zinc finger repeats. In many cases, multiple KRAB-ZFPs could bind to individual families of TEs. Although the majority (~2/3) of human KRAB-ZFPs are enriched on TEs, the majority of TE copies are still not bound by KRAB-ZFPs/TRIM28. The remaining ~1/3 of KRAB-ZFPs, which tend to be more ancient and less likely to be co-occupied by TRIM28, are enriched near transcriptional start sites of genes, leading to speculation that these factors also originally bind to TEs when they evolved, but these target TEs are no longer recognizable as such due to genetic drift.
Evolution of KRAB-ZFPs
A recent study of vertebrate genomes indicated that KRAB-ZFPs likely arose about 420 million years ago in a common ancestor of coelacanth and tetrapods [33]. Interestingly, the KRAB-ZFP genes evolved from a mono-exonic to a multi-exonic configuration. This switch during evolution likely facilitated the independent evolution of KRAB domains and zinc finger arrays. It is believed that all extant KRAB-ZFPs evolved from the PRDM9 or PRDM7 gene [34], both of which contain a KRAB domain and a tandem array of C2H2 zinc fingers separated by a PR-SET domain that catalyzes histone H3K4 and H3K36 methylation (Figure 2) [35–38]. However, unlike most mammalian KRAB-ZFPs that interact with the co-repressor TRIM28, the KRAB domain of PRDM9 does not interact with TRIM28 but instead interacts with CXXC1, a component of the COMPASS complex that activates gene transcription [39]. The PRDM9 gene is responsible for creating an active local chromatin environment that targets meiotic recombination hotspots in germ cells (described in more detail below) [40–42]. A second ancient KRAB-ZFP sub-family, the ZKSCAN genes, contains a SCAN box domain in addition to the KRAB domain and tandem zinc finger repeats, which can mediate homo and hetero-oligomerization with other SCAN-containing proteins [43]. The SCAN domain-containing KRAB-ZFPs have been found in all vertebrates except for frog and chicken [44]. The SCAN domain exhibits striking homology to the C-terminal part of the gag capsid protein from Gypsy/Ty3-like LTR (Long Terminal Repeat) retrotransposons, suggesting this domain may have been created by retrotransposon insertion into a KRAB-ZFP gene during evolution [44]. Of the ZKSCAN genes identified in the human genome, most cannot efficiently bind to TRIM28 through their KRAB domains [45]. Thus, during evolution, changes in the KRAB domain that facilitated binding to TRIM28 and loss of PR-SET or SCAN domains facilitated the creation of the modern family of KRAB-ZFP transcriptional repressors (Figure 2).
Figure 2.
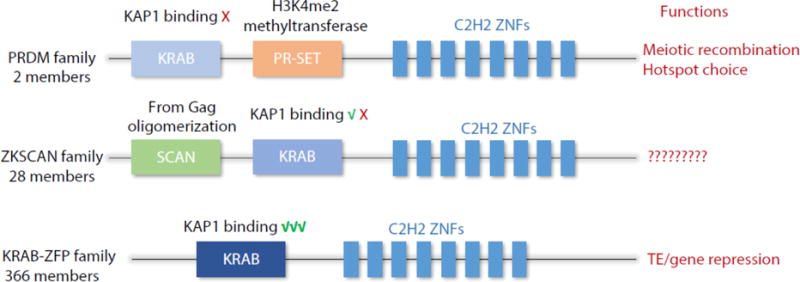
Domain structures, genome content, and likely functions of major KRAB-ZFP subfamilies. The KRAB domain in the PRDM9/7 does not bind to TRIM28, it binds CXXC1, a component of COMPASS coactivator complex. This is consistent with the function of PR-SET domain, which can catalyze histone H3K4me2 methylation. PRDM9 controls meiotic recombination hotspot choice. Some of the ZKSCAN family proteins can bind TRIM28 whereas others do not. The SCAN domain was derived from the Gag capsid protein and can form oligomers with other SCAN domain proteins. However, the biological function is entirely unknown. The newly emerged KRAB-ZFPs without additional domains appear to play a role in epigenetic marking of TEs, imprint control regions, and other genes. Most KRAB domains bind strongly to TRIM28. C2H2 zinc finger domains in all three family proteins are rapidly evolving, suggesting their regulatory repertoire may change during evolution.
An important insight into the evolution and function of KRAB-ZFPs in tetrapods was the finding that the number of tandem zinc finger genes in different species correlated with the LTR transposon load of that species [46]. Since the main function of KRAB-ZFPs is to bind TEs (see Box 1 and 2) that continue to invade and reshape mammalian genomes, KRAB-ZFPs tend to evolve more rapidly than other transcription factor families. Interestingly, most KRAB-ZPFs are found in clusters that overlap segmental duplications in several regions in the genome. For example, in the human genome, there are an estimated 352 KRAB-ZFP genes, 209 of which are located within 6 clusters on Chromosome 19 [47]. In contrast, mice have several large clusters dispersed on different chromosomes containing up to several dozen KRAB-ZFPs [48]. KRAB-ZFP members within a cluster share greater nucleotide and amino acid similarity than KRAB-ZFPs in other clusters [49]. Thus KRAB-ZFP genes are usually born within clusters by local segmental duplication likely caused by unequal crossing over during meiosis. However, new KRAB-ZFPs have also been generated by translocation, retrotransposition, and recombination between zinc finger arrays [48]. Zinc finger arrays have been classified as mini-satellites [48] due to their repeat structure, which are known to be unstable, although there have been no reported systematic measurements of mutation rates of zinc finger arrays or direct measures of array instability. Intriguingly, there are at least two KRAB-ZFPs that bind to repeated motifs found within many zinc finger arrays [33, 50], and this has been suggested to play a role in the deposition of H3K9me3 at the arrays to suppress recombination. Nonetheless, there is a number of evolutionary and molecular forces that allows for an expanding pool of KRAB-ZFP genes that can acquire new DNA binding activities. In support of this notion, amino acid positions that control DNA binding specificity are often under strong positive selection [51–53].
Text Box 2. Co-evolution between KRAB-ZFPs and TEs.
Throughout evolution, waves of retrotransposon insertions have shaped mammalian genomes, including the human genome [93, 94]. A computational analysis of 16 mammalian genomes indicates that the number of KRAB-ZFPs strongly correlates with the number of endogenous LTR elements, suggesting that the KRAB-ZFPs and TEs co-evolved [46]. This led to speculation that TEs are the main drivers of KRAB-ZFP diversification during evolution, (especially the positive selection in the ZNF array fingerprints), and raised the “arms race” model [33, 55]. This model states that the rapidly mutating TEs and germ line invasion of new retroviral families are the main cause of KRAB-ZFP evolution. That is, the dynamic competition between TEs and KRAB-ZFPs drives their selection. So far, several lines of evidence have supported this “arms race” model, indicating that the KRAB-ZFP system may evolve in response to the TEs with increasing diversity. Some good examples include the binding of ZFP809 to the essential PBS-Pro sequence of MuLV and VL30 [61], and the loss of ZNF93 binding site in the newer L1PA subfamilies [68]. Additional support comes from the finding of loss of recognition sites for all ZNF93 paralogs in the newest human-specific LINE-1. Furthermore, a direct prediction of the arms race model is that KRAB-ZFPs should turn over rapidly, as their target TEs decay by genetic drift, and indeed investigation of vertebrate genomes reveals the accumulation of many tandem-ZFP pseudogenes [33]. However, there is also growing evidence suggesting that the arms race between KRAB-ZFPs and TEs cannot be the only driving force of their evolutionary selection. First, some TEs continue to spread by retrotransposition even after the emergence of KRAB-ZFPs that can target these TEs. Second, some KRAB-ZFPs emerge to target TEs that have long since lost the ability to retrotranspose. Third, many TEs are targeted by multiple KRAB-ZFPs, which make KRAB-ZFP evasion by a TE very difficult. Thus, we favor a hybrid model of KRAB-ZFP evolution, whereby KRAB-ZFPs emerge in response to evading TEs, either to limit their retrotranspositional spread, to limit their transcriptional influence on nearby genes in the germ line or in somatic tissues, or to limit their potential to serve as sites of recombination, particularly in the germ line (Figure 3). Regardless of the underlying “purpose” of KRAB-ZFP birth and selection, the outcome for most KRAB-ZFPs is the same, eventual loss from the genome as target TEs decay, unless the KRAB-ZFP has evolved a particularly important function. Examples include Zfp568 and Zfp57, which provide essential functions during embryogenesis that involve regulating the Igf2 gene and genomic imprinting.
Whereas clustered zinc finger proteins are more likely to be species-specific, zinc finger genes located outside the zinc finger clusters tend to be more ancient and conserved during evolution. These ancient KRAB-ZFPs may have simply failed to ever duplicate and create new clusters, or alternatively, these “solo” KRAB-ZFPs might be the remains of ancient clusters that have mostly decayed due to the loss of non-essential KRAB-ZFPs by the same unequal crossing over forces that created modern clusters. This notion is supported by the high number of KRAB-ZFP pseudogenes also found in modern clusters [33, 48]. Sequence analyses have shown that the KRAB domains themselves also evolve rapidly, at a speed comparable with that of the zinc finger domains [54]. Such mutations likely facilitated the initial interaction of the KRAB domain with TRIM28, but also led to loss of TRIM28 interactions in other KRAB-ZFPs that are not repressors.
The question of what fraction of KRAB-ZFPs are true orthologues in mammalian species is not simple to address because zinc fingers and KRAB-domains all share some sequence homology. Putative DNA binding orthologues of modern KRAB-ZFPs can be best traced by comparing the zinc fingerprints of KRAB-ZFPs between species. Fingerprinting analyses have revealed only a few dozen DNA binding orthologues between Mus musculus and Homo sapiens (which diverged ~100 million years ago), with a proportionally higher number of orthologues as species more recently diverged from humans [33, 55] suggesting that the KRAB-ZFP families undergo continuous turnover and amplification by adding new members during evolution [19, 33, 46].
Biological functions of KRAB zinc finger proteins
Previous studies have reported that KRAB-ZFPs are involved in many biological processes including embryonic development, genomic imprinting, cell differentiation, metabolic control, and sexual dimorphism [20, 56–59]. But the major function of KRAB-ZFPs, inferred from genome-wide binding profiles (Box 1) or by direct loss-of-function experiments, is the embryonic silencing of TEs [60, 61]. Genetic ablation of the KRAB-interacting scaffold protein TRIM28 [62] or its partner histone methyltransferase SETDB1 [63] activates multiple TEs, including several classes of endogenous retroviruses (ERVs) in mouse and human embryonic stem cells (ESCs). However, recent studies indicate that KRAB-ZFPs with their corepressors TRIM28 and SETDB1 can transcriptionally repress not only TEs, but also other genes to control their precise expression, which will also be highlighted below.
Repression of TEs
One of the most compelling cases of a KRAB-ZFP that represses TEs expression in vivo is ZFP809. The murine-specific ZFP809 was first identified along with TRIM28 based on its ability to bind and repress the Proline Primer Binding Sequence (Pro-PBS) of Murine Leukemia Viruses (MuLV), which is rapidly transcriptionally silenced upon pro-viral integration in mouse embryonic stem cells [49, 60]. Although several clues have implicated many KRAB-ZFPs in TE silencing during early development, ZFP809 is, so far, the only identified KRAB-ZFP responsible for TE repression supported by genome-wide binding and loss-of-function studies in vivo. ChIP-seq analysis in ESCs and embryonic carcinoma cells revealed that ZFP809 targets several Pro-PBS-containing ERVs by recruiting the TRIM28/SETDB1 repressor complex to these elements. Depletion of ZFP809 during development leads to de-repression of endogenous retrovirus-like elements virus-like 30 (VL30, which are related to MuLV and also utilize the Pro-PBS) during post-implantation development and in most adult organs and tissues [61]. Importantly, deletion of ZFP809 in somatic cells has no effect on VL30 expression, indicating that ZFP809 is involved in epigenetic silencing initiation of VL30 in early development. ZFP809’s role in early development is further supported by the reduced expression of ZFP809 upon differentiation of ESCs [61]. These lines of evidence support the model that KRAB/TRIM28 repression is initiated during early embryo development and heritably maintained in somatic tissues without the continuous requirement for KRAB-ZFPs or corepressor TRIM28 [61]. However, some KRAB-ZFPs not only control the expression of ERVs during early development, but also in adult tissues, including ZFP932 and its paralog Gm15446, which directly bind and regulate different ERVK family subsets [64].
Genome-wide binding analyses of TRIM28 in human ESCs and primary human T lymphocytes [65, 66] indicate that KRAB-ZFPs can not only suppress LTR transposons, but also non-retroviral transposable elements (for a summary of these see [67]) including LINE-1 and several groups of SINE-VNTR-Alu (SVA) families. ZNF91 and ZNF93, two human-specific KRAB-ZFPs, directly target and silence SVA and LINE-1 (L1), respectively [68]. Interestingly, ZNF91/93 have undergone a series of structural changes to accommodate the continuous repression of the rapidly evolving target TEs, providing direct evidence of the evolutionary arms race between KRAB-ZFPs and TEs (Box 2). In the case of L1, novel variants have arisen lacking the ZNF binding site, suggesting TEs can also evade KRAB-ZFP silencing by mutation. However, such mutations may also impact other stages of the L1 retrotransposition cycle.
Repression of neighboring gene expression by KRAB-ZFPs at TE elements
KRAB-ZFPs that bind to TEs not only repress the expression of TEs themselves, but also regulate the expression of nearby genes by suppressing the latent enhancer/promoter activity found within these elements. For example, a nearby VL30 element serves as an ectopic enhancer that activates the Tdrd9 gene in Zfp809 KO but not wild type cells [69], and TRIM28 knockout/knockdown leads to activation of many TE-proximal genes in ESCs and differentiated tissues [64, 70]. In these studies, the loss of KRAB-ZFPs/TRIM28 is accompanied by a loss of the H3K9me3 mark at TEs and an increase in active promoter/enhancer marks (including H3K4me3 and H3K27Ac), which suggests that KRAB-ZFPs actively prevent the binding of activating transcription factors within TEs. Considering the large number of KRAB-ZFPs and TEs in mammalian genomes, it is likely that hundreds, if not thousands, of genes are indirectly regulated by KRAB-ZFPs through TEs proximal effects. These findings have led to speculation that the expansion and selection of KRAB-ZFP binding to TEs was not driven solely by the need to directly suppress TE expression, but was instead an engine driving the domestication of TE derived regulatory sequences (Box 2) [71]. According to this model, continuous innovation and spread of TEs along with their KRAB-ZFPs binders rewires gene expression networks in each species, potentially impacting many aspects of organismal development and physiology.
Despite the large and growing body of evidence that the majority of KRAB-ZFPs bind to TEs to repress their expression and impact transcriptional networks in cells, there has yet to be convincing genetic evidence that this process is essential for host fitness. This can be partly explained by the paucity of KRAB-ZFP knockout models, which are difficult to generate since these genes are largely encoded in large clusters that have undergone recent segmental duplications that are not easily targeted by homologous recombination. Modern gene editing tools should facilitate the development of more KRAB-ZFP knockout models that can be used to assess the in vivo biological functions of this gene family. There has also yet to be a formal demonstration that the KRAB-ZFP/TRIM28 system restricts retro-transposition of endogenous retroelements in vivo. Improved methods of mapping insertions of endogenous retroelements in KRAB-ZFP knockout mice would provide a plausible method to address this question.
Regulation of gene expression by KRAB-ZFPs independent of TE sequence
Approximately one third of human KRAB-ZFPs do not bind preferentially to TEs (Box1). These non-TE binding KRAB-ZFPs are more ancient (evolutionarily speaking) and thus it is possible that some or even most were originally TE binders, but over time their target TE sequences eroded due to genetic drift. Regardless of how such non-TE binding KRAB-ZFPs evolved, there is a growing body of literature into the gene regulatory functions of these proteins, which have been recently extensively reviewed [71]. We will focus attention on three KRAB-ZFPs that are not found in clusters and are implicated in essential vertebrate and mammalian regulatory innovations: ZFP57, the master regulator of genomic imprinting, ZFP568, which represses a placental isoform of the master fetal growth hormone insulin like growth factor 2 (IGF2), and PRDM9, the ancestor of all KRAB-ZFPs and the master regulator of recombination site selection during meiosis.
Genomic Imprinting
Genomic imprinting is the epigenetic process whereby one of two inherited alleles of a gene is selectively silenced in a parent-of-origin specific manner. Genomic imprinting is believed to have initiated in therian mammals with the advent of imprinting of Igf2 [72]. A widely-held hypothesis on the origin of genomic imprinting in mammals and its link with viviparity is the parental conflict hypothesis, which states that the inequality between parental alleles is due to the different interests of each parent as it relates to the evolutionary fitness of their genes. Whereas the paternal genes gain greater evolutionary fitness through success of offspring (potentially at the expense of the mother), the maternal genes gain greater fitness through preserving resources for her own survival and potential future offspring. In support of this theory, maternally imprinted genes, like Igf2, tend to be growth promoting, whereas paternally imprinted genes tend to be growth inhibiting. But the lack of imprinting in monotremes, which also display apparent conflicts over maternal offspring exchange [73], suggests that molecular adaptations in therians that were not present in monotremes were also critical to the establishment of genomic imprinting.
In mice, the differential establishment of heterochromatin and DNA methylation at imprint control regions (ICRs) in the germ line and its maintenance during embryonic development is crucial for imprinted gene regulation [74, 75]. ZFP57 plays the critical role in this process. Loss of maternal-zygotic function of Zfp57 results in a highly penetrant embryonic lethality, and the absence of Zfp57 in oocytes leads to maternal imprinting establishment failure [58]. ZFP57 and its corepressor TRIM28 bind selectively to the DNA-methylated allele of the majority of ICRs in ESCs via the methylated TGCCGC hexanucleotide (utilizing two of its C2H2 zinc fingers) and this binding is necessary for DNA and histone H3K9me3 methylation maintenance [32, 76, 77]. ZFP57 thus protects against imprint erasure during the wave of DNA demethylation that affects most of the mouse genome during early embryogenesis. Studies have also shown that ZFP57 mutation is a shared clinical feature in transient neonatal diabetes patients with DNA hypomethylation at imprinted loci throughout the genome [78] indicating ZFP57 plays a major role in imprinting in humans. Furthermore, the fingerprint of ZFP57 is conserved in eutherian mammals, which suggests its DNA binding properties, and thus its function as a master regulator of imprinting is likely conserved in eutherian mammals [33]. Interestingly, imprinting of Igf2 is also found in marsupials [72] despite the lack of a clear Zfp57 ortholog, suggesting imprint protection may be carried out by a marsupial specific KRAB-ZFP or an entirely distinct mechanism.
Placental Mammal Growth regulation
In addition to ZFP57, which plays a role in genomic imprinting of Igf2, ZFP568 also plays an important role in regulating Igf2 levels in mammals. In mice, Igf2 expression is controlled by three fetal promoters that are expressed in the fetus and placenta and one placental specific promoter called Igf2-P0, all of which are paternally expressed (maternally imprinted). Igf2-P0 accounts for ~10% of placental Igf2 expression [79]. Loss of Igf2-P0 leads to placental growth restriction followed by fetal growth restriction, although mice are viable. Thus Igf2-P0 is thought to be an adaptation that subtly regulates fetal/placental growth balance in mammals [80]. Although Igf2 transcripts are among the most abundant mRNAs in many embryonic tissues, Igf2 levels are very low at implantation. ZFP568 completely controls repression of Igf2-P0 at implantation by binding upstream of the Igf2-P0 promoter and maintaining it in a heterochromatic state specifically in embryonic tissues starting in early development [59]. Silencing of Igf2-P0 is critical for viability of mice, as Zfp568 mutants die at gastrulation, a phenotype that can be reversed by genetic removal of Igf2 [59, 81, 82]. The repression activity of ZFP568 at Igf2 depends on its zinc fingers and intact KRAB domain, as well as the co-factors TRIM28 and SETDB1. Furthermore, mutating the binding site upstream of the Igf2 gene activates Igf2-P0 in ESCs [59]. Although the ZFP568 fingerprint is conserved in mammals, suggesting conservation of Igf2-P0 repressive function, ZNF568 is one of the most rapidly evolving genes since the human-chimpanzee separation [83]. There are three alleles in the human population (designated H, C1 and C2), all of which harbor amino acid changes that could alter or disrupt function, including splicing changes that truncate the KRAB domain (in the C1 and C2 alleles), amino acid substitutions that could alter zinc finger binding activity (in H, C1, and C2 alleles), and a premature stop codon that deletes the final two zinc fingers (in the H allele). Intriguingly, the C alleles are associated with a larger relative head size at birth. Thus, ZNF568 orthologs may have unique functions in the human genome.
Recombination hotspot site determination and mammalian speciation
Speciation genes are those that restrict gene flow between species and related taxa, and cause sterility in hybrids. Prdm9, a histone H3K4 methyltransferase and the ancestor of all KRAB-ZFP genes, is one of the only known mammalian genes that has been demonstrated to cause hybrid sterility in specific F1 hybrid crosses of mice [84, 85]. PRDM9 functions as the major determinant of meiotic recombination hotspots in humans and mice by initiating the formation of localized double strand breaks, which is ultimately determined by the DNA binding properties of its zinc finger array [40, 41]. Intriguingly, the zinc finger arrays of PRDM9 are rapidly evolving in both human and wild mice [86], and there are numerous zinc finger alleles and thus hotspots differences within human and mouse populations [87]. Loss of Prdm9 in mice leads to double strand breaks and crossovers at promoters and other PRDM9-independent H3K4me3 marks, revealing a role for PRDM9 in sequestering double strand break machinery away from functional genomic elements [88]. The hybrid sterility observed in specific F1 crosses of mice correlates with the degree to which distinct PRDM9 variants bind both homologues at double strand breaks, revealing an important role for sub-species specific degradation of PRDM9 binding sites by meiotic drive in hybrid sterility and supporting a wider role of PRDM9 in the early stages of speciation [89]. In addition to the SET domain and zinc finger domains of PRDM9, the KRAB domain, which does not bind to TRIM28, is also essential for hotspot formation and fertility [90, 91]. Although many PRDM9 binding sites are not found within TEs, some alleles of PRDM9 bind to sequence motifs in TEs in both mice and humans [87, 92]. The self-destructive drive against these TE-derived sequences in the human lineage demonstrates the important role that TEs have contributed to hotspot choice in humans [41].
Conclusion and Perspectives
KRAB-ZFPs are expanding and diversifying in the vertebrate genomes that contain them, and growing evidence from loss-of-function studies in the mouse and genome-wide binding studies strongly supports the notion that the need to bind and epigenetically mark TEs is driving KRAB-ZFP evolution. Yet it is unclear if this is due to selective pressure to limit TE’s ability to transpose, their ability to recombine, or to simply block the inherent promoter/enhancer activity that TEs encode (Figure 3, Key Figure). Addressing these questions will require better KRAB-ZFP genetic models (see Outstanding Questions). To date, only a small handful of the several hundred KRAB-ZFP genes have been studied in much detail. Yet several of these KRAB-ZFPs, including Prdm9, Zfp57, and Zfp568, are essential to the mammalian specific adaptations regulating hotspot recombination choice, genomic imprinting, and Igf2 silencing. Thus, KRAB-ZFP genes and their ongoing chase against TEs will likely continue to provide an engine that facilitates mammalian and human adaptations.
Figure 3 Key Figure.
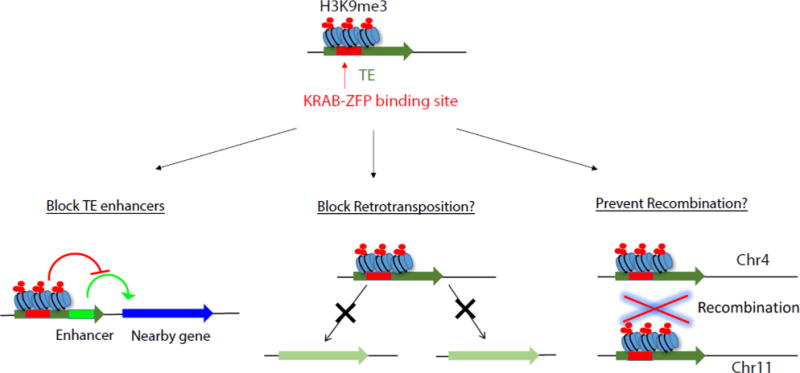
~2/3 of KRAB-ZFPs bind to enhancer regions of TEs and establish H3K9me3 marking at these sites. Such epigenetic marking limits the transcription of TEs and has been shown to be important to block TE enhancers from affecting nearby genes. Yet, whether these effects alone were sufficient to drive their massive expansion in mammals is unknown. Future studies should also illuminate whether KRAB-ZFP based TE marking is critical to prevent the spread of TEs by limiting their retrotransposition or preventing them from serving as sites of non-allelic recombination.
Outstanding questions box.
What is the primary force driving KRAB-ZFP evolution against TEs? Do KRAB-ZFPs limit spread of TEs in the germ line or somatic cells? Do they limit recombination between TEs? Do they limit TE regulatory potential?
Are KRAB-ZFP/TE rewired gene expression networks critical for host fitness or cellular phenotypes?
What is the biological function of most KRAB-ZFP genes? Are conserved KRAB-ZFPs playing important roles in mammalian specific biological processes?
Are there more KRAB-ZFPs like PRDM9, ZFP57, and ZFP568 that may have contributed to speciation?
Are human specific KRAB-ZFPs involved in our separation from great apes and other hominids? Are they involved in the rapid evolution of the human brain?
Are there variant KRAB-ZFPs alleles in addition to PRDM9 and ZNF568 in the human population that contribute to human disease or phenotypes?
How are KRAB-ZFPs regulated at the transcriptional, post-transcriptional, and cluster level? Does KRAB-ZFP regulation contribute to the “gating” of TE enhancers/promoters?
Trends Box.
KRAB-ZFPs make up the largest class of DNA binding transcription factors in mammals. The KRAB-domain is a potent transcriptional repression domain that functions via recruitment of the TRIM28 corepressor, which recruits heterochromatin inducing machinery.
KRAB-ZFPs are rapidly evolving in mammals, primarily at the level of expansion of KRAB-ZFP gene clusters, likely in response to invading waves of TEs.
The majority of KRAB-ZFPs (~2/3 in humans) interact with TEs via specific DNA binding motifs.
Loss-of-function studies of KRAB-ZFP corepressors and individual KRAB-ZFPs demonstrate that KRAB-ZFPs transcriptionally repress target TEs and prevent latent enhancer/promoter activity encoded within TEs from affecting nearby genes.
The continuous cycle of KRAB-ZFP evolution against TEs provides a driving force for new adaptations in mammals.
Additional material – Glossary
- C2H2 Zinc finger
C2H2-type zinc finger proteins (ZFPs) were the first and are the best-characterized class of DNA binding ZFPs. They contain a short β-hairpin and an α-helical fold (ββα structure) with the sequence motif X2-Cys-X2,4-Cys-X12-His-X3,4,5-His, where a single zinc atom is held in place by Cys2His2 (C2H2) amino acid residues in a tetrahedral array. This class of ZFPs can also bind to RNA and mediate protein-protein interactions
- Genomic imprinting
In mammals, somatic cells have two copies of each autosome, one inherited from each parent. Normally, genes from both chromosomes are expressed equally, however, imprinted genes are epigenetically repressed/expressed in a parental-allele specific manner. Imprinting refers to the processes whereby one of the two parental alleles is specifically marked in the male or female germ line, which is maintained in the offspring to control the mono-allelic expression of a gene
- Hybrid sterility
Lack of reproductive capacity of later generation hybrids from parents of different genetic origins. This reproductive mechanism restricts gene flow between taxa in the process of speciation
- KRAB domain
The Kruppel-associated box (KRAB) is a protein domain consisting of ~75 amino acid residues with transcriptional repression activity. The KRAB domain is present in the N-terminal part of about one third of eukaryotic C2H2 zinc finger proteins (which are thus referred to as KRAB-ZFPs). The KRAB domain functions through protein-protein interactions via two amphipathic helices. The most prominent interacting protein is TRIM28
- Retrotransposon
One of two broad classes of transposable elements that can amplify themselves in a genome via a “copy-and-paste” mechanism. These are also called RNA transposons because they involve an RNA intermediate that must be reverse transcribed. Broad classes of retrotransposons including Endogenous Retroviruses (ERVs) or Long Terminal Repeat (LTR) transposons, Long Interspersed Nuclear Elements (LINES), and Short Interspersed Nuclear Elements (SINEs). Since retrotransposons can rapidly increase in copy number they can also increase genome size
- Segmental duplication
Segmental Duplications (SDs) are long DNA segments (usually more than 1kb in length) with high sequence similarity (90–100%). SDs exist in multiple locations in the genome, tandem or interspersed, interchromosomal or intrachromosomal. SDs are believed to give rise to low copy repeats, thus playing a role in chromosomal rearrangement. Many clustered KRAB-ZFP genes overlap SDs
- Transposable elements (TEs)
Selfish genetic elements that have the capacity to move to new locations in the genome and thus change genome content. Types of TEs include DNA transposons that function via a cut-and-paste mechanism and retrotransposons that function via a copy-and-paste mechanism
- Viviparity
“Live-birth”, or the reproductive process that refers to the development of the embryo inside the parental body before live birth, contrary to the reproduction by laying eggs that complete their incubation outside the parental body
- Zinc “fingerprint”
The DNA-contacting amino acids within classic C2H2 zinc finger proteins. Typically, amino acids at positions -1, 2, 3 and 6 of each C2H2 α-helix are considered the fingerprint amino acids that control target specificity
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Peng H, et al. Biochemical analysis of the Kruppel-associated box (KRAB) transcriptional repression domain. J Biol Chem. 2000;275(24):18000–10. doi: 10.1074/jbc.M001499200. [DOI] [PubMed] [Google Scholar]
- 2.Schultz DC, et al. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes & Development. 2002;16(8):919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schultz DC, et al. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15(4):428–43. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macfarlan TS, et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 2011;25(6):594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayyanathan K, et al. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 2003;17(15):1855–69. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lechner MS, et al. Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: Direct chromoshadow domain-KAP-1 corepressor interaction is essential. Molecular and Cellular Biology. 2000;20(17):6449–6465. doi: 10.1128/mcb.20.17.6449-6465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiznerowicz M, et al. The Kruppel-associated box repressor domain can trigger de Novo promoter methylation during mouse early embryogenesis. Journal of Biological Chemistry. 2007;282(47):34535–34541. doi: 10.1074/jbc.M705898200. [DOI] [PubMed] [Google Scholar]
- 8.Rowe HM, et al. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development. 2013;140(3):519–529. doi: 10.1242/dev.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groner AC, et al. KRAB-Zinc Finger Proteins and KAP1 Can Mediate Long-Range Transcriptional Repression through Heterochromatin Spreading. Plos Genetics. 2010;6(3) doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer T, et al. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc Natl Acad Sci U S A. 2009;106(22):8998–9003. doi: 10.1073/pnas.0813063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vissing H, et al. Repression of transcriptional activity by heterologous KRAB domains present in zinc finger proteins. FEBS Lett. 1995;369(2–3):153–7. doi: 10.1016/0014-5793(95)00728-r. [DOI] [PubMed] [Google Scholar]
- 12.Looman C, et al. A novel Kruppel-Associated Box identified in a panel of mammalian zinc finger proteins. Mamm Genome. 2004;15(1):35–40. doi: 10.1007/s00335-003-3022-0. [DOI] [PubMed] [Google Scholar]
- 13.Mark C, et al. Comparative analysis of KRAB zinc finger proteins in rodents and man: evidence for several evolutionarily distinct subfamilies of KRAB zinc finger genes. DNA Cell Biol. 1999;18(5):381–96. doi: 10.1089/104454999315277. [DOI] [PubMed] [Google Scholar]
- 14.Katoh O, et al. ZK1, a novel Kruppel-type zinc finger gene, is induced following exposure to ionizing radiation and enhances apoptotic cell death on hematopoietic cells. Biochem Biophys Res Commun. 1998;249(3):595–600. doi: 10.1006/bbrc.1998.9201. [DOI] [PubMed] [Google Scholar]
- 15.Losson R, Nielsen AL. The NIZP1 KRAB and C2HR domains cross-talk for transcriptional regulation. Biochim Biophys Acta. 2010;1799(5–6):463–8. doi: 10.1016/j.bbagrm.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Yi Z, et al. A novel KRAB zinc-finger protein, ZNF480, expresses in human heart and activates transcriptional activities of AP-1 and SRE. Biochem Biophys Res Commun. 2004;320(2):409–15. doi: 10.1016/j.bbrc.2004.05.182. [DOI] [PubMed] [Google Scholar]
- 17.Hallen L, et al. The KRAB-containing zinc-finger transcriptional regulator ZBRK1 activates SCA2 gene transcription through direct interaction with its gene product, ataxin-2. Hum Mol Genet. 2011;20(1):104–14. doi: 10.1093/hmg/ddq436. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159(3):647–61. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huntley S, et al. A comprehensive catalog of human KRAB-associated zinc finger genes: Insights into the evolutionary history of a large family of transcriptional repressors. Genome Research. 2006;16(5):669–677. doi: 10.1101/gr.4842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biology. 2003;4(10) doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brayer KJ, Segal DJ. Keep your fingers off my DNA: Protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochemistry and Biophysics. 2008;50(3):111–131. doi: 10.1007/s12013-008-9008-5. [DOI] [PubMed] [Google Scholar]
- 22.Pavletich NP, Pabo CO. ZINC FINGER DNA RECOGNITION - CRYSTAL-STRUCTURE OF A ZIF268-DNA COMPLEX AT 2.1-A. Science. 1991;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 23.Pavletich NP, Pabo CO. CRYSTAL-STRUCTURE OF A 5-FINGER GLI-DNA COMPLEX - NEW PERSPECTIVES ON ZINC FINGERS. Science. 1993;261(5129):1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- 24.Patel A, et al. Structural basis for human PRDM9 action at recombination hot spots. Genes Dev. 2016;30(3):257–65. doi: 10.1101/gad.274928.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Najafabadi HS, et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nature Biotechnology. 2015;33(5):555–U181. doi: 10.1038/nbt.3128. [DOI] [PubMed] [Google Scholar]
- 26.Molparia B, et al. ZiF-Predict: a web tool for predicting DNA-binding specificity in C2H2 zinc finger proteins. Genomics Proteomics Bioinformatics. 2010;8(2):122–6. doi: 10.1016/S1672-0229(10)60013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Persikov AV, et al. Predicting DNA recognition by Cys2His2 zinc finger proteins. Bioinformatics. 2009;25(1):22–9. doi: 10.1093/bioinformatics/btn580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Persikov AV, Singh M. De novo prediction of DNA-binding specificities for Cys2His2 zinc finger proteins. Nucleic Acids Res. 2014;42(1):97–108. doi: 10.1093/nar/gkt890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jayakanthan M, et al. ZifBASE: a database of zinc finger proteins and associated resources. BMC Genomics. 2009;10:421. doi: 10.1186/1471-2164-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corsinotti A, et al. Global and Stage Specific Patterns of Kruppel-Associated-Box Zinc Finger Protein Gene Expression in Murine Early Embryonic Cells. Plos One. 2013;8(2) doi: 10.1371/journal.pone.0056721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitges FW, et al. Multiparameter functional diversity of human C2H2 zinc finger proteins. Genome Res. 2016;26(12):1742–1752. doi: 10.1101/gr.209643.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, et al. An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes Dev. 2012;26(21):2374–9. doi: 10.1101/gad.202200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imbeault M, et al. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature. 2017;543(7646):550–554. doi: 10.1038/nature21683. [DOI] [PubMed] [Google Scholar]
- 34.Birtle Z, Ponting CP. Meisetz and the birth of the KRAB motif. Bioinformatics. 2006;22(23):2841–2845. doi: 10.1093/bioinformatics/btl498. [DOI] [PubMed] [Google Scholar]
- 35.Eram MS, et al. Trimethylation of histone H3 lysine 36 by human methyltransferase PRDM9 protein. J Biol Chem. 2014;289(17):12177–88. doi: 10.1074/jbc.M113.523183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powers NR, et al. The Meiotic Recombination Activator PRDM9 Trimethylates Both H3K36 and H3K4 at Recombination Hotspots In Vivo. PLoS Genet. 2016;12(6):e1006146. doi: 10.1371/journal.pgen.1006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu H, et al. Molecular basis for the regulation of the H3K4 methyltransferase activity of PRDM9. Cell Rep. 2013;5(1):13–20. doi: 10.1016/j.celrep.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 38.Grey C, et al. In vivo binding of PRDM9 reveals interactions with noncanonical genomic sites. Genome Res. 2017;27(4):580–590. doi: 10.1101/gr.217240.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parvanov ED, et al. PRDM9 interactions with other proteins provide a link between recombination hotspots and the chromosomal axis in meiosis. Mol Biol Cell. 2017;28(3):488–499. doi: 10.1091/mbc.E16-09-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baudat F, et al. PRDM9 Is a Major Determinant of Meiotic Recombination Hotspots in Humans and Mice. Science. 2010;327(5967):836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Myers S, et al. Drive Against Hotspot Motifs in Primates Implicates the PRDM9 Gene in Meiotic Recombination. Science. 2010;327(5967):876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parvanov ED, et al. Prdm9 Controls Activation of Mammalian Recombination Hotspots. Science. 2010;327(5967):835–835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams AJ, et al. The zinc finger-associated SCAN box is a conserved oligomerization domain. Mol Cell Biol. 1999;19(12):8526–35. doi: 10.1128/mcb.19.12.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emerson RO, Thomas JH. Gypsy and the Birth of the SCAN Domain. Journal of Virology. 2011;85(22):12043–12052. doi: 10.1128/JVI.00867-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Itokawa Y, et al. KAP1-independent transcriptional repression of SCAN-KRAB-containing zinc finger proteins. Biochem Biophys Res Commun. 2009;388(4):689–94. doi: 10.1016/j.bbrc.2009.08.065. [DOI] [PubMed] [Google Scholar]
- 46.Thomas JH, Schneider S. Coevolution of retroelements and tandem zinc finger genes. Genome Research. 2011;21(11):1800–1812. doi: 10.1101/gr.121749.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lukic S, et al. The diversity of zinc-finger genes on human chromosome 19 provides an evolutionary mechanism for defense against inherited endogenous retroviruses. Cell Death and Differentiation. 2014;21(3):381–387. doi: 10.1038/cdd.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kauzlaric A, et al. The mouse genome displays highly dynamic populations of KRAB-zinc finger protein genes and related genetic units. PLoS One. 2017;12(3):e0173746. doi: 10.1371/journal.pone.0173746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolf G, et al. Spotting the enemy within: Targeted silencing of foreign DNA in mammalian genomes by the Kruppel-associated box zinc finger protein family. Mob DNA. 2015;6:17. doi: 10.1186/s13100-015-0050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frietze S, et al. ZNF274 recruits the histone methyltransferase SETDB1 to the 3′ ends of ZNF genes. PLoS One. 2010;5(12):e15082. doi: 10.1371/journal.pone.0015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt D, Durrett R. Adaptive evolution drives the diversification of zinc-finger binding domains. Molecular Biology and Evolution. 2004;21(12):2326–2339. doi: 10.1093/molbev/msh246. [DOI] [PubMed] [Google Scholar]
- 52.Emerson RO, Thomas JH. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009;5(1):e1000325. doi: 10.1371/journal.pgen.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nowick K, et al. Rapid Sequence and Expression Divergence Suggest Selection for Novel Function in Primate-Specific KRAB-ZNF Genes. Molecular Biology and Evolution. 2010;27(11):2606–2617. doi: 10.1093/molbev/msq157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mouse Genome Sequencing, C et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 55.Liu H, et al. Deep Vertebrate Roots for Mammalian Zinc Finger Transcription Factor Subfamilies. Genome Biology and Evolution. 2014;6(3):510–525. doi: 10.1093/gbe/evu030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lupo A, et al. KRAB-Zinc Finger Proteins: A Repressor Family Displaying Multiple Biological Functions. Current Genomics. 2013;14(4):268–278. doi: 10.2174/13892029113149990002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krebs CJ, et al. The KRAB Zinc Finger Protein RSL1 Regulates Sex-and Tissue-Specific Promoter Methylation and Dynamic Hormone-Responsive Chromatin Configuration. Molecular and Cellular Biology. 2012;32(18):3732–3742. doi: 10.1128/MCB.00615-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, et al. A Maternal-Zygotic Effect Gene, Zfp57, Maintains Both Maternal and Paternal Imprints. Developmental Cell. 2008;15(4):547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang P, et al. A placental growth factor is silenced in mouse embryos by the zinc finger protein ZFP568. Science. 2017;356(6339):757–759. doi: 10.1126/science.aah6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458(7242):1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolf G, et al. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev. 2015;29(5):538–54. doi: 10.1101/gad.252767.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rowe HM, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463(7278):237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 63.Matsui T, et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464(7290):927–U149. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 64.Ecco G, et al. Transposable Elements and Their KRAB-ZFP Controllers Regulate Gene Expression in Adult Tissues. Dev Cell. 2016;36(6):611–23. doi: 10.1016/j.devcel.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rowe HM, et al. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res. 2013;23(3):452–61. doi: 10.1101/gr.147678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Turelli P, et al. Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res. 2014;24(8):1260–70. doi: 10.1101/gr.172833.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kazazian HH, Jr, Moran JV. Mobile DNA in Health and Disease. N Engl J Med. 2017;377(4):361–370. doi: 10.1056/NEJMra1510092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jacobs FMJ, et al. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014;516(7530):242–+. doi: 10.1038/nature13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wolf G, et al. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes & Development. 2015;29(5):538–554. doi: 10.1101/gad.252767.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rowe HM, et al. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Research. 2013;23(3):452–461. doi: 10.1101/gr.147678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ecco G, et al. KRAB zinc finger proteins. Development. 2017;144(15):2719–2729. doi: 10.1242/dev.132605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Renfree MB, et al. The origin and evolution of genomic imprinting and viviparity in mammals. Philos Trans R Soc Lond B Biol Sci. 2013;368(1609):20120151. doi: 10.1098/rstb.2012.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Killian JK, et al. Monotreme IGF2 expression and ancestral origin of genomic imprinting. J Exp Zool. 2001;291(2):205–12. doi: 10.1002/jez.1070. [DOI] [PubMed] [Google Scholar]
- 74.Barlow DP. Genomic imprinting: a mammalian epigenetic discovery model. Annu Rev Genet. 2011;45:379–403. doi: 10.1146/annurev-genet-110410-132459. [DOI] [PubMed] [Google Scholar]
- 75.Bartolomei MS, Ferguson-Smith AC. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol. 2011;3(7) doi: 10.1101/cshperspect.a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Quenneville S, et al. In Embryonic Stem Cells, ZFP57/KAP1 Recognize a Methylated Hexanucleotide to Affect Chromatin and DNA Methylation of Imprinting Control Regions. Molecular Cell. 2011;44(3):361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Strogantsev R, et al. Allele-specific binding of ZFP57 in the epigenetic regulation of imprinted and non-imprinted monoallelic expression. Genome Biology. 2015;16 doi: 10.1186/s13059-015-0672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mackay DJG, et al. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nature Genetics. 2008;40(8):949–951. doi: 10.1038/ng.187. [DOI] [PubMed] [Google Scholar]
- 79.Moore T, et al. Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc Natl Acad Sci U S A. 1997;94(23):12509–14. doi: 10.1073/pnas.94.23.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mikaelsson MA, et al. Placental programming of anxiety in adulthood revealed by Igf2-null models. Nat Commun. 2013;4:2311. doi: 10.1038/ncomms3311. [DOI] [PubMed] [Google Scholar]
- 81.Garcia-Garcia MJ, et al. Chato, a KRAB zinc-finger protein, regulates convergent extension in the mouse embryo. Development. 2008;135(18):3053–62. doi: 10.1242/dev.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shibata M, Garcia-Garcia MJ. The mouse KRAB zinc-finger protein CHATO is required in embryonic-derived tissues to control yolk sac and placenta morphogenesis. Dev Biol. 2011;349(2):331–41. doi: 10.1016/j.ydbio.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chien HC, et al. Targeted Disruption in Mice of a Neural Stem Cell-Maintaining, KRAB-Zn Finger-Encoding Gene That Has Rapidly Evolved in the Human Lineage. Plos One. 2012;7(10) doi: 10.1371/journal.pone.0047481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mihola O, et al. A Mouse Speciation Gene Encodes a Meiotic Histone H3 Methyltransferase. Science. 2009;323(5912):373–375. doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- 85.Nowick K, et al. A prominent role of KRAB-ZNF transcription factors in mammalian speciation? Trends Genet. 2013;29(3):130–9. doi: 10.1016/j.tig.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 86.Oliver PL, et al. Accelerated Evolution of the Prdm9 Speciation Gene across Diverse Metazoan Taxa. Plos Genetics. 2009;5(12) doi: 10.1371/journal.pgen.1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Buard J, et al. Diversity of Prdm9 zinc finger array in wild mice unravels new facets of the evolutionary turnover of this coding minisatellite. PLoS One. 2014;9(1):e85021. doi: 10.1371/journal.pone.0085021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brick K, et al. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485(7400):642–5. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Davies B, et al. Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice. Nature. 2016;530(7589):171–176. doi: 10.1038/nature16931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.de Massy B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu Rev Genet. 2013;47:563–99. doi: 10.1146/annurev-genet-110711-155423. [DOI] [PubMed] [Google Scholar]
- 91.Imai Y, et al. The PRDM9 KRAB domain is required for meiosis and involved in protein interactions. Chromosoma. 2017 doi: 10.1007/s00412-017-0631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Myers S, et al. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310(5746):321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 93.Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303(5664):1626–32. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 94.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10(10):691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]


