Abstract
Transposable elements (TEs) are selfish genetic units that typically encode proteins that enable their proliferation in the genome and spread across individual hosts. Here we review a growing number of studies that suggest that TE proteins have often been coopted or ‘domesticated’ by their host as adaptations to a variety of evolutionary conflicts. In particular, TE-derived proteins have been recurrently repurposed as part of defense systems that protect prokaryotes and eukaryotes against the proliferation of infectious or invasive agents, including viruses and TEs themselves. We argue that the domestication of TE proteins may often be the only evolutionary path toward the mitigation of the cost incurred by their own selfish activities.
Keywords: transposable element protein domestication, evolutionary conflicts, adaptation
Domestication of transposable element proteins
Transposable elements (TEs) are selfish genetic elements (see Glossary) that are able to move and amplify within the genome of virtually all walks of life, including prokaryotes, unicellular and multicellular eukaryotes, and even large DNA viruses [1-3]. So-called autonomous TEs encode the enzymatic machinery to promote their own mobilization and propagation (Key Figure, Figure 1) as well as those of related non-autonomous elements, and occasionally unrelated host sequences. The disruptive effects of TEs have been documented extensively, for instance as they integrate into regulatory or coding regions of host genes or when they induce ectopic/non-allelic recombination events [4-6]. As a result, new TE insertions are often deleterious and removed from the population by purifying selection (see Glossary) or they are effectively neutral and fixed through genetic drift [7] (see Glossary). Consistent with the idea that the bulk of TE sequences do not serve host function, the rate and pattern of sequence evolution of TEs that are fixed in a genome generally follows that of unconstrained, neutrally evolving DNA leading to the accumulation of disabled and non-replicative TE ‘skeletons’ throughout genomes [8-10].
Key Figure, figure 1.
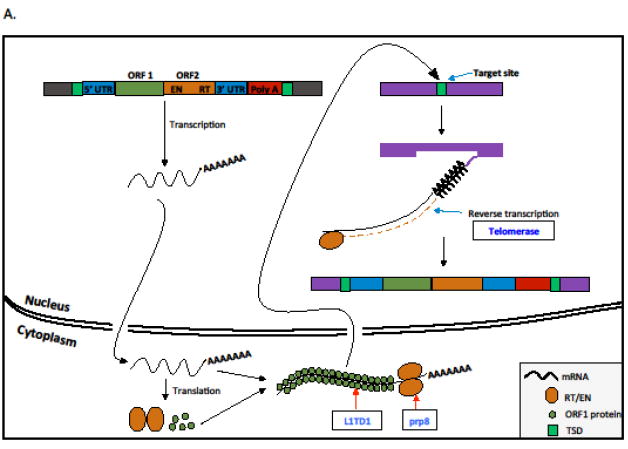
Transposition mechanisms of major types of transposable elements highlighting genes and functions that are involved in conflict. Class I elements or retrotransposons transpose via RNA intermediates (A and B), and Class II elements or DNA transposons mobilize directly as DNA molecules (C). We include retroviruses and endogenous retroviruses under LTR retrotransposons as previously proposed [100]. A. A typical non-LTR retroelement transposition (i.e., LINE element) is initiated by the transcription of the element. The transcript is translated into proteins and they associate with the mRNA and translocate into the nucleus. The retrotranscriptase (RT) protein has endonuclease activity and makes a nick on one of the strands and uses the 3’ end to prime synthesize a cDNA copy and insert into the genome. This process is known as target primed reverse transcription (TPRT). The nicks generated on two DNA strands are generally staggered and this results in target site duplications (TSDs). B. A typical LTR retrotransposon is characterized by long terminal repeats (LTRs) and generally encodes for three major proteins (GAG, POL, and ENV). The transposition is initiated by the transcription of three encoded genes as a single mRNA. The transcript is translated into several protein products. The POL gene is translated to three proteins, integrase (INT), RNAse H (RH), and RT. The GAG forms the capsid protein that encapsulated the LTR mRNA, int, RH, RT into a nucleocapsid virus like particle (VLP). The ENV, which is a glycoprotein, can promote the escape of the VLP from the cell. In the extracellular space, the viral surface ENV glycoprotein can recognize susceptible cells through recognition of the cell receptors and fuse with the cell membrane. Once fused, the nucleocapsid can enter into the cell cytoplasm. Alternatively, the VLP, instead of escaping the cell, can continue the transposition process within a single host cell. In the VLP, the RT reverse transcribes the RNA into cDNA which then associated with the INT. The INT guides the cDNA into the nucleus, where it finds a target site and integrates the element into the genome. Since the INT usually generates a staggered cut, the LTR elements are flanked by TSDs. C. A typical cut and paste DNA TE is flanked by target site duplications (TSDs), Terminal inverted repeats (TIRs) and encodes for at least a transposase. The transposition of the cut and paste element is initiated when the transposase is transcribed and translated. The transposase can either stay as monomer or form multimers. Alternatively, the transposase can also interact with other proteins (either encoded by the TE itself or host proteins). The transposase is then able to translocate into the nucleus where it recognizes and binds the TIRs. After binding to the TIRs the transposase catalyzes the excision of the TE from the donor site. When the TE (bound to the transposase) finds a target site, it makes a staggered cut and inserts itself into the new site. When the staggered cuts are repaired the TE remains flanked by TSDs.
These theoretical and empirical observations are in line with the notion that TEs owe their persistence and extraordinary diversification to their self-replicative and genetically invasive activities [11]. This selfish ‘raison d’être’ does not preclude that, on occasion, parts or whole TE sequences may be coopted to serve cellular function beneficial to the host organism. This process of cooption or ‘molecular domestication’ [12] of TE sequences has become increasingly recognized in recent years as advances in genomics have facilitated the identification, in various organisms, of a growing number of instances of TE-derived sequences that have been repurposed to serve cellular functions. The most robust evidence for such domestication events has come from either (i) comparative genomics whereby particular TE sequences can be inferred to have been immobilized and evolved under functional constraint acting at the level of the host organism for extended periods of time, and/or (ii) genetic evidence whereby mutation or experimental removal of TE sequences have advert effects on cell function and/or host fitness [1, 13-16]. Many of these well-documented examples point to the frequent cooption of TE-derived sequences as noncoding elements modulating host gene expression at the DNA or RNA level [13, 16-20]. By contrast, we herein focus on cases where the coding regions of TEs have been recruited as proteins serving host cell function. Out of the well-studied examples of TE protein domestication (listed in the Supplementary table), a substantial fraction appears to have been driven by the necessity to cope with various evolutionary conflicts and these will be the focus of this review (Table 1). We will highlight specific cases that illustrate both recent and older domestication events and reveal that TE proteins are often domesticated in response to intraspecific and interspecific evolutionary conflicts, as part of an arms race (see Glossary) characterized by ever-changing selective pressures [21]. Interestingly, some of these conflicts have played out repeatedly in multiple lineages and adaptation has occurred through independent TE domestication events, leading to convergent evolutionary innovations. Finally, we will argue that in certain conflicting situations TE domestication might be the sole, inevitable evolutionary resolution.
Table 1.
Compilation of TE protein domestication events highlighted in the text that have been driven by selection to adapt to evolutionary conflicts.
| Gene names | Ancestral TE | TE protein (domain)# | Originally discovered | Taxonomic distribution | Possible conflict | Function related to the conflict | References |
|---|---|---|---|---|---|---|---|
| abp1, cbh1, cbh2 | Tc1/mariner/po go | Transposase (whole) | Schizosacchar omyces pombe | Schizosaccharo mycetale | TE vs. host / Centromere drive | LTR retrotransposons repression; chromatin silencing at centromere. | [44, 45] |
| ALP1 | PIF/Harbinger | Transposase (whole) | Arabidopsis thaliana | Land plants | TE vs. host | Potential role in TE silencing through interaction with Polycomb Repressive Complex 2. | [49] |
| ancHTenv | Gammaretrovir uses | Envelope | Homo sapiens | Hominids | TE vs. host | Restrict viral entry into the host cell. | [42] |
| cag | Tc1/mariner/po go | Transposase (DBD) | Drosophila melanogaster | Unknown | Centromere drive | Predicted to bind centromeric DNA sequence. | [90] |
| Cas1 | Casposon | Transposase (whole) | Bacteria | Archaea and bacteria | Host vs. pathogen | Defense response to virus and maintenance of CRISPR repeat elements. | [33] |
| Cas9 | DNA transposon | tnpB | Bacteria | Archaea and bacteria | Host vs. pathogen | Integration of invading viral DNA into CRISPR locus. | [40] |
| CENPB | Tc1/mariner/pogo | Transposase (DBD) | Homo sapiens | Mammals | Centromere drive | Facilitates mitotic centromere formation, recognizes and binds a 17-bp sequence in the centromeric alpha satellite DNA. | [100, 101] |
| enJSRVenv | Betaretroviruses | Envelope | Sheep | Ovine/unknown | TE vs. host | Restrict viral entry into the host cell. | [41, 102] |
| Fv1 | MuERV-L (gag) (class III) | Capsid Protein | Mus musculus | Mus subgenera | TE vs. host | Murine leukemia virus restriction. | [103] |
| L1TD1 | L1 | ORF1 | Homo sapiens | Mammals | TE vs. host | Potential role in TE control. | [50] |
| MAIL1 | Ty3/gypsy LTR retrotransposo n | Plant mobile domain | Arabidopsis thaliana | Unknown | TE vs. host | Silencing of TEs and genes. Condensation of pericentromeric heterochromatin. | [48] |
| MAIN | Ty3/gypsy LTR retrotransposo | Plant mobile domain | Arabidopsis thaliana | Unknown | TE vs. host | Silencing of TEs and genes. Condensation of pericentromeric heterochromatin. | [48] |
| PiggyMac | piggyBac | Transposase (DBD? + core) | Paramecium tetraurelia | Unknown | TE vs. host | Required for genome rearrangement in Paramecium tetraurelia. | [55] |
| RAG1 | Transib | Transposase (core) | Homo sapiens | Jawed vertebrates | Host vs. pathogen | Important function in V(D)J combination and interacts with RAG2. | [26] |
| RAG2 | Transib | Transposase | Homo sapiens | Jawed vertebrates | Host vs. pathogen | Important function in V(D)J combination and interacts with RAG1. | [26] |
| Refrex-1 | Gammaretrovir uses | Envelope | Domestic cats | Feline/unknown | TE vs. host | Restrict viral entry into the host cell. | [41, 104] |
| Rmcf | Gammaretrovir uses | Envelope | Mus castaneus | Mus subgenera | TE vs. host | Defends against viral infection. | [41, 105] |
| Rmcf2 | Gammaretrovir uses | Envelope | Mus castaneus | Mus subgenera | TE vs. host | Defends against viral infection. | [41, 106] |
| Suppressyn | HERV-F | Envelope | Homo sapiens | Simians | TE vs. host | Regulates syncytins and potential viral restriction into host cells. | [41, 107] |
| Syncytin A, Syncytin B | HERV-F or HERV-H | Envelope | Mus musculus | Murid rodents | Fetus vs. mother | Placenta formation; fusogenic activities ex vivo. | [69] |
| Syncytin 1, Syncytin 2 | HERV-W | Envelope | Homo sapiens | Humans, apes, Old World monkeys | Fetus vs. mother | Cell fusion; placenta formation. | [69] |
| Syncytin-Car1 | CarERV3 (class I) | Envelope | Carnivores | Carnivores | Fetus vs. mother | Placenta-specific expression, fusogenic activities. | [69] |
| Syncytin-Ory1 | Type D retroviruses | Envelope | Rabbits and hares | Leporids: rabbits and hares | Fetus vs. mother | Placenta-specific expression, fusogenic activities. | [69] |
| Syncytin-rum1 | vertebrate retrovirus | Envelope | Bos taurus | Bos taurus | Fetus vs. mother | Placenta-specific expression, fusogenic activities. | [69] |
| TERT | LINE-like retroelement? | Reverse transcriptase | Homo sapiens | Eukaryotes | TE vs. host | RNA-directed DNA polymerase activity; telomerase RNA reverse transcriptase activity. | [108] |
| TPB2 | piggyBac | Transposase (DBD? + core) | Tetrahymena thermophila | Unknown | TE vs. host | Required for genome Rearrangement in Tetrahymena thermophile. | [60] |
| Prp8 | Retroelement | Reverse transcriptase | Homo sapiens | Eukaryotes | mobile introns vs. | Generation of catalytic spliceosome for second transesterification step; mRNA 3’-splice site recognition. | [66] |
DBD: DNA binding domain; Core: Catalytic core
Conflicts between hosts and pathogens
A recurrent theme implicating TE protein domestication is the emergence of adaptive immune systems (see Glossary). V(D)J recombination is a conserved process of jawed vertebrates that creates a virtually infinite repertoire of antibodies in their B and T cells, which in turn allows the recognition and neutralization of a vast diversity of antigens expressed by pathogens [22, 23]. The two crucial components of V(D)J recombination are: (i) the RAG1 and RAG2 proteins, which catalyze the DNA rearrangement reaction, and (ii) their cis-acting DNA sequences, the recombination signal sequences (RSS), which reside at the boundaries of the V (variable), D (diversity), and J (joining) segments that define the specific genomic sequences bound, cleaved, and joined to produce an essentially unlimited diversity of coding sequences [22, 24]. It is now firmly established that the catalytic core of RAG1, the protein that is responsible for cleavage activity, is a domesticated transposase derived from an ancient lineage of DNA transposons dubbed Transib [23, 25]. TE-encoded homologs of RAG1 occur in various invertebrates such as sea urchin and oysters [25, 26]. It is also likely that the RSS motif descends from the terminal inverted repeats (TIRs) of Transib elements, since these sequences and their arrangement are similar in Transib transposons [25]. In addition to RAG1, RAG2 also was shown to have TE origins and several lines of evidence suggested that both RAG1 and RAG2 were domesticated from the same ancestral Transib element [26]. This evolutionary scenario has been solidified by a recent study that functionally characterized an active Transib element from the lancelet, a member of the cephalochordates, which lack V(D)J recombination [27]. This transposon, coined ProtoRAG, encodes both RAG1- and RAG2-like genes arranged just like their domesticated vertebrate homologs and flanked by TIRs that resemble the RSS and is able to undergo TIR-dependent transposition through a mechanism strikingly similar to RAG1/2-mediated DNA rearrangement [27]. These results support the idea that not only RAG1 derives from a transposase, but that RAG2 and RSS also descend from an ancestral transposon related to ProtoRAG.
There is growing evidence that TE domestication was also instrumental to the emergence of another adaptive immune system, but this time of prokaryotes: the CRISPR-Cas system. To defend against the continuous assault of invasive (and often deadly) genetic elements such as plasmids and phages, many bacteria and archaea have evolved a form of adaptive immunity that consists of two minimal components: (i) a core Cas (CRISPR-associated system) protein complex, which has nucleic acid binding and endonuclease activities, and (ii) a guide RNA generated from clustered regularly interspaced short palindromic repeats (CRISPR) loci [28]. CRISPR loci are composed of noncontiguous direct repeats separated by variable spacer sequences that are formed by incorporating DNA bits of invasive genetic elements, thereby providing an heritable record of the cell’s previous exposure to various parasitic elements [29, 30]. If the invader in record enters the cell again, the CRISPR RNA (crRNA) will be used as a guide for the Cas protein complex to recognize and digest the invader’s nucleic acids [31]. Recent studies have uncovered a wide diversity of CRISPR-Cas systems that can be divided into two major classes. Class 1 systems are the most widespread in bacteria and archaea and are defined by multisubunit crRNA effector complexes, whereas class 2 systems represent only about 10% of the CRISPR-Cas loci and are defined by a single subunit crRNA–effector module [32]. Cas1, the most prominent Cas protein found in CRISPR-Cas systems, shares sequence similarity to the transposases encoded by a group of TEs called Casposons [33]. Additionally, Cas1 is able to integrate spacer sequences into the CRISPR locus through a biochemical mechanism that is strikingly similar to transposase [34, 35]. Akin to RSS sequences in V(D)J, it has been proposed that the CRISPR repeats may be derived from the TIRs of Casposon-like elements based on the observation that the TIRs of these transposons are very similar to CRISPR repeats in terms of sequence, size, secondary structure and their predicted ability to be bound and cleaved by Cas1-like transposases [36, 37]. Thus, evidence is mounting that class I CRISPR-Cas systems originated from the domestication of Casposons [33]. Other mobile genetic elements have contributed to the further diversification of Class 1 CRISPR-Cas systems. Notably, reverse transcriptase (RT) sequences derived from mobile group II introns have been coopted repeatedly in bacterial evolution to form Cas1-RT fusion proteins, which enable the acquisition of spacer sequences from parasitic RNA agents [38].
The class 2 CRISPR-Cas system, which is apparently less common, but still remarkably diverse in bacteria is thought to have an independent origin from class 1 CRISPR-Cas system. There is growing support to the notion that class 2 systems were also assembled from parts borrowed from transposons. Cas9-like proteins, which act as effectors in most class 2 systems, show sequence similarity to proteins called TnpB, which are poorly characterized but are commonly found in both autonomous and non-autonomous TEs [39, 40]. Phylogenetic analyses point to multiple domestication events of TnpB-proteins giving rise to different lineages of Cas9-like effectors for several class 2 CRISPR-Cas subtypes [39]. Taken together these findings paint a remarkable picture whereby multiple CRISPR-Cas systems have been assembled independently from the cooption of various types of TE-derived proteins during prokaryotic evolution.
Host-TE conflict resolved through TE domestication
In parallel to the arms race between host and pathogens plays another battle between cells and invasive genetic elements like TEs and retroviral relatives. Cells have evolved ways to overcome these conflicts through pathways and mechanisms that often rely on domesticated TE proteins. Several proteins derived from the Envelope (Env) gene of endogenous retroviruses are known to aid in the protection of the host cell by restricting infection of related retroviruses. Because Env is essential for entry of the virus into the host cell, endogenous Env expression can block viral entry through a competitive process called receptor interference [41]. There are at least six different Env-derived genes identified in species as diverse as mouse, cat, sheep and primates that are capable of protecting against the infection of related retroviruses [41, 42].
In addition to Env proteins, the Gag proteins encoded by endogenous retroviruses can also be coopted for restricting retroviral infection. A classic example is the mouse Fv1 gene which is derived from the Gag gene of a member of the endogenous retrovirus family ERV-L. Fv1 was initially identified as a restriction factor for the murine leukemia virus (which is not directly related to ERV-L), but was subsequently shown to be capable of protecting against a wide variety of retroviruses [43]. Thus, Fv1 may have acquired a broad antiviral function, though the molecular mechanisms by which restriction is achieved remain poorly understood. Interestingly, Fv1 is a rapidly evolving gene with signature of positive selection (see Glossary) diversifying the C-terminal region of the protein, which is known to be important for viral restriction [43], suggesting that this factor has been engaged in an arms race with retroviruses.
In the fission yeast Schizosaccharomyces pombe, three transposase-derived proteins (Abp1, Cbh1 and Cbh2) originated from pogo transposons have taken on partially overlapping function in controlling unrelated retrotransposons called Tf2 elements. These domesticated proteins have been reported to transcriptionally silence Tf2 retrotransposons by tethering histone deacetylases (see Glossary) to the long terminal repeats of these elements, which also prevents the chromosomal integration of Tf2 elements via homologous recombination [44-46]. Thus, proteins derived from one TE class have acquired the ability to silence TEs from a completely different class. TE silencing may not be the sole cellular function of the S. pombe transposase-derived proteins as they are also required for proper chromosome segregation [47]. Therefore, it is unclear whether the TE silencing function of Abp1, Cbh1 and Cbh2 evolved first or emerged secondarily through fortuitous recognition of Tf2 elements. Interestingly, pogo-like transposases have been domesticated in several additional lineages and also in part to serve centromeric function (see Box 2). We speculate that these repeated episodes of transposase domestication might have been promoted to suppress another conflict: the so-called centromere drive (Box 2).
Box 2. Is centromere drive promoting TE domestication?
Centromere binding protein B (CENP-B) is a conserved mammalian factor that is essential for the establishment of centromere identity (Black et al. 2007). CENP-B is derived from the transposase of a pogo-like DNA transposon [87]. Three CENP-B-like proteins (Abp1, Cbh1, and Cbh2) have also been identified in fission yeast by virtue of their sequence and functional similarities to mammalian CENP-B. However, the yeast genes are not orthologous to the mammalian CENP-B and were independently domesticated from a distinct lineage of pogo-like transposons, suggesting a form of convergent evolution [88]. In addition, there are two more reports of independent domestication events of pogo-like transposases: one in lepidopteran species with holocentric chromosomes [89] and one in Drosophila (called CAG), which may also be a centromere-associated protein [89, 90].
The recurrent domestication of pogo-like transposases in evolution is intriguing and might be driven by the ability of these proteins to turn into TE silencers as described in the text for the CENP-B-like proteins of fission yeast [44]. But the association of several CENP-B proteins with centromeric regions suggests that another genetic conflict might be another evolutionary force repeatedly underlying their cooption: the so-called centromere drive model [91]. Unlike mitosis, which produces two identical daughter cells or male meiosis, where all four gametes are produced, in female meiosis only one of the four meiotic products is passed to the next generation through the oocyte. This creates an opportunity for competition between homologous chromosomes to end up in the oocyte. The centromere is positioned in a way that it can effectively orient the chromosome during meiosis I through microtubule attachment and the proper orientation of a chromosome has been shown to be advantageous for its transmission to the next generation [11]. This phenomenon has led to a model dubbed centromere drive, where the centromere that positions the chromosome in the best orientation is selected, and is thought to be responsible for rapid evolution of centromeric DNA as its length and sequence can bias its transmission [92, 93]. Centromere drive is believed not to cause direct negative fertility effects to females, however if it reduces the fertility in males, the centromeric proteins would require to adapt and restore male fertility [94]. The conflict generated could have not only led to the rapid evolution of kinetochore proteins, but in addition, might have also led to the domestication of pogo-like transposase as centromere-binding proteins to adapt to the centromere drive.
In Arabidopsis, two genes MAIL1 and MAIN encode related proteins that appear to be evolutionarily derived from a subset of Ty3/gypsy retrotransposons found in angiosperms [48]. These proteins might have been initially captured from the host by these elements, but appear to have been reclaimed to partake in an epigenetic silencing pathway that transcriptionally represses a broad array of TEs [48]. Genetic loss of these genes resulted in impaired condensation of pericentromeric heterochromatin and upregulation of TE transcription suggesting their protein products are acting as transcriptional repressors. The molecular mechanisms by which MAIL1 and MAIN promote the formation of silent chromatin remain to be characterized, but involve a molecular pathway independent of small-interfering RNAs (see Glossary) and DNA methylation. It is intriguing that proteins related to MAIL1 and MAIN seem to have been acquired by TEs multiple times, including DNA transposons of the Mutator-like superfamily, which may reflect a counter-defense strategy deployed by these elements [48].
ALP1 is another domesticated TE protein identified in Arabidopsis that is involved in yet another epigenetic silencing pathway [49]. ALP1 directly derives from a PIF-like transposase and antagonizes silencing through a direct interaction with the Polycomb Repressor Complex 2 (PRC2), which is known to contribute to TE silencing in Arabidopsis [49]. The authors hypothesized that ALP1’s interaction with PRC2 could be an ancient property of PIF-like transposases that benefited the original transposon as a form of counter-repression [49]. Interestingly, in this case, the domesticated TE protein does not appear to play an effector role in TE repression but exerts a modulatory effect on a TE silencing pathway. The outcome must be beneficial to the host organism since the ALP1 gene display clear signature of evolutionary conservation and purifying selection across diverse land plant species.
Another example of domesticated TE protein that was potentially coopted for TE control is LINE-1 type Transposase Domain-containing 1 (L1TD1 [50]). L1TD1 was coopted in the ancestor of placental mammals from the ORF1 coding region of long interspersed nuclear elements LINE-1 (or L1s), one of the most abundant and persistent TE families in mammalian genomes. While the biochemical and cellular activities of L1TD1 remain to be defined, several observations suggest that it may be engaged in an arms race relationship with L1 elements. First, the evolution of the L1TD1 gene is characterized by bouts of rapid diversification under positive selection in primates and mice, lineages where L1 elements have undergone particularly dramatic bursts of diversification and expansion. Second, the L1TD1 gene has been lost multiple times during mammalian evolution, and at least in one lineage (megabats), its loss correlates with the (otherwise rare) extinction of L1 elements, as if L1TD1 was no longer needed once L1 became extinct. Although human L1TD1 now appears to function as a regulatory protein to maintain embryonic stem cell pluripotency, the authors argue that L1TD1 was initially domesticated to defend against L1 or other TEs [50].
TE proteins co-opted to eliminate TE-derived sequences
Ciliates are single-celled eukaryotes that are unique for harboring dimorphic nuclei [51]. The germline micronucleus (MIC) contains the genomic material that is passed down to the next generation while the somatic macronucleus (MAC) is not passed to the next generation but encodes all the proteins responsible for the organism’s function [51]. Like other typical eukaryotic genomes, the micronucleus contains a large amount of repetitive sequences and TEs interspersed with DNA sequences essential for the host [52, 53]. However, unlike any other genomes, the genes in the MIC are interrupted and sometimes even scrambled by a multitude of nongenic DNA segments, including repetitive elements called Internal Eliminated Sequences or IES that must be excised at the DNA level for correct assembly of the MAC and proper gene expression [54]. To excise IESs some ciliates have co-opted the cleavage properties of transposases. For instance, Paramecium uses PiggyMac (Pgm [55, 56]), a domesticated transposase, while in Tetrahymena at least four other transposase-derived proteins (TPB2, TPB1, TBP6, and Lia5) are required for IES removal [57-60]. While all of these proteins share sequence similarity to the piggyBac superfamily of transposases their evolutionary relationship to one another remains obscure. For example, it is unclear whether the Paramecium and Tetrahymena genes encoding these proteins are orthologous or if each is derived from distinct transposons independently domesticated and/or the product of gene duplication events [54, 58, 61]. The functions of these transposase-derived proteins also appear to vary or to have diverged after they were domesticated. In Paramecium, DNA elimination involves the precise excision of IES, which are located between and within genes, and requires the catalytically active PGM transposase as well as TA dinucleotides at the boundaries of IES. When excised in the MAC only a single TA remains [61]. In Tetrahymena, IES are primarily intergenic and while TPB2 has retained catalytic activities that are essential for IES removal [60], LIA5 appears to have lost its catalytic activity but remain essential for DNA elimination likely through its involvement in chromatin reorganization prior to IES excision [57, 58]. Finally, TPB1 and TPB6 appear to function as catalytically active transposases, but differ from TPB2 in being dedicated to the removal of a small subset of IES that resemble ancient piggyBac transposons [59].
There is growing evidence that IES themselves originated from TEs. In Paramecium, a fraction of IES appears to be derived from recent Tc1/mariner-like element invasions and resemble miniature elements or solitary TIRs, whose sequences appear to have converged to be excised by Paramecium’s PiggyMac leaving no scar behind unlike typical Tc1/mariner transposition [61]. In Tetrahymena, there is also substantial overlap and terminal sequences similarities between IES and TEs, including piggyBac-like elements [53, 59, 62]. These observations and what is known about the biochemistry of IES excision [61] support the idea that the process of IES elimination in Paramecium and Tetrahymena closely resembles the excision of piggyBac transposons, and that this type of elements could have provided both the enzymatic machinery and at least some of the cis-acting sequences now required for the process. Expanding upon this idea, we propose a model in which the dimorphic nuclei system of ciliates fosters the evolution of TEs that are specifically expressed during the transition from MIC to MAC as to be excised during that transition, which would minimize their deleterious impact and lead to their eventual domestication (see Box 1). Another ciliate species, Oxytricha, provides an outstanding model to test the hypothesis. In this species, certain DNA transposons called TBE undergo self-excision during the MIC to MAC transition and TBE transposase expression is essential for IES excision and proper genome unscrambling (Nowacki, et al. 2009). Thus TBE transposons might be at an early step toward domestication [54, 63] (see also Box 1 and Box 3).
Box 1. When TE domestication may be inevitable.
Here we argue that evolutionary conflicts drive TEs to evolve features that initially minimize their deleterious impact on the host, but eventually and inevitably lead to their domestication. We envision that the ciliate MIC/MAC binuclei transition, which involves a step of programmed DNA elimination when the MAC nucleus forms (see main text), might provide such an opportunity. Once this process is in place, natural selection would favor ciliate TEs that are expressed during the transition from MIC to MAC as this behavior would promote their propagation exclusively in the MIC and minimize deleterious effects on somatic development and function in the MAC (Figure I). This system further predicts that TEs capable of excision, such as cut-and-paste DNA transposons, would be favored over those that cannot like retrotransposons. Consistent with this, the TE landscape of Tetrahymena and Oxytricha MIC genomes appears to be dominated by DNA transposons [52, 53] (but maybe less so for Paramecium [83]). Another prediction of this model is that if a transposon lands in a region where its excision becomes essential for proper MAC (somatic) function and fixes in the population, it would impose functional constraint on at least one active transposase gene as well as the cis-acting sequences (TIRs) required for excision of the transposon (Figure I). This situation may progress from a type of mutualism [54, 64], where active TEs capable of excision are maintained by natural selection, to a complete domestication if the transposase becomes physically dissociated from its cis-acting sequences and can only function in trans without increasing the spread of the transposase. Interestingly, the transposases encoded by the TBE family of DNA transposons, which are currently active in Oxytricha, are known to bear the signature of purifying selection [64, 84]. This signature of constraint suggests that TBE transposase activity has to be maintained to preserve host fitness and might reflect an intermediate step toward eventual domestication. A final prediction of the model is that transposons that insert within or close to genes but excise precisely, leaving no molecular ‘footprints’ of their insertion, would be more likely to succeed in colonizing the MIC and therefore be more prone to evolve toward domestication. In this regard, it is notable that all transposases identified to date as domesticated for DNA elimination in ciliates derive from piggyBac transposons, which are known to insert preferentially within or near genes [85] and to produce precise excision events [86].
Box 3. Can whole TEs be domesticated?
A recurrent theme in the examples of TE domestication summarized in this review is that the domesticated TE proteins act in trans even when they act on substrates that resemble TE ends or derive from the same cognate TE (e.g., RAG1/2 in V(D)J recombination). But can a whole TE be domesticated as a unit? Below we argue that domestication is complete only when proteins are separated from rest of the TE sequences as genetic conflict still exist when the whole TE is still replicating even if it is potentially in the trajectory to being domesticated.
In Oxytricha, TEs belonging to Tc1/mariner superfamily excise themselves during the transition from MIC to MAC nucleus and their activity is needed for from MIC to MAC transition [63]. However, the conflict may still be ongoing in this case as the fitness of individuals is predictably lowered by new insertions of the active element that is likely still actively transposing [54] thereby increasing the chances of deleterious TE-excision-disabling mutations as offspring is produced (Figure I).
In Drosophila, the activity of non-LTR retrotransposons elongates telomeres. In these species three non-LTR retrotransposons (Het-A, TART, and TAHRE) retrotranspose to the very ends of the chromosomes using the 3’ end for reverse transcription preventing the telomeres from shortening [95, 96]. These elements preferentially target the end of the chromosomes and are rarely found in other genomic regions [97]. The presence of TEs at the ends of the chromosomes might be viewed as a domestication of whole TEs by the host. However, while the retroelements may have found a “safe heaven” at the ends of the chromosomes, perhaps, after the demise of the telomerase in Drosophila, the genome might actually still be in conflict with the TEs. It should be emphasized that the three non-LTR retroelements that transpose to the ends of the chromosomes in Drosophila have evolved features that differ from their non-LTR relatives such as targeting of the 5’ of other telomeric TEs at the end of chromosomes combined to unusually long UTRs that appear to specialize them toward their genomic niche of telomere targeting [96]. However, they appear to remain selfish elements as given the opportunity, DNA double strand breaks are recognized by these elements and they transpose into other genomic regions as well [96] and the opportunities for selfishness remain [98]. Thus these TEs cannot be considered fully domesticated and although the organism presumably benefits from their insertion at the telomeres there is evidence of ongoing conflict that might only be resolved by a complete domestication whereby the locus producing the template RNA is physically separated from that encoding the reverse transcriptase. It is tempting to speculate that telomerase, itself a reverse transcriptase, might have originated through this evolutionary path from a domesticated retroelement [99].
Parallels have been drawn between the ciliate IES and spliceosomal introns in eukaryotes [64]. Analogously to the TE domestication model for IES removal (see Box 1), it is tempting to speculate that TE proteins might have been ancestrally coopted to ensure spliceosomal intron splicing. There is solid evidence that spliceosomal introns are evolutionarily related and likely derive from sequences resembling group II introns (i.e., a type of class I mobile element). First, group II introns have structural similarity with spliceosomal introns (e.g., several components of the group II intron ribozyme including small RNAs are similar to the small nuclear RNAs of the spliceosome), and they also have a splicing mechanism that is strikingly reminiscent of the removal of spliceosomal introns [65]. Additionally, the prp8 protein, which is an integral component of the spliceosomal complex, displays significant sequence similarity with reverse transcriptases and as such has been proposed to derive from an anciently domesticated retroelement [66]. Since group II introns typically encode a reverse transcriptase that is essential for their insertion (Figure 1), group II and spliceosomal introns are not only similar in structure and excision mechanism, but also in parts of the enzymatic machinery that catalyzes their mobilization [65, 66]. These observations bring credence to the idea that spliceosome and introns could have originated via domestication of TE-encoded proteins and their cis-acting sequences, respectively. This hypothesis is in line with the proposal that the invasion of an ancient group II intron-like mobile element of an early eukaryotic ancestor might have led to the emergence of spliceosomal introns [64, 67]. Insertions of these introns within genes would have been tolerated by natural selection as long as the introns would have been spliced out after transcription, restoring the reading frame, imposing functional constraint on the machinery that ensures their splicing and leading to domestication of the machinery for proper cell function
In summary, TEs evolve ways to replicate and spread in the genome while minimizing their deleterious effects on host fitness, for instance by ensuring their excision at the DNA or RNA level. These processes create a dependence of the host on these enzymatic activities, which leads to the assimilation of TE-encoded proteins to the cellular machinery. Thus, the evolutionary dynamics of host-TE interactions create fertile ground for the domestication of TEs, which in turns add a layer of complexity to the organization and function of the genome.
TE proteins co-opted as a result of conflict between mother and embryo
Syncytins are proteins derived from the Env gene of retroviruses that have been coopted at least nine times during mammal evolution and are thought to play a role in placentation [68-71]. The placenta is a temporary organ that is formed by the fusion of fetal extraembryonic membrane and the maternal uterine tissue, which facilitates metabolic exchanges through the interface between the mother and the fetus [69]. The proposed function of Syncytins in the placenta is based on their restricted or high level of expression in that organ, their ability to mediate cell-to-cell fusion (which is required for the establishment of the syncytiotrophoblast layer at the fetal-maternal interface) and, in some case, also immunosuppressive activities [68]. Knockout studies of two murine-specific Syncytins in the mouse firmly established their critical function in placenta development [72, 73], but it remains unknown whether all Syncytins identified in other mammalian lineages are equally important for placentation. In fact, the evolution of Syncytins presents an evolutionary conundrum, because none are conserved across mammals, but instead each has emerged independently during mammal evolution through cooption events of distinct, lineage-specific retroviral Env sequences [68-71]. Some species like mouse and human even harbor multiple Syncytins in their genomes that originated at different evolutionary time points and there is also evidence that some Syncytins have been lost during evolution [68]. Could the repeated cooption and turnover of Syncytins reflect convergent adaptation to a persistent evolutionary battle? It has been proposed that the interface between the mother and the fetus in the placenta sets the stage for a conflict whereby the fetus selects for the ability to maximize the transfer of nutrients to itself while the mother, in response, adapts to limit the nutrient transfer and maintain overall homeostasis maximizing her offspring number [74]. This conflict is predicted to result in an evolutionary arms race driving rapid placental evolution and potentially Syncytin evolution. The model is supported by several genetic observations, including certain patterns of gene expression (imprinting; that reveals that the conflict might even start as a mother-father conflict [75, 76]) and evolution (positive selection) that are prevalent for placental-specific genes, and at an anatomical level by the remarkable diversification of this organ during mammalian evolution [69, 74, 77, 78].
A placenta feature that might facilitate the recurrent Syncytin cooption for placenta function could be the low level of DNA methylation relative to other tissues, which tends to promote the expression of TEs and endogenous retroviruses in particular in this organ [76, 79, 80]. In addition to the Syncytins, i.e., endogenous retrovirus gene domestication, numerous ERV and other TE sequences have been coopted as cis-regulatory elements to coordinate placental or uterine gene expression during pregnancy [81, 82]. Thus placenta hypomethylation that might facilitate the recurrent recruitment of ERV proteins and placenta-specific regulatory sequences might have influenced how embryos evolved the cellular fusion that started the conflict and might influence how it adapts to the everlasting evolutionary arms race between the fetus and the mother [78].
Concluding remarks
In this review we highlight three major routes by which TE proteins have been domesticated in response to genetic conflicts. First, TE proteins from various classes of elements have been repeatedly coopted to suppress TEs or retroviruses. The recurrence of this phenomenon may be explained by the fact that these TE proteins had pre-existing interactions with cellular machinery and with TEs themselves, which can be readily repurposed for TE suppression. A second, unforeseen route invokes the transition from actively transposing elements toward domestication imposed by their own selfish invasive strategies. This route, which is best exemplified by the process of DNA elimination and the formation of IES in ciliates or possibly the transition from self-splicing to spliceosomal introns, likely contributed to increasing complexity in genome organization and function during evolution. Finally, the last route involves the cooption of TE proteins and sequences for seemingly unexpected novel biological processes such as adaptive immune systems of vertebrates (V(D)J recombination) and prokaryotes (CRISPR-Cas). In those examples where TE proteins are coopted for seemingly completely new functions, the interactions of those proteins, the host cellular machinery and the interactions of TEs they derive from need to be better characterized if we are to obtain a complete picture of how those novel functions evolved.
Lastly, we want to highlight the challenges in assigning function to domesticated TE proteins. These challenges are very similar to assigning functions to any other candidate protein and reverse genetics methods are often used. However, from an evolutionary point of view, there is an interest in exploring the function that initially facilitated the domestication of TE proteins. When a TE protein is domesticated to resolve a conflict, it will most likely be incorporated into host cellular pathways. These pathways may evolve over time to a point that may obscure our understanding of the activity or interaction that initially triggered the domestication event. Thus the present function of a TE-derived protein may not always illuminate the initial process of domestication and, as a consequence, the role of genetic conflicts as the initial driver of TE domestication may remain underestimated.
Supplementary Material
Figure I.
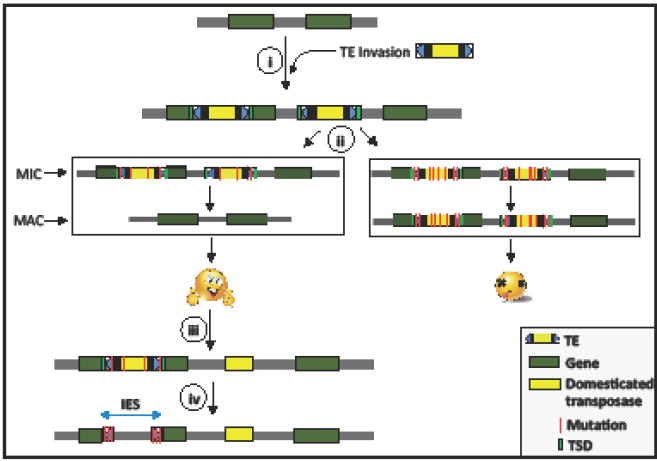
Model of the inevitability of TE domestication using ciliates as example. i) A novel TE invades a naïve ciliate genome and expands in copy number. Upon integration, TEs can potentially disrupt genes and regulatory sequences. ii) Because ciliates have dimorphic nuclei, micronucleus (MIC) and macronucleus (MAC), the organisms in which the TEs evolve to precisely excise during the transformation from the MIC to the MAC will have intact coding regions and regulatory sequences and will survive. These TEs will proliferate undetected by the host. Host with mutated copies of the TEs which cannot excise due to mutations in the TIRs or transposases cannot form intact ORF in the MAC and do not survive. Thus there is purifying selection and organisms that harbor TEs able to precisely excise, provided there are enough active TE copies that provide a source of transposase for excision have higher fitness. These TEs will keep proliferating and the potential for deleterious mutations in the TIRs will increase. iii) Conflict between TE and host is resolved by domesticating a TE protein to excise related TEs during the transition to the MAC nucleus. iv) Overtime the TE-related sequences accumulate mutations beyond recognition. However, the regions of TE (generally parts of TIRs) are under purifying selection since these sequences are important for the excision of disruptive sequences and gives rise to IESs.
Trends Box.
-
Transposable elements are selfish DNA elements That are able to increase in copy number by Xploiting host cellular functions.
Domestication of TE sequences by the host for cellular function is an evolutionary process that has been unexpectedlycommon.
Proteins encoded by transposable elements (TEs) are often repurposed to Perform host functions as part of novel protein---coding genes. Domesticated TE proteins are frequently coopted to mitigate evolutionary conflicts, especially in defense against pathogens and invasive genetic elements.
For certain TE conflicts, domestication might be an inevitable outcome.
Outstanding Questions Box.
--- Why do DNA transposon proteins appear to be more prone to domestication than retroelement proteins?
--- What types of interaction interfaces between host and TEs facilitate TE domestication?Are some interfaces more likely to be preserved and coopted?
--- How often does TE domestication lead to convergent molecular innovations?
--- How are TE genes put under the regulatory control of the host? Does it involve the replacement or modification of TE’s ancestral regulatory properties? Does the local genomic environment near a TE’s landing site play a major role?
--- To what extent is the population genetics of a species affecting the propensity and path toward TE domestication?
--- Are species accumulating more TEs in their genome more likely to co---opt TEs?
Acknowledgments
EB would like to acknowledge the support from the National Institute of General Medical Sciences of the National Institutes of Health (GM071813). CF is supported by grants GM112972, GM059290, HG009391, from the National Institutes of Health. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Glossary
- Adaptive immune systems
Antigen specific response system that involves antigen-specific recognition and neutralization.
- Arms race
In this publication, an evolutionary conflict that involves continuous competition between interacting species or genetic elements to adapt to the ever-changing interacting partner.
- Genetic drift
Evolutionary process leading to the chance change in allele frequencies due to the random effects caused by sampling in populations of finite population size.
- Histone deacetylases
Enzymes whose activity involves the removal of acetyl groups of histones. This histone modification most often leads to chromatin condensation.
- Positive selection
Evolutionary process leading to the increase in frequency of new beneficial alleles. When it occurs in the context of an arms race it is recurrent and leaves behind a signature of fast protein evolution.
- Purifying selection
Evolutionary process leading to the decrease in frequency of new deleterious alleles.
- Reverse transcription
Protein activity that leads to the synthesis of DNA using RNA as a template.
- Selfish genetic element
DNA sequences that have evolved the ability to propagate at a cost to the host.
- Small-interfering RNAs
In this publication, short cellular RNAs complementary to mRNAs that prompt targeted mRNA degradation and, often, targeted gene silencing through chromatin remodeling.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annual Review of Genetics. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curcio MJ, Derbyshire KM. The outs and ins of transposition: from mu to kangaroo. Nat Rev Mol Cell Biol. 2003;4(11):865–77. doi: 10.1038/nrm1241. [DOI] [PubMed] [Google Scholar]
- 3.Sun C, et al. DNA transposons have colonized the genome of the giant virus Pandoravirus salinus. BMC Biol. 2015;13:38. doi: 10.1186/s12915-015-0145-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hancks DC, Kazazian HH., Jr Roles for retrotransposon insertions in human disease. Mob DNA. 2016;7:9. doi: 10.1186/s13100-016-0065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beauregard A, et al. The take and give between retrotransposable elements and their hosts. Annu Rev Genet. 2008;42:587–617. doi: 10.1146/annurev.genet.42.110807.091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 2011;12(9):615–27. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lynch M. The origins of genome architecture. Sinauer Assocs., Inc; Sunderland, MA: 2007. [Google Scholar]
- 8.Wacholder AC, et al. Inference of transposable element ancestry. PLoS Genet. 2014;10(8):e1004482. doi: 10.1371/journal.pgen.1004482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carr M, et al. Evolutionary genomics of transposable elements in Saccharomyces cerevisiae. PLoS One. 2012;7(11):e50978. doi: 10.1371/journal.pone.0050978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Koning AP, et al. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7(12):e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burt A, Trivers R. Genes in conflict: The biology of selfish genetic elements. The Belknap Press of Harvard University Press; 2006. [Google Scholar]
- 12.Miller WJ, et al. Molecular domestication of mobile elements. Genetica. 1997;100(1-3):261–70. [PubMed] [Google Scholar]
- 13.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9(5):397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joly-Lopez Z, et al. Phylogenetic and Genomic Analyses Resolve the Origin of Important Plant Genes Derived from Transposable Elements. Mol Biol Evol. 2016;33(8):1937–56. doi: 10.1093/molbev/msw067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joly-Lopez Z, et al. A gene family derived from transposable elements during early angiosperm evolution has reproductive fitness benefits in Arabidopsis thaliana. PLoS Genet. 2012;8(9):e1002931. doi: 10.1371/journal.pgen.1002931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chuong EB, et al. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet. 2017;18(2):71–86. doi: 10.1038/nrg.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapusta A, Feschotte C. Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends Genet. 2014;30(10):439–52. doi: 10.1016/j.tig.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bourque G. Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr Opin Genet Dev. 2009;19(6):607–12. doi: 10.1016/j.gde.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 19.Rebollo R, et al. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet. 2012;46:21–42. doi: 10.1146/annurev-genet-110711-155621. [DOI] [PubMed] [Google Scholar]
- 20.Elbarbary RA, et al. Retrotransposons as regulators of gene expression. Science. 2016;351(6274):aac7247. doi: 10.1126/science.aac7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rice WR. Intergenomic conflict, interlocus antagonistic coevolutiona dn evolution of reproductive isolation. In: Howard DJ, Berlocher SH, editors. In Endless forms : species and speciation. Oxford University Press; 1998. pp. 261–270. [Google Scholar]
- 22.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–32. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 23.Carmona LM, Schatz DG. New insights into the evolutionary origins of the recombination-activating gene proteins and V(D)J recombination. FEBS J. 2016 doi: 10.1111/febs.13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oettinger MA, et al. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248(4962):1517–23. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 25.Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3(6):e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapitonov VV, Koonin EV. Evolution of the RAG1-RAG2 locus: both proteins came from the same transposon. Biol Direct. 2015;10:20. doi: 10.1186/s13062-015-0055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang S, et al. Discovery of an Active RAG Transposon Illuminates the Origins of V(D)J Recombination. Cell. 2016;166(1):102–14. doi: 10.1016/j.cell.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(5962):167–70. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 29.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 30.van der Ploeg JR. Analysis of CRISPR in Streptococcus mutans suggests frequent occurrence of acquired immunity against infection by M102-like bacteriophages. Microbiology. 2009;155(6):1966–1976. doi: 10.1099/mic.0.027508-0. [DOI] [PubMed] [Google Scholar]
- 31.Jiang F, Doudna JA. CRISPR-Cas9 Structures and Mechanisms. Annu Rev Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 32.Makarova KS, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13(11):722–36. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krupovic M, et al. Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity. BMC Biol. 2014;12:36. doi: 10.1186/1741-7007-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nunez JK, et al. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature. 2015;519(7542):193–8. doi: 10.1038/nature14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hickman AB, Dyda F. The casposon-encoded Cas1 protein from Aciduliprofundum boonei is a DNA integrase that generates target site duplications. Nucleic Acids Res. 2015;43(22):10576–87. doi: 10.1093/nar/gkv1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koonin EV, Krupovic M. Evolution of adaptive immunity from transposable elements combined with innate immune systems. Nat Rev Genet. 2015;16(3):184–92. doi: 10.1038/nrg3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beguin P, et al. Casposon integration shows strong target site preference and recapitulates protospacer integration by CRISPR-Cas systems. Nucleic Acids Res. 2016;44(21):10367–10376. doi: 10.1093/nar/gkw821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silas S, et al. Direct CRISPR spacer acquisition from RNA by a natural reverse transcriptase-Cas1 fusion protein. Science. 2016;351(6276):aad4234. doi: 10.1126/science.aad4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shmakov S, et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15(3):169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kapitonov VV, et al. ISC, a Novel Group of Bacterial and Archaeal DNA Transposons That Encode Cas9 Homologs. J Bacteriol. 2015;198(5):797–807. doi: 10.1128/JB.00783-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malfavon-Borja R, Feschotte C. Fighting fire with fire: endogenous retrovirus envelopes as restriction factors. J Virol. 2015;89(8):4047–50. doi: 10.1128/JVI.03653-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blanco-Melo D, et al. Co-option of an endogenous retrovirus envelope for host defense in hominid ancestors. Elife. 2017:6. doi: 10.7554/eLife.22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yap MW, et al. Evolution of the retroviral restriction gene Fv1: inhibition of non-MLV retroviruses. PLoS Pathog. 2014;10(3):e1003968. doi: 10.1371/journal.ppat.1003968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cam HP, et al. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature. 2008;451(7177):431–6. doi: 10.1038/nature06499. [DOI] [PubMed] [Google Scholar]
- 45.Johansen P, Cam HP. Suppression of Meiotic Recombination by CENP-B Homologs in Schizosaccharomyces pombe. Genetics. 2015;201(3):897–904. doi: 10.1534/genetics.115.179465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murton HE, et al. Restriction of Retrotransposon Mobilization in Schizosaccharomyces pombe by Transcriptional Silencing and Higher-Order Chromatin Organization. Genetics. 2016;203(4):1669–78. doi: 10.1534/genetics.116.189118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baum M, Clarke L. Fission yeast homologs of human CENP-B have redundant functions affecting cell growth and chromosome segregation. Mol Cell Biol. 2000;20(8):2852–64. doi: 10.1128/mcb.20.8.2852-2864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ikeda Y, et al. Arabidopsis proteins with a transposon-related domain act in gene silencing. Nat Commun. 2017;8:15122. doi: 10.1038/ncomms15122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang SC, et al. Kicking against the PRCs – A Domesticated Transposase Antagonises Silencing Mediated by Polycomb Group Proteins and Is an Accessory Component of Polycomb Repressive Complex 2. PLoS Genetics. 2015;11(12):e1005660. doi: 10.1371/journal.pgen.1005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McLaughlin RN, Jr, et al. Positive selection and multiple losses of the LINE-1-derived L1TD1 gene in mammals suggest a dual role in genome defense and pluripotency. PLoS Genet. 2014;10(9):e1004531. doi: 10.1371/journal.pgen.1004531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58(2):233–67. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X, et al. The architecture of a scrambled genome reveals massive levels of genomic rearrangement during development. Cell. 2014;158(5):1187–98. doi: 10.1016/j.cell.2014.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hamilton EP, et al. Structure of the germline genome of Tetrahymena thermophila and relationship to the massively rearranged somatic genome. Elife. 2016:5. doi: 10.7554/eLife.19090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vogt A, et al. Transposon domestication versus mutualism in ciliate genome rearrangements. PLoS Genet. 2013;9(8):e1003659. doi: 10.1371/journal.pgen.1003659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baudry C, et al. PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes Dev. 2009;23(21):2478–83. doi: 10.1101/gad.547309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dubois E, et al. Multimerization properties of PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements. Nucleic Acids Res. 2017;45(6):3204–3216. doi: 10.1093/nar/gkw1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vogt A, Mochizuki K. A domesticated PiggyBac transposase interacts with heterochromatin and catalyzes reproducible DNA elimination in Tetrahymena. PLoS Genet. 2013;9(12):e1004032. doi: 10.1371/journal.pgen.1004032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shieh AW, Chalker DL. LIA5 is required for nuclear reorganization and programmed DNA rearrangements occurring during tetrahymena macronuclear differentiation. PLoS One. 2013;8(9):e75337. doi: 10.1371/journal.pone.0075337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng CY, et al. The piggyBac transposon-derived genes TPB1 and TPB6 mediate essential transposon-like excision during the developmental rearrangement of key genes in Tetrahymena thermophila. Genes Dev. 2016;30(24):2724–2736. doi: 10.1101/gad.290460.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng CY, et al. A domesticated piggyBac transposase plays key roles in heterochromatin dynamics and DNA cleavage during programmed DNA deletion in Tetrahymena thermophila. Mol Biol Cell. 2010;21(10):1753–62. doi: 10.1091/mbc.E09-12-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arnaiz O, et al. The Paramecium germline genome provides a niche for intragenic parasitic DNA: evolutionary dynamics of internal eliminated sequences. PLoS Genet. 2012;8(10):e1002984. doi: 10.1371/journal.pgen.1002984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fass JN, et al. Genome-Scale Analysis of Programmed DNA Elimination Sites in Tetrahymena thermophila. G3 (Bethesda) 2011;1(6):515–22. doi: 10.1534/g3.111.000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nowacki M, et al. A functional role for transposases in a large eukaryotic genome. Science. 2009;324(5929):935–8. doi: 10.1126/science.1170023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Witherspoon DJ, et al. Selection on the protein-coding genes of the TBE1 family of transposable elements in the ciliates Oxytricha fallax and O. trifallax. Mol Biol Evol. 1997;14(7):696–706. doi: 10.1093/oxfordjournals.molbev.a025809. [DOI] [PubMed] [Google Scholar]
- 65.Lambowitz AM, Belfort M. Mobile Bacterial Group II Introns at the Crux of Eukaryotic Evolution. Microbiol Spectr. 2015;3(1) doi: 10.1128/microbiolspec.MDNA3-0050-2014. MDNA3-0050-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dlakic M, Mushegian A. Prp8, the pivotal protein of the spliceosomal catalytic center, evolved from a retroelement-encoded reverse transcriptase. RNA. 2011;17(5):799–808. doi: 10.1261/rna.2396011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006;440(7080):41–5. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- 68.Esnault C, et al. Differential evolutionary fate of an ancestral primate endogenous retrovirus envelope gene, the EnvV syncytin, captured for a function in placentation. PLoS Genet. 2013;9(3):e1003400. doi: 10.1371/journal.pgen.1003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lavialle C, et al. Paleovirology of ‘syncytins’, retroviral env genes exapted for a role in placentation. Philos Trans R Soc Lond B Biol Sci. 2013;368(1626):20120507. doi: 10.1098/rstb.2012.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cornelis G, et al. Retroviral envelope gene captures and syncytin exaptation for placentation in marsupials. Proc Natl Acad Sci U S A. 2015;112(5):E487–96. doi: 10.1073/pnas.1417000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cornelis G, et al. Retroviral envelope syncytin capture in an ancestrally diverged mammalian clade for placentation in the primitive Afrotherian tenrecs. Proc Natl Acad Sci U S A. 2014;111(41):E4332–41. doi: 10.1073/pnas.1412268111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dupressoir A, et al. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc Natl Acad Sci U S A. 2011;108(46):E1164–73. doi: 10.1073/pnas.1112304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dupressoir A, et al. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci U S A. 2009;106(29):12127–32. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haig D. Placental growth hormone-related proteins and prolactin-related proteins. Placenta. 2008;29(Suppl A):S36–41. doi: 10.1016/j.placenta.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 75.Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 1991;7(2):45–9. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- 76.Haig D. Transposable elements: Self-seekers of the germline, team-players of the soma. Bioessays. 2016;38(11):1158–1166. doi: 10.1002/bies.201600125. [DOI] [PubMed] [Google Scholar]
- 77.Chuong EB, et al. Maternal-fetal conflict: rapidly evolving proteins in the rodent placenta. Mol Biol Evol. 2010;27(6):1221–5. doi: 10.1093/molbev/msq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chuong EB. Retroviruses facilitate the rapid evolution of the mammalian placenta. Bioessays. 2013;35(10):853–61. doi: 10.1002/bies.201300059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haig D. Going retro: Transposable elements, embryonic stem cells, and the mammalian placenta (retrospective on DOI 10.1002/bies.201300059) Bioessays. 2015;37(11):1154. doi: 10.1002/bies.201500114. [DOI] [PubMed] [Google Scholar]
- 80.Schroeder DI, et al. Early Developmental and Evolutionary Origins of Gene Body DNA Methylation Patterns in Mammalian Placentas. PLoS Genet. 2015;11(8):e1005442. doi: 10.1371/journal.pgen.1005442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lynch VJ, et al. Ancient transposable elements transformed the uterine regulatory landscape and transcriptome during the evolution of mammalian pregnancy. Cell Rep. 2015;10(4):551–61. doi: 10.1016/j.celrep.2014.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chuong EB, et al. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat Genet. 2013;45(3):325–9. doi: 10.1038/ng.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guerin F, et al. Flow cytometry sorting of nuclei enables the first global characterization of Paramecium germline DNA and transposable elements. BMC Genomics. 2017;18(1):327. doi: 10.1186/s12864-017-3713-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen X, Landweber LF. Phylogenomic analysis reveals genome-wide purifying selection on TBE transposons in the ciliate Oxytricha. Mob DNA. 2016;7:2. doi: 10.1186/s13100-016-0057-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gogol-Doring A, et al. Genome-wide Profiling Reveals Remarkable Parallels Between Insertion Site Selection Properties of the MLV Retrovirus and the piggyBac Transposon in Primary Human CD4+ T Cells. Mol Ther. 2016;24(3):592–606. doi: 10.1038/mt.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mitra R, et al. piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 2008;27(7):1097–109. doi: 10.1038/emboj.2008.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smit AF, Riggs AD. Tiggers and DNA transposon fossils in the human genome. Proc Natl Acad Sci U S A. 1996;93(4):1443–8. doi: 10.1073/pnas.93.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Casola C, et al. Convergent domestication of pogo-like transposases into centromere-binding proteins in fission yeast and mammals. Mol Biol Evol. 2008;25(1):29–41. doi: 10.1093/molbev/msm221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.d’Alencon E, et al. Characterization of a CENP-B homolog in the holocentric Lepidoptera Spodoptera frugiperda. Gene. 2011;485(2):91–101. doi: 10.1016/j.gene.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 90.Mateo L, Gonzalez J. Pogo-like transposases have been repeatedly domesticated into CENP-B-related proteins. Genome Biol Evol. 2014;6(8):2008–16. doi: 10.1093/gbe/evu153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Henikoff S, et al. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293(5532):1098–102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 92.McLaughlin RN, Jr, Malik HS. Genetic conflicts: the usual suspects and beyond. J Exp Biol. 2017;220(Pt 1):6–17. doi: 10.1242/jeb.148148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Malik HS. Mimulus finds centromeres in the driver’s seat. Trends Ecol Evol. 2005;20(4):151–4. doi: 10.1016/j.tree.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 94.Roach KC, et al. Rapid evolution of centromeres and centromeric/kinetochore proteins. In: Singh RS, et al., editors. In Rapidly Evolving Genes and Genetic Systems. Oxford University Press; 2012. pp. 83–93. [Google Scholar]
- 95.Pardue ML, Debaryshe P. Adapting to life at the end of the line: How Drosophila telomeric retrotransposons cope with their job. Mob Genet Elements. 2011;1(2):128–134. doi: 10.4161/mge.1.2.16914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pardue ML, DeBaryshe PG. Retrotransposons that maintain chromosome ends. Proc Natl Acad Sci U S A. 2011;108(51):20317–24. doi: 10.1073/pnas.1100278108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Servant G, Deininger PL. Insertion of Retrotransposons at Chromosome Ends: Adaptive Response to Chromosome Maintenance. Front Genet. 2016;6:358. doi: 10.3389/fgene.2015.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee YCG, et al. Recurrent innovation at genes required for telomere integrity in Drosophila. Mol Biol Evol. 2016;34(2):467–482. doi: 10.1093/molbev/msw248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.de Lange T. T-loops and the origin of telomeres. Nat Rev Mol Cell Biol. 2004;5(4):323–9. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- 100.Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8(12):973–82. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


