Abstract
Proteins are constantly challenged by environmental stress conditions that threaten their structure and function. Especially problematic are oxidative, acid, or severe heat stress, which induce very rapid and widespread protein unfolding and generate conditions that make canonical chaperones and/or transcriptional responses inadequate to protect the proteome. Here, we review recent advances in identifying and characterizing stress-activated chaperones, which are inactive under non-stress conditions but become potent chaperones under specific protein-unfolding stress conditions. We discuss the posttranslational mechanisms by which these chaperones sense stress and consider the role intrinsic disorder plays in their regulation and function. We examine their physiological roles under both non-stress and stress conditions, their integration into the cellular proteostasis network, and their potential as novel therapeutic targets.
Keywords: proteostasis, molecular chaperone, oxidative stress, protein unfolding, protein aggregation
Stress-Activated Chaperones
Cells and organisms are constantly challenged by a variety of intracellular and extracellular stress conditions. Some of the most vulnerable organisms are bacteria and unicellular parasites, which are exposed not only to rapidly changing environmental conditions, but also encounter extremely fast acting stress conditions upon entry into the host. These stress conditions, which have evolved as part of the mammalian host defense, include sudden changes in temperature as encountered during host invasion and/or inflammation, sudden drops in pH as experienced when ingested bacteria pass through the mammalian stomach, or exposure to very high levels of reactive oxygen species (ROS) or reactive chlorine species (RCS) at the epithelia of the gut or during the oxidative burst in macrophages [1-3]. All of these stress conditions cause significant cellular damage in a matter of seconds to minutes, simultaneously affecting the structure and stability of many different proteins [4, 5] (see Box 1). To counteract these sudden-onset protein-damaging conditions, organisms have evolved a subgroup of molecular chaperones that are posttranslationally activated under the very specific stress conditions that demand their activity (see Table 1). To achieve this, these chaperones have developed elaborate regulatory mechanisms to rapidly sense the respective stress conditions and respond with the specific activation of their chaperone function. Once activated, the chaperones typically serve as ATP-independent chaperone holdases (see Glossary) that apparently form stable complexes with the unfolding client proteins and prevent them from irreversibly aggregating as long as stress conditions persist. Stress-induced transcriptional upregulation of these chaperones increases their levels and likely contributes to prolonged protection [6]. Once the stress subsides, for instance when ingested bacteria reach the intestine or a parasite leaves its mammalian host, the stress-activated chaperones return to their original, chaperone-inactive state. The client proteins are released and can either refold on their own or use canonical chaperone foldases (Table 1) for refolding. This mechanism provides a unique ability to deal with stress conditions that would otherwise inactivate and kill the organisms. Although stress-activated chaperones have primarily been identified in bacteria, recent work suggests that they might also serve important protective functions in eukaryotic cells. In this review, we discuss the various strategies that stress-activated chaperones employ to sense and respond to fast acting stress conditions, including oxidative, acid, and heat stress, and consider the physiological roles that these important first line of defense proteins play in cell survival.
Box 1. Effects of Oxidative, Acid, or Heat Stress on Protein Structure and Function.
Proteins are the major targets of endogenous and exogenous stress. Elevated temperatures and low pH are known to destabilize a protein's tertiary structure by disrupting hydrogen bonds and electrostatic interactions, causing loss of secondary structure and inducing unfolding [4, 5]. Exposure of hydrophobic surfaces, normally buried in the interior of the protein, can cause non-specific protein-protein interactions and lead to irreversible protein aggregation. Reactive oxygen and chlorine species cause a variety of side chain modifications, including the oxidation of sulfur-containing cysteine and methionine, chlorination of side-chain amines, and oxidation of histidines and tryptophans [14, 91, 92]. These modifications, when affecting functionally or structurally important residues, can lead to loss of protein function, increased destabilization, protein unfolding, fragmentation, and aggregation. In some proteins, posttranslational modifications, especially thiol oxidation, are reversible processes and have regulatory functions [15, 93].
Table 1. Stress-Activated Chaperones.
| Prokaryotes | In vitro Activating Conditions | Stress-Sensitive Phenotypes* | Ref |
|---|---|---|---|
| Hsp33 | H2O2 + Temp > 43°C HOCl or HOBr Bilesalt + H2O2 | H2O2 + heat shock HOCl or HOBr Bilesalt | [20] [28, 101] [30] |
| RidA | HOCl | HOCl | [60] |
| HdeA / HdeB | pH 2 / pH 4 | pH 2-3 / pH 4 | [65, 66, 73] |
| Eukaryotes | In vitro Activating Conditions | Stress-Sensitive Phenotypes* | Ref |
| Get3 | H2O2 + Cu2+ | Cu2+ | [41, 55] |
| Hsp26 | Temp > 43°C | - | [76] |
| HspB5/HspB1 | pH 6.5-7.5 | n.d. | [77, 78] |
| Tsa1 | H2O2 Temp > 43°C | H2O2 Heat shock | [85] |
| SmPrxI | pH 4.2 H2O2 | n.d. | [86] |
| mTXPNx/C2C-Prx1 | Temp > 40°C | Heat shock | [87, 88, 90] |
only stress-related phenotypes of the respective deletion mutants are listed; n.d. not determined.
Redox-Regulated Chaperones: First Defenders Against Oxidative Stress
Aerobically growing cells contain an assortment of antioxidant enzymes (e.g., peroxiredoxins, catalases) and small redox-buffering systems (e.g., glutathione) that work together to maintain redox homeostasis and protect cells against the toxic accumulation of ROS (for an overview, see Ref. [7]). However, as nearly always in life, systems can fail. In humans, for instance, aging, age-related neurodegenerative diseases, and certain metabolic diseases (e.g., diabetes) can disturb the cellular redox homeostasis, causing a condition termed oxidative stress [8-10]. Moreover, UV light, gamma rays, and X-rays, as well as various pollutants can promote oxidative stress in humans [11]. Bacteria, too, are exposed to high levels of oxidative stress when they invade their hosts. Neutrophils, part of the innate immune defense system, contain myeloperoxidase, which converts peroxide into the extremely reactive hypochlorous acid (HOCl, bleach) that acts as a very powerful natural disinfectant [1].
The expression of most known molecular chaperones is under heat shock control and is triggered by the accumulation of protein unfolding intermediates [12]. Although this response works well for stress conditions that slowly increase the pool of unfolded proteins, some ROS, such as hydroxyl radicals and RCS are, by definition, extremely reactive. They can very rapidly react with protein side chains, with rate constants typically in the 106-107 M-1 s-1 range, and cause irreversible damage (see Box 1) long before cells can respond with transcriptional upregulation and de novo synthesis of new chaperones, which can take up to an hour in bacteria [13, 14]. Moreover, many proteins involved in transcription and translation are themselves highly oxidation-sensitive, causing a decrease in general protein transcription and translation during severe oxidative stress [15, 16]. Finally, oxidative stress triggers a sharp decrease in cellular ATP levels (Figure 1). This not only contributes to the downregulation of many essential processes during oxidative stress [17], but also reduces the activity of some canonical chaperones, such as heat shock protein (Hsp) 70 (DnaK) [18]. It has long been thought that the drop in intracellular ATP levels is due to the oxidative inactivation of glyceraldehyde dehydrogenase and other players of glycolysis and oxidative phosphorylation [17]. Recent studies in Escherichia coli, however, demonstrated that the observed decline in ATP levels upon HOCl stress is at least in part caused by the active re-routing of ATP into long chains of inorganic phosphate, which themselves function as molecular chaperones that protect proteins against oxidative unfolding (Figure 1) [19].
Figure 1. Stress-Activated Chaperones Ensure Bacterial Survival.
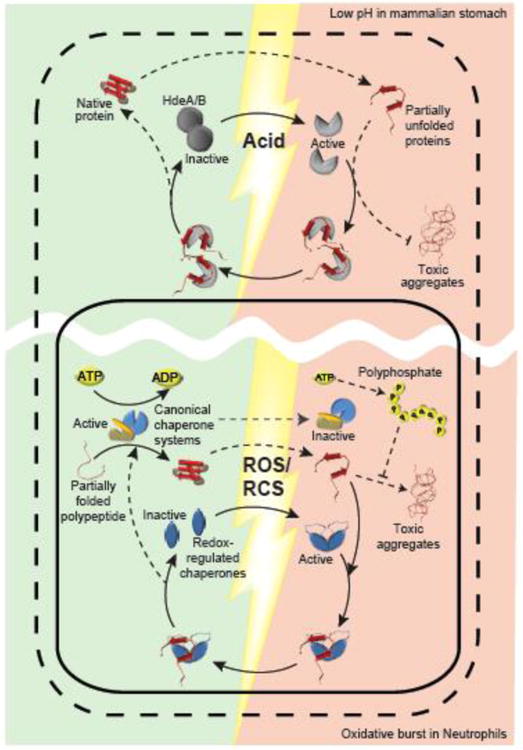
Bacteria are exposed to extremely fast acting stress conditions that have evolved as part of the mammalian host defense. Ingested gram-negative bacteria that colonize the intestine need to survive the highly acidic environment of the stomach. Their porous outer membrane (dashed line) allows the free passage of small molecules such as protons causing the sudden acid-induced unfolding of periplasmic proteins (red shaded area). To deal with these stress conditions, Gram-negative bacteria employ the periplasmic chaperones HdeA and HdeB (grey), which undergo rapid acid-induced unfolding and dissociation, activating their ATP-independent chaperone function. Chaperone-active HdeA/B bind unfolding proteins (red), maintain their solubility and prevent the accumulation of toxic protein aggregates. Upon return to neutral pH (green shaded area), HdeA mediates client protein refolding concomitant with its own refolding. Another mammalian host defense involves the production of high levels of reactive oxygen or chlorine species (ROS/RCS), such as H2O2 and HOCl, respectively at gut epithelia and in macrophages. ROS/RCS-mediated side chain modifications cause protein unfolding and aggregation in the cytosol (red shaded area within solid line). In addition, ROS/RCS cause a decline in cellular ATP levels, and directly inhibit canonical ATP-dependent chaperones. In bacteria, ATP is largely rerouted to polyphosphate, which functions as chaperone itself. To cope with this stress, bacteria employ redox-regulated ATP-independent chaperone such as Hsp33 and RidA (blue), which sense ROS/RCS through disulfide bond formation or N-chlorination of amino acid side chains, respectively. Chaperone-active Hsp33/RidA bind unfolding proteins, prevent their aggregation and maintain their solubility. Once reducing, non-stress conditions and ATP levels are restored, canonical chaperones release and refold the unfolded client proteins bound to Hsp33. So far it remains unclear whether RidA can transfer its substrate to canonical chaperones for refolding.
Over the past few years, several chaperones have been identified whose chaperone function is activated as a direct consequence of ROS/RCS-mediated posttranslational modifications, either through oxidation of redox sensitive cysteines (e.g., prokaryotic Hsp33, eukaryotic Get3) or chlorination of amino acid side chains (e.g., E. coli RidA). Deletion of these genes renders the respective organisms highly sensitive to in vivo protein aggregation and causes a significant increase in oxidative stress sensitivity, suggesting that one mechanism by which physiological oxidants damage cells and kill organisms is through the oxidative unfolding and aggregation of proteins.
Hsp33: A Paradigm for Stress-Activated Chaperones
The first report of a redox-regulated chaperone came in 1999 [20], only a few months after the discovery that transient oxidation-induced disulfide bond formation can take place within the reducing environment of the E. coli cytosol and is used for the activation of the oxidative stress transcriptional regulator OxyR [21]. Hsp33, a 33 kDa heat shock protein that is highly conserved in bacteria, several eukaryotic parasites such as Leishmania, and green algae, was found to use oxidation of four absolutely conserved cysteines to convert from an inactive chaperone under non-stress conditions to an active chaperone under oxidative stress conditions [20] (Figure 1). The finding that cytosolic proteins, such as OxyR and Hsp33, use reversible disulfide bond formation to actively control their protein function, contributed to a paradigm shift in the field. Now, almost 20 years later and fostered by the development of highly quantitative redox proteomic techniques, hundreds of proteins are known to use redox regulation as a posttranslational mechanism to control their protein activity in response to oxidative stress [22-24].
The stress sensor in Hsp33 is composed or four absolutely conserved cysteine residues that reside in Hsp33's C-terminal redox switch domain. Under non-stress conditions, all four cysteines are engaged in the high-affinity binding of one zinc ion (Figure 2). Zinc binding appears to serve at least two purposes: it primes the cysteines for rapid oxidation by coordinating them in a highly reactive thiolate anion form, but, at the same time, it stabilizes them and prevents their premature oxidation [25]. Over the years, many more proteins have been identified that use zinc-coordinating cysteine motifs as functional redox switches, which directly communicate increased ROS levels to physiological changes in organisms [26].
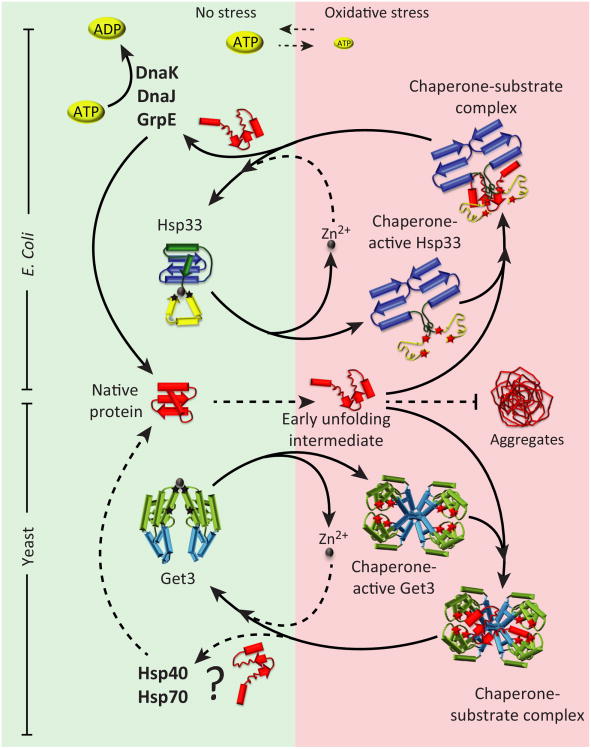
Figure 2. Analogies between pro-and eukaryotic redox-regulated chaperones.
Hsp33 in bacteria and the structurally unrelated tail-anchored protein insertion factor Get3 in yeast turn into effective ATP-independent chaperones during oxidative stress conditions. Under reducing non-stress conditions, both proteins are chaperone-inactive. Hsp33, a monomer when reduced, contains four redox sensitive cysteines (indicated with stars), which are located in the C-terminal redox-sensing domain (yellow). The cysteines are arranged in a C-X-C and C-X-Y-C motif and coordinate one zinc ion. Zinc binding confers stability to the C-terminus as well as an adjacent flexible linker region (green), which nestles onto the N-terminal domain of Hsp33 (blue). Get3, a dimer when reduced, also contains four redox sensitive cysteines arranged in a C-X-C and C-X-Y-C motif (stars) and coordinates zinc (grey sphere). In contrast to Hsp33, however, only the C-X-C motifs of the two monomers coordinate zinc, which stabilizes the reduced dimer. The two other cysteines are located in close vicinity to the ATP binding pocket of the ATPase domain (light green) of Get3. Upon exposure to oxidative stress, each protein forms two intramolecular disulfide bonds, connecting the two neighboring cysteines (red star) within the respective motifs. This oxidative disulfide bond formation causes zinc release in both proteins, and massive structural rearrangements. Oxidative activation of Hsp33 involves significant unfolding of the C-terminal redox-sensing domain and the flexible linker region, and is accompanied by dimerization. In Get3, structural rearrangements appear to affect the ATPase domain and alpha-helical subdomain (blue) and leads to the formation of tetramers and higher oligomers with no significant ATPase activity. Both oxidized Hsp33 and Get3 bind tightly to unfolding protein intermediates and prevent their aggregation in an ATP-independent manner. Upon return to non-stress condition, thiols in both chaperones become reduced and the structural rearrangements are reversed. Unfolded substrate proteins bound to Hsp33 are transferred to the DnaK/DnaJ/GrpE system, which promotes refolding to their native structures. The fate of Get3's client proteins remains to be elucidated.
The mechanism by which Hsp33 activates its chaperone function appears to be precisely tailored to oxidative stress conditions that induce protein unfolding. For example, neither peroxide alone nor heat alone causes Hsp33 activation. This makes physiological sense because peroxide alone leads to cellular ATP decline but does not cause protein unfolding in bacteria [18], and heat-induced aggregation is taken care off by canonical chaperones [27]. Peroxide stress in combination with heat stress, however, causes protein aggregation, rapidly inactivates canonical chaperones such as DnaK within minutes, and, most importantly, activates Hsp33 [18]. By contrast, HOCl, which oxidizes and unfolds proteins within seconds at any temperature, activates Hsp33 at any temperature [28]. Analysis of Hsp33's three-dimensional structure revealed the answer as to how Hsp33's redox switch can be so precisely tuned to these specific stress conditions. Hsp33 consists of two domains, a compactly folded N-terminal domain and a C-terminal redox-sensing domain, which is equally compactly folded as long as the four cysteines are reduced and zinc coordinated [25] (Figure 2). The two domains are connected by a highly charged ∼50 aa linker region, which folds into three helices and covers a largely hydrophobic four-stranded β-sheet platform in the N-terminal domain [25, 29]. In the presence of kinetically slow (non-protein unfolding) oxidants, such as peroxide, the distal C-X-X-C pair engages in a disulfide bond, which causes zinc release and the partial unfolding of Hsp33's C-terminal domain. This unfolding destabilizes the upstream linker region and converts it into a thermolabile folding sensor, ready to unfold should unfolding conditions (e.g., elevated temperatures, bile salts) be encountered as well [30, 31]. Once the linker region unfolds, the proximal C-X-C pair forms the second disulfide bond, leading to the constitutive unfolding of the linker region and the constitutive activation of Hsp33 as a chaperone [32]. Kinetically fast, protein-unfolding oxidants, such as HOCl, rapidly form both disulfide bonds in Hsp33 and activate the chaperone within the mixing time of in vitro experiments (T1/2 <1 min) [28]. Once disulfide bonded and partially unfolded, two oxidized Hsp33's monomers associate and form a chaperone-active dimer [28] (Figure 2).
Proteins with regions of intrinsic disorder have long been known to exist (Box 2) [33]. However, intrinsic disorder was generally associated with the inactive form of the protein, and refolding was thought to be required to attain cellular function. The discovery that Hsp33 must lose its structure and convert into an intrinsically disordered protein to gain activity suggested a second paradigm shift in biology. Several other so-called conditionally disordered chaperones are also known to activate by partial protein unfolding [34].
Box 2. Role of Intrinsic Disorder in Chaperone Function.
Stress-activated chaperones that are activated by partial unfolding are termed conditionally disordered because they can adopt a folded or an intrinsically disordered (ID) state, depending on the environmental conditions [34]. Regions of intrinsic disorder are typically rich in polar charged and uncharged amino acids and poor in hydrophobic amino acids [94], and many excellent prediction programs have been developed to identify these regions in proteins [95]. Due to a lack in hydrophobic core, ID regions are unable to fold into defined secondary or tertiary structures unless stabilized by other proteins, lipids, or membranes [96]. In reduced Hsp33, the linker region, which is predicted to be intrinsically disordered and unable to fold in isolation [32], is stabilized by the nearby zinc-containing redox switch. When the zinc binding domain is destabilized by disulfide bond formation and zinc release, however, the linker region converts into an intrinsically disordered structure [35]. Disordered regions appear to play several important roles in chaperone function. They provide: (1) wide-range electrostatic interactions, which appear to be involved in the initial recognition and binding of unfolding intermediates [35, 97] and which are followed by closer-range hydrophobic interactions that stabilize the complex; (2) flexibility and plasticity, which are necessary to bind and interact with a variety of different protein-unfolding intermediates [35, 67]; and (3) solubility of the client-chaperone complex, which is necessary to prevent co-aggregation [32]. Potential refolding of the disordered region upon client binding might contribute to increased stability of the complex, and has been proposed to generate an entropy transfer reaction that destabilizes the bound client protein and allows it to re-enter the folding pathway [34, 35, 98]. This new chaperone category includes yeast Hsp26, bacterial Hsp33, yeast Get3, and HdeA [35, 67, 99]. Both sHsps and Prx have been shown to also undergo structural changes consistent with unfolding, but the regions involved appear less extensive [87, 100]. Nevertheless, the role of these ID regions in chaperone function is likely the same.
The initial finding that unfolding of the linker region exposes hydrophobic surfaces that are otherwise buried in reduced Hsp33 provided a straightforward model by which Hsp33 and potentially other intrinsically disordered chaperones use partial unfolding as a mode of activation (Box 2) [31]. More recent studies revealed that chaperone-active Hsp33 uses its own disordered, hydrophilic linker region to directly interact with early protein-unfolding intermediates [35]. Binding of these partially structured intermediates appears to stabilize and potentially refold the linker region, contributing to the stabilization of the chaperone-client complex [35]. Indeed, a recent study using unnatural amino acids incorporated into Hsp33 to enable either in vivo crosslinking or site-directed 19F nuclear magnetic resonance spectroscopy, revealed a composite client-binding site in Hsp33 that consists of polar residues from the flexible linker region and nonpolar residues from the N-terminal β-sheet platform surface of Hsp33 [36].
The oxidative activation of Hsp33's chaperone function is fully reversible (Figure 1). Upon returning to non-stress-reducing and protein-refolding conditions, oxidized Hsp33 dimers are reduced by the cellular glutaredoxin and thioredoxin system yet remain bound to their client proteins [37]. Upon restoration of the cellular ATP levels, ATP-dependent chaperone foldases composed of the bacterial DnaK/DnaJ/GrpE (Hsp70) system mediate the release of Hsp33's client proteins and trigger its full inactivation [35, 37]. Subsequently, the KJE system supports the refolding of client proteins to the native state (Figure 1). By using this controlled inactivation mechanism, client proteins are released only when redox homeostasis is restored and the canonical proteostasis network is present to support their refolding [37].
Several questions regarding Hsp33's working mechanism and in vivo roles remain open, including one relating Hsp33 function under non-stress conditions. Indeed, a recently conducted study in E. coli reported that overexpression of Hsp33 rescued the synthetically lethal phenotype of a strain lacking the canonical chaperones trigger factor (TF) and DnaK [38]. The study suggested that Hsp33, by sequestering the essential elongation factor-Tu (EF-TU) for degradation to the Lon protease, slows down de novo protein synthesis, alleviating the need for TF and DnaK. Since the authors did not investigate the redox status of Hsp33 under these conditions, it remains to be tested whether this activity represents a specific function of reduced Hsp33, or whether it relates to the previously reported interaction between Hsp33 and EF-Tu under oxidative stress conditions [39].
Get3: The Link to Redox-Regulated Chaperones in Eukaryotes
Except for Clamydomonas and some members of the Trypanosoma family, no Hsp33 homologues have been found in any eukaryotes. This observation raised the question as to how eukaryotic cells cope with oxidative protein unfolding. Recent evidence suggests that the highly conserved eukaryotic ATPase Get3 (TRC40 in mammals) moonlights as a redox-regulated chaperone under ATP-depleting stress conditions, such as oxidative stress [40, 41]. Like Hsp33, Get3 exhibits no significant chaperone activity under reducing non-stress conditions. In contrast to Hsp33, however, reduced cytosolic Get3 has a well-known job—it serves as the central player of the Guided Entry of Tail-anchored proteins (GET) pathway in yeast, where it mediates the posttranslational integration of tail-anchored (TA) proteins into the membrane of the endoplasmic reticulum (ER) [42-44]. Under these non-stress conditions, dimeric Get3 shuttles TA proteins from the ribosome to the ER membrane [45, 46] using ATP-binding and hydrolysis to control the binding and release of TA proteins [47, 48].
As would be expected for a protein that is involved in the global targeting of TA proteins [49], deletion of the mammalian Get3 homologous TRC40 or its partners is embryonically lethal [50]. It was therefore surprising that mutants of get3 in Saccharomyces cerevisiae or Arabidopsis thaliana did not show any global growth defects [51]. Moreover, while many different TA proteins were found to employ the Get3/TRC40 system for successful membrane targeting in vitro, in vivo studies in yeast or tissue-specific knockout mice failed to identify more than a handful of proteins that show targeting defects [52]. This is likely because at least two functional backup systems exist that take over the targeting of TA proteins in the absence of the GET pathway [53, 54]. This result, however, raised questions regarding the observed stress-associated phenotypes of a get3 deletion strain, which included increased copper and oxidative stress sensitivity as well as decreased survival during glucose starvation [55, 56]. Indeed, recent studies revealed that the copper sensitive phenotype of a get3 deletion strain is in fact not due to a defect in TA targeting but due to the lack of a second function of Get3 as an oxidative stress-activated, ATP-independent general chaperone [41].
Although no sequence homology exists between Hsp33 and Get3, the two proteins show some intriguing similarities: both coordinate zinc via a conserved CXXC-motif, release zinc upon oxidative disulfide bond formation, and undergo partial oxidative unfolding that results in an alternative structure that is active as a molecular chaperone [41] (Figure 2). These structural rearrangements, which can be monitored on secondary, tertiary, and quarternary structure levels, cause inactivation of Get3's ATPase activity, exposure of hydrophobic surfaces, and formation of higher oligomeric species, with a tetramer being the smallest chaperone-active unit [41]. While we hypothesize that the newly exposed hydrophobic residues will likely serve as interaction sites for unfolded substrate, high-resolution structures of oxidized Get3 tetramers in the presence of client proteins are still lacking. Most importantly, all of the conformational and functional changes in Get3 are fully reversible, allowing Get3 to return to its ATPase-dependent functions once stress conditions no longer exist [41] (Figure 2).
The discovery that Get3 has two distinct functions in vivo is in excellent agreement with previous findings in Caenorhabditis elegans, where deletion of the Get3 homologue ASNA-1/TRC40 was found to cause a severe growth defect and increased sensitivity to the oxidative stress-inducing anti-cancer drug cisplatin [57, 58]. Intriguingly, while expression of wild-type ASNA-1 rescued both phenotypes, a mutant variant of ASNA-1 lacking two of the conserved cysteines rescued the growth defect but not the cisplatin sensitivity [59]. These results suggest that ASNA/TRC40 also works as a dual-function protein in higher eukaryotes. Questions to be addressed in the near future include the client specificity of oxidized Get3 and the role of the other components of the GET pathway in Get3's chaperone function. It is exciting to think that the chaperone function of Get3/TRC40 might be the physiologically more relevant activity of this conserved class of proteins. Further studies are required to address this important question.
N-chlorination as Mechanism of Oxidative Chaperone Activation
More recently, another intriguing redox-mediated activation mechanism for oxidative stress-activated chaperones was discovered in E. coli. The protein RidA, a member of the highly conserved yet functionally diverse YjgF/YER057c/UK114 protein family [60], was found to specifically undergo reversible N-chlorination reactions, which turn the enzyme into a highly active chaperone that protects bacteria specifically against HOCl stress-mediated protein aggregation [60].
Salmonella enterica RidA has long been known for its ability to speed up the release of ammonia from enamine/imine intermediates of the pyridoxal 5′-phosphate-dependent threonine dehydratase (IlvA) [61]. In the presence of RCS, such as HOCl or monochloramine, however, RidA was found to strongly inhibit the activity of IlvA [60]. This surprising result raised the question as to whether RidA might function as a protein-binding chaperone holdase that inhibits IlvA's catalytic activity by forming a tight complex. Indeed, in vitro aggregation studies with unfolded IlvA and other model substrates showed that HOCl-treated RidA prevents protein aggregation, whereas untreated RidA shows no significant chaperone activity [60]. Deletion of RidA increased E. coli's sensitivity to HOCl, suggesting that RidA is an integral component that protects E. coli cells against RCS [60].
Only incubation of RidA with RCS activated the chaperone function, whereas incubation with other tested oxidants, such as peroxide or diamide, had no effect [60]. Subsequent structural studies revealed that RidA's activation is accompanied by an increase in surface hydrophobicity and the formation of higher oligomers, hallmarks of other stress-activated chaperones [41, 60]. The observation that RCS activated a cysteine-free RidA variant excluded cysteine oxidation as possible regulation mechanism [60]. However, the authors found that HOCl-treated RidA showed substantially decreased levels of free amino groups and up to seven different N-chlorination sites. These results led to the conclusion that N-chlorination of lysine and/or arginine side chains is likely responsible for the activation of RidA's chaperone activity [60]. These results agree well with earlier studies on alpha-2 macroglobulin, which also shows an increase in chaperone activity likely due to the chlorination of positively charged side chains [62]. So far, it is unclear whether one or more N-chlorinations are responsible for the observed effects.
Note that RidA's chaperone function appears to be relevant in other organisms as well. However, although N-chlorination of RidA's amino acid side chains was fully reversible by DTT, ascorbic acid, or the physiological redox systems Trx or GSH, activation of the Drosophila melanogaster homologue DUK114 appears to be irreversible [60]. The next step is to determine whether chaperone activation is mediated by the N-chlorination of specific residues or due to a general increase in surface hydrophobicity, triggered by the neutralization of positive side chains. If the latter is true, many more proteins might undergo HOCl-mediated conversion into molecular chaperones than previously anticipated, bolstering the defense system against oxidative stress.
Acid-Activated Chaperones: Defense Against Low pH Stress
One of the most extreme and rapid changes in environmental pH that organisms encounter is in the mammalian stomach, where up to 2 L of 10 mM hydrochloric acid (HCl) (pH 2.0) are produced per day to aid in food digestion and protect the host against foodborne pathogens [63]. This raises the obvious question as to how enterobacteria survive this voyage, which is necessary to colonize the intestinal tract. Due to the porous nature of the outer membrane, the periplasm of Gram-negative bacteria is in free exchange with small molecules in the extracellular environment, making periplasmic proteins particularly vulnerable to acidic pH [64]. Moreover, since the periplasm lacks ATP, none of the canonical chaperones are present to protect and refold proteins. To deal with the proteotoxic effects of highly acidic pH, Gram-negative bacteria contain the small periplasmic proteins HdeA and HdeB (∼10 kDa), which undergo rapid acid-induced activation as molecular chaperones [65, 66] (Figure 1).
At neutral pH, HdeA is compactly folded, dimeric, and chaperone-inactive. When exposed to acidic environments, however, HdeA partially unfolds and within seconds dissociates into chaperone-active monomers, exposing a hydrophobic dimer interface in the process, which appears to be crucial for client binding [67]. Intramolecular FRET studies revealed that the partial unfolding is critical for HdeA to adopt different conformations, an apparently necessary prerequisite for its broad client specificity [67].
Its small size and unique mode of activation have made HdeA a popular candidate for structural studies and molecular dynamics simulations. Constant-pH molecular dynamics (CPHMD) together with mutational studies revealed key residues that appear to be responsible for pH sensing [68]. Indeed, mutation of only two key acidic residues was sufficient to convert HdeA into a constitutively active chaperone at near neutral pH [69]. Recent NMR studies revealed that while the dimer structure of HdeA is maintained until pH 3, increasing charge neutralization of Asp and Glu residues causes a loosening of the tertiary structure and the eventual dissociation into chaperone-active monomers at pH <3 [70]. Upon return to neutral pH, HdeA appears to mediate the refolding of its client proteins in an ATP-independent manner concomitant with the return into its dimeric chaperone-inactive conformation. It is thought that HdeA exploits a “slow-release mechanism” to keep the concentration of unbound aggregation-prone substrate species low and hence facilitate efficient client refolding [71] (Figure 1).
More recently, HdeB, which is part of the hdeA-hdeB acid stress operon and displays structural similarities to HdeA, was shown to have chaperone activity under low pH as well [72]. However, while HdeA is chaperone-active under severe acid stress (e.g., pH <3), HdeB exhibits its highest chaperone activity at pH 4 [73]. Moreover, unlike HdeA, HdeB remains dimeric and apparently folded in its chaperone-active state, suggesting two rather distinct activation mechanisms for HdeA and HdeB [73]. Recently conducted pH-dependent NMR studies suggest that changes in HdeB's intrinsic dynamic properties could contribute to its chaperone function at pH 4 [74]. The differential regulation of chaperone activity by pH for HdeA and HdeB shows how bacteria have adapted to survive varying degrees of acid stress-induced protein-unfolding conditions.
Multi-Stress Sensing Chaperone Families
At least two protein families have been described in which individual family members sense different stress conditions yet undergo similar conformational changes that lead to the specific activation of their chaperone function. One of these is the small heat shock proteins (sHsps), a diverse group of chaperones that play important roles in maintaining protein homeostasis in all kingdoms of life [75]. Stress-specific activation of their chaperone function has been observed for several members. Yeast Hsp26, for instance, undergoes local unfolding events upon exposure to elevated temperatures [76], whereas mammalian HspB5/HSBP1 appears to use the protonation state of specific His residues to sense low pH conditions [77, 78]. Although the sensing mechanisms are different, the structural events that follow are quite similar. The respective sHsps, which typically form large oligomeric structures that are considered to be inactive storage forms of the chaperones [76], dissociate into chaperone-active dimers with high client binding activity [76, 79]. Upon binding their client proteins, which effectively prevents their aggregation, the sHsps reassemble into higher molecular weight chaperone-client complexes. Once non-stress conditions have been restored, the client proteins can then be extracted and refolded by members of the Hsp70/Hsp104-system [27, 80]. Outstanding, very recent reviews are available that provide deeper mechanistic and regulatory insights into the chaperone function of sHsps [75, 81]
A second group of multi-stress sensing chaperones includes members of the large family of peroxiredoxins (Prxs), highly conserved and extremely abundant proteins [82]. In their “day job,” Prxs function as true enzymes, using the presence of a highly reactive cysteine in their active site to effectively detoxify peroxide and other reactive oxygen and nitrogen species (for review, see [83]). In their role as peroxidases, Prxs are directly involved in cellular H2O2 signaling and protein oxidation [83, 84]. In addition, however, certain members of the Prx family reversibly lose their peroxidase function and switch into effective ATP-independent chaperones (Figure 3). Stress conditions that trigger chaperone activation of Prxs include severe peroxide stress (e.g., yeast Tsa1, mammalian hPrxII), which is mediated by the overoxidation of the active site cysteine [84, 85], low pH (e.g., SmPrxI from the trematode Schistosoma mansoni) [86], or elevated temperatures (e.g., mitochondrial mTXNPx from Leishmania infantum, C2C-Prx1 from Chinese cabbage) (Figure 3) [87, 88]. Because we discussed the mechanisms used to sense elevated ROS levels and acidic pH in some detail, we will focus this final discussion on the temperature sensing mechanism of mitochondrial mTXNPx from the parasite L. infantum. Recent studies revealed that the temperature-mediated switch from peroxidase to chaperone function allows parasites to rapidly adapt to the elevated temperatures experienced during their transition from the insect to the mammalian host [87].
Figure 3. Peroxiredoxins - A Multi-stress Sensing Dual-Function Protein Family.
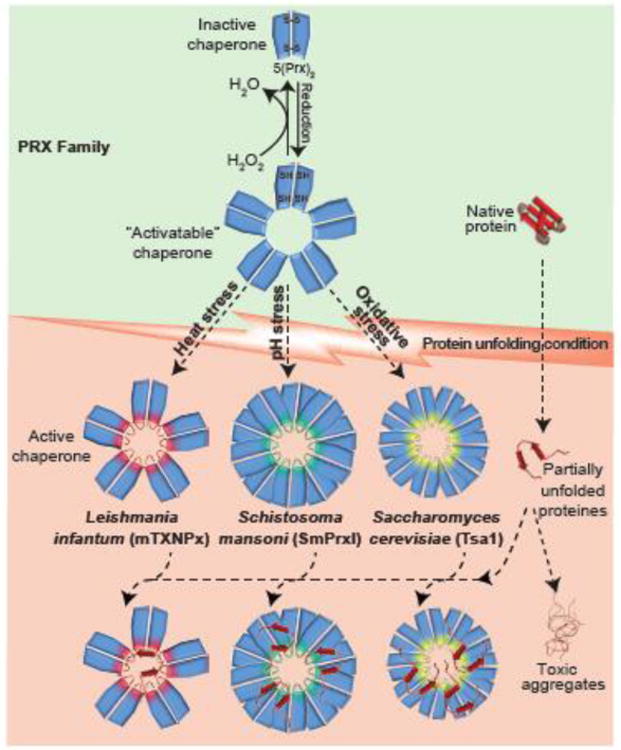
Members of the peroxiredoxin (PRX) family (blue) effectively detoxify peroxide and other reactive oxygen and nitrogen species (green shaded area). As part of the oxidation/reduction process, PRX cycles between reduced decamers and oxidized dimers. Members of the PRX family have been found to switch into potent ATP-independent chaperones once specific protein-unfolding stress conditions are encountered. Leishmania infantum peroxiredoxin mTXNPx, for instance, specifically senses heat stress, whereas Schistosoma mansoni SmPrxI's responds to pH stress and Tsa1 from Saccharomyces cerevisiae is activated by high levels of ROS. The mechanism of stress sensing depends on the respective stress conditions (please see text for details) but the structural changes that the PRX members undergo appear to be similar. They include the formation of chaperone-active decamers and higher oligomeric states, which show increased exposure of hydrophobic surfaces, and decreased peroxiase activity. The chaperone-active peroxiredoxins bind unfolding proteins and hence prevent the formation of cytotoxic protein aggregates. While the binding of unfolding client proteins in the center of the ring-like structure has been shown for mTXNPx, it is unclear where the other PRXs bind their client proteins. mTXNPx interacts with canonical chaperone systems to release the client proteins and support their refolding. Stress-specific peroxiredoxins in parasites appear to be necessary for parasitic organisms to successfully propagate in the mammalian host.
Prxs typically cycle between a reduced, decameric ring-like conformation and an oxidized dimeric conformation [83]. In vitro studies showed that at elevated temperatures, reduced mTXNPx within minutes undergoes significant structural rearrangements, which increase its structural flexibility, expose previously buried hydrophobic patches, and trigger ATP-independent chaperone activity. When present in the oxidized, dimeric conformation, however, none of these structural changes occur, and mTXNPx remains inactive as a chaperone. Further studies into this dual function of reduced mTXNPx revealed that as the chaperone activity increases with temperature, the peroxidase activity decreases. These results suggest that reduced mTXNPx exists in two alternative conformations that appear to be functionally exclusive and are determined by the environmental temperature [87]. Transmission electron microscopy of mTXNPx decamers incubated with client proteins at elevated temperatures indicated that binding of the client proteins occurs in the center of each decameric ring. Reference-free class averages showing top-down views also revealed the presence of up to five spoke-like structures that extend from mTXNPx into the interior of the ring. These structures likely represent the highly flexible N-termini of Prx, which are known to face the interior of the ring but have so far remained unresolved in X-ray structures [89]. Indeed, when the N-termini of mTXNPx were modified by His-tags, the stress-induced chaperone function of mTXNPx was lost, but the peroxidase activity remained [87].
In vivo studies showed that raising the temperature to 37°C leads to extensive protein unfolding and aggregation in L. infantum mutants lacking mTXNPx, and reduces survival of the parasites in mammalian hosts. Whereas expression of wild-type mTXNPx or a peroxidase-inactive variant of mTXNPx fully rescued both effects [90], expression of the N-terminally tagged chaperone-inactive, peroxidase-active variant failed to do so [87]. These results clearly demonstrate that the stress-induced chaperone function of reduced mTXNPx and not its peroxidase activity is crucial for parasitic adaptation and survival in the mammalian host.
Concluding Remarks and Future Perspectives
Over the past decade, an increasing number of stress-activated chaperones have been discovered. They share at least three major features: (1) they are activated in a matter of seconds to minutes upon exposure to specific stress conditions, making them ideal first responders to protect cells against conditions that lead to sudden protein unfolding and aggregation (oxidative stress, heat stress, and/or acid stress); (2) they undergo highly stress-specific structural rearrangements during their activation, ensuring that they are only activated when truly needed; and (3) they function ATP-independently, which allows them to prevent protein aggregation under stress conditions or in cellular compartments that are devoid of ATP. Of course, one might wonder why nature didn't want these chaperones to be always active, given their highly effective chaperoning activity. However, since their protein binding activity, which is of very high affinity and low specificity, cannot be regulated by any other means (i.e., ATP binding), activation under non-stress conditions would likely interfere with de novo protein folding processes and proteostasis. Indeed, bacteria expressing a constitutively active variant of Hsp33 suffered severe growth disadvantages and accumulated large amounts of cellular proteins in insoluble aggregates [32]. The fact that many of these proteins have a second, typically mutually exclusive function under non-stress conditions likely ensures that the respective genes are not lost during evolution even in those organisms that do not experience these particular stress conditions over many generations.
Another commonality among stress-activated chaperones is their prevalence in pathogenic bacteria and parasites. While it simply might require more research to find additional stress-activated chaperones in eukaryotes, it is enticing that several of the stress-activated chaperones identified in pathogens so far (e.g., HdeA, HdeB, Hsp33) have no clear eukaryotic homolog, making them potentially very attractive new drug targets. Given that these chaperones all use partial protein unfolding as their mode of activation, it is tempting to speculate that small compounds that stabilize the chaperones could be used to specifically prevent their activation, and hence might serve as suitable new therapeutics in combating bacterial and parasitic diseases.
Finally, it will be exciting to learn how many more proteins that have been characterized as enzymes, targeting factors, or co-factors turn out to convert into chaperones under extreme stress conditions (see Outstanding Questions). Partial unfolding comes naturally to proteins—all that is needed to become an effective buffer for other unfolding proteins is the right arrangement of hydrophobic surfaces to foster interactions with them, and hydrophilic regions to prevent self-aggregation and keep client-chaperone complexes soluble. Once the stress conditions are over and energy supplies have been restored, the canonical chaperones can take over, reactivating unfolded proteins and restoring protein homeostasis in the cell.
Outstanding Questions.
For stress-activated chaperones with dual functions, are both functions equally relevant in vivo or is one more important than the other?
What proportion, if any, of stress-activated chaperones is chaperone-active even under non-stress conditions?
How many more proteins are yet to be discovered that undergo stress-induced unfolding and convert into protein-binding chaperones under specific stress conditions?
Trends.
Cells require a fast reacting and flexible proteostasis network to quickly respond and adapt to sudden environmental changes under energy limiting conditions.
Stress-induced activation of chaperone function involves conversion into a partially disordered conformation (i.e., conditional disorder), which appears to be directly involved in client interaction.
Many stress-activated chaperones serve as dual-function proteins with distinct, mutually exclusive enzymatic or regulatory activities under non-stress conditions.
N-chlorination of amino acid side chains serves as a novel mechanism to activate chaperones in response to HOCl stress.
Acknowledgments
We thank Drs. J. Bardwell, L. Leichert, B. Schwappach and F. Stull, for critically reading select portions of this manuscript. Grants by the National Institutes of Health (R35-GM-122506-01) and the Deutsche Forschungsgemeinschaft supported this work.
Glossary
- Canonical chaperones
The term canonical chaperone is used to describe members of the Hsp60/10 family (GroEL/ES in bacteria), the Hsp70/Hsp40 family (DnaK/DnaJ in bacteria) and Hsp90 (HtpG in bacteria) that represent the most common properties among molecular chaperones
- Chaperone foldases
Chaperone foldases are a specific subgroup of molecular chaperones that assist in folding nascent polypeptide chains and/or preventing protein aggregation, and in refolding unfolded proteins and/or extracting proteins from aggregates. Binding and release of client proteins is typically regulated by ATP binding and hydrolysis. Repeated cycles of client binding and release ensure proper client folding. Typical ATP-dependent foldases include members of the Hsp60 and Hsp70 chaperone families
- Chaperone holdases
Most ATP-independent chaperones work as chaperone holdases. They tightly bind to unfolding client proteins and prevent their aggregation. Some chaperone holdases are specifically active under distinct stress conditions (e.g., Hsp33), whereas others, like Hsp40, are constantly active under normal conditions. Stress-activated chaperone holdases often depend on ATP-dependent foldases for client refolding once stress conditions subside. The more recently discovered bacterial chaperones Spy and HdeA, which reside in the bacterial periplasm, show both ATP-independent holdase and foldase function
- Heat shock protein (Hsp)
Heat shock proteins are a highly conserved group of protein families whose expression is induced by the accumulation of protein-unfolding intermediates. Protein unfolding can be caused by a variety of stress conditions, including elevated temperatures (i.e., heat shock), oxidative stress, nutrient starvation, and viral infections. During non-stress conditions, most Hsps are constitutively expressed and participate in the folding and trafficking of nascent polypeptides, the assembly of multi-protein complexes, and/or the coordinated degradation of proteins. Under stress conditions, the increased expression of Hsps protects cells against the accumulation of aggregated proteins and/or promotes disassembly, refolding or degradation of unfolded, aggregated proteins
- Proteostasis
This blend of the words “protein” and “homeostasis” describes a network of chaperones and proteases that support and regulate de novo protein folding, trafficking, and degradation of proteins. The interconnected and competitive pathways of the network maintain a correctly folded proteome and the health of the organism itself. The proteostasis network is highly flexible, using posttranslational regulation of protein activity to adjust to changing environments within minutes, as well as transcriptional responses to ensure long-term adaptation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Klebanoff SJ, et al. Myeloperoxidase: a front-line defender against phagocytosed microorganisms. J Leukoc Biol. 2013;93(2):185–98. doi: 10.1189/jlb.0712349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee CT, Repasky EA. Opposing roles for heat and heat shock proteins in macrophage functions during inflammation: a function of cell activation state? Front Immunol. 2012;3:140. doi: 10.3389/fimmu.2012.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grasberger H, et al. Dual oxidases control release of hydrogen peroxide by the gastric epithelium to prevent Helicobacter felis infection and inflammation in mice. Gastroenterology. 2013;145(5):1045–54. doi: 10.1053/j.gastro.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goto Y, et al. Acid-induced folding of proteins. Proc Natl Acad Sci U S A. 1990;87(2):573–7. doi: 10.1073/pnas.87.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weijers M, et al. Heat-induced denaturation and aggregation of ovalbumin at neutral pH described by irreversible first-order kinetics. Protein Sci. 2003;12(12):2693–703. doi: 10.1110/ps.03242803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morano KA, et al. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics. 2012;190(4):1157–95. doi: 10.1534/genetics.111.128033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marengo B, et al. Redox Homeostasis and Cellular Antioxidant Systems: Crucial Players in Cancer Growth and Therapy. Oxid Med Cell Longev. 2016;2016:6235641. doi: 10.1155/2016/6235641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Y, Zhao B. Oxidative stress and the pathogenesis of Alzheimer's disease. Oxid Med Cell Longev. 2013;2013:316523. doi: 10.1155/2013/316523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maritim AC, et al. Diabetes, oxidative stress, and antioxidants: a review. J Biochem Mol Toxicol. 2003;17(1):24–38. doi: 10.1002/jbt.10058. [DOI] [PubMed] [Google Scholar]
- 10.Knoefler D, et al. About the dangers, costs and benefits of living an aerobic lifestyle. Biochem Soc Trans. 2014;42(4):917–21. doi: 10.1042/BST20140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aseervatham GS, et al. Environmental factors and unhealthy lifestyle influence oxidative stress in humans--an overview. Environ Sci Pollut Res Int. 2013;20(7):4356–69. doi: 10.1007/s11356-013-1748-0. [DOI] [PubMed] [Google Scholar]
- 12.Verghese J, et al. Biology of the heat shock response and protein chaperones: budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol Mol Biol Rev. 2012;76(2):115–58. doi: 10.1128/MMBR.05018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dukan S, et al. Hypochlorous acid activates the heat shock and soxRS systems of Escherichia coli. Appl Environ Microbiol. 1996;62(11):4003–8. doi: 10.1128/aem.62.11.4003-4008.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pattison DI, Davies MJ. Absolute rate constants for the reaction of hypochlorous acid with protein side chains and peptide bonds. Chem Res Toxicol. 2001;14(10):1453–64. doi: 10.1021/tx0155451. [DOI] [PubMed] [Google Scholar]
- 15.Brandes N, et al. Thiol-based redox switches in eukaryotic proteins. Antioxid Redox Signal. 2009;11(5):997–1014. doi: 10.1089/ars.2008.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shenton D, et al. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J Biol Chem. 2006;281(39):29011–21. doi: 10.1074/jbc.M601545200. [DOI] [PubMed] [Google Scholar]
- 17.Colussi C, et al. H2O2-induced block of glycolysis as an active ADP-ribosylation reaction protecting cells from apoptosis. FASEB J. 2000;14(14):2266–76. doi: 10.1096/fj.00-0074com. [DOI] [PubMed] [Google Scholar]
- 18.Winter J, et al. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol Cell. 2005;17(3):381–92. doi: 10.1016/j.molcel.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 19.Gray MJ, et al. Polyphosphate is a primordial chaperone. Mol Cell. 2014;53(5):689–99. doi: 10.1016/j.molcel.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakob U, et al. Chaperone activity with a redox switch. Cell. 1999;96(3):341–52. doi: 10.1016/s0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- 21.Zheng M, et al. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279(5357):1718–21. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 22.Leichert LI, et al. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci U S A. 2008;105(24):8197–202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J, et al. The Expanding Landscape of the Thiol Redox Proteome. Mol Cell Proteomics. 2016;15(1):1–11. doi: 10.1074/mcp.O115.056051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindemann C, et al. Redox proteomics uncovers peroxynitrite-sensitive proteins that help Escherichia coli to overcome nitrosative stress. J Biol Chem. 2013;288(27):19698–714. doi: 10.1074/jbc.M113.457556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janda I, et al. The crystal structure of the reduced, Zn2+-bound form of the B. subtilis Hsp33 chaperone and its implications for the activation mechanism. Structure. 2004;12(10):1901–7. doi: 10.1016/j.str.2004.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maret W. Zinc coordination environments in proteins as redox sensors and signal transducers. Antioxid Redox Signal. 2006;8(9-10):1419–41. doi: 10.1089/ars.2006.8.1419. [DOI] [PubMed] [Google Scholar]
- 27.Haslbeck M, et al. Disassembling protein aggregates in the yeast cytosol. The cooperation of Hsp26 with Ssa1 and Hsp104. J Biol Chem. 2005;280(25):23861–8. doi: 10.1074/jbc.M502697200. [DOI] [PubMed] [Google Scholar]
- 28.Winter J, et al. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell. 2008;135(4):691–701. doi: 10.1016/j.cell.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vijayalakshmi J, et al. The 2.2 A crystal structure of Hsp33: a heat shock protein with redox-regulated chaperone activity. Structure. 2001;9(5):367–75. doi: 10.1016/s0969-2126(01)00597-4. [DOI] [PubMed] [Google Scholar]
- 30.Cremers CM, et al. Bile salts act as effective protein-unfolding agents and instigators of disulfide stress in vivo. Proc Natl Acad Sci U S A. 2014;111(16):E1610–9. doi: 10.1073/pnas.1401941111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ilbert M, et al. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat Struct Mol Biol. 2007;14(6):556–63. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cremers CM, et al. Unfolding of metastable linker region is at the core of Hsp33 activation as a redox-regulated chaperone. J Biol Chem. 2010;285(15):11243–51. doi: 10.1074/jbc.M109.084350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cortese MS, et al. Intrinsic disorder in scaffold proteins: getting more from less. Prog Biophys Mol Biol. 2008;98(1):85–106. doi: 10.1016/j.pbiomolbio.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bardwell JC, Jakob U. Conditional disorder in chaperone action. Trends Biochem Sci. 2012;37(12):517–25. doi: 10.1016/j.tibs.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reichmann D, et al. Order out of disorder: working cycle of an intrinsically unfolded chaperone. Cell. 2012;148(5):947–57. doi: 10.1016/j.cell.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Groitl B, et al. Protein unfolding as a switch from self-recognition to high-affinity client binding. Nat Commun. 2016;7:10357. doi: 10.1038/ncomms10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffmann JH, et al. Identification of a redox-regulated chaperone network. EMBO J. 2004;23(1):160–8. doi: 10.1038/sj.emboj.7600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruel N, et al. Hsp33 controls elongation factor-Tu stability and allows Escherichia coli growth in the absence of the major DnaK and trigger factor chaperones. J Biol Chem. 2012;287(53):44435–46. doi: 10.1074/jbc.M112.418525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wholey WY, Jakob U. Hsp33 confers bleach resistance by protecting elongation factor Tu against oxidative degradation in Vibrio cholerae. Mol Microbiol. 2012;83(5):981–91. doi: 10.1111/j.1365-2958.2012.07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Powis K, et al. Get3 is a holdase chaperone and moves to deposition sites for aggregated proteins when membrane targeting is blocked. J Cell Sci. 2013;126(Pt 2):473–83. doi: 10.1242/jcs.112151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voth W, et al. The protein targeting factor Get3 functions as ATP-independent chaperone under oxidative stress conditions. Mol Cell. 2014;56(1):116–27. doi: 10.1016/j.molcel.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borgese N, et al. The tale of tail-anchored proteins: coming from the cytosol and looking for a membrane. J Cell Biol. 2003;161(6):1013–9. doi: 10.1083/jcb.200303069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuldiner M, et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134(4):634–45. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shao S, Hegde RS. Membrane protein insertion at the endoplasmic reticulum. Annu Rev Cell Dev Biol. 2011;27:25–56. doi: 10.1146/annurev-cellbio-092910-154125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mariappan M, et al. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 2010;466(7310):1120–4. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mateja A, et al. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 2009;461(7262):361–6. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stefer S, et al. Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex. Science. 2011;333(6043):758–62. doi: 10.1126/science.1207125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suloway CJ, et al. Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc Natl Acad Sci U S A. 2009;106(35):14849–54. doi: 10.1073/pnas.0907522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128(6):1147–59. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 50.Mukhopadhyay R, et al. Targeted disruption of the mouse Asna1 gene results in embryonic lethality. FEBS Lett. 2006;580(16):3889–94. doi: 10.1016/j.febslet.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 51.Xing S, et al. Loss of GET pathway orthologs in Arabidopsis thaliana causes root hair growth defects and affects SNARE abundance. Proc Natl Acad Sci U S A. 2017;114(8):E1544–E1553. doi: 10.1073/pnas.1619525114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rivera-Monroy J, et al. Mice lacking WRB reveal differential biogenesis requirements of tail-anchored proteins in vivo. Sci Rep. 2016;6:39464. doi: 10.1038/srep39464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aviram N, et al. The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature. 2016;540(7631):134–138. doi: 10.1038/nature20169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rabu C, et al. A precursor-specific role for Hsp40/Hsc70 during tail-anchored protein integration at the endoplasmic reticulum. J Biol Chem. 2008;283(41):27504–13. doi: 10.1074/jbc.M804591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen J, et al. The Saccharomyces cerevisiae Arr4p is involved in metal and heat tolerance. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine. 2003;16(3):369–78. doi: 10.1023/a:1022504311669. [DOI] [PubMed] [Google Scholar]
- 56.Metz J, et al. The yeast Arr4p ATPase binds the chloride transporter Gef1p when copper is available in the cytosol. The Journal of biological chemistry. 2006;281(1):410–7. doi: 10.1074/jbc.M507481200. [DOI] [PubMed] [Google Scholar]
- 57.Tsutsumishita Y, et al. Involvement of H2O2 production in cisplatin-induced nephrotoxicity. Biochem Biophys Res Commun. 1998;242(2):310–2. doi: 10.1006/bbrc.1997.7962. [DOI] [PubMed] [Google Scholar]
- 58.Marullo R, et al. Cisplatin induces a mitochondrial-ROS response that contributes to cytotoxicity depending on mitochondrial redox status and bioenergetic functions. PLoS One. 2013;8(11):e81162. doi: 10.1371/journal.pone.0081162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hemmingsson O, et al. ASNA-1 activity modulates sensitivity to cisplatin. Cancer research. 2010;70(24):10321–8. doi: 10.1158/0008-5472.CAN-10-1548. [DOI] [PubMed] [Google Scholar]
- 60.Muller A, et al. Activation of RidA chaperone function by N-chlorination. Nat Commun. 2014;5:5804. doi: 10.1038/ncomms6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lambrecht JA, et al. Conserved YjgF protein family deaminates reactive enamine/imine intermediates of pyridoxal 5′-phosphate (PLP)-dependent enzyme reactions. J Biol Chem. 2012;287(5):3454–61. doi: 10.1074/jbc.M111.304477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wyatt AR, et al. Hypochlorite-induced structural modifications enhance the chaperone activity of human alpha2-macroglobulin. Proc Natl Acad Sci U S A. 2014;111(20):E2081–90. doi: 10.1073/pnas.1403379111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith JL. The role of gastric acid in preventing foodborne disease and how bacteria overcome acid conditions. J Food Prot. 2003;66(7):1292–303. doi: 10.4315/0362-028x-66.7.1292. [DOI] [PubMed] [Google Scholar]
- 64.Koebnik R, et al. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol. 2000;37(2):239–53. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- 65.Hong W, et al. Periplasmic protein HdeA exhibits chaperone-like activity exclusively within stomach pH range by transforming into disordered conformation. J Biol Chem. 2005;280(29):27029–34. doi: 10.1074/jbc.M503934200. [DOI] [PubMed] [Google Scholar]
- 66.Gajiwala KS, Burley SK. HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J Mol Biol. 2000;295(3):605–12. doi: 10.1006/jmbi.1999.3347. [DOI] [PubMed] [Google Scholar]
- 67.Tapley TL, et al. Structural plasticity of an acid-activated chaperone allows promiscuous substrate binding. Proc Natl Acad Sci U S A. 2009;106(14):5557–62. doi: 10.1073/pnas.0811811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang BW, et al. Probing pH-dependent dissociation of HdeA dimers. J Am Chem Soc. 2011;133(48):19393–8. doi: 10.1021/ja2060066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Foit L, et al. Chaperone activation by unfolding. Proc Natl Acad Sci U S A. 2013;110(14):E1254–62. doi: 10.1073/pnas.1222458110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garrison MA, Crowhurst KA. NMR-monitored titration of acid-stress bacterial chaperone HdeA reveals that Asp and Glu charge neutralization produces a loosened dimer structure in preparation for protein unfolding and chaperone activation. Protein Sci. 2014;23(2):167–78. doi: 10.1002/pro.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tapley TL, et al. Protein refolding by pH-triggered chaperone binding and release. Proc Natl Acad Sci U S A. 2010;107(3):1071–6. doi: 10.1073/pnas.0911610107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kern R, et al. Escherichia coli HdeB is an acid stress chaperone. J Bacteriol. 2007;189(2):603–10. doi: 10.1128/JB.01522-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dahl JU, et al. HdeB functions as an acid-protective chaperone in bacteria. J Biol Chem. 2015;290(1):65–75. doi: 10.1074/jbc.M114.612986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ding J, et al. HdeB chaperone activity is coupled to its intrinsic dynamic properties. Sci Rep. 2015;5:16856. doi: 10.1038/srep16856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carra S, et al. The growing world of small heat shock proteins: from structure to functions. Cell Stress Chaperones. 2017;22(4):601–611. doi: 10.1007/s12192-017-0787-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haslbeck M, et al. Hsp26: a temperature-regulated chaperone. EMBO J. 1999;18(23):6744–51. doi: 10.1093/emboj/18.23.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rajagopal P, et al. A conserved histidine modulates HSPB5 structure to trigger chaperone activity in response to stress-related acidosis. Elife. 2015;4 doi: 10.7554/eLife.07304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clouser AF, Klevit RE. pH-dependent structural modulation is conserved in the human small heat shock protein HSBP1. Cell Stress Chaperones. 2017;22(4):569–575. doi: 10.1007/s12192-017-0783-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stengel F, et al. Quaternary dynamics and plasticity underlie small heat shock protein chaperone function. Proc Natl Acad Sci U S A. 2010;107(5):2007–12. doi: 10.1073/pnas.0910126107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ungelenk S, et al. Small heat shock proteins sequester misfolding proteins in near-native conformation for cellular protection and efficient refolding. Nat Commun. 2016;7:13673. doi: 10.1038/ncomms13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Treweek TM, et al. Small heat-shock proteins: important players in regulating cellular proteostasis. Cell Mol Life Sci. 2015;72(3):429–51. doi: 10.1007/s00018-014-1754-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Harper AF, et al. An Atlas of Peroxiredoxins Created Using an Active Site Profile-Based Approach to Functionally Relevant Clustering of Proteins. PLoS Comput Biol. 2017;13(2):e1005284. doi: 10.1371/journal.pcbi.1005284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Poole LB, et al. Overview of peroxiredoxins in oxidant defense and redox regulation. Curr Protoc Toxicol. 2011;Chapter 7 doi: 10.1002/0471140856.tx0709s49. Unit7 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rhee SG, Woo HA. Multiple functions of peroxiredoxins:peroxidases, sensors and regulators of the intracellular messenger H(2)O(2), and protein chaperones. Antioxid Redox Signal. 2011;15(3):781–94. doi: 10.1089/ars.2010.3393. [DOI] [PubMed] [Google Scholar]
- 85.Jang HH, et al. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117(5):625–35. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 86.Saccoccia F, et al. Moonlighting by different stressors: crystal structure of the chaperone species of a 2-Cys peroxiredoxin. Structure. 2012;20(3):429–39. doi: 10.1016/j.str.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Teixeira F, et al. Mitochondrial peroxiredoxin functions as crucial chaperone reservoir in Leishmania infantum. Proc Natl Acad Sci U S A. 2015;112(7):E616–24. doi: 10.1073/pnas.1419682112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim SY, et al. Oligomerization and chaperone activity of a plant 2-Cys peroxiredoxin in response to oxidative stress. Plant Science. 2009;177(3):227–232. [Google Scholar]
- 89.Wang X, et al. Structural insights into the peroxidase activity and inactivation of human peroxiredoxin 4. Biochem J. 2012;441(1):113–8. doi: 10.1042/BJ20110380. [DOI] [PubMed] [Google Scholar]
- 90.Castro H, et al. Leishmania mitochondrial peroxiredoxin plays a crucial peroxidase-unrelated role during infection: insight into its novel chaperone activity. PLoS Pathog. 2011;7(10):e1002325. doi: 10.1371/journal.ppat.1002325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stadtman ER, Levine RL. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 2003;25(3-4):207–18. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- 92.Winterbourn CC, Kettle AJ. Redox reactions and microbial killing in the neutrophil phagosome. Antioxid Redox Signal. 2013;18(6):642–60. doi: 10.1089/ars.2012.4827. [DOI] [PubMed] [Google Scholar]
- 93.Antelmann H, Helmann JD. Thiol-based redox switches and gene regulation. Antioxid Redox Signal. 2011;14(6):1049–63. doi: 10.1089/ars.2010.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Uversky VN. The intrinsic disorder alphabet. III. Dual personality of serine. Intrinsically Disord Proteins. 2015;3(1):e1027032. doi: 10.1080/21690707.2015.1027032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Deng X, et al. A comprehensive overview of computational protein disorder prediction methods. Mol Biosyst. 2012;8(1):114–21. doi: 10.1039/c1mb05207a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hsu WL, et al. Intrinsic protein disorder and protein-protein interactions. Pac Symp Biocomput. 2012:116–27. [PubMed] [Google Scholar]
- 97.Koldewey P, et al. Forces Driving Chaperone Action. Cell. 2016;166(2):369–79. doi: 10.1016/j.cell.2016.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18(11):1169–75. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 99.Haslbeck M, et al. Some like it hot: the structure and function of small heat-shock proteins. Nat Struct Mol Biol. 2005;12(10):842–6. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- 100.Basha E, et al. Small heat shock proteins and alpha-crystallins: dynamic proteins with flexible functions. Trends Biochem Sci. 2012;37(3):106–17. doi: 10.1016/j.tibs.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Groitl B, et al. Pseudomonas aeruginosa defense systems against microbicidal oxidants. Mol Microbiol. 2017 doi: 10.1111/mmi.13768. [DOI] [PMC free article] [PubMed] [Google Scholar]