Abstract
Background
Recently, the nature of the lipid-ligand of Pru p 3, one of the most common plant food allergens in Southern Europe, has been identified as a derivative of the alkaloid camptothecin bound to phytosphingosine. However, the origin of its immunological activity is still unknown.
Objective
We sought to evaluate the role of the Pru p 3 lipid-ligand in the immunogenic activity of Pru p 3.
Methods
In vitro cultures of different cell types (monocyte-derived dendritic cells (moDCs), PBMCs and epithelial and iNKT-hybridoma cell lines) have been used to determine the immunological capacity of the ligand, by measuring cell proliferation, maturation markers and cytokine production. To study the capacity of the lipid-ligand to promote sensitization to Pru p 3 in vivo, a mouse model of anaphylaxis to peach has been produced and changes in the humoral and basophil responses have been analyzed.
Results
The lipid-ligand of Pru p 3 induced maturation of moDCsc and proliferation of PBMCs. Its immunological activity resided in the phytosphingosine tail of the ligand. The adjuvant activity of the ligand was also confirmed in vivo, where the complex of Pru p 3-ligand induced higher levels of IgE than Pru p 3 alone. The immunological capacity of the Pru p 3 ligand was mediated by CD1d, as maturation of moDCs was inhibited by anti-CD1d antibodies and Pru p 3-ligand co-localized with CD1d on epithelial cells. Finally, Pru p 3-ligand presented by CD1d was able to interact with iNKTs.
Conclusions & Clinical Relevance
The Pru p 3 lipid-ligand could act as an adjuvant to promote sensitization to Pru p 3, through its recognition by CD1d receptors. This intrinsic adjuvant activity of the accompanying lipid cargo could be a general essential feature of the mechanism underlying the phenomenon of allergenicity.
Keywords: Peach allergy, lipid transfer proteins, Pru p 3, iNKT, CD1d
Introduction
The prevalence of allergy is increasing in Europe where it has been predicted that over 50% of the population will have some form of allergy by 20501,2, with a most dramatic increase in children whose quality of life is severely affected3. The annual cost of allergic diseases for national health systems is as high as for seasonal influenza4, and it has been estimated that the annual cost of food allergy exceeds thousands of millions of euros in Europe alone5.
Even though the role of proteins in allergic sensitization has been extensively studied6,7, the reasons why a specific protein may have allergenic activity are not yet well understood. Several hundred protein allergens have been described (www.allergen.org) and a large number of their epitopes identified8. However, only 1% of known protein families include allergens and besides, not all the members of those families are allergenic6,7,8. Although no biochemical feature common to allergenic proteins has been found, more than 50% of allergens have been shown to be lipid carriers, including Bet v 1 (the major allergen of birch pollen) and 2S allergens, which contain a large hydrophobic pocket able to harbour and store many lipid compounds and flavonoids as well8,9,10. Recently, the ligand of Pru p 3, the peach allergenic lipid transfer protein (LTP), has been identified as a derivative of the alkaloid camptothecin bound to phytosphingosine (Figure 1a), a lipidtail which is inserted into the Pru p 3 cavity9. Phytosphingosine is a structural component in membranes10, while camptothecin is widely used as a chemotherapeutic agent for its ability to inhibit the enzymatic activity of topoisomerase I11.
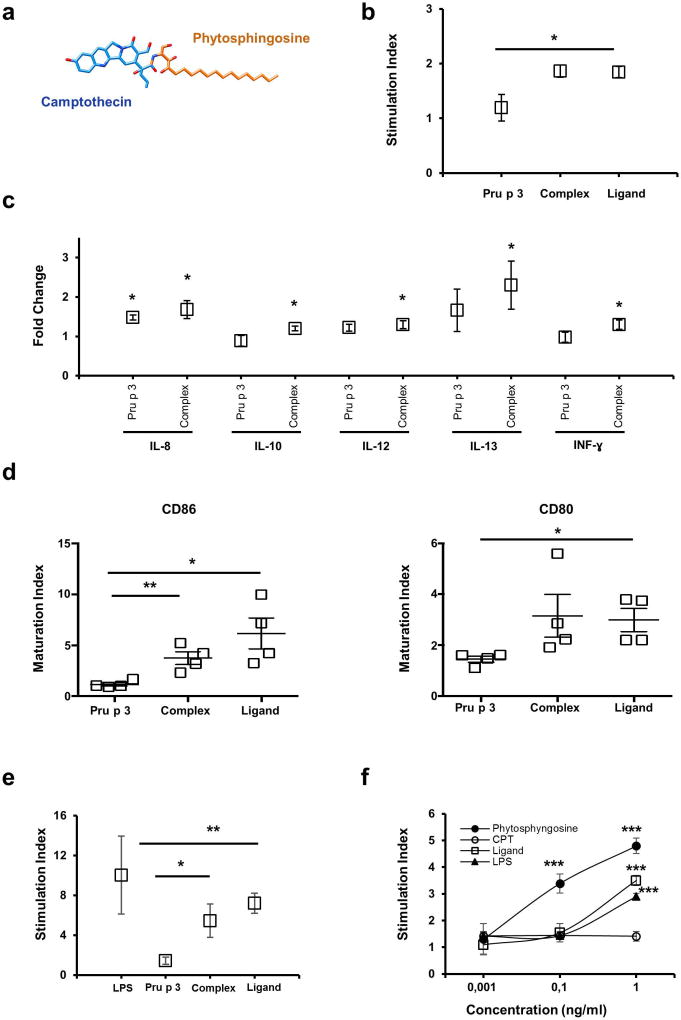
Figure 1. Characterization of immunogenic activity of the lipid-ligand of Pru p 3.
(a) Structural formula of the lipid-ligand of Pru p 3 (10-hydroxy-camptothecin linked to phytosphingosine) (b) Proliferation assays of human PBMCs (n=6) stained with CSFE were incubated with Pru p 3 and Complex (Pru p 3; 10 µg/ml and the lipid-ligand 1 µg/ml). After 5 days, the proliferation of PBMCs was measured by loss of CSFE staining. Stimulation index (SI) was calculated as (% CSFEneg-PBMCs + antigen)/(% CSFEneg-PBMCs – antigen). Means and SE (bars) are shown. *p < 0.05. (c) Cytokine produced by PMBCs by Bioplex system. The results are expressed as the fold change between the amount (pg/mL) produced in the presence of the stimulus (Pru p 3, Complex), and the amount without stimulus. Means and SE (bars) are shown. *p < 0.001. (d) Phenotype of human monocyte-derived DCs (moDC) induced by Pru p 3 and its lipid-ligand. Changes in the expression of costimulatory molecules (CD80 and CD86) in monocyte-derived DCs from healthy donors, after stimulation with Pru p 3 alone (Pru p 3; 10 µg/mL), with the lipid-ligand (Complex; 10 µg/ml), and with the lipid-ligand only (Ligand; 1 µg/mL). Maturation indices of CD80 and CD86 expression are shown (p<0.05). (e, f) NF-κB/AP-1 activation using the transfected cell line THP1-XBlue™. Cells (1×106 cells/ml) were incubated with Pru p 3 (Pru p 3; 5 µg/ml), Pru p 3 with lipid-ligand (Complex; 5 µg/ml) and only lipid-ligand (Ligand; 1 µg/ml) in (e), or with the lipid-ligand (Ligand), phytosphingosine (Phytosphingosine) and campthotecin (CPT) in (f). In both, LPS was used as positive control. Activation of NF-κB/AP-1 was revealed with QUANTI-Blue™. *p < 0.05 **p < 0.01 ***p<0.001.
Lipid ligands carried by allergens, or exposed together, might influence the host response to the specific allergen. Accordingly, immune-regulatory lipids could significantly contribute to the allergenicity of several proteins12. All of these molecules are attractive candidates as non-self structures recognized by the innate immune system as being capable of inducing Th2-skewed responses that contribute to allergy in susceptible individuals13. Lipids from the pollen coat and furry animals as well as the so-called pollen-associated lipid mediators are co-delivered together with the allergens and can modulate the immune responses of predisposed subjects by interacting with the innate immune system and invariant natural killer T (iNKT) cells13.
iNKT cells are innate-like T lymphocytes with important immunomodulatory properties that can be damaging (e.g. allergic inflammation) or protective (e.g. maintenance of transplant tolerance, inhibition of autoimmunity)14. They express glycolipid-reactive αβ T cell receptors (TCRs) together with several characteristic markers of NK cells and they are “CD1-restricted”15.The CD1-receptor family includes monomorphic major histocompatibility complex (MHC) class I-like glycoproteins that can present lipid antigens. The CD1 family in humans has five members, namely CD1a–e, while rodents only express CD1d16. Given the nonpolar nature of lipids, their mobilization across an aqueous endocytic environment likely requires extraction from membranes and, in most of the cases, transport across aqueous biological buffers into the lipid-binding grooves of CD1 molecules.
The results presented in the current report suggest that the lipid-ligand carried by the peach allergen Pru p 3 could act as an adjuvant with the ability to modulate the immune system towards a Th2 response. Pru p 3 could release its lipid ligand to CD1 receptors present in the epithelial and dendritic cells, which in turn would present it to iNKT cells, ultimately resulting in activation of the immune response. Activation of the mucosal immune system by allergen-associated lipids may provide an important missing link in our understanding about the genesis of food allergic responses.
Materials and Methods
Isolation of the lipid-ligand of Pru p 3
To carry out the assays, the recombinant Pru p 3 protein was used. In this way, the absence of pollutants (lipid-ligand residues) in the purified protein was ensured. As “complex”, it is referred to the mix of Pru p 3 and its purified ligand (10:1). The ratio was determined by binding assays between Pru p 3 and the lipid-ligand as previously described9.The recombinant Pru p 3 was produced in P Pastoris and isolated using the methods describe by Diaz-Perales et al. (2002)17. The presence of endotoxin in the protein-samples was measured using the Pierce LAL Chromogenic Endotoxin Quantitation Kit (Thermo Scientific, USA), following manufacturer’s instructions.
The lipid-ligand of Pru p 3 was extracted form peach peel. Briefly, peach peel extract was performed in PBS with 0.5 M NaCl and separated by cation-exchange chromatography on a Waters AccellTM Plus CM SepPakR cartridge (Waters Corp, Milford, MA, USA) with 20 mM formic acid, pH 4.0. The cationic retained fraction was separated by size-exclusion LH-20 chromatography using methanol:water (70:30) as the mobile phase. Fractions were checked using thin layer chromatography (TLC) on silica gel–coated plates (Merck, USA), and developed with methanol:water (70:30 v/v) in a saturated chromatography chamber for about 20 min. The plates were visualized under UV light (254 nm) and stained with vanillin (1% in ethanol) and dichlorofluorescein (0.1% in ethanol-NaOH 2.5 mM) (Sigma-Aldrich, Germany). Fractions containing fluorescence were pooled and re-purified by HPLC as previously described18 with slight modifications. Isocratic analytical HPLC was performed using an RP-C18 column Nucleosil 120 C18 5 µm 250×4mm (Scharlab, Spain). The mobile phase for alkaloid elution was acetonitrile: water (85:15) at a flow rate 0.5 ml/min for 150 µl of sample. Sample peaks were detected using UV light at 254 nm. The standard was prepared using methanol 80% further diluted in 20% acetonitrile. All reagents were HPLC grade.
Lymphocyte transformation test (LTT) with peripheral blood mononuclear cells (PMBCs)
Buffy coat (cell-enriched plasma) samples were obtained from 8 healthy donors at the Transfusion Centre (Madrid, Spain) for isolation of PBMCs. The study was approved by the ethics committees of the Transfusion Centre (PO.DlS.09; Madrid, Spain) and the Technical University of Madrid (PIC 52-2013; Madrid, Spain).
PBMCs isolated using a density gradient centrifugation on Lymphoprep (Axis-Shield, Oslo, Norway) were stained with CSFE stained following the manufacturer’s instructions (Bdbioscience, CA, USA). Then, cultures (5×105 cells per well) were established in the presence of 5 µg/ml of Pru p 3 or Complex (Pru p 3 with its ligand) in triplicate in 96-well plates (Costar, NY, USA) for proliferation analysis in a total volume of 200 µl of RPMI media (Invitrogen), supplemented with 10% (v/v) of fetal calf serum (Invitrogen), 0.02 mM mercaptoethanol, 2 mM of glutamine, and 10 mM HEPES, for 5 days at 37°C, in a 5% CO2 humidified atmosphere. The stimulation index (SI) was calculated as the ratio between counts of antigen-stimulated cultures and counts without any activator. Supernatants were obtained to measure cytokines.
Statistically significant differences were analysed by GraphPad6 using Mann-Whitney test. P-values<0.05 were considered significant for all assays.
Quantification of the cytokines produced by PMBCs
To measure the PMBCs cytokines, Bio-Plex Pro™ Assays (Bio-Rad 10014905) for 27 different proteins detection powered by Luminex xMAP technology was used following the instructions at www.bio-rad.com/bio-plex. The final analysis was performed with the Bio-Plex Manager™ Software using xMAP technology for analysis of multiplex data. Cytokines were quantified according to a calibration curve in pg/mL (Supplementary Table 1). The statistical analysis was performed using the two-way ANOVA with corrections for multiple comparisons. P values<0.001 were considered significant. Then, cytokines with statistically significant difference were also represented as fold change calculated as the ratio between the amount of cytokines present in the culture with stimulus and the cytokine content of the control without stimulation.
Generation of human antigen-presenting cells (moDC)
Monocytes were purified from human PBMCs by positive selection using CD14 Dynabeads (Invitrogen, Norway), following the manufacturer’s protocol. Purity was assessed by flow cytometry (BD Accuri Cytometers, USA) and found to be higher than 95%. Dendritic cells (DCs) were derived from monocytes (moDC) by culturing the CD14+ fraction in complete medium RPMI (Invitrogen, Norway) with L-glutamine (brand, country), 10% heat-inactivated fetal bovine serum (FBS, Lonza, Germany), 100 mg/ml antibiotics (streptomycin and penicillin; Invitrogen, Norway), 200 ng/ml rhIL4, and 100 ng/ml rhGM-CSF (Immunotools, Germany) for 4–5 days at 5% CO2 and 37°C, as described previously19. Immature DCs (iDCs) derived from monocytes were incubated in complete medium at 5 × 105 cells/ml in 24-well plates (Falcon BD Labware, France) with Pru p 3 at 20 µg/ml, with or without lipid-ligand (2 µg/ml). LPS at 10 µg/ml (Sigma-Aldrich, USA) was used as a positive control. After 72 h stimulation at 37°C in 5% CO2, treated DCs were recovered, and maturation was assessed by up-regulation of CD80 and CD86 (Immunotools, Germany) in an Accuri cytometer (BD Accuri Cytometers, USA). Untreated DCs were used as a control. In order to determine the receptor involved in molecular recognition, cells were previously incubated with 1 ug of polyclonal antibodies anti TLR2, TLR4 (Invivogen, France) and anti CD1d (Thermo Fisher). Results were expressed as the activation of CD80 or/and C86 and the maturation index was calculated as the ratio between stimulated and non-stimulated DCs. The statistical analysis was performed using the Kruskal-Wallis test, and p-values <0.05 were considered positive.
NF-κB/AP-1 activation
The transfected cell line THP1-XBlue™ (Invivogen, France) derived from the human monocytic THP-1 cell line was used according to the manufacturer’s instructions. In brief, THP1-XBlue cells were seeded at a density of 1×106 cells/ml in cRPMI medium (RPMI 1640, 2 mM L-glutamine, 10 mM HEPES, 10% heat-inactivated FBS, PenStrep cocktail (penicillin 50 µg/ml and streptomycin 50 µg/ml) and 200 µg/ml of Zeocin. One microgram of sample/per well (5 µg/µl) was added to the cells for 24 h. LPS (Sigma-Aldrich, USA) and PBS were used as positive and negative controls, respectively. Twenty microliters of cell suspension (~200,000 cells) per well was added to a flat-bottom 96-well plate containing 180 µl of QUANTI-Blue™ per well and incubated for 18–24 h. Soluble embryonic alkaline phosphatase (SEAP) levels were determined using a spectrophotometer at 620 nm. The statistical analysis was performed using the Kruskal-Wallis test, and p-values <0.05 were considered positive.
Epicutaneous sensitization of mice and Pru p 3 challenge
C3H/HeOuJ mice, which present normal TLR4, were obtained from Jackson Laboratories (Bar Harbor, ME) and used with the permission of the Institutional Animal Care and Use Committee (IACUC) (number LA11-00273). Six- to eight-week-old female mice were anesthetized, and abdominal fur was removed with depilatory cream (Veet, Reckitt Benckiser, Parsippany, NJ), immediately followed by exposure to Pru p 3 (100 ug), with or without lipid-ligand (10ug) in 50 ul of PBS spread on the abdominal skin to dry.
Mice were exposed weekly for a total of 6 exposures and challenged a week after the last exposure. Mice were challenged by intraperitoneal injection with increasing doses of Pru p 3. Body temperature was measured before and 30 minutes after challenge by rectal thermometer (WPI Instruments, Sarasota, FL).
Antibody measurement
Pru p 3-specific IgE and IgG1 were measured in serum obtained prior to challenge, by directly coating Pru p 3 on the plate and detecting with biotinylated anti-IgE or IgG1 (BD Biosciences, Franklin Lakes, NJ), followed by Avidin-HRP and TMB reagent (eBiosciences, San Diego, CA). A SpectraMax plate reader with background subtraction was used to measure absorbance (450 nm). The statistical analysis was performed using SPSSS 17.0 and two ways ANOVA with corrections for multiple comparisons. P values<0.05 or p<0.01 were considered positives.
Basophil activation tests
Basophil activation tests were performed as previously described20. Blood was diluted in RPMI and incubated at 37°C for 90 minutes with Pru p 3 at different concentrations. Red blood cells were lysed, and cells were stained with CD49b and IgE to detect the population of basophils, after gating out T and B cells with CD3/CD19 staining. CD200R was used as basophil activation marker (all antibodies were from eBioscience). The statistical analysis was performed using SPSSS 17.0 and two ways ANOVA with corrections for multiple comparisons. P values <0.05 or <0.01 were considered positive.
Epithelial cell culture
The Caco-2 cell line was purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) and grown following the manufacturer’s instructions. Cells were seeded in 24-well Transwell® culture plates (0.4 mm pore diameter, Corning Inc., NY, USA) at a density of 8×104 cells per well and grown for 18–21 days, replacing the medium every 2 days. The integrity of the Caco-2 cell monolayer was checked by measuring the transepithelial electrical resistance (TEER) using a Millicell-ERS device (Millipore, Bedford, MA, USA). Cell monolayers were used in transport studies when values of TEER reached a plateau exceeding 300 Wcm2.
Immunolocalization of Pru p 3 and innate immune receptors (TLR2, TLR4 and CD1d)
Monolayers of Caco 2 cells were cultured as described above. Pru p 3, without or with its lipid-ligand (Complex), was added in the apical chamber during short times (5 min, 10 min). Cells were then fixed with 4% formaldehyde in PBS, pH 7.4, at 4°C for 15 min. After washing with PBS, samples were permeabilized by freeze-thaw cycles. After additional washing with PBS, specific anti-Pru p 3 (1:50; Jimenez Díaz Foundation21), anti-TLR2 (Invivogen, France), anti-TLR4 (Invivogen, France) and anti-CD1d antibodies (Thermo Fisher) were incubated overnight at 4°C. The binding was revealed using secondary antibodies produced in the appropriate host: anti-rabbit Alexa 647-conjugated antibodies for detected Pru p 3 and anti-mouse Alexa 488-conjugated antibodies for detected the innate receptors (Molecular Probes). The specimens were mounted with glycerol:PBS 1:1 and observed with a Leica TCS-SP8 confocal microscope using the 488-nm/567-nm laser excitation.
Generation of CD1d-based Artificial Antigen Presenting cell (aAPC)
Dynabeads® M-450 epoxy beads (Invitrogen) were coated with dimeric murine CD1d:Ig fusion Protein (BD Biosciences) and with Leaf Tm purified anti-mouse CD28 clone CD28.2 (Biolegend) as costimulatory molecule at 4°C with rotation during 48 h. Subsequently, 5×107 beads/ml were incubated with α-Galactosylceramide (5 ug; Sigma) as a positive control, the complex (Pru p 3+lipid-ligand; 5 µg) or lipid-ligand alone (0.5 ug) during 48 h at 37 °C. Then, beads were co-cultured with a murine iNKT cell line (DN32.D3 NKT cell hybridoma was kindly provided by Prof. Tonya Webb, University of Maryland School of Medicine, Baltimore, USA.) in 1:1 ratio in a 96 well flat bottom. The supernatant was collected after 24 h and the production of IL-2 was measured by ELISA assays.
CD1d-Dextramer loaded with the lipid-ligand of Pru p 3
A standard protocol recommended by the company including some modifications according to the stability of the lipid-ligand was used to load the CD1d/unloaded Dextramers®(Immudex TF1037.01) with the lipid-ligand of Pru p 3. The lipid was dissolved in methanol at 1 mg/ml and dissolved completely at room temperature. Dextramer with the lipid was always kept at 2–8°C in the dark until its use with cultured cells. The binding to experimental cells was performed at 37°C for 10 min, and then analysed by flow cytometry (BD-Accuri C6, BD-Biosciences, USA) or confocal microscopy (Leica TCS-SP8)
Statistical analyses
Statistical analyses were performed using SPSS 17.0 and GraphPad 6. The t test, Kruskal-Wallis test, one-way ANOVA, and two-way ANOVA with corrections for multiple comparisons were used when applicable. p-values <0.05 were considered significant for all assays.
Structural analyses
For Pru p 3, the chain B of the crystal structure PDB id. 2B5S22 was used. For saposins, the crystal structures from the following PDB id's were selected: 2DOB (saposin A), 1N69 (saposin B), 2GTG (saposin C), and 2RB3 (saposin D). Identification of secondary structure in Pru p 3 and saposins was performed with DSSP23. Three methods to determine structural alignments were employed to compare the structures of Pru p 3 and saposins: (i) the rigid (no twists) version of FATCAT (Flexible structure AlignmenT by Chaining Aligned fragment pairs allowing Twists) algorithm24, (ii) TM (Template Modelling)-align superposition algorithm25 and (iii) CE (Combinatorial Extension) algorithm26. According to the prescriptions given in the references for these methods, aligned structures with FATCAT are considered to have structural relationship if the alignment p-value is < 0.01, with lower values indicating higher similarity. Structural alignments obtained with TM-align are scored with a “TM-score” in the (0.0–1.0) scale, with values closer to 1.0 indicating better similarities and values between 0.40 and 0.50 suggesting structural relationship. For CE superpositions, Z-scores between 3.0 and 4.0 suggest some structural similarity and values > 4.0 indicate a stronger structural resemblance. Molecular graphics were prepared and rendered with PyMOL 1.8.2 (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC).
Results
The lipid-ligand of Pru p 3 is responsible for the activation of antigen-presenting cells
The immunological activity of Pru p 3 with or without its lipid-ligand was investigated using human PBMCs obtained from four healthy donors. PBMCs were stained with CSFE and after that, Pru p 3 and the complex were added. On the 5th day, PBMCs proliferation was quantified by flow cytometer, with strong activation in the presence of the ligand (Figure 1b; p≤ 0.001). This activation was also measured as the production of pro-inflammatory cytokines (Supplementary Table 1). Significant differences were only observed in the production of IL8, IL10, INFγ, but especially in the production of IL13 in the presence of ligand (Figure 1c).
In the same way, moDCs derived from peripheral blood of the same donors were also incubated with Pru p 3 and its ligand. As shown in Figure 1d, the presence of the lipid-ligand gave rise to significant induction of the maturation markers CD80 and CD86, while no changes were observed when only Pru p 3 was utilized (p≤ 0.001).
Moreover, the immunological activity of the lipid-ligand was also confirmed using THP1-XBlue™ cells (human monocyte cell line). The lipid-ligand, alone or in complex with Pru p 3, was able to stimulate NF-κB/AP-1 activation in these cells (Figure 1e; p=0.01). To identify the fraction of the lipid-ligand that had immunological activity, THP1-XBlueTM cells were again incubated in the presence of increasing amounts of the lipid-ligand, camptothecin and phytosphingosine. Thus, it was phytosphingosine the only one to be capable of inducing a similar response as that produced by the lipid-ligand (Figure 1f), indicating that the immunological activity was associated to the phytosphingosine hydrophobic moiety and not to the camptothecin polar head of the lipid-ligand (Figure 1a).
The lipid-ligand of Pru p 3 increases the capacity to sensitize mice through the epicutaneous route
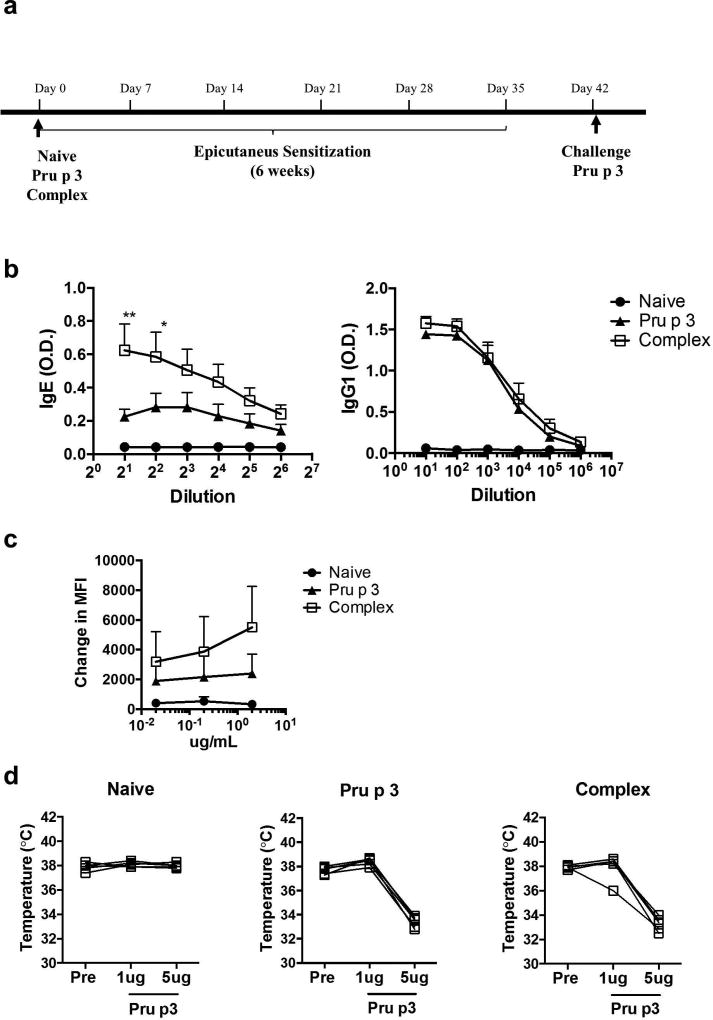
To determine the role of the lipid-ligand of Pru p 3 in inducing allergic sensitization, mice were epicutaneously exposed to Pru p 3, with or without its lipid-ligand, once a week for six weeks, following a previously described protocol27 (Figure 2a). Briefly, hair was removed with depilatory cream, but no tape stripping was performed and the antigen was added topically in PBS without occlusive bandages. After six weeks of sensitization, blood was obtained for basophil activation test and specific immunoglobulin measurement. Mice exposed to Pru p 3 plus its lipid-ligand (Complex in Figure 2b) developed significantly more Pru p 3-specific IgE than Pru p 3 alone (Pru p 3, Figure 2b), while IgG1 levels were similar in both cases. Similarly, basophil activation was increased in mice sensitized with the complex, as reflected by a higher increase in CD200R MFI (Figure 2c). Despite the differences in IgE induction, when mice were intraperitoneally challenged with Pru p 3, no differences were observed in terms of drop in body temperature during anaphylactic responses (Figure 2d).
Figure 2. Impact of the lipid-ligand of Pru p 3 on peach allergy.
(a) Mice were left naïve or topically exposed to 100 ug of Pru p 3 alone (Pru p 3 label) or Pru p 3 together with 10 ug of its lipid-ligand (Complex) once per week for six weeks. After sensitization, intraperitoneal challenge was performed with Pru p 3. (b) Antigen-specific IgE and IgG were measured prior to allergen challenge. Data are mean ± SEM. (c) Pru p 3-induced basophil activation was measured by up-regulation of CD200R. Change in MFI (median fluorescence intensity) was calculated with respect to the stimulation with media alone. Data are mean ± SEM. (d) Mice were challenged with increasing doses of Pru p 3 by intraperitoneal challenge. Body temperature prior to the challenge and 30 minutes after each ip injection are shown. n = 5 mice per group. *p < 0.05, **p < 0.01.
Pru p 3 is bound by CD1d in the presence of the lipid-ligand
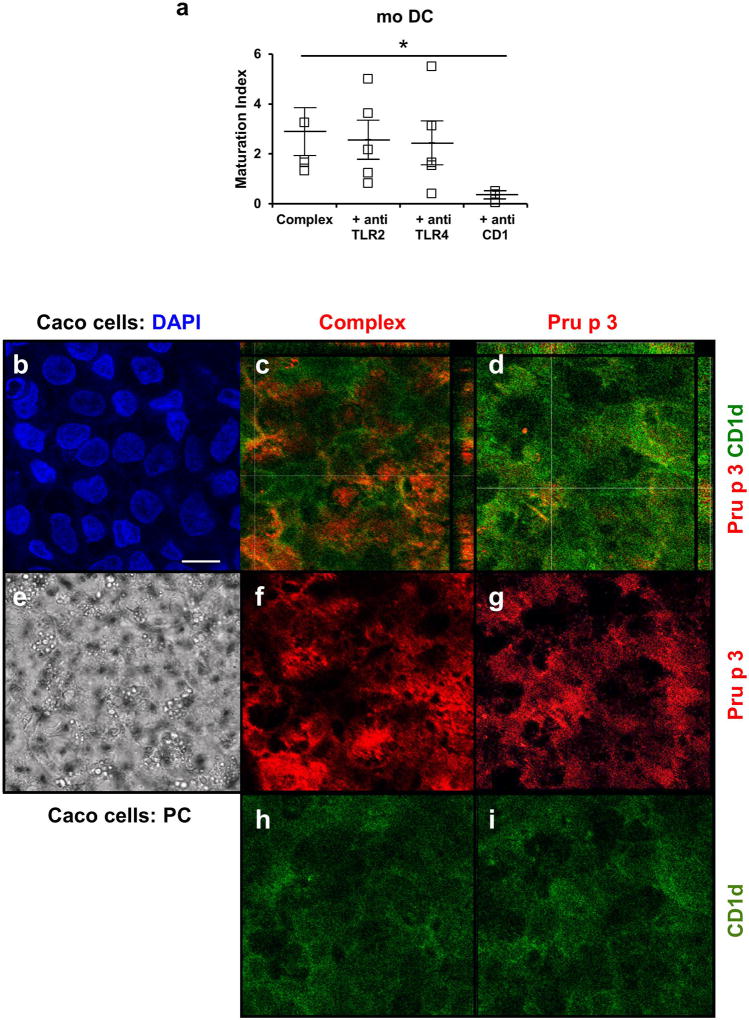
To determine which receptor mediated the recognition of the lipid-ligand, specific antibodies to TLR2, TLR4 and CD1d (receptors that recognize lipid ligands) were added to Pru p 3-moDCs cultures and DC maturation was studied. Maturation of human moDC could be only inhibited in presence of anti-CD1d (Figure 3a), but not with polyclonal antibodies anti-TLR2 or anti-TLR4 (p<0.001).
Figure 3.
(a) Inhibitory effect in the activated phenotype (positive CD80 and/or CD86) of moDCs, after stimulation with Pru p 3 (Pru p 3; 10 µg/mL) or Pru p 3 with its lipid-ligand (Complex; 10 µg/ml) and specific antibodies against TLR2, TLR4 and CD1d. (*p < 0.05). (b–i)) Immunolocalization of Pru p 3 (Red) or Complex (Red), and CD1d surface proteins (Green) in polarized CaCo 2 monolayer after 5 minutes incubation. (b) DAPI staining (e) Phase contrast. (c, d) Co-localization was just found between CD1 protein expression and Pru p 3. Bar 20 microns.
In the same way, polarized epithelial monolayers (Caco2 cells, Figure 3b) were also incubated with Pru p 3 (Pru p 3) and complex (Complex), during short times (5 min). After fixing cells, immune receptors (PRR) such as TLR2, TLR4 and CD1d were localized using specific antibodies. In the presence of the ligand, Pru p 3 colocalized with CD1d-type receptors (Figure 3c) but not when Pru p 3 was alone (Figure 3d). In the case of TLR2 and TLR4, no differences in localization were observed in the presence of absence of the lipid-ligand (Supplementary Figure 1).
The lipid-ligand presented by CD1d could stimulate iNKTs
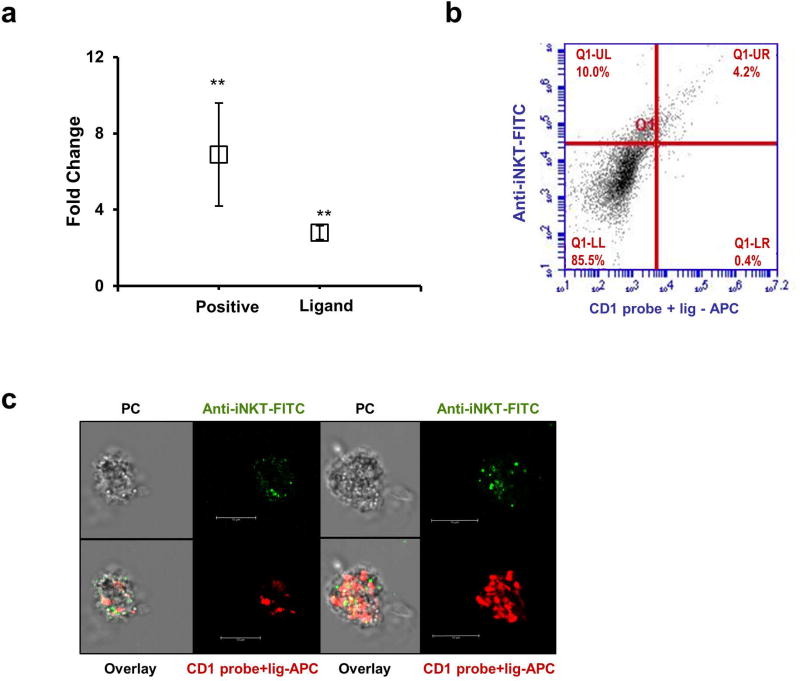
According to the results above described, we studied the role of the lipid-ligand in the activation of iNKT cells through CD1d receptor. iNKTs have been described as specialized cells in the recognitions of glycolipid antigens. Using a CD1d-based artificial antigen-presenting cell system (aAPC)28 and the murine DN32.D3 iNKT cell hybridoma, a clear increase in the secretion of IL-2 was observed in the presence of the lipid-ligand (Ligand) and the positive control α-Galactosylceramide (Positive), indicating the activation of iNKTs (Figure 4a).
Figure 4.
(a) CD1d-based artificial antigen-presenting cells (aAPC) were incubated with a positive control (α-Galactosylceramide; 5 µgr), and with the lipid-ligand (0.5 ug) during 48h at 37 °C. After that, aAPC were co-cultured with iNKT cells at a 1:1 ratio in a 96 well flat bottom. The supernatant was collected at 24h and the production of IL-2 was measured by ELISA assays. (b) Cyc5-labeled CD1d-tetramers (red) were loaded with the lipid-ligand of Pru p 3 and incubated with healthy volunteer’s PBMCs during 2 h. iNKTs were detected with a monoclonal antibody specific for the TCR of iNKTs labelled FITC (Vα24-Jα18 combined with Vβ11). (c) Confocal microscopy image of CD1d-tetramers-lipid-ligand probe (red) and TCR iNKTs (green) and phase contrast image of two different examples.
To further demonstrate that the lipid-ligand could interact with iNKTs through the CD1d receptor, the lipid-ligand was incubated with a CD1d unloaded human dextramer according to the manufacturer's instructions. This probe is capable of binding to the TCR present on the surface of iNKTs provided it has the appropriate lipid. As can be seen in Figure 4b, the fluorescent probe loaded with the lipid-ligand (labelled with APC) was able to interact with iNKT cells (labelled with FITC). In fact, most of the lipid-ligand-CD1d probes bound specifically to iNKTs (Figure 4b). In the figure 4c, it could be observed two examples of iNKT cell surrounded by CD1d-tetramers-lipid-ligand probe labelled with APC (red).
Pru p 3 could play a putative role as an external saposin
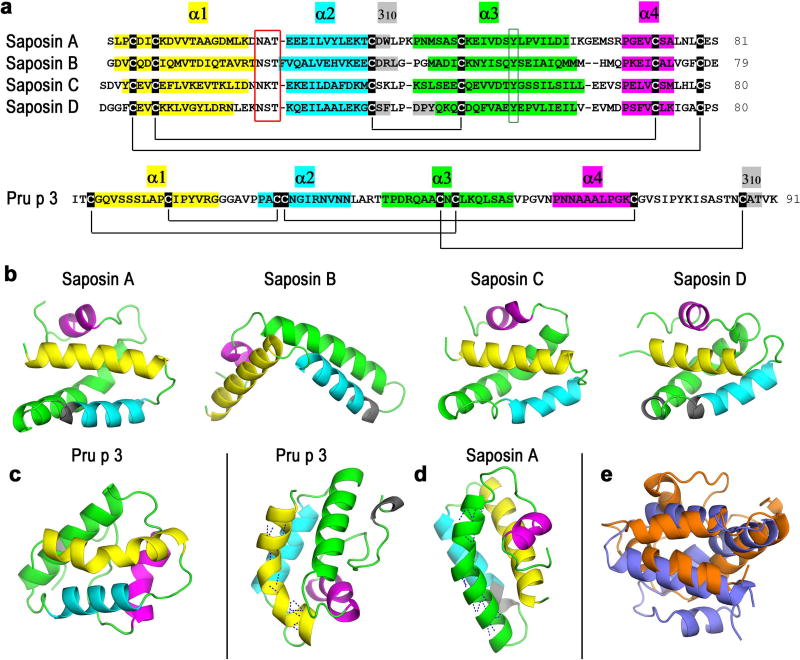
The experimental structure of Pru p 3 shows similar features to those of saposins, small non-enzymatic lipid transfer proteins that assist the loading of lipids onto CD1d. There are four saposins A, B, C, and D with pairwise sequence identity not higher than 39% that show a highly conserved structure with four α-helices and three disulphide bridges. Although not essential for their activity, they have a conserved N-glycosylation site (Figure 5a). The spatial arrangement of the four α-helices is nearly identical (Figure 5b). However, while in the absence of lipids, saposins adopt a compact form (saposins A, C and D in Figure 5b), in the presence of lipids they undergo a conformational change adopting an open form (saposin B in figure 5b). The architecture of saposins also present some particular features: (i) the longest helix has a highly localized kink associated with a local disruption of two main-chain hydrogen bonds around a conserved Tyr54 residue which is not the classic example of disruption caused by a proline, (ii) the shortest α4 helix starts at a conserved proline, and (iii) with the exception of saposin C, they also have short 310 helices (Figure 5a and 5b).
Figure 5. Structures of saposins and Pru p 3.
(a) Alignment of sequences of saposins A–D and sequence of Pru p 3. Colours labelling the residues that form α and 310 helices are also used in (b)–(d) below. Black lines connecting marked cysteines represent disulphide bridges. Red box indicates conserved N-linked glycosylation sites and green box indicates conserved tyrosines at the kink of helix α3 shown in (d) for saposin A. (b) X-ray crystal structures of saposins A–D. (c) X-ray crystal structure of Pru p 3. (d) Kink in the longest helix α1 in Pru p3 and α3 in saposin A. Dashed lines in these helices represent main chain hydrogen bonds. (e) Structural superposition of saposin A (blue) and Pru p 3 (orange) computed with FATCAT rigid.
The structure of Pru p 3 (Figure 5c) presents significant structural similarities to saposins. Not only Pru p 3 has a similar architecture with four α helices, one 310 helix and disulfide bonds (four instead of three in saposins), but it also displays particular structural features analogous to those indicated above for saposins. In fact, Pru p 3 presents a kink in the longest α helix (although the disruption of main-chain hydrogen bonds is caused by a proline), its shortest α4 helix starts also at a proline and it exhibits also a three-residue 310 helix (Figure 5a and 5d). Besides these structural characteristics, Pru p 3 is similar to saposins in size, existence of an inner cavity and lipid transfer abilities that are known to span diverse lipidligands29. Three procedures used to obtain structural alignments based on different approaches (FATCAT, TM-Align, and CE: see Materials and Methods) found that the best structural similarity is actually that existing between Pru p 3 and saposin A (Figure 5e). Superposition scores for these three methods were the following. FATCAT (rigid algorithm) significance p-value score was 0.00196 for Pru p 3/saposin A while it ranged between 0.00348 and 0.0101 for the remaining saposins. TM-Align TM-score was 0.403 for Pru p 3/saposin A while it ranged between 0.358 and 0.392 for the remaining saposins and CE significance Z-score was 4.07 for Pru p 3/saposin A while it ranged between 1.99 and 3.50 for the remaining saposins. Within the confidence levels conveyed by those scores (see Materials and Methods), it can be concluded that Pru p 3 and saposin A have a significant structural relationship.
Discussion
According to our own results and evidence reported in the literature in recent years, we hypothesized that allergic sensitization is enhanced by the recognition of lipid-ligands carried by some allergens. These lipid-ligands could be exposed to immune cells at the time of first contact, inducing the production of specific IgE against the protein carrier associated with the ligands. In a later encounter, the interaction between the allergen and specific IgE bound to the high-affinity receptor (FcεRI) on the surface of effector cells (mast cells and basophils) would induce the symptoms associated with allergy.
In the case of Pru p 3, our results reported here suggest that the lipid-ligand of this LTP is a critical collaborator for activation of dendritic cells, CD1 presentation and iNKT cells, associated with enhanced induction of antigen-specific IgE in mouse models.
The lipid-ligand carried by Pru p 3 seems to act as an adjuvant with the ability to modulate the immune system toward a Th2 response associated with class-switching towards IgE production, which is characteristic of allergy. Although Pru p 3 in the absence of the lipid-ligand was able to sensitize mice, the Pru p 3-lipid-ligand complex induced a much more robust IgE production. This result suggests that the presence of the lipid-ligand could increase susceptibility to peach allergy and decrease the exposure level to Pru p 3 required to induce sensitization to this allergen, thus acting as an adjuvant. In addition, we have shown for the first time that Pru p 3 is able to sensitize through the skin in a mouse model. The results presented here and the fact that the majority of peach allergic patients experience their first allergic reaction the first time they consume this food, together suggest that epicutaneous exposure is the main route of sensitization to peach, as it has been proposed for other allergens27,30,31.
In this sense, Pru p 3 in the presence of its lipid-ligand was presented by CD1d receptors in epithelial and dendritic cells. CD1 proteins are expressed on professional APCs such as Langerhans cells, dendritic cells, monocytes, B lymphocytes, intestinal epithelium, smooth muscle and endothelial blood vessels. They can present lipids and glycolipids to T-cell subpopulations, which in turn mediate effector functions including cytokine secretion16,32. CD1 lipid loading seems to occur in endosomal compartments of many antigen presenting cells such as epithelial mucosa. In their trafficking and binding to lipid antigens for recognition of T-cell receptors, CD1 molecules are assisted by saposins, small lipid transfer proteins that facilitate non-enzymatically the hydrolysis of a variety of glycosphingolipids in lysosomes,33,34 although the mechanism of this transfer and whether there is a direct CD1-saposin binding is still unknown. As CD1 molecules recycle through cellular compartments, they encounter in addition to saposins other proteins such as Niemann-Pick type C2 protein35 or apolipoprotein E36 that are also involved in the loading of lipids onto CD1.
Based upon their similar structural features, we propose that there could also be a functional relationship between Pru p 3 and saposins. There are four homologous saposins A, B, C, and D that derive from a unique prosaposin precursor protein upon endosomal proteolytic cleavage. Saposins are cysteine-rich proteins with four α-helices that exist in both isolated soluble and lipid-bound states. Although their sequences have pairwise identities not higher than 39%37, their topology is conserved and their architecture is rather similar. Structural alignments determined with three different algorithms reveal a significant relationship between Pru p 3 and saposin A. Given these structural resemblances together with the similarities in size, cavity and lipid transfer abilities, we conjecture that Pru p 3 could play a role in the trafficking of molecules with lipidmoieties that are eventually loaded onto CD1d receptors.
In any event, we have shown in this work that the lipid-ligand of Pru p 3, perhaps especially its phytosphingosin domain, is presented to iNKT cells by CD1 receptors. Although it is likely that this is not the only innate immune pathway involved in the recognition of Pru p 3, our results reveal that it is active in moDC and epithelial mucosa. This is not the first report associating iNKTs to allergen activation. iNKTs can be activated by phospholipids, particularly phosphatidylethanolamine and phosphatidylcholine present on the surface of cypress nuts16. A recent study analyzed the general ability of olive pollen lipids to activate DCs and stimulate iNKT cells and demonstrated that polar lipids isolated from olive pollen grains upregulated CD1d on DCs, which then activated iNKT cells in co-cultures38. In the same way, the 2S albumin from Brazil nut, Ber e 1, was not sufficient to cause IgE or IgG production in mice39. In contrast, the co-administration of the allergen with polar lipid fractions isolated from the same nut induced the production of Ber e 1-specific IgE and IgG1. In the absence of iNKT cells, the specific IgE levels were lower40. Lastly, it has been suggested that the activation of iNKT cells induce Th2-mediated inflammation to cow’s milk-derived sphingolipids41.
In summary, we have shown that the lipid-ligand of Pru p 3 enhances the allergic activity and participates actively in the triggering of antigen-presenting cells. Pru p 3 would tend to be targeted by innate immune response because of its auto-adjuvant lipid-ligand. Moreover, Pru p 3 has structural features that support its possible involvement in trafficking and loading of lipid compounds onto CD1d receptors. The fact that other allergens have been described as lipid binding proteins, suggest that intrinsic adjuvant activity by such proteins and their accompanying lipid cargo could be a general essential feature of the mechanisms underlying the phenomenon of allergenicity.
Supplementary Material
Supplementary figure 1: Immunolocalization of Caco 2 monolayer after 5 minutes of incubation with Pru p 3 or Complex. Pru p 3 was detected by polyclonal antibodies in red (Alexa 647) and TLR2 and TLR4 were detected in green (Alexa 488). From left to right: TLR2 (green), Pru p 3 (red), DAPI, phase contrast, triple combination (TLR/Pru p 3/DAPI) and colocalization of Pru p 3 and TLR. Bar 20 microns.
Acknowledgments
This project was supported by grant project BIO2013-41403R from Ministerio de Ciencia e Innovación (Spain) and Thematic Networks and Cooperative Research Centers: RIRAAF (RD12/0013/0014) and NIAID R21AI124062 (CB). LT was supported in part by the Robin Chemers Neustein Postdoctoral Fellowship. NCB was also supported in part by STSM Grant from COST Action FA1402.
Abbreviations
- ELISA
enzyme-linked immunosorbent assay
- FBS
fetal bovine serum
- GM-CSF
granulocyte macrophage-colony stimulating factor
- HPLC
high-performance liquid chromatography
- Ip
Intraperitoneal
- LPS
lipopolysaccharide
- moDCs
monocyte-derived dendritic cells
- MFI
median fluorescence intensity
- NF-κB
nuclear factor kappa-light-chain enhancer of activated B cells
- nsLTPs
non-specific lipid transfer proteins
- PALMs
pollen-associated lipid mediators
- PAMPs
pathogen-associated molecular patterns
- PBMCs
peripheral blood mononuclear cells
- PBS
phosphate-buffered saline
- PDB
Protein Data Bank
- RPMI
Roswell Park Memorial Institute
- SEAP
secreted embryonic alkaline phosphatase
- TLC
thin layer chromatography
- TLR4
toll-like receptor 4
Footnotes
Conflict of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Pawankar R, Canonica GW, Holgate ST, Lockey RF. Allergic diseases and asthma: a major global health concern. Current opinion in allergy and clinical immunology. 2012;12:39–41. doi: 10.1097/ACI.0b013e32834ec13b. [DOI] [PubMed] [Google Scholar]
- 2.Pawankar R. The unmet global health need of severe and complex allergies: meeting the challenge. The World Allergy Organization journal. 2012;5:20–21. doi: 10.1097/WOX.0b013e31824a5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hedlin G, et al. Problematic severe asthma in children, not one problem but many: a GA2LEN initiative. The European respiratory journal. 2010;36:196–201. doi: 10.1183/09031936.00104809. [DOI] [PubMed] [Google Scholar]
- 4.Muraro A, et al. The management of the allergic child at school: EAACI/GA2LEN Task Force on the allergic child at school. Allergy. 2010;65:681–689. doi: 10.1111/j.1398-9995.2010.02343.x. [DOI] [PubMed] [Google Scholar]
- 5.Fox M, et al. Health sector costs of self-reported food allergy in Europe: a patient-based cost of illness study. European journal of public health. 2013;23:757–762. doi: 10.1093/eurpub/ckt010. [DOI] [PubMed] [Google Scholar]
- 6.Breiteneder H, Radauer C. A classification of plant food allergens. The Journal of allergy and clinical immunology. 2004;113:821–830. doi: 10.1016/j.jaci.2004.01.779. [DOI] [PubMed] [Google Scholar]
- 7.Hoffmann-Sommergruber K. Plant allergens and pathogenesis-related proteins. What do they have in common? International archives of allergy and immunology. 2000;122:155–166. doi: 10.1159/000024392. [DOI] [PubMed] [Google Scholar]
- 8.Aalberse RC, Crameri R. IgE-binding epitopes: a reappraisal. Allergy. 2011;66:1261–1274. doi: 10.1111/j.1398-9995.2011.02656.x. [DOI] [PubMed] [Google Scholar]
- 9.Cubells-Baeza N, Gómez-Casado C, Tordesillas L, Ramírez-Castillejo C, Garrido-Arandia M, González-Melendi P, Herrero M, Pacios LF, Díaz-Perales A. Identification of the ligand of Pru p 3, a peach LTP. Plant Mol Biol. 2017 doi: 10.1007/s11103-017-0590-z. first online 15 Mar 2017. [DOI] [PubMed] [Google Scholar]
- 10.Fischer CL, et al. The roles of cutaneous lipids in host defense. Biochimica et biophysica acta. 2014;1841:319–322. doi: 10.1016/j.bbalip.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lorence A, Nessler CL. Camptothecin, over four decades of surprising findings. Phytochemistry. 2004;65:2735–2749. doi: 10.1016/j.phytochem.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Thomas WR. Allergen ligands in the initiation of allergic sensitization. Current allergy and asthma reports. 2014;14:432. doi: 10.1007/s11882-014-0432-x. [DOI] [PubMed] [Google Scholar]
- 13.Bublin M, Eiwegger T, Breiteneder H. Do lipids influence the allergic sensitization process? The Journal of allergy and clinical immunology. 2014;134:521–529. doi: 10.1016/j.jaci.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szabo PA, Anantha RV, Shaler CR, McCormick JK, Haeryfar SM. CD1d- and MR1-Restricted T Cells in Sepsis. Front Immunol. 2015;6:401. doi: 10.3389/fimmu.2015.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann SC, et al. Invariant natural killer T cells are enriched at the site of cutaneous inflammation in lupus erythematosus. J Dermatol Sci. 2013;71:22–28. doi: 10.1016/j.jdermsci.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 16.Agea E, et al. Human CD1-restricted T cell recognition of lipids from pollens. J Exp Med. 2005;202:295–308. doi: 10.1084/jem.20050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diaz-Perales A, et al. cDNA cloning and heterologous expression of the major allergens from peach and apple belonging to the lipid-transfer protein family. Clinical and experimental allergy. 2002;32:87–92. doi: 10.1046/j.0022-0477.2001.01257.x. [DOI] [PubMed] [Google Scholar]
- 18.Kulkarni AV, Patwardhan AA, Lele U, Malpathak NP. Production of camptothecin in cultures of Chonemorpha grandiflora. Pharmacognosy research. 2010;2:296–299. doi: 10.4103/0974-8490.72327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tordesillas L, et al. Transport of Pru p 3 across gastrointestinal epithelium - an essential step towards the induction of food allergy? Clinical and experimental allergy. 2013;43:1374–1383. doi: 10.1111/cea.12202. [DOI] [PubMed] [Google Scholar]
- 20.Leonard SA, et al. Dietary baked egg accelerates resolution of egg allergy in children. The Journal of allergy and clinical immunology. 2012;130:473–480. doi: 10.1016/j.jaci.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palacin A, et al. Graph based study of allergen cross-reactivity of plant lipid transfer proteins (LTPs) using microarray in a multicenter study. PloS one. 2012;7:e50799. doi: 10.1371/journal.pone.0050799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasquato N, et al. Crystal structure of peach Pru p 3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens. Journal of molecular biology. 2006;356:684–694. doi: 10.1016/j.jmb.2005.11.063. [DOI] [PubMed] [Google Scholar]
- 23.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 24.Ye Y, Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19(Suppl 2):ii246–255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 27.Tordesillas L, et al. Skin exposure promotes a Th2-dependent sensitization to peanut allergens. J Clin Invest. 2014;124:4965–4975. doi: 10.1172/JCI75660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.East JE, Sun W, Webb TJ. Artificial antigen presenting cell (aAPC) mediated activation and expansion of natural killer T cells. J Vis Exp. 2012 doi: 10.3791/4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pacios LF, et al. Computational study of ligand binding in lipid transfer proteins: Structures, interfaces, and free energies of protein-lipid complexes. Journal of computational chemistry. 2012;33:1831–1844. doi: 10.1002/jcc.23012. [DOI] [PubMed] [Google Scholar]
- 30.Benede S, Blazquez AB, Chiang D, Tordesillas L, Berin MC. The rise of food allergy: Environmental factors and emerging treatments. EBioMedicine. 2016;7:27–34. doi: 10.1016/j.ebiom.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noti M, et al. Exposure to food allergens through inflamed skin promotes intestinal food allergy through the thymic stromal lymphopoietin-basophil axis. The Journal of allergy and clinical immunology. 2014;133:1390–1399. doi: 10.1016/j.jaci.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stenger S, et al. Differential effects of cytolytic T cell subsets on intracellular infection. Science. 1997;276:1684–1687. doi: 10.1126/science.276.5319.1684. [DOI] [PubMed] [Google Scholar]
- 33.Leon L, et al. Saposins utilize two strategies for lipid transfer and CD1 antigen presentation. Proc Natl Acad Sci USA. 2012;109:4357–4364. doi: 10.1073/pnas.1200764109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vartabedian VF, Savage PB, Teyton L. The processing and presentation of lipids and glycolipids to the immune system. Immunol Rev. 2016;272:109–119. doi: 10.1111/imr.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barral DC, Brenner MB. CD1 antigen presentation: how it works. Nat Rev Immunol. 2007;7:929–941. doi: 10.1038/nri2191. [DOI] [PubMed] [Google Scholar]
- 36.Zajonc DM, Girardi E. Recognition of Microbial Glycolipids by Natural Killer T Cells. Front Immunol. 2015;6:400. doi: 10.3389/fimmu.2015.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahn VE, Leyko P, Alattia JR, Chen L, Prive GG. Crystal structures of saposins A and C. Protein Sci. 2006;15:1849–1857. doi: 10.1110/ps.062256606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abos-Gracia B, et al. Olea europaea pollen lipids activate invariant natural killer T cells by upregulating CD1d expression on dendritic cells. The Journal of allergy and clinical immunology. 2013;131:1393–1399. doi: 10.1016/j.jaci.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 39.Dearman RJ, Alcocer MJ, Kimber I. Influence of plant lipids on immune responses in mice to the major Brazil nut allergen Ber e 1. Clinical and experimental allergy. 2007;37:582–591. doi: 10.1111/j.1365-2222.2007.02689.x. [DOI] [PubMed] [Google Scholar]
- 40.Mirotti L, et al. Lipids are required for the development of Brazil nut allergy: the role of mouse and human iNKT cells. Allergy. 2013;68:74–83. doi: 10.1111/all.12057. [DOI] [PubMed] [Google Scholar]
- 41.Jyonouchi S, et al. Invariant natural killer T cells in children with eosinophilic esophagitis. Clinical and experimental allergy. 2014;44:58–68. doi: 10.1111/cea.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1: Immunolocalization of Caco 2 monolayer after 5 minutes of incubation with Pru p 3 or Complex. Pru p 3 was detected by polyclonal antibodies in red (Alexa 647) and TLR2 and TLR4 were detected in green (Alexa 488). From left to right: TLR2 (green), Pru p 3 (red), DAPI, phase contrast, triple combination (TLR/Pru p 3/DAPI) and colocalization of Pru p 3 and TLR. Bar 20 microns.