Abstract
Influenza therapeutics with new targets and mechanisms of action are urgently needed to combat potential new pandemics, emerging viruses, and constantly mutating circulating strains. We report here on design and structural characterization of potent peptidic inhibitors against influenza hemagglutinin (HA). The peptide design was based on complementarity determining region (CDR) loops of anti-HA human broadly neutralizing antibodies, FI6v3 and CR9114. The optimized peptides exhibit nanomolar affinity and neutralization against group 1 influenza A viruses including the 2009 H1N1 pandemic and avian H5N1 strains. The peptide inhibitors bind to the highly conserved stem epitope and block the low pH-induced conformational rearrangements associated with membrane fusion. These peptidic compounds and their advantageous biological properties should accelerate development of novel small molecule and peptide-based therapeutics against influenza virus.
Current influenza drug therapies that are available worldwide target only two viral proteins, the M2 channel and neuraminidase (NA), both of which function at critical stages of the virus lifecycle. M2 is involved in proton conducting activity in both early and late stages of replication and NA in the release of nascent virions (1, 2). However, escape mutations have resulted in drug-resistant viruses that diminish or ablate their therapeutic effects (3, 4). Therefore, new therapeutics with novel mechanisms of action are urgently required to combat the persistent global threat imposed by influenza virus. Therapeutic strategies aimed at targeting the highly conserved functional regions on influenza hemagglutinin (HA) that are involved in viral entry should be highly effective and may reduce the likelihood of generating escape mutants.
Infection by influenza virus is initiated by HA receptor binding at the cell surface and then by HA-mediated fusion of viral and host cellular membranes in the low pH of endosomal compartments (5). HA is the major glycoprotein on the influenza virus surface and is a homotrimer with each monomer composed of two subunits, HA1 and HA2, linked by a single disulfide bond (6). The HA1 subunit forms the membrane-distal globular head that contains the receptor binding site (RBS), as well the highly variable immunodominant regions that surround the RBS. HA2, as well as N- and C-terminal regions of HA1, form the highly conserved, membrane-proximal stem. HA1 interacts first with sialylated receptors on the host-cell epithelial surface, after which the bound virus is internalized by endocytosis. At the low pH of endosomes, the fusion potential of HA is activated, and HA dramatically rearranges to release its fusion peptide that anchors to the endosomal membrane, thereby triggering events that lead to fusion of the viral and host membranes (7).
Recent breakthroughs in isolation and characterization of human broadly neutralizing antibodies (bnAbs), reviewed (8, 9), against HA have raised hopes for a more universal vaccine as well as the possibility for designing therapeutics that target this region and may be less prone to resistance. Such bnAbs target the HA membrane-proximal stem region and block the large conformational rearrangements associated with membrane fusion, thereby neutralizing the virus. Structural characterization of their epitopes identified a highly conserved site of vulnerability at the HA1/HA2 interface in the HA stem that is present on influenza A group 1 and group 2 serotypes that represent 18 HA subtypes (H1-H18), as well as on both influenza B lineages (10).
This relatively recent structural information from bnAb-HA complexes has already inspired and guided the design of non-immunoglobulin protein scaffolds against the HA stem (11, 12). Novel small proteins, such as HB80 and HB36, were engineered de novo and based on the paratope of bnAb CR6261 by selecting scaffolds where amino-acid side chains were designed to occupy the conserved hydrophobic stem pockets and displayed in the appropriate configuration and conformation on scaffold proteins. These small proteins show comparable affinity, mode of binding, and neutralization breadth to CR6261, and inhibit the low pH conformational change in HA. Improved variants of HB36 also protect mice against lethal challenge from the 2009 H1N1 pandemic virus (13).
The success of these effective protein scaffolds encouraged us to explore the feasibility of engineering even smaller ligands against influenza virus. Smaller peptidic ligands present a promising alternative to biologics by retaining high affinity, selectivity and overcoming challenges such as cell permeability, low bioavailability, and high production cost (14). Furthermore, passive immunotherapy against influenza currently requires hospitalization for intravenous treatment (15). Strategies to develop peptides based on complementarity determining region (CDR) loops have been reported for other viral targets (16–18). However, the absence of any structural insights into their mechanism of action hindered further development. The collective structural information derived from interactions of several hydrophobic pockets in the influenza HA stem with heavy-chain CDR loops and framework region (FR3) of bnAbs FI6v3 and CR9114 guided our approach. Here we report on the successful development and structural characterization of potent cyclic peptides as influenza HA fusion inhibitors.
bnAb-guided peptide design
A rich compendium of structural and functional information is available on HA stem-targeting bnAbs encoded by germline genes VH1-69 [CR9114 (10), CR6261 (19), F10 (20) and A06 (21)] and VH3-30 [FI6v3 (22), 3.1 (23) and 39.29 (24)]. BnAbs CR6261, F10, A06, and 3.1 are specific against group 1 influenza A viruses, whereas FI6v3, 39.29 and CR9114 show pan influenza reactivity against group 1 and 2, as well as influenza B viruses (CR9114). All of these bnAbs interact with a highly conserved hydrophobic groove at the HA1/ HA2 interface in the HA stem. For CR9114, the key interacting residues are derived from HCDRs 2, 3 and FR3, whereas in FI6v3, HCDR3 serves as the dominant interacting motif (Fig. 1A). Therefore, to recapitulate most of these interactions within a single peptide with easier feasibility of cyclization, the HCDR3 loop of FI6v3 was chosen as the starting point for construction of peptidic ligands (Fig. 1, B and C).
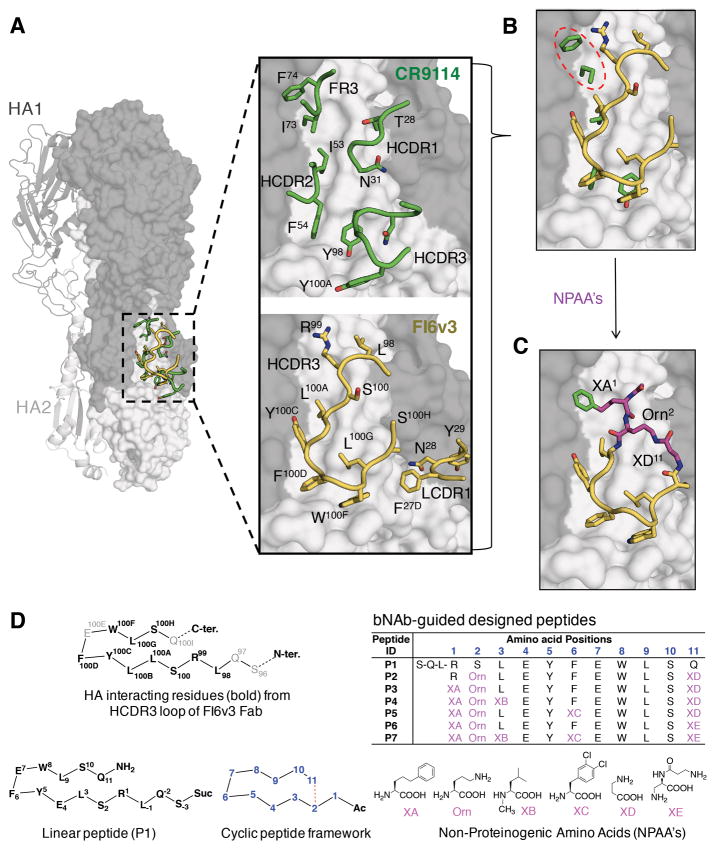
Fig. 1. bnAb-based cyclic peptide design strategy.
(A) Overlay of structures of influenza hemagglutinin from group 1 A/Puerto Rico/8/1934 (H1N1) strain (H1/PR8) (PDB 1RU7) (grey) with HA-interacting loop residues from the Fabs of HA-Fab complexes of CR9114 (PDB 4FQI) (green) and FI6v3 (PDB 3ZTN) (yellow). One HA protomer of H1/PR8 is represented as a cartoon and the two protomers in surface representation. HA1 is in dark grey and HA2 in light grey. Loop residues from the complementarity determining regions of the heavy (H) and light (L) chain (CDR 1, 2, 3) and framework region 3 (FR3) of each antibody that interact with the hydrophobic groove in the HA1/HA2 interface are in stick and ribbon representation. (B) Overlay of the hydrophobic groove interacting side chains from CR9114 HCDR2, HCDR3 and FR3 (green) with the HCDR3 loop from FI6v3 (yellow). CR9114 FR3 residues occupying one of the hydrophobic cavities on the HA stem not occupied by FI6v3 are highlighted with a dashed red oval. (C) Blueprint of the peptides constructed by merging features from the HCDR3 loop of FI6v3 (yellow) with FR3 of CR9114 (green). Non-proteinogenic amino acids (NPAAs) (magenta) used to link features derived from the two antibodies are shown in magenta. (D) Representative peptides in this study. Amino-acid sequences are shown for HCDR3 of FI6v3, linear peptide P1, and cyclized peptides P2-P7, with the chemical structures of corresponding NPAAs illustrated below.
A series of linear peptides varying in length, terminal-capping moieties, and mutations (table S1; fig. S1), was synthesized, characterized by LC-MS, and tested in an AlphaLISA competition binding assay against a panel of HAs from viruses representing influenza A group 1 [H1N1 A/California/07/2009 (H1/Cal), H1N1 A/New Caledonia/20/1999 (H1/NCa) and H5N1 A/Vietnam/1203/2004 (H5/Viet)], group 2 [H3N2 A/Brisbane/10/2007 (H3/Bris) and H7N7 A/Netherlands/219/2003 (H7/Neth)], and influenza B [B/Florida/4/2006) (B/Flo)] (SI-text S1). The linear sequence variant P1 displayed weak binding competition on group 1 H1 and H5 HAs (Figs. 1D and 2A; fig. S2). Peptide P1 is identical to FI6v3 HCDR3 except at position 4, where Leu100B in FI6v3 was replaced by Glu4 to improve peptide solubility (n.b. peptide residues are indicated in bold throughout) (Fig. 1D).
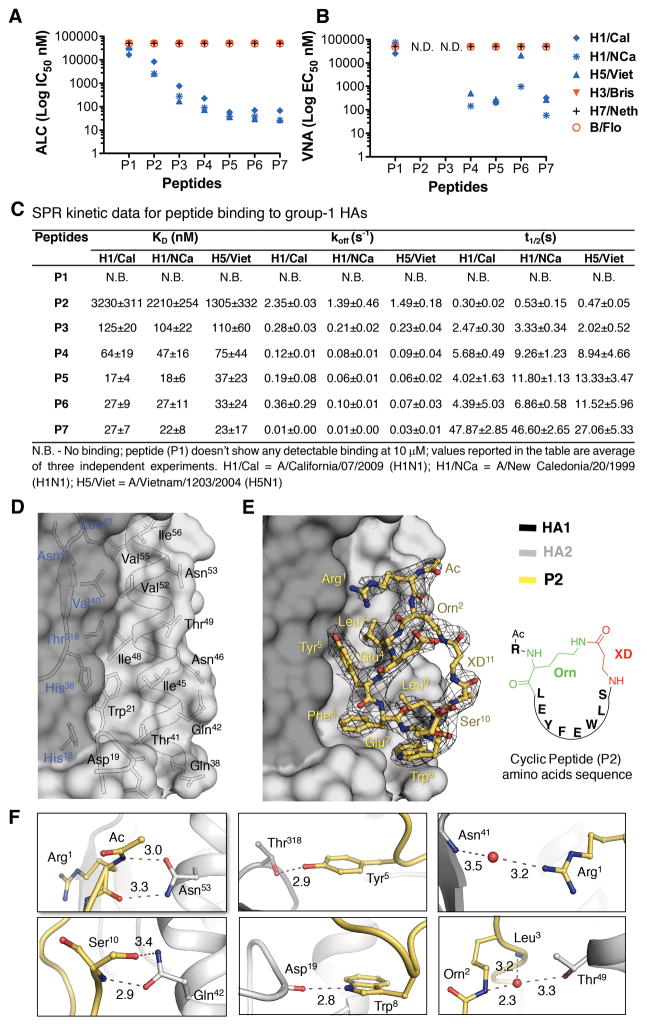
Fig. 2. In vitro binding, neutralization and crystal structure of the peptides.
(A) Peptide-mediated inhibition of HAs from different virus strains determined using an AlphaLISA competition (ALC) binding assay, reported as IC50. Peptides were tested against group 1 H1 HAs (H1/Cal = A/California/07/2009 (H1N1); H1/NCa = A/New Caledonia/20/1999 (H1N1)); H5 HA (H5/Viet = A/Vietnam/1203/2004 (H5N1)); group 2 H3 HA (H3/Bris = A/Brisbane/10/2007 (H3N2); H7 HA (H7/Neth = A/Netherlands/219/2003 (H7N7)) and an HA from influenza B virus (B/Flo = B/Florida/4/2006). Either CR9114 Fab or small protein HB80.4 was used as a positive control. (B) Virus neutralization potential of peptides determined in the virus neutralization assay (VNA), reported as EC50. N.D. = not determined. (C) SPR kinetic data for the peptides, as determined for group 1 H1 and H5 HAs. (D–F) Crystal structure of peptide P2 in complex with H1/PR8 HA. (D) P2 binding residues on the molecular surface of H1/PR8 HA (HA1-dark grey and HA2-light grey). (E) P2 (yellow sticks) in complex with H1/PR8 HA. A 2Fo-Fc map contoured at 1σ is displayed around P2. Residues of the peptide are numbered as in Fig. 1D. (F) Non-covalent interactions of P2 with H1/PR8 HA. Peptide is shown in yellow sticks, HA in grey and waters as red spheres. The distances are in Å.
In vitro affinity maturation of stem-targeting peptides
Starting from the sequence of P1, the peptide conformation was constrained by cyclization. To determine optimal ring size, a library of cyclic peptides of varying lengths was constructed incorporating non-proteinogenic amino acids (NPAAs) for lactam formation and using various cyclization strategies i.e. head-to-tail, sidechain-to-sidechain and sidechain-to-tail. In the latter constructs, an ornithine (Orn2) sidechain was fused with the carboxyl terminus of β-alanine (XD11) (Fig. 1, C and D). Synthesis and screening of this peptide library (table S2 and SI-text S2) led to P2 with micromolar binding and potency against H1/Cal, H1/NCa and H5/Viet HAs (KD’s of 1–3 μM by SPR, IC50’s of 2–8 μM in AlphaLISA); however, no binding was observed to group 2 or influenza B HAs (Figs. 1D and 2, A to C, figs. S1 to S3). Thus, constraining the peptide backbone via cyclization improved the performance of the peptidic ligands against group 1 HAs.
In the next round, further optimization was aided by our co-crystal structure of P2 with H1 HA from A/Puerto Rico/8/1934 (H1N1) (H1/PR8) (Fig. 2, D to F). The Arg1 guanidinium moiety was found to have unfavorable interactions with a hydrophobic cavity in the stem. However, the corresponding stem cavity is occupied by Ile73 and Phe74 of FR3 in CR9114 (Fig. 1, A and B). Thus, Arg1 of P2 was substituted by the hydrophobic 5-phenyl-norvaline (XA1) in P3 (Fig. 1D), which improved binding by 11–26 fold (KD=104–125 nM) with 9–15 fold improvement in potency (IC50=177–766 nM), compared to P2 (Fig. 2, A and C). The P3-HA complex is also slightly more stable (koff ≈0.2 s−1) than P2-HA (koff >1 s−1) (Fig. 2C and fig. S3).
Subsequent peptides P4, P5 and P6 (Fig. 1D) were then constructed based on P2 and P3, which suggested that rigidification of the peptide macrocycle and optimization of the HA-peptide interactions through incorporation of NPAAs can improve stability of the complex and enhance binding and neutralization. All three peptides demonstrate improved binding (KD), complex stability (koff), and potency (IC50) compared to P3 (Fig 2, A and C). While P4 showed modest improvement of 2–3 times (x) (KD=47–75 nM, IC50=76–228 nM), P5 and P6 exhibited 3–13x higher affinity and potency (KD=17–37 nM, IC50=30–70 nM) against all three HAs tested (Fig. 2, A and C, figs. S2 and S3). Furthermore, these peptides neutralize H1N1 and H5N1 viruses (Fig. 2B).
To investigate if binding and neutralization could be further improved, the enhancing features of P4, P5 and P6 were incorporated into a single peptide P7 (Fig. 1D). P7 shows similar affinity, potency and virus neutralization as P5 and P6 (Fig. 2, A to C, and figs. S2 and S3), but the HA-P7 complex is more stable with 2–25x lower koff (0.01–0.03 s−1) and 2–12x higher t1/2 (27–48 s) (Fig. 2C). Further, P7 demonstrates broad group 1 specificity when tested against a panel of HAs from the influenza A (group 1 and 2) and B viruses (figs. S4 and S5, and table S3).
Structural analysis of cyclic peptide binding to HA
Crystal structures of cyclic peptides (P2 to P7) in complex with H1/PR8 HA were determined at 2.28–3.10 Å resolutions (Figs. 2 to 4, figs. S6 to S9, and tables S4 and S5). All peptides recognize the highly conserved hydrophobic stem groove (Fig. 2, D and E, fig. S6). The peptides contact HA1 residues 18, 38–42 and 318, HA2 A-helix (residues 38–56), and a turn encompassing HA2 19–21 (Fig. 2D) that are remarkably similar to the epitopes of the stem-targeting bnAbs (CR9114, FI6v3, CR6261, and F10) (Fig. 3, A and B).
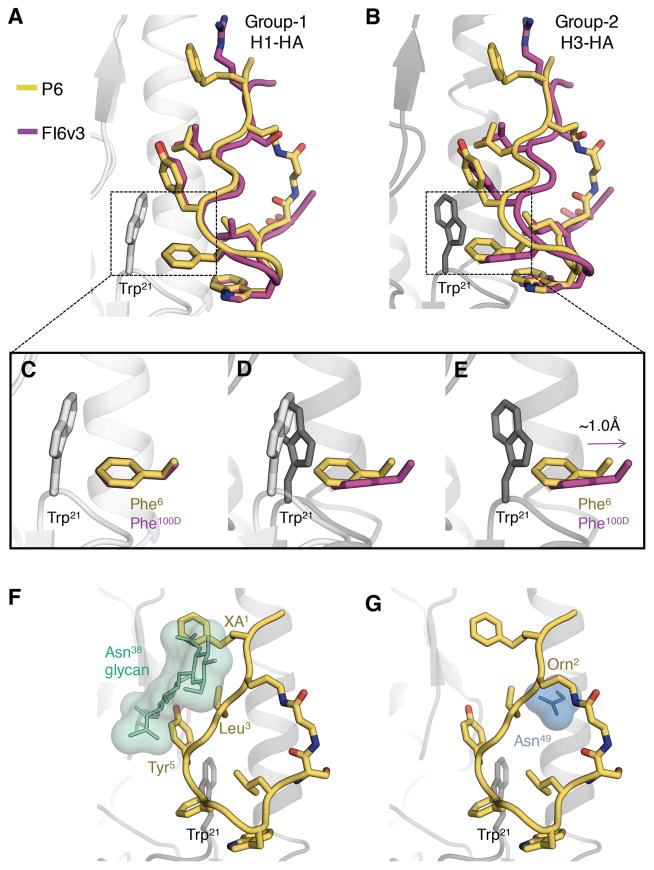
Fig. 4. Structural elucidation of the group 1 HA specificity of the peptides.
(A) Superimposition of H1 HA bound to FI6v3 HCDR3 (PDB 3ZTN, magenta) versus H1/PR8 HA bound to P6 (yellow). Only peptide P6 is shown from the PR8 complex. (B) Superimposition of H3 HA bound to FI6v3 HCDR3 (PDB 3ZTJ, magenta) versus H1/PR8 HA bound to P6 (yellow). (C–E) Depiction of potential steric clashes between Phe6 from P6 and Trp21 from group 2 HA. (C) Zoomed in view of Phe100D/Phe6 interactions with Trp21 from group 1 H1 HA. (D) Change in orientation of Trp21 in group 1 versus group 2 HAs. (E) Displacement of Phe100D to avoid a steric clash with Trp21 from group 2 HA and the predicted clash of Phe6 with Trp21 from group 2 HA. (F) Potential steric clash between XA1, Leu3 and Tyr5 from P6 and the glycosylated Asn38 (green) in group 2 HAs and (G) Potential steric clash between ornithine (Orn2) from P6 and Asn49 (light blue) in group 2 HAs, from an overlay of the H1/PR8 bound P6 with H3 HA.
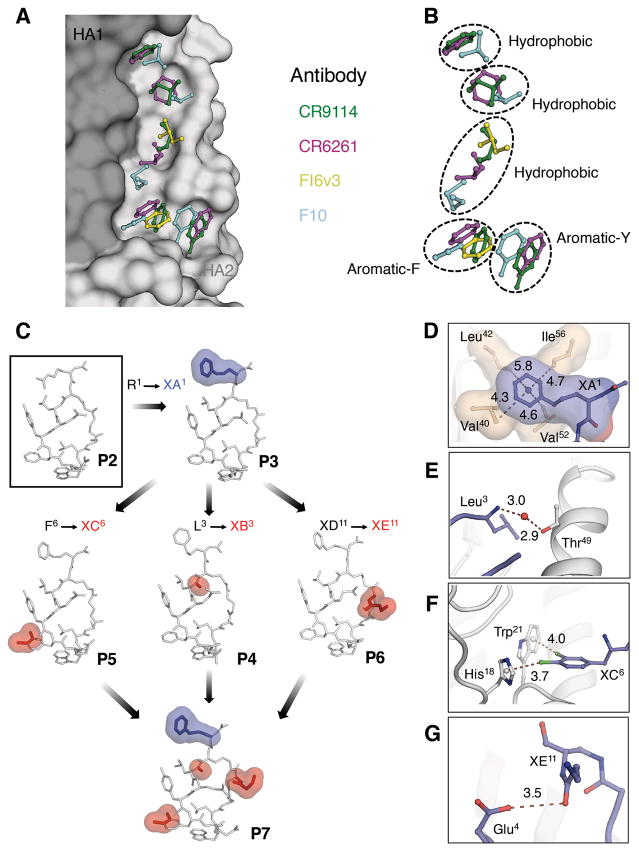
Fig. 3. bnAb guided affinity maturation of the peptides.
(A, B) The stem epitope of bnAbs on H1 HA from A/Puerto Rico/8/1934 (H1N1) (H1/PR8) is shown in surface representation with HA1 in dark grey and HA2 in light grey (see Fig. 1 for overall location). Antibodies in complex with HA [CR9114 Fab (PDB 4FQI), FI6v3 Fab (PDB 3ZTN), CR6261 Fab (PDB 3GBN) and F10 Fab (PDB 3FKU)] were superimposed on H1/PR8 HA. The side chains of hydrophobic residues from HCDR 1, 2, 3 and FR3 of each antibody that occupy pockets in the stem hydrophobic groove are represented in ball-and-stick and colored (CR9114 - green, CR6261 - magenta, FI6v3 - yellow and F10 - cyan). (C) Sequential changes introduced into the peptides during the process of affinity maturation are highlighted with blue and red surfaces on the crystal structures of the respective peptides P2 to P7 in complex with H1/PR8 HA. (D–G). Interactions of peptides with PR8 HA introduced by the sequential changes shown in C. (D) In P3, non-proteinogenic amino acid XA1 occupies a hydrophobic cavity in the stem epitope. The distance between the centroid of the phenyl ring and the surrounding hydrophobic residues is given in Å. (E) Water-mediated H-bond interaction of the backbone amide of Leu3 from P5 with Thr49 in HA2 A-helix. (F) Sigma hole-π interactions between the non-proteinogenic amino acid XC6 from P5 with His18 and Trp21 from HA1 and HA2, respectively. The distance between the centroid of aromatic ring and the chlorine atoms of XC6 is reported in Å. (G) Intramolecular H-bonds between Glu4 and XE11 in P6. The H-bond distance is in Å.
The nonpolar contacts made by the hydrophobic residues in the peptide macrocycles can be grouped into two distinct regions on the HA stem (Fig. 2D). The first region, consisting of HA1 Val40, Leu42 and Thr318 and Ile48, Val52, Val55 and Ile56 on the HA2 A-helix, is contacted by peptide residues Arg1 or XA1, Leu3 or XB3, and Tyr5 (Fig. 2, D and E; peptide residue abbreviations are in Fig. 1D). Phe6 or XC6, Trp8 and Leu9 contact the second region consisting of His18, His38, Trp21, Gln38, Thr41, Gln42 and Ile45 (Fig. 2, D and E). Polar interactions include eleven hydrogen bonds, where six are direct and five are mediated through water molecules (Fig. 2F). Four hydrogen bonds are involved in recognition of the A-helix at Asn53 and Gln42 by the peptide macrocycle backbone carbonyls of Arg1 (or XA1) and Ser10 (Fig. 2F). The Tyr5 and Trp8 side chains hydrogen bond with Thr318 and the Asp19 main-chain carbonyl (Fig. 2F). The cyclic peptides bury ~544–593 Å2 on the HA surface, which is fairly comparable to the surface area buried by the anti-stem Fabs in their HA complexes (~630–680 Å2) (10, 19, 22).
Next, we elucidated the structural basis for improvement of the optimized peptides compared to P2. The rationale behind substituting Arg1 of P2 with XA1 in P3 was to mimic interactions made by FR3 Ile73 and Phe74 in bNAb CR9114 (Figs. 1 and 3, A and B). Indeed, the significant improvement in affinity and potency of P3 can be explained by gain of hydrophobic contacts of XA1 with a small hydrophobic cavity formed by Val40, Leu42, Val52, and Ile56 (Fig. 3, C and D). N-methylation (Leu3 → XB3) rigidifies the peptide backbone in P4 and also mediates entropy gain by displacing a conserved water molecule bound to the A-helix (Fig. 3, C and E). Whereas incorporation of di-chloro phenylalanine (Phe6 → XC6) in P5 resulted in sigma hole-pi interactions with His18 and Trp21, respectively (Fig. 3, C and F). Such interactions in halogen-aromatic systems can contribute up to ~2.0 Kcal/mol binding energy (25). In P6, XE11 rigidifies the peptide macrocycle by forming an intra-molecular H-bond with Glu4 (Fig. 3, C and G).
Group 1 HA binding specificity
Despite the presence of interacting residues from pan influenza antibodies FI6v3 and CR9114, and targeting the same epitope on the surface of HA, the peptides neutralize group 1 and not group 2 or influenza B viruses (Fig. 2B). This activity can be rationalized by comparing the structures of apo and bnAb-complexed HAs from group 1 and 2 versus the peptide-HA complexes determined here. The key differences between the peptide epitope on H1 HA and the corresponding region on H3 HA are the orientation of HA2 Trp21, a glycosylation site at HA1 Asn38 in group 2 HA, and Thr49 in the A-helix of group 1 H1 HAs to compared Asn49 in group 2 H3 HAs (26). A conserved Phe (from different locations) in group-specific, as well as more bnAbs, make edge-to-face aromatic interactions with Trp21. However, only bnAbs FI6v3 and CR9114 appear to demonstrate sufficient flexibility in their key interacting CDR loops, containing Phe100D (HCDR3) and Phe54 (HCDR2) respectively, to accommodate the different orientations of Trp21 in the two HA groups (10, 22). Comparison of peptide P6 versus FI6v3 bound to group 1 H1 HA demonstrates a perfect overlay of Phe6 and Phe100D (Fig. 4, A and C) whereas, when FI6v3 is bound to group 2 H3 HA, Phe100D is displaced ~1Å outwardly compared to Phe6 (Fig. 4, B and E). Consequently, while FI6v3 can interact favorably with Trp21 of H3 HA, P6 and the other peptides evidently are more constrained and, hence, cannot avoid steric clashes with H3 HA Trp21 (Fig. 4D). Similarly, steric clashes between the corresponding Phe of group 1 specific antibodies, i.e. CR6261 and F10, and Trp21 of group 2 HA probably contribute to their group 1 specificity.
The second difference pertains to a glycosylation site at HA1 Asn38 in group 2 HAs. In unliganded (apo) group 2 HAs, the Asn38 glycan projects toward the HA2 A-helix of the same HA subunit such that it would overlap with the expected footprint of the stem-targeting antibodies and the peptides. Notwithstanding, when bnAbs FI6v3 and CR9114 bind group 2 HAs, they are able to reorient the glycan to insert their heavy chain CDRs into the corresponding hydrophobic groove and acquire cross-group reactivity. The peptides do not appear to be capable of reorienting the glycan and, therefore, would experience steric clashes of XA1, Leu3 and Tyr5 with the Asn38 glycan on group 2 HAs (Fig. 4F).
The third key difference arises at position 49 in the A-helix, where the bulkier Asn49 in H3 HAs would clash with the ornithine (Orn2) introduced into the peptides for cyclization (Fig. 4G). Overall, the crystal structures indicate that the cumulative effect of these differences account for why the peptides do not interact with group 2 HAs.
Mechanism of viral fusion inhibition
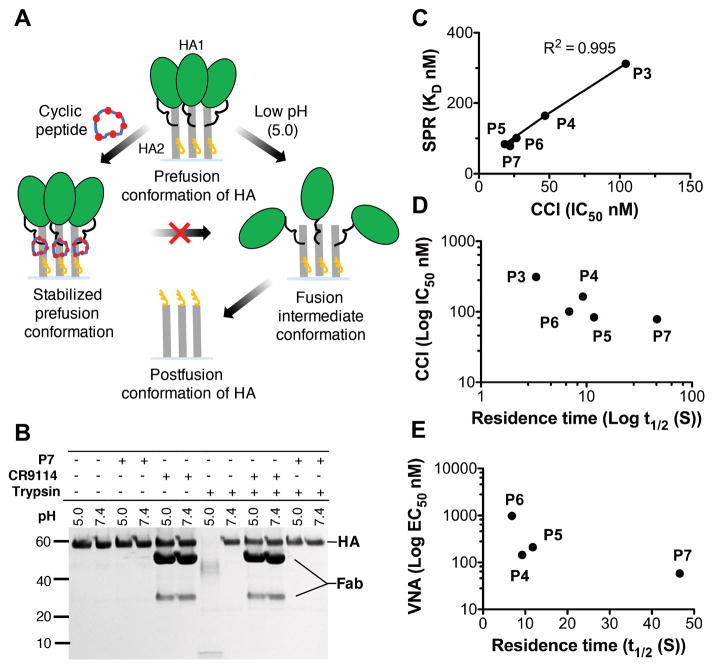
Viral fusion with the host cell membrane is mediated by a large conformational change in HA that is triggered by the low pH of the endosome. Stem-targeting bnAbs, such as CR9114, FI6v3 and CR6261, inhibit viral fusion by stabilizing the trimer even at low pH (10, 19, 22). Since the peptides mimic the binding modes of these antibodies, it was expected that they would have a similar mechanism of action. The first piece of evidence came from crystallization of the P3, P4, P5 and P7 peptide-HA complexes under low pH conditions (pH 4.0–5.0), which is well below the pH of membrane fusion (table S4) (7). The H1/PR8 HA structure in complex with various peptides is essentially identical to the prefusion apo HA (fig. S9). Therefore, the peptides appear to stabilize the prefusion state of HA and prevent the pH-dependent conformational rearrangements that lead to membrane fusion (Fig. 5A).
Fig. 5. Mechanism of HA conformational change inhibition by the peptides.
(A) Cartoon representation of the mechanism of conformational change inhibition by peptides. (B) The trypsin susceptibility assay establishes that P7 inhibits the low pH-induced conformational changes in H1/PR8 HA. Exposure to low pH renders the H1/PR8 HA sensitive to trypsin digestion (lanes 7 vs 8), but peptide P7 prevents its conversion to a trypsin-susceptible conformation (lanes 11 vs 12). The mechanism is similar to that of fusion inhibiting CR9114 Fab (lanes 9 vs 10). (C to E) A correlation of IC50 and EC50 was derived from conformational change inhibition (CCI) and virus neutralization assays (VNA), with SPR-derived constants KD and t1/2.
To directly assess inhibition of the HA rearrangements that are associated with fusion activation by the peptides, conformational change inhibition (CCI) and trypsin susceptibility (TS) assays (19, 27) were performed (Fig. 5, B to D, figs. S10 and S11, and table S6). All of the cyclic peptides prevented the conformational change at low pH in the CCI assay (table S6) and the trend demonstrated by CCI-derived IC50 and virus neutralization assay derived EC50 values correlates well with kinetic parameters derived from SPR (Fig. 5, C to E and SI-text S3). The TS assay with representative peptide P7 corroborated the CCI assay results. Therefore, the reported peptides are not only strong HA binders, but also effective inhibitors of the low pH conformation change. These data imply that, like the bnAbs, the cyclic peptides indeed function by stabilizing the HA trimer throughout endosomal entry and subsequent trafficking to late endosomes (28).
In vitro stability and in vivo pharmacokinetic profiling of peptide
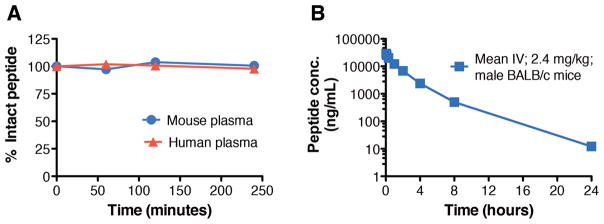
To investigate the translational potential of the cyclic peptides identified in this study, in vitro stability and in vivo pharmacokinetic studies were performed (Fig. 6). In vitro stability of peptide P7 was assessed in human and mouse plasma. P7 (~2 μg/ml) was incubated with the plasma at 37ºC and measured by LC-MS. P7 does not show degradation during the entire time course, i.e. ~4 hours (Fig. 6A), implying that cyclization and incorporation of NPAAs endows the peptide with resistance to proteolytic cleavage. In vivo pharmacokinetic profiling was then used to assess the stability and clearance of P7 after intravenous administration in BALB/c mice. P7 has a half-life of ~2.7 hours and is cleared from plasma in ~24 hours (Fig. 6B). Furthermore, the peptides do not show any significant cytotoxic effects in the human lung derived Calu-3 cell line (fig. S12). These translational data demonstrate that the peptidic fusion inhibitors discovered here are highly promising for development of influenza therapeutics.
Fig. 6. Plasma stability and in vivo pharmacokinetic profile of peptide P7.
(A) Stability of P7 in BALB/c mouse (blue circles) and human (red triangles) plasma at 37º C. (B) Plasma concentration of P7 following intravenous injection at 2.4 mg/kg in male BALB/c mice (n=3/group). Peptide concentrations from the plasma samples were analyzed by LC-MS/MS and individual plasma concentration-time profiles were subjected to non-compartmental pharmacokinetic analysis.
Implications for novel therapeutics against influenza virus
Here, we report on the development of effective peptidic inhibitors of influenza virus that neutralize by inhibiting the HA conformational rearrangements at low pH. Extensive data from virus neutralization, AlphaLISA, SPR, TS, CCI assays, X-ray crystallography, in vitro and in vivo stability studies, and their lack of cytotoxicity, provide validation that these peptide fusion inhibitors have potential to translate into the clinic. The peptides were designed based on how the antigen binding loops of pan influenza A (and B) antibodies interact with diverse HAs. Constraining the peptide macrocycle by cyclization and addition of non-proteinogenic amino acids led to improvement in the affinity and potency of peptides P4, P5, P6 and P7. Structural characterization vividly illustrated that the peptides do indeed recognize the highly conserved HA stem epitope and explain their specificity for group 1 versus group 2 influenza viruses. Thus, the peptides developed here should accelerate the field of small molecule therapeutics against influenza HA (27). This approach could be further applied to other sites of vulnerability on the HA and other pathogenic viruses such as HIV-1, Ebola, Zika etc.
Supplementary Material
Summary.
Potent peptidic inhibitors that target the stem fusion region of influenza hemagglutinin provide valuable insights on development of novel therapeutics against influenza virus.
Acknowledgments
We thank P. Raboisson for useful discussions on peptide design, M. Jongeneelen and A. van Eijgen for execution of AlphaLISA and virus neutralization assays, S. Blokland for production and site-specific modification of hemagglutinin and setup and executing of conformational change inhibition assays, M. Seijsener-Peeters for scientific guidance of assay execution and data analysis, F. Jahouh for designing the analytical methodology for peptide stability studies, H. Tien for help in setting up automated crystallization screens, W. Koudstaal and J.P. Verenini for help with manuscript formatting, and R. Stanfield and X. Dai for discussions on refinement. This work is supported in part by NIH grants R56 AI117675 and R56 AI127371 (to I.A.W.). R.U.K. is grateful to the Swiss National Science Foundation for early mobility post-doctoral fellowship. X-ray data sets were collected at the Advanced Photon Source, Argonne National Laboratory (beamline 23 ID-D), the Advanced Light Source (beamline 5.0.3) and the Stanford Synchrotron Radiation Lightsource (beamline 12-2). GM/CA CAT is funded in whole or in part with federal funds from the National Cancer Institute (Y1-CO-1020) and NIGMS (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy (DOE), Basic Energy Sciences, Office of Science, under contract no. DE-AC02-06CH11357. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under contract no. DE-AC02-05CH11231. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS, NIAID or NIH. All data and code to understand and assess the conclusions of this research are available in the main text, supplementary materials and via the following repositories: Protein Data Bank accession codes 5W6U, 5W6I, 5W5U, 5W5S, 5W6R and 5W6T. This is manuscript 29451 from The Scripps Research Institute. A patent application related to this work has been filed (application number WO2016EP60438; publication number WO2016180826). Sharing of materials described in this patent application will be subject to standard material transfer agreements.
Footnotes
Material and Methods
References (29–39)
REFERENCES AND NOTES
- 1.Das K, Aramini JM, Ma LC, Krug RM, Arnold E. Structures of influenza A proteins and insights into antiviral drug targets. Nat Struct Mol Biol. 2010;17:530–538. doi: 10.1038/nsmb.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Clercq E. Antiviral agents active against influenza A viruses. Nat Rev Drug Discov. 2006;5:1015–1025. doi: 10.1038/nrd2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moscona A. Global transmission of oseltamivir-resistant influenza. N Engl J Med. 2009;360:953–956. doi: 10.1056/NEJMp0900648. [DOI] [PubMed] [Google Scholar]
- 4.Sheu TG, et al. Dual resistance to adamantanes and oseltamivir among seasonal influenza A(H1N1) viruses: 2008–2010. J Infect Dis. 2011;203:13–17. doi: 10.1093/infdis/jiq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 6.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 7.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 8.Julien JP, Lee PS, Wilson IA. Structural insights into key sites of vulnerability on HIV-1 Env and influenza HA. Immunol Rev. 2012;250:180–198. doi: 10.1111/imr.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee PS, Wilson IA. Structural characterization of viral epitopes recognized by broadly cross-reactive antibodies. Curr Top Microbiol Immunol. 2015;386:323–341. doi: 10.1007/82_2014_413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dreyfus C, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleishman SJ, et al. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitehead TA, et al. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat Biotechnol. 2012;30:543–548. doi: 10.1038/nbt.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koday MT, et al. A Computationally designed hemagglutinin stem-binding protein provides in vivo protection from influenza independent of a host immune response. PLoS Pathog. 2016;12:e1005409. doi: 10.1371/journal.ppat.1005409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Craik DJ, Fairlie DP, Liras S, Price D. The future of peptide-based drugs. Chem Biol Drug Des. 2013;81:136–147. doi: 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]
- 15.Sparrow E, Friede M, Sheikh M, Torvaldsen S, Newall AT. Passive immunization for influenza through antibody therapies, a review of the pipeline, challenges and potential applications. Vaccine. 2016;34:5442–5448. doi: 10.1016/j.vaccine.2016.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casset F, et al. A peptide mimetic of an anti-CD4 monoclonal antibody by rational design. Biochem Biophys Res Commun. 2003;307:198–205. doi: 10.1016/s0006-291x(03)01131-8. [DOI] [PubMed] [Google Scholar]
- 17.Williams WV, Kieber-Emmons T, VonFeldt J, Greene MI, Weiner DB. Design of bioactive peptides based on antibody hypervariable region structures. Development of conformationally constrained and dimeric peptides with enhanced affinity. J Biol Chem. 1991;266:5182–5190. [PubMed] [Google Scholar]
- 18.Levi M, et al. A complementarity-determining region synthetic peptide acts as a miniantibody and neutralizes human immunodeficiency virus type 1 in vitro. Proc Natl Acad Sci USA. 1993;90:4374–4378. doi: 10.1073/pnas.90.10.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kashyap AK, et al. Protection from the 2009 H1N1 pandemic influenza by an antibody from combinatorial survivor-based libraries. PLoS Pathog. 2010;6:e1000990. doi: 10.1371/journal.ppat.1000990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 23.Wyrzucki A, et al. Alternative recognition of the conserved stem epitope in influenza A virus hemagglutinin by a VH3–30-encoded heterosubtypic antibody. J Virol. 2014;88:7083–7092. doi: 10.1128/JVI.00178-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura G, et al. An in vivo human-plasmablast enrichment technique allows rapid identification of therapeutic influenza A antibodies. Cell Host Microbe. 2013;14:93–103. doi: 10.1016/j.chom.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 25.Imai YN, Inoue Y, Nakanishi I, Kitaura K. Cl-pi interactions in protein-ligand complexes. Protein Sci. 2008;17:1129–1137. doi: 10.1110/ps.033910.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russell RJ, et al. H1 and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology. 2004;325:287–296. doi: 10.1016/j.virol.2004.04.040. [DOI] [PubMed] [Google Scholar]
- 27.Kadam RU, Wilson IA. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc Natl Acad Sci USA. 2017;114:206–214. doi: 10.1073/pnas.1617020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brandenburg B, et al. Mechanisms of hemagglutinin targeted influenza virus neutralization. PLoS One. 2013;8:e80034. doi: 10.1371/journal.pone.0080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.