Abstract
Background
We previously demonstrated that chronic alcohol ingestion augments TGFβ1 expression in the lung fibroblast and increases the risk of fibroproliferative disrepair in a mouse model of acute lung injury. The effect of alcohol on TGFβ1 is mitigated by treatment with sulforaphane, which can activate nuclear factor (erythroid-derived 2)-like 2 (Nrf2). However, the mechanisms by which alcohol amplifies, or SFP attenuates, TGFβ1 expression in the fibroblast are not known. MicroRNA (miR)-21 has been shown to inhibit Smad7, a TGFβ1 signaling inhibitor. In the present study, we hypothesized that alcohol augments TGFβ1 expression through up-regulation of miR-21, which subsequently inhibits Smad7.
Methods
Primary mouse lung fibroblasts were cultured ± alcohol ± SFP and assessed for gene expression of miR-21, and gene and/or protein expression of Nrf2, Nrf2-regulated anti-oxidant enzymes, Smad7, STAT3, and TGFβ1. NIH 3T3 fibroblasts were transfected with a miR-21 inhibitor and cultured ± alcohol. α-SMA, Smad7, and TGFβ1 protein expression were then assessed. In parallel, NIH 3T3 lung fibroblasts were transfected with Nrf2 silencing RNA (siRNA) and cultured ± alcohol ± SFP. Gene expression of miR-21, Nrf2, Smad7, and TGFβ1 were assessed.
Results
MiR-21 gene expression was increased by 12-fold at 48 hours and Smad7 gene and protein expression were reduced by ∼30% in alcohol-treated fibroblasts. In parallel, inhibition of miR-21 attenuated alcohol-mediated decrease in Smad7 and increase in TGFβ1 and alpha-smooth muscle actin protein expression. Treatment with SFP mitigated the effect of alcohol on miR-21, Smad7, and total and phosphorylated STAT3, and restored Nrf2-regulated antioxidant gene expression. Silencing of Nrf2 prevented the effect of SFP on miR-21, Smad7, and TGFβ1 gene expression in alcohol-treated NIH 3T3 fibroblasts.
Conclusions
Alcohol treatment increases TGFβ1 in fibroblasts, at least in part, through augmentation of miR-21, which then inhibits Smad7 expression. These effects can be attenuated by activation of Nrf2 with SFP.
Keywords: Alcohol, Smad7, miR-21, TGFβ1, fibroblast
Introduction
Chronic alcohol abuse is an international health concern, and has been estimated to directly contribute to 3.2-5.9% of overall global mortality (Rehm and Imtiaz, 2016). Furthermore, long-term alcohol ingestion is associated with significant organ dysfunction, and increased burden of mental disorders, lung disease, cancer, infection, and cirrhosis (Rehm and Imtiaz, 2016, Kershaw and Guidot, 2008). Our group previously demonstrated an increase in the pro-fibrotic cytokine, TGFβ1, in lung fibroblasts of alcohol-fed mice and rats, which translated to an increased risk of fibrotic disrepair following acute lung injury in chronic alcohol-exposed animals (Bechara et al., 2004, Sueblinvong et al., 2014b, Sueblinvong et al., 2014a). In addition, we showed that in alveolar epithelial cells and fibroblasts, alcohol exposure decreased the expression and activity of nuclear factor (erythroid-derived 2)-like 2 (Nrf2), a master transcription factor that regulates the anti-oxidant response element and promotes expression of anti-oxidant enzymes such as heme oxygenase-1 (HO-1), glutamate-cysteine ligase catalytic subunit (GCLC), glutamate-cysteine ligase modifier subunit (GCLM), and glutathione S-transferase (GST)(Jensen et al., 2013, Sueblinvong et al., 2014b, Zhang and Forman, 2009). We also determined that treatment with sulforaphane (SFP), a Nrf2 activator, attenuated TGFβ1 expression in alcohol-treated lung fibroblasts (Sueblinvong et al., 2014b). Moreover, in alcohol-treated fibroblasts, inhibition of TGFβ1 expression or signaling restored Nrf2 activity (Sueblinvong et al., 2014b). These data suggest that alcohol exposure may lead to chronic oxidative stress in the lung by suppressing Nrf2 and activating TGFβ1 in a vicious positive feedback loop. Despite these important discoveries, there are still several questions left to be answered. Importantly, the pathway by which alcohol augments TGFβ1 in the murine primary lung fibroblast remains unknown. Additionally, it is unclear if the effect of SFP can be ascribed to Nrf2 activation or to another regulatory pathway (Clarke et al., 2008).
TGFβ1 is secreted from the cell bound to the latency-associated peptide (LAP), which together form latent, or inactive, TGFβ; however, latent TGFβ cannot bind to cell surface receptors due to steric hindrance from the LAP (Aschner and Downey, 2016). Active TGFβ is formed after the LAP is cleaved from latent TGFβ1 due to a variety of mechanisms (Aschner and Downey, 2016). The canonical TGFβ1 signaling cascade is a complex series of events that begins in the extracellular microenvironment when active TGFβ1 ligand binds to the TGFβ receptor (TβR) II, promoting heterotetramerization with TβRI (Aschner and Downey, 2016). Downstream signaling by TβRI through phosphorylation of regulatory Smad (R-Smad) transcription factors promotes fibroblast-to-myofibroblast differentiation, with an increase in myofibroblastic markers such as alpha-smooth muscle actin (α-SMA), augmentation in TGFβ1 expression, and ultimately, fibrogenesis (Aschner and Downey, 2016, Phan, 2008). Activation of the canonical pathway is negatively controlled by Smad7, an inhibitory Smad (I-Smad) which binds to the intracellular domain of TβRI and prevents downstream activation of R-Smads (Aschner and Downey, 2016). It was recently shown that Smad7 was suppressed, with an associated increase in TGFβ1 pro-fibrotic signaling, in a mouse and rat model of bleomycin-induced acute lung injury and fibrosis (Hayashi et al., 1997, Venkatesan et al., 2004). In light of these findings, it is reasonable to speculate that alcohol may increase TGFβ1 by inhibiting Smad7.
MicroRNAs (miRs) are small, non-coding RNAs that inhibit protein expression by binding to the 3′ untranslated region (UTR) of target gene transcripts, increasing mRNA degradation, and/or inhibiting protein translation (Ha and Kim, 2014). There are over 2500 miRs discovered in humans, and it is estimated that the majority of human protein coding genes have at least 1 miR binding site (Ha and Kim, 2014). MiRs have been implicated in a number of disease processes including idiopathic pulmonary fibrosis (IPF), and serve as potential therapeutic targets for disease (Pandit et al., 2011). MiR-21 is up-regulated in myofibroblasts from patients with IPF, and it's expression is under positive control by several transcription factors, including signal transducer and activator of transcription 3 (STAT3)(Krichevsky and Gabriely, 2009, Liu et al., 2010). Recently, miR-21 was found to be increased in a mouse model of bleomycin-induced pulmonary fibrosis, and is thought to promote fibrosis by augmenting TGFβ1 signaling through direct inhibition of Smad7 protein expression (Liu et al., 2010); however, no study to date has evaluated the interplay between alcohol, miR-21, and Smad7.
In the present study, we aim to determine how alcohol mediates an increase in TGFβ1 in the murine lung fibroblast, and to determine how SFP attenuates TGFβ1. Additionally, we explore whether alcohol-mediated TGFβ1 augmentation is associated with an increase in the myofibroblastic marker, α-SMA. We hypothesize that alcohol induces miR-21 gene expression, which subsequently inhibits Smad7 protein expression, leading to a relative gain of function in the TβRI with resultant increase in TGFβ1 signaling and expression. Furthermore, we believe that SFP ultimately inhibits TGFβ1 expression by activating Nrf2.
Materials and Methods
Cell culture and treatment
Primary lung fibroblasts (PLFs) were isolated from the lungs of C57Bl/6 (wild-type, Jackson Laboratories, Bar Harbor, ME) mice as previously described (Roman et al., 2005). PLFs (passage 3-7) and NIH 3T3 mouse lung fibroblasts (ATCC, Manassas, VA) were cultured in DMEM with 4.5 g/L glucose supplemented with 20% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 U/ml streptomycin. Treatments included alcohol (60 mM) and sulforaphane (SFP, 5 μM).
Nrf2 silencing RNA (siRNA) transfection
NIH 3T3 cells were seeded in 6-well plates and transfected the next day with 5nM rat Nrf2 siRNA vector (Invitrogen, Carlsbad, CA) or the same concentration of scrambled control vector (Invitrogen, Carlsbad, CA) using HiPerFect (Qiagen, Valencia, CA) as described in the manufacturer protocol. At 24 hours post-transfection, some groups were exposed to alcohol at 60 mM, and all groups were cultured for an additional 48 hours. Cells were then harvested for microRNA analysis as described below.
Down-regulation of miR-21
To confirm the role of miR-21 in alcohol-mediated Smad7 expression, NIH 3T3 cells were transfected with Lipofectamine 3000 containing miR-21 inhibitor (5′UAGCUUAUCAGACUGAUGUUGA; 20 nM at the final concentration) or anti-miR negative control (20 nM) according to the manufacturer's protocol (Qiagen, Valencia, CA). Six hours after transfection, serum-free medium was replaced with complete culture medium, and cells were incubated overnight. Cells were treated with alcohol (60 mM) 24 hours after transfection, and then collected for gene and protein expression analysis at 48 hours and 72 hours respectively, as described below.
RNA and microRNA isolation, reverse transcription and quantitative PCR
The mirVana miRNA Isolation Kit was used to extract and purify messenger RNA (mRNA) and microRNA from mouse PLFs or NIH 3T3 lung fibroblasts according to the manufacturer's protocol (Life technologies, Grand Island, NY). In brief, following extraction and purification, mRNA and microRNA fractions were eluted into separate vials, and RNA concentrations and purity were determined. cDNA was synthesized from 250 ng microRNA per sample using the miScript II RT kit (Qiagen, Valencia, CA). cDNA was synthesized from 1 μg mRNA as previously described (Sueblinvong et al., 2014a). Quantitative PCR (qPCR) for mRNA expression was performed with primer sets for 18s, GCLC, GCLM, GSTT2, HO-1, Nrf2, Smad7, and TGFβ1 (Table 1) using iQ SYBR Green Supermix (Bio-Rad). qPCR for SNORD95 and miR-21 (5′UAGCUUAUCAGACUGAUGUUGA) gene expression was performed with primers sets from Qiagen using QuantiTect SYBR Green PCR Kit (Qiagen, Valencia, CA). The iCycler sequence detection system (Bio-Rad) was used for the real-time PCR analysis. The level of target mRNA or microRNA expression was normalized to 18s or SNORD95 housekeeping genes, and relative target mRNA or microRNA levels were determined according to the comparative cycle threshold method (Applied Biosystems 7900HT Sequence Detection System, User Bulletin No. 2; Applied Biosystems). Relative expression values were calculated as previously described (Sueblinvong et al., 2014a).
Table 1. Primers sequences.
| Genes | Forward Primer | Reverse Primer |
|---|---|---|
| m18s | 5′ GGA CCA GAG CGA AAG CA 3′ | 5′ ACC CAC GGA ATC GAG AAA 3′ |
| mGCLC | 5′ GCA CATCTA CCA CGC AGT CAA 3′ | 5′ ACA TCG CCT CCA TTC AGT AAC 3′ |
| mGCLM | 5′ CGG GAA CCT GCT CAA CTG 3′ | 5′ TCT TCT CTT TCA TCG GGA TTT A 3′ |
| mGST | 5′ CTC AAA GGG CAG CAC ATG AG 3′ | 5′ GTG GTC TGC CAC CTG GTA CT 3′ |
| mHO-1 | 5′ TGC TCG AAT GAA CAC TCT GG 3′ | 5′ TCC TCT GTC AGC ATC ACC TG 3′ |
| mNrf2 | 5′ TCA GTC TTC ACT GCC CCT CA 3′ | 5′ AGC TCC TGC CAA ACT TGC TC 3′ |
| mSmad7 | 5′ CAA ACC AAC TGC AGG CTG TC 3′ | 5′ CCCC CAG GGG CCA GAT AAT TC 3′ |
| mTGFβ1 | 5′ CCG TGG CTT CTA GTC CTG AC 3′ | 5′ GAV TGG CGA GCC TTA GTT TG 3′ |
Protein isolation and analysis
Total protein from PLF or NIH 3T3 cell lysates were isolated as previously described (Sueblinvong et al., 2012). Equal amounts of protein samples were separated on 4-20% gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, and transferred to nitrocellulose membranes. The blots were blocked in blocking buffer (5% non-fat dry milk, 0.1% Tween 20 in TBS, pH 7.4) for 1 hour, then incubated with mouse anti-mouse Smad7 (R&D, 1:1,000), rabbit anti-mouse α-SMA (Abcam, 1:1,000), mouse anti-mouse STAT3 (Santa Cruz, 1:200), mouse anti-mouse Tyr 705 p-STAT3 at (1:200, Santa Cruz), rabbit anti-mouse GAPDH (Sigma, 1:50,000), and rat anti-mouse TGFβ1 (BD Pharmingen, 1:500) at 4°C overnight in blocking buffer or 5% bovine serum albumin in 0.1% Tween 20 in TBS (for phosphorylated STAT3). They were then washed and incubated for 1 hour at room temperature with an appropriate horseradish peroxidase–conjugated secondary antibody (Amersham Biosciences, Pittsburgh, PA), washed again, and visualized via enzyme-linked chemiluminescence using the SuperSignal West Pico kit (Pierce Biotechnology, Rockford, IL).
Statistical analysis
Unpaired two tailed t-tests or one-way ANOVA were used for comparisons between groups using GraphPad Prism and GraphPad InStat version 4. Post-test analysis using Dunnett's correction method was performed if statistical significance was reached by one-way ANOVA. GraphPad Prism and GraphPad InStat version 5 were used to calculate statistics. Significant differences were accepted at a P level of < 0.05.
Results
Alcohol exposure attenuates Smad7 gene and protein expression in primary lung fibroblasts
We first sought to determine if alcohol inhibits Smad7 in lung fibroblasts. We found that alcohol treatment initially increased Smad7 gene expression at 24 hours (not statistically significant as compared to untreated cells); however, at 48 hours, Smad7 gene expression was suppressed by ∼50%, and by approximately 90% at 72 hours as compared to untreated cells (P<0.05)(Figure 1A). Figure 1B highlights an approximate 30% reduction in Smad7 protein (P<0.05) expression after 72 hours of treatment with alcohol. These results suggest that alcohol may amplify TGFβ1 signaling and expression by down-regulating the TGFβ1 signaling inhibitor, Smad7.
Figure 1. Alcohol exposure in vitro decreased Smad7 gene and protein expression in murine primary lung fibroblasts.
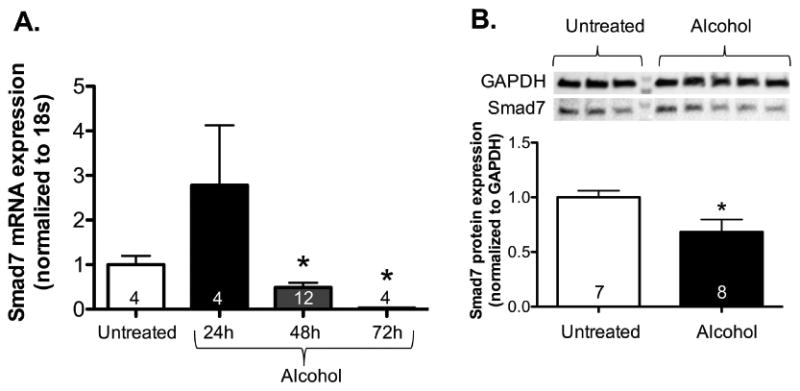
Primary lung fibroblasts (PLFs) were isolated from the lungs of C57BL/6 mice, and after 3-7 passages, cells were cultured ± alcohol (60 mM) for up to 72 hours, messenger RNA was isolated at 24, 48, and 72 hours and total protein was isolated at 72 hours. (A) At 24 hours of treatment, PLFs showed a trend toward an increase in Smad7 gene expression; however, gene expression was reduced by approximately 30% at 48 hours and nearly eliminated at 72 hours (as reflected by Smad7 mRNA levels, normalized to 18s). (B) There was an approximate 30% reduction of Smad7 protein expression at 72 hours as determined by Western immunoblot analyses (normalized to GAPDH; representative gels shown above summary data). *P<0.05 compared to untreated groups.
Alcohol exposure increases miR-21 expression in primary lung fibroblasts
MiR-21 has previously been shown to inhibit Smad7 expression in a variety of cells (Pandit et al., 2011). We speculated that alcohol might suppress Smad7 expression in lung fibroblasts by up-regulating miR-21. Lung fibroblasts were cultured with or without alcohol, and harvested for miR-21 gene expression analysis at 12, 24, and 48 hours. Figure 2 demonstrates a slight increase in miR-21 expression at 12 and 24 hours, with a significant increase by ∼12-fold at 48 hours (P<0.05). These findings suggest that in alcohol-treated murine PLFs, heightened miR-21 gene expression may directly suppress Smad7 with resultant increase in TGFβ1 expression.
Figure 2. Alcohol exposure in vitro increased microRNA-21 gene expression in murine primary lung fibroblasts.
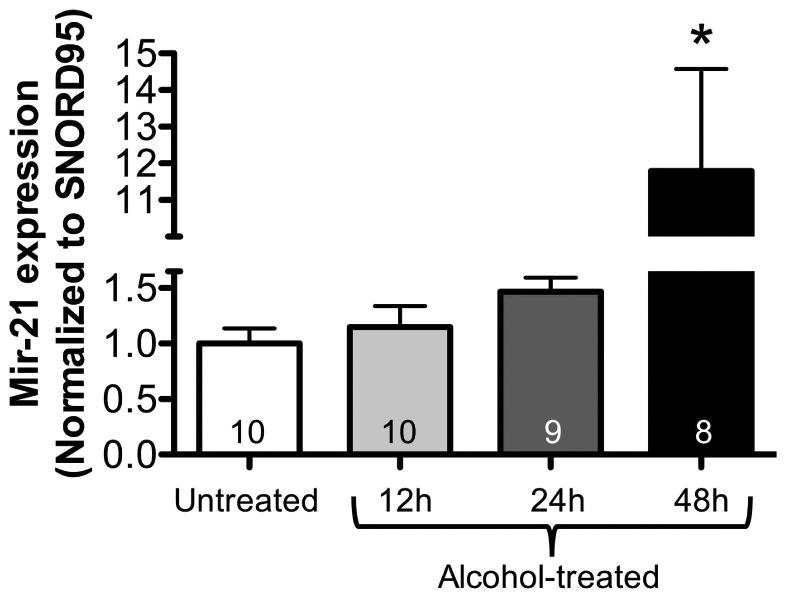
Murine primary lung fibroblasts (PLFs) were cultured ± alcohol (60 mM) for 12-48 hours and small RNA was isolated at 12, 24, and 48 hours. MicroRNA-21 (miR-21) expression was analyzed, and alcohol-treated PLFs showed a trend toward increased miR-21 expression at 12 and 24 hours. MiR-21 expression was significantly increased at 48 hours as compared to untreated cells (data normalized to a housekeeping gene SNORD95). *P<0.05 compared to untreated groups.
Alcohol exposure suppresses Nrf2 and augments TGFβ1 gene expression in primary lung fibroblasts
We performed a time-course study in primary lung fibroblasts to compare the effect of alcohol on TGFβ1 and Nrf2 gene expression with the temporal changes in miR-21 and Smad7 expression. There was a progressive fall in Nrf2 expression at 24, 48 and 72 hours, with a complementary rise in TGFβ1 expression at 24 and 48 hours in alcohol-treated murine primary lung fibroblasts compared to untreated controls as shown in Figures 3A and B, respectively. These results correlate with the results depicted in Figures 1 and Figure 2, and do not exclude the possibility that alcohol induces miR-21, which subsequently suppresses Smad7, leading to augmentation of TGFβ1 and inhibition of Nrf2.
Figure 3. Alcohol exposure in vitro attenuated Nrf2 and augmented TGFβ1 gene expression in murine primary lung fibroblasts.
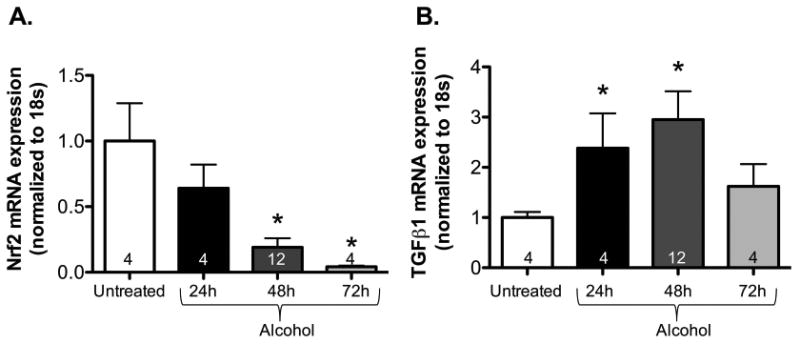
Murine primary lung fibroblasts (PLFs) were cultured ± alcohol (60 mM) for 24-72 hours and messenger RNA was isolated at 24, 48, and 72 hours. (A) Consistent with our previous findings, alcohol treatment suppressed Nrf2 gene expression starting at 48 hours, and gene expression remained suppressed at 72 hours (as reflected by Nrf2 mRNA levels, normalized to 18s) compared to untreated cells. (B) Similarly, alcohol treatment augmented TGFβ1 gene expression at 24 and 48 hours compared to untreated cells. However, there was a decrease in TGFβ1 gene expression at 72 hours. *P<0.05 compared to untreated groups.
Inhibition of miR-21 normalizes Smad7, and partially suppresses latent and active TGFβ1 protein expression in alcohol-treated NIH 3T3 fibroblasts
To confirm that alcohol-mediated augmentation of TGFβ1 expression was indeed secondary to inhibition of Smad7 by miR-21, we transfected NIH 3T3 lung fibroblasts with miR-21 inhibitor and treated with or without alcohol. Consistent with our findings in PLFs in Figure 1B, Smad7 protein expression was suppressed and both latent and active TGFβ1 protein expression were induced (P<0.05) in alcohol-treated cells transfected with negative control compared to untreated cells transfected with negative control (Figures 4A and 4B). Additionally, miR-21 inhibitor alone had no effect on Smad7 or TGFβ1 protein expression. Alcohol failed to suppress Smad7 protein expression in cells transfected with miR-21 inhibitor (P<0.05, Figure 4A). Along the same line, alcohol-induced latent and active TGFβ1 protein expression was partially attenuated in cells transfected with miR-21 inhibitor (Figure 4B). These data confirm that alcohol-mediated induction of miR-21 attenuates Smad7, and miR-21-mediated suppression of Smad7 contributes to an increase in TGFβ1 expression and activation.
Figure 4. In vitro inhibition of miR-21 restored Smad7 and partially attenuated TGFβ1 protein expression in alcohol-treated NIH 3T3 fibroblasts.
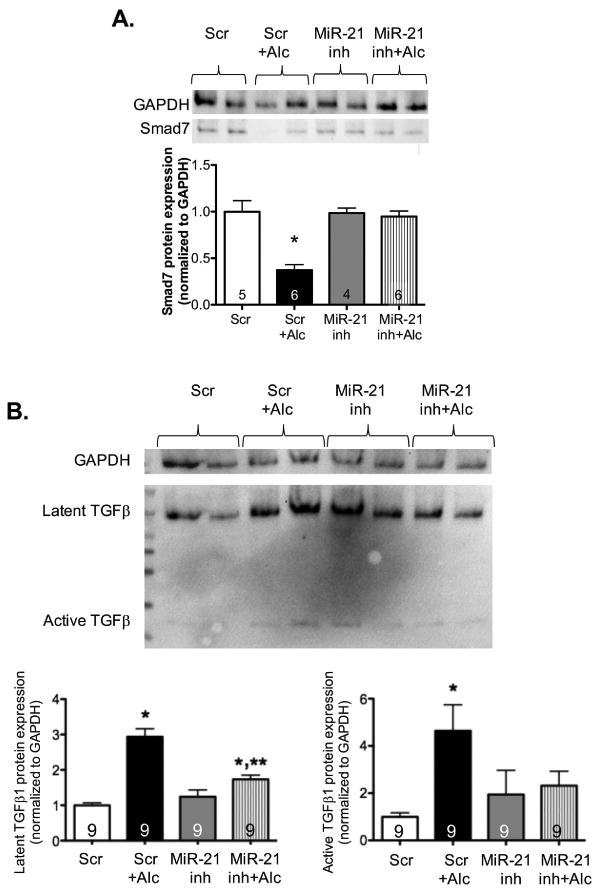
NIH 3T3 fibroblasts were transfected with miR-21 inhibitor (miR-21 inh; 10μM) or anti-miR negative control (scramble) and treated ± alcohol (Alc; 60 mM) starting at 24 hours after transfection. At 72 hours, cells were collected for protein expression analyses by Western immunoblot. (A) Consistent with the findings in PLFs, alcohol-treated NIH 3T3 cells transfected with negative control showed a decrease in Smad7 protein expression. Inhibition of miR-21 attenuated alcohol-mediated Smad7 suppression (normalized to GAPDH; representative gels shown above summary data). (B) Similarly, alcohol-treated NIH 3T3 cells transfected with negative control showed a significant increase in both latent and active TGFβ1 protein expression. Inhibition of miR-21 partially attenuated alcohol-induced latent and active TGFβ1 protein expression (normalized to GAPDH; representative gels shown above summary data). *P<0.05 compared to negative control (scramble RNA) transfected group. **P<0.05 compared to negative control (scramble RNA) + alcohol treatment group.
Alcohol augments protein expression of α-SMA, which can be partially attenuated by miR-21 inhibition in NIH 3T3 cells
To determine the role of alcohol and miR-21 in fibroblast-to-myofibroblast transdifferentiation, we assessed protein expression of the myofibroblast marker, α-SMA, in alcohol-treated NIH 3T3 cell. Figure 5 demonstrates that alcohol-treated cells transfected with negative control had a trend toward increased α-SMA expression compared to untreated cells transfected with negative control (P=0.05). Furthermore, this effect appeared to be partially attenuated in alcohol-treated cells transfected with miR-21 inhibitor.
Figure 5. Alcohol treatment trended toward induction of α-SMA protein expression in NIH 3T3 cells, which can be partially attenuated by miR-21 inhibition.
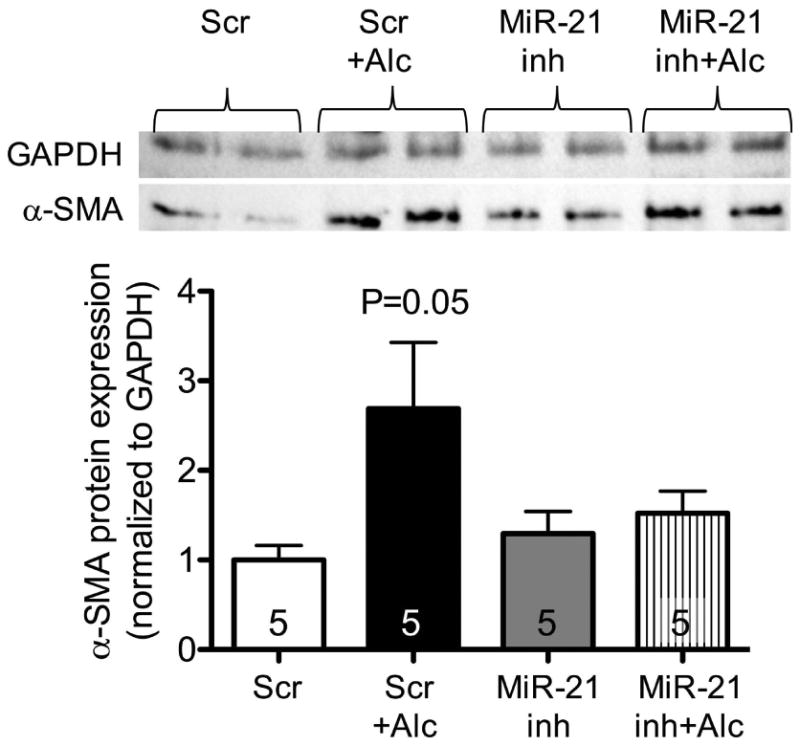
NIH 3T3 fibroblasts were transfected with miR-21 inhibitor (miR-21 inh; 10μM) or anti-miR negative control (scramble) and treated ± alcohol (Alc; 60 mM) starting at 24 hours after transfection. At 72 hours, cells were collected and analyzed for α-SMA protein expression by Western immunoblot.
Treatment with SFP attenuates miR-21 gene expression and restores Smad7 protein expression in primary lung fibroblasts
We have previously shown that treatment with the Nrf2 activator, SFP, could attenuate alcohol-mediated increase in TGFβ1 expression (Sueblinvong et al., 2014b). In the present study, we hypothesized that SFP achieves this effect by down-regulating miR-21 and/or up-regulating Smad7. We first confirmed that SFP could activate Nrf2 in alcohol-treated PLFs by evaluating gene expression of known Nrf2-dependent antioxidant enzymes. We show in Figure 6A that SFP increased Nrf2-dependent enzyme gene (e.g. GCLC, GCLM, GST, and HO-1) expression in alcohol-treated cells. In light of this, we assessed the effect of SFP on the expression of miR-21 and it's downstream target, Smad7. Figure 6B demonstrates that SFP mitigated alcohol-induced miR-21 gene expression by ∼50% (P<0.05) at 48 hours as compared to cells treated with alcohol alone. In parallel, Smad7 gene and protein expression were augmented following co-treatment of PLFs with alcohol and SFP as compared to alcohol alone (Figure 7A and 7B). These data suggest that SFP attenuates alcohol-induced TGFβ1 by modifying the miR-21-Smad7 axis.
Figure 6. Sulforaphane attenuated miR-21 gene expression in alcohol-treated murine primary lung fibroblasts.
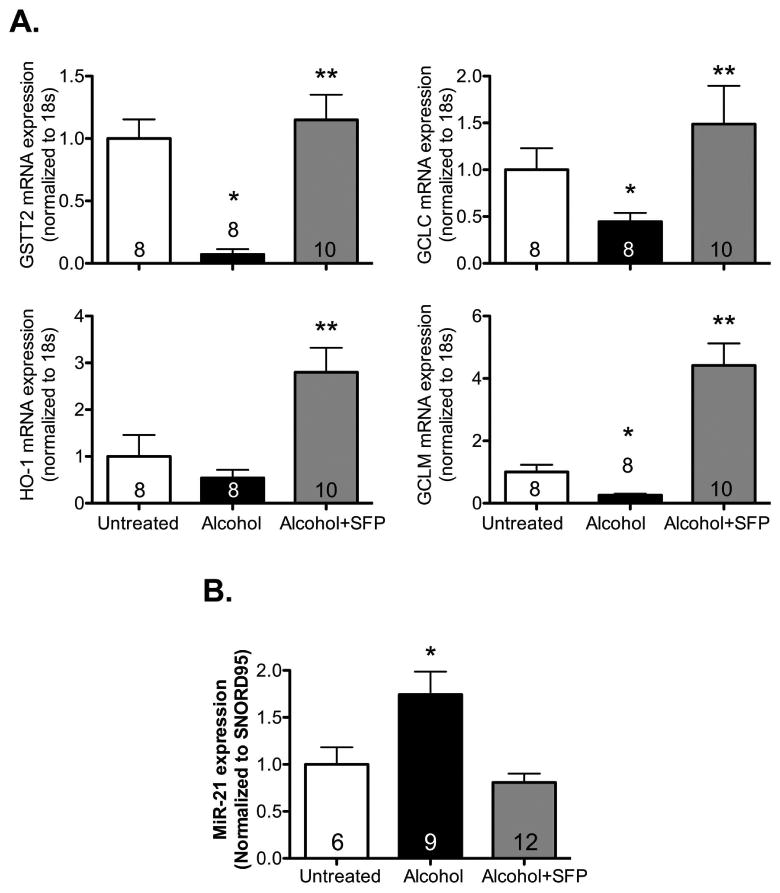
Murine primary lung fibroblasts (PLFs) were treated in vitro ± alcohol (Alc; 60 mM) ± sulforaphane (SFP; 5μM). Large and small RNA and protein were isolated at 48 hours and 72 hours, respectively. (A) Alcohol treatment significantly suppressed some of the Nrf2-dependent enzymes including glutamate-cysteine ligase catalytic subunit (GCLC), glutamate-cysteine ligase modifier subunit (GCLM), and glutathione S-transferase theta 2 (GSTT2) gene expression, but not heme oxygenase-1 (HO-1). Addition of SFP significantly increased gene expression of all Nrf2-dependent enzymes (as reflected by mRNA levels, normalized to 18s). (B) At 48 hours, alcohol treatment increased miR-21 gene expression while an addition of SFP attenuated the effect of alcohol on miR-21 in PLFs (normalized to SNORD95). *P<0.05 compared to untreated groups.
Figure 7. In parallel, sulforaphane restored Smad7 gene and protein expression alcohol-treated murine primary lung fibroblasts in vitro.
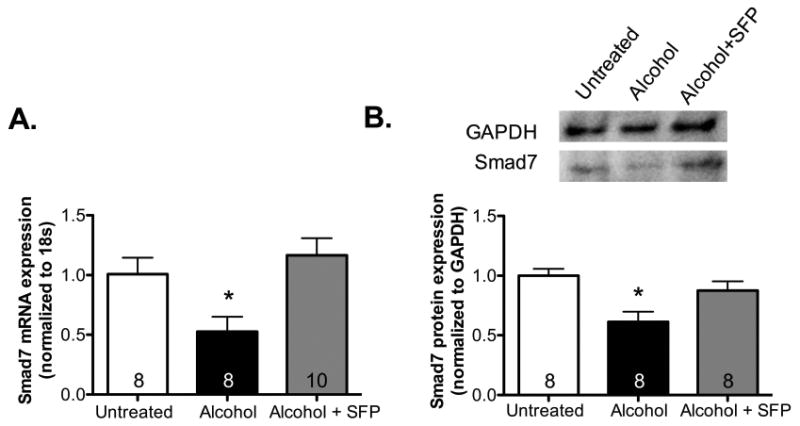
Murine primary lung fibroblasts (PLFs) were treated in vitro ± alcohol (Alc; 60 mM) ± sulforaphane (SFP; 5μM), messenger RNA and protein were isolated at 48 hours and 72 hours, respectively. Sulforaphane treatment restored (A) Smad7 gene expression (as reflected by Smad7 mRNA levels, normalized to 18s) and (B) Smad7 protein expression in alcohol-treated PLFs (normalized to GAPDH; representative gels shown above summary data). *P<0.05 compared to untreated groups.
Alcohol induces and sulforaphane suppresses STAT3 expression and activation in primary lung fibroblasts
We speculated that miR-21 induction might be secondary to an alcohol-mediated increase in STAT3 expression and activation in primary lung fibroblasts. In order to determine this, we treated primary lung fibroblasts with alcohol ± SFP, and measured protein expression of STAT3 and phosphorylated STAT3. Figure 8 demonstrates ∼3.5-fold increase in total STAT3 (Figure 8A) and ∼4-fold increase in pSTAT3 (Figure 8B) expression in cells treated with alcohol compared to untreated cells (P<0.05). The effect of alcohol on STAT3 was attenuated by treatment with SFP.
Figure 8. Alcohol increased STAT3 protein expression and activation, which is attenuated by sulforaphane in primary lung fibroblasts.
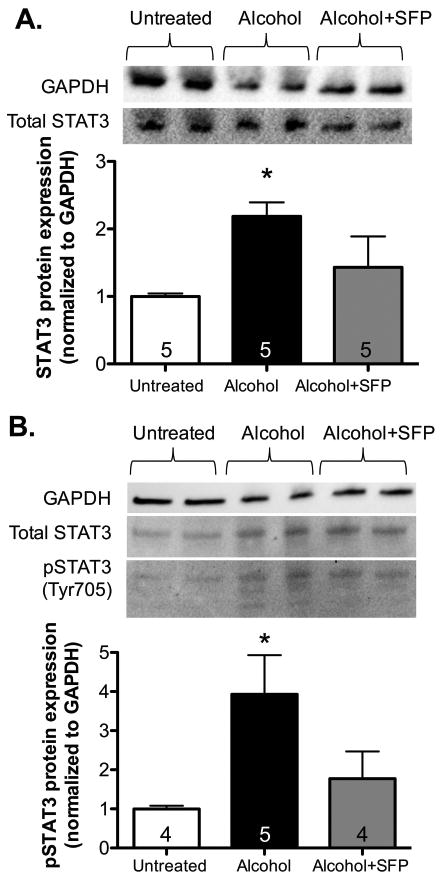
Murine primary lung fibroblasts (PLFs) were treated in vitro ± alcohol (Alc; 60 mM) ± sulforaphane (SFP; 5μM), total protein was isolated at 72 hours for Western immunoblot analyses. (A) Alcohol treatment increased total STAT3 protein expression, and addition of SFP normalized STAT3 expression. (B) Alcohol treatment increased Tyr 705 phosphorylated STAT3, while SFP treatment suppressed it. Protein expression was normalized to GAPDH; representative gels shown above summary data. *P<0.05 compared to untreated groups.
Treatment with SFP attenuates miR-21 gene expression through a Nrf2-dependent pathway in NIH 3T3 fibroblasts
To determine if SFP affects the miR-21/Smad7 pathway by it's known ability to activate Nrf2 rather than another mechanism of action, we transfected NIH 3T3 fibroblasts with Nrf2 silencing RNA (Nrf2 RNAi) or negative control RNA (scramble RNAi), then treated with or without SFP and/or alcohol. We first confirmed that transfection with Nrf2 siRNA was successful. Figure 9A shows ∼80% suppression of Nrf2 gene expression in cells transfected with Nrf2 RNAi as compared to cells transfected with scramble RNAi. We then assessed miR-21 expression, and as expected, alcohol induced miR-21 gene expression in cells transfected with scramble RNAi as compared to untreated cells transfected with scramble RNAi. Furthermore, SFP treatment attenuated alcohol-induced miR-21 expression in cells transfected with scramble RNAi (Figure 9B). In parallel, silencing of Nrf2 gene expression did not alter miR-21 expression. Addition of SFP to alcohol-treated cells transfected with Nrf2 RNAi failed to suppress miR-21 gene expression to a level comparable to cells transfected with scramble RNAi + alcohol + SFP (Figure 9B). Finally, we assessed Smad7 and TGFβ1 gene expression in cells transfected with Nrf2 RNAi, and as expected, we found that alcohol suppressed Smad7 and augmented TGFβ1 expression, while SFP attenuated these effects in cells transfected with scramble RNAi (Figure 10A and 10B). Interestingly, sulforaphane treatment significantly suppressed Smad7 gene expression in cells transfected with Nrf2 RNAi (P<0.05, Figure 10A). Co-treatment with alcohol and SFP had no significant effect on Smad7 gene expression in cells transfected with Nrf2 RNAi (Figure 10A). As previously shown by our group, Nrf2 RNAi transfection alone with or without SFP treatment did not significantly alter TGFβ1 gene expression (Sueblinvong et al., 2014b). Treatment with alcohol and SFP significantly accentuated TGFβ1 gene expression in cells transfected with Nrf2 RNAi (P<0.05, Figure 10B). These data suggest SFP mitigates the effects of alcohol on miR-21, Smad7, and TGFβ1 gene expression through the Nrf2 signaling pathway.
Figure 9. Sulforaphane treatment failed to attenuate miR-21 gene expression in NIH 3T3 cells transfected with Nrf2 silencing RNA.
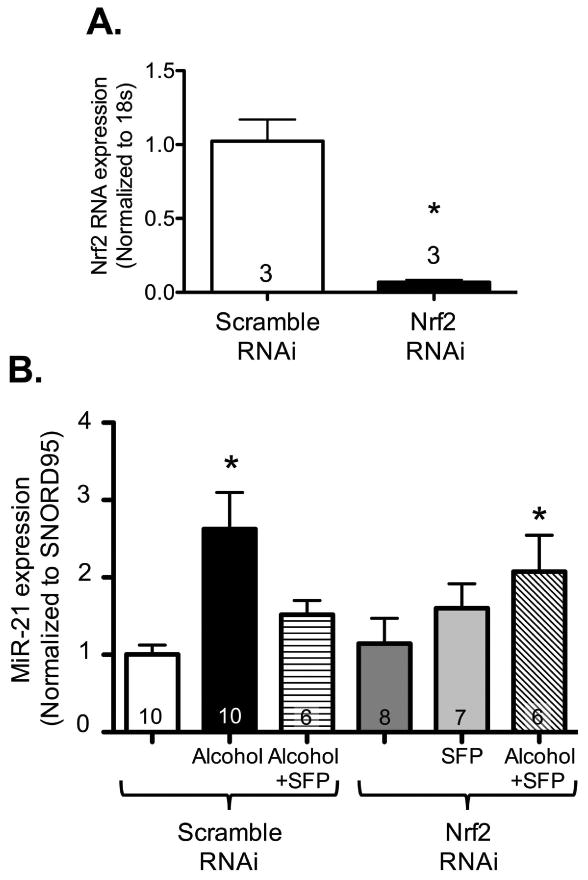
NIH 3T3 fibroblasts were transfected with Nrf2 siRNA (Nrf2 RNAi; 5nM) or negative control (scramble RNAi; 5nm) and treated ± alcohol (Alc; 60 mM) ± sulforaphane (SFP; 5μM) starting at 24 hours after transfection. (A) At 24 hours following Nrf2 siRNA transfection, Nrf2 gene expression was analyzed by quantitative PCR to assess for transfection efficacy. Cells transfected with Nrf2 siRNA showed a significant reduction (∼80%) in Nrf2 gene expression compared to cells transfected with negative controls (as reflected by Nrf2 mRNA levels, normalized to 18s). (B) At 48 hours following alcohol ± sulforaphane treatment, small RNA was isolated and miR-21 gene expression was analyzed. As shown above, alcohol treatment significantly increased miR-21 gene expression while SFP normalized it in scramble RNAi transfected groups. Nrf2 RNAi transfection alone did not alter miR-21 gene expression; however, SFP failed to suppress miR-21 gene expression in alcohol-treated cells transfected with Nrf2 RNAi. *P<0.05 compared to untreated groups transfected with negative control (scramble siRNA).
Figure 10. In parallel, sulforaphane failed to attenuate the effect of alcohol on Smad7 and TGFβ1 gene expression in NIH 3T3 cells transfected with Nrf2 silencing RNA.
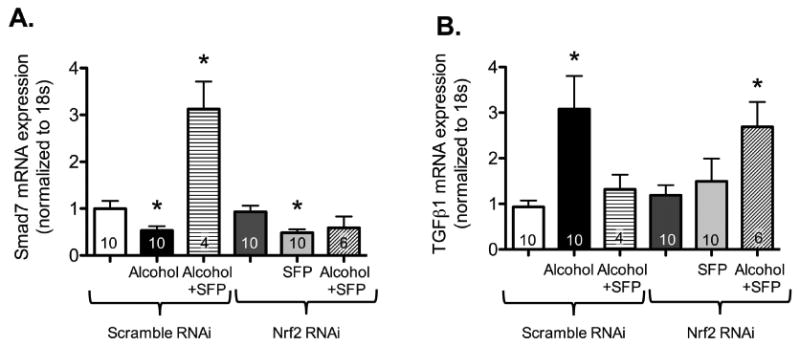
NIH 3T3 fibroblasts were transfected with Nrf2 siRNA (Nrf2 RNAi; 5nM) or negative control (scramble RNAi; 5nm) and treated ± alcohol (Alc; 60 mM) ± sulforaphane (SFP; 5μM) starting at 24 hours after transfection. At 48 hours following Nrf2 siRNA transfection, mRNA was isolated and Smad7 and TGFβ1 gene expression were analyzed by quantitative PCR. (A) In scramble RNAi-transfected cells, alcohol treatment suppressed Smad7 gene expression while addition of SFP significantly increased it. In Nrf2 RNAi-transfected cells, SFP treatment significantly decreased Smad7 gene expression while co-treatment of these cells with SFP and alcohol had no significant effect on Smad7 gene expression. (B) As expected, alcohol treatment significantly increased TGFβ1 gene expression in cells transfected with scramble RNAi, while SFP suppressed it. However, in Nrf2 RNAi transfection, SFP treatment alone did not have any significant effect on TGFβ1 gene expression, while alcohol + SFP treatment significantly increased it. *P<0.05 compared to untreated groups transfected with negative control (scramble siRNA).
Discussion
In this study, we determined that Smad7 protein expression is suppressed and miR-21 gene expression is increased in murine primary lung fibroblasts following alcohol exposure. Expanding upon this, alcohol-induced miR-21 appeared to directly suppress Smad7, as evidenced by restoration of Smad7 expression in alcohol-treated cells transfected with miR-21 inhibitor. Additionally, inhibition of miR-21 partially attenuated alcohol-induced latent and active TGFβ1 protein expression. Alcohol treatment also amplified expression and activation of the transcription factor STAT3 which is known to activate transcription of miR-21. Furthermore, alcohol-treated lung fibroblasts had increased α-SMA protein expression which can be attenuated by inhibition of miR-21. Finally, we showed that SFP inhibits miR-21 and restores Smad7 expression in alcohol-treated lung fibroblasts, which is mediated by activation of Nrf2. Taken together, these results suggest that alcohol augments TGFβ1 expression in murine lung fibroblasts, at least in part, by increasing miR-21 which suppresses Smad7 expression, leading to an increase in downstream signaling and self-induction of TGFβ1; however, there may be other mechanisms by which alcohol increases active TGFβ1. Alcohol-mediated increase in α-SMA suggests that augmentation of TGFβ1 by alcohol promotes an increase in fibroblast-to-myofibroblast differentiation in vitro. Lastly, activation of Nrf2 signaling by SFP limits miR-21 expression and subsequent Smad7 augmentation, and serves as a potential therapeutic option to prevent alcohol-mediated fibroproliferative disrepair following acute lung injury.
Our group previously showed that chronic alcohol exposure leads to oxidative stress in the lung, as reflected by alveolar depletion of the critical antioxidant glutathione, representing a fundamental mechanism underlying alcohol-induced lung dysfunction (Holguin et al., 1998, Gauthier et al., 2005, Moss et al., 1999). We further identified TGFβ1 as a proximal and critical mediator of alcohol-induced oxidative stress in the lung which increases susceptibility to acute injury and subsequent fibroproliferative disrepair (Sueblinvong et al., 2014b, Bechara et al., 2004). To identify a potential mechanism by which alcohol induces TGFβ1 expression and amplifies TGFβ1 signaling, we assessed the natural TGFβ1 inhibitor, Smad7, and found that alcohol exposure suppresses Smad7 protein expression by ∼30%. Smad7 inhibits canonical TGFβ1 signaling by binding to the TβRI which subsequently inhibits fibroblast-to-myofibroblast transition and self-induction of TGFβ1 activation and expression (Nakao et al., 1997, Hayashi et al., 1997, Aschner and Downey, 2016). Our results are in accordance with previous studies demonstrating lower levels of Smad7 in a bleomycin model of pulmonary fibrosis (Venkatesan et al., 2004, Liu et al., 2010). Furthermore, overexpression of Smad7 was shown to prevent pulmonary fibrosis following bleomycin exposure and renal fibrosis after obstructive uropathy (Chung et al., 2013, Nakao et al., 1999). These results are also in accordance with extra-pulmonary models of fibrosis, as knocking out Smad7 gene expression has previously been associated with increased risk of liver injury and fibrosis after an acute insult (Zhu et al., 2011, Hamzavi et al., 2008). However, this is the first study directly linking alcohol exposure with inhibition of Smad7 in the primary lung fibroblast. Given our findings, alcohol-mediated inhibition of Smad7 is a novel mechanism to explain downstream amplification of TGFβ1 and the increased risk of fibroproliferative disrepair following acute lung injury noted previously in our lab (Sueblinvong et al., 2014a, Sueblinvong et al., 2014b). Further studies are warranted to determine if augmentation of Smad7 can serve as a therapeutic target for management of end organ complications seen in chronic alcohol use.
To clarify the mechanism by which alcohol suppresses Smad7, we assessed miR-21, a known mediator in the TGFβ1 signaling pathway (Liu et al., 2010). In this study, we found that miR-21 gene expression was significantly increased by ∼12-fold in alcohol-treated lung fibroblasts. This finding is not unsurprising, as miR-21 expression was shown to be greater in the livers of alcohol-fed compared to control-fed rats following partial hepatectomy (Dippold et al., 2012). Prior literature clearly demonstrates Smad7 mRNA has a 3′ UTR binding site for miR-21 (Lin et al., 2014, Li et al., 2013, Liu et al., 2010). These data led us to hypothesize that alcohol-induced miR-21 expression attenuates Smad7 expression in lung fibroblasts. To evaluate this, we transfected NIH 3T3 cells with miR-21 inhibitor. We found that there was restoration of Smad7 protein expression in alcohol-treated cells transfected with miR-21 inhibitor as compared to alcohol-treated cells transfected with negative control. This confirmed our suspicion that alcohol-mediated increase in miR-21 directly inhibits Smad7 expression in the murine lung fibroblast. These results are similar to the study by Liu et al where bleomycin-mediated attenuation of Smad7 was prevented in fibroblasts transfected with anti-miR-21 probes (Liu et al., 2010). To understand if alcohol augments miR-21 directly, or if it acts upstream of miR-21, we measured expression of the transcription factor, STAT3, which has been shown to promote miR-21 expression in a variety of cell types (Krichevsky and Gabriely, 2009). We found that alcohol treatment increased both total and phosphorylated STAT3 protein expression in murine PLFs, suggesting that alcohol promotes miR-21 expression indirectly through induction and activation of the STAT3 signaling pathway; however, additional studies are required before confirming this mechanism.
In our present study, inhibition of miR-21 with resultant restoration in Smad7 led to partial attenuation of latent and active TGFβ1 in alcohol-treated cells. These data confirm our hypothesis that alcohol mediates an increase in miR-21 which attenuates Smad7, leading to a relative increase in TGFβ1 signaling and amplification in TGFβ1 expression in the murine fibroblast. However, given that both latent and active TGFβ1 protein expression were not completely normalized following transfection with miR-21 inhibitor, alcohol may activate TGFβ1 through an additional pathway independent of miR-21 and Smad7. Alcohol-mediated suppression of Nrf2 with resultant increase in oxidative stress provides one explanation for this finding, as TGFβ1 is known to be activated by reactive oxygen species (ROS)(Barcellos-Hoff and Dix, 1996, Sullivan et al., 2008, Aschner and Downey, 2016, Jensen et al., 2013). The increase in oxidative stress is likely independent of miR-21, and hence, would not be affected by miR-21 inhibition. As previously shown, we found that activation of Nrf2-ARE signaling pathway lead to partial attenuation of TGFβ1 expression, and vice versa, activation of TGFβ1 led to suppression of Nrf2-ARE signaling (Sueblinvong et al., 2014b). Taken together, these data suggest that alcohol not only induces TGFβ1 expression by suppressing Smad7, but also by inhibiting Nrf2 with resultant increased oxidative stress in a ‘vicious cycle’. Future studies exploring both therapeutic and preventative measures targeting the miR-21-Smad7 axis and Nrf2 signaling are essential to prevent the detrimental effect of alcohol on lung repair.
As mentioned above, we and others previously showed that alcohol-mediated suppression of Nrf2 activity and increase in TGFβ1 expression are linked in a feedback loop (or ‘vicious cycle’), and that activation of Nrf2 with SFP suppresses TGFβ1 (Sullivan et al., 2008, Sueblinvong et al., 2014b). To determine if the inhibitory effect of SFP on TGFβ1 expression was mediated through a miR-21/Smad7 dependent pathway, we treated fibroblasts with alcohol and SFP, and found that SFP restored mir-21 and Smad7 expression to the level expressed in untreated cells. Interestingly, SFP also reduced total STAT3 and pSTAT3 expression in alcohol-treated fibroblasts. These data suggest that SFP exerts it's effect upstream of the STAT3/miR-21/Smad7 axis, potentially by restoring Nrf2 activity and mitigating alcohol-induced oxidative stress.
SFP is a naturally occurring isothiocyanate found in cruciferous vegetables, and is believed to have numerous potential benefits including chemoprevention, neuroprotection, and relief from oxidative stress (Tarozzi et al., 2013, Clarke et al., 2008). The effect of SFP can be attributed to a variety of mechanisms including HDAC inhibition, cell cycle arrest, and inhibition of cytochrome P450 enzymes (Clarke et al., 2008). Our group is particularly interested in the ability of SFP to activate Nrf2, a master transcription factor that positively regulates the anti-oxidant response element (ARE) in response to oxidative stress (Hybertson et al., 2011). We previously demonstrated that alcohol promotes oxidative stress in the lung by inhibiting Nrf2, and that SFP restores Nrf2 function in the alveolar epithelium and fibroblast (Jensen et al., 2013, Sueblinvong et al., 2014b). Given the above, our current study findings are not unexpected. We again confirmed that SFP could activate Nrf2 and restore Nrf2-dependent enzyme expression in murine PLFs. To determine if SFP restored Smad7 expression through a Nrf2-dependent pathway rather than another mechanism previously mentioned, we transfected NIH 3T3 fibroblasts with Nrf2 siRNA. In cells transfected with Nrf2 siRNA and treated with alcohol, co-treatment with SFP failed to decrease miR-21 gene expression to a level comparable to untreated and alcohol + SFP-treated cells transfected with scramble siRNA. In parallel, we also showed that addition of SFP to cells transfected with Nrf2 siRNA ± alcohol did not significantly alter Smad7 or TGFβ1 gene expression. These data imply that SFP affects the miR-21/Smad7/TGFβ1 pathway through a Nrf2-dependent mechanism.
Taken together, we have outlined a pathway describing that alcohol augments TGFβ1 by increasing miR-21, which suppresses Smad7 (Figure 11). While not specifically addressed in this study, this model is almost certainly circular with multiple feedback loops. Supporting evidence for this comes from a prior study demonstrating how TGFβ1 and miR-21 augment one another in an amplifying circuit in human primary lung fibroblasts (Liu et al., 2010). In this study, we used two different fibroblast cell lines; primary lung fibroblasts from C57BL/6 mice and NIH 3T3 fibroblasts. NIH 3T3 fibroblasts were used because they were easier to transfect with siRNA and the miR-21 inhibitor. While the results may be theoretically different in each cell line, the differences would likely be minimal given both cell lines are from the same species. Murine fibroblast rather than human fibroblast was chosen in order to draw comparisons to prior studies; however, future studies are warranted in human fibroblasts to determine if the outcomes remain the same. Lastly, all of the experiments were done in vitro to ensure we were measuring the response to alcohol from the fibroblast rather than other cell types in the lung. Future in vivo studies are warranted to ensure the outcomes remain the same.
Figure 11. Proposed pathway outlining how alcohol augments TGFβ1 expression in the murine lung fibroblast.
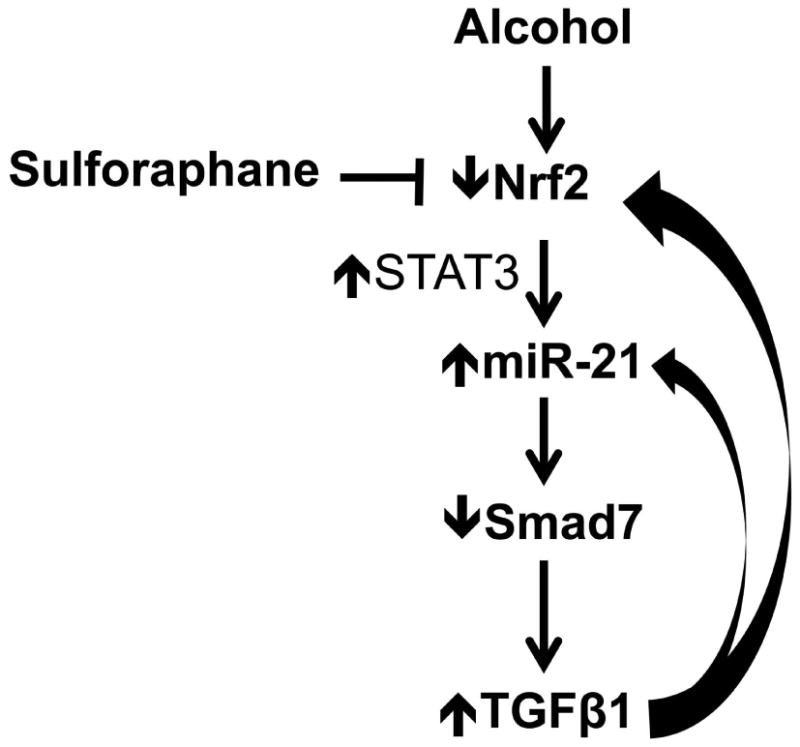
Alcohol exposure increases miR-21. Smad7 protein expression is then suppressed by miR-21. Inhibition of Smad7 leads to an increase in TGFβ1 signaling and augmentation of TGFβ1 gene and protein expression. TGFβ1 then amplifies miR-21 gene expression via positive feedback and inhibits Nrf2 (as previously shown by our group). MiR-21 and Smad7 expression can be normalized by the Nrf2 activator, SFP, which diminishes TGFβ1 and TGFβ1-mediated amplification of miR-21.
In conclusion, we determined for the first time that alcohol exposure augments miR-21, which in turn, suppresses Smad7 expression in the murine primary lung fibroblast. This is one novel mechanism to explain how alcohol amplifies TGFβ1 in the lung fibroblast, and provides a potential new target for prevention of pulmonary fibroproliferative disrepair secondary to chronic alcohol ingestion. SFP inhibits alcohol-mediated augmentation of miR-21 by activating Nrf2, and serves as a potential therapy for prevention or treatment of pulmonary fibroproliferative disrepair secondary to chronic alcohol use. While this study focused on the miR-21/Smad7 pathway, there are potentially other mechanisms by which alcohol augments active TGFβ1 expression in the fibroblast, including alcohol-mediated increase in ROS. Future studies are warranted aimed at identifying these pathways with a goal of establishing a multi-targeted approach to therapy. Follow-up in vivo studies would be helpful to confirm our most recent findings; in particular, it is important to determine if miR-21 and Smad7 are normalized by SFP in the whole lung, and if SFP or another Nrf2 activator can prevent lung disrepair following acute lung injury in people who chronically ingest alcohol.
Acknowledgments
Research support: NIH K08 AA021404-01 for VS, NIH T32 HL116271 for LTM, and the Veterans Affairs Career Development -02 (1 IK2 BX001707-01A1) for DEG.
References
- Aschner Y, Downey GP. Transforming Growth Factor-beta: Master Regulator of the Respiratory System in Health and Disease. Am J Respir Cell Mol Biol. 2016;54:647–55. doi: 10.1165/rcmb.2015-0391TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcellos-Hoff MH, Dix TA. Redox-mediated activation of latent transforming growth factor-beta 1. Mol Endocrinol. 1996;10:1077–83. doi: 10.1210/mend.10.9.8885242. [DOI] [PubMed] [Google Scholar]
- Bechara RI, Brown LA, Roman J, Joshi PC, Guidot DM. Transforming growth factor beta1 expression and activation is increased in the alcoholic rat lung. Am J Respir Crit Care Med. 2004;170:188–94. doi: 10.1164/rccm.200304-478OC. [DOI] [PubMed] [Google Scholar]
- Chung AC, Dong Y, Yang W, Zhong X, Li R, Lan HY. Smad7 suppresses renal fibrosis via altering expression of TGF-beta/Smad3-regulated microRNAs. Mol Ther. 2013;21:388–98. doi: 10.1038/mt.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JD, Dashwood RH, Ho E. Multi-targeted prevention of cancer by sulforaphane. Cancer Lett. 2008;269:291–304. doi: 10.1016/j.canlet.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dippold RP, Vadigepalli R, Gonye GE, Hoek JB. Chronic ethanol feeding enhances miR-21 induction during liver regeneration while inhibiting proliferation in rats. Am J Physiol Gastrointest Liver Physiol. 2012;303:G733–43. doi: 10.1152/ajpgi.00019.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier TW, Ping XD, Harris FL, Wong M, Elbahesh H, Brown LA. Fetal alcohol exposure impairs alveolar macrophage function via decreased glutathione availability. Pediatr Res. 2005;57:76–81. doi: 10.1203/01.PDR.0000149108.44152.D3. [DOI] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–24. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Hamzavi J, Ehnert S, Godoy P, Ciuclan L, Weng H, Mertens PR, Heuchel R, Dooley S. Disruption of the Smad7 gene enhances CCI4-dependent liver damage and fibrogenesis in mice. J Cell Mol Med. 2008;12:2130–44. doi: 10.1111/j.1582-4934.2008.00262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, Richardson MA, Topper JN, Gimbrone MA, JR, JL Wrana, Falb D. The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell. 1997;89:1165–73. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- Holguin F, Moss I, Brown LA, Guidot DM. Chronic ethanol ingestion impairs alveolar type II cell glutathione homeostasis and function and predisposes to endotoxin-mediated acute edematous lung injury in rats. J Clin Invest. 1998;101:761–8. doi: 10.1172/JCI1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hybertson BM, Gao B, Bose SK, Mccord JM. Oxidative stress in health and disease: the therapeutic potential of Nrf2 activation. Mol Aspects Med. 2011;32:234–46. doi: 10.1016/j.mam.2011.10.006. [DOI] [PubMed] [Google Scholar]
- Jensen JS, Fan X, Guidot DM. Alcohol causes alveolar epithelial oxidative stress by inhibiting the nuclear factor (erythroid-derived 2)-like 2-antioxidant response element signaling pathway. Am J Respir Cell Mol Biol. 2013;48:511–7. doi: 10.1165/rcmb.2012-0334OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw CD, Guidot DM. Alcoholic lung disease. Alcohol Res Health. 2008;31:66–75. [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhang D, Wang Y, Sun P, Hou X, Larner J, Xiong W, Mi J. MiR-21/Smad 7 signaling determines TGF-beta1-induced CAF formation. Sci Rep. 2013;3:2038. doi: 10.1038/srep02038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Gan H, Zhang H, Tang W, Sun Y, Tang X, Kong D, Zhou J, Wang Y, Zhu Y. MicroRNA21 inhibits SMAD7 expression through a target sequence in the 3′ untranslated region and inhibits proliferation of renal tubular epithelial cells. Mol Med Rep. 2014;10:707–12. doi: 10.3892/mmr.2014.2312. [DOI] [PubMed] [Google Scholar]
- Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207:1589–97. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss M, Steinberg KP, Guidot DM, Duhon GF, Treece P, Wolken R, Hudson LD, Parsons PE. The effect of chronic alcohol abuse on the incidence of ARDS and the severity of the multiple organ dysfunction syndrome in adults with septic shock: an interim and multivariate analysis. Chest. 1999;116:97S–98S. [PubMed] [Google Scholar]
- Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, Ten Dijke P. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389:631–5. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- Nakao A, Fujii M, Matsumura R, Kumano K, Saito Y, Miyazono K, Iwamoto I. Transient gene transfer and expression of Smad7 prevents bleomycin-induced lung fibrosis in mice. J Clin Invest. 1999;104:5–11. doi: 10.1172/JCI6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit KV, Milosevic J, Kaminski N. MicroRNAs in idiopathic pulmonary fibrosis. Transl Res. 2011;157:191–9. doi: 10.1016/j.trsl.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Phan SH. Biology of fibroblasts and myofibroblasts. Proc Am Thorac Soc. 2008;5:334–7. doi: 10.1513/pats.200708-146DR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm J, Imtiaz S. A narrative review of alcohol consumption as a risk factor for global burden of disease. Subst Abuse Treat Prev Policy. 2016;11:37. doi: 10.1186/s13011-016-0081-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman J, Ritzenthaler JD, Bechara R, Brown LA, Guidot D. Ethanol stimulates the expression of fibronectin in lung fibroblasts via kinase-dependent signals that activate CREB. Am J Physiol Lung Cell Mol Physiol. 2005;288:L975–87. doi: 10.1152/ajplung.00003.2004. [DOI] [PubMed] [Google Scholar]
- Sueblinvong V, Kerchberger VE, Saghafi R, Mills ST, Fan X, Guidot DM. Chronic alcohol ingestion primes the lung for bleomycin-induced fibrosis in mice. Alcohol Clin Exp Res. 2014a;38:336–43. doi: 10.1111/acer.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueblinvong V, Neujahr DC, Mills ST, Roser-Page S, Ritzenthaler JD, Guidot D, Rojas M, Roman J. Predisposition for disrepair in the aged lung. Am J Med Sci. 2012;344:41–51. doi: 10.1097/MAJ.0b013e318234c132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueblinvong V, Tseng V, Smith T, Saghafi R, Mills ST, Neujahr DC, Guidot DM. TGFbeta1 mediates alcohol-induced Nrf2 suppression in lung fibroblasts. Alcohol Clin Exp Res. 2014b;38:2731–42. doi: 10.1111/acer.12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan DE, Ferris M, Pociask D, Brody AR. The latent form of TGFbeta(1) is induced by TNFalpha through an ERK specific pathway and is activated by asbestos-derived reactive oxygen species in vitro and in vivo. J Immunotoxicol. 2008;5:145–9. doi: 10.1080/15476910802085822. [DOI] [PubMed] [Google Scholar]
- Tarozzi A, Angeloni C, Malaguti M, Morroni F, Hrelia S, Hrelia P. Sulforaphane as a potential protective phytochemical against neurodegenerative diseases. Oxid Med Cell Longev. 2013;2013:415078. doi: 10.1155/2013/415078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan N, Pini L, Ludwig MS. Changes in Smad expression and subcellular localization in bleomycin-induced pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2004;287:L1342–7. doi: 10.1152/ajplung.00035.2004. [DOI] [PubMed] [Google Scholar]
- Zhang H, Forman HJ. Signaling pathways involved in phase II gene induction by alpha, beta-unsaturated aldehydes. Toxicol Ind Health. 2009;25:269–78. doi: 10.1177/0748233709102209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Wang L, Wang X, Luo X, Yang L, Zhang R, Yin H, Xie D, Pan Y, Chen Y. Hepatic deletion of Smad7 in mouse leads to spontaneous liver dysfunction and aggravates alcoholic liver injury. PLoS One. 2011;6:e17415. doi: 10.1371/journal.pone.0017415. [DOI] [PMC free article] [PubMed] [Google Scholar]


