Abstract
Recent developments in automated optical sectioning microscope systems have enabled researchers to conduct high resolution, three-dimensional (3D) microscopy at the scale of millimeters in various types of tissues. This powerful technology allows the exploration of tissues at an unprecedented level of detail, while preserving the spatial context. By doing so, such technology will also enable researchers to explore cellular and molecular signatures within tissue and correlate with disease course. This will allow an improved understanding of pathophysiology and facilitate a precision medicine approach to assess the response to treatment. The ability to perform large-scale imaging in 3D cannot be realized without the widespread availability of accessible quantitative analysis. In this review, we will outline recent advances in large-scale 3D imaging, and discuss the available methodologies to perform meaningful analysis and potential applications in translational research.
Introduction
The diagnostic approach to human disease frequently relies on a pathological assessment of a biopsy from an organ involved in disease. This standard approach not only can define a disease, but also can help determine optimal therapy and provide prognostic information. In a few instances, such as protocol biopsies in solid organ transplant settings, repeated histopathology evaluation can also be useful to evaluate response to therapy [1, 2]. With advancements in our understanding of molecular and cellular pathogenesis of several diseases, it has become apparent that the information obtained from a biopsy can be used to further individualize diagnosis and therapy; a process referred to as precision medicine. For instance, the presence of a specific cellular marker may imply a more aggressive disease and/or determine responsiveness to therapy. The field of oncology offers several illustrations where a form of precision medicine is already implemented in clinical use. For example, the adoption of specific tumor stains (estrogen receptor, human epidermal growth factor 2 receptor, HER2) [3–5] and gene expression profiling [6] to determine prognosis and shape therapy has now become a standard of care in the management of breast cancer.
Before implementing a promising new tool on tissue specimens for clinical use, such a tool will require validation in preclinical models and testing of its utility in clinical studies. Because of the finite amount of the sampled tissue, especially in clinical settings, an ideal state-of-the-art tissue-interrogation technology should: 1) allow maximum extraction of the information from specimens of all sizes; 2) be amenable to standardization and reproducibility; 3) enable the production of quantitative analysis that can be easily performed in non-specialized centers. The use of immuno-histochemical or immuno-fluorescence techniques to study protein expression in situ have been a cornerstone in clinical and research histopathology evaluation. Despite the widespread use of these techniques, the ability to translate quantitative observations into objective data points has been challenging. In addition to problems with reproducibility, the inherent bias from sampling a small area of tissue can be limiting.
To address some of these limitations, several researchers have adopted the use of whole slide scanning to capture the entire area of available tissue and minimize sampling bias. In addition, many digital pathology software tools are being developed and implemented for use in clinical research studies. These tools may allow better standardization and objectivity. However, in the best-case scenario of optimal use, these technical advances still limit tissue examination in a 2-dimensional (2D) plane. A major shortcoming in such approach is the inability to capture the spatial characteristics and structures within complex organs. For example, a glomerulus or an entire physiological unit of kidney, the nephron, extends the full depth of the kidney. Further, complex cellular interactions, such as immune cells, do not limit their interactions to 2D planes. In addition, when phenotyping cells using multiple labels, the uneven distribution of many cell surface markers reduces the accuracy of a simple 2D survey. Therefore, the characteristics of morphologically complex organs and the interactions between various types of cells in situ is better captured using three-dimensional (3D) imaging.
Recent development in optical sectioning microscopes have allowed researchers to perform high resolution 3D microscopy at the millimeter scale in various tissues-mesoscale imaging. With the implementation of multiple laser lines and methods of spectral unmixing, it is possible to characterize up to 8 different labels within tissue slices. Microscopy of this magnitude offers several advantages over conventional approaches to tissue histology: first, mesoscale images support the holistic interrogation of hundreds of thousands of cells, minimizing sampling bias and improving the detection of rare events; second, mesoscale images capture tissue structures necessary to interpret pathology-the distribution of cells throughout the tissue and their possible activities; third, the characteristics of morphologically complex cells (such as neurons, dendritic cells, etc.) and their interactions can be captured.
In this review, we will outline recent advances in mesoscale 3D imaging, its evolving methodology and discuss the available tools to perform quantitative analysis, and its potential applications in translational research1. The implementation of mesoscale multi-fluorescence 3D imaging and analysis will provide a powerful tool that allows unbiased high-resolution molecular and cellular mapping of tissue specimen in health and disease states.
Enabling Technologies
In the past 10 years, we have witnessed an explosion in the tools and technologies to enable mesoscale 3D imaging. These advancements have included progress in tissue clearing and tissue staining, the availability of automated microscopes with rediscovered and new optical sectioning modalities, and the development of new analytical approaches. The clinical setting imposes several restrictions on the adoption of new technologies including diagnostic value that can be delivered in a timely fashion for responsive interventions and efficient use of often limiting amounts of sample or tissue. Current practice in tissue section imaging includes immuno-fluorescence and small fluorescent-molecule staining to identify cellular markers and tissue structure. As 3D mesoscale imaging is a volumetric extension of thin section imaging, it can rely on the multitude of antibodies and reagents developed for thin-section staining. Thus, the adoption of mesoscale 3D imaging for clinical application will require that 1) there be additional or improved information gained, such as more accurate and/or additional parameters for quantitation and diagnostics, 2) samples must be processed, imaged and quantified on the order of hours and days not weeks, 3) sample processing must be efficient, as human biopsy or tissue samples are available in limited amounts; approaches that are compatible with multiple rounds of analysis would be ideal, 4) sample processing must be compatible with existing practice (e.g. immunofluorescence, etc.) and not require expensive instrumentation or extraordinary expertise to use. The additional information gained from 3D mesoscale imaging will enable the interrogation of hundreds of thousands of cells, the precise quantitation of spatial relationships in 3D and the potential to develop new models and parameters for diagnostics. Thus, we would argue that the translational potential of new tools, techniques and approaches for 3D mesoscale imaging, hinges on speed, efficiency and compatibility with existing practice.
Tissue Preparation
Several methodologies can be used to prepare a tissue section for volumetric imaging. In its simplest form, a reasonably sized tissue is fixed using 4% paraformaldehyde immediately after harvesting, and subsequently sectioned using a vibrotome into slices of 50 to 100 μm thick [7–9]. This process applies to solid organ samples, such as the kidney, as we described previously. In our experience, a tissue section of 50 μm will allow uniform penetration and staining with antibodies and small molecules [7, 8, 10]. For small specimen or soft organs, tissue sectioning is only feasible after freezing with cryoprotection. This process also applies to small human specimens, such as core samples from a kidney biopsy. Additionally, we found that human biopsies preserved in Michel’s solution prior to freezing remain well suited for processing and fluorescence 3D imaging [10]. When imaging a large surface area like a section from a core biopsy (4–6 mm2), the magnitude of data obtained from 50 μm sections is tremendous, >10 GB with multiple channels (Figure 1). However, it may also be necessary to image deeper into tissues, up to hundreds of microns and millimeters, to assess whole glomeruli or the entirety of a nephron[11]. This is now possible with advancements in tissue preparation and the use of “tissue clearing”. The opaqueness of organs is due to inhomogeneous scattering of light in the tissue. This inhomogeneous scattering is a result of variation in refractive index (RI) in the tissue due to anatomical structure, different cell types in tissues, the variety in the organization of organelles in these cells and the RI differences between membranes, the cytosol and protein. More than a hundred years ago, anatomists recognized the inhomogeneity could be minimized and began developing tissue clearing protocols[12, 13]. The goal in tissue clearing is to remove this RI inhomogeneity. When effective, tissue clearing uncovers mesoscale 3D structure from normally opaque tissue (Figure 2) [14–17].
Figure 1. Mosaic of 80 image volumes collected from a mouse kidney thick section.
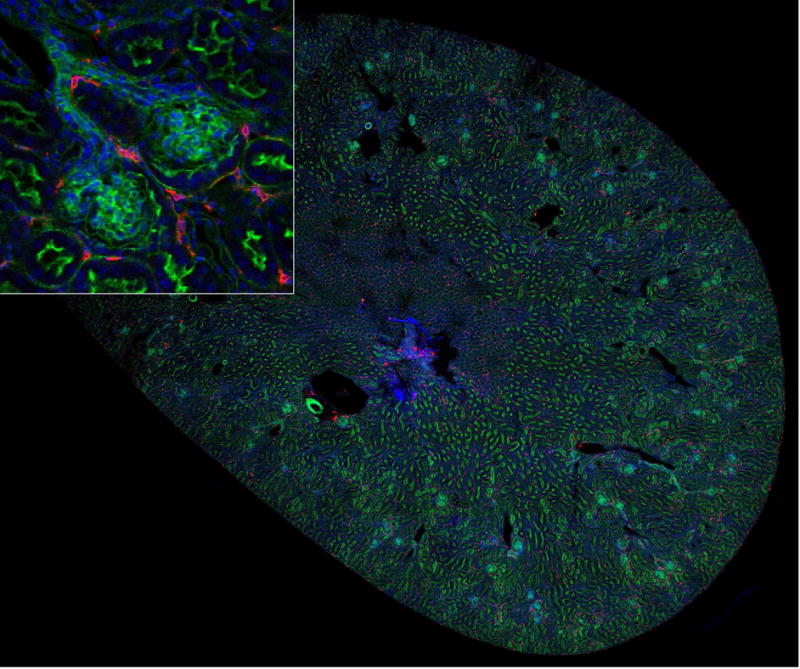
A 50 μm section of mouse kidney was cut with a vibratome and stained for nuclei (DAPI,blue), F-actin (green), and with an antibody against MHCII (red). Image volumes were collected using a 20X, NA 0.75 Leica objective on a Leica SP8 confocal microscope and mosaic merged in Fiji. Image field is 6 mm across. Inset – magnified region showing MHCII cells adjacent to two glomeruli.
Figure 2. Tissue clearing and imaging of mesoscale 3D specimen generates complex, high resolution volumes.
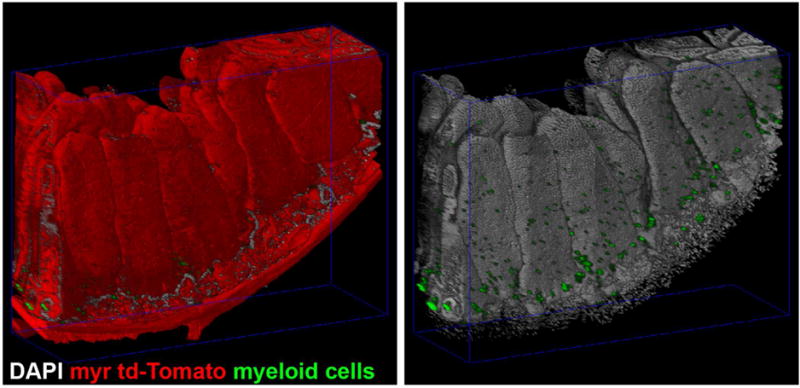
Intestine from FLT3Cre+; ROSA26mTmG/mTmG mice expressing myristoylated-td-Tomato(myr td-Tomato, red) and GFP in myeloid cells(myeloid cells, green) were dissected, stained for nuclei(DAPI, gray) and cleared with Triton X-100 in phosphate buffered saline. Multiple overlapping volumes were imaged by confocal laser scanning microscopy and mosaic merged. At left, DNA and GFP are shown to demonstrate the number and complexity of the enterocytes and intercalated myeloid cells.
There are at least 18 unique approaches to clearing tissue (as reviewed in Richardson and Lichtman)[13]. These approaches have been developed for a variety of purposes including but not limited to clearing specific organs (i.e. brain), to stabilize membranes and lipid dyes or fluorescent proteins, or to be compatible with and extend existing staining protocols (i.e. immunofluorescence and repeated rounds of immunofluorescence). Taking into consideration clinical restrictions as described above, we can focus on protocols with relatively short durations (within days vs. weeks), that support immunostaining. This leaves ten of the eighteen techniques reviewed: iDISCO, BABB, Sucrose, FocusClear, ClearT2, CUBIC, Clarity, PARS, SWITCH and ACT-PRESTO[13]. Papers using BABB, Sucrose, FocusClear, ClearT2, CUBIC and iDISCO all demonstrate immunostaining. iDISCO is unique in that it was specifically developed to support rapid immunostaining and clearing[18]. Pushing the state-of-the-art, SWITCH is especially exciting as 20 successive rounds of immunostaining were demonstrated on large volumes[19]. This is ideal for extracting the maximum amount of information from the limited amount of tissue provided by human biopsies. While it is possible that other protocols based upon hydrogel embedding are also compatible with multiple rounds of immunostaining (e.g, Clarity, PARS, and ACT-PRESTO[20–23]) adoption of these techniques is complicated by their reliance on specialized equipment and procedures for electrophoretic tissue clearing.
While it is difficult to recommend a particular protocol, perhaps the best criterion is the degree to which a protocol has been adopted for research studies. A non-rigorous survey of the primary literature in PubMed demonstrates that the iDISCO and Clarity approaches are, to date, amongst the most widely applied when considering the number of years since publication. The choice of technique will also depend upon the outcome of systematic validations in which results obtained from mesoscale analysis of cleared tissues are compared with those obtained from conventional pathology approaches. These validations are critical as there is no guarantee that tissue or organ structure will not be altered by clearing, as tissue shrinkage[16] and tissue expansion and/or protein loss[20] have been observed. With clinical samples tissue clearing may be further complicated by the impact of different pathophysiology on a protocol’s efficacy. It seems reasonable to assume that the best protocol will need to be empirically validated for different types of clinical samples[24].
Staining and multispectral imaging
The ability to clear tissue is only useful if there are complimentary methods for labeling tissue structures of interest-including cellular and molecular markers. Fortunately, there are well-established techniques for staining of tissue and thin sections with dyes and by immunofluorescence[25, 26]. The accessibility of antibodies to structures buried in the specimen is one of the major challenge in labeling thicker tissues. To circumvent this problem, small molecules can be used to label structures. For instance, there are fluorescent toxins, nucleic acid binding dyes and sugar binding lectins for labeling cell-surface proteins. Some examples include, fluorescent-taxol for microtubules, fluorescent phallotoxins for filamentous actin (f-actin), DAPI ((4′,6-diamidino-2-phenylindole) which bind DNA and lectins for labeling sugar modified surface proteins. Vector Labs supplies kits that include a wide variety of fluorescently conjugated lectins that bind a variety sugar chains and can provide labeling of tissue structures. These small molecules have the advantage of size when staining large specimens. A weakness of using some small molecule dyes are their dependence on specific structures that may be poorly preserved during fixation or damaged by tissue clearing. For instance, in our hands f-actin has proven difficult to stain with some aqueous tissue clearing protocols. As fluorescence-labeled phallotoxin binds between actin monomers in f-actin, we hypothesize that f-actin is labile in the protocols we’ve tried[27].
Immunofluorescence staining allows for a greater variety of specific and targeted labeling. Antibodies may bind to protein conformations or “discontinuous epitopes” or short peptide sequences[28]. Linear peptide antibodies may be less sensitive to tissue preparations that include denaturation of higher order protein structure[29, 30]. Unfortunately, due to their size, antibodies may have limited penetrance into tissue. Antibody protease digestions products, Fab and Fab2 fragment, may partially alleviate this size restriction. More exciting, great potential exists in the application of naturally small antibodies or nanobodies[31]. Furthermore, mechanical and electrophoretic methods to increase the penetration and staining of the antibodies are being explored[19, 22, 32].
With standard multi-channel imaging on a laser scanning confocal 3–4 spectrally separable dyes can be imaged. Starting with Alexa dyes (ThermoFisher), there has been an ever-growing repertoire of high quality fluorophores across the visible light spectrum with excitations tuned to the major laser excitation wavelengths of confocal microscopes. Further, distinct emission wavelengths have enabled off-the-shelf four channel imaging on commercially available microscopes. With linear unmixing multiple fluorophores can be combined with overlapping emission in an imaging experiment. This allows for the imaging up to eight markers in a sample (Figure 3 and 5). Linear unmixing separates the contribution of single fluorophores in a multiple fluorophore labeled sample using a reference spectra(Figure 3, left panel)[33]. An exciting possibility is combining both spectral unmixing and a fluorophore’s fluorescent lifetime. Lifetime is a characteristic time that it takes a fluorophore to emit a photon after excitation. If lifetimes are distinct, this information could be used to separate spectrally identical species, further increasing the number of molecular targets that could be imaged[34]. The use of multiphoton fluorescence lifetime imaging microscopy to image intrinsic fluorophores and metabolic state is also an interesting prospect[35–37]. For instance, imaging intrinsic fluorophores with fluorescence lifetime imaging microscopy could provide a method for rapid assessment of clinical samples[38].
Figure 3. Spectral unmixing of seven labels in a human kidney biopsy volume.
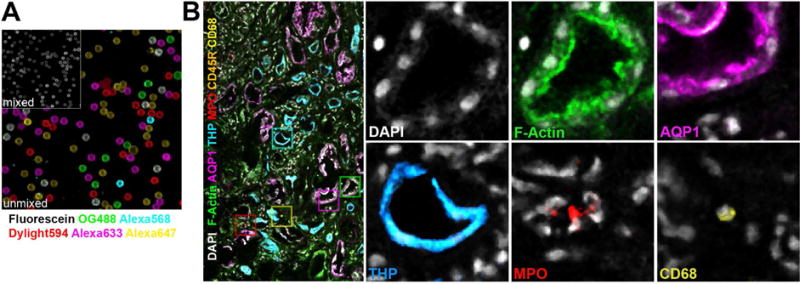
Beads and the biopsy were labeled with seven fluorophores. Multiple overlapping confocal volumes of the human biopsy were collected and mosaic merged. Spectra from the independently labeled beads were used to spectrally unmix the seven fluorophores with LASX(Leica). (A) Secondary antibodies independently absorbed to latex beads were combined and then spectrally unmixed with reference spectra. Reference spectra were collected from individually labeled beads. (B) Human biopsies were stained for nuclei (DAPI, gray), F-actin (phalloidin-Oregon Green 488, green) and species specific secondary antibodies labeled with Dylight594, Alexa647, Fluorescein, Alexa633 and Alexa568 were used to label primary antibodies against myeloperoxidase (MPO, red), macrophages (CD68, yellow), B-cells (CD45R, orange), Aquaporin-1(AQP1, magenta) and Tamm-Horsfall Protein (THP, cyan).
Figure 5. Six channel 3D mesoscale imaging and analysis of a human nephrectomy section by tissue cytometry.
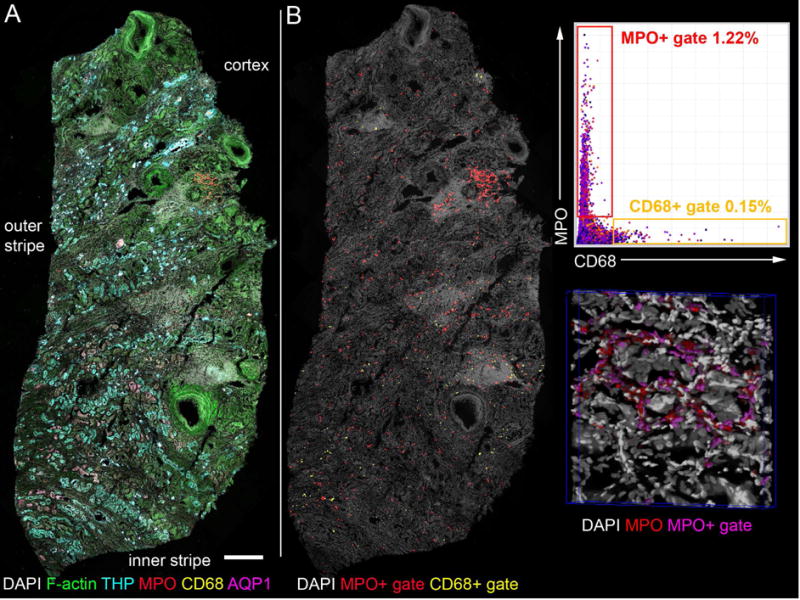
(A) Human nephrectomy was stained for nuclei (DAPI, gray), F-actin (phalloidin-Oregon Green 488, green), myeloperoxidase (MPO, red), macrophages (CD68, yellow), Aquaporin-1(AQP1, magenta) and Tamm-Horsfall Protein (THP, cyan) and with species specific secondary antibodies labeled with Dylight594, Alexa647, Fluorescein, Alexa633 and Alexa568 respectively. Multiple overlapping volumes were imaged by confocal laser scanning microscopy, spectrally unmixed and mosaic merged. Spectral unmixing was performed with single fluorophore labeled beads as references. (B) Merged and unmixed image was analyzed by VTEA and CD68 and MPO positive cells were quantified. At right, gates were drawn on scatter plot in VTEA to identify CD68+ and MPO+ cells (yellow and red gates respectively). Left, these cells were mapped to the original volume, a maximum projection is shown (as indicated in yellow, CD68+ and red, MPO+). At lower right, a portion of the volume is rendered by Voxx in 3D with nuclei (gray), myeloperoxidase (MPO, red) and gated nuclei/cells (magenta). Scale bar = 500 μm. This image was used with permission from the Journal of the American Society of Nephrology (Cover July 2017).
The degree to which cellular and molecular markers are present in specimens is going to be affected by both tissue preparation and underlying pathologies. As tissue clearing can impact the availability of markers, staining with new reagents and new tissues will need to be validated experimentally. Tissue preparation and staining is thus a careful balance between the homogenization of the RIs in a specimen while retaining the salient markers.
Imaging Platforms
One impetus driving research into tissue clarification has been the concomitant flourishing of optical sectioning microscopes that support three-dimensional imaging. Optical sectioning microscopes minimize out-of-plane fluorescence thereby increasing the contrast of the in-focus image[25, 39]. Optical sectioning microscopes have the advantage over serial sectioning approaches as the tissue is not destroyed in the process[40, 41]. This allows the sample to be restained or imaged by complementary approaches such as micro-computed tomography. The confocal microscope is the workhorse of mesoscale high-resolution imaging and is commonly used to image large specimens[20–22, 42, 43]. Confocal microscopes, which are decades old in design, have become highly automated and easy to use and capable of scanning millimeter scale samples labeled with multiple fluorophores. Further, spectral imaging is easily implemented and available on most commercial confocal microscopes.
An alternative approach to confocal microscopy is to use an optical sectioning microscope that only excites the imaging focal plane. There are two approaches that accomplish this, 1) multiphoton excitation microscopy and 2) the many variations of light-sheet microscopy (LSM). These modalities have advantages in 3D mesoscale imaging but they also have unique complications. Multiphoton fluorescence microscopy depends upon the simultaneous absorption of two photons, which occurs only at the focus of the objective lens(Reviewed in [44]). Primarily used to image deep into scattering biological tissues, multiphoton microscopy has also been used to image large cleared tissue at depth[14, 18, 45, 46]. Light sheet microscopy, like confocal microscopy, is a technology that was developed decades ago[47–49] (Reviewed in [50]). In its simplest form, LSM is based upon stimulation of fluorescence by a thin sheet of light, which is imaged by a camera placed orthogonal to the light sheet. LSM’s major advantages are 1) its adaptability in imaging large specimens and 2) its speed (Reviewed in Power and Huisken, 2017)[51]. Recently, it has seen a resurgence in imaging naturally clear embryos [52, 53]. Over the past two and half decade’s light sheet microscopy has also been applied to imaging other large specimens such as cleared tissue and organs[15, 16, 18, 21–23, 43, 48, 54].
Confocal microscopes are available from the four major manufacturers of microscopes, including Zeiss, Leica, Nikon and Olympus. All can be configured for multiphoton excitation at additional cost of one to two hundred thousand dollars in laser and beam routing optics. It should be noted that light sheet microscopes are also commercially available from three manufacturers, Leica, Zeiss and LaVisionBiotec. Of these, LaVisionBiotec’s, Ultramicroscope has been use in tissue clearing publications[15, 18, 55, 56].
For the purposes discussed here, confocal microscopy has significant advantages over multiphoton and light sheet microscopy. First, multiphoton and LSM are typically limited to no more than 4 or 5 different fluorophores, recent designs of confocal microscopes support imaging upwards of eight different probes. Second, confocal microscopy is also not limited to specific research groups with expertise in building and running microscopes, thanks to the abundance of commercial easy to use platforms. For instance, after a few hours of training a non-expert researcher can be imaging on these platforms.
The combination of tissue clearing, fluorescence labeling of multiple markers (more than four or five) and imaging of large tissues and organs by confocal microscopy forms the pillars of a holistic approach to the analysis of millimeter scale specimens (Figure 2 and 5). We can reasonably imagine applying these techniques to human samples and investigate key cellular players and structural changes in cancer biopsies, follow the progress of transplant rejection and examine and characterize tissue pathophysiology all in mesoscale 3D volumes. Further, with appropriate vetting and validation, models of pathophysiology could be developed which may prove useful in a clinical setting. Thus, the software tool(s) that enable researchers to develop such models could become central to defining the translation potential of this combination.
Exploring and analyzing mesoscale 3D volumes
In general, any approach to analyzing mesoscale 3D volumes needs to be 1) robust and able to support large and complex datasets (Figure 2 and 5), 2) provide an intuitive bidirectional workflow integrating image processing to analysis with 3D visualization to facilitate discovery and 3) provide simple extensibility for testing and use of new methods of image processing and analysis (Figure 4B). Currently tools available for analyzing mesoscale datasets fall into two broad categories, commercial and open source (Figure 4A). Unfortunately, the number of tools that are specifically tailored to analyzing mesoscale 3D datasets is limited. In addition, the ability to perform meaningful analysis on large datasets frequently requires the combination of commercial and custom software whose cost and/or sophistication is beyond the reach of many biomedical researchers. Fortunately, there are many active efforts to improve image analysis approaches in 3D, including solutions for visualization, segmentation and analysis[57–62]. As we discuss below, our own efforts have focused on combining efficient exploration and quantitative analysis of mesoscale 3D image datasets into a single platform that will provide wide accessibility of this powerful approach to the research community.
Figure 4. Available tools with integrated 3D tissue cytometry workflow.
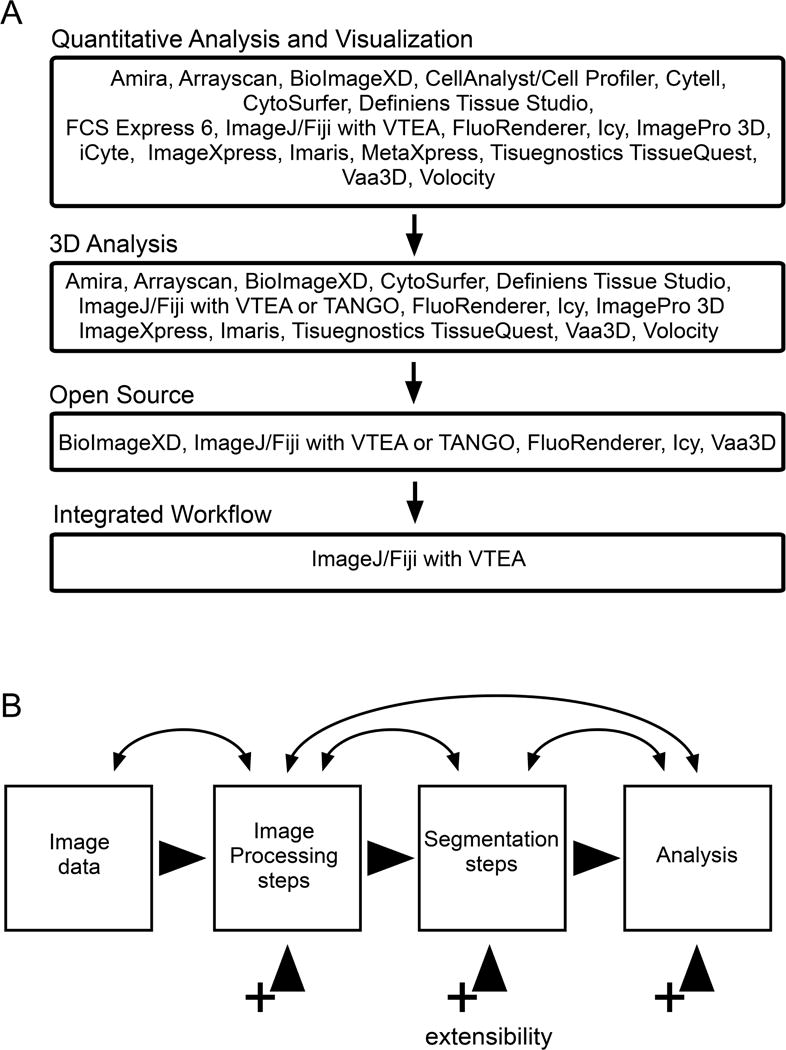
(A) VTEA provides the most comprehensive and easy-to-use open source tool for tissue cytometry. (B) VTEA was designed to support extensibility and a bidirectional workflow. A bidirectional workflow supports and encourages an iterative refinement of image processing and segmentation to facilitate accurate quantitative analyses. A common interface and workflow for tissue cytometry will enable reproducible user interaction and results. Extensibility is available or planned for the three major components of VTEA: image processing, segmentation and analysis (visualization). This will allow for the incorporation of new approaches in the developing field of image processing and segmentation and to address new challenges.
Existing commercial software capable of 3D analysis are targeted towards research and/or image based high content screening systems for drug discovery. No software or platforms has been designed specifically for mesoscale 3D image analysis. Research focused commercial packages include Amira(ThermoFisher), MetaExpress/Metamorph (Molecular Devices), Imaris (Bitplane/Andor) and Volocity (Perkin-Elmer) are stalwarts in image processing and 3D visualization and analysis. These packages provide researchers with solutions for visualization and segmentation out-of-the-box. Further, Imaris includes methods to facilitate the use of sophisticated image analysis tools via ImageJ/FIJI and MATLAB[59]. Although previously focused on research applications Perkin Elmer has extended Volocity to support its portfolio of imaging high content screening (IHCS) systems. There are number of additional companies that are making IHCS systems including GE Lifesciences and Tissuegnostics. Tissuegnostic’s TissueQuest software was the first to provide researchers with a tool that provided a flow cytometry like interface to analyze single cell phenotypes from 2D image data[63, 64]. Recently, both GE Lifesciences and TissueGnostics have brought to market confocal based IHCS systems to support 3D imaging. Whether these novel systems will play a role in large-scale or mesoscale 3D imaging is yet to be seen. Lastly, Definiens’ TissueStudio, ImageMiner and XD fill a unique role in the image processing software landscape. Their software integrates segmentation tools, with a well-developed machine learning engine, “Cognition Network Technology” and the capacity for customization by the end user. This customization will prove powerful. For instance, being able to extend segmentation algorithms will make meeting the challenges of segmenting different nuclei possible (Figure 2 vs. Figure 5).
The alternative to commercial software is the active community of open source and freeware software projects centered around ImageJ/FIJI and Icy [59, 65]. The absence of a financial barrier to use the software can make them easier for developers and user to adopt. Open source and freeware software can also facilitate the adoption of new techniques in a field, through extensibility, without the restrictions imposed by commercial solutions, including closed software or limiting license agreements. A disadvantage of open source and freeware software is that on-going support of plugins and tools may be difficult with changing developers and interests. FIJI and Icy are both general purpose image processing applications that can be easily extended and improved by the community through plugins. There are a handful of plugins that facilitate 3D image analysis (Figure 4A)[60, 66–68]. Beyond FIJI and Icy there are additional open source and/or freeware for 3D rendering including Vaa3D [57, 61, 62, 69]. Vaa3D has also implemented a plugin architecture for image processing and analysis. Although Vaa3D, FIJI and Icy may provide disparate tools required in image processing and some analysis tools, none of the software provides an easy to use bidirectional interface that integrates image processing, segmentation and exploration.
We recently developed volumetric tissue exploration and analysis (VTEA) software in response to the need for an integrated and powerful, yet user-friendly, solution to analyze large 3D image volumes and mesoscale datasets of tissue[10]. VTEA was designed around an intuitive interface with a bi-directional workflow to facilitate discovery and provide a framework for extensibility and reproducibility. The unique value of VTEA rests on the following features: 1) it requires mastery of a single software system, 2) the components are organized into an intuitive image analysis workflow accessible to researchers without expertise in image analysis, 3) the various components of the image and data analysis workflow are dynamically interlinked in a bi-directional workflow and 4) has been designed and will include extensible image processing, segmentation, visualization and data interrogation components. VTEA is available for download via the FIJI updater as described at, https://imagej.net/VTEA.
The uniqueness of VTEA is laid-out in Figure 4A. Software packages that can analyze 3D data are a mix of open source projects and commercial software. Although some commercial software provides mechanisms for customization and addition of functionality-notably Imaris and Tissue Studio, they are expensive. If we exclude these packages and focus on open source the remaining software include several general image analysis packages (Icy and FIJI) that may be full featured but require significant expertise to use[59, 65] or visualization software which may include some or limited image processing and object segmentation tools (FluoRender, BioImageXD and Vaa3D)[57, 61, 69]. The two remaining tools, VTEA and TANGO a FIJI plugin for analyzing nuclear organization, both integrate multiple aspects of image processing, segmentation and visualization into a workflow. As such the user is not required to keep track of analyses, the processing, segmentation and objects and their associated measurements-the plugins do the bookkeeping. Importantly, the various aspects of either tool are extensible to accommodate new techniques and approaches. How VTEA differentiates itself from TANGO is the inclusion of quantitative visualization and analysis tools currently in the form of dynamically linked scatter plots and images. This is in part due to the underlying intended utility of the two tools. TANGO was developed to aid in identifying and characterizing nuclear morphology[67] and VTEA was developed to aid in identifying, characterizing hundreds of thousands of cells and enable 3D tissue cytometry and exploratory analysis[10].
Application of Large-scale quantitative 3D tissue imaging and analysis in research settings
The ability to interrogate intact tissues by performing quantitative surveys of specific cells, based on a surface marker label, is an immediate application of this technology. This process, referred to as 3D tissue cytometry (3DTC) is the equivalent of performing flow cytometry to intact tissues, except that the spatial context is preserved, and no bias is introduced through tissue digestion and homogenization.
Examples of such an approach applied on a limited-scale were demonstrated in the works of Gerner et al. [70] or Moreau et al. [71], who used multiple software packages in a complex analytical pipeline to distinguish and localize several immune cell populations in mouse tissue. Liarski and colleagues used a similar approach to measure distances between lymphocytes in human kidney biopsies [72]. Recently, we showed that the streamlined approach of VTEA can be easily used to characterize various resident immune cell populations in the mouse kidney [10]. Furthermore, we showed that VTEA analysis can be easily implemented on large-scale volumes, to quantitate immune cell distribution in entire thick sections of human kidney core biopsies. This approach is again demonstrated in Figure 5, where immune cell distribution using VTEA is shown in a nephrectomy thick section, imaged using a large-scale confocal microscopy approach. Because of the 3D nature of the datasets analyzed by VTEA, both the proportion of cells and their spatial density can be measured. This approach is implemented using a region-of-interest based analysis, where VTEA determines the density of specific immune cells in relation to specific structures. In an example from a human kidney biopsy (Figure 6), VTEA can distinguish a difference in the density of neutrophils around proximal tubules, based on a spatial parameter specific to these tubules. We also used this approach to measure dendritic cell clustering around glomeruli of mouse kidney [10]. Because measurements are normalized to the imaged volume, density-based measurements will extend the usefulness of VTEA by allowing comparisons not only within a specimen, and but also between different conditions or treatments. Through the extensibility feature in VTEA, the implementation of more complex, machine learning-based algorithms (such as cell clustering and shape recognition) will further enhance the automation of analysis. Furthermore, these algorithms will yield rich, “Big-data” style outputs for the hundreds of thousands of cells it already can classify.
Figure 6. Quantifying the spatial density of infiltrating neutrophils.
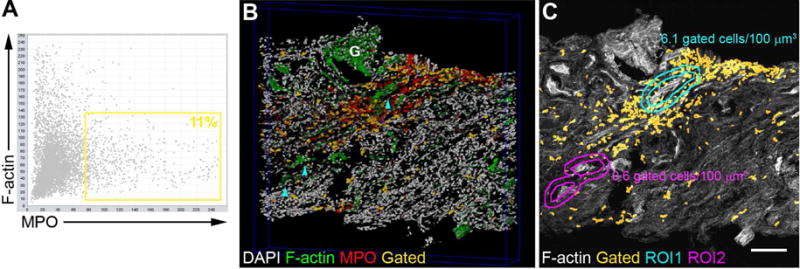
3D imaging and analysis of a biopsy stained for nuclei (DAPI, gray), F-actin (phalloidin,green) and myeloperoxidase (MPO, red). (A) Scatter plot showing all cells with their associated F-actin and MPO staining. A gate for MPO positive cells is indicated by the yellow rectangle. (B) Representative 3D rendering with image data for DAPI, F-actin, MPO and an overlay (yellow) mapping the nuclei of MPO+ cells gated in A directly on the image volume. Proximal tubules (cyan arrowheads) and a glomerulus (G) are indicated. (C) Region-of-interest (ROI) interrogation of the image volume determines neutrophil density around different tubule segments. In this example, there was ten times more neutrophils around PT contiguous to a glomerulus (<100 μm distance) vs. non-contiguous PT (ROI1 vs ROI2, respectively). Scale bar = 100 μm.
It is also important to emphasize that, because of the advances in intracellular labeling techniques, and the increasing repertoire of antibodies that can detect the activation of specific pathways, it is now possible to perform 3D quantitative analysis to determine specific pathway activation and signaling in a tissue. For example, through the specific detection of phosphorylated form of cJUN, VTEA can determine the activation of c-Jun-N-terminal kinase (JNK) signaling in a specific cell type (Figure 7). The functional quantitative data extracted from a tissue, complements the cytometry analysis and provides a molecular context to studying tissue from a specific disease. We can envision the power and complexity of the data obtained from a tissue simultaneously probed for various cell and pathway activation markers, which will allow a precise determination of the cell type(s) activated in a disease process.
Figure 7. Kidney injury induced signaling quantified in 3D by tissue cytometry.
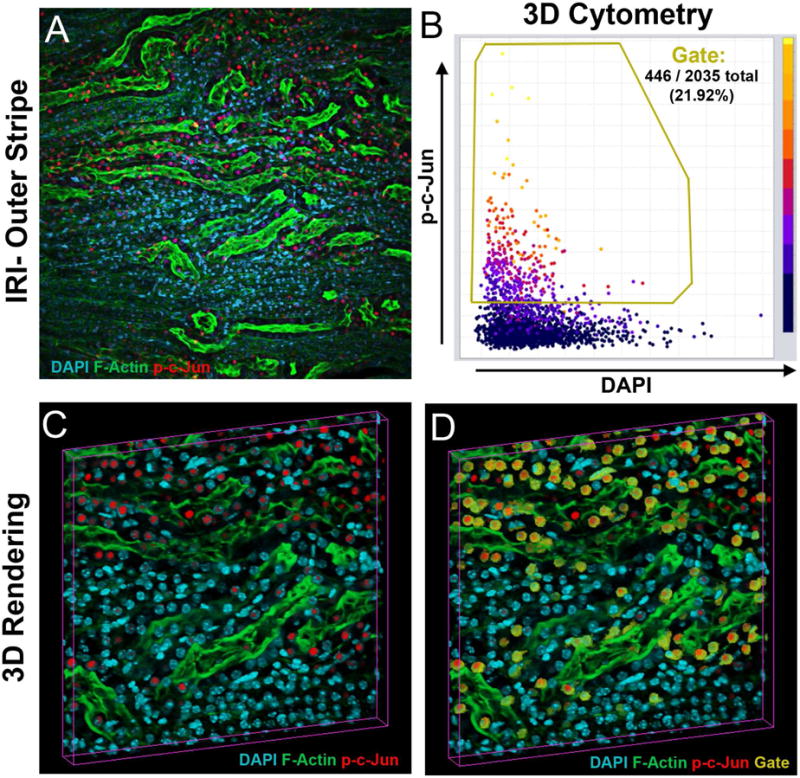
(A)-Z-projection of a 50μm-thick image volume from the kidney of a mouse 6 hrs after ischemia-reperfusion injury, stained for DAPI (cyan), F-Actin (green) and p-c-Jun (red). (B) VTEA scatterplot of nucleated cells, with a gate (in yellow) for cells with nuclear (activated) c-Jun. (C) Volume rendering of panel A. (D) Volume rendering with gated cells as given in B, indicated in yellow.
Translational implications
Although large-scale 3D quantitative imaging of tissue is not yet used in clinical applications, this approach is being implemented in various venues of translational and clinical research. We are using this approach to aid on-going clinical research by our group in the fields of kidney disease, urology and endocrinology. We expect our approaches will be more widely applicable. Essential to the adoption of this approach are the availability of banked human tissue samples, and the development of a corresponding longitudinal clinical database. In diabetes research, we are implementing large-scale 3D quantitative imaging on archived kidney biopsy tissue to understand the immune cell signatures of patients who progress to end stage renal disease. We are using a similar approach to understand the characteristics of human acute kidney injury, and identify patients who are at risk of progression to chronic kidney disease. Another example of application is in the field of human urolithiasis. Determining the different signatures of inflammatory cells in human kidney papillary biopsies for various type of kidney stones will unravel key pathways to understand the pathogenesis, and potentially offer a personalized approach for prevention and treatment of this heterogeneous disease. We envision a similar approach in characterizing tissue from other organs and diseases. For example, a better quantitative approach for tumor markers within a biopsy can better characterize various types of cancer, and complement gene expression studies [6] to characterize prognosis and responsiveness to therapy.
Concluding Remarks
We are witnessing the dawn of a new era in tissue interrogation, through implementing important scientific advancements in the field, such as novel clearing techniques, multiple markers to identify subtypes of cells, large scale 3D imaging, and the use of innovative software, such as VTEA, for investigating and exploring mesoscale 3D imaging data. This synergistic approach will likely enhance our understanding of disease pathophysiology across organs, and facilitate the move to clinical application. In clinical studies, large-scale 3D quantitative imaging could be adapted to query human tissue biopsies at the molecular level, thereby providing a tool that could empower personalized medicine, especially when combined with genomics and transcriptomic data. Undoubtedly, these efforts need to be expanded. Furthermore, the application of novel 3D analytic methodologies to cellular and molecular pathology is still an evolving field. Nevertheless, it is very likely that the observed progress in studying tissue specimens will adapt advancements in the fields of image processing and machine learning to unlock the third dimension of mesoscale 3D imaging.
Acknowledgments
Funding agencies:
NIH/NIDDK DK076169 Diacomp (TME and PCD)
VA Merit Award, NIH-NIDDK Program Project Grant (P01DK056788) TME.
Indiana Clinical and Translational Sciences Institute, funded in part by grant #UL1 TR001108 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award.
National Institutes of Health O’Brien Center for Advanced Renal Microscopic Analysis (NIH-NIDDK P30 DK079312).
Microscopy studies were conducted at the Indiana Center for Biological Microscopy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
All data presented in this manuscript conform to the ethical guidelines for human and animal research and has been approved by the Indiana University Institutional Review Board and the Institutional Animal Care and Use Committee.
Conflict of Interest statement: The authors declare no conflict of interests, and all authors have read the journal’s policy on disclosure of potential conflicts of interest.
The authors have read the journal’s authorship agreement; the manuscript has been reviewed and approved by all authors.
References
- 1.Furness PN, Philpott CM, Chorbadjian MT, Nicholson ML, Bosmans JL, Corthouts BL, et al. Protocol biopsy of the stable renal transplant: a multicenter study of methods and complication rates. Transplantation. 2003;76:969–73. doi: 10.1097/01.TP.0000082542.99416.11. [DOI] [PubMed] [Google Scholar]
- 2.Hiemann NE, Wellnhofer E, Knosalla C, Lehmkuhl HB, Stein J, Hetzer R, et al. Prognostic impact of microvasculopathy on survival after heart transplantation: evidence from 9713 endomyocardial biopsies. Circulation. 2007;116:1274–82. doi: 10.1161/CIRCULATIONAHA.106.647149. [DOI] [PubMed] [Google Scholar]
- 3.Bartlett JM, Brookes CL, Robson T, van de Velde CJ, Billingham LJ, Campbell FM, et al. Estrogen receptor and progesterone receptor as predictive biomarkers of response to endocrine therapy: a prospectively powered pathology study in the Tamoxifen and Exemestane Adjuvant Multinational trial. J Clin Oncol. 2011;29:1531–8. doi: 10.1200/JCO.2010.30.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17:1474–81. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- 5.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 6.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El-Achkar TM, McCracken R, Liu Y, Heitmeier MR, Bourgeois S, Ryerse J, et al. Tamm-Horsfall protein translocates to the basolateral domain of thick ascending limbs, interstitium, and circulation during recovery from acute kidney injury. American journal of physiology Renal physiology. 2013;304:F1066–75. doi: 10.1152/ajprenal.00543.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Achkar TM, McCracken R, Rauchman M, Heitmeier MR, Al-Aly Z, Dagher PC, et al. Tamm-Horsfall protein-deficient thick ascending limbs promote injury to neighboring S3 segments in an MIP-2-dependent mechanism. American journal of physiology Renal physiology. 2011;300:F999–1007. doi: 10.1152/ajprenal.00621.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Achkar TM, Wu XR, Rauchman M, McCracken R, Kiefer S, Dagher PC. Tamm-Horsfall protein protects the kidney from ischemic injury by decreasing inflammation and altering TLR4 expression. American journal of physiology Renal physiology. 2008;295:F534–44. doi: 10.1152/ajprenal.00083.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winfree S, Khan S, Micanovic R, Eadon MT, Kelly KJ, Sutton TA, et al. Quantitative Three-Dimensional Tissue Cytometry to Study Kidney Tissue and Resident Immune Cells. J Am Soc Nephrol. 2017 doi: 10.1681/ASN.2016091027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olson E, Levene MJ, Torres R. Multiphoton microscopy with clearing for three dimensional histology of kidney biopsies. Biomed Opt Express. 2016;7:3089–96. doi: 10.1364/BOE.7.003089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haust MD. Tetrahydrofuran (THF) for routine dehydration, clearing, and infiltration. Am J Clin Pathol. 1959;31:357–61. doi: 10.1093/ajcp/31.4_ts.357. [DOI] [PubMed] [Google Scholar]
- 13.Richardson DS, Lichtman JW. Clarifying Tissue Clearing. Cell. 2015;162:246–57. doi: 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ke MT, Fujimoto S, Imai T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci. 2013;16:1154–61. doi: 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
- 15.Susaki EA, Tainaka K, Perrin D, Yukinaga H, Kuno A, Ueda HR. Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat Protoc. 2015;10:1709–27. doi: 10.1038/nprot.2015.085. [DOI] [PubMed] [Google Scholar]
- 16.Hama H, Hioki H, Namiki K, Hoshida T, Kurokawa H, Ishidate F, et al. ScaleS: an optical clearing palette for biological imaging. Nat Neurosci. 2015;18:1518–29. doi: 10.1038/nn.4107. [DOI] [PubMed] [Google Scholar]
- 17.Erturk A, Bradke F. High-resolution imaging of entire organs by 3-dimensional imaging of solvent cleared organs (3DISCO) Exp Neurol. 2013;242:57–64. doi: 10.1016/j.expneurol.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 18.Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Murray E, Cho JH, Goodwin D, Ku T, Swaney J, Kim SY, et al. Simple, Scalable Proteomic Imaging for High-Dimensional Profiling of Intact Systems. Cell. 2015;163:1500–14. doi: 10.1016/j.cell.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–7. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen CK, Lubeck E, et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 2014;158:945–58. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee E, Choi J, Jo Y, Kim JY, Jang YJ, Lee HM, et al. ACT-PRESTO: Rapid and consistent tissue clearing and labeling method for 3-dimensional (3D) imaging. Sci Rep. 2016;6:18631. doi: 10.1038/srep18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat Protoc. 2014;9:1682–97. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolesova H, Capek M, Radochova B, Janacek J, Sedmera D. Comparison of different tissue clearing methods and 3D imaging techniques for visualization of GFP-expressing mouse embryos and embryonic hearts. Histochem Cell Biol. 2016;146:141–52. doi: 10.1007/s00418-016-1441-8. [DOI] [PubMed] [Google Scholar]
- 25.Pawley JB. Handbook of biological confocal microscopy. 3rd. New York, NY: Springer; 2006. [Google Scholar]
- 26.Bacallao R, Stelzer EH. Preservation of biological specimens for observation in a confocal fluorescence microscope and operational principles of confocal fluorescence microscopy. Methods Cell Biol. 1989;31:437–52. doi: 10.1016/s0091-679x(08)61621-0. [DOI] [PubMed] [Google Scholar]
- 27.Oda T, Namba K, Maeda Y. Position and orientation of phalloidin in F-actin determined by X-ray fiber diffraction analysis. Biophys J. 2005;88:2727–36. doi: 10.1529/biophysj.104.047753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Getzoff ED, Tainer JA, Lerner RA, Geysen HM. The chemistry and mechanism of antibody binding to protein antigens. Adv Immunol. 1988;43:1–98. doi: 10.1016/s0065-2776(08)60363-6. [DOI] [PubMed] [Google Scholar]
- 29.Brown MC, Joaquim TR, Chambers R, Onisk DV, Yin F, Moriango JM, et al. Impact of immunization technology and assay application on antibody performance–a systematic comparative evaluation. PLoS One. 2011;6:e28718. doi: 10.1371/journal.pone.0028718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forsstrom B, Axnas BB, Rockberg J, Danielsson H, Bohlin A, Uhlen M. Dissecting antibodies with regards to linear and conformational epitopes. PLoS One. 2015;10:e0121673. doi: 10.1371/journal.pone.0121673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 2013;82:775–97. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 32.Liu H, Kao WW. A novel protocol of whole mount electro-immunofluorescence staining. Mol Vis. 2009;15:505–17. [PMC free article] [PubMed] [Google Scholar]
- 33.Garini Y, Young IT, McNamara G. Spectral imaging: principles and applications. Cytometry A. 2006;69:735–47. doi: 10.1002/cyto.a.20311. [DOI] [PubMed] [Google Scholar]
- 34.Pelet S, Previte MJ, Kim D, Kim KH, Su TT, So PT. Frequency domain lifetime and spectral imaging microscopy. Microsc Res Tech. 2006;69:861–74. doi: 10.1002/jemt.20361. [DOI] [PubMed] [Google Scholar]
- 35.Winfree S, Hato T, Day RN. Intravital microscopy of biosensor activities and intrinsic metabolic states. Methods. 2017 doi: 10.1016/j.ymeth.2017.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hato T, Winfree S, Day R, Sandoval RM, Molitoris BA, Yoder MC, et al. Two-Photon Intravital Fluorescence Lifetime Imaging of the Kidney Reveals Cell-Type Specific Metabolic Signatures. J Am Soc Nephrol. 2017 doi: 10.1681/ASN.2016101153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stringari C, Cinquin A, Cinquin O, Digman MA, Donovan PJ, Gratton E. Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue. Proc Natl Acad Sci U S A. 2011;108:13582–7. doi: 10.1073/pnas.1108161108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giacomelli MG, Sheikine Y, Vardeh H, Connolly JL, Fujimoto JG. Rapid imaging of surgical breast excisions using direct temporal sampling two photon fluorescent lifetime imaging. Biomed Opt Express. 2015;6:4317–25. doi: 10.1364/BOE.6.004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyde A. Memoir on Inventing the Confocal Scanning Microscope. Scanning. 10:128–38. [Google Scholar]
- 40.Ragan T, Kadiri LR, Venkataraju KU, Bahlmann K, Sutin J, Taranda J, et al. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nature methods. 2012;9:255–8. doi: 10.1038/nmeth.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amato SP, Pan F, Schwartz J, Ragan TM. Whole Brain Imaging with Serial Two-Photon Tomography. Front Neuroanat. 2016;10:31. doi: 10.3389/fnana.2016.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee E, Sun W. ACT-PRESTO: Biological Tissue Clearing and Immunolabeling Methods for Volume Imaging. J Vis Exp. 2016 doi: 10.3791/54904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–39. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 44.Ustione A, Piston DW. A simple introduction to multiphoton microscopy. Journal of microscopy. 2011;243:221–6. doi: 10.1111/j.1365-2818.2011.03532.x. [DOI] [PubMed] [Google Scholar]
- 45.Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci. 2011;14:1481–8. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- 46.Hou B, Zhang D, Zhao S, Wei M, Yang Z, Wang S, et al. Scalable and DiI-compatible optical clearance of the mammalian brain. Front Neuroanat. 2015;9:19. doi: 10.3389/fnana.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siedentopf HZR. Über Sichtbarmachung und Groessenbestimmung ultra mikroskopischer Teilchen, mit besonderer Anwendung auf Goldrubinglaesern. Annalen der Physik. 1903;10:1–39. [Google Scholar]
- 48.Dodt HU, Leischner U, Schierloh A, Jahrling N, Mauch CP, Deininger K, et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nature methods. 2007;4:331–6. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- 49.Voie AH, Burns DH, Spelman FA. Orthogonal-plane fluorescence optical sectioning: three-dimensional imaging of macroscopic biological specimens. Journal of microscopy. 1993;170:229–36. doi: 10.1111/j.1365-2818.1993.tb03346.x. [DOI] [PubMed] [Google Scholar]
- 50.Santi PA. Light sheet fluorescence microscopy: a review. J Histochem Cytochem. 2011;59:129–38. doi: 10.1369/0022155410394857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Power RM, Huisken J. A guide to light-sheet fluorescence microscopy for multiscale imaging. Nature methods. 2017;14:360–73. doi: 10.1038/nmeth.4224. [DOI] [PubMed] [Google Scholar]
- 52.Huisken J, Stainier DY. Selective plane illumination microscopy techniques in developmental biology. Development. 2009;136:1963–75. doi: 10.1242/dev.022426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–9. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 54.Erturk A, Becker K, Jahrling N, Mauch CP, Hojer CD, Egen JG, et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc. 2012;7:1983–95. doi: 10.1038/nprot.2012.119. [DOI] [PubMed] [Google Scholar]
- 55.Epp JR, Niibori Y, Liz Hsiang HL, Mercaldo V, Deisseroth K, Josselyn SA, et al. Optimization of CLARITY for Clearing Whole-Brain and Other Intact Organs(1,2,3) eNeuro. 2015:2. doi: 10.1523/ENEURO.0022-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobosz M, Ntziachristos V, Scheuer W, Strobel S. Multispectral fluorescence ultramicroscopy: three-dimensional visualization and automatic quantification of tumor morphology, drug penetration, and antiangiogenic treatment response. Neoplasia. 2014;16:1–13. doi: 10.1593/neo.131848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peng H, Bria A, Zhou Z, Iannello G, Long F. Extensible visualization and analysis for multidimensional images using Vaa3D. Nat Protoc. 2014;9:193–208. doi: 10.1038/nprot.2014.011. [DOI] [PubMed] [Google Scholar]
- 58.Haubold C, Schiegg M, Kreshuk A, Berg S, Koethe U, Hamprecht FA. Segmenting and Tracking Multiple Dividing Targets Using ilastik. Adv Anat Embryol Cell Biol. 2016;219:199–229. doi: 10.1007/978-3-319-28549-8_8. [DOI] [PubMed] [Google Scholar]
- 59.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nature methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Royer LA, Weigert M, Gunther U, Maghelli N, Jug F, Sbalzarini IF, et al. ClearVolume: open-source live 3D visualization for light-sheet microscopy. Nature methods. 2015;12:480–1. doi: 10.1038/nmeth.3372. [DOI] [PubMed] [Google Scholar]
- 61.Wan Y, Otsuna H, Chien CB, Hansen C. FluoRender: An Application of 2D Image Space Methods for 3D and 4D Confocal Microscopy Data Visualization in Neurobiology Research. IEEE Pac Vis Symp. 2012:201–8. doi: 10.1109/pacificvis.2012.6183592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clendenon JL, Phillips CL, Sandoval RM, Fang S, Dunn KW. Voxx: a PC-based, near real-time volume rendering system for biological microscopy. Am J Physiol Cell Physiol. 2002;282:C213–8. doi: 10.1152/ajpcell.2002.282.1.C213. [DOI] [PubMed] [Google Scholar]
- 63.Ecker RC, Steiner GE. Microscopy-based multicolor tissue cytometry at the single-cell level. Cytometry A. 2004;59:182–90. doi: 10.1002/cyto.a.20052. [DOI] [PubMed] [Google Scholar]
- 64.Ecker RC, Rogojanu R, Streit M, Oesterreicher K, Steiner GE. An improved method for discrimination of cell populations in tissue sections using microscopy-based multicolor tissue cytometry. Cytometry A. 2006;69:119–23. doi: 10.1002/cyto.a.20219. [DOI] [PubMed] [Google Scholar]
- 65.de Chaumont F, Dallongeville S, Chenouard N, Herve N, Pop S, Provoost T, et al. Icy: an open bioimage informatics platform for extended reproducible research. Nature methods. 2012;9:690–6. doi: 10.1038/nmeth.2075. [DOI] [PubMed] [Google Scholar]
- 66.Ollion J, Cochennec J, Loll F, Escude C, Boudier T. Analysis of nuclear organization with TANGO, software for high-throughput quantitative analysis of 3D fluorescence microscopy images. Methods Mol Biol. 2015;1228:203–22. doi: 10.1007/978-1-4939-1680-1_16. [DOI] [PubMed] [Google Scholar]
- 67.Ollion J, Cochennec J, Loll F, Escude C, Boudier T. TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics. 2013;29:1840–1. doi: 10.1093/bioinformatics/btt276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rizk A, Paul G, Incardona P, Bugarski M, Mansouri M, Niemann A, et al. Segmentation and quantification of subcellular structures in fluorescence microscopy images using Squassh. Nat Protoc. 2014;9:586–96. doi: 10.1038/nprot.2014.037. [DOI] [PubMed] [Google Scholar]
- 69.Kankaanpaa P, Paavolainen L, Tiitta S, Karjalainen M, Paivarinne J, Nieminen J, et al. BioImageXD: an open, general-purpose and high-throughput image-processing platform. Nature methods. 2012;9:683–9. doi: 10.1038/nmeth.2047. [DOI] [PubMed] [Google Scholar]
- 70.Gerner MY, Kastenmuller W, Ifrim I, Kabat J, Germain RN. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity. 2012;37:364–76. doi: 10.1016/j.immuni.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moreau HD, Lemaitre F, Terriac E, Azar G, Piel M, Lennon-Dumenil AM, et al. Dynamic in situ cytometry uncovers T cell receptor signaling during immunological synapses and kinapses in vivo. Immunity. 2012;37:351–63. doi: 10.1016/j.immuni.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 72.Liarski VM, Kaverina N, Chang A, Brandt D, Yanez D, Talasnik L, et al. Cell distance mapping identifies functional T follicular helper cells in inflamed human renal tissue. Sci Transl Med. 2014;6:230ra46. doi: 10.1126/scitranslmed.3008146. [DOI] [PMC free article] [PubMed] [Google Scholar]


