Abstract
Although patients with primary insomnia experience sleep disruption, they are able to maintain normal performance on a variety of cognitive tasks. This observation suggests that insomnia may be a condition where predisposing factors simultaneously increase the risk for insomnia and also mitigate against the deleterious consequences of waking. To gain insight into processes that might regulate sleep and buffer neuronal circuits during sleep loss, we manipulated three genes, fat facet (faf), highwire (hiw) and the GABA receptor Resistance to dieldrin (Rdl), that were differentially modulated in a Drosophila model of insomnia. Our results indicate that increasing faf and decreasing hiw or Rdl within wake-promoting large ventral lateral clock neurons (lLNvs) induces sleep loss. As expected, sleep loss induced by decreasing hiw in the lLNvs results in deficits in short-term memory and increases of synaptic growth. However, sleep loss induced by knocking down Rdl in the lLNvs protects flies from sleep-loss induced deficits in short-term memory and increases in synaptic markers. Surprisingly, decreasing hiw and Rdl within the Mushroom Bodies (MBs) protects against the negative effects of sleep deprivation (SD) as indicated by the absence of a subsequent homeostatic response, or deficits in short-term memory. Together these results indicate that specific genes are able to disrupt sleep and protect against the negative consequences of waking in a circuit dependent manner.
Keywords: sleep, plasticity, learning, memory, homeostasis, Drosophila, ubiquitin, GABA-A receptors
Introduction
In humans, sleep disruption has debilitating consequences on daytime functioning and health (Banks and Dinges, 2007). As a consequence, insomnia has an important economic burden with both direct (medical) and indirect (e.g., absenteeism, accidents) costs (Ozminkowski et al., 2007). Unfortunately the neuronal mechanisms underlying insomnia and their relationship to cognitive deficits are poorly understood. For example, patients with primary insomnia experience sleep disruption, for 4 years or more (Chevalier et al., 1999) and while substantially impaired (Hauri, 1997; Drake et al., 2003; Buysse et al., 2007; Edinger et al., 2008) are able to maintain normal performance on a variety of tasks (Riedel and Lichstein, 2000; Orff et al., 2007). The ability to maintain normal performance is surprising given the well-established negative impact of inadequate sleep on a variety of cognitive tests (Krause et al., 2017).These observations suggest that insomnia may be a condition where predisposing factors simultaneously increase the risk for insomnia and also mitigate against the deleterious consequences of waking. Understanding how patients with insomnia can maintain cognitive abilities during sleep loss may provide novel insights into the relationship between sleep and plasticity.
Using a laboratory selection strategy, we have isolated a population of flies (insomniac-like flies referred to as ins-l flies) that share many features with human insomnia, including difficulty falling asleep, difficulty staying asleep, inadequate sleep duration and poor quality sleep as evidenced by daytime cognitive impairments (Seugnet et al., 2009c). Whole genome profiling identified a large set of genes (~1000) that were differentially regulated in ins-l flies (Seugnet et al., 2009c). Among the genes differentially expressed in ins-l flies, 30 genes are involved in synaptic transmission and constituted a significantly over-represented biological process as defined by Gene Ontology. These genes represent a possible link between synaptic activity, sleep regulation and behavioral performance. Indeed, two of these genes, highwire (hiw) an E3 ubiquitin ligase (DiAntonio and Hicke, 2004), and Resistant to dieldrin (Rdl), a GABA-A receptor (Henderson et al., 1993), enhance memory under some conditions (Liu et al., 2007; Huang et al., 2012). Given their role in both memory, and sleep (see below) the ubiquitin-proteasome system and GABA signaling represent two pathways that are uniquely situated to not only modulate sleep time but also buffer neuronal circuits during sleep loss.
The ubiquitin-proteasome system plays an important role in activity- dependent plasticity and has recently been implicated in regulating sleep (Stavropoulos and Young, 2011; Freeman et al., 2012; Pfeiffenberger and Allada, 2012; Jarome and Helmstetter, 2013). Specifically, flies mutant for an adaptor for the Cullin-3 ubiquitin ligase complex, insomniac, display dramatic reductions in total sleep time (~400 min/day). Moreover, total sleep is also reduced when either insomniac (inc) or Cullin-3 (Cul3) are knocked down using RNAi. While these data strongly implicate the ubiquitin-proteasome complex as regulators of sleep time, changes in sleep were only observed when inc and Cul-3 were knocked down pan-neuronally throughout development but did not reduce sleep when they were manipulated in adults (Pfeiffenberger and Allada, 2012). In comparison, data from human patients with insomnia indicate that distinct neuronal circuits contribute to various insomnia phenotypes in adults (Nofzinger et al., 2004). Thus, a closer evaluation of the circuits impacted by the ubiquitin-proteasome system is warranted.
GABA signaling has also been shown to modulate sleep in flies (Agosto et al., 2008; Parisky et al., 2008; Chung et al., 2009). The role of the GABA-A receptor Rdl in regulating sleep onset was first identified by demonstrating that the wake-promoting effects of the anticonvulsant, carbamazepine, were reduced in Rdl mutants (Agosto et al., 2008). Based upon the immunohistochemical localization of Rdl, genetic studies localized the sleep-regulating properties of Rdl to a subset of clock neurons, the ventral lateral neurons (LNvs; Parisky et al., 2008; Chung et al., 2009). Although subsequent studies have begun to investigate molecular mechanisms regulating the Rdl receptor in the LNvs (Li et al., 2013; Liu et al., 2014), little is known about how modulating the Rdl receptor in the LNvs, or other brain regions, impacts behavioral plasticity.
As mentioned above, insomnia patients perform well on a variety of cognitive tasks despite experiencing inadequate sleep (Riedel and Lichstein, 2000). Of the genes differentially expressed in insomnia-like flies, the only genes that both disrupt sleep and enhance memory are those involved in the ubiquitin-proteasome system (hiw) and Rdl. Although hiw and Rdl do not appear to be functionally related, human studies have identified a number of unrelated genes that are associated with insomnia phenotypes (for review see Lind and Gehrman, 2016). Moreover, whether distinct genes influence insomnia is predicted to be differentially influenced by changes in the environment (Lind and Gehrman, 2016). That is, the literature predicts that genes with independent function will likely influence insomnia phenotypes in different ways. With that in mind, we asked whether hiw and Rdl could independently influence sleep and cognitive impairments during sleep loss. Surprisingly, our results indicate that the ability of faf, hiw or Rdl to modulate sleep and protect against cognitive impairments during waking is circuit dependent. These data provide additional insight into potential mechanisms that allow patients with insomnia to maintain cognitive ability during sleep disruption.
Materials and Methods
Fly Stocks, Sleep Monitoring and Sleep Deprivation
Cs flies were obtained from the Bloomington Drosophila stock center. UAS-hiwRNAi lines were obtained from the Vienna Drosophila RNAi center (VDRC; Dietzl et al., 2007). We obtained fafEP3520, UAS-hiw∆RING (UAS-hiwDN)stocks from A. DiAntonio (Washington University in St.Louis, MO, USA), UAS-hiwRNAi from the Vienna Drosophila Resource Center. GAL4 lines were selected based upon their expression throughout the brain or in neuronal populations known to be involved in sleep and plasticity. Drivers expressing broadly throughout the brain include elav-GAL4, MJ85B-GAL4, Cha-GAL4 and TH-GAL4. The Mushroom Bodies (MBs) play a role in sleep regulation and memory; drivers that express in the MBs include 247-Gal4, 30Y-GAL4, c309-GAL4, and 201y. The central complex drivers have been implicated in plasticity and include c232-GAL4 and c205-GAL4. The pars intercerebralis drivers include 50y-GAL4, c687-GAL4, Jan191-GAL4 and Feb194-Gal4. Finally, we obtained drivers expressing the circadian clock including cry39-GAL4, cry16-GAL4, cry-Gal80 stocks from M. Rosbash (Brandeis University), PDF-GAL4 flies from P. Taghert (Washington University in St.Louis, MO, USA). UAS-Rdl RNAi stocks and other GAL4 drivers were obtained from Bloomington stock center, P. Taghert, A. DiAntonio, S. Birman (Université de la Mediterannée), R. Greenspan (Neuroscience Institute, San Diego, CA, USA), A. Sehgal (University of Pennsylvania), and R. Allada (Northwestern University). For GAL4:UAS experiments, parental lines were outcrossed to Cs flies. Flies were cultured at 25°C, 50% to 60% humidity, in 12 h:12 h Light:Dark cycle, on a standard food containing yeast, dark corn syrup, molasses, dextrose and agar. Newly eclosed female adult flies were collected from culture vials daily under CO2 anesthesia. Three day old flies were then individually placed into 65 mm glass tubes so that sleep parameters can be continuously evaluated using the Trikinetics activity monitoring system as previously described (Shaw et al., 2000)1. Flies were sleep deprived using an automated SD apparatus that has been found to produce waking without nonspecifically activating stress responses (Shaw et al., 2002). Flies were sleep deprived using the SNAP from ZT 12 (beginning of the dark phase) to ZT 0 (beginning of the light phase). Unless otherwise stated, at least 16 flies were analyzed for each experimental condition. Differences in sleep time were assessed using either a Student’s t-test or analysis of variance (ANOVA) which were followed by planned comparisons using a Modified Bonferroni correction.
Locomotor Rhythms
Flies were individually placed into 5 × 65 mm tubes with regular food and placed into constant conditions for 10 days. Locomotor activity was continuously recorded in 30 min bins using the Trikinetics system. Locomotor rhythms were analyzed for 6 or 9 days using MATLAB (Mathworks, Natick, MA, USA) based computational tools designed by Levine et al. (2002). At least 30 flies were analyzed for each genotype.
Learning
The APS performance test was performed as previously described (Le Bourg and Buecher, 2002). Flies are individually tested in a T-maze where they are allowed to choose between a dark and a lighted vial. Adult flies are phototaxic and choose the lighted alley in the absence of reinforcer. During the test a filter paper soaked with a quinine solution is placed in the lighted vial to provide an aversive association. In the course of 16 trials through the maze flies learn to make more frequent choices to the dark vial (photonegative choices). The number of photonegative choices is tabulated during four successive blocks of four trials and the performance score is the % of photonegative choices made in the last block of four trials. At least eight flies were evaluated for each condition. For each experiment, learning was evaluated by the same experimenter who was blind to genotype and condition. All flies were tested in the morning between ZT0 and ZT4. Flies were sleep deprived using the SNAP from ZT 12 (beginning of the dark phase) to ZT 0 (beginning of the light phase) and until each flies was tested for learning. Learning scores are normally distributed (Seugnet et al., 2009b). Differences between scores were assessed using either a Student’s t-test or ANOVA which were followed by planned comparisons using a Modified Bonferroni correction.
Photosensitivity
Photosensitivity was evaluated using the T-maze with no filter paper. The average proportion of choices to the lighted vial during 10 trials was calculated for each individual fly. The phototaxis index (PI) is the average of the scores obtained for at least 5 flies ± SEM.
Quinine/Humidity Sensitivity
Sensitivity to quinine/humidity was evaluated as Le Bourg and Buecher (2002) with the following modifications: each fly was individually placed in a 14 cm transparent cylindrical tube covered with filter paper, uniformly lighted and maintained horizontal. In one half of the apparatus the filter paper is soaked with quinine solution while the other half is kept dry. The quinine/humidity sensitivity index (referred to as Quinine Sensitivity Index or QSI) was determined by calculating the time in seconds that the fly spent on the dry side of the tube when the other side had been wetted with quinine, during a 5 min period.
Immunohistochemistry
Brains were removed from the head casing and fixed in 4% paraformaldehyde in phosphate buffer solution (PBS; 1.86 mM NaHPO, 8.41 mM NaHPO, and 175 mM NaCl) for 1 h and washed in PBS. Following a 2-h pre-incubation in 3% normal goat serum in PBS-TX (PBS containing 0.3% Triton X-100), brains were washed in PBS-TX. Primary antibodies were Rabbit anti-GFP (1:1000; Sigma) and mouse anti-PDF (1:50; DSHB, University of Iowa), washed in PBS-TX and incubated in the appropriate fluorescent secondary antibodies.
Confocal Microscopy
Confocal images with a 1 μm slice thickness were collected on an Olympus microscope provided by the Washington University Center for Cellular Imaging in St. Louis, MO, USA. Confocal stacks of PDF terminals were quantified as described previously (Donlea et al., 2009). Briefly, immuno-positive terminals were counted using the ImageJ binary thresholding algorithm. The number of synaptic terminals for the GAL4/+ parental control used to generate a mean. The mean of the GAL4/+ was used to normalize each individual UAS/+ and GAL4>UAS brain. The individual normalized values were then used to calculate the mean and standard error for the group. The normalized values for each group were then evaluated using a one-way ANOVA and a modified Bonferroni test.
Statistics
All comparison were done using a Student’s T-test or, if appropriate, ANOVA and subsequent modified Bonferroni comparisons unless otherwise stated. All statistically different groups are defined as p < 0.05.
Results
Circuit-Dependent Regulation of Sleep by faf and hiw
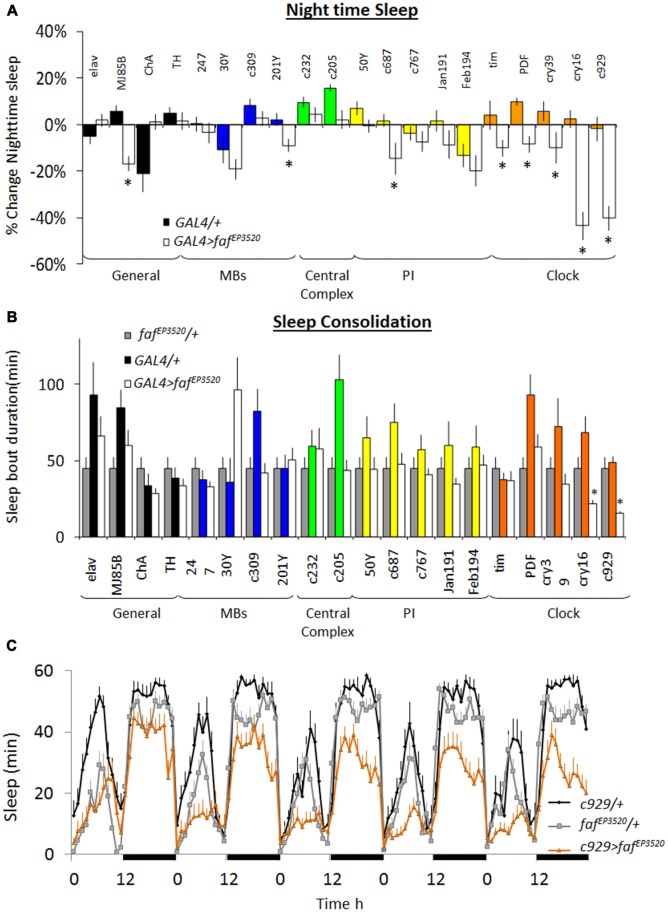
As mentioned, whole genome profiling revealed that faf was increased in ins-l flies compared to normal sleeping background controls identifying it as a potential contributor to insomnia phenotypes (Seugnet et al., 2009c). The increase of faf in ins-l flies was confirmed in an independent cohort using QPCR (data not shown). Given this expression profile, we hypothesized that over-expressing faf would recapitulate important features of the insomnia phenotype such as reduced sleep time and sleep fragmentation. The P-element line, fafEP3520, contains a UAS that can be used to induce functional faf (DiAntonio et al., 2001). Thus, we conducted a mini-brain screen to express faf in circuits known to modulate sleep and waking in flies (Joiner et al., 2006; Pitman et al., 2006; Foltenyi et al., 2007; Sheeba et al., 2008; Donlea et al., 2011). Although previous reports demonstrate that knocking-down inc and Cul-3 with the pan-neuronal elav-GAL4 driver substantially reduced sleep time (Stavropoulos and Young, 2011; Pfeiffenberger and Allada, 2012), elav-GAL4>fafEP3520 did not reduce night-time sleep compared to genetic controls (Figure 1A, left). However, we observed a ~20% reduction in total sleep time when we expressed fafEP3520 using MJ85b-GAL4, a stronger driver that expresses broadly throughout the brain (Joiner and Griffith, 1997; Dubnau et al., 2001); no changes in sleep were observed when faf was expressed in cholinergic or dopaminergic neurons (Figure 1A). Surprisingly, expressing faf in other sleep/wake centers (e.g., Mushroom Bodies (MBs), Fan Shaped body (FB), Central Complex, Pars Intercerebralis (PI)) did not consistently alter sleep parameters (Figure 1A). In contrast, all five GAL4 drivers that express in clock neurons significantly reduced night-time sleep when compared to both parental controls. Indeed, the only drivers that reduced night-time sleep and also disrupted sleep consolidation when expressing fafEP3520 were cry16-GAL4 and c929-GAL4 (Figures 1A,B). Since insomnia tends to become more prevalent with age in humans, we asked whether expressing faf in clock cells would disrupt sleep from the first day of life, similar to the results seen with inc and Cul-3, or whether the changes in sleep would develop over time in adults. In wild-type flies, sleep reaches a stable level by 3-days of age (Shaw et al., 2000). We did not observe differences in sleep in young c929-GAL4>fafEP3520 and their parental controls during their first 2 days of adult life (data not shown). Similarly, c929-GAL4/+ (black), fafEP3520/+ (gray) and c929-GAL4>fafEP3520 (orange) flies slept similarly on day 3 (Figure 1C, left). However, while sleep remained stable over days in c929-GAL4/+ and fafEP3520/+ controls, sleep progressively declined in mature c929-GAL4>fafEP3520 flies and stabilized by 7–8 days of age (Figure 1C). Thus, disrupting the ubiquitin-proteasome system in clock cells (see below) by expressing a gain-of-function allele of faf disrupts sleep in mature adults, and recapitulates two key features seen in ins-l flies.
Figure 1.
Over-expression of fat facet (faf) modulates sleep in a circuit dependent manner. (A) Change in night-time sleep following faf over-expression with GAL4 drivers that target different areas of the brain. Nighttime sleep is expressed as %change from fafEP3520/+ at day 8; (n > 14/group; *p < 0.05). (B) Average night bout duration at day 8. Gray bars: fafEP3520/+; colored bars: GAL4/+; white bars: GAL4>fafEP3520 combinations (n > 14/group; *p < 0.05). (C) Sleep is reduced in c929-GAL4/+; fafEP3520/+ flies compared to parental controls. Data is presented as minutes of sleep/h starting on day 3; black bars indicate lights-off (one of three replicates of n > 16/group).
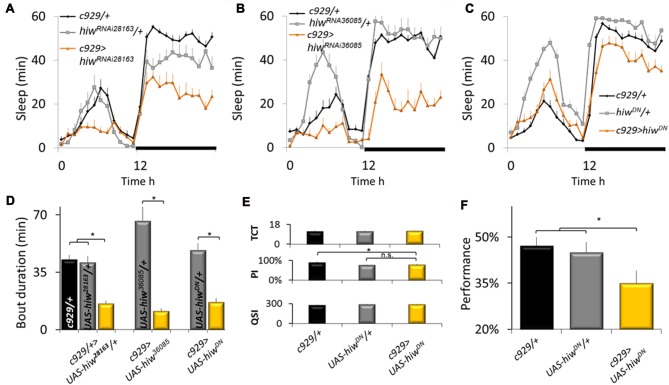
To further evaluate the role of ubiquitination pathways for sleep regulation, we obtained loss-of-function alleles for the E3 ubiquitin ligase highwire (hiw). As mentioned above, loss-of-function alleles for hiw phenocopy gain-of-function alleles for faf. As seen in Figures 2A,B knockdown of hiw in c929-GAL4 expressing neurons reduced sleep time using two independent RNAi lines that target a different portion of the gene. Reduced sleep was also observed when expressing a dominant negative form of highwire (hiwDN; Figure 2C). As with gain-of-function alleles for faf, all three loss-of-function alleles for hiw also substantially disrupted sleep consolidation at night (Figure 2D). These phenotypes were not observed when expressing wild-type hiw (UAS-hiwWT) with c929-GAL4 (data not shown).
Figure 2.
Disrupting highwire (hiw) increases waking. (A–C) Reducing hiw in c929 expressing cells using hiwRNAi28163, hiwRNA36085, or UAS-hiwDN disrupts sleep. A Genotype (3) X Time (24) repeated measures analysis of variance (ANOVA) revealed a significant Genotype × Time interaction, ANOVA F(46,11058) = 4.96; p = 9.9E-16, F(46,1196) = 5.97; p = 9.9E-16, and F(46,996) = 5.26; p = 9.9E-16, respectively. (D) Disrupting hiw reduces average sleep bout duration at night, one-way ANOVA for Genotype F(6,93) = 6.58, p = 7.97E-006; *p < 0.05 Modified Bonferroni test. (E) c929-GAL4>UAS-hiwDN flies exhibit normal values for the Time to complete Test (TCT), the Photosensitivity Index (PSI) and the Quinine Sensitivity Index (QSI) that are not statistically different from both c929-GAL4/+ and UAS-hiwDN/+ parental controls, one-way ANOVA F(2,26) = 0.22, p = 0.8, ANOVA F(2,14) = 6.3, p = 0.01, ANOVA F(2,14) = 0.73, p = 0.9, respectively; *p < 0.05 modified Bonferroni test). (F) Baseline learning is reduced in flies expressing UAS-hiwDN in c929 expressing cells one-way ANOVA F(2,26) = 3.54, p = 0.042; *p < 0.05 modified Bonferroni test).
Sleep loss impairs performance on many cognitive tasks including those that require short-term memory and response-inhibition (Frey et al., 2004; Chuah et al., 2006 Dissel and Shaw, 2013). Thus, to determine whether the reduction in sleep seen in c929-GAL4>hiwDN flies is the result of a failed ability to generate adequate sleep or the consequence of a reduction in sleep need, we evaluated performance using Aversive Phototaxic Suppression (APS) assay. The APS is an established short-term memory assay that is extremely sensitive to sleep disruption (Seugnet et al., 2008, 2009a,c, 2011a,b; Thimgan et al., 2010, 2015; Dissel et al., 2015a,b,c, 2017). In the APS, flies are individually placed in a T-maze and allowed to choose between a lighted and darkened chamber over 16 trials. Flies that do not display phototaxis during the first block of four trials are excluded from further analysis (Le Bourg and Buecher, 2002; Seugnet et al., 2009a). During 16 trials, flies learn to avoid the lighted chamber that is paired with an aversive stimulus (quinine/humidity). The performance index is calculated as the percentage of times the fly chooses the dark vial during the last four trials of the 16 trial test and short term memory (STM) is defined as selecting the dark vial on two or more occasions during Block 4. Before being tested for STM, flies are first examined to ensure that they exhibit normal photosensitivity and quinine sensitivity (Le Bourg and Buecher, 2002; Seugnet et al., 2008, 2009b). This step is important since changes to sensory thresholds could confound the ability to detect true changes in associate learning (Kahsai and Zars, 2011; Dubnau and Chiang, 2013; Dissel et al., 2015a). As seen in Figure 2E, photosensitivity and quinine sensitivity for c929-GAL4>UAS-hiwDN flies fall within the range seen in wild-type flies (Seugnet et al., 2008, 2009b) and are not statistically different from both parental controls. Importantly, c929-GAL4>UAS-hiwDN flies exhibit performance deficits while c929-GAL4/+ and UAS-hiwDN/+ parental controls display wild-type STM (Figure 2F). Thus, disrupting the ubiquitin-proteasome system in clock cells interferes with the ability of the flies to obtain needed sleep.
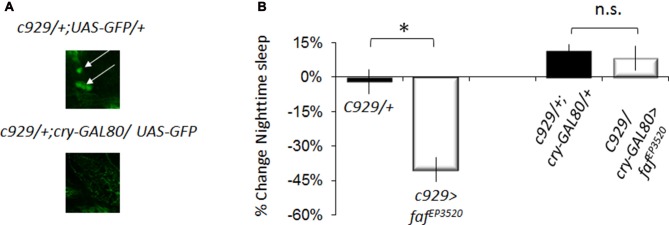
The pattern of expression for pdf-GAL4, cry16-GAL4 and c929-GAL4 all include the large ventral lateral clock neurons (lLNvs) indicating that the effects of faf and hiw are likely mediated through the large ventral lateral neurons (lLNvs; Grima et al., 2004; Stoleru et al., 2004). However, c929-GAL4 is expressed in other peptidergic neurons (Park et al., 2008). To determine whether faf over-expression in the lLNvs was responsible for the sleep reduction phenotype, we combined c929-GAL4 with cry-Gal80, which targets the GAL4 inhibitor GAL80 to all clock neurons (Stoleru et al., 2004). cry-GAL80 effectively removed GAL4 mediated induction in the lLNvs as assessed with a UAS-GFP reporter (Figure 3A bottom) and abolished the sleep reduction phenotype induced by c929-GAL4 with faf EP3520 (Figure 3B). Thus, expression of faf in the lLNvs regulates night-time sleep.
Figure 3.
Disrupting hiw in clock cells increases waking. (A) Anti-GFP immunostaining for c929-GAL4/+ ;UAS -GFP/+ and c929/+ ;UAS-GFP/cryGal80 showing expression in the pars intercerebralis (arrowhead) and the lLNv clock neurons (arrows). (B) Blocking expression of faf in lLNvs prevents the reduction in night-time sleep (%change fafEP3520/+; one of two replicates of n < 16/group shown; F(3,58) = 23.42, p = 4.67E-10, *p < 0.05).
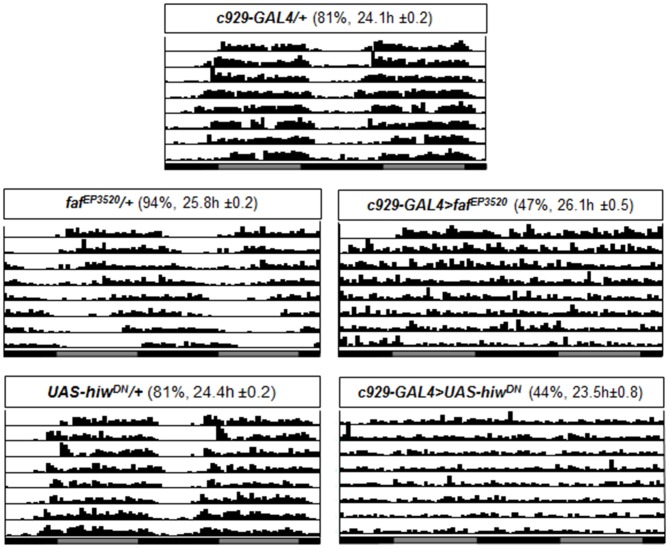
Neither inc, nor Cul3 alter free running circadian rhythms when disrupted in clock cells. To determine whether faf, and hiw would alter free running rhythms, we combined fafEP3520, and UAS-HiwDN, with c929-GAL4 and PDF-GAL4. As seen in Figure 4, >40% of c929-GAL4 >fafEP3520 and c929-GAL4 >UAS-hiwDN where arrhythmic in free running conditions (Figure 4). Thus, in contrast to inc and Cul3, expression of faf in the lLNvs disrupts circadian locomotor rhythms.
Figure 4.
Circadian rhythms are disrupted when fafEP3520 or UAS-hiwDN, are expressed using c929-GAL4. Representative single fly actograms from DD1 to DD8. Values in parenthesis (%) refer to the percentage of rhythmic flies and the period τ ± SEM. 16–32 flies were evaluated for each group.
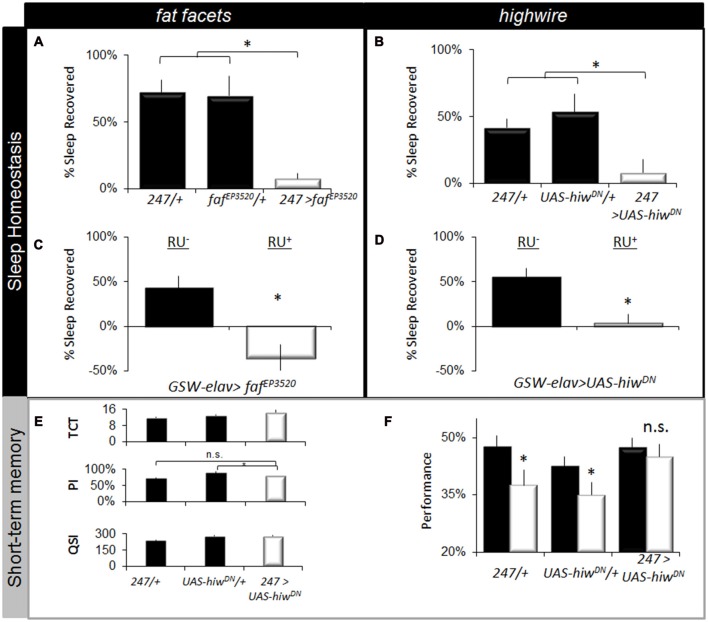
To determine whether disrupting the ubiquitin-proteasome system would alter other aspects of sleep regulation, we evaluated sleep homeostasis following 12 h of SD. Previous studies have shown that the MB are important for sleep regulation (Joiner et al., 2006; Pitman et al., 2006). Moreover, performance in the APS involves MB neurons which are particularly sensitive to sleep loss (Seugnet et al., 2008, 2009b). As seen in Figures 5A,B, expressing fafEP3520 or UAS-hiwDN in the MBs using 247-GAL4 significantly attenuated sleep rebound compared to both parental controls. Previous reports suggest that sleep phenotypes are only observed when the ubiquitin-proteasome system is disrupted during development (Pfeiffenberger and Allada, 2012). Thus, we used the inducible GeneSwitch system (Osterwalder et al., 2001) to express fafEP3520 or UAS-hiwDN in adults. As seen in Figures 5C,D, adult GSW-elav>fafEP3520 and GSW-elav>UAS-hiwDN flies fed RU486 (RU+) exhibited a significantly reduced sleep rebound compared to genetically identical, vehicle fed siblings (RU−). It should be noted that we and others have consistently reported that RU does not influence a variety of phenotypes including lifespan, sleep, sleep homeostasis, short-term memory, short-term memory following SD, olfactory conditioning, phototaxis, geotaxis, locomotion, the escape response (Mao et al., 2004; Seugnet et al., 2008; Vanderheyden et al., 2013; Dissel et al., 2015c). Thus, disrupting the ubiquitin-proteasome system in adults attenuates sleep homeostasis.
Figure 5.
Disrupting fafEP3520 or hiw attenuates the negative effects of waking. (A,B) Expressing fafEP3520 and hiwDN in the Mushroom Bodies (MBs) using 247-GAL4 reduces homeostasis. Parental lines are shaded (n > 26/group; F(4,195) = 18, p = 1.41E-12, *p < 0.05, modified Bonferroni correction). (C,D) Expressing fafEP3520 and hiwDN in the adult brain using GeneSwitch (GSW-elav) reduced sleep homeostasis (% of sleep recovered in 24 h following a 12 h sleep deprivation (SD); n > 20/group). Flies were fed RU486 (RU+) or vehicle (RU−); genotype designations are inset (t test, *p < 0.05). (E) 247-GAL4 >UAS-hiwDN flies exhibit normal values for the TCT, PSI and QSI that are not statistically different from both 247-GAL4/+ and UAS-hiwDN/+ parental controls, *p < 0.05 modified Bonferroni test. (F) Learning is maintained following SD when hiwDN is expressed in the MBs (n > 8/group, *p < 0.05, modified Bonferroni test).
The lack of a homeostatic response following SD may represent either an adaptation that allows animals to better withstand the negative effects of waking, or it may simply indicate that the flies have lost their capacity to respond normally to sleep loss (Seugnet et al., 2011a; Donlea et al., 2012; Dissel et al., 2015b). To distinguish between these two possibilities, we evaluated STM. If flies have reduced sensitivity to sleep loss, they should maintain their ability to learn after SD (Dissel et al., 2015b). However, if they are simply unable to generate the needed compensatory response they should be learning impaired (Dissel et al., 2015b; Dissel and Shaw, 2017). Unfortunately, learning could not be evaluated in faf over-expressing flies due to deficits in phototaxis. A previous study has found that expressing UAS-hiwDN in MBs throughout development does not result in alterations in morphology (Huang et al., 2012). Thus we evaluated the effects of SD on learning in 247-GAL4/UAS-hiwDN flies. As above, we first evaluated photosensitivity and quinine sensitivity to rule out the possibility that changes in behavior could be due to non-associative cues. As seen in Figure 5E, photosensitivity, quinine sensitivity and time to complete the task were similar between genotypes. Importantly, flies expressing hiwDN in the MB maintain normal learning after SD while the outcrossed parental lines (247-GAL4/+ and UAS-hiwDN/+) are significantly impaired (Figure 5F). The magnitude of the learning deficit observed in the parental lines following SD is similar to that previously reported for sleep deprived wild-type flies and flies lacking MBs (Seugnet et al., 2008). Moreover the deficits in learning following sleep loss in flies are within the range of effect sizes observed following sleep loss in humans and rodents across a number of cognitive domains (Frey et al., 2007; Fu et al., 2007; Pierard et al., 2007). Thus, reducing hiw function in the MB confers resistance to SD as measured by both sleep homeostasis and STM.
Circuit-Dependent Regulation of Sleep by Rdl
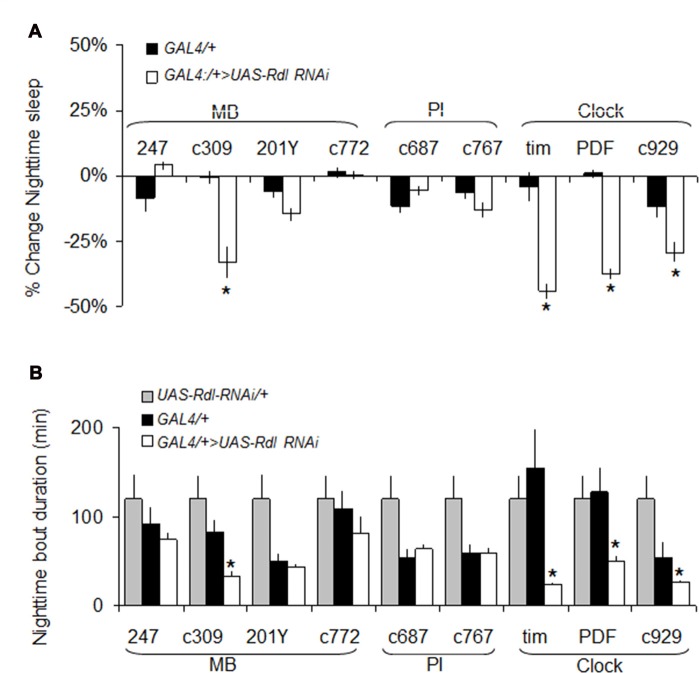
As mentioned above, we have hypothesized that Rdl may play a role in insomnia-phenotypes since Rdl is downregulated in ins-l flies (Seugnet et al., 2009c), is broadly expressed in the brain, including in neurons that impact sleep and waking (Liu et al., 2007; Parisky et al., 2008), and can enhance memory under some circumstances (Liu et al., 2007). While knocking down Rdl in lLNvs has been shown to increase waking, its role in other sleep wake-circuits and its impact on sleep homeostasis remains unclear (Agosto et al., 2008; Parisky et al., 2008; Chung et al., 2009). Thus, we conducted a mini-brain screen to express RdlRNAi in brain structures associated with sleep regulation. Consistent with previous reports, knocking down Rdl in clock neurons reduced night-time sleep and increased sleep fragmentation (Figures 6A,B).
Figure 6.
RdlRNAi screen. (A) Changes in nighttime sleep following UAS-Rdl RNAi expression using GAL4 drivers that target different areas of the brain involved in sleep regulation (MB, Mushroom Bodies, PI: pars intercerebralis, Clock: clock neurons). Nighttime sleep is presented as % change from UAS-Rdl RNAi/+ at age day 8 (n > 14/group; *p < 0.05). (B) Average nighttime sleep bout duration for the GAL4>UAS-Rdl RNAi combinations shown in (A) compared to both GAL4/+ and UAS-Rdl RNAi /+ (n > 14/group, *p < 0.05).
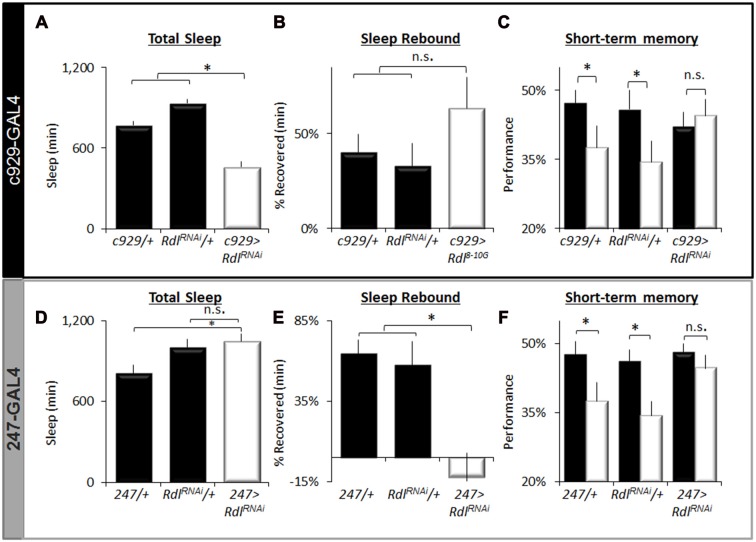
To further characterize the effects of knocking down Rdl on sleep homeostasis and STM, we focused on c929-GAL4 and 247-GAL4. As seen in Figure 7A, knocking down Rdl in c929-GAL4 expressing cells significantly reduces total sleep time compared to c929-GAL4/+ and UAS-RdlRNAi/+ parental controls as previously described. Similar results were observed with two other RNAi line (Rdli4-5 and Rdl31286; data not shown). Although c929-GAL4>UAS-RdlRNAi flies are short sleepers, 12 h of additional sleep loss is not accompanied by a larger sleep rebound compared to parental controls (Figure 7B). Importantly, short-sleeping c929-GAL4>UAS-RdlRNAi flies exhibit intact STM which is not adversely affected by an additional 12 h of SD (Figure 7C). Note that while performance in c929-GAL4>UAS-RdlRNAi flies appears to be slightly lower than parental lines, the difference is not significant and well within the range observed for wild-type flies (Seugnet et al., 2008). Thus, in contrast to the learning impairments seen following disrupting faf and hiw in lLNvs, knocking down Rdl seems to protect flies from the deleterious effects of waking as measured by both a reduced sleep rebound and intact STM.
Figure 7.
Regulation of sleep by Rdl. (A) Total Sleep is reduced in c929-GAL4>UAS-RdlRNAi flies compared to c929-GAL4/+ and UAS-RdlRNAi/+ parental controls, one-way ANOVA for genotype F(2,68) = 39, p = 4.86E-12, *p < 0.05, Modified Bonferroni correction. (B) Sleep homeostasis in c929-GAL4>UAS-RdlRNAi is not statistically different from both c929-GAL4/+ and UAS-RdlRNAi/+ parental controls (n > 14/group; *p < 0.05 modified Bonferroni correction). (C) Learning under baseline (black) and following a 12 h SD (white) in c929-GAL4>UAS-RdlRNAi and c929-GAL4/+, UAS-RdlRNAi/+ (n > 8/group; main effect for Condition F(1,50) = 3.85, p = 0.05; *p < 0.05 Modified Bonferroni correction). (D) 247-GAL4 <UAS-RdlRNAi flies exhibit normal levels of Total Sleep that are not statistically different from both 247-GAL4/+ and UAS-RdlRNAi/+ parental controls, one-way ANOVA for genotype F(2,40) = 4.7, p = 0.01, *p < 0.05, modified Bonferroni correction. (E) Sleep homeostasis (% of sleep recovered 24 h following a 12 h SD) in 247-GAL4/+, UAS-Rdl RNAi/+ and 247-GAL4>UAS-RdlRNAi flies (n > 14/group; F(2,48) = 5.23, p = 0.008, *p < 0.05, modified Bonferroni correction). (F) Learning under baseline (black) and following a 12 h SD (white) in 247-GAL4/+, UAS-Rdl RNAi/+ and 247-GAL4>UAS-RdlRNAi (n > 8/group; Genotype X Condition (baseline vs. sleep deprived) interaction F(1,83) = 3.90, p = 0.024; *p < 0.05).
To determine if knocking down Rdl in the MBs would impact sleep homeostasis or STM, we expressed UAS-RdlRNAi using 247-GAL4; knocking down Rdl in the MBs throughout development does not result in obvious morphological defects (Liu et al., 2007). As seen in Figure 7D, knocking down Rdl in the MBs did not affect total sleep time. However, knocking down Rdl in the MBs altered sleep regulation as evidenced by a significantly reduced sleep rebound compared to 247-GAL4/+ and UAS-RdlRNAi/+ parental controls (Figure 7E). Although both 247-GAL4/+ and UAS-RdlRNAi/+ parental controls displayed impaired STM following SD 247-GAL4>UAS-RdlRNAi flies maintained normal STM both before and after sleep loss (Figure 7F). No changes were seen in photosensitivity of quinine sensitivity indicating that the observed results were not due to changes in sensory thresholds (data not shown). Together these data suggest that knocking down Rdl both in clock neurons (lLNvs) and in the MBs can protect flies from the deleterious effects of SD.
Differential Modulation of Synaptic Plasticity by hiw and Rdl
Previous studies have reported that SD increases synaptic markers (Gilestro et al., 2009; Bushey et al., 2011). Notably, the impact of sleep loss on synaptic plasticity can be readily examined by quantifying pigment dispersing factor (PDF) positive terminals in projections from the lLNvs (Vanderheyden et al., 2013). As noted above, behavioral plasticity is disrupted in short-sleeping c929/+>UAS-hiwRNAi/+ flies but is preserved in short sleeping c929-GAL4/+>UAS-RdlRNAi/+ flies (Figures 2A,F, 7A,C). These data lead to the hypothesis that c929/+>UAS-hiwRNAi/+ flies should exhibit an increase in the number of PDF-positive terminals typical of sleep deprived flies while c929-GAL4/+>UAS-RdlRNAi/+ should be unaffected. As seen in Figures 8A–C, short-sleeping c929/+>UAS-hiwRNAi/+ flies display an increase in the number and size of PDF positive punctae compared to both of their normal sleeping parental controls (c929/+, UAS-hiwRNAi/+). In contrast, the number, size and intensity of PDF-positive terminals is preserved in short-sleeping c929-GAL4/+>UAS-RdlRNAi/+ flies compared to their normal sleeping parental controls (c929-GAL4/+>UAS-RdlRNAi/+; Figure 8D). Thus, knocking down Rdl in lLNvs protects flies from the negative impact of waking as measured both by STM and by examining structural plasticity. In contrast, the increase in synaptic number and morphology induced by knocking down hiw in the lLNvs is deleterious to behavioral and structural plasticity.
Figure 8.
Knocking down Rdl in the lLNvs protects flies from sleep-loss induced increases in synaptic markers. (A) Representative images of PDF immunohistochemistry of c929-GAL4/+>UAS-hiwRNAi/+ flies reveals an increase in the number (upper panels) and size (lower panels) of varicosities compared to c929-GAL4/+ and UAS-hiwRNAi/+ parental controls. (B) Quantification of terminal number expressed as a percentage of GAL4/+ (upper panel) revealed a significant One-way ANOVA for genotype F(2,30) = 6.0, p = 0.006, n = 9–12 each group; *p < 0.05 modified Bonferroni correction. Quantification of bouton size (lower panel) revealed a significant one-way ANOVA for genotype F(2,30) = 17.6, p = 1.07E-05, n = 9–12 each group; *p < 0.05 modified Bonferroni correction. (C) Representative images of PDF immunohistochemistry of c929-GAL4/+>UAS-RdlRNAi/+ flies does not show any change in the number (upper) or size (lower) of PDF positive punctae compared to c929-GAL4/+ and UAS-RdlRNAi/+ parental controls. (D) Quantification of terminal number and size did not reveal a significant one-way ANOVA for number or size F(2,17) = 0.2, p = 0.79 and F(2,17) = 1.9, p = 0.18, respectively; n = 7 each group.
Discussion
The de-ubiquitination enzyme faf and the E3 ubiquitin ligase hiw are evolutionary conserved synaptic proteins that regulate similar targets across phyla (Chen et al., 2000; Burgess et al., 2004; Nakata et al., 2005). The role of these two genes in the context of sleep is unknown. By manipulating faf and hiw, the present results provide evidence for an important role of specific ubiquitination pathways in the regulation of sleep homeostasis, the vulnerability to sleep loss and the regulation of sleep quotas. Interestingly, the impact of faf and hiw in conferring vulnerability or resilience to sleep loss is circuit dependent. Previous studies have found that the pan-neuronal disruption of inc and Cul3, two genes that are also involved in protein degradation pathways, reduces sleep time and attenuates sleep rebound (Stavropoulos and Young, 2011; Pfeiffenberger and Allada, 2012). However, in contrast to faf and hiw, inc and Cul3 only impact sleep if they are disrupted during development and do not seem to alter sleep when expressed in clock cells. Thus, these data confirm and extend the previous observations by demonstrating that novel ubiquitin pathways can be used in distinct circuits to modulate sleep and the response to sleep loss in adults.
As mentioned above, patients with primary insomnia are able to maintain normal performance on a variety of tasks even though they experience substantial sleep disruption (Riedel and Lichstein, 2000; Orff et al., 2007). These data suggest that insomnia may be a condition where predisposing genetic factors increase the risk for insomnia while simultaneously mitigating against at least some of the deleterious consequences of waking. Although the pan-neuronal expression of fafEP3520 did not increase waking, sleep was substantially disrupted when fafEP3520 was expressed in clock cells. The role of the ubiquitin-proteasome system within clock cells was confirmed by expressing loss-of-function alleles of the E3 ubiquitin ligase UAS-hiwDN. Importantly, the waking associated with disrupting the ubiquitin-proteasome system within clock cells was associated with STM impairments thereby recapitulating several key features of ins-l flies. In addition to disrupting STM, knocking down hiw also resulted in quantitative increases in the number and size of PDF-positive punctae similar to that observed during SD. In contrast to clock cells, however, when the ubiquitin-proteasome system was disrupted in the MBs, flies became resistant to SD. Interestingly, a previous report suggests that the ability of hiwDN to improve memory when expressed in the MBs was not due to its impact on Kenyon Cell morphology (Huang et al., 2012). These data suggest that the activity of a gene in one circuit may disrupt sleep and increase vulnerability to sleep loss while the activity of that same gene in a separate neuronal circuit may protect the animal from the negative consequences of waking. Understanding how hiwDN protects the MBs during SD will be the focus of future studies.
It is important to emphasize that inducing fafEP3520 and UAS-hiwDN in the adult fly brain using the conditional GeneSwitch GSW-elav driver results in a reduction of the homeostatic response following SD. As the induction of the transgene occurred in adult flies, this result indicates that faf can impact sleep regulation in the absence of developmental defects. This observation is consistent with the observation that disrupting hiw throughout development does not result in obvious structural MB deficits (Huang et al., 2012). Interestingly, knockdown of inc and Cul3 in adults using GSelav did not result in short-sleeping flies indicating that inc and Cul3 primarily influence sleep by disrupting neuronal circuitry during development. Indeed, both inc and Cul3 mutants are known to have morphological defects in the MBs (Zhu et al., 2005; Pfeiffenberger and Allada, 2012). Nonetheless, only 20%–30% of inc flies were found to have gross morphological defects in their MBs as identified by anti-FASCICLINII (FASII) immunohistochemistry. Since 90% of inc flies exhibit sleep disruption, it is possible that disrupted MB morphology may not fully explain the sleep phenotype in inc and Cul3 mutants (Pfeiffenberger and Allada, 2012). While the reduced sleep seen in inc flies may not be exclusively due to changes in gross MB morphology, many of the expected morphological defects such as defective re-elaboration of MB dendrites during metamorphosis, and short dendrites that fail to form typical claw-like structure are more readily detected using mosaic analysis (Zhu et al., 2005). Moreover, it is difficult to exclude the possibility that inc and Cul3 may have altered the development of additional neuronal groups that could disrupt circuits which could directly, or indirectly, influence sleep. It is interesting to note that STM remained intact, both during baseline and following SD, in flies expressing UAS-hiwDN in the MBs indicating that the effects were not due to deficits in MB structure (Seugnet et al., 2008). Indeed, disrupting hiw in the adult brain using GSW-elav can facilitate long-term memory (Huang et al., 2012). Taken together, these results suggest that disrupting the ubiquitin proteasome system by manipulating faf and hiw modulates the sensitivity to sleep loss by its effect on the buildup of target proteins, rather than by disrupting neuronal circuits during development.
It is possible that the phenotypes observed after manipulating faf and hiw with specific GAL4 drivers could be the result of ectopic expression, unrelated to the endogenous faf and hiw functions. While this possibility cannot be formally excluded we consider it unlikely for three main reasons: First, if altering faf and hiw resulted in a non-specific disruption of neuronal function, we would expect to observe sleep phenotype following expression in brain regions previously identified as playing an important role in sleep regulation, such as the MB, the fan shaped body, the pars intercerebralis or dopaminergic neurons. None of these cells types produced a change in sleep time upon faf over-expression. In fact only a few GAL4 drivers resulted in a change of sleep when used to over-express faf, and all of these drivers produced a reduction of nighttime sleep. Second, all the phenotypes described here for faf over-expression could be phenocopied by a down-regulation of hiw function. In the case of c929-GAL4 expressing cells, this was achieved by both a dominant negative form of hiw and RNAi constructs. It is unlikely that these two very different ways of reducing gene activity would result in the same phenotype if they were not specific and directly targeted to endogenous gene function. Finally, the hiwDN construct specifically blocks the ubiquitin ligase activity of the hiw protein (Wu et al., 2005), suggesting that the phenotypes observed in this study are linked to ubiquitination and not to other aspects of hiw function. Indeed, careful studies of temporal requirement for hiw function suggest that hiw may be maintaining synaptic transmission efficacy throughout the larval life of the animal, independently of its role in morphological plasticity (Wu et al., 2005).
Although it is not yet clear how the ubiquitin-proteasome system could protect flies from the negative effects of waking in specific circuits, a growing body of evidence has emphasized the important role that the ubiquitin system plays in neurons (Kaang and Choi, 2012). As mentioned, both faf and hiw regulate synaptogenesis and the elimination of synapses at the larval NMJ (DiAntonio et al., 2001). Moreover, the ubiquitin system can be regulated by neuronal activity, and seems to modulate several aspects of presynaptic and postsynaptic neurotransmission. Indeed, ubiquitin pathways not only regulate both excitatory and inhibitory receptors, it appears that the ubiquitin-proteasome system can also impact activity-dependent structural remodeling as evidenced by ubiquitin-dependent changes in spine morphology, size and density (Kaang and Choi, 2012). Interestingly, sleep and waking are also known to modulate activity-dependent changes at the synapse (Maret et al., 2011) suggesting that sleep and waking may rely upon the ubiquitin proteasome system to carry out some of their functions.
Given that, in clock neurons, faf and hiw increase the vulnerability to waking while in MB neurons faf and hiw confer resistance to waking, we asked whether modulating the activity of these circuits using an independent molecular pathway would reveal similar outcomes. The expectation that independent molecular pathways can influence insomnia phenotypes seems reasonable given the number and diversity of genes associated with human insomnia (for review see Lind and Gehrman, 2016).
Interestingly, knocking down Rdl both in clock cells and in the MB seemed to protect flies from the negative effects of extended waking. In the case of reducing Rdl in clock cells, flies maintained their ability to form STM despite being very short sleepers. In fact, short sleeping c929-GAL4>UAS-RdlRNAi flies maintained wild-type STM even when they were exposed to an additional 12 h of SD. This latter observation is interesting given that c929-GAL4/+>UAS-RdlRNAi/+ flies respond to 12 h of SD with a normal sleep rebound indicating that they have accrued additional sleep debt. The ability to acquire STM during a sleep rebound reinforces previous conclusions that impairments in APS cannot be attributed to increased sleep drive (Seugnet et al., 2008). It should be emphasized that the phenotype observed when UAS-hiwDN or UAS-RdlRNAi are expressed in clock cells looks identical when considering baseline sleep alone. Moreover, one might infer that c929-GAL4/+>UAS-hiwDN/+ are resistant to sleep loss and that c929-GAL4/+>UAS-RdlRNAi/+ are vulnerable to sleep loss based solely upon the absence or presence of a sleep rebound, respectively. However, when one considers STM, a phenotype that is known to be highly responsive to sleep loss in humans, rodents and flies, it becomes clear that the phenotype observed in c929-GAL4/+>UAS-hiwDN/+ is very different from that observed c929-GAL4/+>UAS-RdlRNAi/+ flies. Indeed, c929-GAL4/+>UAS-hiwDN/+ flies differ not only in behavioral plasticity but also structural plasticity. Identifying the difference in resilience/vulnerability to sleep loss would have been difficult, if not impossible, if we had only used sleep metrics as both the independent and dependent variable as is typically the case (Sehgal and Mignot, 2011; Dissel and Shaw, 2013).
In recent years greater attention has been given to elucidating mechanisms regulating sleep homeostasis (Joiner, 2016). Although genetic evidence has implicated R2 ring neurons of the Ellipsoid body and a subset of neurons in the dorsal Fan Shaped body as integrating signals conveying sleep need (Donlea et al., 2014; Liu et al., 2016; Pimentel et al., 2016), little is known about how or where these signals originate. Our data implicate the MBs as a likely input, into the circuitry underlying sleep homeostasis. It is worth noting that not all manipulations that increase waking are compensated by a subsequent homeostatic response (Joiner, 2016; Allada et al., 2017). For example, sleep rebound is absent or reduced following short periods of starvation or following heat shock during SD (Shaw et al., 2002; Thimgan et al., 2010, 2015; Donlea et al., 2012). Similarly, a recent study has found that activation of acetylcholine neurons induces episodes of waking that are followed by a homeostatic response while wake induced by activating octopaminergic neurons fails to induce a sleep rebound (Seidner et al., 2015). These data suggests the existence of independent arousal circuits that have differential access to homeostatic circuitry (Seidner et al., 2015). However, whereas waking induced by starvation does not disrupt learning and memory (Thimgan et al., 2010), waking induced by the activation of cholinergic and octopaminergic neurons results in learning deficits (Seidner et al., 2015). Thus, while arousal circuits may have differential access to the sleep homeostat, their ability to impair neurons involved in learning and memory may not be as restricted. Indeed, our data indicate that it is also possible for SD to activate homeostatic circuitry while simultaneously protecting against sleep-loss induced cognitive impairments (e.g., sleep deprived c929-GAL4/+>UAS-RdlRNAi flies). That is, under some circumstances, sleep homeostasis and cognitive deficits are dissociable. The organization of arousal, homeostatic and cognitive circuits may provide the architecture whereby insomnia-related genes can differentially result in sleep loss and protect flies from the negative effects of waking. Understanding how sleep deprived c929-GAL4/+>UAS-RdlRNAi flies can maintain normal cognitive abilities may reveal organizational insights into how extended waking impacts adaptive behavior during insomnia.
Author Contributions
LS, SD and PJS designed the experiments and analyzed data. LS, SD and MT completed the behavioral and genetic experiments. SD and LC conducted confocal imaging. LS, SD, MT and PJS wrote this manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Krishna Melnattur for helpful comment and suggestions.
Funding. This study was funded in part by the National Institute of Health R01NS051305-13 and 2 R01 NH076980-06 (PJS), Neuroscience Blueprint Core grant P30 NS057105 to the Bakewell Neuroimaging Laboratory.
References
- Agosto J., Choi J. C., Parisky K. M., Stilwell G., Rosbash M., Griffith L. C. (2008). Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat. Neurosci. 11, 354–359. 10.1038/nn2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R., Cirelli C., Sehgal A. (2017). Molecular mechanisms of sleep homeostasis in flies and mammals. Cold Spring Harb. Perspect. Biol. 9:a027730. 10.1101/cshperspect.a027730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks S., Dinges D. F. (2007). Behavioral and physiological consequences of sleep restriction. J. Clin. Sleep Med. 3, 519–528. [PMC free article] [PubMed] [Google Scholar]
- Burgess R. W., Peterson K. A., Johnson M. J., Roix J. J., Welsh I. C., O’Brien T. P. (2004). Evidence for a conserved function in synapse formation reveals Phr1 as a candidate gene for respiratory failure in newborn mice. Mol. Cell. Biol. 24, 1096–1105. 10.1128/mcb.24.3.1096-1105.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey D., Tononi G., Cirelli C. (2011). Sleep and synaptic homeostasis: structural evidence in Drosophila. Science 332, 1576–1581. 10.1126/science.1202839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buysse D. J., Thompson W., Scott J., Franzen P. L., Germain A., Hall M., et al. (2007). Daytime symptoms in primary insomnia: a prospective analysis using ecological momentary assessment. Sleep Med. 8, 198–208. 10.1016/j.sleep.2006.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Overstreet E., Wood S. A., Fischer J. A. (2000). On the conservation of function of the Drosophila fat facets deubiquitinating enzyme and fam, its mouse homolog. Dev. Genes Evol. 210, 603–610. 10.1007/s004270000109 [DOI] [PubMed] [Google Scholar]
- Chevalier H., Los F., Boichut D., Bianchi M., Nutt D. J., Hajak G., et al. (1999). Evaluation of severe insomnia in the general population: results of a European multinational survey. J. Psychopharmacol. 13, S21–S24. 10.1177/026988119901304s04 [DOI] [PubMed] [Google Scholar]
- Chuah Y. M., Venkatraman V., Dinges D. F., Chee M. W. (2006). The neural basis of interindividual variability in inhibitory efficiency after sleep deprivation. J. Neurosci. 26, 7156–7162. 10.1523/JNEUROSCI.0906-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung B. Y., Kilman V. L., Keath J. R., Pitman J. L., Allada R. (2009). The GABAA receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr. Biol. 19, 386–390. 10.1016/j.cub.2009.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiAntonio A., Haghighi A. P., Portman S. L., Lee J. D., Amaranto A. M., Goodman C. S. (2001). Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature 412, 449–452. 10.1038/35086595 [DOI] [PubMed] [Google Scholar]
- DiAntonio A., Hicke L. (2004). Ubiquitin-dependent regulation of the synapse. Annu. Rev. Neurosci. 27, 223–246. 10.1146/annurev.neuro.27.070203.144317 [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., Fellner M., et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Dissel S., Angadi V., Kirszenblat L., Suzuki Y., Donlea J., Klose M., et al. (2015a). Sleep restores behavioral plasticity to Drosophila mutants. Curr. Biol. 25, 1270–1281. 10.1016/j.cub.2015.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissel S., Melnattur K., Shaw P. J. (2015b). Sleep, performance, and memory in flies. Curr. Sleep Med. Rep. 1, 47–54. 10.1007/s40675-014-0006-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissel S., Seugnet L., Thimgan M. S., Silverman N., Angadi V., Thacher P. V., et al. (2015c). Differential activation of immune factors in neurons and glia contribute to individual differences in resilience/vulnerability to sleep disruption. Brain Behav. Immun. 47, 75–85. 10.1016/j.bbi.2014.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissel S., Klose M., Donlea J., Lijuan C., English D., Winsky-Sommerer R., et al. (2017). Sleep can be used as a therapeutic to reverse memory impairment and the underlying pathology in Drosophila models of Alzheimer’s disease. Neurobiol. Sleep Circadian Rhythms 2, 15–26. 10.1016/j.nbscr.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissel S., Shaw P. J. (2013). “Drosophlia model systems for genetic sleep research,” in The Genetic Basis of Sleep and Sleep Disorders, eds Shaw P. J., Tafti M., Thorpy M. (New York, NY: Cambridge University Press; ), 43–53. [Google Scholar]
- Dissel S., Shaw P. J. (2017). “Sleep and memory formation in Drosophila,” in Learning and Memory: A Comprehensive Reference, ed. Byrne J. H. (Cambridge, MA: Academic Press; ), 179–198. [Google Scholar]
- Donlea J., Leahy A., Thimgan M. S., Suzuki Y., Hughson B. N., Sokolowski M. B., et al. (2012). Foraging alters resilience/vulnerability to sleep disruption and starvation in Drosophila. Proc. Natl. Acad. Sci. U S A 109, 2613–2618. 10.1073/pnas.1112623109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea J. M., Pimentel D., Miesenbock G. (2014). Neuronal machinery of sleep homeostasis in Drosophila. Neuron 81, 860–872. 10.1016/j.neuron.2013.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea J. M., Ramanan N., Shaw P. J. (2009). Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science 324, 105–108. 10.1126/science.1166657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea J. M., Thimgan M. S., Suzuki Y., Gottschalk L., Shaw P. J. (2011). Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science 332, 1571–1576. 10.1126/science.1202249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake C. L., Roehrs T., Roth T. (2003). Insomnia causes, consequences, and therapeutics: an overview. Depress. Anxiety 18, 163–176. 10.1002/da.10151 [DOI] [PubMed] [Google Scholar]
- Dubnau J., Chiang A. S. (2013). Systems memory consolidation in Drosophila. Curr. Opin. Neurobiol. 23, 84–91. 10.1016/j.conb.2012.09.006 [DOI] [PubMed] [Google Scholar]
- Dubnau J., Grady L., Kitamoto T., Tully T. (2001). Disruption of neurotransmission in Drosophila mushroom body blocks retrieval but not acquisition of memory. Nature 411, 476–480. 10.1038/35078077 [DOI] [PubMed] [Google Scholar]
- Edinger J. D., Means M. K., Carney C. E., Krystal A. D. (2008). Psychomotor performance deficits and their relation to prior nights’ sleep among individuals with primary insomnia. Sleep 31, 599–607. 10.1093/sleep/31.5.599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltenyi K., Greenspan R. J., Newport J. W. (2007). Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat. Neurosci. 10, 1160–1167. 10.1038/nn1957 [DOI] [PubMed] [Google Scholar]
- Freeman A., Pranski E., Miller D., Radmard S., Bernhard D., Jinnah H., et al. (2012). Sleep fragmentation and motor restlessness in a Drosophila model of restless legs syndrome. Curr. Biol. 22, 1142–1148. 10.1016/j.cub.2012.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey D. J., Badia P., Wright K. P., Jr. (2004). Inter- and intra-individual variability in performance near the circadian nadir during sleep deprivation. J. Sleep Res. 13, 305–315. 10.1111/j.1365-2869.2004.00429.x [DOI] [PubMed] [Google Scholar]
- Frey D. J., Fleshner M., Wright K. P., Jr. (2007). The effects of 40 hours of total sleep deprivation on inflammatory markers in healthy young adults. Brain Behav. Immun. 21, 1050–1057. 10.1016/j.bbi.2007.04.003 [DOI] [PubMed] [Google Scholar]
- Fu J., Li P., Ouyang X., Gu C., Song Z., Gao J., et al. (2007). Rapid eye movement sleep deprivation selectively impairs recall of fear extinction in hippocampus-independent tasks in rats. Neuroscience 144, 1186–1192. 10.1016/j.neuroscience.2006.10.050 [DOI] [PubMed] [Google Scholar]
- Gilestro G. F., Tononi G., Cirelli C. (2009). Widespread changes in synaptic markers as a function of sleep and wakefulness in Drosophila. Science 324, 109–112. 10.1126/science.1166673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B., Chelot E., Xia R., Rouyer F. (2004). Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431, 869–873. 10.1038/nature02935 [DOI] [PubMed] [Google Scholar]
- Hauri P. J. (1997). Cognitive deficits in insomnia patients. Acta Neurol. Belg. 97, 113–117. [PubMed] [Google Scholar]
- Henderson J. E., Soderlund D. M., Knipple D. C. (1993). Characterization of a putative γ-aminobutyric acid (GABA) receptor β subunit gene from Drosophila melanogaster. Biochem. Biophys. Res. Commun. 193, 474–482. 10.1006/bbrc.1993.1648 [DOI] [PubMed] [Google Scholar]
- Huang C., Zheng X., Zhao H., Li M., Wang P., Xie Z., et al. (2012). A permissive role of mushroom body α/β core neurons in long-term memory consolidation in Drosophila. Curr. Biol. 22, 1981–1989. 10.1016/j.cub.2012.08.048 [DOI] [PubMed] [Google Scholar]
- Jarome T. J., Helmstetter F. J. (2013). The ubiquitin-proteasome system as a critical regulator of synaptic plasticity and long-term memory formation. Neurobiol. Learn. Mem. 105, 107–116. 10.1016/j.nlm.2013.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner W. J. (2016). Unraveling the evolutionary determinants of sleep. Curr. Biol. 26, R1073–R1087. 10.1016/j.cub.2016.08.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner W. J., Crocker A., White B. H., Sehgal A. (2006). Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441, 757–760. 10.1038/nature04811 [DOI] [PubMed] [Google Scholar]
- Joiner M. A., Griffith L. C. (1997). CaM kinase II and visual input modulate memory formation in the neuronal circuit controlling courtship conditioning. J. Neurosci. 17, 9384–9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaang B. K., Choi J. H. (2012). Synaptic protein degradation in memory reorganization. Adv. Exp. Med. Biol. 970, 221–240. 10.1007/978-3-7091-0932-8_10 [DOI] [PubMed] [Google Scholar]
- Kahsai L., Zars T. (2011). Learning and memory in Drosophila: behavior, genetics, and neural systems. Int. Rev. Neurobiol. 99, 139–167. 10.1016/b978-0-12-387003-2.00006-9 [DOI] [PubMed] [Google Scholar]
- Krause A. J., Simon E. B., Mander B. A., Greer S. M., Saletin J. M., Goldstein-Piekarski A. N., et al. (2017). The sleep-deprived human brain. Nat. Rev. Neurosci. 18, 404–418. 10.1038/nrn.2017.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourg E., Buecher C. (2002). Learned suppression of photopositive tendencies in Drosophila melanogaster. Anim. Learn. Behav. 30, 330–341. 10.3758/bf03195958 [DOI] [PubMed] [Google Scholar]
- Levine J. D., Funes P., Dowse H. B., Hall J. C. (2002). Signal analysis of behavioral and molecular cycles. BMC Neurosci. 3:1. 10.1186/1471-2202-3-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhou Z., Zhang X., Tong H., Li P., Zhang Z. C., et al. (2013). Drosophila neuroligin 4 regulates sleep through modulating GABA transmission. J. Neurosci. 33, 15545–15554. 10.1523/JNEUROSCI.0819-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind M. J., Gehrman P. R. (2016). Genetic pathways to insomnia. Brain Sci. 6:E64. 10.3390/brainsci6040064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Lamaze A., Liu Q., Tabuchi M., Yang Y., Fowler M., et al. (2014). WIDE AWAKE mediates the circadian timing of sleep onset. Neuron 82, 151–166. 10.1016/j.neuron.2014.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Liu Q., Tabuchi M., Wu M. N. (2016). Sleep drive is encoded by neural plastic changes in a dedicated circuit. Cell 165, 1347–1360. 10.1016/j.cell.2016.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Krause W. C., Davis R. L. (2007). GABAA receptor RDL inhibits Drosophila olfactory associative learning. Neuron 56, 1090–1102. 10.1016/j.neuron.2007.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z., Roman G., Zong L., Davis R. L. (2004). Pharmacogenetic rescue in time and space of the rutabaga memory impairment by using Gene-Switch. Proc. Natl. Acad. Sci. U S A 101, 198–203. 10.1073/pnas.0306128101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maret S., Faraguna U., Nelson A. B., Cirelli C., Tononi G. (2011). Sleep and waking modulate spine turnover in the adolescent mouse cortex. Nat. Neurosci. 14, 1418–1420. 10.1038/nn.2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K., Abrams B., Grill B., Goncharov A., Huang X., Chisholm A. D., et al. (2005). Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell 120, 407–420. 10.1016/j.cell.2004.12.017 [DOI] [PubMed] [Google Scholar]
- Nofzinger E. A., Buysse D. J., Germain A., Price J. C., Miewald J. M., Kupfer D. J. (2004). Functional neuroimaging evidence for hyperarousal in insomnia. Am. J. Psychiatry 161, 2126–2128. 10.1176/appi.ajp.161.11.2126 [DOI] [PubMed] [Google Scholar]
- Orff H. J., Drummond S. P., Nowakowski S., Perils M. L. (2007). Discrepancy between subjective symptomatology and objective neuropsychological performance in insomnia. Sleep 30, 1205–1211. 10.1093/sleep/30.9.1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder T., Yoon K. S., White B. H., Keshishian H. (2001). A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. U S A 98, 12596–12601. 10.1073/pnas.221303298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozminkowski R. J., Wang S., Walsh J. K. (2007). The direct and indirect costs of untreated insomnia in adults in the United States. Sleep 30, 263–273. 10.1093/sleep/30.3.263 [DOI] [PubMed] [Google Scholar]
- Parisky K. M., Agosto J., Pulver S. R., Shang Y., Kuklin E., Hodge J. J., et al. (2008). PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron 60, 672–682. 10.1016/j.neuron.2008.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D., Veenstra J. A., Park J. H., Taghert P. H. (2008). Mapping peptidergic cells in Drosophila: Where DIMM fits in. PLoS One 3:e1896. 10.1371/journal.pone.0001896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffenberger C., Allada R. (2012). Cul3 and the BTB adaptor insomniac are key regulators of sleep homeostasis and a dopamine arousal pathway in Drosophila. PLoS Genet. 8:e1003003. 10.1371/journal.pgen.1003003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierard C., Liscia P., Philippin J. N., Mons N., Lafon T., Chauveau F., et al. (2007). Modafinil restores memory performance and neural activity impaired by sleep deprivation in mice. Pharmacol. Biochem. Behav. 88, 55–63. 10.1016/j.pbb.2007.07.006 [DOI] [PubMed] [Google Scholar]
- Pimentel D., Donlea J. M., Talbot C. B., Song S. M., Thurston A. J., Miesenbock G. (2016). Operation of a homeostatic sleep switch. Nature 536, 333–337. 10.1038/nature19055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman J. L., McGill J. J., Keegan K. P., Allada R. (2006). A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature 441, 753–756. 10.1038/nature04739 [DOI] [PubMed] [Google Scholar]
- Riedel B. W., Lichstein K. L. (2000). Insomnia and daytime functioning. Sleep Med. Rev. 4, 277–298. 10.1053/smrv.1999.0074 [DOI] [PubMed] [Google Scholar]
- Sehgal A., Mignot E. (2011). Genetics of sleep and sleep disorders. Cell 146, 194–207. 10.1016/j.cell.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidner G., Robinson J. E., Wu M., Worden K., Masek P., Roberts S. W., et al. (2015). Identification of neurons with a privileged role in sleep homeostasis in Drosophila melanogaster. Curr. Biol. 25, 2928–2938. 10.1016/j.cub.2015.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L., Galvin J. E., Suzuki Y., Gottschalk L., Shaw P. J. (2009a). Persistent short-term memory defects following sleep deprivation in a Drosophila model of Parkinson disease. Sleep 32, 984–992. 10.1093/sleep/32.8.984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L., Suzuki Y., Stidd R., Shaw P. J. (2009b). Aversive phototaxic suppression: evaluation of a short-term memory assay in Drosophila melanogaster. Genes Brain Behav. 8, 377–389. 10.1111/j.1601-183x.2009.00483.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L., Suzuki Y., Thimgan M., Donlea J., Gimbel S. I., Gottschalk L., et al. (2009c). Identifying sleep regulatory genes using a Drosophila model of insomnia. J. Neurosci. 29, 7148–7157. 10.1523/JNEUROSCI.5629-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L., Suzuki Y., Donlea J. M., Gottschalk L., Shaw P. J. (2011a). Sleep deprivation during early-adult development results in long-lasting learning deficits in adult Drosophila. Sleep 34, 137–146. 10.1093/sleep/34.2.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L., Suzuki Y., Merlin G., Gottschalk L., Duntley S. P., Shaw P. J. (2011b). Notch signaling modulates sleep homeostasis and learning after sleep deprivation in Drosophila. Curr. Biol. 21, 835–840. 10.1016/j.cub.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L., Suzuki Y., Vine L., Gottschalk L., Shaw P. J. (2008). D1 receptor activation in the mushroom bodies rescues sleep-loss-induced learning impairments in Drosophila. Curr. Biol. 18, 1110–1117. 10.1016/j.cub.2008.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P. J., Cirelli C., Greenspan R. J., Tononi G. (2000). Correlates of sleep and waking in Drosophila melanogaster. Science 287, 1834–1837. 10.1126/science.287.5459.1834 [DOI] [PubMed] [Google Scholar]
- Shaw P. J., Tononi G., Greenspan R. J., Robinson D. F. (2002). Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature 417, 287–291. 10.1038/417287a [DOI] [PubMed] [Google Scholar]
- Sheeba V., Fogle K. J., Kaneko M., Rashid S., Chou Y. T., Sharma V. K., et al. (2008). Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr. Biol. 18, 1537–1545. 10.1016/j.cub.2008.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavropoulos N., Young M. W. (2011). insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron 72, 964–976. 10.1016/j.neuron.2011.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D., Peng Y., Agosto J., Rosbash M. (2004). Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431, 862–868. 10.1038/nature02926 [DOI] [PubMed] [Google Scholar]
- Thimgan M. S., Seugnet L., Turk J., Shaw P. J. (2015). Identification of genes associated with resilience/vulnerability to sleep deprivation and starvation in Drosophila. Sleep 38, 801–814. 10.5665/sleep.4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimgan M. S., Suzuki Y., Seugnet L., Gottschalk L., Shaw P. J. (2010). The perilipin homologue, lipid storage droplet 2, regulates sleep homeostasis and prevents learning impairments following sleep loss. PLoS Biol. 8:e1000466. 10.1371/journal.pbio.1000466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderheyden W. M., Gerstner J. R., Tanenhaus A., Yin J. C., Shaw P. J. (2013). ERK phosphorylation regulates sleep and plasticity in Drosophila. PLoS One 8:e81554. 10.1371/journal.pone.0081554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Wairkar Y. P., Collins C. A., DiAntonio A. (2005). Highwire function at the Drosophila neuromuscular junction: spatial, structural, and temporal requirements. J. Neurosci. 25, 9557–9566. 10.1523/jneurosci.2532-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Perez R., Pan M., Lee T. (2005). Requirement of Cul3 for axonal arborization and dendritic elaboration in Drosophila mushroom body neurons. J. Neurosci. 25, 4189–4197. 10.1523/jneurosci.0149-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]