Abstract
We present the genomic and expressed myosin II sequences from the giant waterbug, Lethocerus indicus. The intron rich gene appears relatively ancient and contains six regions of mutually exclusive exons that are alternatively spliced. Alternatively spliced regions may be involved in the asymmetric myosin dimer structure known as the interacting heads motif, as well as stabilizing the interacting heads motif within the thick filament. A lack of negative charge in the myosin S2 domain may explain why Lethocerus thick filaments display a perpendicular interacting heads motif, rather than one folded back to contact S2, as is seen in other thick filament types such as those from tarantula.
Electronic supplementary material
The online version of this article (doi:10.1007/s10974-017-9476-6) contains supplementary material, which is available to authorized users.
Keywords: Myosin II, Insect flight muscle, Alternative splicing, Interacting heads motif
Introduction
The indirect flight muscles of the giant waterbug, Lethocerus spp., have long held a special place in muscle research, thanks to their conveniently large fiber size and exquisitely well-ordered filament lattice. Key evidence that informed the early swinging crossbridge theory of contraction (Huxley and Brown 1967; Huxley 1969) was the observation of tilted myosin heads attached to actin in rigor Lethocerus muscle (Reedy et al. 1965). The first time-resolved X-ray diffraction of actively contracting muscle took advantage of the oscillatory contraction mode of Lethocerus muscle (Tregear and Miller 1969). The subsequent development of modern synchrotron X-ray sources was initially driven by the problem of muscle (Holmes and Rosenbaum 1998), and the first synchrotron X-ray pattern was recorded from Lethocerus muscle (Rosenbaum et al. 1971). The first cryo-EM images of isolated myosin filaments were also from Lethocerus (Menetret et al. 1988).
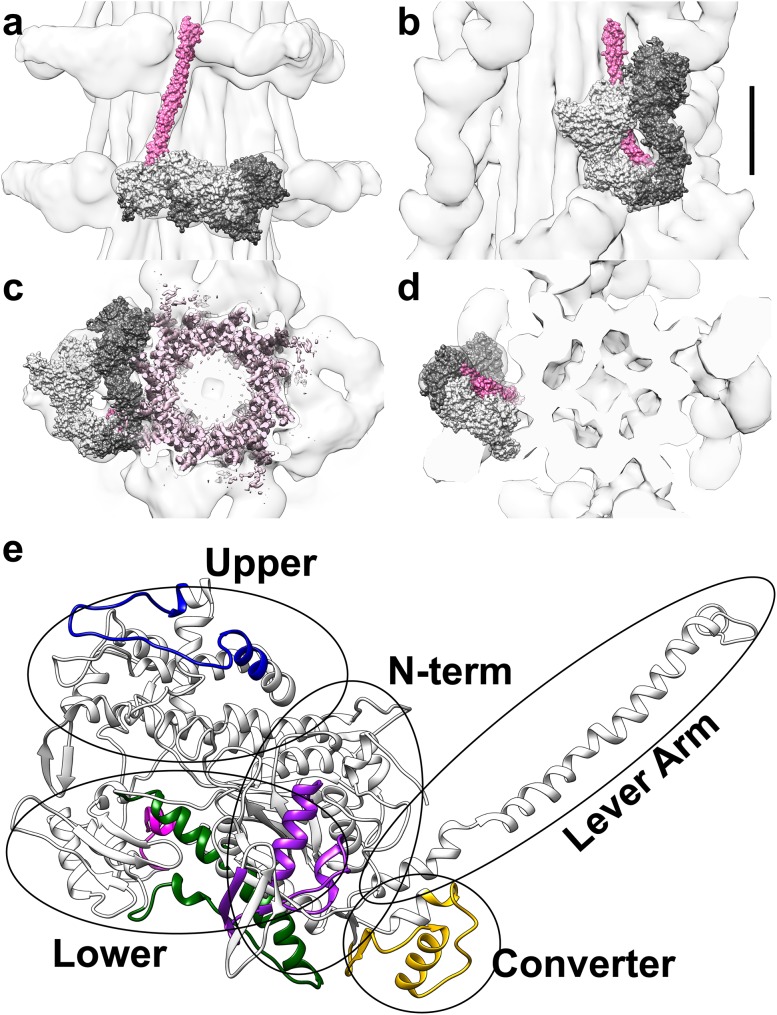
A recent three-dimensional reconstruction of the myosin-containing thick filament from Lethocerus yielded several surprises (Hu et al. 2016). The myosin heads formed an interacting heads motif (IHM), which had previously been observed in smooth muscle myosin crystals (Wendt et al. 1999) and in other striated muscle thick filament types (Woodhead et al. 2005, 2013; Zoghbi et al. 2008; Zhao et al. 2009). But in the Lethocerus thick filament, the IHM was oriented in a unique way, perpendicular to the filament axis (Fig. 1a–d). Other insects with asynchronous flight muscle may have the same orientation of the IHM in the relaxed thick filament. Resolution in the latest reconstruction was especially good within the backbone of the thick filament (~5 Å), revealing the twists and turns of the α-helical coiled-coil rod in its native environment, as well as extra non-myosin densities embedded within the backbone (Hu et al. 2016). With this data we can begin to construct an atomic model of the entire 160-nm long myosin rod domain; in turn making it urgent that we obtain the correct amino acid sequence for Lethocerus myosin, which was previously unknown.
Fig. 1.
a–d Thick filament reconstructions from Lethocerus (Hu et al. 2016) on the left (a, c) and tarantula (Alamo et al. 2016) on the right (b, d) are shown in longitudinal (a, b) and cross-section views (c, d). A space-filling model of the myosin IHM, PDB 3JBH (Alamo et al. 2016), is fit within both maps. Although myosin is a dimer, the two heads of the IHM are not equivalent. One head (light gray) is called blocked, because its actin-binding domain contacts the back of its partner, called the free head (dark gray). In b and c, the IHMs are oriented similarly, although the filaments are perpendicular in these two views. Thus, the IHM is perpendicular to the S2 domain (pink) in Lethocerus (a); whereas it folds back to lie on top of the S2 domain in tarantula (b). The thick filament bare zone is towards the top of the page (a, b) or below the plane of the page (c, d). Scale bar 100 Å. e Ribbon diagram of myosin S1 head from the tarantula model, residues 1-838 of PDB 3JBH.g (Alamo et al. 2016), shows the five regions expected to be alternatively spliced in Lethocerus, color coded purple, blue, dark green, magenta, and yellow for MXEs 1, 5, 7, 8 and 10, respectively. A sixth alternatively spliced region is expected within the helical rod domain (not shown), near the junction between S2 and light meromyosin (LMM). The N-terminal, upper and lower 50 kD, converter, and lever arm domains of the S1 head are circled and labeled
In many species, the myosin gene shows clusters of mutually exclusive exons (MXEs) that are alternatively spliced to give different protein isoforms (Bernstein et al. 1986; Wassenberg et al. 1987; George et al. 1989; Odronitz and Kollmar 2008; Kollmar and Hatje 2014). Evolutionary analysis shows eleven potential MXE clusters that code for specific regions of the molecule, ten within the S1 myosin head (Fig. 1e) and one within the helical rod domain (Odronitz and Kollmar 2008; Kollmar and Hatje 2014). The ancestral arthropod gene is predicted to be intron rich, with 42 exons that are typically short (Kollmar and Hatje 2014). The exon numbering and which MXE clusters are present vary among different taxa due to variable intron/exon loss. For example, the Drosophila myosin gene retains MXE clusters 1, 5, 7, 10 and 11, but has single exons for the remaining potential MXE clusters, whereas Hemiptera like Lethocerus retain the same five MXE clusters plus cluster 8 (Kollmar and Hatje 2014). Additionally, a short or long C-terminus is encoded by either inclusion or exclusion of the penultimate exon, which has an early stop codon (Bernstein et al. 1986; Odronitz and Kollmar 2008; Kollmar and Hatje 2014). The generally accepted view is that the alternative splicing fine-tunes the biophysical properties of myosin as needed for different muscle types (Bernstein and Milligan 1997). We propose here that the alternative splicing may also affect the stability of the IHM and the structural differences seen in thick filaments from different muscle types.
Results
We sought the expressed myosin sequence by cloning Lethocerus cDNA and initially retrieved 52 partially overlapping clones (Supplemental Information, Methods). Using the partial clones to design new primers, we retrieved and report here eight unique full-length myosin clones, two unique partial clones, and the 5′ and 3′ untranslated regions, termed clones X1–X12 (GenBank Accession #s MF071206-MF071217). Simultaneously we initiated whole genome shotgun sequencing from Lethocerus DNA. Genome annotation is still in progress, but the scaffold containing the muscle myosin sequence has been identified and analyzed (GenBank Accession # MF078003).
The gene structure and amino acid sequences of Lethocerus muscle myosin show the expected six MXE clusters that are alternatively spliced as well as the short/long C-termini (Fig. 2). The gene has 39 exons that are typically short, similar to other Hemiptera and in contrast to Drosophila myosin with only 19 exons that are necessarily longer (Kollmar and Hatje 2014). The location of 34 of the introns exactly match those predicted for the ancestral arthropod myosin gene [compare Fig. 2 to Fig. 5 of Odronitz and Kollmar (2008)], suggesting that the Lethocerus myosin gene is relatively ancient and undergone little intron loss. Similar to other species, the first exon lacks a start codon and is untranslated, and protein expression begins with the second exon (George et al. 1989; Odronitz and Kollmar 2008; Kollmar and Hatje 2014). At least one MEF2 promoter sequence and a number of E-box sequences are found either before the first exon or within the first intron, consistent with Drosophila myosin (Hess et al. 2007). We also identified a 3′ polyadenylation site that was 233 bases downstream of the stop codon in the last exon. In the genomic sequence, this site includes both upstream and downstream polyadenylation signals, 20 and 10 base pairs away respectively, as expected (Retelska et al. 2006). We found clones expressing almost all of the possible variants for each MXE cluster, with the exception of exons 10a, 14b, and 20c, which must still be considered putative variants (Fig. 2b, italics).
Fig. 2.
a In the Lethocerus myosin gene structure, light gray bars represent introns and taller boxes represent exons. Numbers above indicate the MXE cluster according to Kollmar and Hatje (2014); numbers below indicate the exon number within the Lethocerus myosin gene. Dark gray boxes represent single exons. MXE clusters 1, 5, 7, 8, 10 and 11 are outlined, numbered and color-coded. Unnumbered white boxes represent MXE clusters 2–4, 6 and 9, for which Lethocerus has single exons but some species have alternatively spliced clusters of MXEs. b The protein sequence is arranged by exon, with MXE clusters outlined and color-coded as in a. Exons 10a, 14b, and 20c are italicized to indicate they are putative exons for which no clone has been found to date
Our initial cloning used cDNA prepared from mixed muscle types, so we expected multiple clones. We were most interested in knowing which myosin isoforms are expressed in the dorsal longitudinal muscles, which are the ones usually used in muscle research. Therefore we prepared cDNA from mRNA separately isolated from the dorsal longitudinal, the oblique, and the dorsal ventral indirect flight muscles, and then used PCR to screen for the presence or absence of every exon variant in each of the three muscles (Supplemental Fig. 1). All three muscles tested negative for expression of exons 10a, 14b, and 20c, confirming our initial cloning that these variants are not expressed at detectable levels in adult flight muscles. Surprisingly, the dorsal ventral muscles tested positive for all of the remaining exon variants except 14c; therefore nine of the ten unique clones we obtained appear to be expressed in dorsal ventral muscles. The dorsal ventral muscles are difficult to dissect and it is possible that our sample was contaminated with other muscle types, such as direct flight, skeletal, or somatic muscles, which may explain the apparent multiplicity of variants expressed. In contrast, the dorsal longitudinal and the oblique muscles are easy to isolate, and they expressed only exons 3b, 10c, 14a, 17a/b, 20a and 29a, as well as both the short and long C-termini. Thus we found that only clones X1, X2 and X4 are expressed in the dorsal longitudinal and oblique flight muscles (Supplemental Information, Table 1).
When clone X2 is used to query the known Drosophila melanogaster myosin sequences in a BLAST search (Altschul et al. 1997), it is 83–85% similar to all Drosophila myosins, but most similar to isoform K, the adult flight-muscle specific isoform (Zhang and Bernstein 2001). Clone X1 is identical to X2 except for having the long C-terminus, rather than the short one. In Drosophila indirect flight muscle, there is evidence of sequential expression of a long C-terminal myosin early in sarcomere development and a short C-terminal myosin later (Orfanos and Sparrow 2013). In contrast, our results indicate concurrent expression of both the long and the short versions in Lethocerus flight muscle. Clone X4 is identical to X1, except for expressing exon 17a, instead of 17b. Exon 17 corresponds to MXE 8 and codes for a variable region of the myosin motor domain known as loop 2 (Kollmar and Hatje 2014). We used quantitative PCR to estimate the relative levels in the dorsal longitudinal muscles and found that exon 17b was expressed slightly more than exon 17a (mean ratio = 4 ± 2, from three cDNA samples independently prepared from different insects).
Discussion
To date, the most detailed model of the IHM is PDB 3JBH, which was flexibly fit into a 20-Å resolution map of the tarantula thick filament. 3JBH revealed conserved residues that may be involved in specific contacts that stabilize the IHM (Alamo et al. 2016). We note here that many of those proposed contacts involve the alternatively spliced MXEs as can be seen when the MXEs are mapped onto the IHM (Fig. 3). Intramolecular contacts between the two motor domains include blocked-head MXEs 5 and 6 with free-head MXEs 7 and 10 (Fig. 3a). Although we do not yet have a flexibly fit model of the Lethocerus IHM, similar contacts would be expected. Likewise, similar contacts would be expected when the IHM is observed in isolated myosin molecules (Jung et al. 2008). The head–head interactions of the IHM are thought to give rise to the very low fibrillar ATPase rate known as the super-relaxed state (Hooijman et al. 2011). We suggest that alternative splicing of the MXEs, in addition to its widely studied effect on acto-myosin interactions (Bernstein and Milligan 1997), may also affect the stability of the IHM, and therefore affect species-specific differences in the kinetics of ATP exchange in the super-relaxed state (Naber et al. 2011).
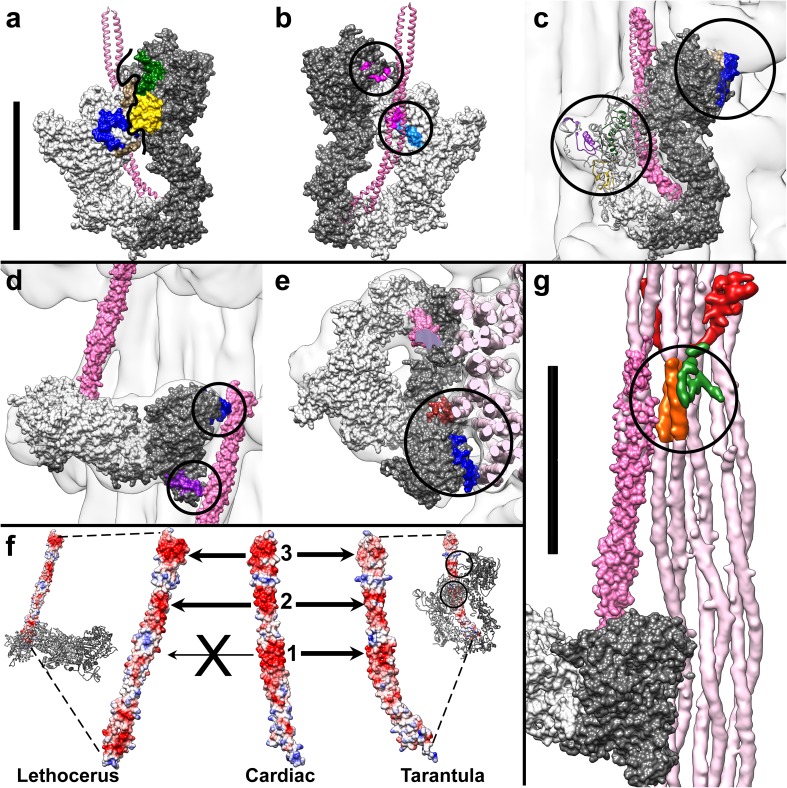
Fig. 3.
Color-coded MXEs are shown in context of the IHM and thick filament structures of tarantula (a–c) and Lethocerus (d, e, g). a Surface representation of the tarantula IHM shows blocked-head MXEs 5 and 6 (blue and tan) contacting free-head MXEs 7 and 10 (green and yellow). The heavy black line separates the blocked head (light gray) from the free head (dark gray). Ribbon representation of the S2 domain is shown in pink passing behind the IHM. b The view flipped 180° shows the back of the IHM where blocked-head MXEs 8 and 9 (magenta and light blue) contact the pink S2 domain (lower circle). In the upper circle, free-head MXE 8 (magenta) does not directly contact S2, but may be necessary to position the part that does contact S2. This region is a flexible loop known as loop 2 (Rayment et al. 1993). c Same view as a, with the transparent thick filament structure overlaid. In the upper circle, free-head MXEs 5 and 6 (blue and tan) contact an adjacent IHM. In the lower circle blocked-head MXEs 1, 7 and 10 (purple, green and yellow) contact the thick filament backbone. For clarity, the blocked head is shown as a ribbon diagram, whereas the free head is shown as a surface diagram. d In the Lethocerus IHM, free-head MXEs 1 and 5 (purple and blue) contact a neighboring S2 (pink). e A cross section shows the contacts of MXE 2 and 5 (brown and blue) to the thick filament backbone, where the α-helical coiled-coils can be seen (light pink). This view is flipped 180° relative to Fig. 1c. Because this is a rigid-body fitting of 3JBH to the Lethocerus thick filament structure, the three contacts shown here (d, e) should be considered speculative. In contrast, more confidence can be assigned to the tarantula contacts (a–c) because 3JBH was flexibly fit to the tarantula structure. MXE clusters 1, 5, 7, 10 and 11 (a–e purple, blue, green and yellow, and g orange) are alternatively spliced in both arachnids and Lethocerus; whereas clusters 2 and 9 (e brown, and b light blue) are alternatively spliced in arachnids but not Lethocerus, cluster 8 (b magenta) is alternatively spliced in Lethocerus but not arachnids, and cluster 6 (a, c tan) is not alternatively spliced in either but is in other species such as scallops (Kollmar and Hatje 2014). f N-terminal S2 structures from tarantula myosin (3JBH), cardiac myosin (2FXM), and Lethocerus myosin (homology model based on 2FXM) are color coded by electrostatic potential from negative (red) to neutral (white) to positive (blue). The three red rings of negative charge seen in the cardiac structure are numbered. Lethocerus myosin does not appear to have Ring 1. g In the Lethocerus structure, MXE 11 (orange) is within the thick filament backbone made of coiled-coil rod domains (light pink) and contacts two extra proteins (green and red) and the S2 domain (dark pink) of a neighboring molecule. Scalebars = 100 Å
In the folded-back IHM structure seen in tarantula thick filaments, blocked-head MXEs 8 and 9 make an intramolecular contact with their own S2 tail (Fig. 3b). Additionally, free-head MXEs 5 and 6 are involved in intermolecular contacts with the adjacent IHM to the right and one level up, while blocked-head MXEs 1, 7 and 10 are involved in intermolecular contacts with the S2 arising from the IHM to the left and one level down (Fig. 3c). All of these interactions are totally absent or distinct in the Lethocerus thick filament structure. In the Lethocerus thick filament, only MXEs 1 and 5 contact the S2 domain coming from the IHM to the right and one level down (Fig. 3d), and MXE 2 contacts the thick filament backbone (Fig. 3e). In particular, MXE 8 is not positioned to make any potential contacts within the Lethocerus IHM, unlike the tarantula IHM in which blocked-head MXE 8 contacts S2 (Fig. 3b). This structural difference suggests some difference in S2 (and/or MXE 8, discussed later). A crystal structure of this portion of S2 from human cardiac myosin revealed three rings of concentrated negative charge, termed Rings 1–3. The contact between blocked-head MXE 8 and S2 seen in the tarantula structure involves Ring 1 (Blankenfeldt et al. 2006). We built a homology model of S2 using the Lethocerus sequence and it reveals that Lethocerus myosin has much less negative charge at Ring 1, compared to cardiac or tarantula S2 (Fig. 3f). Therefore, lacking the Ring 1 negative charge, Lethocerus myosin may be unable to stabilize the folded-back orientation and thus prefers the perpendicular orientation of the IHM.
Alternatively spliced MXE 11 is located within the α-helical coiled-coil rod domain of myosin, near what is known as the S2-LMM junction. The S2-LMM junction is susceptible to proteolysis and the location of a bend in the rod domain in solubilized molecules, and is therefore thought to act as a hinge (Elliott and Offer 1978; Suggs et al. 2007; Miller et al. 2009). However, the Lethocerus thick filament structure shows this site to be well embedded within the thick filament backbone (Hu et al. 2016) and unlikely to serve as a hinge in this context. MXE 11 is, nevertheless, the site of several interesting interactions (Fig. 3g). It makes contacts with two extra, presumably non-myosin, densities (Hu et al. 2016). It also contacts the S2 domain of the adjacent molecule where that molecule joins the backbone. Previous efforts to pull the entire length of S2 free from the thick filament backbone by swelling rigor muscles failed, and showed only the first 11 nm of S2 (Liu et al. 2006), indicating that the contact between S2 and MXE 11 must be fairly strong.
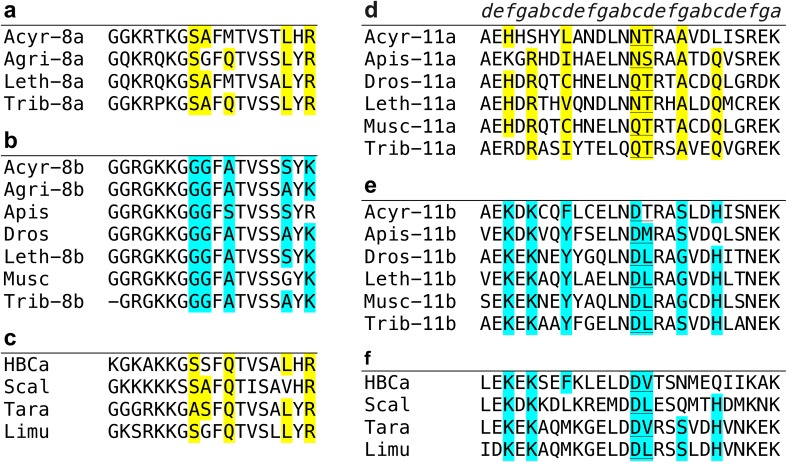
MXEs 8 and 11 typically have two variants if they are alternatively spliced, or a single exon otherwise (Odronitz and Kollmar 2008; Kollmar and Hatje 2014). Lethocerus dorsal longitudinal muscle expresses primarily MXE 8b. Drosophila, Apis, and Musca have a single exon for MXE 8, and they are more 8b-like when compared with insects that retain MXE 8 (Fig. 4a, b). All of these insects have asynchronous flight muscles, similar to Lethocerus. In contrast, sequences from human β-cardiac, scallop, tarantula, and Limulus myosins are more 8a-like (Fig. 4c). All four of these myosins have thick filaments with the folded-back IHM. Similarly, comparison of MXE 11 sequences shows that human β-cardiac, scallop, tarantula, and Limulus myosins are more 11b-like (Fig. 4d–f), in contrast to Lethocerus and Drosophila flight muscles which express MXE 11a (Miller et al. 2009; Suggs et al. 2007; Collier et al. 1990). Therefore, we suggest that this may be a general trend. Thick filaments will show the folded-back IHM if their myosin sequences in these regions are more similar to MXEs 8a and 11b, whereas they will show the perpendicular IHM if their sequences are more similar to 8b and 11a. This idea is complicated by our observation that Lethocerus dorsal longitudinal muscles express both MXE 8b and 8a. However, the Lethocerus IHM structure was based on a subset of thick filaments that excluded the filament ends and selected for perpendicular heads (Hu et al. 2016), so it is possible the reconstruction excluded heads with 8a sequence.
Fig. 4.
Sequence alignments of MXEs 8 and 11 show residues, highlighted in yellow and cyan, that are conserved but different in the two versions of each MXE. a, b MXE 8 sequences from seven insect species, Acyrthosiphon pisum, Agrilus planipennis, Apis mellifera, Drosophila melanogaster, Lethocerus indicus, Musca domestica, and Tribolium castaneum. c Sequences from human β-cardiac, scallop, tarantula, and Limulus myosins are more like the insect 8a versions. d–f In MXE 11, the sequences from human β-cardiac, scallop, tarantula, and Limulus myosins are more like the insect 11b versions. Most notable is a pair of residues at positions 14–15 (underlined). In the 11a versions this pair is polar and uncharged, asparagine or glutamine paired with serine or threonine. In contrast, in the 11b versions it is negatively charged aspartate paired with an aliphatic residue. The canonical heptad repeat for the α-helical coiled-coil is shown at the top of d
Further analysis awaits an atomic model of the rod and a flexible fitting of the Lethocerus IHM structure, which we are currently pursuing.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. Michael Reedy for project inspiration, specimen collection, and dissection. We thank Dr. Belinda Bullard for Lethocerus cDNA, the myosin regulatory light chain sequence, and helpful comments on the manuscript. This work was supported by the United States National Institute of Health, Grant AR014317, and the National High Technology Research and Development Program of China (863), Grant 2015AA020105.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s10974-017-9476-6) contains supplementary material, which is available to authorized users.
References
- Alamo L, Qi D, Wriggers W, Pinto A, Zhu J, Bilbao A, Gillilan RE, Hu S, Padrón R. Conserved intramolecular interactions maintain myosin interacting-heads motifs explaining tarantula muscle super-relaxed state structural basis. J Mol Biol. 2016;428(6):1142–1164. doi: 10.1016/j.jmb.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein SI, Milligan RA. Fine tuning a molecular motor: the location of alternative domains in the Drosophila myosin head. J Mol Biol. 1997;271(1):1–6. doi: 10.1006/jmbi.1997.1160. [DOI] [PubMed] [Google Scholar]
- Bernstein SI, Hansen CJ, Becker KD, Wassenberg DR, 2nd, Roche ES, Donady JJ, Emerson CP., Jr Alternative RNA splicing generates transcripts encoding a thorax-specific isoform of Drosophila melanogaster myosin heavy chain. Mol Cell Biol. 1986;6(7):2511–2519. doi: 10.1128/MCB.6.7.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenfeldt W, Thoma NH, Wray JS, Gautel M, Schlichting I. Crystal structures of human cardiac beta-myosin II S2-Delta provide insight into the functional role of the S2 subfragment. Proc Natl Acad Sci USA. 2006;103(47):17713–17717. doi: 10.1073/pnas.0606741103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier VL, Kronert WA, O’, Donnell PT, Edwards KA, Bernstein SI. Alternative myosin hinge regions are utilized in a tissue-specific fashion that correlates with muscle contraction speed. Genes Dev. 1990;4(6):885–895. doi: 10.1101/gad.4.6.885. [DOI] [PubMed] [Google Scholar]
- Elliott A, Offer G. Shape and flexibility of the myosin molecule. J Mol Biol. 1978;123(4):505–519. doi: 10.1016/0022-2836(78)90204-8. [DOI] [PubMed] [Google Scholar]
- George EL, Ober MB, Emerson CP., Jr Functional domains of the Drosophila melanogaster muscle myosin heavy-chain gene are encoded by alternatively spliced exons. Mol Cell Biol. 1989;9(7):2957–2974. doi: 10.1128/MCB.9.7.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess NK, Singer PA, Trinh K, Nikkhoy M, Bernstein SI (2007) Transcriptional regulation of the Drosophila melanogaster muscle myosin heavy-chain gene. Gene Expression Patterns GEP 7(4):413–422. doi:10.1016/j.modgep.2006.11.007 [DOI] [PMC free article] [PubMed]
- Holmes KC, Rosenbaum G. How X-ray Diffraction with Synchrotron Radiation Got Started. J Synchrotron Radiat. 1998 doi: 10.1107/S0909049597018578. [DOI] [PubMed] [Google Scholar]
- Hooijman P, Stewart MA, Cooke R. A new state of cardiac myosin with very slow ATP turnover: a potential cardioprotective mechanism in the heart. Biophys J. 2011;100(8):1969–1976. doi: 10.1016/j.bpj.2011.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Taylor DW, Reedy MK, Edwards RJ, Taylor KA. Structure of myosin filaments from relaxed Lethocerus flight muscle by cryo-EM at 6 A resolution. Sci Adv. 2016;2(9):e1600058. doi: 10.1126/sciadv.1600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley HE. The mechanism of muscular contraction. Science. 1969;164(886):1356–1365. doi: 10.1126/science.164.3886.1356. [DOI] [PubMed] [Google Scholar]
- Huxley HE, Brown W. The low-angle X-ray diagram of vertebrate striated muscle and its behaviour during contraction and rigor. J Mol Biol. 1967;30(2):383–434. doi: 10.1016/S0022-2836(67)80046-9. [DOI] [PubMed] [Google Scholar]
- Jung HS, Komatsu S, Ikebe M, Craig R. Head-head and head-tail interaction: a general mechanism for switching off myosin II activity in cells. Mol Biol Cell. 2008;19(8):3234–3242. doi: 10.1091/mbc.E08-02-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmar M, Hatje K. Shared gene structures and clusters of mutually exclusive spliced exons within the metazoan muscle myosin heavy chain genes. PLoS One. 2014;9(2):e88111. doi: 10.1371/journal.pone.0088111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wu S, Reedy MC, Winkler H, Lucaveche C, Cheng Y, Reedy MK, Taylor KA. Electron tomography of swollen rigor fibers of insect flight muscle reveals a short and variably angled S2 domain. J Mol Biol. 2006;362(4):844–860. doi: 10.1016/j.jmb.2006.07.084. [DOI] [PubMed] [Google Scholar]
- Menetret JF, Hofmann W, Lepault J. Cryo-electron microscopy of insect flight muscle thick filaments. An approach to dynamic electron microscope studies. J Mol Biol. 1988;202(1):175–178. doi: 10.1016/0022-2836(88)90530-X. [DOI] [PubMed] [Google Scholar]
- Miller MS, Dambacher CM, Knowles AF, Braddock JM, Farman GP, Irving TC, Swank DM, Bernstein SI, Maughan DW. Alternative S2 hinge regions of the myosin rod affect myofibrillar structure and myosin kinetics. Biophys J. 2009;96(10):4132–4143. doi: 10.1016/j.bpj.2009.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naber N, Cooke R, Pate E. Slow myosin ATP turnover in the super-relaxed state in tarantula muscle. J Mol Biol. 2011;411(5):943–950. doi: 10.1016/j.jmb.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odronitz F, Kollmar M. Comparative genomic analysis of the arthropod muscle myosin heavy chain genes allows ancestral gene reconstruction and reveals a new type of ‘partially’ processed pseudogene. BMC Mol Biol. 2008;9:21. doi: 10.1186/1471-2199-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orfanos Z, Sparrow JC. Myosin isoform switching during assembly of the Drosophila flight muscle thick filament lattice. J Cell Sci. 2013 doi: 10.1242/jcs.110361. [DOI] [PubMed] [Google Scholar]
- Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261(5117):50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Reedy MK, Holmes KC, Tregear RT. Induced changes in orientation of the cross-bridges of glycerinated insect flight muscle. Nature. 1965;207(5003):1276–1280. doi: 10.1038/2071276a0. [DOI] [PubMed] [Google Scholar]
- Retelska D, Iseli C, Bucher P, Jongeneel CV, Naef F. Similarities and differences of polyadenylation signals in human and fly. BMC Genomics. 2006;7:176. doi: 10.1186/1471-2164-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum G, Holmes KC, Witz J. Synchrotron Radiation as a source for X-Ray diffraction. Nature. 1971;230(5294):434–437. doi: 10.1038/230434a0. [DOI] [Google Scholar]
- Suggs JA, Cammarato A, Kronert WA, Nikkhoy M, Dambacher CM, Megighian A, Bernstein SI. Alternative S2 hinge regions of the myosin rod differentially affect muscle function, myofibril dimensions and myosin tail length. J Mol Biol. 2007;367(5):1312–1329. doi: 10.1016/j.jmb.2007.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tregear RT, Miller A. Evidence of crossbridge movement during contraction of insect flight muscle. Nature. 1969;222(5199):1184–1185. doi: 10.1038/2221184a0. [DOI] [Google Scholar]
- Wassenberg DR, 2nd, Kronert WA, O’Donnell PT, Bernstein SI. Analysis of the 5′ end of the Drosophila muscle myosin heavy chain gene. Alternatively spliced transcripts initiate at a single site and intron locations are conserved compared to myosin genes of other organisms. J Biol Chem. 1987;262(22):10741–10747. [PubMed] [Google Scholar]
- Wendt T, Taylor D, Messier T, Trybus KM, Taylor KA. Visualization of head-head interactions in the inhibited state of smooth muscle myosin. J Cell Biol. 1999;147(7):1385–1390. doi: 10.1083/jcb.147.7.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhead JL, Zhao FQ, Craig R, Egelman EH, Alamo L, Padron R. Atomic model of a myosin filament in the relaxed state. Nature. 2005;436(7054):1195–1199. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- Woodhead JL, Zhao FQ, Craig R. Structural basis of the relaxed state of a Ca2+-regulated myosin filament and its evolutionary implications. Proc Natl Acad Sci USA. 2013;110(21):8561–8566. doi: 10.1073/pnas.1218462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Bernstein SI. Spatially and temporally regulated expression of myosin heavy chain alternative exons during Drosophila embryogenesis. Mech Dev. 2001;101(1–2):35–45. doi: 10.1016/S0925-4773(00)00549-9. [DOI] [PubMed] [Google Scholar]
- Zhao FQ, Craig R, Woodhead JL. Head-head interaction characterizes the relaxed state of Limulus muscle myosin filaments. J Mol Biol. 2009;385(2):423–431. doi: 10.1016/j.jmb.2008.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi ME, Woodhead JL, Moss RL, Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc Natl Acad Sci U S A. 2008;105(7):2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.