Abstract
In higher plants, miR156 regulates the vegetative phase change via the target SBP/SPL genes. The regulation of miR156 during ontogenetic processes is not fully understood. In the apple genome, of 31 putative MdMIR156 genes that encode pre-miR156, seven were dominantly expressed. However, the transcript levels of only MdMIR156a5 and MdMIR156a12 decreased significantly during the vegetative phase change, which was consistent with the mature miR156 level, indicating that miR156 is under transcriptional regulation. Leaf H2O2 content was higher in the adult phase than in the juvenile phase because of excess H2O2 accumulation in chloroplasts. When in vitro shoots were treated with menadione, diphenyleneiodonium, L-2-oxothiazolidine-4-carboxylic acid or buthionine sulphoximine, the expressions of MdMIR156a5, MdMIR156a12, and as well miR156 were coordinated with reduced glutathione (GSH) contents and glutathione/glutathione disulfide ratio but not H2O2 contents. Alteration of miR156 expression level by MdMIR156a6-overexpressing or miR156-mimetic transgenic Nicotiana benthamiana did not cause a corresponding change in reactive oxygen species or GSH status. Collectively, the results indicate that the vegetative phase change in apple is controlled by the MdMIR156a5 and MdMIR156a12 transcriptional regulatory network in response to the plastid–nucleus redox signals, such as GSH.
Introduction
Perennial plants undergo several developmental transitions during the life cycle, including the transition from an embryonic to post-embryonic mode of growth, the vegetative phase change (juvenile-to-adult vegetative transition), and the floral transition (vegetative-to-reproductive transition)1,2. In apple seedlings, the vegetative phase change occurs at approximately the 80th node, and natural floral induction is initiated at about the 120th node3. The vegetative phase change usually occurs once in a life cycle, but this process is reversible (defined as rejuvenation) under certain circumstances, under which the shoot meristem regains juvenility and most juvenile traits after reproductive maturity is attained4–6. The vegetative phase change is targeted by plant breeders to shorten the breeding cycle, whereas rejuvenation is critical for plant nurseries and propagators of many perennial woody plants to achieve rapid vegetative growth, to improve rooting ability and to realize efficient proliferation7.
The microRNA miR156 regulates the vegetative phase change via inhibition of the target SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) genes8,9. In Malus species, during phase change, miR156 expression declines at about the 80th node and attains a minimum level at about the 140th node; on the other hand, during rejuvenation, miR156 levels are recovered in rejuvenated in vitro shoots after several subculture cycles6,10. The miRNAs can be overexpressed in transformants driven by 35 S promoter or otherwise can be successfully knocked-down by transgenic plants expressing a miRNA-resistant version of a miRNA target or modifying the sequence of the AtIPS1 gene11–15. Overexpression of miR156 in transgenic Populus × canadensis reduces the transcripts of miR156-targeted SPL genes and drastically prolongs the juvenile phase16. The expressions of NbSPLs decreased or increased at least five-fold changes in 35 S:MdMIR156a6 or MIM156 transgenic tobacco (Nicotiana benthamiana) lines, respectively17.
MicroRNAs can be regulated at transcription, post-transcriptional cleavage or both stages. The processing of miRNAs from long primary transcripts (pri-miRNAs) requires the activity of several proteins, including DICER-LIKE 1 (DCL1), the double stranded RNA-binding protein HYPONASTIC LEAVES 1 (HYL1), and the zinc-finger protein SERRATE (SE)18–23. Loss-of-function mutants of HYL1, SE or ARGONAUTE 1 (AGO1) cause defects in the timing of the juvenile phase in Arabidopsis 24–28.
The ontogenetic signal upstream of miR156 is proposed to originate from leaf primordia. Defoliation disrupts miR156 expression, which implies that a mobile signal(s) is (are) derived from the pre-existing leaves29,30. One potential mobile signal that triggers the juvenile-to-adult transition is sugar. Recent study reveals that trehalose-6-phosphate (T6P) pathway regulates flowering at two sites in plant. In the leaves, TREHALOSE-6-PHOSPHATE SYNTHASE 1 (TPS1) activity is required for the induction of the florigen FT, even under inductive photoperiod31. In addition, the T6P pathway affects the expression of important flowering-time and flower-patterning genes at the shoot apical meristem independently of the photoperiod pathway31,32. The loss of TPS1 causes Arabidopsis thaliana to flower extremely late33,34. Sucrose inhibits pri-miR156 transcription and processing. Supplying Arabidopsis plants with exogenous sugar reduces the levels of MIR156A and MIR156C transcripts by 40%, whereas sugar deprivation increases their expression35,36. However, the Arabidopsis mutant gin2-1, which lacks HEXOKINASE 1 (HXK1), is only slightly precocious in the transition to the adult phase, and thus sugar may not be the only factor that regulates miR156 expression35.
Redox signaling hub interacting with phytohormone signaling network controls growth, differentiation, development and environmental response in plants37. Redox homeostasis is well maintained by the precisely regulated generation and scavenging of H2O2, superoxide (O2 −), singlet oxygen (1O2) and other types of reactive oxygen species (ROS)38. H2O2, the most abundant ROS in plants, is produced mainly in chloroplast, apoplast and mitochondria and H2O2 itself is an inorganic second messenger acts on many redox sensitive regulation processes39,40,41. Under none stressful conditions, the dominant H2O2 production sites in plant are chloroplast and peroxisome, and chloroplastic H2O2 induces early signal responses42.
Glutathione (GSH) and ascorbate (ASC) coupling is a central hub of redox regulation, and GSH is considered as an important redox buffer in plants43,44. GSH concentration, GSH/glutathione disulfide (GSSG) ratio and the cellular/subcellular compartmentation are tightly regulated45,46,47.
Several changes in ROS generation and scavenging were age related in some organisms. In Drosophila melanogaster, Musca domestica and Mus musculus, production of GSH declines and that of ROS increases with age48,49. In apple seedlings, the expression of miR156 decreased gradually during ontogenesis. H2O2 contents, glutathione reductase activity and the expressions of some MdGR gene family members increased remarkably. However, the GSH content and GSH to GSSG ratio declined10,50. When H2O2 concentrations of in vitro shoots of an apple seedling are manipulated with menadione (MED) or diphenyleneiodonium (DPI) treatment, the concentrations of GSH was observed extremely lower in MED treated in vitro shoots than in untreated control, and the relative expression of miR156 decreased to a significantly low level. In DPI treated in vitro shoots, a slight but statistically significant increase in GSH was detected, a drastic increase in miR156 expression was observed. When GSH contents were altered with L-2-oxothiazolidine-4-carboxylic acid (OTC) or buthionine sulphoximine (BSO) application, miR156 expression varies concomitantly, but no substantial changes in H2O2 concentrations were detected in OTC and BSO treated in vitro shoots compared to the control10. It seems that the H2O2 levels are not a direct factor than GSH affecting miR156 expression, but the exact relationship between ROS, GSH and miR156 remains unclear.
Cellular ROS can be formed in almost all kinds of organelles. In non-photosynthetic organisms and mammalian cells, more than 90% of cellular ROS is produced in the mitochondria as a result of untimely spontaneous transfer of electrons to oxygen, primarily from complexes I and III of the respiratory chain51,52. In animals, age-related ROS accumulate in mitochondria and the endoplasmic reticulum52. Recent studies have identified respiratory burst oxidase homologues (Rboh), plant homologues of the catalytic subunit of phagocyte NADPH oxidase (gp91phox), as a source of ROS during the apoplastic oxidative burst53. In green plants, excitation of pigments and photosynthetic electron transfer reactions in an oxygen-rich environment lead to the production of ROS54. In higher plants, the site of the dramatically higher H2O2 content in the reproductive phase compared with the juvenile phase is presently unknown.
To understand whether the phase change-associated decrease in miR156 expression level is due to transcriptional regulation by redox homeostasis in plants, in this study on apple (Malus domestica) we first confirmed the ontogenesis-related members of the MdMIR156 gene family. The response of MdMIR156 transcription to the changes in redox status was evaluated using in vitro shoots, and the site of the phase change-associated ROS generation was investigated by transmission electron microscopy.
Results
miR156 is under transcriptional regulation during phase change
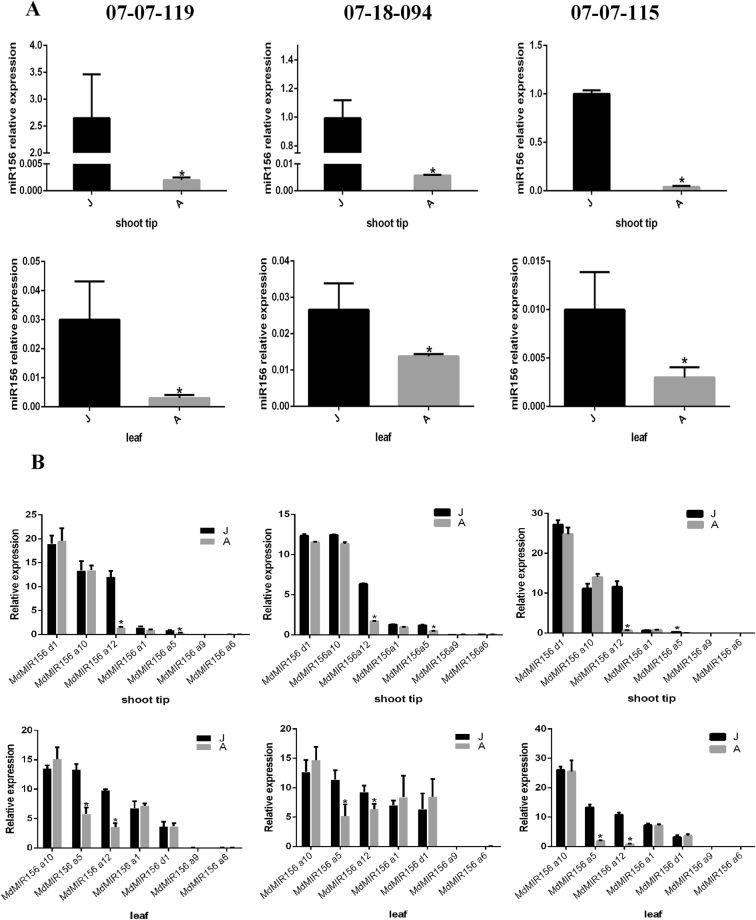
In both leaf and shoot tip samples, the expression level of mature miR156 in the adult phase was dramatically lower than that in the juvenile phase (Fig. 1A).
Figure 1.
Quantitative expression of miR156 (A) and major members of the MdMIR156 gene family (B) in shoot tips and leaves in the juvenile (J) and adult (A) phase of three individuals of Malus asiatica ‘Zisai Pearl’ × M. domestica ‘Red Fuji’. Error bars represent the SD of three experimental replicates. Asterisk represents p < 0.05 (Duncan’s multiple-range test).
Twenty-seven gene family members containing SPL-box were identified in the apple genome, 14 of which were putative targets of miR156 (MdSPL3, 4, 6, 7, 10, 11, 12, 18, 20, 21, 22, 23, 24, 26). MdSPL6 did not express and the transcripts of MdSPL10&11 or MdSPL21&22 were identical to each other. Six MdSPL members (MdSPL26, 24, 3, 20, 18 and 23) were dominantly expressed in the three hybrids (Fig. S1). The reduced levels of miR156 during phase change were also concomitant with significantly and robustly increased expressions of its targets, MdSPL26, MdSPL23, and MdSPL10&11 (Fig. S1).
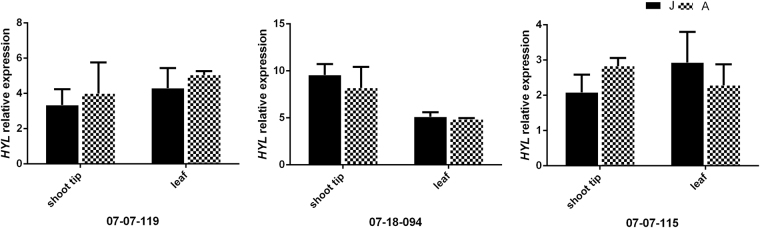
In the apple genome, there are 31 putative MdMIR156 genes encode pre-miR156 (Fig. S5, Table S7). Of these 31 genes, seven were dominantly expressed in shoot tip tissue and the leaf (MdMIR156d1, a10, a12, a1, a5, a9 and a6). Despite the transcripts of MdMIR156d1, MdMIR156a10 and MdMIR156a1 were slightly higher at adult phase than juvenile phase, but their differences were not statistically significant. The transcripts of MdMIR156a5 and MdMIR156a12 exhibited significantly lower levels in the adult phase than in the juvenile phase, which was closely correlated with mature miR156 expression (Fig. 1B). No notable changes in the expression of MdHYL1, which encodes the double-stranded RNA-binding protein HYL1, were detected between the juvenile and adult phases in either shoot tip or leaf samples (Fig. 2). These data indicate that miR156 expression is, at least partially, under transcriptional regulation, and that MdMIR156a5 and MdMIR156a12 are key family members responsive to ontogenetic cues.
Figure 2.
Quantitative expression of MdHYL1 in shoot tips and leaves in juvenile (J) and adult phase (A) of three individuals of Malus asiatica ‘Zisai Pearl’ × M. domestica ‘Red Fuji’. Error bars represent the SD of three experimental replicates.
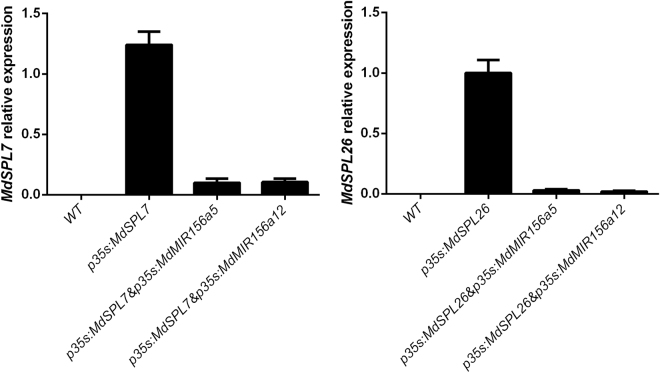
In transiently transformed tobacco, quantitative RT-PCR analysis showed that MdSPL7 and MdSPL26 transcript levels were reduced significantly in both MdMIR156a5 and MdMIR156a12 co-transformed lines (Fig. 3). These results indicated that transcripts of MdMIR156a5 and MdMIR156a12 may degrade MdSPLs, the target of miR156, and thus are indicated to be precursor genes of mature miR156.
Figure 3.
Quantitative expression of MdSPL7 and MdSPL26 in wild and transgenic lines of Nicotiana benthamiana. The expression constructs MdMIR156 (a5 or a12) and MdSPL (MdSPL7 or MdSPL26) were mixed and transformed simultaneously into N. benthamiana by injection. The experiment was performed with three replicates injected with five plaques. The pBI121 vector harboring the GUS reporter gene was also injected to confirm the conversion efficiency. GUS was used as the reference gene. Error bars represent the SD of three experimental replicates.
miR156 is regulated downstream of GSH and ROS
Given that in vitro shoots derived from adult-phase explants undergo rejuvenation, the juvenility of the in vitro shoot changes with subculture cycles6. In the preliminary experiments, in suspension cells derived from apple ‘Orin’ leaves, mature miR156 expression level and redox parameters remained constant and robust with successive subculture cycles. Therefore, suspension cells were an ideal system to use for redox treatments (Figs S2–S4).
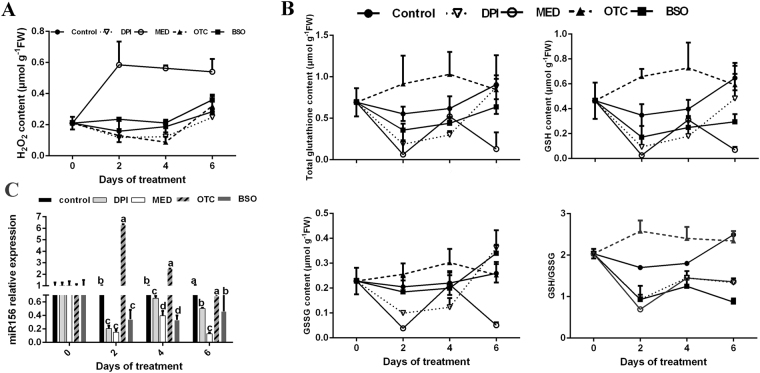
The H2O2 concentration of suspension cells on MED-supplemented medium increased significantly as expected on days 2–6 of treatment (Fig. 4A). In contrast, H2O2 concentration in DPI-treated suspension cells was lower than that of the untreated control on days 2–4 (Fig. 4A). No significant changes in H2O2 were detected in response to OTC and BSO treatment compared with the untreated control (Fig. 4A).
Figure 4.
Changes in hydrogen peroxide (H2O2) concentration (A), glutathione (GSH) content, glutathione/glutathione disulfide (GSH/GSSG) (B) and relative expression of mature miR156 (C) of suspension cells of apple ‘Orin’ leaf treated with redox-modulating chemicals. The suspension cells were cultured for six days in medium supplemented with either 50 μM menadione (MED), 5 μM diphenyleneiodonium (DPI), 50 μM L-2-oxothiazolidine-4-carboxylic acid (OTC) or 0.5 mM buthionine sulphoximine (BSO). Error bars represent the SD of three biological replicates.
The concentrations of GSH, GSSG, and GSH + GSSG, and the GSH/GSSG ratio decreased significantly on days 2–6 in the MED treatment (Fig. 4B). Compared with the untreated control, in the DPI treatment no obvious changes in GSSG concentrations occurred throughout the experimental period, whereas a significant decline in GSH, GSH + GSSG and GSH/GSSG ratio were detected on days 2–6 (Fig. 4B). On days 2–4 of OTC treatment, a dramatic increase in concentration of reduced GSH was observed and consequently the GSH/GSSG ratio was elevated (Fig. 4B). In the suspension cells cultured in BSO-supplemented medium, significant depletions in reduced GSH and total GSH + GSSG concentrations were observed on days 2–6 of treatment. However, the GSSG concentration did not change, resulting in a low GSH/GSSG ratio throughout the experiment (Fig. 4B).
The relative expression level of miR156 in the MED, DPI, and BSO treatments were significantly decreased on days 2–6, whereas a dramatic increase in miR156 expression level was observed in the OTC treatment except on day 6 (Fig. 4C). These results prompted us to examine the expressions of miR156 targets MdSPL26, MdSPL23, and MdSPL10&11, which responded to phase change. qRT-PCR analysis revealed that the expressions of MdSPL26 was elevated in BSO treatment but decreased in response to OTC treatment (Fig. S6). Thus the impacts of GSH on miR156 expression were established.
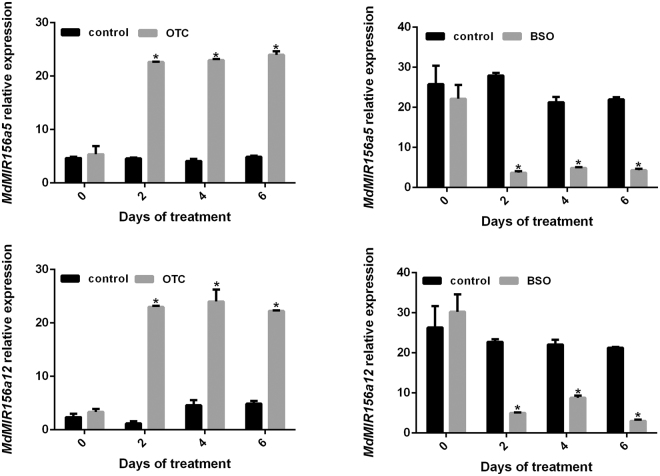
We examined the transcript levels of MdMIR156a5 and MdMIR156a12 in response to OTC and BSO treatment. The transcript levels of MdMIR156a5 and MdMIR156a12 increased by up to 20-fold in response to OTC treatment, but decreased significantly in response to BSO treatment (Fig. 5). These changes were consistent with the changes in miR156 expression level.
Figure 5.
The relative expression of MdMIR156a5 and MdMIR156a12 of suspension cells of apple ‘Orin’ leaf treated with redox-modulating chemicals. The suspension cells were cultured for six days in medium supplemented with either 50 μM L-2-oxothiazolidine-4-carboxylic acid (OTC) or 0.5 mM buthionine sulphoximine (BSO). Error bars represent the SD of three biological replicates. Asterisk represents p<0.05 (Ducan's multiple-range test).
Thus, when suspension cells were treated with exogenous redox modulators, the changes in transcript levels of MdMIR156a5, MdMIR156a12, and mature miR156 were consistent with the changes in reduced GSH concentration and the GSH/GSSG ratio, and showed no direct correspondence with H2O2 concentration.
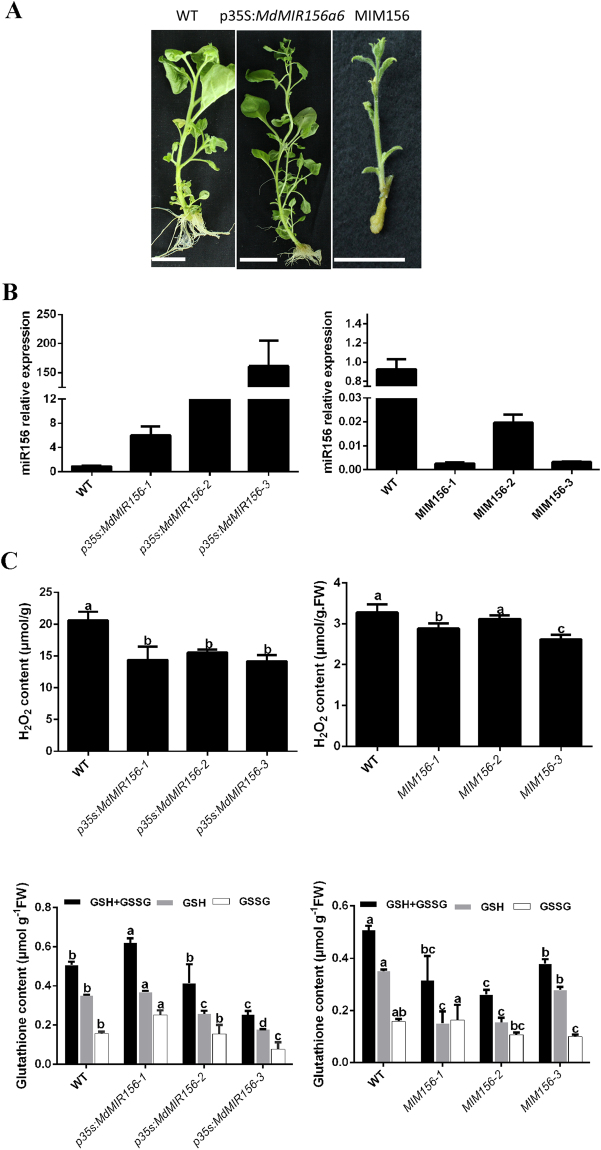
To test whether the changes in miR156 expression level may affect redox homeostasis, an expression construct containing the miR156 precursor MdMIR156a6 under the control of the Cauliflower mosaic virus 35 S promoter, and a target mimic, consisting of a non-cleavable RNA that formed a non-productive interaction with a complementary miR156 (MIM156) to inhibit the activity of miR156, were genetically transformed into Nicotiana benthamiana. The transgenic plants showed multiple morphological changes; 35 S:MdMIR156a6-overexpressing plants showed increased number of leaves (double that of the wild type [WT]), shorter internode length, and inhibition of flowering, whereas the transgenic MIM156-overexpressing plants flowered profusely (Fig. 6A).
Figure 6.
Nicotiana benthamiana transformant in vitro shoots constitutively expressing MIM156 or MdMIR156a6. (A) Phenotypes of Nicotiana benthamiana transformant. (B) Expression analysis of miR156 in Nicotiana benthamiana transformant in vitro shoots constitutively expressing MIM156 or MdMIR156a6 compared with the wild type (WT). 5S was used as a loading control. (C) Changes in H2O2 concentration and glutathione concentration in Nicotiana benthamiana transformant in vitro shoots constitutively expressing MIM156 or MdMIR156a6 compared with the wild type (WT). Error bars represent the SD of three biological replicates. Different letters indicate statistical significance (p < 0.05; Duncan’s multiple-range test).
Three independent MdMIR156a6 overexpression transgenic lines (p35S:MdMIR156-1, p35S:MdMIR156-2, and p35S:MdMIR156-3) and three miR156 mimetic transgenic lines (MIM156-1, MIM156-2, and MIM156-3) were chosen for further analysis. MdMIR156a6 overexpression transgenic plants accumulated much higher quantities of mature miR156 than the WT plants, whereas miR156 expression was greatly inhibited in the MIM156 transgenic plants (Fig. 6B).
The H2O2 and GSH concentrations showed no regular differences between the MdMIR156a6-overexpressing and miR156-mimetic transgenic lines, and also between the WT and transgenic lines (Fig. 6C). These data indicated that the change in miR156 expression level did not affect H2O2 and glutathione concentrations constantly. Taken together, these results indicated that miR156 was regulated downstream of ROS and GSH.
Changes in sugar metabolism during phase change
To determine whether sugar is involved in the phase change in apple seedlings, we examined the contents of glucose, fructose, the activity of TPS and the expression of MdHXK gene family members. The glucose content was significantly higher in adult phase than those in juvenile phase in 07-07-115, but no significant differences were detected between juvenile and adult tissue in 07-07-119 and 07-18-094, and no significant variation in TPS activity was found between the he juvenile and adult phases (Fig. S7). Of the ten members of MdHXK gene in apple genome, eight (except MdHXK6 and MdHXK10) were actively expressed in leaf tissue in apple. To get better convincing data, nine hybrid trees were used for analyzing the variations in MdHXKs expression patterns between juvenile and adult phase and to consider if MdHXKs were co-expressed with miR156. The data showed obviously that the miR156 expressions were significantly higher in the juvenile phase, which was consistent among all the nine hybrids (Fig. S8). The expressions of MdHXK5 and MdHXK8 were higher in juvenile than in adult leaves in 07-07-115, and 07-07-119, respectively, however, the expressions of MdHXK1, MdHXK2 and MdHXK5 were significantly higher in the adult phase in 07-05-017, 07-18-094, 07-06-140 and 07-07-119, respectively (Fig. S8). Together, no substantial and consistent variations in sugar metabolism can be concluded between the juvenile and the adult phase.
The site of phase-related elevation in ROS concentration
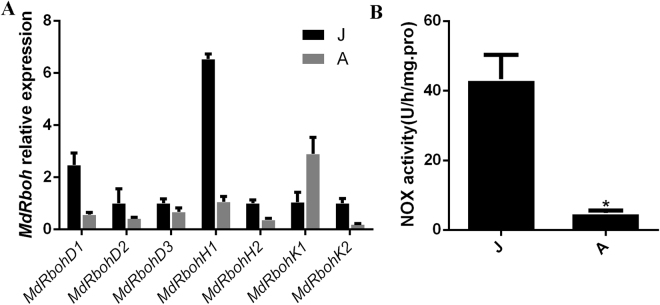
To confirm whether or not the phase-related changes in ROS concentration are generated from the apoplast, NADPH oxidase (NOX) activity and expression of Rboh gene family members in leaf samples of apple hybrid trees was measured. The apple genome contains seven members in MdRboh gene family, of which six (D1, D2, D3, H1, H2, and K2) showed more active transcription in the juvenile phase, especially MdRboh H1 (Fig. 7A). Only the transcript level of MdRboh K1 was higher in the adult phase than in the juvenile phase (Fig. 7A). Consistently, NOX activity was significantly higher in the juvenile phase than in the adult phase (Fig. 7B). Changes in NOX activity and MdRboh gene transcription were not consistent with H2O2 dynamics during the phase change and therefore the ontogenetic-specific elevation in ROS concentration was not generated in the apoplast.
Figure 7.
Quantitative expression of members of the MdRboh gene family (A) and NADPH oxidase (NOX) activity (B) in the juvenile (J) and adult (A) phase of Malus asiatica ‘Zisai Pearl’ × M. domestica ‘Red Fuji’. Error bars represent the SD of three biological replicates. Asterisk represents p < 0.05 (Duncan’s multiple-range test).
Subsequently we measured the concentration of H2O2, GSH miR156 and miR156-targeted AtSPL genes (AtSPL3 and AtSPL9) expression in Atrbohd and Atrbohf mutant. In Atrbohd mutant, the concentration of H2O2, GSH, expressions of miR156 and AtSPLs did not differ significantly from the wild-type, in Atrbohf mutant, however, there was a significant decrease of GSH content, GSH/GSSG ratio and miR156 transcripts, the expression of AtSPL9 correspondingly increased (Figs S9, S10). These data from Arabidopsis further support that GSH, but not H2O2, affects miR156 expression.
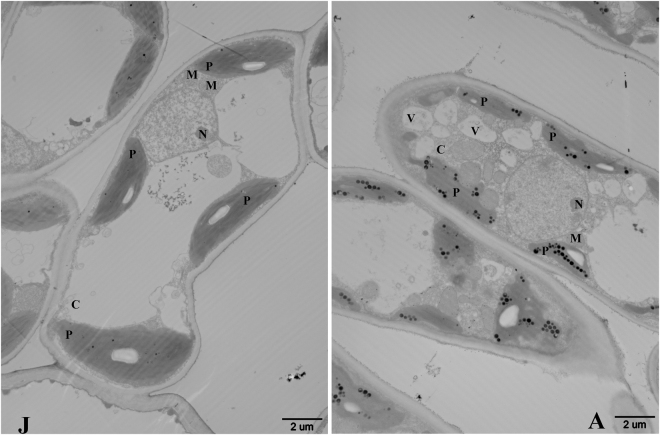
To further confirm if the phase-related ROS are produced in plastids, subcellular H2O2 compartmentation was visualized using CeCl3 staining and transmission electron microscopy. Accumulation of a large amount of H2O2 was clearly observed in the chloroplast in leaf samples of the adult phase, in contrast, deposits of CeCl3 were barely detected in leaf samples of the juvenile phase (Fig. 8). No obvious differences in CeCl3 deposition were observed in the cytosol, nucleus, and other subcellular compartments between leaf samples of the juvenile and adult phases. These data indicated that the phase-related ROS are generated and accumulated in plastids in cells of the adult phase.
Figure 8.
Cytochemical detection of H2O2 accumulation in the juvenile (left) and adult phase (right) of Malus asiatica ‘Zisai Pearl’ × M. domestica ‘Red Fuji’. H2O2 was visualized by transmission electron microscopy at the subcellular level using CeCl3 staining. P: plastid, M: mitochondrion, N: nucleus, V: vacuole, C: cytoplasm.
Discussion
The microRNA 156 control of the vegetative phase change in plants is under transcriptional regulation. In the apple genome, 31 MdMIR156 genes were identified by RNA library sequencing and bioinformatic analysis55. Prior to the current study, only one MdMIR156 precursor (MdMIR156h) has been experimentally verified to be processed into mature miR15656. Then, we have previously validated that MdMIR156a6 is one of the coding genes for miR156 precursor, because several NbSPLs were down-regulated in 35 S:MdMIR156a6 transgenic tobacco lines, but increased in 35 S:MIM156 lines17. We confirmed in this study MdMIR156a5 and MdMIR156a12 can also be transcribed and processed into mature miR156 by using the same approaches.
The present results showed that of the seven family members predominantly expressed in leaf and shoot tip tissues, only transcript abundance of MdMIR156a5 and MdMIR156a12 were significantly higher in juvenile than adult phases, which is consistent with the dynamics of miR156 expression. In leaf or stem samples of some seedlings, the expressions of MdMIR156a1, MdMIR156a10 and MdMIR156d1 were slightly higher in adult phase, but the variations were not statistically significant and were inconsistent among seedling individuals. Given that not all the members of the MdMIR156 gene family responded to ontogenetic signals, additional functions of miR156 may be unexplored. In Arabidopsis thaliana, for example, auxin induces expression of MIR156B and MIR156D in roots and subsequently increases miR156 expression during lateral-root growth57. When the H2O2 or GSH concentrations of suspension cells were altered by BSO or OTC treatment, the levels of MdMIR156a5 and MdMIRa12 transcripts as well as the mature miR156 expression level changed correspondingly. Consequently, the changes in miR156 levels were concomitant with the expression of the dominant ontogenesis responsive miR156 target, MdSPL26, which is highly consistent with our previous findings in M. xiaojinensis 17.
The present results are strongly supported by an analogous observation that the developmental stage associated decline in miR156 expression level is partially mediated by sugar at the transcriptional level of MIR156A and MIR156C in Arabidopsis 35. MdHYL1, a homolog of HYL1 in Arabidopsis thaliana involved in processing of primary miRNA precursors, did not vary in transcription level between the juvenile and adult phases in leaf and shoot tip tissue. This finding is insufficient to exclude the contribution of post-transcriptional processing of primary miR156 precursors on the decline in mature miR156, but it does not conflict with the hypothesized transcriptional regulation of miR156 during the vegetative phase change.
MdMIR156 transcription is regulated downstream of redox signals. In our previous experiments, in which in vitro shoots were treated with redox-modulating chemicals, miR156 transcript levels varied with GSH concentration and GSH/GSSG ratio but not with H2O2 concentration10. In the present study, we used suspension cells to avoid variation in miR156 expression with successive subculture cycles. In response to OTC or BSO treatment, the transcription of MdMIR156a5, MdMIR156a12 and thus mature miR156 varied depending on GSH concentration and GSH/GSSG ratio. Conversely, whereas miR156 transcript level was modulated by transformation with MIM156 and MdMIR156 overexpression, the H2O2 and GSH concentration in transgenic N. benthamiana plants did not change consistently, which suggested that H2O2 and GSH are involved upstream of the miR156 transcriptional regulatory network.
The adult-phase-specific ROS are generated in plastids. During the adult phase, the H2O2 concentration increases dramatically, as does the concentrations or activities of ROS scavengers10. These data indicate that the accumulation of H2O2 is not because of decreased scavenging capacity, but because of elevation in H2O2 production. Both NOX activity and MdRboh transcript levels were higher (except MdRboh K1) in the juvenile phase than in the adult phase, therefore the adult-phase-related H2O2 is not produced in the apoplast. In Atrbohd mutant, the H2O2 and GSH contents did not change, RbohD could not be the crucial member in the redox pathways. In Atrbohf mutants, the H2O2 content did not change at the tissue level, but the apoplast H2O2 decreased52. Because the activity of apoplast γ- glutamyltransferase, which degrades oxidized GSH adducts including GSSG for cysteine recycling, is suppressed when apoplast redox environment gets more reductive, the GSH biosynthesis in chloroplast will be suppressed too due to restricted cysteine availability58,59,60. Therefore, both GSH concentration and thus miR156 transcript level declined significantly (Fig. S11).
Because sugar is once proposed as a mobile signal associated with the vegetative phase change in Arabidopsis 35,36. TPS1 activity is necessary for floral induction31–34. To date, there are no data illustrating the relationship among sugar, GSH and miR156 expression. In this study, changes in GSH level and miR156 expression were closely correlated, but we did not find significant and consistent variations in sugar contents and TPS enzyme activity between the juvenile and the adult phase in three apple hybrid trees. In further, none of the eight MdHXK gene family members exhibited significant and robust variation between the juvenile and the adult phase. We thus also suspected that the variations in MdHXKs expressions between ontogenetic phases were caused by the factors independent with miR156 levels, which agreed to the postulation by Yang and the colleagues35.
Recently, in Arabidopsis, miR159 has been found to modulate vegetative phase change upstream of miR156 through MYB3361. Though the expressions of both pri-MIR156A and mature miR156 are much higher during growth in mir159ab mutant than in wild type Col-0, but they still decline with days after planting, which indicates an independent mechanism is more powerful on regulating vegetative phase change61. Guo and the co-workers proposed that miR159-MYB33 pathway acts on vegetative phase change may be dependent on light61. In photosynthetic plants, chloroplast is not only the major subcellular light sensor and receptor, but also one of the major ROS generating sites39–41,42. Transmission electron microscopic observations revealed that in the adult phase H2O2 accumulated only in chloroplasts, indicating that plastids are the main site of phase-related ROS production. No differences in H2O2 concentration in subcellular compartments other than chloroplasts were detected between the juvenile and adult phases, although H2O2 is permeable across membranes62. The other forms of ROS, such as singlet oxygen or superoxide anion radicals, are considered to be impermeable63. Although ROS molecules are established as modulators of gene expression, it is widely accepted that ROS themselves cannot act as signaling molecules in the nucleus because of their high reactivity64. Hence, we propose that subsequent retrograde redox signals, rather than chloroplast ROS per se, may act on MdMIR156 transcription. During phase change in apple, the contents of H2O2 and GSH varied significantly within a considerable extent, in chemically-treated apple in vitro shoots, concentrations of H2O2 and GSH changed drastically, even to an extreme level10,65,66,67. But in all cases, miR156 levels varied with GSH concentration and GSH/GSSG ratio but not directly with H2O2 concentration10. In the present experiment, the H2O2 concentration of suspension cells was either enhanced in response to MED or inhibited by DPI treatment, whereas both GSH concentration and miR156 transcript level declined significantly. MED and DPI alter apoplast H2O2 production and the apoplast redox state may affect GSH regeneration activity58,59,60. On the other hand, GSH levels were altered by OTC or BSO treatment, MdMIR156s and mature miR156 transcription varied with GSH concentration, but H2O2 concentration did not change throughout the experimental period. In Atrbohf mutant, the miR156 transcription also changed with GSH concentration. These findings implied that GSH may act downstream of ROS in the MdMIR156 transcription regulatory network. We propose that GSH may be an element of the plastid-nucleus retrograde redox signals, because GSH nucleus sequestration functions as a retrograde signal closely associated with developmental signals68,69. Nuclear GSH may create suitable redox environments for DNA synthesis and repair70. As a result, GSH stimulates cell proliferation and cell differentiation in meristematic tissues71,72. In future studies on the ontogenesis in plants, we recommend that greater attention should be paid to GSH.
Conclusions
During the vegetative phase change in apple, ROS accumulate and miR156 transcription declined. The phase-related ROS were generated and accumulated in plastids. In the apple genome, of the 31 putative genes that encode precursors of miR156, MdMIR156a5 and MdMIR156a12 responded to ontogenetic regulation, and these two genes were transcriptionally regulated downstream of redox signals such as GSH.
Methods
Plant materials and chemical treatments
Eight-year-old trees raised from hybrid seeds of the cross Malus asiatica ‘Zisai Pearl’ × M. domestica ‘Red Fuji’ were used. ‘Zisai Pearl’ is a Chinese domestic cultivar originating from Hebei. Given the hyper-heterozygosity of the parents and strong segregation among the hybrids, to estimate the genetic variation between individuals, three intact seedlings grown on their own roots, namely 07-07-115, 07-07-119 and 07-18-094, were sampled as biological replicates. To validate the expression patterns of some genes in a larger population, six additional seedlings were chosen randomly (Fig. S7). Young leaves and unlignified shoot tips were sampled from 1-year-old suckers and annual branches. The samples taken from the 1st to the 80th nodes were defined as the juvenile phase, and those from the 120th node to the canopy top as the adult phase73. The leaves or the shoot tips collected from the same ontogenetic phase were respectively mixed and divided into three experimental replicates, frozen immediately and stored at −80 °C. The transgenic plants 35 S:MdMIR156a6 and 35 S:MIM156 were described previously15 , 17.
To validate the changes in miR156 in response to GSH or redox homeostasis alteration, an ideal experimental system with constant and robust miR156 expression is necessary, because in plant the ROS or GSH levels and miR156 expression varied with the process of vegetative phase change and as well subculture cycles during rejuvenation6,10. As porcine granulosa cells were used in several studies in mammalian74,75, developmentally de-differentiated suspension cells derived from the leaf of M. domestica cultivar ‘Orin’ were treated by addition of redox modulating chemicals to the culture medium. In ‘Orin’ suspension cells, the redox parameters and the expression of miR156 remained constant and robust with successive subculture cycles (Figs S2–S4). The concentrations used in these experiments were optimized in preliminary tests (Supplementary Tables S1–S4). MED (50 µM) (Sigma-Aldrich, Beijing, China) and 5 µM DPI (Sigma-Aldrich) were used as an inducer and inhibitor of H2O2, respectively, and 50 µM OTC (Sigma-Aldrich) and 0.5 mM BSO (Sigma-Aldrich) were used as a precursor and inhibitor of GSH biosynthesis, respectively. Suspension cells cultured on medium containing no redox-altering reagent were used as the control. The experiment employed a completely randomized experimental design with three replicates. The sampling time points were on days 0, 2, 4 and 6 of the respective treatments.
The Atrbohd and Atrbohf mutants in Arabidopsis thaliana were obtained from College of Biological Sciences, China Agricultural University. The seeds were surface-sterilized with 2.5% NaClO then sown onto plates containing Murashige and Skoog media containing 3% sucrose and 0.75% agar. The measurements were done using two-week-old seedlings. The qRT-PCR primers for AtSPL3, AtSPL9 and UBQ10 were referenced to Yang et al.35.
Relative expression levels of miR156, MdHYL1, MdMIR156 and MdSPL family members
The expression levels of mature miR156 were analyzed using the method described by Du et al.10. From 500 mg of leaf sample, microRNA was extracted using the RNAiso for Small RNA kit (9753Q, Takara, Dalian, China) following the manufacturer’s instructions. For reverse transcription, 2 μg microRNA extract was diluted in 12 μL diethyl pyrocarbonate (DEPC) water plus 1 μL of dNTPs (10 mMol·L-1, Takara, Dalian, China) and 1 μL stem-loop primer (10 μM). The mixture was incubated at 70 °C for 5 min, and then immediately put on ice for 5 min. Afterwards, 1.25 μL dNTPs (10 mMol·L-1, Takara), 2.5 μL buffer (5 × ) (Takara), 0.6 μL RNasin, and 1 μL M-MLV (5U μL−1, Takara) were added to the extracted solution. Reverse transcription was accomplished at 42 °C for 1 h, followed by 70 °C for 10 min. RT-qPCR of miR156 was performed as follows:10 μL miRcute miRNA Premix (2×), 0.4 μL upstream primers, 0.4 μL downstream primers and 2 μL template (cDNA), and DEPC water to 20 μL. Program: 94 °C for 2 min, and 40 cycles of 94 °C for 20 s, and 60 °C for 34 s. RT-qPCR was carried out using the ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The primer sequences used for miR156 and 5S rRNA are listed in Supplementary Table S5.
For the MdMIR156 expression assay, total RNA was extracted using a modified cetyltrimethylammonium bromide method76. The sequences of all genes were obtained from Xia et al.55. The mature sequences of microRNAs are designated with ‘miR’ as prefix in the database, whereas miRNA genes are given names of the form such as ath-MIR166a. Lettered suffixes describe distinct loci expressing all related mature miRNAs77,78. The sequences of MdHYL1 (MDP0000304933) and MdSPLs were obtained from the apple genome website (http://genomics.research.iasma.it/) by means of a basic local alignment search tool (BLAST) search. β-Actin was used as the reference gene. The primer pairs used are listed in Supplementary Table S6.
Plasmid construction and genetic transformation
To verify that MdMIR156 genes can be processed into mature miR156, the expression constructs MdMIR156 (a5 or a12) and MdSPL (MdSPL7 - MDP0000170630 or MdSPL26- MDP0000142582) were mixed and transformed simultaneously into Nicotiana benthamiana by injection79. The experiment was performed with three replicates injected with five plaques. The pBI121 vector containing the β-glucuronidase (GUS) reporter gene was also injected to confirm the conversion efficiency. The transient expression of the transgenes was detected by quantitative RT-PCR. GUS was used as the reference gene.
Assay of redox homeostasis
Concentrations of H2O2 and glutathione were determined using methods described previously10,80.
NOX activity and relative expression of MdRboh gene family members
The activity of NOX was measured following the method of Yang et al.81. The sequences of Rbohs were obtained from the apple genome website (http://genomics.research.iasma.it/) by means of a BLAST search. Based on the BLASTP search results for the proteins encoded by the examined genes in the National Center of Biotechnology Information database (http://www.ncbi.nlm.nih.gov/), primers were designed using the Primer Express 5.0 software (AuGCT, Beijing, China). The Rboh gene members were named in accordance with Darius et al.82. The primer pairs used are listed in Supplementary Table S8.
Analysis of sugar contents, TPS activity and HXK gene expression
Leaves samples of juvenile and adult phase of three hybrid trees were used in these experiments as biological replicates. Soluble sugars were extracted and the concentrations of glucose and fructose were separated with a high performance liquid chromatography (Agilent Technologies, CA, USA) and were determined with a refractive index detector (Agilent Technologies, CA, USA) according to the protocols described by Liang et al.83. The TPS enzyme activity was assayed by using the method of Garg et al.84. The sequences of HXKs were obtained from the apple genome website (http://genomics.research.iasma.it/) by means of a BLAST search. The primer pairs for HXK gene members were designed with Primer 5.0 and the sequences are listed in Supplementary Table S9.
Cytochemical detection of H2O2
Hydrogen peroxide was visualized by transmission electron microscopy at the subcellular level using CeCl3 staining81. Tissue samples (2 mM × 2 mM) were excised from the leaves of the juvenile or adult phases, then vacuum infiltrated with freshly prepared 5 mM CeCl3 in 50 mM 3-(N-morpholino)-propane-sulfonic acid at pH 7.2 for 30 min. The tissue samples were then fixed in 2.5% (v/v) glutaraldehyde in 0.2 M sodium phosphate buffer (PBS), pH 7.2, for 1 h at room temperature and kept overnight at 4 °C. After fixation, the tissue samples were washed twice for 10 min in PBS and dehydrated in a graded acetone series (30, 50, 70, 80, 90 and 100% [v/v]), progressively embedded in increasing concentrations of acetone–resin mixtures, and incubated over two 24 h replacements of pure epoxy resin before polymerization at 60 °C for 48 h. Ultrathin sections were cut, using a diamond knife mounted on nickel grids, and examined without staining with a transmission electron microscope (JEM-1230, JEOL, JAPAN) at an accelerating voltage of 80 kV.
Statistical analysis
All experimental data were subjected to analysis of variance and Duncan’s multiple-range test. Differences were defined as significant at p < 0.05.
Data Availability
All data generated or analysed during this study are included in this published article and its Supplementary Information files.
Electronic supplementary material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC) [Grant no. 31372020; 31672107]; the earmarked fund for China Agriculture Research System (CARS-27); Key Laboratory of Biology and Genetic Improvement of Horticultural Crops (Nutrition and Physiology), Ministry of Agricultural, and also PXM2017_014207_000043. We would like to thanks Prof. Shuhua Yang for kindly providing us Atrbohd and Atrbohd mutants.
Author Contributions
Xinzhong Zhang conceived and designed the experiments, Xiaolin Jia, Xiaozhao Xu and Fei Shen conducted the experiments, Xiaolin Jia, Ya Kun Chen, Qing Bo Zheng, Zhen Du and Xinzhong Zhang analysed the data, wrote the manuscript and coordinated its revision. Yi Wang, Ting Wu, Xue Feng Xu and Zhen Hai Han read and provided helpful discussions, and approved the final version.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-14671-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Poethig RS. Phase change and the regulation of developmental timing in plants. Science. 2003;301:334–346. doi: 10.1126/science.1085328. [DOI] [PubMed] [Google Scholar]
- 2.Poethig RS. Vegetative phase change and shoot maturation in plants. Curr Top Dev Biol. 2013;105:125–152. doi: 10.1016/B978-0-12-396968-2.00005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang XZ, et al. Potential polyphenol markers of phase change in apple (Malus domestica) J Plant Physiol. 2007;164:574–580. doi: 10.1016/j.jplph.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Hackett W. P. Juvenility, maturation, and rejuvenation in woody plants. Horti Rev p:109–56 (1985).
- 5.Zimmerman RH, Hackett WP, Pharis RP. Springer. 1985. Hormonal aspects of phase change and precocious flowering, in hormonal regulation of development III; pp. 79–115. [Google Scholar]
- 6.Xiao ZF, et al. The lose of juvenility elicits adventitious rooting recalcitrance in apple rootstocks. Plant Cell Tiss Organ Cult. 2014;119:51–63. doi: 10.1007/s11240-014-0513-5. [DOI] [Google Scholar]
- 7.Meilan R. Floral induction in woody angiosperms. New Forests. 1997;14:179–202. doi: 10.1023/A:1006560603966. [DOI] [Google Scholar]
- 8.Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu G, et al. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138:750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du Z, et al. Redox homeostasis and reactive oxygen species scavengers shift during ontogenetic phase changes in apple. Plant Sci. 2015;236:283–294. doi: 10.1016/j.plantsci.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Jones-Rhoades MW, Bartel DP, Bartel. B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 12.Schwab R, et al. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8:517–27. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 13.Kim J, et al. MicroRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. 2005;42:84–94. doi: 10.1111/j.1365-313X.2005.02354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mallory AC, et al. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J. 2004;23:3356–64. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franco-Zorrilla JM, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 16.Wang JW, et al. MiRNA control of vegetative phase change in trees. PLoS Genet. 2011;7:e1002012. doi: 10.1371/journal.pgen.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu XZ, et al. High miR156 expression is required for Auxin-induced adventitious root formation via MxSPL26 independent of PINs and ARFs in Malus xiaojinensis. Front Plant Sci. 2017;8:1059. doi: 10.3389/fpls.2017.01059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiraguri A, et al. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol Biol. 2005;57:173–188. doi: 10.1007/s11103-004-6853-5. [DOI] [PubMed] [Google Scholar]
- 19.Kurihara Y, Takashi Y, Watanabe Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006;12:206–212. doi: 10.1261/rna.2146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J. SERRATE: a new player on the plant microRNA scene. Embo Rep. 2006;7:1052–1058. doi: 10.1038/sj.embor.7400806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong Z, Han MH, Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. P Natl Acad Sci USA. 2008;105:9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang SW, et al. Structure of Arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing. Structure. 2010;18:594–605. doi: 10.1016/j.str.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Machida S, Chen HY, Yuan YA. Molecular insights into miRNA processing by Arabidopsis thaliana SERRATE. Nucleic Acids Res. 2011;39:7828–7836. doi: 10.1093/nar/gkr428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McConnell JR, Barton MK. Effect of mutations in the PINHEAD gene of Arabidopsis on the formation of shoot apical meristems. Dev Genet. 1995;16:358–366. doi: 10.1002/dvg.1020160409. [DOI] [Google Scholar]
- 25.Clarke JH, Tack DT, Findlay K, Van Montagu M, Van Lijsebettens M. The SERRATE locus controls the formation of the early juvenile leaves and phase length in Arabidopsis. Plant J. 1999;20:493–501. doi: 10.1046/j.1365-313x.1999.00623.x. [DOI] [PubMed] [Google Scholar]
- 26.Liu Q, et al. The ARGONAUTE10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. Plant J. 2009;58:27–40. doi: 10.1111/j.1365-313X.2008.03757.x. [DOI] [PubMed] [Google Scholar]
- 27.Ji L, et al. ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet. 2011;7:e1001358. doi: 10.1371/journal.pgen.1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li SX, Yang X, Wu FJ, He YK. HYL1 controls the miR156-mediated juvenile phase of vegetative growth. J Exp Bot. 2012;63:2787–2798. doi: 10.1093/jxb/err465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashby A. Studies on the morphogenesis of leaves I. An essay on leaf shape. New Phytol. 1948;47:153–176. doi: 10.1111/j.1469-8137.1948.tb05098.x. [DOI] [Google Scholar]
- 30.Yang L, Conway SR, Poethig RS. Vegetative phase change is mediated by a leaf-derived signal that represses the transcription of miR156. Development. 2011;138:245–249. doi: 10.1242/dev.058578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu S, Lian H, Wang JW. Plant developmental transitions: the role of microRNAs and sugars. Curr Opin Plant Biol. 2015;27:1–7. doi: 10.1016/j.pbi.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Wahl V, et al. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science. 2013;339:704–707. doi: 10.1126/science.1230406. [DOI] [PubMed] [Google Scholar]
- 33.Van Dijken AJ, Schluepmann H, Smeekens SC. Arabidopsis trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiol. 2004;135:969–977. doi: 10.1104/pp.104.039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gomez LD, Gilday A, Feil R, Lunn JE, Graham IA. AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. Plant J. 2010;64:1–13. doi: 10.1111/j.1365-313X.2010.04312.x. [DOI] [PubMed] [Google Scholar]
- 35.Yang L, Xu ML, Koo Y, He YJ, Poethig RS. Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. Elife. 2013;2:e00260. doi: 10.7554/eLife.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu S, et al. Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. Elife. 2013;2:e00269. doi: 10.7554/eLife.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Considine MJ, Foyer CH. Redox regulation of plant development. Antioxid Redox Sign. 2014;21:1305–1326. doi: 10.1089/ars.2013.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foyer CH, Noctor G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell. 2005;17:1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrov V. D. & Van Breusegem, F. Hydrogen peroxide—a central hub for information flow in plant cells. AoB Plants, pls014 (2012). [DOI] [PMC free article] [PubMed]
- 40.Mittler R, Vanderauwera S, Gollery M, Breusegem FV. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 41.Li SW, Xue L, Xu S, Feng H, An A. Hydrogen peroxide acts as a signal molecule in the adventitious root formation of mung bean seedlings. Environ Exp Bot. 2009;65:63–71. doi: 10.1016/j.envexpbot.2008.06.004. [DOI] [Google Scholar]
- 42.Sewelam N, et al. Spatial H2O2 signaling specificity: H2O2 from chloroplasts and peroxisomes modulates the plant transcriptome differentially. Mol Plant. 2014;7:1191–1210. doi: 10.1093/mp/ssu070. [DOI] [PubMed] [Google Scholar]
- 43.De Tullio MC, Jiang K, Feldman LJ. Redox regulation of root apical meristem organization: Connecting root development to its environment. Plant Physiol Bioch. 2010;48:328–336. doi: 10.1016/j.plaphy.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 44.Schnaubelt D, et al. Low glutathione regulates gene expression and the redox potentials of the nucleus and cytosol in Arabidopsis thaliana. Plant Cell Environ. 2015;38:266–279. doi: 10.1111/pce.12252. [DOI] [PubMed] [Google Scholar]
- 45.Ballatori N, Krance SM, Marchan R, Hammond CL. Plasma membrane glutathione transporters and their roles in cell physiology and pathophysiology. Mol Aspects Med. 2009;30:13–28. doi: 10.1016/j.mam.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noctor G, et al. Glutathione in plants: an integrated overview. Plant Cell Environ. 2012;35:454–484. doi: 10.1111/j.1365-3040.2011.02400.x. [DOI] [PubMed] [Google Scholar]
- 47.Koffler BE, Bloem E, Zellnig G, Zechmanna B. High resolution imaging of subcellular glutathione concentrations by quantitative immunoelectron microscopy in different leaf areas of Arabidopsis. Micron. 2013;45:119–128. doi: 10.1016/j.micron.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cochemé HM, et al. Measurement of H2O2 within living Drosophila during aging using a ratiometric mass spectrometry probe targeted to the mitochondrial matrix. Cell Metab. 2011;13:340–350. doi: 10.1016/j.cmet.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sohal RS, Orr WC. The redox stress hypothesis of aging. Free Radical Biol Med. 2012;52:539–555. doi: 10.1016/j.freeradbiomed.2011.10.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ji N, Wang Y, Wu T, Zhang XZ, Han ZH. Correlation analysis of miR156 expression and redox status during the phase change of Malus xiaojinensis seedlings. J China Agri Uni. 2016;21:59–64. [Google Scholar]
- 51.Møller IM. Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Mol Biol. 2001;52:561–591. doi: 10.1146/annurev.arplant.52.1.561. [DOI] [PubMed] [Google Scholar]
- 52.Ferguson M, Mockett RJ, Shen Y, Orr WC. Age-associated decline in mitochondrial respiration and electron transport in Drosophila melanogaster. Biochem J. 2005;390:501–511. doi: 10.1042/BJ20042130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sagi M, Fluhr R. Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol. 2006;14:336–340. doi: 10.1104/pp.106.078089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dietz KJ, Turkan I, Krieger-Liszkay A. Redox- and reactive oxygen species-dependent signaling into and out of the photosynthesizing chloroplast. Plant Physiol. 2016;171:1541–1550. doi: 10.1104/pp.16.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia R, Zhu H, An YQ, Eric PB, Liu ZR. Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 2012;13:R47. doi: 10.1186/gb-2012-13-6-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun C, Zhao Q, Liu DD, You CX, Hao YJ. Ectopic expression of the apple Md-miRNA 156h gene regulates flower and fruit development in Arabidopsis. Plant Cell Tiss Organ Cult. 2013;112:343–351. doi: 10.1007/s11240-012-0241-7. [DOI] [Google Scholar]
- 57.Yu N, Niu QW, Ng KH, Chua NH. The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J. 2015;83:673–685. doi: 10.1111/tpj.12919. [DOI] [PubMed] [Google Scholar]
- 58.Martin MN, Saladores PH, Lambert E, Hudson AO, Leustek T. Localization of members of the γ-glutamyl transpeptidase family identifies sites of glutathione and glutathione S-conjugate hydrolysis. Plant Physiol. 2007;144:1715–1732. doi: 10.1104/pp.106.094409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang HQ, Forman HJ. Redox regulation of γ-glutamyl transpeptidase. Am J Resp Cell Mol. 2009;41:509–515. doi: 10.1165/rcmb.2009-0169TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Noctor G, Mhamdi A, Queval G, Foyer CH. Regulating the redox gatekeeper: vacuolar sequestration puts glutathione disulfide in its place. Plant Physiol. 2013;163:665–671. doi: 10.1104/pp.113.223545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo CK, et al. Repression of miR156 by miR159 regulates the timing of the juvenile-to-adult transition in Arabidopsis. Plant Cell. 2017;29:1293–1304. doi: 10.1105/tpc.16.00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petrov VD, Breeusegem FV. Hydrogen peroxide—a central hub for information flow in plant cells. AoB Plants. 2012;1:pls014. doi: 10.1093/aobpla/pls014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Noctor G, Foyer CH. Intracelluar redox compartmentation and ROS-related communication in regulation and signaling. Plant Physiol. 2016;171:1581–1592. doi: 10.1104/pp.16.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miret JA, Munne-Bosch S. Redox signaling and stress tolerance in plants: a focus on vitamin E. Ann NY Acad Sci. 2015;1340:29–38. doi: 10.1111/nyas.12639. [DOI] [PubMed] [Google Scholar]
- 65.Chen J, Zhu C, Li LP, Sun ZY, Pan XB. Effects of exogenous salicylic acid on growth and H2O2-metabolizing enzymes in rice seedlings under lead stress. J Environ Sci. 2007;19:44–49. doi: 10.1016/S1001-0742(07)60007-2. [DOI] [PubMed] [Google Scholar]
- 66.Zhang XL, Jia XF, Yu B, Gao Y, Bai JG. Exogenous hydrogen peroxide influences antioxidant enzyme activity and lipid peroxidation in cucumber leaves at low light. Sci Hortic. 2011;129:656–662. doi: 10.1016/j.scienta.2011.05.009. [DOI] [Google Scholar]
- 67.Amini SA, Shabani L, Afghani L, Jalalpour Z, Sharifi-tehrani M. Squalestatin-induced production of taxol and baccatin in cell suspension culture of yew (Taxus baccata L.) Turk J Biol. 2014;8:528–536. doi: 10.3906/biy-1401-47. [DOI] [Google Scholar]
- 68.Voehringer DW, McConkey DJ, McDonnell TJ, Brisbay S, Meyn RE. Bcl-2 expression causes redistribution of glutathione to the nucleus. P Natl Acad Sci USA. 1998;95:2956–2960. doi: 10.1073/pnas.95.6.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vivancos PD, et al. Recruitment of glutathione into the nucleus during cell proliferation adjusts whole-cell redox homeostasis in Arabidopsis thaliana and lowers the oxidative defence shield. Plant J. 2010;64:825–838. doi: 10.1111/j.1365-313X.2010.04371.x. [DOI] [PubMed] [Google Scholar]
- 70.Garcia-Gimenez JL, et al. Nuclear glutathione. Biochim Biophys Acta. 2013;1830:3304–3316. doi: 10.1016/j.bbagen.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 71.Ogawa K. Glutathione-associated regulation of plant growth and stress responses. Antiox Rad Signal. 2005;7:973–981. doi: 10.1089/ars.2005.7.973. [DOI] [PubMed] [Google Scholar]
- 72.Pasternak T, Asard H, Potters G, Jansen MAK. The thiol compounds glutathione and homoglutathione differentially affect cell development in alfalfa (Medicago sativa L.) Plant Physiol Biochem. 2014;74:16–23. doi: 10.1016/j.plaphy.2013.10.028. [DOI] [PubMed] [Google Scholar]
- 73.Gao Y, et al. Differences in gene expression and regulation during ontogenetic phase change in apple seedlings. Plant Mol Biol Rep. 2013;32:357–371. doi: 10.1007/s11105-013-0648-2. [DOI] [Google Scholar]
- 74.Zhu L, et al. Zearalenone induces apoptosis and necrosis in porcine granulosa cells via a caspase-3- and caspase-9-dependent mitochondrial signaling pathway. J Cell Physiol. 2012;227:1814–1820. doi: 10.1002/jcp.22906. [DOI] [PubMed] [Google Scholar]
- 75.Duda M, Knet M, Tabarowski Z, Slomczynska M. Luteal macrophage conditioned medium affects steroidogenesis in porcine granulosa cells. Reprod Biol. 2011;11:117–34. doi: 10.1016/S1642-431X(12)60049-7. [DOI] [PubMed] [Google Scholar]
- 76.Zhang YG, Chen JH, Han ZH, Xu XF, Li TZ. Comparison of methods for total RNA isolation from Malus xiaojinensis and cDNA LD-PCR amplification. Biotech Bull. 2004;4:50–53. [Google Scholar]
- 77.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright A. J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Griffiths-Jones S, Saini HK, van Dongen S, Enright A. J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xing LB, et al. Genome-wide identification of vegetative phase transition-associated microRNAs and targetpredictions using degradome sequencing in Malus hupehensis. BMC Genomics. 2014;15:1125. doi: 10.1186/1471-2164-15-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mukherjee SP, Choudhuri MA. Implication of hydrogen peroxide–ascorbate system on membrane permeability of water stressed vigna seedlings. New Phytol. 1985;99:355–360. doi: 10.1111/j.1469-8137.1985.tb03663.x. [DOI] [Google Scholar]
- 81.Yang W, et al. Hydrogen peroxide is a second messenger in the salicylic acid-triggered adventitious rooting process in mung bean seedlings. PLoS ONE. 2013;8:e84580. doi: 10.1371/journal.pone.0084580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Darius C, et al. Characterization of apple NADPH oxidase genes and their expression associated with oxidative stress in shoot culture in vitro. Plant Cell Tiss Organ Cult. 2016;124:621–633. doi: 10.1007/s11240-015-0920-2. [DOI] [Google Scholar]
- 83.Liang ZC, et al. Inheritance of sugar and acid contents in the ripe berries of a tetraploid × diploid grape cross population. Euphytica. 2011;182:251–259. doi: 10.1007/s10681-011-0487-x. [DOI] [Google Scholar]
- 84.Garg N, Pandey R. High effectiveness of exotic arbuscular mycorrhizal fungi is reflected in improved rhizobial symbiosis and trehalose turnover in Cajanus cajan genotypes grown under salinity stress. Fungal Ecology. 2016;21:57–67. doi: 10.1016/j.funeco.2016.04.001. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its Supplementary Information files.