Abstract
Increased knowledge of the molecular differences between indolent and aggressive prostate cancer is needed for improved risk stratification and treatment selection. Secreted frizzled-related protein 4 (SFRP4) is a modulator of the cancer-associated Wnt pathway, and previously suggested as a potential marker for prostate cancer aggressiveness. In this study, we investigated and validated the association between SFRP4 gene expression and aggressiveness in nine independent cohorts (n = 2157). By differential expression and combined meta-analysis of all cohorts, we detected significantly higher SFRP4 expression in cancer compared with normal samples, and in high (3–5) compared with low (1–2) Grade Group samples. SFRP4 expression was a significant predictor of biochemical recurrence in six of seven cohorts and in the overall analysis, and was a significant predictor of metastatic event in one cohort. In our study cohort, where metabolic information was available, SFRP4 expression correlated significantly with the concentrations of citrate and spermine, two previously suggested biomarkers for aggressive prostate cancer. SFRP4 immunohistochemistry in an independent cohort (n = 33) was not associated with aggressiveness. To conclude, high SFRP4 gene expression is associated with high Grade Group and recurrent prostate cancer after surgery. Future studies investigating the mechanistic and clinical usefulness of SFRP4 in prostate cancer are warranted.
Introduction
Prostate cancer is the second most common cancer and the fifth leading cause of cancer related death in men worldwide1. The lack of accurate markers to separate aggressive from non-aggressive prostate cancer at an early time point is causing considerable overtreatment of indolent cancers2. Discovery of new biomarkers of aggressiveness, as well as improved understanding of differences between indolent and aggressive prostate cancer, are therefore highly needed.
The family of secreted frizzled-related proteins (SFRP1–5) are extracellular inhibitors of Wnt signalling, a pathway identified for its role in carcinogenesis3. The SFRPs are in general regarded as tumour suppressors, however, oncogenic properties have also been suggested due to biphasic modulation of Wnt signalling4,5 and interactions with other signalling pathways4. SFRP4 is the largest and the most structurally different of the family members6. In several types of cancer, SFRP4 follows a tumour suppressor pattern with epigenetic silencing and reduced gene expression, as reviewed by Pohl et al.7. However, for prostate cancer, increased gene expression of SFRP4 has been observed8,9, and shown to be a predictor of recurrent disease10. Additionally, SFRP4 has been included in different gene expression signatures linked to prostate cancer aggressiveness and recurrence10,11, including our previously published signature for non-canonical Wnt pathway and epithelial-to-mesenchymal transition (NCWP-EMT) markers12. Protein levels of SFRP4 measured by immunohistochemistry is discordant in prostate cancer; Horvath et al.13,14 reported increased expression of membranous SFRP4 staining to be associated with good prognosis, while Mortensen et al.10 reported cytoplasmic expression to be linked to worse prognosis. Overall SFRP4 appears to be a potential biomarker candidate for prostate cancer aggressiveness, and there is a need to validate and clarify the role of SFRP4 in prostate cancer.
Reprogramming of metabolism is one of the hallmarks of cancer development15. For prostate cancer, the metabolites citrate and spermine have shown promise as biomarkers and are found in lower concentrations in aggressive compared to indolent cancers16,17. Our NCWP-EMT gene expression signature was associated with reduced concentrations of these metabolites12, but the correlation between SFRP4 gene and protein expression levels, and citrate and spermine has not previously been investigated in prostate cancer. Our previously published method for integration of gene expression levels with metabolic data and histopathology of the exact same samples, gives an excellent opportunity to examine this18.
The overall aim of this study was to investigate and validate SFRP4 gene expression in prostate cancer, and its relation to cancer aggressiveness. The results were validated in eight independent, publically available gene expression prostate cancer cohorts with patient follow-up data. Furthermore, SFRP4 protein expression was assessed using immunohistochemistry in a separate cohort. Our approach of including several independent patient cohorts gave increased statistical power, and improved the accuracy and generalisation of the results.
Results
Our study cohort consisted of 156 prostate tissue samples from 41 patients, of which 116 were cancer tissue samples19. Eight independent prostate cancer validation cohorts were downloaded from Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA), giving a total number of 2157 samples from 1884 patients. Five of the validation cohorts included normal samples as well as cancer samples. Our additional patient cohort for immunohistochemistry analysis, termed the IHC cohort12, included prostate cancer samples from 40 patients. Clinical and histopathological data for all patient cohorts included in the study are listed in Table 1.
Table 1.
Clinical and histopathological variables of all ten cohorts.
| Clinical variables | Study cohort | IHC cohort | Erho et al. | TCGA-PRAD | CAM Ross-Adams et al. |
|---|---|---|---|---|---|
| Samples (patients) | 156 (41) | 40 (40) | 545 (545) | 549 (497) | 186 (163) |
| Cancer samples (patients) | 116 (41) | 40 (40) | 545 (545) | 497 (497) | 112 (112) |
| Age at diagnosis, years (median, range) | 64 (48–69) | 61 (48–73) | 65.3 ± 6.4 | 61 (41–78) | 61 (41–73) |
| PSA before surgery, ng/mL (median, range) | 9.1 (4.0–45.8) | 8.85 (5.2–18) | — | 7.4 (0.7–107) | 7.8 (3.2–23.7) |
| Grade Groups | |||||
| Low (1–2) | 60 (52%) | 19 (47.5%) | 334 (61%)a | 207 (42%) | 82 (73%) |
| High (3–5) | 56 (48%) | 21 (52.5%) | 211 (39%)a | 289 (58%) | 30 (27%) |
| Pathological T stage | |||||
| pT1 | — | — | — | — | — |
| pT2 | 70 (60%) | 27 (68%) | 219 (40%) | 187 (38%) | 33 (29%) |
| pT3 | 40 (35%) | 12 (30%) | 253 (47%) | 293 (59%) | 74 (66%) |
| pT4 | — | — | 9 (2%) | 1 (1%) | |
| No data | 6 (5%) | 1 (2%) | 73 (13%) | 8 (1%) | 4 (4%) |
| Follow-up | |||||
| Endpoint | BCR | BCR | Metastasis | BCR | Recurrence |
| Occurred | 13 (32%) | 16 (40%) | 212 (39%)b | 91 (18%) | 19 (17%) |
| Not occurred | 21 (51%) | 21 (53%) | 333 (69%)b | 399 (80%) | 93 (83%) |
| No data | 7 (17%) | 3 (8%) | — | 7 (2%) | — |
| Clinical variable | STK Ross-Adams et al. | Wang et al. | Sboner et al. | Taylor et al. | Mortensen et al. |
| Samples (patients) | 94 (94) | 136 (82) | 281 (281) | 160 (131) | 50 (50) |
| Cancer samples (patients) | 94 (94) | 65 (56) | 281 (281) | 131 (131) | 36 (36) |
| Age years (median, range) | 63 (43–77) | 74 (51–91) | 58 (37–73) | 63 (46–71) | |
| PSA before surgery, ng/mL (median, range) | 7.95 (1.5–117) | 6.62 (1.0–75) | 5.92 (1.0–46) | 16 (5.0–43) | |
| Grade Groups | |||||
| Low (1–2) | 60 (64%) | 50 (77%) | 162 (58%) | 107 (82%) | 32 (89%)a |
| High (3–5) | 34 (36%) | 15 (23%) | 119 (42%) | 24 (18%) | 4 (11%)a |
| Pathological T-stage | |||||
| pT1 | — | 1 (2%) | 281c (100%) | — | — |
| pT2 | 48 (51%) | 32 (57%) | — | 85 (65%) | 19 (53%) |
| pT3 | 42 (45%) | 20 (35%) | — | 40 (30%) | 17 (47) |
| pT4 | — | 1 (2%) | — | 6 (5%) | — |
| No data | 4 (4%) | 2 (2%) | — | — | — |
| Follow-up | |||||
| Endpoint | Recurrence | BCR | PCa-death | BCR | BCR |
| Occurred | 45 (48%) | 29 (52%) | 165 (59%) | 27 (21%) | 22 (61%) |
| Not occurred | 48 (51%) | 27 (48%) | 116 (41%) | 104 (79%) | 14(39%) |
| No data | 1 (1%) | — | — | — | — |
Abbreviations: BCR – biochemical recurrence, PCa-death – prostate cancer-specific death.
aIn Erho et al. and Mortensen et al.: Low Grade Group 1–3 and high Grade Group 4–5 (due to lack of information to separate Grade Group 2 and 3).
bIn Erho et al. metastatic progression at 10-year patient follow-up.
cClinical T-stage.
SFRP4 expression in cancer
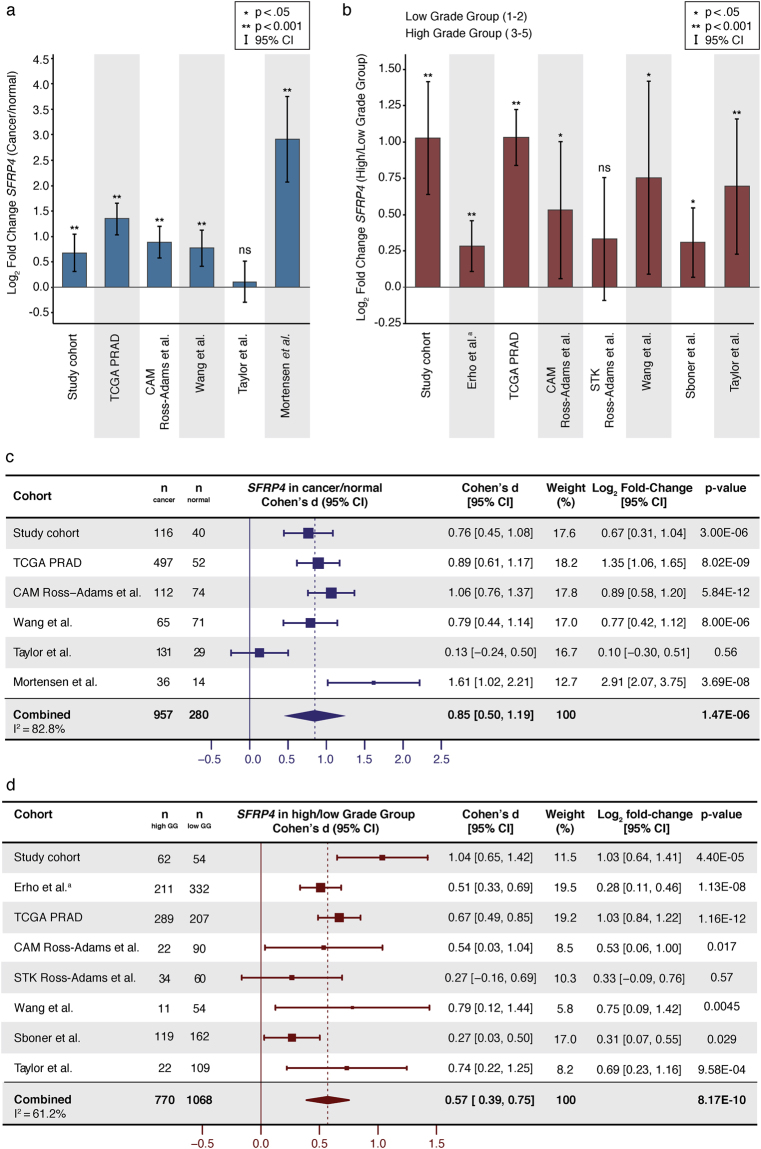
In our study cohort, there was significantly higher SFRP4 expression in cancer samples compared with normal samples (t-test p < 0.001, Fig. 1a). This was also true for four of the five independent validation cohorts which included expression data from both cancer and normal samples (Fig. 1a). Meta-analysis of all the cohorts gave a significant combined Cohen’s d of 0.85 (p < 0.001, Fig. 1c). This is considered a large effect-size and indicates a considerable difference in mean SRFP4 expression between normal and cancer tissue. Together, this clearly shows significant upregulation of SFRP4 in prostate cancer compared with normal prostate tissue.
Figure 1.
SFRP4 gene expression in prostate cancer. (a) Log2 fold change of SFRP4 expression in cancer compared with normal samples (b) Log2 fold change of SFRP4 expression in high Grade Group (3–5) compared with low Grade Group (1–2) samples. (c) Forest plot and meta-analysis of SFRP4 expression in prostate cancer compared with normal prostate samples. (d) Forest plot and meta-analysis of SFRP4 expression in high Grade Group compared with low Grade Group prostate cancer samples. Abbreviations: ns - not significant, GG – Grade Group, CI – confidence interval. aIn the Erho et al. cohort, low Grade Group included GG 1–3, and high Grade Group included GG 4–5.
SFRP4 expression in cancer with high Grade Group
In our study cohort, there was significantly higher SFRP4 expression in high Grade Groups (3–5) compared with low Grade Groups (1–2) cancer samples (t-test p < 0.001, Fig. 1b), and this was confirmed in six of the seven validation cohorts (Fig. 1b). Meta-analysis of all the analysed cohorts further strengthened this finding, giving a significant combined Cohen’s d of 0.57 (p < 0.001, Fig. 1d). The Mortensen et al. cohort was excluded from differential expression analysis between high and low Grade Groups due to the low number of high Grade Group samples (n = 4).
SFRP4 expression and pathological T-stage and preoperative PSA value
SFRP4 expression was significantly higher in samples from patients with a high pathological T-stage (≥T3a) compared with low T-stage samples (≤T2c) in six out of the seven cohorts that included information on T-stage (Supplementary Table S1). SFRP4 expression was not correlated with preoperative PSA in any of the cohorts (Supplementary Table S1).
SFRP4 and patient follow-up
In our study cohort, the continuous value of SFRP4 expression was a significant predictor of biochemical recurrence (PSA ≥ 0.2 ng/mL) after radical prostatectomy by univariate Cox proportional hazards analysis (p = 0.007, Fig. 2). This was further confirmed in five of the six validation cohorts with biochemical recurrence as endpoints (Fig. 2). Meta-analysis of the six cohorts with microarray based gene expression gave a significant combined SFRP4 standardised hazard ratio (HR) of 1.70 for prediction of biochemical recurrence (p < 0.001, Fig. 2). Continuous SFRP4 expression was not a predictor of prostate cancer-specific death in the watchful waiting Sboner et al. cohort (HR 1.0, p = 0.96, Fig. 2). Logistic regression analysis showed SFRP4 expression to be a predictor of metastases after radical prostatectomy in the Erho et al. cohort (odds ratio 2.34, p < 0.001, Fig. 2).
Figure 2.
Univariate Cox proportional hazard analysis of SFRP4 expression and follow-up endpoints. SFRP4 gene expression was used as a continuous variable in the analyses. Meta-analysis was performed on the cohorts with microarray based SFRP4 gene expression data and biochemical recurrence (PSA ≥ 0.2 ng/mL) as endpoint. For the cohorts with multiple samples per patients (study and Wang et al. cohort), one sample per patient was selected by random. Abbreviations: CI – confidence interval, HR – hazard ratio, BCR – biochemical recurrence. aThe Erho et al. cohort was analysed by logistic regression, with odds ratio as the effect size.
SFRP4 expression and metabolism
In our study cohort, the SFRP4 expression level was negatively correlated with concentrations of citrate (Pearson’s r = −0.53, p < 0.001) and the polyamine spermine (Pearson’s r = −0.49, p < 0.001) (Fig. 3). These were the strongest correlations to citrate and spermine of all the genes in our previously published NCWP-EMT gene expression signature12 (Supplementary Table S2).
Figure 3.
Correlations with metabolism. Linear Pearson correlations between SFRP4 gene expression and citrate and spermine in our study cohort. All variables are log2 transformed.
SFRP4 immunohistochemistry
In our IHC cohort, seven of the 40 samples were excluded from further analysis due to insufficient or lack of tumour cells in the immunohistochemically stained sections. We did not detect membranous SFRP4 staining of prostate cancer cells in any samples. However, cytoplasmic SFRP4 staining of different intensities was identified and categorised into four different scores (Fig. 4). Proportion of positive cancer cells were also scored and multiplied with staining intensity to create a staining index (Supplementary Table S3). Full immunohistochemistry scoring of each sample along with clinical, histopathological and metabolic data is shown in Supplementary Table S4.
Figure 4.
Immunohistochemistry of SFRP4. Examples of staining intensities 0 to 3 in our IHC cohort.
There was no relationship between Grade Groups and SFRP4 staining index (Fisher’s exact p = 1.0). This was also the case when looking at staining intensity and proportion separately (Fisher’s exact p = 0.80 and p = 0.82, respectively). Furthermore, no significant associations between SFRP4 staining and biochemical recurrence (Log-rank: staining index p = 0.87, intensity p = 0.82, proportion p = 0.95), nor any significant correlation between SFRP4 staining index and citrate and spermine concentrations (Pearson’s r = 0.13 p = 0.47 and Pearson’s r = 0.18 p = 0.32, respectively) were detected.
Discussion
In this study, we performed analyses of SFRP4 gene expression and validated the results in eight independent prostate cancer cohorts. We showed SFRP4 expression to be increased in prostate cancer, and further increased in high Grade Group (3–5) compared with low Grade Group (1–2) cancers. Additionally, SFRP4 gene expression was found to be a predictor of worse outcome in prostatectomy-treated prostate cancer patients. Furthermore, the SFRP4 expression level was negatively correlated with the concentrations of citrate and spermine in the samples. Together, these results underpin SFRP4 as a biomarker candidate of prostate cancer aggressiveness.
We showed SFRP4 gene expression to be increased in prostate cancer compared with normal tissue in five of six cohorts, and in the combined meta-analysis of all cohorts. This is in agreement with Luo et al.8 and Wissmann et al.9, who investigated matched tumour and normal tissue samples from 16 and 56 prostate cancer patients, respectively. Contradictory, García-Tobilla et al.20 did not find significantly different expression levels of SFRP4 between normal and prostate cancer tissue, however, this study suffered from small sample size (normal n = 4, cancer n = 11). In a previous paper, we also showed increased SFRP4 expression in prostate cancer when balancing for stroma content in the samples12. Interestingly, two studies have shown increased SFRP4 expression in prostate cancer tissue compared with benign prostate hyperplasia20,21, but this approach was not possible to pursue in our study. To summarise, previous studies have in general reported increased SFRP4 gene expression in prostate cancer compared with normal prostate tissue, but they have been carried out on small cohorts. The result of the present study adds substantial validation to SFRP4 expression being increased in prostate cancer.
We showed increased expression of SFRP4 in high Grade Group compared with low Grade Group cancer samples, as well as an association between SFRP4 expression and risk of biochemical recurrence and metastasis after radical prostatectomy. SFRP4 gene expression has previously been linked to more aggressive prostate cancer; Luo et al.8 showed increased expression of SFRP4 in tissue samples from prostate cancer patients with pathological stage T3a-b compared with pathological stage T2b. Mortensen et al.10 discovered SFRP4 to be a part of two aggressive gene expression clusters, as well as being an independent predictor of recurrence after prostatectomy in an independent cohort. Our previously published NCWP-EMT gene expression signature, which included SFRP4 as one of 15 genes, was associated with biochemical recurrence and metastasis after prostatectomy12. Furthermore, Oncotype DX® Prostate Cancer, a commercially available gene expression signature, includes SFRP4 as one of its 17 genes, and has been associated with clinical recurrence of prostate cancer after prostatectomy11. Our analyses of multiple independent cohorts in the current study, further support high SFRP4 expression to be associated with more aggressive prostate cancer. To conclude, several studies8,10–12, including the current study, support SFRP4 gene expression to be upregulated in aggressive compared with less aggressive prostate cancer.
SFRP4 is classified as an inhibitor of Wnt signalling, a pathway implicated in carcinogenesis3. Consequently, SFRP4 is expected to be a tumour suppressor, and to be downregulated in aggressive cancer. As reviewed by Pohl et al., DNA hypermethylation of the SFRP4 promotor and reduced SFRP4 gene expression is observed in many types of cancers, including endometrial, ovarian, bladder, and oesophageal cancer7. Although SFRP4 expression in prostate cancer tissue seems to deviate from this, a few prostate cell line studies have supported tumour suppressor properties of SFRP4 also in prostate cancer; Horvath et al.13,14 showed that PC3 and LNCaP cell lines modified to overproduce SFRP4 proteins had reduced cellular proliferation compared to controls. Furthermore, García-Tobilla et al.20 showed reduced gene expression of SFRP4 in prostate cancer cell lines (LNCaP, PC3, DU145 and 22Rv1) compared with control cells (PREC). However, they did not detect DNA hypermethylation at the SFRP4 promotors that could explain this downregulation in any of the cell lines20. Absence of SFRP4 hypermethylation was also shown in both prostate cancer cell lines and in tumour tissue in a study by Perry et al.21. In contrast to the study by García-Tobilla et al.20, and in coherence with human prostate cancer tissue studies, Perry et al.21 also detected upregulation of SFRP4 in all prostate cancer cell lines (LNCaP, PC3, DU145 and 22Rv1) compared with controls (PWR-1 and RWPE1). Interestingly, in the two latter mentioned studies20,21, DNA hypermethylation of SFRP2, SFRP3, and SFRP5 was detected in both cell lines and human prostate cancer tissues20,21. This is in agreement with findings in colorectal cancer, where Suzuki et al.22 suggested that SFRP4 may not be an important inhibitor of the Wnt signalling pathway due to lower frequency of DNA hypermethylation and weaker Wnt signalling inhibition compared with other SFRP family members. This may be translatable to prostate cancer, and could explain why SFRP4 is not downregulated in cancer. However, studies of how SFRP4 regulates the Wnt signalling pathway and other pathways in prostate cancer are necessary before a conclusion can be drawn.
In the current study, we detected an association between SFRP4 expression and development of metastases after prostatectomy in the Erho et al. cohort. Bones are the most frequent site for haematogenous metastases in prostate cancer23, and, interestingly, SFRP4 has been suggested to have an important role in bone homeostasis24,25. However, to our knowledge, the function of SFRP4 in bone metastases has not been investigated. A hypothesis to explain the association between SFRP4 gene expression and high Grade Groups, as well as recurrence and metastasis after prostatectomy, could therefore be that SFRP4 increases the cancer cell’s ability to metastasise to bone. Future studies investigating the role of SFRP4 in prostate cancer bone metastases would consequently be of interest.
For patient follow-up in this study, we used the surrogate endpoints of biochemical recurrence and metastases, in all except one cohort, Sboner et al., in which prostate cancer-specific death was used. Such surrogate endpoints are commonly used in prostate cancer studies, due to a natural long survival time of patients. Although recurrence after prostatectomy in general represents more aggressive disease, we also recognise that the use of surrogate endpoints is a limitation to the clinical value of the results, as only a minority of patients with biochemical recurrence will experience systemic progression or prostate cancer-specific death26. In the Sboner et al. cohort, we did not see any association between SFRP4 gene expression and cancer-specific death. This cohort did, however, differ substantially from the other analysed cohorts, where all patients included had incidental prostate cancer discovered by trans-urethral resection of the prostate (TURP), and were classified as stage T1a-T1b, NX, and M0 disease. The samples used for gene expression were from the TURP procedure. Although most prostate cancers arise from the peripheral zone, resection performed by TURP represents the transition zone, and is likely to detect a higher rate of transition zone prostate cancers. Substantial differences in gene expression between tumours of different zonal origin has previously been observed27. This may limit future clinical use of SFRP4 expression for risk stratification in patients with transitional zone prostate cancers, and potentially also in patients with very early stage prostate cancer, and this should be further investigated.
Changes in metabolism is regarded as one of the hallmarks of cancer15. In prostate cancer, the concentrations of the metabolites citrate and spermine are shown to be reduced in cancer compared with normal tissue28,29, and further reduced in high Gleason score prostate cancer16. A recent study has also shown citrate and spermine to be predictors of prostate cancer biochemical recurrence in three independent cohorts17. The high negative correlation between SFRP4 expression and spermine and citrate in our study cohort further supports SFRP4 expression to be associated with aggressive cancer. One of the normal functions of prostate cells is production of citrate and spermine for the prostatic fluid, and reduced concentration of these metabolites may signify loss of normal prostate function. However, whether these metabolic mechanisms are directly related to SFRP4 expression was not investigated in the current study.
We did not find any association between immunohistochemical staining of SFRP4 and histopathological, metabolic, or follow-up data in our IHC cohort in this study. Our cohort only included tissue samples from 33 patients, as it was originally part of a demanding integrated analysis of metabolomics, histopathology, and patient follow-up12,30. This small sample size limits the interpretation of our immunohistochemistry results. There are only four previous studies including immunohistochemistry of SFRP4 in prostate cancer, and there are no standardised protocols for staining or scoring. Three of these studies were based on the same SFRP4 stained cohort of tissue microarray (TMA) samples from 229 radical prostatectomy patients13,14,31, where membranous SFRP4 staining was found to be associated with good prognosis13. In the current study, we did not observe any membranous staining of SFRP4. The lack of membranous staining is in accordance with a previous study by Mortensen et al.10, which included TMA sections from 517 radical prostatectomy patients. Our IHC cohort was stained by the same commercially available antibody and in the same dilution as used in the Mortensen et al. study10, which may explain the similar staining pattern. The use of different antibodies compared with the Horvath et al. study13 may be a possible cause of the observed disparity in membranous staining. In addition, the relatively weak staining of SFRP4 in the current study (Fig. 4) may have been insufficient to demonstrate membrane staining. In contrast to the TMA sections used in both the Mortensen et al.10 and Horvath et al.13 studies, our IHC cohort consisted of sections from needle biopsy samples. These biopsy sections were larger than the standard TMA sections, and we observed staining intensity heterogeneity within each sample. Consequently, we experienced some challenges when determining the immunohistochemistry intensity score of the samples. As mentioned, there are limitations of the immunohistochemistry evaluation of SFRP4 in the current study, especially with regard to the small sample size, and as a consequence, no certain conclusion can be made based on our results. Nevertheless, we have demonstrated a few issues that are important to address before immunohistochemistry of SFRP4 can have a role in prostate cancer risk stratification, including the lack of standardised staining and evaluation protocols, and the uncertain impact of staining heterogeneity.
In the current study, we did not directly investigate possible clinical applications of SFRP4 expression, and this should be examined in future studies. Absolute quantification of SFRP4 mRNA by real time PCR in biopsies may have a role for risk stratification and treatment selection for prostate cancer patients, including selection of patients for active surveillance and adjuvant treatment. Another interesting possibility for further studies, is investigation of the SFRP4 gene and protein expression levels in less invasive liquid biopsies such as serum, urine, and prostatic and seminal fluid.
In this study, we have validated the presence of increased SFRP4 gene expression in prostate cancer tissue, and we detected and validated higher SFRP4 gene expression in high Grade Group compared with low Grade Group cancer. We further showed that the SFRP4 gene expression level was as a predictor of recurrence and metastasis after prostatectomy. Finally, we showed a negative correlation between SFRP4 gene expression and the indolent metabolic markers, citrate and spermine. To conclude, SFRP4 expression is associated with more aggressive disease, and is a biomarker candidate for risk stratification of prostate cancer patients. SFRP4 may therefore potentially be useful in the selection of candidates for active surveillance as well as for patients in need of adjuvant or more aggressive treatment, and SFRP4 deserves further attention in prostate cancer studies.
Methods
Ethics statement
The study was approved by and carried out in accordance to the regulations of the Regional Committee for Medical and Health Research Ethics, Central Norway (identifiers 4.2007.1890 and 4.2007.1654). All patients signed a written informed consent.
Patients and samples
The samples in our study and IHC cohort were donated from patients diagnosed with localised or locally advanced prostate cancer and treated with radical prostatectomy between 2007 to 2010 at St. Olav’s Hospital, Trondheim University Hospital. None of the patients had received any prostate cancer treatment prior to surgery. Samples in the study cohort were harvested from fresh-frozen prostatectomy specimens with a highly standardised method as previously described by Bertilsson et al.18. The samples in the IHC cohort were collected as needle biopsies within approximately two minutes after the prostatectomy, and were snap frozen.
Follow-up
At least five-year follow-up data were collected for the patients in our study and IHC cohort. Biochemical recurrence was defined as serum PSA levels of at least 0.2 ng/mL in two independent measurements, or, in case of missing PSA values/time-point, onset of salvage therapy.
Histopathology
For histopathological evaluation, a cryosection from each fresh frozen tissue sample in our study cohort and two formalin-fixed paraffin-embedded sections of each sample in our IHC cohort were used. All sections were evaluated by an experienced pathologist as previously described12. The reproducibility of the histopathological evaluation has previously been assessed in our study cohort, by an independent pathologist blinded for previous evaluation, where high interrater agreement (κ = 0.66) was reported12. Post-operative Gleason score was obtained from the whole-mount prostate sections according to the clinical criteria for prostate cancer. The reported Gleason scores were directly converted to Grade Groups according to the new grading system for prostate cancer32. For statistical analyses, the samples and patients were divided into two groups: low Grade Group (1–2) and high Grade Group (3–5).
Metabolomics
The samples in the study cohort and IHC cohort were analysed by proton high-resolution magic angle spinning magnetic resonance spectroscopy (HR-MAS MRS) using a Bruker Avance DRX600 Spectrometer (Bruker Biopsin, Germany). LCModel was applied for absolute quantification of 23 metabolites from the spectra. More details on the HR-MAS MRS acquisition and metabolite quantification have been described by Giskeødegård et al. for the study cohort16 and Hansen et al. for the IHC cohort30.
Microarray gene expression
Gene expression analysis was performed on the tissue samples in the study cohort after HR-MAS MRS. Illumina Human HT-12v4 Expression Bead Chip (Illumina) were used to measure relative gene expression as previously described by Bertilsson et al.19.
Immunohistochemistry
In the IHC cohort, immunohistochemistry was performed using 4 μm thick, formalin fixed, paraffin embedded tissue sections. Rabbit polyclonal antibody against SFRP4 (Protein Tech catalogue: 15328-1-AP) was used in a 1:200 dilution at pH 9. The sections were counterstained with haematoxylin. Every section was evaluated for SFRP4 staining location (membranous or cytoplasmic). The most common staining intensity of each sample was scored from 0 to 3 (Fig. 4) based on the staining intensities described by Mortensen et al.10. Additionally, the proportion of positive cancer cells was scored from 0 to 3, and was multiplied by the intensity score to obtain a staining index (0–9). For statistical analyses, the staining index was categorised into three groups (0, 1–3, and 4–9). Further details on the scoring procedure are given in Supplementary Table S3. Two independent readings were performed by one pathologist experienced in immunohistochemistry and one physician. When scoring differed, consensus was reached by re-evaluation of the sections together.
Validation cohorts
For validation, the following seven prostate cancer cohorts with available microarray gene expression and follow-up data were downloaded from GEO: Erho et al. (GSE46691)33,34, CAM (Cambridge) Ross-Adams et al. (GSE70768)35, STK (Stockholm) Ross-Adams et al. (GSE70769)35, Wang et al. (GSE8218)36–38, Sboner et al. (GSE16560)39, Taylor et al. (GSE21035/32)40, and Mortensen et al. (GSE46602)10. In addition, a RNA sequencing (RNA Seq) cohort of prostate adenocarcinomas, TCGA PRAD, was downloaded from TCGA41,42. Cancer samples for all cohorts were from radical prostatectomy specimens, except Sboner et al. which was from a watchful waiting patient cohort of incidental prostate cancer discovered by transurethral resection of the prostate. Normal samples in Mortensen et al. were from surgical prostate specimens from patients with bladder cancer, four of the normal prostate samples in Wang et al. were autopsy samples from normal subjects, the rest and the other cohorts were adjacent normal prostate tissue from prostatectomy specimens. According to histopathology, the samples were divided into the same two groups as our cohorts: low Grade Group (1–2) and high Grade Group (3–5). The Erho et al. and Mortensen et al. cohorts did not included information to separate Grade Group 2 and 3, and for these cohorts the low and high Grade Groups were defined as Grade Group 1–3 and 4–5, respectively. Biochemical recurrence was the follow-up endpoint in Wang et al., Taylor et al., Mortensen et al., and TCGA PRAD. Biochemical recurrence and/or salvage treatment were the recurrence endpoints in the CAM and STK Ross-Adams et al. cohorts. For the Erho et al. cohort, metastasis was the endpoint, and prostate cancer-specific death was the endpoint in the Sboner et al. cohort. Clinical and histopathological data of the cohorts are listed in Table 1, and an overview table of the cohorts is included as Supplementary Table S5.
Statistical analysis
When more than one probe for SFRP4 existed in a cohort, the probe with the highest variance was chosen for statistical analyses. For all analyses, SFRP4 gene expression data were log2 transformed if not previously performed. For the gene expression cohorts, independent sample t-tests (two-tailed) were used for comparisons between two groups. Q-Q plots were used to check the normality assumption; small deviations were accepted due to the robustness of the test. Equal variance assumption was tested by Levene’s test, and corrected for when applicable. Fieller’s method was used to obtain pooled confidence interval for the log2 fold changes. To obtain Cohen’s d, a standardised effect size for meta-analyses, the difference between two means (cancer and normal, and high and low Grade Group) were divided by their pooled standard deviation. Meta-analyses by random-effect model were performed using the metafor package43 in R44.
In the two cohorts with multiple samples per patients (our study cohort and the Wang et al. cohort), one sample per patient was randomly selected for survival analyses. Univariate Cox proportional hazard regression analyses were performed on the continuous SFRP4 expression. The proportional hazard assumption was tested using the survival package45 in R44. Standardised hazard ratios were obtained by multiplying the natural logarithm of the hazard ratio (beta) by its standard deviation46. Cohorts with microarray based gene expression data and biochemical recurrence as endpoint were included in a random-effect model meta-analysis, which was performed in R44 using the metafor package43. Due to unavailable time-point of the events in the Erho et al. cohort, logistical regression was used for the follow-up analyses of this cohort.
A two-tailed t-test was performed on SFRP4 expression between high pathological T-stage (≥T3a) and low T-stage (≤T2c) for the seven cohorts that included information about T-stage. The two cohorts excluded from this analysis were the Erho et al. cohort where T-stage information was only available for the whole cohort and not for the individual samples, and the Sboner et al. cohort which only included clinical T-stage (all T1c). Pearson correlation was performed to calculate the correlation coefficient (Pearson’s r) between preoperative PSA and SFRP4 expression for the same cohorts.
Pearson correlation coefficients (two-tailed) were used to test the correlations between gene expression and log2 transformed concentrations of the metabolites citrate and spermine in our study and IHC cohort. Fisher exact tests (two-tailed) were used to examine the relationship between immunohistochemistry staining and histopathological Grade Groups, and log-rank statistics were used to investigate the relationship between SFRP4 staining and time to biochemical recurrence.
For all statistical tests the significant level was set at p = 0.05. When mentioned, analyses were performed in R44, all other analyses were performed in SPSS47.
Electronic supplementary material
Acknowledgements
The study was funded by grants from the Norwegian University of Science and Technology (NTNU), the Norwegian Cancer Society, and the Liaison Committee for education, research and innovation in Central Norway. The tissue samples in our study cohort and IHC cohort were collected and stored by Biobank1, St. Olav’s Hospital, and the MR Cancer Group, NTNU, respectively. HR-MAS MRS was performed at the MR Core Facility, NTNU. Histological staining was performed by the Cellular & Molecular Imaging Core Facility (CMIC), NTNU. The microarray service was provided by the Genomics Core Facility, NTNU, and Norwegian Microarray Consortium (NMC). The authors thank Trond Viset for histopathological evaluation, Alan J. Wright and Ailin F. Hansen for LCModel quantification of the metabolites in our study and IHC cohort, respectively, and Øyvind Salvesen for assistance with statistical meta-analyses.
Author Contributions
E.S., M.K.A., F.D., T.F.B., M.B.R., and M.-B.T. contributed to the conception and design of the study. E.S., H.B., M.B.R. and M.-B.T. developed methods and performed experiments. E.S. and A.M.B. performed immunohistochemistry scoring. Data analysis was performed by E.S. with consult from M.B.R. and M.-B.T. The paper was written by E.S., and all authors edited and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Morten B. Rye and May-Britt Tessem contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-14622-3.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Elise Sandsmark, Email: elise.sandsmark@gmail.com.
May-Britt Tessem, Email: may-britt.tessem@ntnu.no.
References
- 1.Torre LA, et al. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Loeb S, et al. Overdiagnosis and overtreatment of prostate cancer. European urology. 2014;65:1046–1055. doi: 10.1016/j.eururo.2013.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nusse R, Varmus HE. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 4.Bovolenta P, Esteve P, Ruiz JM, Cisneros E, Lopez-Rios J. Beyond Wnt inhibition: new functions of secreted Frizzled-related proteins in development and disease. Journal of cell science. 2008;121:737–746. doi: 10.1242/jcs.026096. [DOI] [PubMed] [Google Scholar]
- 5.Uren A, et al. Secreted frizzled-related protein-1 binds directly to Wingless and is a biphasic modulator of Wnt signaling. The Journal of biological chemistry. 2000;275:4374–4382. doi: 10.1074/jbc.275.6.4374. [DOI] [PubMed] [Google Scholar]
- 6.Jones SE, Jomary C. Secreted Frizzled-related proteins: searching for relationships and patterns. BioEssays: news and reviews in molecular, cellular and developmental biology. 2002;24:811–820. doi: 10.1002/bies.10136. [DOI] [PubMed] [Google Scholar]
- 7.Pohl S, Scott R, Arfuso F, Perumal V, Dharmarajan A. Secreted frizzled-related protein 4 and its implications in cancer and apoptosis. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:143–152. doi: 10.1007/s13277-014-2956-z. [DOI] [PubMed] [Google Scholar]
- 8.Luo JH, et al. Gene expression analysis of prostate cancers. Molecular carcinogenesis. 2002;33:25–35. doi: 10.1002/mc.10018. [DOI] [PubMed] [Google Scholar]
- 9.Wissmann C, et al. WIF1, a component of the Wnt pathway, is down-regulated in prostate, breast, lung, and bladder cancer. The Journal of pathology. 2003;201:204–212. doi: 10.1002/path.1449. [DOI] [PubMed] [Google Scholar]
- 10.Mortensen MM, et al. Expression profiling of prostate cancer tissue delineates genes associated with recurrence after prostatectomy. Scientific reports. 2015;5:16018. doi: 10.1038/srep16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein EA, et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. European urology. 2014;66:550–560. doi: 10.1016/j.eururo.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Sandsmark, E. et al. A novel non-canonical Wnt signature for prostate cancer aggressiveness. Oncotarget. 8(6), 9572–9586, 10.18632/oncotarget.14161 (2016). [DOI] [PMC free article] [PubMed]
- 13.Horvath LG, et al. Membranous expression of secreted frizzled-related protein 4 predicts for good prognosis in localized prostate cancer and inhibits PC3 cellular proliferation in vitro. Clinical cancer research: an official journal of the American Association for Cancer Research. 2004;10:615–625. doi: 10.1158/1078-0432.CCR-0707-03. [DOI] [PubMed] [Google Scholar]
- 14.Horvath LG, et al. Secreted frizzled-related protein 4 inhibits proliferation and metastatic potential in prostate cancer. The Prostate. 2007;67:1081–1090. doi: 10.1002/pros.20607. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 16.Giskeodegard GF, et al. Spermine and citrate as metabolic biomarkers for assessing prostate cancer aggressiveness. PloS one. 2013;8:e62375. doi: 10.1371/journal.pone.0062375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braadland, P. R. et al. Ex vivo metabolic fingerprinting identifies biomarkers predictive of prostate cancer recurrence following radical prostatectomy. British Journal of Cancer. 10.1038/bjc.2017.346 (2017). [DOI] [PMC free article] [PubMed]
- 18.Bertilsson H, et al. A new method to provide a fresh frozen prostate slice suitable for gene expression study and MR spectroscopy. The Prostate. 2011;71:461–469. doi: 10.1002/pros.21260. [DOI] [PubMed] [Google Scholar]
- 19.Bertilsson H, et al. Changes in gene transcription underlying the aberrant citrate and choline metabolism in human prostate cancer samples. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:3261–3269. doi: 10.1158/1078-0432.CCR-11-2929. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Tobilla P, et al. SFRP1 repression in prostate cancer is triggered by two different epigenetic mechanisms. Gene. 2016;593:292–301. doi: 10.1016/j.gene.2016.08.030. [DOI] [PubMed] [Google Scholar]
- 21.Perry AS, et al. Gene expression and epigenetic discovery screen reveal methylation of SFRP2 in prostate cancer. International journal of cancer. 2013;132:1771–1780. doi: 10.1002/ijc.27798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki H, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nature genetics. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 23.Bubendorf L, et al. Metastatic patterns of prostate cancer: an autopsy study of 1,589 patients. Human pathology. 2000;31:578–583. doi: 10.1053/hp.2000.6698. [DOI] [PubMed] [Google Scholar]
- 24.Haraguchi R, et al. sFRP4-dependent Wnt signal modulation is critical for bone remodeling during postnatal development and age-related bone loss. Scientific reports. 2016;6:25198. doi: 10.1038/srep25198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simsek Kiper PO, et al. Cortical-Bone Fragility–Insights from sFRP4 Deficiency in Pyle’s Disease. The New England journal of medicine. 2016;374:2553–2562. doi: 10.1056/NEJMoa1509342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boorjian SA, et al. Long-term risk of clinical progression after biochemical recurrence following radical prostatectomy: the impact of time from surgery to recurrence. European urology. 2011;59:893–899. doi: 10.1016/j.eururo.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 27.Sinnott JA, et al. Molecular differences in transition zone and peripheral zone prostate tumors. Carcinogenesis. 2015;36:632–638. doi: 10.1093/carcin/bgv051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swanson MG, et al. Quantitative analysis of prostate metabolites using 1H HR-MAS spectroscopy. Magnetic resonance in medicine. 2006;55:1257–1264. doi: 10.1002/mrm.20909. [DOI] [PubMed] [Google Scholar]
- 29.van der Graaf M, et al. Proton MR spectroscopy of prostatic tissue focused on the detection of spermine, a possible biomarker of malignant behavior in prostate cancer. Magma (New York, N.Y.) 2000;10:153–159. doi: 10.1007/BF02590640. [DOI] [PubMed] [Google Scholar]
- 30.Hansen AF, et al. Presence of TMPRSS2-ERG is associated with alterations of the metabolic profile in human prostate cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.9817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yip PY, et al. Low AZGP1 expression predicts for recurrence in margin-positive, localized prostate cancer. The Prostate. 2011;71:1638–1645. doi: 10.1002/pros.21381. [DOI] [PubMed] [Google Scholar]
- 32.Epstein JI, et al. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. The American journal of surgical pathology. 2016;40:244–252. doi: 10.1097/PAS.0000000000000530. [DOI] [PubMed] [Google Scholar]
- 33.Erho N, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PloS one. 2013;8:e66855. doi: 10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao SG, et al. The Landscape of Prognostic Outlier Genes in High-Risk Prostate Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2016;22:1777–1786. doi: 10.1158/1078-0432.CCR-15-1250. [DOI] [PubMed] [Google Scholar]
- 35.Ross-Adams H, et al. Integration of copy number and transcriptomics provides risk stratification in prostate cancer: A discovery and validation cohort study. EBioMedicine. 2015;2:1133–1144. doi: 10.1016/j.ebiom.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, et al. In silico estimates of tissue components in surgical samples based on expression profiling data. Cancer research. 2010;70:6448–6455. doi: 10.1158/0008-5472.CAN-10-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia Z, et al. Diagnosis of prostate cancer using differentially expressed genes in stroma. Cancer research. 2011;71:2476–2487. doi: 10.1158/0008-5472.CAN-10-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen X, et al. An accurate prostate cancer prognosticator using a seven-gene signature plus Gleason score and taking cell type heterogeneity into account. PloS one. 2012;7:e45178. doi: 10.1371/journal.pone.0045178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sboner A, et al. Molecular sampling of prostate cancer: a dilemma for predicting disease progression. BMC medical genomics. 2010;3:8. doi: 10.1186/1755-8794-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.The Molecular Taxonomy of Primary Prostate Cancer. Cell163, 1011–1025, 10.1016/j.cell.2015.10.025 (2015). [DOI] [PMC free article] [PubMed]
- 42.TheCancerGenomeAtlas(TCGA), http://www.cbioportal.org/study?id=prad_tcga-clinical Accessed: 05.10.2016.
- 43.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw. 2010;36:1–48. doi: 10.18637/jss.v036.i03. [DOI] [Google Scholar]
- 44.R Core Team. R: A language and environment for statistical computing, v3.3. 1, http://www.r-project.org/. R Foundation for Statistical Computing, Vienna, Austria (2013).
- 45.Therneau, T. A Package for Survival Analysis in S. R package version 2.38, https://cran.r-project.org/package=survival. (2015).
- 46.Crager MR. Generalizing the standardized hazard ratio to multivariate proportional hazards regression, with an application to clinical~ genomic studies. Journal of Applied Statistics. 2012;39:399–417. doi: 10.1080/02664763.2011.594034. [DOI] [Google Scholar]
- 47.IBM Corp. IBM SPSS Statistics for Macintosh, v24.0. Armonk, NY: IBM Corp. (Released 2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.