Abstract
The Melanoma-associated Antigen gene family (MAGE) generally encodes for tumour antigens. We had identified that one of the MAGE gene members, Mageb16 was highly expressed in undifferentiated murine embryonic stem cells (ESCs). While the role of Mageb16 in stemness and differentiation of pluripotent stem cells is completely unknown, here, in our current study, we have demonstrated that Mageb16 (41 kDa) is distributed in cytosol and/or in surface membrane in undifferentiated ESCs. A transcriptome study performed at differentiated short hairpin RNA (shRNA)-mediated Mageb16 knockdown (KD) ESCs and scrambled control (SCR) ESCs until a period of 22 days, revealed that Mageb16 KD ESCs mainly differentiated towards cells expressing mesodermal and cardiovascular lineage - gene markers. Gene markers of other mesoderm-oriented biological processes such as adipogenesis, osteogenesis, limb morphogenesis and spermatogenesis were also significantly enriched in the differentiated Mageb16 KD ESCs. The expression levels of contractile genes were higher in differentiated Mageb16 KD ESCs when compared to differentiated SCR and wild ESCs, suggesting a higher cardiomyogenic potential of Mageb16 depleted ESCs. Further analysis indicates that regulative epigenetic networks and nucleocytoplasmic modifications induced by the depletion of Mageb16, may play a probable role in differentiation.
Introduction
Melanoma-associated Antigen family (MAGE) of genes are located in X chromosome and encodes for tumour antigens which normally bind to T-lymphocytes in cancer patients1. The MAGE family genes are classified into type I and type II. Type I MAGE genes are further divided into subfamilies MAGE-A, -B and -C which are silent in almost all normal tissues except in testes and placenta. Interestingly, the type I MAGE genes are also highly expressed in rapidly proliferating cells such as tumour cells and in developing embryos2. The type II MAGE genes (MAGE D1-D4, E, F, G and H) are highly conserved and expressed in many adult tissues as well as in many tissues during embryonic development3,4. Homologous genes have also been identified in human, mice, and zebrafish2. Human MAGEB16 belongs to the MAGE B subfamily (GenBank accession No.: NM_001099921.1) which have been identified based on the sequence and the isoelectric points of analogous proteins5. When verified for protein similarities between murine Mageb16 and human MAGEB16, only 52% residue identity has been observed. Among the MAGE B subfamily, subgroups of genes- B1, B2, B3 and B4 share 70–80% of nucleotide identity and 49–68% of amino acid residue identity5. Recently, it has been reported that amongst 12 different human and mice tissues, the expression level of MAGEB16 was very high only in the testis6. Authors have also reported very high expression levels of Mageb16 in testis of 3 to 18 day postnatal mice embryos and though slightly decreased, the expression level was still high in testis of 4 month old adult mice6. Authors proposed that the testis-specific expression of MAGEB16 in human and Mageb16 in mice is regulated by the CpG methylation status of their promoter regions6.
In order to identify novel gene networks controlling fundamental biological processes of pluripotency and differentiation in murine ESCs, we recently performed microarray analysis of undifferentiated, early and late differentiated ESCs. Our transcriptome data showed that the most critical early differentiation processes occurs at day 2 and 3 of differentiation7. Besides monitoring well-annotated genes involved in regulation of pluripotency, germ layer formation and late differentiation processes toward somatic cell lines, Mageb16 has been identified showing a similar time kinetic of expression like the pluripotency factors Oct4 and Nanog 7. The expression level of Mageb16 was very high in undifferentiated ESCs which considerably declined immediately after differentiation7.
Interestingly, siRNA knockdown of Mageb16 in undifferentiated ESCs cultured under monolayer conditions resulted in a remarkable upregulation of mesodermal, ectodermal and endodermal marker genes after 48 h of differentiation as compared to untreated and control scrambled (SCR)-oligonucleotide treated ESCs7. These results suggested a crucial key role of Mageb16 for maintenance of pluripotency and differentiation of ESCs. In the present study, this hypothesis has been evaluated by generating a transgenic mESC line in which Mageb16 was permanently silenced by knockdown (KD) of Mageb16 using a shRNA directed to Mageb16 mRNA. Moreover, for a detailed study of its functional role in controlling pluripotency and differentiation Mageb16 KD ESCs were randomly differentiated for different time points using the embryoid body (EB) methodology. Signal transduction pathways affected by the depletion of Mageb16 were identified by detailed transcriptome and bioinformatics analysis. Additionally, the cellular localization of Mageb16 has been determined in ESCs.
Results
Knockdown efficiency of Mageb16 in ESCs
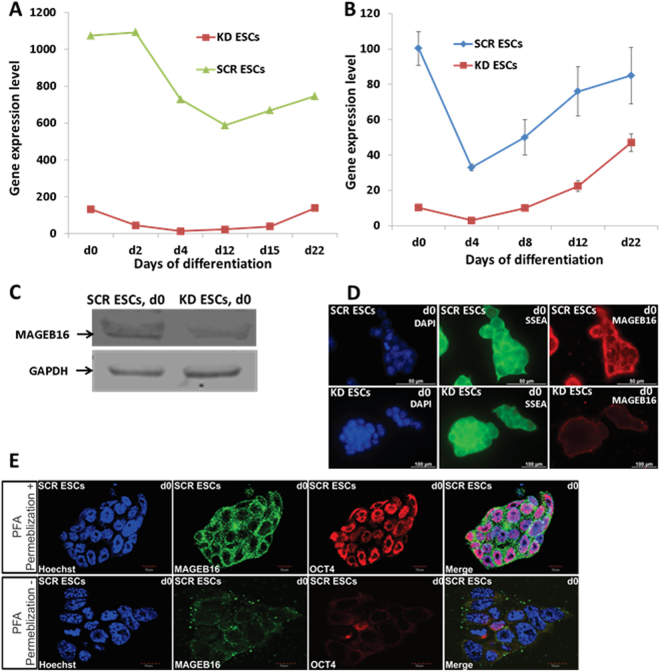
Maximal expression of Mageb16 has been observed in undifferentiated SCR ESCs and the expression started declining at 4-day old embryoid bodies (EBs) (Fig. 1A) (Microarray data). The expression of Mageb16 was drastically reduced in KD ESCs and remained low until 22-days in EBs (Fig. 1A). The efficiency of the Mageb16 knockdown has been additionally confirmed by qPCR and western blot (independent experiments from microarray experiments). As indicated in Fig. 1B, the expression level of Mageb16 in the SCR ESCs was very high (=100%) and declined by 30, 50 and 75% at 4-, 8-, and 22-days in SCR EBs, respectively. In comparison to SCR EBs, the MageB16 mRNA expression level was reduced by more than 90% in KD ESCs and the expression level remained significantly lower in 4-, 8-, and 12-days KD EBs. Knockdown of Mageb16 has also been confirmed at the protein level as shown by western blot analysis (Fig. 1C). MAGEB16 was distributed in cytosol and the cell surface of undifferentiated control SCR ESCs but not in the nucleus (Fig. 1D). No visible Mageb16 was observed in KD ESCs (Fig. 1D). As expected, Ssea1 a cell surface protein associated with undifferentiated ESCs8, was expressed in both undifferentiated SCR and KD ESCs. Confocal imaging of permeabilized and non-permeablized SCR ESCs, further confirmed the localization of MAGEB16 in cytosol and the cell plasma membrane (supplementary Figure 1E).
Figure 1.
Mageb16 expression in differentiated KD and SCR ESCs. (A) Mageb16 expression in undifferentiated and differentiated KD and SCR ESCs (microarray data). (B) qPCR analysis of Mageb16 expression in undifferentiated and differentiated KD and SCR ESCs (2 to 22 day EBs). (C) MAGEB16 protein expression in the KD and SCR ESCs. After preparation of the protein lysates, 10 µg protein was analysed by western blotting. Chemiluminescence detection of MAGEB16 has been performed using MAGEB16 polyclonal antibodies (1:250) and GAPDH has been detected using the anti-GAPDH antibody (1:2500) dilutions. (D) Cellular localization of MAGEB16 and SSEA1 in SCR and KD ESCs. Immunocytochemistry has been performed using primary anti-SSEA1 andibodies (1:50) (green colour) and anti-MAGEB16 antibodies (1:50) (red colour) and goat anti mouse IgM-alexa fluor 488 secondary antibody (1:1000) and goat anti rabbit IgG-alexa fluor 568 as secondary antibody (1: 1000). Cells were co-stained with nuclear marker Hoechst 33342. The overlay of nuclear and MAGEB16 staining reveals that the presence of MAGEB16 is restricted to cytosol and/or surface membrane (scale bar: 100 µm). (E) Confocal microscopy (upper panel+: permebilized cells; lower panel−: non-permebilized). Immunocytochemistry was performed using primary anti-OCT4 antibodies (1:250) (red color) and anti-MAGEB16 antibodies (1:50) (green color) and goat anti rabbit IgM-alexa fluor 488 secondary antibody (1:1000) and goat anti mouse IgG-alexa fluor 568 as secondary antibody (1:1000). Cells were co-stained with nuclear marker Hoechst 33342. The overlay of nuclear, OCT4 and MAGEB16 staining reveals the presence of MAGEB16, which is restricted to cytoplasmic domains of the ESCs (scale bar: 10 µm).
Gene expression modulations regulated by the depleted Mageb16 during differentiation
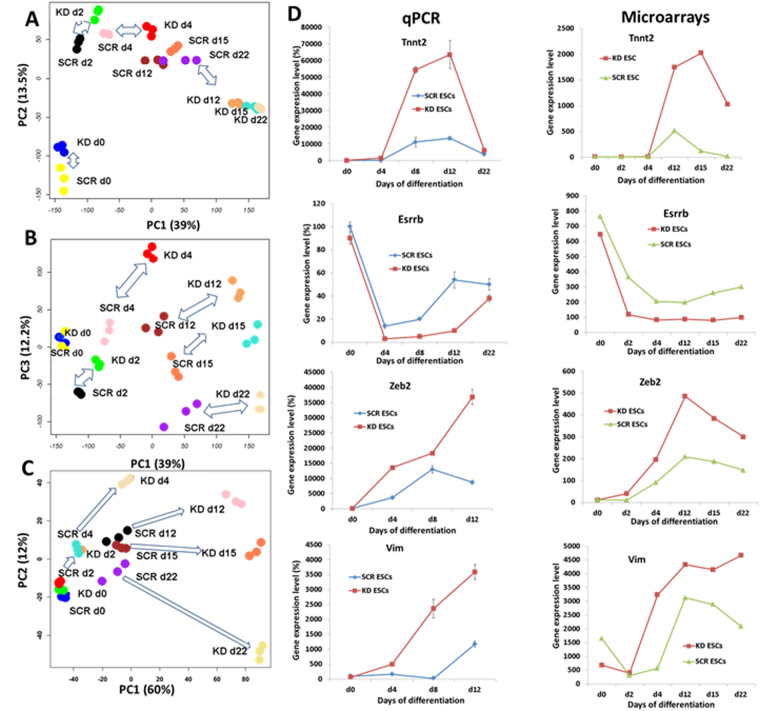
Principal component analysis (PCA) of the transcriptomes was performed for- undifferentiated Mageb16 KD ESCs, SCR ESCs and differentiated (2-, 4-, 12-, 15- and 22 days) EBs of both the populations (Fig. 2). The PCA plot of the analysis of the entire transcriptome of the undifferentiated and differentiated ESC populations and EBs of both Mageb16 depleted and SCR cell types indicates a 39% variance in the principal component 1 PC1,13.5% variance in PC2 and 12.2% in PC3 (Fig. 1A and B). A Clear separation between undifferentiated SCR ESCs and undifferentiated Mageb16 KD ESCs was obvious in PC2 (Fig. 2A). The significant distance between the different differentiation time points of both the populations were seen clearly at PC3 with 12.3% of variance. Using the significance of change FDR corrected P value < 0.05 and size of change at least 2 fold of the gene expression part of statistical correction, we determined 4499 statistically significant expressed probe sets doing statistical extensive comparisons of different cell populations of our study. Among the probe sets there were 174 non-annotated, 225 Riken cDNAs and 3060 annotated genes. As shown in Fig. 2C, significant transcriptome of differentiated KD ESCs and EBs shows clear variance of its earlier expression foldaway compared to control SCR ESCs and EBs. The main difference of a 60% variance was observed in PC 1 whereas 12% variance has been observed in PC2 (in total 72% variance). Validation of microarray data has been performed by choosing five genes (including Mageb16, see Fig. 1A, B) applying qPCR. As shown in Fig. 2D, qPCR data for expression of Tnnt2, Esrrb, Zeb2 and Vim were consistent with microarray gene expression data (To be considered is that the qPCR validation of Vim and Zeb2 has been performed for undifferentiated SCR and KD ESCs and 4-, 8- and 12-day SCR and KD EBs).
Figure 2.
Principal Component Analysis of the entire normalized array datasets. After normalization of the entire transcriptome dataset obtained from both of the undifferentiated and differentiated SCR and Mageb16 KD ESCs (2-day to 22-day EBs), they were analysed by principal component analysis (PCA). Each sphere represents individual sample from a color-coded triplicate sample. (A) PC1 shows the main variability among the transcriptome differences and PC2 shows the second largest variability. (B) PCA plot with PC1 and PC3 to monitor a clear separation at the third largest variance (PC3). (C) PCA of the significantly differential expressed genes (at least 2 fold; FDR corrected P value < 0.05) between undifferentiated and differentiated SCR or undifferentiated and differentiated KD ESCs. A PC1 and PC2 variance of 60% and 12%, respectively, has been obtained indicating a clear separation between the transcriptomes of the differentiated SCR and KD ESCs. (D) Gene expression of representative genes determined by qPCR analysis. The gene expression data of triplicates for each experimental condition are expressed as mean ± SD (*P < 0.05 for KD 12-day versus control SCR 12-day EBs). The expression of Mabeb16 determined by qPCR is shown in Fig. 1B.
K-means clusters and GO analysis
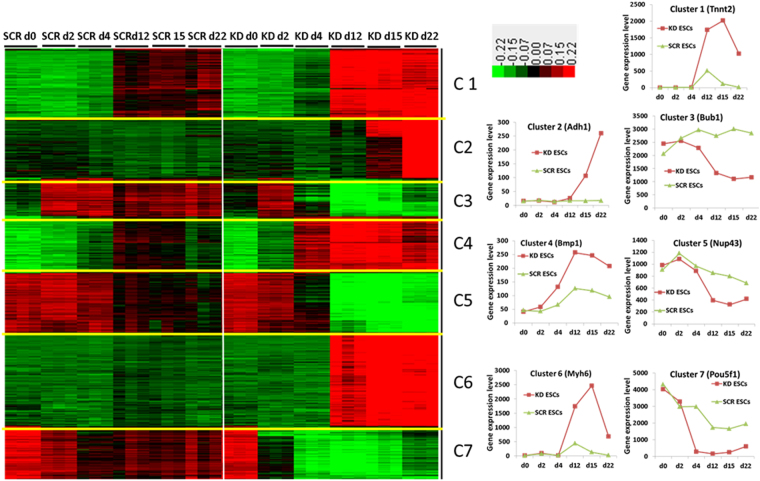
Part of Gene set enrichment analysis, applying the k-means clustering, significantly altered genes (exhibiting at least 2-fold regulation in each condition) were clustered in seven specific gene clusters. The clustering was based on their expression pattern over the different differentiation time points (Fig. 3). Also illustrated on Fig. 3 (right) are the expression pattern of representative genes from the different clusters. To identify specific Gene Ontology (GO), biological processes (BPs), molecular function (MF) and cellular component (CC) along with KEGG pathways, the differentially expressed genes were analysed using Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/) tool. The most biologically significant “parent” terms and more specialized “child” terms of the biological processes of 7 clusters are shown in Table 1. The complete GO analysis is shown in the Supplementary Dataset File.
Figure 3.
Visualization of k-means clustering of differentially expressed probe sets with Euclidean distance measurement and k = 7 group clusters. Replicates are displayed in the vertical axis and genes in the horizontal axis. Log2 transformed signal intensities are depicted in colour code. The heatmap indicates high expression levels in red, intermediate expression level in dark grey and low expression levels in green. Representative diagrams for the gene expression pattern of the different clusters are shown on the right.
Table 1.
Specific GO and KEGG pathways of the cluster specific genes indicated in Fig. 3.
| Term | Gene Nr. | PValue |
|---|---|---|
| Cluster 1 | ||
| GO:0009888~tissue development | 49 | 7.76E-12 |
| GO:0001568~blood vessel development | 24 | 5.57E-08 |
| GO:0001944~vasculature development | 24 | 8.69E-08 |
| GO:0048706~embryonic skeletal system development | 14 | 1.43E-07 |
| GO:0060348~bone development | 15 | 1.58E-06 |
| GO:0051216~cartilage development | 12 | 3.86E-06 |
| GO:0007420~brain development | 23 | 3.90E-06 |
| GO:0001649~osteoblast differentiation | 9 | 2.02E-05 |
| GO:0007417~central nervous system development | 25 | 2.20E-05 |
| GO:0030900~forebrain development | 15 | 8.57E-05 |
| mmu05414:Dilated cardiomyopathy | 11 | 2.16E-04 |
| GO:0060485~mesenchyme development | 8 | 2.41E-04 |
| GO:0043009~chordate embryonic development | 24 | 4.31E-04 |
| GO:0030324~lung development | 10 | 0.0019 |
| GO:0050767~regulation of neurogenesis | 10 | 0.0062 |
| GO:0001654~eye development | 11 | 0.0065 |
| GO:0030902~hindbrain development | 7 | 0.0067 |
| Cluster 4 | ||
| GO:0009888~tissue development | 46 | 1.87E-13 |
| GO:0001568~blood vessel development | 27 | 4.74E-12 |
| GO:0001944~vasculature development | 27 | 8.28E-12 |
| GO:0007507~heart development | 22 | 5.64E-09 |
| GO:0060429~epithelium development | 23 | 3.70E-08 |
| GO:0030324~lung development | 13 | 2.71E-06 |
| GO:0060485~mesenchyme development | 9 | 7.00E-06 |
| GO:0048732~gland development | 16 | 1.27E-05 |
| GO:0007517~muscle organ development | 15 | 1.51E-05 |
| GO:0009952~anterior/posterior pattern formation | 14 | 1.52E-05 |
| GO:0014032~neural crest cell development | 7 | 4.80E-05 |
| mmu04510:Focal adhesion | 14 | 6.91E-05 |
| GO:0022008~neurogenesis | 26 | 1.07E-04 |
| GO:0060348~bone development | 11 | 1.51E-04 |
| GO:0001656~metanephros development | 8 | 1.70E-04 |
| GO:0030326~embryonic limb morphogenesis | 9 | 8.25E-04 |
| GO:0007420~brain development | 16 | 8.65E04 |
| Cluster 2 | ||
| mmu00982:Drug metabolism | 23 | 3.08E-18 |
| mmu00980:Metabolism of xenobiotics by cytochrome P450 | 19 | 1.90E-14 |
| mmu00830:Retinol metabolism | 19 | 3.39E-14 |
| GO:0009888~tissue development | 34 | 1.14E-07 |
| GO:0060429~epithelium development | 21 | 1.92E-07 |
| mmu00140:Steroid hormone biosynthesis | 10 | 1.50E-06 |
| mmu00983:Drug metabolism | 10 | 2.66E-06 |
| GO:0008202~steroid metabolic process | 14 | 1.08E-05 |
| mmu00590:Arachidonic acid metabolism | 10 | 2.43E-04 |
| GO:0008544~epidermis development | 10 | 5.85E-04 |
| GO:0007398~ectoderm development | 10 | 9.15E-04 |
| Cluster 3 | ||
| GO:0007126~meiosis | 11 | 5.73E-07 |
| GO:0005634~nucleus | 89 | 7.74E-06 |
| GO:0000279~M phase | 15 | 6.85E-05 |
| GO:0004089~carbonate dehydratase activity | 5 | 1.02E-04 |
| mmu00270:Cysteine and methionine metabolism | 5 | 7.46E-04 |
| GO:0000775~chromosome, centromeric region | 8 | 8.26E-04 |
| GO:0007283~spermatogenesis | 10 | 0.0125 |
| GO:0006006~glucose metabolic process | 7 | 0.0166 |
| mmu00480:Glutathione metabolism | 4 | 0.0279 |
| Cluster 5 | ||
| GO:0005634~nucleus | 208 | 2.83E-40 |
| GO:0006259~DNA metabolic process | 57 | 1.26E-27 |
| GO:0000279~M phase | 46 | 1.66E-25 |
| GO:0005730~nucleolus | 37 | 1.60E-16 |
| mmu04110:Cell cycle | 17 | 2.62E-09 |
| mmu03030:DNA replication | 8 | 4.07E-06 |
| GO:0006310~DNA recombination | 11 | 7.53E-06 |
| GO:0007126~meiosis | 11 | 3.16E-05 |
| GO:0051327~M phase of meiotic cell cycle | 11 | 3.16E-05 |
| mmu04115:p53 signaling pathway | 9 | 5.43E-05 |
| GO:0005654~nucleoplasm | 28 | 3.18E-04 |
| GO:0034470~ncRNA processing | 12 | 0.001 |
| GO:0040029~regulation of gene expression, epigenetic | 7 | 0.005 |
| GO:0006913~nucleocytoplasmic transport | 8 | 0.006 |
| GO:0046930~pore complex | 6 | 0.022 |
| GO:0007498~mesoderm development | 5 | 0.045 |
| mmu00051:Fructose and mannose metabolism | 5 | 0.005 |
| Cluster 6 | ||
| mmu04610:Complement and coagulation cascades | 31 | 8.09E-21 |
| GO:0002526~acute inflammatory response | 26 | 2.41E-17 |
| GO:0007596~blood coagulation | 22 | 1.83E-14 |
| GO:0019724~B cell mediated immunity | 16 | 5.99E-09 |
| GO:0043292~contractile fiber | 18 | 1.85E-08 |
| GO:0060429~epithelium development | 28 | 1.35E-06 |
| GO:0001944~vasculature development | 24 | 2.98E-05 |
| GO:0019752~carboxylic acid metabolic process | 35 | 1.00E-04 |
| GO:0048732~gland development | 17 | 0.0018 |
| GO:0050873~brown fat cell differentiation | 6 | 0.0032 |
| GO:0007398~ectoderm development | 12 | 0.0079 |
| GO:0031016~pancreas development | 5 | 0.041 |
| Cluster 7 | ||
| GO:0019827~stem cell maintenance | 9 | 2.69E-09 |
| GO:0048864~stem cell development | 9 | 4.07E-09 |
| GO:0005634~nucleus | 115 | 1.94E-07 |
| GO:0010468~regulation of gene expression | 73 | 5.93E-06 |
| GO:0007283~spermatogenesis | 16 | 6.50E-05 |
| GO:0007498~mesoderm development | 6 | 0.004 |
| GO:0007398~ectoderm development | 6 | 0.095 |
| GO:0007492~endoderm development | 3 | 0.096 |
As indicated in Table 1, in comparison to the differentiated SCR ESCs, terms associated with tissue development were significantly enriched in Mageb16 depleted 12-, 15- and 22-day EBs (Fig. 3, Cluster 1). The most prominent BPs were mainly associated with development of mesoderm derivatives such as GO:0001568~blood vessel development, GO:0001944~vasculature development, GO:0048706~embryonic skeletal system development, GO:0051216~cartilage development and GO:0001649~osteoblast differentiation. The second set of most prominent BPs were associated with development of ectodermal derivatives such as GO:0007420~brain development, GO:0007417~central nervous system development and GO:0060429~epithelium development. Similarly, highly significant developmental GOs participating in the development of mesoderm derivatives have also been observed by the analysis of Cluster 4 genes indicating higher expression levels in 4-, 12-, 15-, and 22 days KD EBs in comparison to the differentiated SCR ESCs. This is followed by development of ectoderm derivatives such as GO:0007420~brain development, GO:0007417~central nervous system development and gland development. Analysis of the cluster 2 genes which were highly upregulated in the 22 days KD EBs in comparison to 22 days SCR EBs, indicated metabolism as the main KEGG (mmu00982: Drug metabolism) with the enriched biological GO, GO:0070330~aromatase activity. It included several “child” metabolism associated processes such as GO:0006631~fatty acid metabolic process, mmu00590:Arachidonic acid metabolism and other. Additionally, epithelium-, epidermis-, ectoderm-, gland-, brown fat cell- and lung development have also been identified in the Cluster 2 genes. In comparison to the 12-, 15- and 22-day SCR EBs, cluster 3 genes indicated a low expression level in the appropriate 12-, 15- and 22-day KD EBs. A further analysis indicated that BPs associated with the process of meiosis (GO:0007126~meiosis) and spermatogenesis (Dnmt3a, Rec8, Tex15, Sycp3, Tcfl5, Hsf2, Nr6a1, Mael, Rad18, Nlrp14) were significantly repressed (see Table 1 and Supplementary Dataset File).
Cluster 5 genes showed similar expression levels in KD ESCs, 2-, and 4-day KD EBs in comparison to the SCR ESCs, 2-, and 4-day SCR EBs. In this context, expression of genes belonging to the GO:0006259~DNA metabolic process, GO:0000279~M phase, GO:0005634~nucleus and GO:0005730~nucleolus were significantly repressed in the differentiated 12-, 15- and 22-day KD EBs in comparison to 12-, 15- and 22-day SCR EBs.
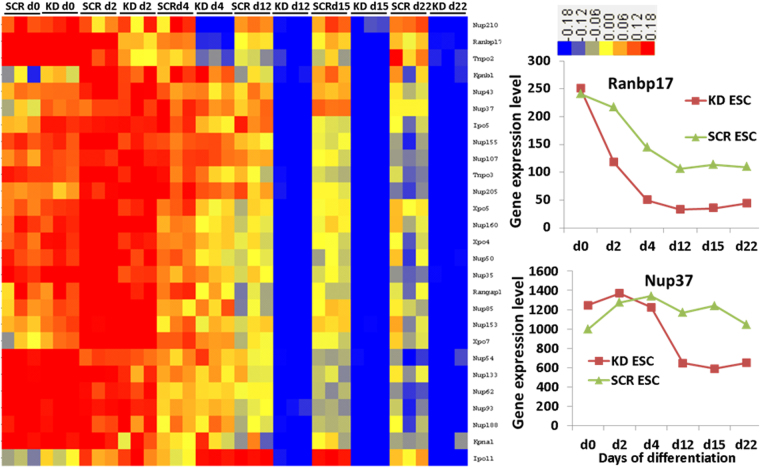
Interestingly, genes belonging to the GO:0007498~mesoderm, GO:0031080~Nup107–160 complex, GO:0006913~nucleocytoplasmic transport (8 genes) and GO:0034470~ncRNA processing, GOTERM_BP_5,GO:0040029~regulation of gene expression and genes involved in epigenetics (Xist, Hat1, Lin28a, Dnmt3b, Baz2a, Brca1, Hells) are also included in cluster 5 genes (Fig. 3, Table 1). The expression pattern of nucleocytoplasmic transport and nuclear pore genes in cluster 5, and in addition, other well annotated genes belonging to these GOs, are shown in Fig. 4. As indicated, gene expression level remained high during differentiation of the KD ESCs and later time points of differentiation in comparison to the SCR EBs. In comparison to SCR ESCs and EBs, several nucleocytoplasmic transport and nuclear pore genes were downregulated after 12-days of differentiation in KD ESCs and EBs (Fig. 4). Some of the genes identified among them are Ranbp17, Nup43, Nup133, Nup37, Kpnb1 and Kpna1. The absolute expression levels of Ranbp17 and Nup37 at the different differentiation time points are also shown in Fig. 4(right).
Figure 4.
Visualization of differentially expressed genes participating in nucleocytoplasmic trafficking. Replicates are displayed in the vertical axis and genes in the horizontal axis. Log2 transformed signal intensities are depicted in colour code. The heatmap indicates high expression levels in red, intermediate expression level in yellow and low expression levels in blue. Representative diagrams on the right show the gene expression of the Ranbp17 and Nup37.
A similar gene expression pattern to cluster 5 has been observed in cluster 7 genes. The differences between the expression level of differentiated KD and SCR ESCs were obvious at 4-day EBs becoming more prominent with increasing time of differentiation. Interestingly, genes belonging to the GO:0019827~stem cell maintenance and GO:0007498~mesoderm development indicated lower expression level in 4 to 22-day KD EBs in comparison to the 4 to 22-day SCR EBs. The comparative expression of a few select genes from these 3 clusters- cluster 3, 5 and 7 is illustrated in the Supplementary Information, Figure S1.
Analysis of cluster 6 genes, which possess higher expression, levels in 12-, 15- and 22 days KD EBs as compared to SCR EBs identified mainly GOs and KEGGs associated with development of mesoderm derivatives. Among them were mmu04610:Complement and coagulation cascades, GO:0001568~blood vessel development, GO:0050873~brown fat cell differentiation, GO:0048732~gland development, GO:0031016~pancreas development and those responsible for functional processes of the heart have been identified.
Expression of key genes associated with the contractility of cardiomyocytes
In order to figure out whether Mageb16 depletion resulted in impaired cardiomyogenesis, (a process that is driven by the mesoderm) we plotted the expression values of key contractility genes such as Tnnt2 (Troponin T2), Actc1 (Alpha-Cardiac Actin), Myl2 (Cardiac Ventricular Myosin Light Chain 2), Myl3, (Cardiac Myosin Light Chain 1), Myh6 (Myosin Heavy Chain, Cardiac Muscle Alpha Isoform) and Myh7 (Myosin Heavy Chain, Cardiac Muscle Beta Isoform). As shown in Figure S2, there were no significant differences in the time kinetic in differentiating ESCs and SCR. However, the expression levels of the contractility genes in KD ESCs were significantly higher in differentiating KD ESCs. In addition, we monitored the contractile foci of 13-days SCR and KD EBs, which exhibit more contractile foci in 12-days KD EBs (see representative videos).
Discussion
The discovery of novel molecular mechanisms controlling embryogenesis under in vivo conditions is problematic, cost-intensive and a time-consuming procedure requiring comprehensive animal studies. Nevertheless the developmental network can be partly recapitulated in vitro by the use of cultured ESCs which can be applied to recognize critical developmental processes. Recently, we have identified Mageb16 as a novel regulator of pluripotency and differentiation in ESCs7. In this context, we have demonstrated that siRNA knockdown of Mageb16- a highly expressed gene marker in the pluripotent state of ESCs, resulted in a high expression of gene markers for all three germ layer- T bra (mesoderm), FGF5 (ectoderm), and HNF4a (endoderm) after 48 h. According to Uniprot database (http://www.uniprot.org/uniprot/Q9CWV4), Mageb16 encodes for a protein of 363 amino acids (41 kDa). The molecular weight of MAGEB16 protein was verified using Western blotting methodology (Fig. 1). According to the Cell-PLoc 2.0 bioinformatics tool for predicting subcellular localization of proteins in different organisms9, MAGEB16 is predicted to be located in the cytosol. We confirmed this prediction using confocal imaging (Fig. 1), wherein, MAGEB16 has been detected in cell surface and/or cytosol.
To identify the mechanism by which MAGEB16 regulates the onset of differentiation in ESCs, we generated KD ESCs in which Mageb16 is silenced. Applying the classical hanging drop protocol, we differentiated the cells until day 22 and performed microarray studies at different time points of differentiation. In summary, DAVID analysis of our data showed that the level of key pluripotency markers such as Pou5f1, Nanog and Sox2 declined faster during differentiated KD ESCs as compared to both SCR and WT ESCs (Figure S1). Based on the GOs and KEGG pathways enriched in Mageb16 deficient differentiated KD ESCs, Mageb16 mainly regulated the development of mesodermal directed lineages such as blood-, blood vessel-, heart-, muscle-, bone-, cartilage-, metanephros-, epithelial- and kidney development. In addition, the enriched KEGG pathways such as ECM-receptor interaction, PPAR signalling pathway (crucial for maintenance of hematopoietic stem cells), linoleic metabolism (associated with mesenchymal stem cells), hypertrophic and dilated cardiomyopathy suggest mesoderm related lineage specification too. However, enrichment of some ectoderm associated biological processes such as neurogenesis, lung development and axonogenesis suggest regulation of ectoderm and epithelial lineages also. Notably, the time kinetic of expression of classic contractility genes (Figure S2) was very similar in differentiating ESCs and SCR. However, the expression values were higher in differentiating KD ESCs which could be correlated to a higher beating activity in 13-days differentiating KD ESCs. These findings suggest a stronger orientation towards cardiomyogenic differentiation in differentiating Mageb16 KD ESCs.
Notably, analysis of microarray data indicated a faster downregulation of pluripotency genes such as Pou5f1, Nanog and Sox2 as compared to SCR ESCs and ESCs (Cluster 7, Figure S1). However, the expression of Pou5f1, Nanog and Sox2 did not completely downregulated in 12-, 15-, and 22-days ESC- and SCR EBs. These results suggest that these factors regulate self-renewal but they also are involved in the differentiation processes of ESCs. In this context, it has been shown that Pou5f1, Nanog and Sox2 not only play an important role in maintaining pluripotency but are also involved in differentiation depending on the presence and expression level of other differentiation factors10,11. As demonstrated earlier, a certain amount of Pou5f1 also induces differentiation of ESCs to mesodermal and primitive endoderm cells10.
Interestingly, while the expression level of pluripotency factors was very similar in undifferentiated KD ESCs and SCR ESCs, significant differences were observed in 12, 15 and 22-day EBs (Fig. 3, cluster 7, Figure S1). Although Mageb16 expression level was very low in KD ESCs, the expression of pluripotency genes identified in Cluster 5 (including Pou5f1, Nanog and Sox2), remained very high and similar to that of undifferentiated SCR ESCs. From these findings, we may conclude that Mageb16 is not directly involved in regulating expression of pluripotency genes but rather acting through fine-tuning the differentiation program of ESCs when differentiation is initiating.
Interestingly, the expression of genes such as Hat1, Lin28a and Brca1 participating in the epigenetic regulation of gene expression, as well as in nucleocytoplasmic transport and ncRNA processing indicated the characteristic cluster 4 expression pattern with significant lower expression levels in 12, 15 and 22-day KD EBs in comparison to the corresponding SCR-or the WT EBs (Figure S1). HAT1 plays an important epigenetic role in cellular chromatin assembly12. Epigenetic DNA modifications including chromatin rearrangements by histone modifications are involved in mammalian developmental processes and are characteristic of differentiation of ESCs toward somatic cells13. Hat1 has been found to be overexpressed in adult stem cells such as hematopoietic stem cells (HSCs)14 and neural stem cells15.
LIN28 is an RNA-binding protein promoting pluripotency of ESCs via regulation of the microRNA let-7 regulates transition between pluripotency and committed cell lineages16. LIN28 is highly upregulated in undifferentiated cells and its expression declined during differentiation16. Our findings also demonstrated high expression levels in SCR ESCs and KD ESCs and the expression declined with increasing time of differentiation with significantly lower levels at 12- to 22-day in KD EBs as compared to corresponding SCR EBs.
There is a clear indication that depletion of Mageb16 is crucial for differentiation of ESCs specifically towards mesoderm depended adipogenesis, osteogenesis, cardiovascular lineages development, limb morphogenesis along with epithelial differentiation and neurogenesis (Cluster 1 and 4). These crucial developmental processes are well orchestrated by the participation of the highly conserved Hox genes (Hoxd9, Hoxc8 and Hoxa9). In this context, Hox genes were found in Cluster 1 and 4 showing a high expression level in 12- to 22-day Mageb16 deficient KD EBs in comparison to the corresponding Mageb16 expressing SCR EBs. Additionally, Tbx4 (cluster 4) and Itga4 which regulate osteoblast differentiation17 as well as Shh and Igfbp5 (cluster 1) which are participating on the development of cardiovascular system18 are also highly upregulated in 12- to 22-day Mageb16-deficient KD ESCs in comparison to the corresponding control EBs. In addition, mesodermal genes were significantly faster downregulated in the differentiated Mageb16 deficient KD ESCs compared to the SCR ESCs (Cluster 7, Table 1) correlating with the prominent formation of mesodermal derivatives.
Interestingly we found in Cluster 5, genes that encode for nuclear pore proteins which are essential for exchanging mRNAs and ncRNA between nucleus and cytosol. The expression level of these genes becomes lower during differentiation of Mageb16 deficient KD ESCs in comparison to Mageb16 expressing differentiated SCR ESCs and ESCs. There is increasing evidence that the process of nucleocytoplasmic trafficking (NCT), including nuclear pore complex proteins regulate ESCs differentiation19–21. Among those genes showing low expression values with progressive differentiation of Mageb16 KD ESCs, Ranbp17, Nup43, Nup133 and Nup37 have been identified. Xpo4, Xpo5 and Xpo7, which belong to the exportin gene family mediating the transport of molecules from nucleus to cytosol, also exhibited low expression levels. Interestingly, importins such as Ipo5 and Ipo11 and exportins such as Xpo4, Xpo5 and Xpo7 are also critical for the regulation of differentiation in ESCs21–23. Normally, their expression is tightly regulated during lineage commitment of ESCs21–23. Moreover, the expression levels of Kpnb1, Kpna1, Tnpo2 and Tnpo3 which are crucial for nuclear protein export21–23 were lower in Mageb16 deficient differentiated KD ESCs. In conclusion, the rapid downregulation of importins and exportins during differentiation of Mageb16 deficient KD ESCs correlates well with faster progression of differentiation. In this context, the opposite phenomenon has been observed by Strip2 deficient KD ESCs. In this context, we recently demonstrated for the first time that Strip2 is essential for the onset of the differentiation in ESCs24,25.
Until now, the functional role of Mageb16 is unknown. More recently it has been demonstrated that among different tissues, MAGEB16 was highly upregulated only in human and mice testis6. Furthermore, authors reported that the CpG islands of the 5′ upstream region of MAGEB16 was highly demethylated in the testis in striking contrast to highly methylated MAGEB16 expressed in other tissues6. The authors have also suggested that MAGEB16 is a critical regulator of spermatogenesis. This report6 strongly supports our findings, demonstrating that genes involved in the process of meiosis and spermatogenesis (Dnmt3a, Rec8, Tex15, Sycp3, Tcfl5, Hsf2, Nr6a1, Mael, Rad18, Nlrp14) were downregulated at later time points of differentiation in the Mageb16 KD ESCs in comparison to Mageb16 expressing SCR ESCs (Cluster 3 genes, Table 1 and Table S1). Accordingly, among the spermatogenesis genes, DNMT3A was reported to induce epigenetic modification of DNA via CpG island methylation, which again suggests the important role of MAGEB16 in spermatogenesis. One of the key regulators of spermatogenesis is Mael (Supplementary Information, Figure S1) which is expressed in male germ cells and testis but under pathological conditions also in cancer cells26. Demethylation of CpG islands in Mael is also associated with its expression26. Moreover, in the absence of MAGEB16, spermatogenesis genes become downregulated during formation of spermatocytes. From these findings, we may suggest that Mageb16 is a master regulator of spermatogenesis. Our results additionally demonstrate that MAGEB16 is a key regulator of differentiation processes in ESCs via regulation of the expression of pluripotency factors. In the absence of Mageb16, expression level of pluripotency factors declined more rapidly during differentiation of ESCs and their expression level remained very low as compared to MAGEB16 expressing ESCs. Our findings give strong evidences that Mageb16 could be critical for the expression of genes participating in nucleocytoplasmic trafficking of proteins and RNAs between cytosol and nucleus (and vice versa), which in turn may regulate the expression of pluripotency factors such as Pou5f1 and Nanog. However, the cellular mechanisms by which Mageb16 affect the nucleocytoplasmic trafficking of factors essential for differentiation of ESCs remain to be elucidated.
Methods
Murine ESC culture and Mageb16 KD stable cell line generation
Permanent knockdown of Mageb16 has been performed using RNA interference technology that employs a short hairpin RNA targeting Mageb16. Culturing of the murine CGR8 ESCs (ECACC 95011018) was performed as reported previously7. Four sets of shRNA Plasmid vector (pGFP-V-RS_HuSH TM Origene) targeting Mageb16 are used. These plasmids are retroviral silencing plasmids of Origene (Rockville, MD, USA) with GFP plus screening for transfection efficiency and puromycin resistance for stable polyclonal cell line generation. The shMageb16 target sequence on its mRNA was 5′-CATTCCACCAGATCAGTCAGATAGCACAG-3′ from the exon region (ENSMUSE00000697770) and the sequence is common to both Mageb16 transcript variants 1 and 2. Non-targeting scrambled shRNA plasmid (TR30013) was also purchased from Origene. Transfection complex of Lipofectamine® LTX with Plus™ reagent (Life Technologies Cn: 15338–100) and shRNA plasmid targeting Mageb16 was prepared and transfected into CGR8 as per the protocol suggested by the producer. A similar transfection complex with shRNA plasmid of non-target was prepared and transfected into CGR8 for Scrambled. The selection process was initiated immediately after 24 hours of transfection with 3.5 µg/ml puromycin to have stable knockdown clones. After 12 days of transfection, the surviving and stably proliferating cells were selected for further investigations.
Differentiation of ESCs using hanging drop protocol
Random differentiation of Mageb16 KD ESCs (KD ESCs), scrambled ESCS (SCR ESCs) and wild type ESCs (WT ESCs) was performed using hanging drop method for forming embryoid bodies (EBs) as described previously7. EBs from all three-cell populations were collected at different time intervals- 2-, 4-, 12, 15- and day 22 for microarray studies.
RNA isolation and microarray dataset generation
Total RNA was isolated from ESCs and EBs generated with KD-, SCR- and WT ESCs using RNeasy mini kit (Qiagen, Hilden, Germany). Then, 100 ng total RNA was used for amplified RNA (aRNA) amplification with GeneChip 3′ IVT Express Kit (Affymetrix, Santa Clara, CA, USA). After 16 h of biotinylated in vitro transcription, aRNA was purified and 15 μg of purified aRNA was fragmented with fragmentation buffer. Next, 12.5 μg of fragmented aRNA was hybridized with Mouse Genome 430 2.0 arrays (Affymetrix) for 16 h at 45 °C. Arrays were washed and stained with phycoerythrin with Affymetrix Fluidics Station 450 and scanned using Affymetrix Gene-Chip Scanner 3000 7 G (Affymetrix).
Transcriptome analysis and Gene Ontology (GO) enrichment Analysis
The quality control matrices were confirmed with Affymetrix GCOS software. The raw data were background corrected, summarized and normalized using RMA algorithm executed by R bioconductor packages27. A PCA was performed to observe the transcriptome variability between the different time points of differentiation. Significantly regulated transcripts were determined by empirical Bayes linear model applied using the LIMMA package in R28. The significance of the change was calculated correcting P-value of t-score with false discovery rate using Benjamini–Hochberg method at P < 0.05 and a 2-fold size of change was calculated. Furthermore significantly differential expressed probe sets were analysed by k-mean cluster analysis after a transcript-wise normalization of signal values to a mean of 0 and standard deviation (SD) of 1 using Euclidian distance measurement and k = 7, using the Cluster 3.0 tool. To determine the biological significance of differentially expressed transcripts (DETs), the differential expressed genes were analysed by the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics tool (http://david.abcc.ncifcrf.gov/). DAVID provides biological processes (BPs), molecular function (MF) and cellular component (CC) for the differential expressed transcripts (DET) with an EASE enrichment score at P < 0.01. For filtering the highly significant enriched Biological Processes and KEGG pathways, adjusted P value with Benjamini correction was used with the threshold p value < 0.05.
Validation of microarray data sets with qRT-PCR
ABI 7500 FAST Detection System (Applied Biosystems) was used to read the gene profile generated at quantitative real-time-polymerase chain reaction. Total RNA from KD- SCR- and WT ESCs and from their EBs at day 2-, 4-, 12-, 15- and 22-day old EBs was reverse transcribed using SuperScript VILO cDNA synthesis kit with random hexamers as suggested (Invitrogen GmbH). qRT-PCR was performed in triplicates for every sample using QuantiFast SYBER Green PCR assay (Qiagen) and TaqMan Gene Expression Assay (Life Technologies). QuantiFast SYBER Green PCR assay reactions were performed with the following conditions, beginning with Taq activation at 95 °C for 5 min, thereafter 40 cycles of 10 sec denaturation at 95 °C, 30 sec of combined annealing and extension at 60 °C and finally ending with a melting curve acquisition. Primers used are shown in Table S2.
Western blotting and immunohistochemistry analysis
Western blot analysis was performed with 10 µg total protein. Total protein extracts were separated using 4–12% Bis-Tris Plus precast polyacrylamide gels (Thermo Fisher, Karlsruhe, Germany) by electrophoresis and blotted onto polyvinylidene fluoride membranes (Thermo Fisher, Karlsruhe, Germany). After blocking the membranes with 5% non-fat milk suspended in T-PBS (0.1% Tween 20, Sigma–Aldrich), the membranes were incubated with primary antibodies in 1% non-fat milk at 4 °C overnight. The proteins were visualized using the ECL Pierce Fast Western Blot system (Thermo Fisher Scientific, 35050).
Polyclonal antibodies against MAGEB16 were generated by Pineda Antibody services by immunization of rabbits with the peptide Mageb16-3: NH2-CQGSPVIPPDQSDSTDL-CONH2 (17 amino acids). Anti-GAPDH (Abcam, ab 9485) was purchased from Abcam (Abcam, Cambridge, UK). For immunocytochemistry analysis, undifferentiated KD and SCR ESCs were plated on gelatine-coated coverslips for 2 days. On day 2, the ESCs were fixed with 99% methanol (Roth, Germany) at −20 °C for 10 min. Thereafter cells were permeabilized with 0.3% triton X-100 (Sigma-Aldrich, Germany) at room temperature (RT) for 20 min. Cells were blocked with 5% bovine serum albumin (BSA) (PAA, Austria) and stained with anti-MAGEB16 (1:50) and anti SSEA-1 (Santa cruz, sc-101462, 1:50). Primary antibodies were detected with species matched Alexa-488 conjugated secondary antibodies (Invitrogen, USA). Alexa-568 conjugated secondary antibodies and DAPI (Invitrogen, USA) were used to stain cell nuclei as recommended by the manufacturer’s protocol. Images were captured with an Axiovert 200 microscope and Axiovision 4.3 software (Carl Zeiss, Germany).
We further visualized the expression of MAGEB16 using confocal microscopy. For this aim immunocytochemistry was performed with 4% PFA fixed undifferentiated SCR ESCs on day2. To visualize MAGEB16, staining was performed with and without permeabilization. Briefly, ESCs were fixed with 4% PFA (Roth, Germany) at room temperature for 10 min. For permeabilization, cells were permeabilized with 0.3% triton X-100 (Sigma-Aldrich, Germany) at room temperature (RT) for 20 min and some cells were preceded without permeabilization to next step (non permeabilized). The cells were then blocked with 5% bovine serum albumin (BSA) (PAA, Austria) and stained with anti-MAGEB16 (1:50) and anti OCT4 (Santa cruz, sc-5279, 1:250). Primary antibodies were detected with species matched Alexa-488 conjugated secondary antibodies (Invitrogen, USA). Alexa-568 conjugated secondary antibodies and Hoechst (Invitrogen, USA) were used to stain cell nuclei as recommended by the manufacturer’s protocol. Images were captured with an Olympus FluoView1000 confocal system (Olympus).
Data availability statement
The microarray data were submitted to the GEO database with the association number for the full mRNA expression dataset GSE103615.
Electronic supplementary material
Acknowledgements
The present work has been supported by the DFG (SA 568/17-2). We would like to thank Ms. Susan Romani for contributing for the microarray experiments.
Author Contributions
J.A.D., performed the experiments, contributes to the analysing of the microarray data and contributed to the writing of the manuscript. A.S., supervised the study, wrote the manuscript and prepared the figures and tables. S.P.S., performed the Western blotting and the qPCR experiments. P.S., and J.H., carefully read the manuscript and improved the language quality of the manuscript. M.X.D., contributed to the analysing of the microarray data. S.P performed the confocal microscopy with an Olympus FluoView1000 confocal system.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
John Antonydas Gaspar and Sureshkumar Perumal Srinivasan contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-14561-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.van der Bruggen P, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 2.Zhang G, Zhou H, Xue X. Complex roles of NRAGE on tumor. Tumour Biol. 2016 doi: 10.1007/s13277-016-5084-0. [DOI] [PubMed] [Google Scholar]
- 3.Barker PA, Salehi A. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J Neurosci Res. 2002;67:705–712. doi: 10.1002/jnr.10160. [DOI] [PubMed] [Google Scholar]
- 4.Lopez-Sanchez N, Gonzalez-Fernandez Z, Niinobe M, Yoshikawa K, Frade JM. Single mage gene in the chicken genome encodes CMage, a protein with functional similarities to mammalian type II Mage proteins. Physiol Genomics. 2007;30:156–171. doi: 10.1152/physiolgenomics.00249.2006. [DOI] [PubMed] [Google Scholar]
- 5.Chomez P, et al. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 2001;61:5544–5551. [PubMed] [Google Scholar]
- 6.Liu Y, et al. Demethylation of CpG islands in the 5′ upstream regions mediates the expression of the human testis-specific gene MAGEB16 and its mouse homolog Mageb16. BMB Rep. 2014;47:86–91. doi: 10.5483/BMBRep.2014.47.2.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaspar JA, et al. Gene expression signatures defining fundamental biological processes in pluripotent, early, and late differentiated embryonic stem cells. Stem Cells Dev. 2012;21:2471–2484. doi: 10.1089/scd.2011.0637. [DOI] [PubMed] [Google Scholar]
- 8.Muramatsu T. Embryoglycan: a highly branched poly-N-acetyllactosamine in pluripotent stem cells and early embryonic cells. Glycoconj J. 2016 doi: 10.1007/s10719-016-9673-3. [DOI] [PubMed] [Google Scholar]
- 9.Chou KC, Shen HB. A new method for predicting the subcellular localization of eukaryotic proteins with both single and multiple sites: Euk-mPLoc 2.0. PLoS One. 2010;5:e9931. doi: 10.1371/journal.pone.0009931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Oron E, Nelson B, Razis S, Ivanova N. Distinct Lineage Specification Roles for NANOG, OCT4, and SOX2 in Human Embryonic Stem Cells. Cell Stem Cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 12.Parthun MR. Hat1: the emerging cellular roles of a type B histone acetyltransferase. Oncogene. 2007;26:5319–5328. doi: 10.1038/sj.onc.1210602. [DOI] [PubMed] [Google Scholar]
- 13.Golob JL, Paige SL, Muskheli V, Pabon L, Murry CE. Chromatin remodeling during mouse and human embryonic stem cell differentiation. Dev Dyn. 2008;237:1389–1398. doi: 10.1002/dvdy.21545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park IK, et al. Differential gene expression profiling of adult murine hematopoietic stem cells. Blood. 2002;99:488–498. doi: 10.1182/blood.V99.2.488. [DOI] [PubMed] [Google Scholar]
- 15.Lim DA, et al. In vivo transcriptional profile analysis reveals RNA splicing and chromatin remodeling as prominent processes for adult neurogenesis. Mol Cell Neurosci. 2006;31:131–148. doi: 10.1016/j.mcn.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Tsialikas J, Romer-Seibert J. LIN28: roles and regulation in development and beyond. Development. 2015;142:2397–2404. doi: 10.1242/dev.117580. [DOI] [PubMed] [Google Scholar]
- 17.Seifert A, Werheid DF, Knapp SM, Tobiasch E. Role of Hox genes in stem cell differentiation. World J Stem Cells. 2015;7:583–595. doi: 10.4252/wjsc.v7.i3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein D, Benchellal M, Kleff V, Jakob HG, Ergun S. Hox genes are involved in vascular wall-resident multipotent stem cell differentiation into smooth muscle cells. Sci Rep. 2013;3:2178. doi: 10.1038/srep02178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D’Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, Hetzer MW. A change in nuclear pore complex composition regulates cell differentiation. Dev Cell. 2012;22:446–458. doi: 10.1016/j.devcel.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacinto FV, Benner C, Hetzer MW. The nucleoporin Nup153 regulates embryonic stem cell pluripotency through gene silencing. Genes Dev. 2015;29:1224–1238. doi: 10.1101/gad.260919.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yasuhara N, Yoneda Y. Importins in the maintenance and lineage commitment of ES cells. Neurochem Int. 2017;105:32–41. doi: 10.1016/j.neuint.2017.01.020. [DOI] [PubMed] [Google Scholar]
- 22.Okada N, et al. Importins and exportins in cellular differentiation. J Cell Mol Med. 2008;12:1863–1871. doi: 10.1111/j.1582-4934.2008.00437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J, et al. Gating pluripotency via nuclear pores. Trends Mol Med. 2014;20:1–7. doi: 10.1016/j.molmed.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 24.Sabour D, et al. STRIP2 Is Indispensable for the Onset of Embryonic Stem Cell Differentiation. Mol Ther Methods Clin Dev. 2017;5:116–129. doi: 10.1016/j.omtm.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagh V, et al. Fam40b is required for lineage commitment of murine embryonic stem cells. Cell Death Dis. 2014;5:e1320. doi: 10.1038/cddis.2014.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao L, et al. Identification of a novel human cancer/testis gene MAEL that is regulated by DNA methylation. Mol Biol Rep. 2010;37:2355–2360. doi: 10.1007/s11033-009-9741-x. [DOI] [PubMed] [Google Scholar]
- 27.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 28.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data were submitted to the GEO database with the association number for the full mRNA expression dataset GSE103615.