Abstract
Aim:
Analysis reconstruction networks from two diseases, IBD and NASH and their relationship, based on systems biology methods.
Background:
IBD and NASH are two complex diseases, with progressive prevalence and high cost for countries. There are some reports on co-existence of these two diseases. In addition, they have some similar risk factors such as age, obesity, and insulin resistance. Therefore, systems biology approach can help to discover their relationship.
Methods:
DisGeNET and STRING databases were sources of disease genes and constructing networks. Three plugins of Cytoscape software, including ClusterONE, ClueGO and CluePedia, were used to analyze and cluster networks and enrichment of pathways. Based on degree and Betweenness, hubs and bottleneck nodes were defined.
Results:
Common genes between IBD and NASH construct a network with 99 nodes. Common genes between IBD and NASH were extracted and imported to STRING database to construct PPI network. The resulting network contained 99 nodes and 333 edges. Five genes were selected as hubs: JAK2, TLR2, TP53, TLR4 and STAT3 and five genes were selected as bottleneck including: JAK2, TP53, AGT, CYP3A4 and TLR4. These genes were hubs in analysis network that was constructed from hubs of NASH and IBD networks.
Conclusion:
Systems biology methods, specifically PPI networks, can be useful for analyzing complicated related diseases. Finding Hub and bottleneck proteins should be the goal of drug designing and introducing disease markers.
Key Words: Inflammatory bowel diseases (IBD), Non-alcoholic steatohepatitis (NASH), Protein-protein interaction (PPI) network analysis, Hub-bottlenecks, Protein clusters
Introduction
Non-alcoholic steatohepatitis (NASH) is a subtype of Non-alcoholic fatty liver disease (NAFLD) which has the potential to progress to cirrhosis, hepatic failure or hepatocellular carcinoma (1-3). This disease is a liver inflammation caused by fat accumulation in the liver. NASH and NAFLD are common diseases in industrial countries (4) and have been reported in Australia, India, Japan, the Middle East, New Zealand, North America, South America, northern Europe, southern Europe and South East Asia (4). In fact nowadays, NASH is the second liver disease in the United States and will be a gold standard for liver transplant in a few years (5). Biopsy of liver is the only way for diagnosing NASH, but it is a costly and error-prone method (6) with some risks (7). So, reducing the cost of early diagnosis of this disease is very important (8).
Inflammatory bowel disease (IBD) is a chronic gastrointestinal disorder that consists primarily of two types: ulcerative colitis and Crohn's disease. Both usually involve severe diarrhea, pain, fatigue and weight loss. This disease is caused by dysregulated immune response to host intestinal microflora (9). IBD can be debilitating and sometimes leads to life-threatening complications (10). IBD patients may develop some complicated diseases such as sclerosing cholangitis and autoimmune hepatitis. Most occurrences of IBD are seen in North America, resulting in high costs of health care measures (11). IBD patients are more likely to show overweight and obesity (12-14). Interestingly, these obese persons are at risk of NASH (15). Some scientist report the incidence of NASH in IBD in range of 6.2% to 40% (16-18). Gisbert et al analyzed 786 IBD patients (49% CD and 51% UC subgroup) and reported 40.8% prevalence of NASH in these patients (16). In a study by Sourianarayanane et al, the same investigation was performed in 928 patients (53% CD and 47% UC subgroup) to find an incidence of only 8.2%. Indeed, some NASH risk factors were reported in IBD population, such as small bowel surgery (13), hypertension (13), obesity (13), steroid use (13), active disease (19), duration of IBD (19), prior IBD surgery (19) and anti-TNFα use (19). However, the pathogenesis of NASH in IBD patients is a mystery because of disease-specific risk factors, such as chronic inflammation, drug-induced hepatotoxicity, steroid exposure, malnutrition and gut dysbiosis that is shared between both diseases (20, 21). Systems biology methods can be helpful to provide a new perspective of shared molecular mechanisms in related diseases such as IBD and NASH (22-25). Protein-protein interaction (PPI) network analysis is one of the major fields in systems biology in which analyzed complex interactome of proteins as a main source of data (26). Using systems biology method such as comparison between gene sets of diseases, constructing PPI network and pathway enrichment can be helpful to decipher the shared mechanism of IBD and NASH. In this study, we reported seven important shared proteins between these diseases that can be used not only as markers of disease, but also as targets for drug designing.
Methods
DisGeNET is a discovery platform containing one of the largest publicly available collections of genes and variants associated with human diseases (27). The related genes of IBD and NASH were exported from DisGeNET database and used to construct PPI network. The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING), a database for predicted protein-protein interactions at EMBL clusters the extracted results from many protein-protein interactions databases, like Mint, BioGrid, etc. It also uses the information from KEGG pathways and reactome to provide the best annotations for the interactions of one protein (28). We constructed IBD and NASH networks by submitting gene list to STRING database and analyzed the networks by Cytoscape software (29).
A network is composed of nodes (e.g., genes or proteins) and edges/links (e.g., co-expression relationships or physical interactions). In network biology terms, degree, and Betweenness are important centrality parameters that are useful for analysis network topology. Edges/links of a node are called the degree of that node. Nodes with high degree are called hubs and nodes that achieve top-ten or top-five percent of betweenness centrality are called bottlenecks (both based on researcher’s definition) (30). So, nodes that are simultaneously hubs and bottlenecks are named hub-bottlenecks (31). Average degree (A.D) and standard deviation (SD) of degrees were calculated and nodes with degree above two*SD + A.D were selected as hub proteins in each network. Also, the top five percent of betweenness centrality measures were selected as bottleneck proteins. Shared genes, hubs and bottleneck proteins of these two networks were extracted and used for further analysis. The common network was constructed by importing shared genes in STRING database and clustered by ClusterONE plugin of Cytoscape software (32) that finds overlapping protein complexes in a protein interaction network loaded into Cytoscape. (overlap threshold = 1, node penalty = 0, haircut threshold = 0) (33). Pathway enrichment and the relation between pathways were accomplished using ClueGO and CluePedia plugins of Cytoscape software (34, 35).
Results
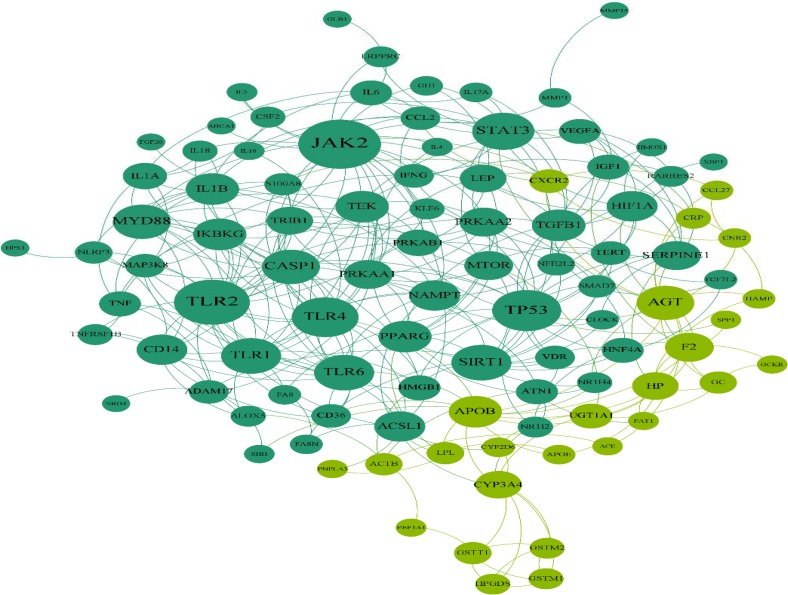
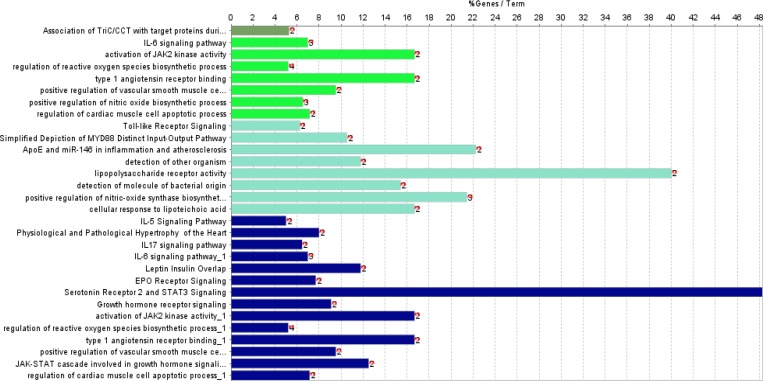
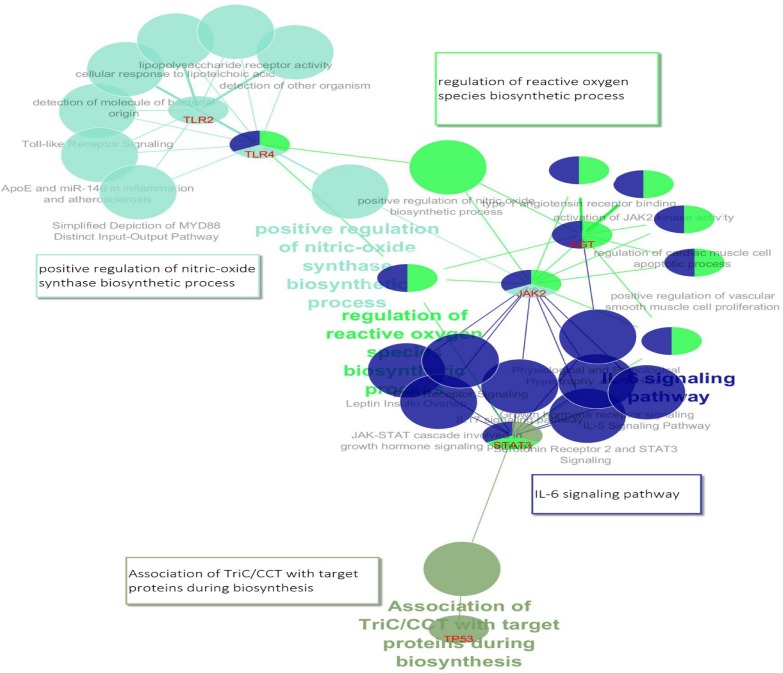
From DisGeNET, 838 and 331 genes were extracted for IBD and NASH, respectively. Totally, 113 genes were shared between the two lists and were named common genes. The common genes network that was constructed using STRING database, has 99 nodes and 333 edges and two clusters (figure 1). Five genes were selected as hubs: JAK2, TLR2, TP53, TLR4 and STAT3; and five genes were selected as bottleneck including: JAK2, TP53, AGT, CYP3A4 and TLR2 (Table 1). Therefore, in this network, we have three hub- bottleneck genes: JAK2, TLR2 and TP53. Also clustering yielded two cluster: cluster one with 73 nodes (p-value: 0.00004) and cluster two with 23 nodes (p-value: 0.0032) (figure 1). In addition, IBD and NASH networks were constructed by STRING database. The IBD network from STRING analysis consists of 653 nodes and 4110 edges, while the NASH network comprises 257 nodes and 965 edges. Analyzing IBD and NASH networks showed that 54 hubs and three bottlenecks were common between these two networks (Table 2). All hubs and bottlenecks in the common gene network appeared in the list of common hubs from IBD and NASH networks (Table2). So these seven genes were used for pathways enrichment and gene ontology. Gene ontology results showed 24 pathways that can be classified in four clusters (Table 3 and figure 2). These clusters were named based on the main cluster’s pathway: 1. Association of TriC/CCT with target proteins during biosynthesis; 2. IL-6 signaling pathway; 3. Regulation of reactive oxygen species biosynthetic process; and 4. Positive regulation of nitric oxide biosynthetic process. Due to shared mechanism, some pathways appeared in more than one cluster. CluePedia analysis showed that IL-6 signaling pathway group was the central group that connects other groups (Figure3).
Figure 1.
Common gene network containing 99 nodes and 333 edges. This network includes two modules that are highlighted by dark green (cluster one) and light green (cluster two
Table 1.
Shared Hub and Bottleneck genes of IBD and NASH networks. Hub-bottleneck genes that are the same as table 1 are marked with asterisks
| Hubs | Degree | Bottlenecks | B.N |
|---|---|---|---|
| GMPS | 3 | SIRT1 | 0.116186 |
| PRKAG2 | 12 | TP53** | 0.100492 |
| MLST8 | 9 | HIF1A | 0.086405 |
| MTOR | 18 | ||
| MAP3K7 | 22 | ||
| TRAF6 | 19 | ||
| PRKAG1 | 12 | ||
| PRKAB1 | 13 | ||
| PRKAB2 | 12 | ||
| ACTB | 3 | ||
| ACTG1 | 4 | ||
| PRKAA2 | 14 | ||
| SERPINC1 | 5 | ||
| F2 | 7 | ||
| RPTOR | 18 | ||
| TLR4** | 17 | ||
| IKBKG | 18 | ||
| BIRC3 | 12 | ||
| TRAF3 | 11 | ||
| TICAM1 | 11 | ||
| UBE2N | 12 | ||
| BIRC2 | 14 | ||
| MYD88 | 15 | ||
| IKBKB | 21 | ||
| SERPIND1 | 5 | ||
| FGG | 11 | ||
| PRKAA1 | 16 | ||
| TP53** | 20 | ||
| CREBBP | 22 | ||
| MDM4 | 5 | ||
| HIF1A | 10 | ||
| CD14 | 12 | ||
| TLR2** | 18 | ||
| AGT** | 9 | ||
| TERT | 6 | ||
| STAT3** | 17 | ||
| JAK2** | 20 | ||
| HMGB1 | 9 | ||
| SIRT1 | 9 | ||
| PPARG | 9 | ||
| VEGFA | 8 | ||
| LEP | 11 | ||
| IL6 | 3 | ||
| IGF1 | 6 | ||
| IL1B | 8 | ||
| TGFB1 | 11 | ||
| IFNG | 4 | ||
| SERPINE1 | 7 | ||
| IL4 | 3 | ||
| CSF2 | 4 | ||
| APOB | 7 | ||
| GH1 | 3 | ||
| TEK | 11 | ||
| CYP3A4** | 2 |
Table 2.
Hub and Bottleneck genes of Common network genes. Hub-bottleneck genes are bolded
| Hubs | Degree | Bottlenecks | Betweenness score |
|---|---|---|---|
| JAK2 | 25 | JAK2 | 0.160941 |
| TLR4 | 22 | TP53 | 0.10348 |
| TP53 | 19 | AGT | 0.102813 |
| TLR2 | 18 | CYP3A4 | 0.083948 |
| STAT3 | 16 | TLR4 | 0.083942 |
Table 3.
Enriched pathways list, based on seven genes that are shared between table 1 and table 2
| GO Term | Ontology Source | Adj-PValue | Associated Genes Found | GO Groups |
|---|---|---|---|---|
| Association of TriC/CCT with target proteins during biosynthesis | REACTOME | 160.0E-6 | STAT3, TP53 | 0 |
| IL-6 signaling pathway | WikiPathways | 7.1E-6 | AGT, JAK2, STAT3 | 1 |
| activation of JAK2 kinase activity | GO_BiologicalProcess | 100.0E-6 | AGT, JAK2 | |
| regulation of reactive oxygen species biosynthetic process | GO_BiologicalProcess | 180.0E-9 | AGT, JAK2, STAT3, TLR4 | |
| type 1 angiotensin receptor binding | GO_MolecularFunction | 100.0E-6 | AGT, JAK2 | |
| positive regulation of vascular smooth muscle cell proliferation | GO_BiologicalProcess | 210.0E-6 | AGT, JAK2 | |
| positive regulation of nitric oxide biosynthetic process | GO_BiologicalProcess | 8.3E-6 | AGT, JAK2, TLR4 | |
| regulation of cardiac muscle cell apoptotic process | GO_BiologicalProcess | 210.0E-6 | AGT, JAK2 | |
| Toll-like Receptor Signaling | WikiPathways | 170.0E-6 | TLR2, TLR4 | 2 |
| Simplified depiction of MYD88 distinct input-output pathway | WikiPathways | 190.0E-6 | TLR2, TLR4 | |
| ApoE and miR-146 in inflammation and atherosclerosis | WikiPathways | 62.0E-6 | TLR2, TLR4 | |
| detection of other organism | GO_BiologicalProcess | 170.0E-6 | TLR2, TLR4 | |
| lipopolysaccharide receptor activity | GO_MolecularFunction | 18.0E-6 | TLR2, TLR4 | |
| detection of molecule of bacterial origin | GO_BiologicalProcess | 110.0E-6 | TLR2, TLR4 | |
| positive regulation of nitric-oxide synthase biosynthetic process | GO_BiologicalProcess | 220.0E-9 | JAK2, TLR2, TLR4 | |
| cellular response to lipoteichoic acid | GO_BiologicalProcess | 100.0E-6 | TLR2, TLR4 | |
| IL-5 Signaling Pathway | WikiPathways | 90.0E-6 | JAK2, STAT3 | 3 |
| Physiological and Pathological Hypertrophy of the Heart | WikiPathways | 240.0E-6 | AGT, STAT3 | |
| IL17 signaling pathway | WikiPathways | 210.0E-6 | JAK2, STAT3 | |
| IL-6 signaling pathway | WikiPathways | 7.1E-6 | AGT, JAK2, STAT3 | |
| Leptin Insulin Overlap | WikiPathways | 170.0E-6 | JAK2, STAT3 | |
| EPO Receptor Signaling | WikiPathways | 220.0E-6 | JAK2, STAT3 | |
| Serotonin Receptor 2 and STAT3 Signaling | WikiPathways | 11.0E-6 | JAK2, STAT3 | |
| Growth hormone receptor signaling | REACTOME | 210.0E-6 | JAK2, STAT3 | |
| activation of JAK2 kinase activity | GO_BiologicalProcess | 100.0E-6 | AGT, JAK2 | |
| regulation of reactive oxygen species biosynthetic process | GO_BiologicalProcess | 180.0E-9 | AGT, JAK2, STAT3, TLR4 | |
| type 1 angiotensin receptor binding | GO_MolecularFunction | 100.0E-6 | AGT, JAK2 | |
| positive regulation of vascular smooth muscle cell proliferation | GO_BiologicalProcess | 210.0E-6 | AGT, JAK2 | |
| JAK-STAT cascade involved in growth hormone signaling pathway | GO_BiologicalProcess | 160.0E-6 | JAK2, STAT3 | |
| regulation of cardiac muscle cell apoptotic process | GO_BiologicalProcess | 210.0E-6 | AGT, JAK2 |
Figure 2.
Enriched pathways by seven genes that are shared between table 1 and table 2. Pathways are colored based on classification
Figure 3.
CluePedia map of enriched pathways, main pathways and their relation to genes are shown. Four groups of GO in table 3 are shown in this figure schematically. Group 0: Association of TriC/CCT with target proteins during biosynthesis, dark green; Group 1: IL-6 signaling pathway, dark blue; Group 2: regulation of reactive oxygen species biosynthetic process, light green; positive regulation of vascular smooth muscle cell proliferation, light blue
Discussion
Systems biology methods such as PPI network analysis and pathway enrichment have been used broadly to discover main proteins and pathways underlay complex diseases (36). Different types of cancers, various kinds of neurodegenerative diseases and disorders and also many cellular condition are analyzed via protein-protein interaction method (37-42) The co-existence of NAFLD with IBD is becoming increasingly recognized (43). In this study, we used the complete genes list of the two diseases (IBD and NASH) that may have shared mechanism based on risk factors and previous studies (44). According to network analysis of the common network (see table 1) and also findings in table 2, it is indicated that seven key genes are related to the two diseases. The comparative study of several diseases has shown that there are common informatics biomarker panels in the studied cases (38, 39) . Five of the seven selected genes (JAK2, TLR2, TP53, TLR4 and STAT3) were in one cluster in Common genes network. The Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling cascade is a central pathway whose regulation is important for a variety of biological processes and whose disruption can cause progressive NASH (45). Also, Janus kinase (JAK) inhibitors have been developed as a new small molecule therapy for autoimmune disease such as IBD (46). On the other hand, Interleukin-6 (IL-6) is among the many cytokines that activate JAK/STAT signaling. Knockout of IL-6 gene affects mice with obesity and NASH (47). Interestingly, the role of IL-6 in IBD immunopathogenesis and its clinical relevance in IBD therapy and diagnostics are well studied (48). Also, TLR2 and TP53 have important roles in both IBD and NASH. TLRs are key mediators of innate host defense in the intestine, involved in maintaining mucosal as well as commensal homeostasis (49). Findings in diverse murine models of colitis have revealed the vital role of TLR dysfunction in IBD pathogenesis (50). Cengiz et al showed that serum TLR4 levels were elevated in NASH patients in comparison with healthy controls. Moreover, in NASH patients, serum level of TLR4 was able to predict liver fibrosis (51). TP53, encoding p53 protein, triggers apoptosis in NASH (52) and IBD patients (53). The finding (see table 3) indicates that JAK2 is involved in 20 biological processes among 30 GO terms (about 70% attributions). STAT3 and AGT, the other two genes, are related to about 40% of terms and TP53 the common genes in many cancer diseases is involved only in one term. Involvement of the key genes in the cluster of biological processes is shown in the figure 3. As depicted in this figure, the central role of TLR4, STAT3 and JAK2 is highlighted. It seems that JAK2, STAT3 and AGT are the most important genes that are connected closely to the two compared diseases.
These results showed that application of systems biology methods unravels the secret behind common mechanism of IBD and NASH diseases. The real impact of IBD therapies on co-existing NAFLD also needs to be further assessed. Also, appropriate screening tools and strategies for the management of co-existing diseases in IBD patients are lacking. Clarification of these issues may enhance early intervention and improve patient outcomes.
Conflict of interests
The authors declare that they have no conflict of interest.
References
- 1.Ratziu V, Bellentani S, Cortez-Pinto H, Day C, Marchesini G. A position statement on NAFLD/NASH based on the EASL 2009 special conference. J Hepatol. 2010;53:372–84. doi: 10.1016/j.jhep.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Beaton MD. Current treatment options for nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Can J Gastroenterol . 2012;26:353–7. doi: 10.1155/2012/725468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matteoni CA, Younossi ZM, Gramlich T, Boparai N, Liu YC, McCullough AJ. Nonalcoholic fatty liver disease: a spectrum of clinical and pathological severity. Gastroenterology. 1999;116:1413–9. doi: 10.1016/s0016-5085(99)70506-8. [DOI] [PubMed] [Google Scholar]
- 4.McCullough AJ. The epidemiology and risk factors of NASH. Fatty liver disease. In: Farrell GC, George J, Hall PM, McCullough AJ, editors. NASH and related disorders. Blackwell Publishing Ltd; 2005. pp. 23–37. [Google Scholar]
- 5.Ahmed A, Wong RJ, Harrison SA. Nonalcoholic Fatty Liver Disease Review: Diagnosis, Treatment, and Outcomes. Clin Gastroenterol Hepatol. 2015;13:2062–70. doi: 10.1016/j.cgh.2015.07.029. [DOI] [PubMed] [Google Scholar]
- 6.Ratziu V, Charlotte F, Heurtier A, Gombert S, Giral P, Bruckert E, et al. Sampling variability of liver biopsy in nonalcoholic fatty liver disease. Gastroenterology. 2005;128:1898–906. doi: 10.1053/j.gastro.2005.03.084. [DOI] [PubMed] [Google Scholar]
- 7.Cadranel JF, Rufat P, Degos F. Practices of liver biopsy in France: results of a prospective nationwide survey For the Group of Epidemiology of the French Association for the Study of the Liver (AFEF) Hepatology. 2000;32:477–81. doi: 10.1053/jhep.2000.16602. [DOI] [PubMed] [Google Scholar]
- 8.Younossi ZM, McCullough AJ, Ong JP, Barnes DS, Post A, Tavill A, et al. Obesity and non-alcoholic fatty liver disease in chronic hepatitis C. J Clin Gastroenterol. 2004;38:705–9. doi: 10.1097/01.mcg.0000135372.10846.2a. [DOI] [PubMed] [Google Scholar]
- 9.Asadzadeh-Aghdaee H, Shahrokh S, Norouzinia M, Hosseini M, Keramatinia A, Jamalan M, et al. Introduction of inflammatory bowel disease biomarkers panel using protein-protein interaction (PPI) network analysis. Gastroenterol Hepatol Bed Bench. 2016;9:S8–S13. [PMC free article] [PubMed] [Google Scholar]
- 10.Chao CY, Battat R, Al Khoury A, Restellini S, Sebastiani G, Bessissow T. Co-existence of non-alcoholic fatty liver disease and inflammatory bowel disease: A review article. World J Gastroenterol. 2016;22:7727–34. doi: 10.3748/wjg.v22.i34.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rocchi A, Benchimol EI, Bernstein CN, Bitton A, Feagan B, Panaccione R, et al. Inflammatory bowel disease: a Canadian burden of illness review. Can J Gastroenterol. 2012;26:811–7. doi: 10.1155/2012/984575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steed H, Walsh S, Reynolds N. A brief report of the epidemiology of obesity in the inflammatory bowel disease population of Tayside, Scotland. Obes Facts. 2009;2:370–2. doi: 10.1159/000262276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geerling BJ, Badart-Smook A, Stockbrügger RW, Brummer RJ. Comprehensive nutritional status in patients with long-standing Crohn disease currently in remission. Am J Clin Nutr. 1998;67:919–26. doi: 10.1093/ajcn/67.5.919. [DOI] [PubMed] [Google Scholar]
- 14.Long MD, Crandall WV, Leibowitz IH, Duffy L, del Rosario F, Kim SC, et al. ImproveCareNow Collaborative for Pediatric IBD. Prevalence and epidemiology of overweight and obesity in children with inflammatory bowel disease. Inflamm Bowel Dis. 2011;17:2162–8. doi: 10.1002/ibd.21585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong RJ, Ahmed A. Obesity and non-alcoholic fatty liver disease: Disparate associations among Asian populations. World J Hepatol. 2014;6:263–73. doi: 10.4254/wjh.v6.i5.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gisbert JP, Luna M, González-Lama Y, Pousa ID, Velasco M, Moreno-Otero R, et al. Liver injury in inflammatory bowel disease: long-term follow-up study of 786 patients. Inflamm Bowel Dis. 2007;13:1106–14. doi: 10.1002/ibd.20160. [DOI] [PubMed] [Google Scholar]
- 17.Sourianarayanane A, Garg G, Smith TH, Butt MI, McCullough AJ, Shen B. Risk factors of non-alcoholic fatty liver disease in patients with inflammatory bowel disease. J Crohns Colitis. 2013;7:e279–85. doi: 10.1016/j.crohns.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 18.Bargiggia S, Maconi G, Elli M, Molteni P, Ardizzone S, Parente F, et al. Sonographic prevalence of liver steatosis and biliary tract stones in patients with inflammatory bowel disease: study of 511 subjects at a single center. J Clin Gastroenterol. 2003;36:417–20. doi: 10.1097/00004836-200305000-00012. [DOI] [PubMed] [Google Scholar]
- 19.Bessissow T, Le NH, Rollet K, Afif W, Bitton A, Sebastiani G. Incidence and Predictors of Nonalcoholic Fatty Liver Disease by Serum Biomarkers in Patients with Inflammatory Bowel Disease. Inflamm Bowel Dis. 2016;22:1937–44. doi: 10.1097/MIB.0000000000000832. [DOI] [PubMed] [Google Scholar]
- 20.Bringiotti R, Ierardi E, Lovero R, Losurdo G, Di Leo A, Principi M. Intestinal microbiota: The explosive mixture at the origin of inflammatory bowel disease? World J Gastrointest Pathophysiol. 2014;5:550–9. doi: 10.4291/wjgp.v5.i4.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miele L, Valenza V, La Torre G, Montalto M, Cammarota G, Ricci R, et al. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology. 2009;49:1877–87. doi: 10.1002/hep.22848. [DOI] [PubMed] [Google Scholar]
- 22.Bosley J, Boren C, Lee S, Grøtli M, Nielsen J, Uhlen M, et al. Improving the economics of NASH/NAFLD treatment through the use of systemsbiology. Drug Discov Today. 2017 doi: 10.1016/j.drudis.2017.07.005. pii: S1359-6446(17)30025-9. [DOI] [PubMed] [Google Scholar]
- 23.van Koppen A, Verschuren L, Nagabukuro H, Costessi A, Morrison MC, Salic K, et al. Systems biology approach to identify processes and early markers for fibrosis in metabolically-induced NASH in mice. Hepatology. 2016;64:S853. [Google Scholar]
- 24.Moco S, Candela M, Chuang E, Draper C, Cominetti O, Montoliu I, et al. Systems biology approaches for inflammatory bowel disease: emphasis on gut microbial metabolism. Inflamm Bowel Dis. 2014;20:2104–14. doi: 10.1097/MIB.0000000000000116. [DOI] [PubMed] [Google Scholar]
- 25.Palmieri O, Mazza T, Castellana S, Panza A, Latiano T, Corritore G, et al. Inflammatory Bowel Disease Meets Systems Biology: A Multi-Omics Challenge and Frontier. OMICS. 2016;20:692–8. doi: 10.1089/omi.2016.0147. [DOI] [PubMed] [Google Scholar]
- 26.Safaei A, Arefi Oskouie A, Mohebbi SR, Rezaei-Tavirani M, Mahboubi M, Peyvandi M, et al. Metabolomic analysis of human cirrhosis, hepatocellular carcinoma, non-alcoholic fatty liver disease and non-alcoholic steatohepatitis diseases. Gastroenterol Hepatol Bed Bench. 2016;9:158–73. [PMC free article] [PubMed] [Google Scholar]
- 27.Piñero J, Bravo À, Queralt-Rosinach N, Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45(D1):D833–9. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–8. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Safaei A, Rezaei Tavirani M, Arefi Oskouei A, Zamanian Azodi M, Mohebbi SR, Nikzamir AR. Protein-protein interaction network analysis of cirrhosis liver disease. Gastroenterol Hepatol Bed Bench. 2016;9:114–23. [PMC free article] [PubMed] [Google Scholar]
- 31.Zamanian-Azodi M, Mortazavi-Tabatabaei SA, Mansouri V, Vafaee R. Metabolite-protein interaction (MPI) network analysis of obsessive-compulsive disorder (OCD) from reported metabolites. Arvand Journal of Health and Medical Sciences. 2016;1:112–20. [Google Scholar]
- 32.Wu H, Gao L, Dong J, Yang X. Detecting overlapping protein complexes by rough-fuzzy clustering in protein-protein interaction networks. PLoS One. 2014;9:e91856. doi: 10.1371/journal.pone.0091856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhosle A, Chandra N. Structural analysis of dihydrofolate reductases enables rationalization of antifolate binding affinities and suggests repurposing possibilities. FEBS J. 2016;283:1139–67. doi: 10.1111/febs.13662. [DOI] [PubMed] [Google Scholar]
- 34.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–3. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–3. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hunter DJ. Gene-environment interactions in human diseases. Nat Rev Genet. 2005;6:287–98. doi: 10.1038/nrg1578. [DOI] [PubMed] [Google Scholar]
- 37.Safari-Alighiarloo N, Taghizadeh M, Rezaei-Tavirani M, Goliaei B, Peyvandi AA. Protein-protein interaction networks (PPI) and complex diseases. Gastroenterol Hepatol Bed Bench. 2014;7:17–31. [PMC free article] [PubMed] [Google Scholar]
- 38.Zamanian-Azodi M, Rezaei-Tavirani M, Rahmati-Rad S, Hasanzadeh H, Rezaei Tavirani M, Seyyedi SS. Protein-Protein Interaction Network could reveal the relationship between the breast and colon cancer. Gastroenterol Hepatol Bed Bench. 2015;8:215–24. [PMC free article] [PubMed] [Google Scholar]
- 39.Rezaei-Tavirani M, Zamanian-Azodi M, Rajabi S, Masoudi-Nejad A, Rostami-Nejad M, Rahmatirad S. Protein Clustering and Interactome Analysis in Parkinson and Alzheimer's Diseases. Arch Iran Med. 2016;19:101–9. [PubMed] [Google Scholar]
- 40.Safari-Alighiarloo N, Taghizadeh M, Tabatabaei SM, Shahsavari S, Namaki S, Khodakarim S, et al. Identification of new key genes for type 1 diabetes through construction and analysis of protein-protein interaction networks based on blood and pancreatic islet transcriptomes. J Diabetes. 2017;9:764–77. doi: 10.1111/1753-0407.12483. [DOI] [PubMed] [Google Scholar]
- 41.Zali H, Rezaei Tavirani M. Meningioma protein-protein interaction network. Arch Iran Med. 2014;17:262–72. [PubMed] [Google Scholar]
- 42.Abbaszadeh HA, Peyvandi AA, Sadeghi Y, Safaei A, Zamanian-Azodi M, Khoramgah MS, et al. Er: YAG Laser and Cyclosporin A Effect on Cell Cycle Regulation of Human Gingival Fibroblast Cells. J Lasers Med Sci. 2017;8:143–9. doi: 10.15171/jlms.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaidos JKJ, Fuchs M. Increased Prevalence of NAFLD in IBD Patients. Dig Dis Sci. 2017;62:1362. doi: 10.1007/s10620-017-4552-8. [DOI] [PubMed] [Google Scholar]
- 44.McGowan CE, Jones P, Long MD, Barritt AS 4th. Changing shape of disease: nonalcoholic fatty liver disease in Crohn's disease-a case series and review of the literature. Inflamm Bowel Dis. 2012;18:49–54. doi: 10.1002/ibd.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riordan JD, Nadeau JH. Modeling progressive non-alcoholic fatty liver disease in the laboratory mouse. Mamm Genome. 2014;25:473–86. doi: 10.1007/s00335-014-9521-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boland BS, Sandborn WJ, Chang JT. Update on Janus kinase antagonists in inflammatory bowel disease. Gastroenterol Clin North Am. 2014;43:603–17. doi: 10.1016/j.gtc.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matthews VB, Allen TL, Risis S, Chan MH, Henstridge DC, Watson N, et al. Interleukin-6-deficient mice develop hepatic inflammation and systemic insulin resistance. Diabetologia. 2010;53:2431–41. doi: 10.1007/s00125-010-1865-y. [DOI] [PubMed] [Google Scholar]
- 48.Mudter J, Neurath MF. Il-6 signaling in inflammatory bowel disease: pathophysiological role and clinical relevance. Inflamm Bowel Dis. 2007;13:1016–23. doi: 10.1002/ibd.20148. [DOI] [PubMed] [Google Scholar]
- 49.Cario E. Toll-like receptors in inflammatory bowel diseases: a decade later. Inflamm Bowel Dis. 2010;16:1583–97. doi: 10.1002/ibd.21282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rachmilewitz D, Katakura K, Karmeli F, Hayashi T, Reinus C, Rudensky B, et al. Toll-like receptor 9 signaling mediates the anti-inflammatory effects of probiotics in murine experimental colitis. Gastroenterology. 2004;126:520–8. doi: 10.1053/j.gastro.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 51.Cengiz M, Ozenirler S, Elbeg S. Role of serum toll-like receptors 2 and 4 in non-alcoholic steatohepatitis and liver fibrosis. J Gastroenterol Hepatol. 2015;30:1190–6. doi: 10.1111/jgh.12924. [DOI] [PubMed] [Google Scholar]
- 52.Farrell GC, Larter CZ, Hou JY, Zhang RH, Yeh MM, Williams J, et al. Apoptosis in experimental NASH is associated with p53 activation and TRAIL receptor expression. J Gastroenterol Hepatol. 2009;24:443–52. doi: 10.1111/j.1440-1746.2009.05785.x. [DOI] [PubMed] [Google Scholar]
- 53.Goretsky T, Dirisina R, Sinh P, Mittal N, Managlia E, Williams DB, et al. p53 mediates TNF-induced epithelial cell apoptosis in IBD. Am J Pathol. 2012;181:1306–15. doi: 10.1016/j.ajpath.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]