Abstract
Aim:
In the present study, we investigated the prevalence of E. coli pathotypes and Shigella sero-groups and their antimicrobial profiles among diarrheic children in Nairobi city, Kenya.
Background:
Although diarrheagenic E. coli pathotypes and Shigella sero-groups are leading causes of diarrhea in children under five years in developing countries, their distribution and antimicrobial resistance vary from place to place and over time in a given region.
Methods:
In a cross-sectional study, we enrolled diarrheic children (n=354) under five years seeking treatment at Mbagathi Hospital, Nairobi city, Kenya,. Stool samples were collected from all children for bacterial culture. Bacterial isolation and identification was performed by conventional microbiological methods. Polymerase chain amplification was used to detect aspU, aggR, andpcvd432 for EAEC, est and elt for ETEC, eae for EPEC, stx for EHEC, and ipaH for EIEC and Shigella species. Antimicrobial profile was determined by disk diffusion method.
Results:
The prevalence of EAEC, ETEC, EPEC (eae), EIEC (ipaH) was 21.2%, 10.5%, 4.5%, and 0.6%, respectively, while that of mixed infection was 0.6%for ETEC/EAEC and 0.3%for EAEC/EPEC/ETEC. No EHEC strain was isolated. Pathogenetic analysis for EAEC showed that5.9% carried aspU,8.2% possessed both aspU and aggR and 7.1% had a combination of aspU, aggR andpcvd432 while that of ETEC was 2.3% for elt, 6.5% for both elt and est and 1.7% for est. The combination of aspU with aggR, elt and est, and pcvd432 with aggR, aspU and est was 0.3% for each case of ETEC/EAEC mixed infection. The aspU gene co-existed with aggR, pcvd432, eae and elt in the EAEC/EPEC/ETEC mixed infection. The prevalence of S. boydii, S. dysenteriae, S. flexneriand, S. sonnei was 0.8%, 0.6%, 1.7%, and 0.8%, respectively. No E. coli pathotype and shigella co-infection was detected. In addition, both E. coli pathotypes and Shigella species were resistant to ampicillin, trimethoprim/sulfamethoxazole, streptomycin, chloramphenicol and tetracycline while gentamycin and kanamycin resistance occurred in diarrheagenic E. coli.
Conclusion:
E. coli pathotypes and Shigella sero-groups harboring virulent genes are important causes of diarrhea in children in Kenya. The increasing spectrum of antibiotic resistance in diarrheagenic E. coli and Shigella species necessitates the development of antimicrobial stewardship education-programs to influence prescribing behavior as well as optimizing the use of effective antimicrobials in Kenya.
Key Words: E. coli pathotypes, Shigella sero-groups, Antimicrobial profile.
Introduction
Diarrheal diseases are the second leading cause of mortality in children under five years accounting for 1.7 billion episodes and more than 0·5 million deaths globally, most of which occur in developing countries (1). Shigella sero-groups and Escherichia coli pathotypes are important etiology of diarrhea mostly in children younger than 5 years in developing countries (2, 3). It is estimated that Shigella causes 164 million cases of bloody diarrhea and more than one million deaths while E. coli pathotypes accounts for about 56 million diarrhea episodes and more than 0.2 million deaths annually, the majority of which occur in children less than five years of age (2-5).
There are six diarrheagenic E. coli (DEC) pathotypes which are differentiated from intestinal flora E. coli by the presence of virulence genes. This include aafII, ast, aggR, aaspU and pcvd432 genes for enteroaggregative E. coli (EAEC), eae and bfp genes for enteropathogenic E. coli (EPEC), stx1 and stx2 genes for enterohemorrhagic E. coli (EHEC), heat stable (est) and labile (elt) toxin genes for enterotoxigenic E. coli (ETEC), virf, ipaH and ipaL genes for enteroinvasive E. coli (EIEC), and daaE gene for diffusely adherent E. coli (DAEC) (6-8). Specific virulent genes for the seventh E. coli pathotype, crohn's disease-associated adherent-invasive E. coli (AIEC), have not been discovered to date. AIEC phenotypic identification is based on their ability to replicate within intestinal epithelial cells and macrophages (9, 10). In Kenya, conflicting reports on predominant diarrheagenic E. coli pathotype in adults and children (11-15) suggest that the prevalence of diarrheagenic E. coli pathotypes as etiology of diarrhea vary from region to region and even between adults and children in a country (16, 17). Previous study in Nairobi city, Kenya, identified EPEC as the major cause of diarrhea in children (15). However, this finding (15) is unreliable since one virulence gene was used to identify EAEC despite the heterogeneous nature of EAEC virulence factors (8, 18).
Shigella species is represented by four sero-groups namely Shigella dysenteriae, S. flexneri, S. boydii and S. sonnei. Shigella sero-groups are further classified based on structural difference of O-antigens polysaccharide into at least 15 serotypes of S. dysenteriae, 19 serotypes of S. flexneri, 20 serotypes of S. boydii and one serotype of S. sonnei (19) indicating the effectiveness of vaccine-induced serotype-specific protection (20). The severity of shigellosis vary within the sero-groups (21) and is linked to the expression of several virulence genes such as the invasion-associated locus (ial), the invasion plasmid antigen H (ipaH), ipaBCD, Shigella enterotoxin 1 and 2 (ShET-1 and ShET-2), ipgD, icsA and virA genes associated with colonization, invasion, intracellular survival and toxin-mediated disease (22). IpaH gene is carried by all four Shigella sero-groups and is used for the molecular diagnosis of shigellosis (22, 23). Variation in the prevalence of Shigella sero-groups among adults and children in Kenya (2, 24-27) suggest that the distribution of Shigella sero-group is heterogeneous over time and place in a country (21). To our knowledge, however, the distribution of Shigella sero-groups among diarrheic children under five years in Nairobi city, Kenya, has not been reported.
Antimicrobial resistance in Shigella species and E. coli has been reported globally and increasing level of resistance is a growing concern (28). In Kenya, the vast majority of antibiotics prescriptions are made based on empirical diagnosis driving resistance among enteric pathogens (25) at this time when development of new antibiotics has gone down (29). Previous studies determined antimicrobial susceptibly patterns of Shigella and E. coli in Kenya (24, 25, 27, 30-32). Since antimicrobial resistance of E. coli and Shigella vary by region and over time (33), continuous antimicrobial surveillance is key in preventing emergence of antimicrobial resistant strains as well as guiding effective treatment (28). Therefore, this study determined diarrheagenic E. coli pathotypes and Shigella sero-groups and their antimicrobial susceptibility patterns in diarrheic children in Nairobi city, Kenya.
Methods
Study site, design and population
This cross-sectional study was conducted in Nairobi city, Kenya. Participants were diarrheic children under five years, seeking treatment at Mbagathi hospital, Nairobi city, Kenya. Diarrhea was defined, according World Health Organization (WHO) guidelines as the occurrence of three or more loose, liquid, or watery stools in a 24-hour period (34). A questionnaire was used to obtain information on age, gender, travel history, nausea, vomiting, abdominal pain, and diarrhea history of the children from the parents/guardians. Additional information on occupation of the guardian and water source and treatment was recorded on questionnaire. Clinical features such as body temperature, and dehydration signs were collected by clinicians. Stool specimens from diarrheic children were collected using wide mouthed, dry, leak-proof, sterile plastic containers and microbiology laboratory analysis performed within two hours of collection. Samples from children who had received antibiotics were excluded from the study.
Bacteriological procedures
Stool samples were plated on MacConckey agar (MCA), Xylose lysine deoxycholate agar (XLD), and Sorbitol MacConkey agar (SMAC) and incubated at 37°C overnight for the isolation and identification of E.coli and Shigella species. Identification of E.coli and Shigella species was performed by following the WHO recommendations (35). Shigella sero-goups were determined using serotyping kit (Denka Seiken Co. Ltd., Tokyo, Japan). The E. coli and Shigella isolates were further characterized for the presence of virulence genes.
Detection of diarrheagenic E. coli pathotype and Shigella virulence genes
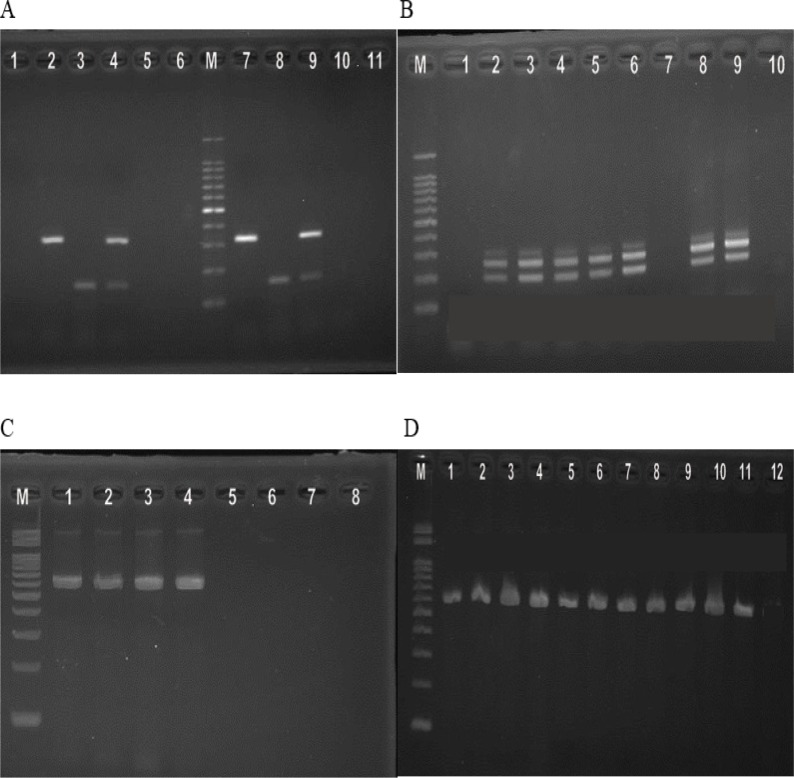
DNA from cultured isolates of E. coli and Shigella species were extracted using QIAamp® DNA Mini Kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer’s recommendations. Initially, multiplex PCR to detect ETEC was performed using virulence gene-specific primers to detect est and elt for ETEC (Table 1) (6). This was followed by multiplex PCR with primers to detect aspU, aggR, and pcvd432 for EAEC, eae for EPEC, stx for EHEC, and ipaH for EIEC (figure 1) (6). Singleplex PCR to detect ipaH-gene of Shigella sero-group isolates was performed using previously published primers (Table 1) (6). PCR reaction was carried out with 2.5 μL of the template DNA added to 47.5 μL mix containing, DreamTaq Green PCR Master Mix, nuclease free water and 1.0 μM of each primer (Thermo Fisher Scientific Inc., Waltham, Massachusetts, USA). Cycling conditions were initial denaturation at 95°C for 2 minutes, followed by 35 cycles of denaturation at 95°C for 30 seconds, annealing at 55°C for 30 seconds, extension at 72°C for 1 minute and final extension at 72°C for 10 minutes. The amplified DNA products were visualized by agarose gel electrophoresis method.
Table 1.
PCR primers used in this study
| Primers, Genes and Sequence (5'- 3') | Amplicon size (bp) | Target gene |
|---|---|---|
| SK1 CCCGAATTCGGCACAAGCATAAGC SK2 CCCGGATCCGTCTCGCCAGTATTCG |
881 | eae |
| VTcom-u: GAGCGAAATAATTTATTATGTG VTcom-d: TGATGATGGCAATTCAGTAT |
518 | stx1 and stx2 |
| AL65: TTAATAGCACCCGGTACAAGCAGG AL125: CCTGACTCTTCAAAAGAGAAAATTAC |
147 | est |
| LT1: TCTCTATGTGCATACGGAGC LTr: CCATACTGATTGCCCGCAAT |
322 | eltB |
| ipaIII: GTTCCCTTGACCGCCTTTCCGATACCGTC ipaIV: GCCGGTCAGCCACCCTCTGAGAGTAC |
600 | IpaH |
| aggRks1: GTATACACAAAAGAAGGAAGC aggRks2: ACAGAATCGTCAGCATCAGC |
254 | aggR |
| Eaggfp: AGACTCTGGCGAAAGACTGTATC Eaggbp: ATGGCTGTCTGTAATAGATGAGAAC |
194 | Pcvd432 |
| aspU-3: GCCTTTGCGGGTGGTAGCGG aspU-2: AACCCATTCGGTTAGAGCAC |
282 | aspU |
Figure 1.
A) Multiplex PCR for detecting ETEC pathotype. Lane M: molecular weight marker. Lanes 1 and 11: Negative control. Lane 2 and 7: elt (322 bp). Lanes 3 and 8: est (147 bp). Lanes 4 and 9: elt (322 bp) and est (147 bp). Lane 5, 6 and 10: elt and est-gene negative E. coli isolates. (B)Multiplex PCR for detecting EAEC, EIEC, and EPEC pathotypes. Lanes 2,3,4,5,6,8 and 9: EAEC-aggR (254 bp) and aspU (282 bp). Lanes: 1 and 7: aggR, ipaH, and aspU-gene negative samples.Lane 10: Negative control. (C) Multiplex PCR for detecting EAEC, EIEC, and EPEC pathotypes. Lane M: Molecular weight marker. Lanes 1, 2, 3, 4:EIEC-ipaH-gene (600 bp). Lane 5,6 and 7: ipaH-gene negative sample. Lane 8: Negative control. (D) Singleplex PCR for detecting shigellaipaH-gene. Lane M: Molecular weight marker. Lanes 1,2,3,4,5,6,7,8, 9, 10 and 11: ipaH(619 bp). Lane 12: Negative control
Antimicrobial susceptibility profile
The bacterial isolates were tested for antibiotic resistance by the disk diffusion method according to established standard operating procedures (36). The antibiotics tested included ampicillin, trimethoprim/sulfamethoxazole, ceftriaxone, streptomycin, amoxicillin/clavulanic acid, gentamycin, kanamycin, ciprofloxacin, chloramphenicol, erythromycin, nalidixic acid and tetracycline.
Data analysis
Statistical analyses were performed using SPSS version 19.0 for Windows (IBM SPSS Statistics for Windows, Version 19.0. Armonk, NY: IBM Corp.). Descriptive statistics were used to analyze the data and the results expressed as frequency and percentage.
Ethical considerations
This study was ethically approved by Kenyatta National Hospital/University of Nairobi (KNH-UoN) Ethics and Research Committee and was conducted according to Helsinki declarations. A consent form was read and signed by either parent or guardian of each child. Diarrheic children were treated by clinicians according to World Health Organization guidelines for treatment of diarrhea in children (34). All study participants' information and test results were confidentially kept.
Results
Demographic and clinical information of study participants
The demographic and clinical information of the diarrheic children under five years of age in Nairobi, Kenya, is presented in table 2. A total of 354 children with diarrhea were included in the study. The age distribution showed that 242 (68.4%) children were within the age group 1 and 30 months and 112 (31.6%) children were between 31 and 60 months. The overall gender distribution was 170 (48%) female and 184 (52%) male. Guardians of 351 (99.2%) and 3 (0.8%) children reported using piped and borehole water, respectively. In addition, guardians of 206 (58.6%) and 10 (2.8%) children, respectively, reported treating drinking and having travelled to their rural homes two weeks prior to the start of the study. Occupation distribution showed that two (0.6%), 17 (4.8%), 5 (1.4%), 14 (4.0%), and 178 (50.3%) of the guardians were in the health care practitioner, office administrative support, construction/installation/repair, education/training, and sales, respectively, while 138 (39.0%) were unemployed.
Table 2.
Demographic and clinical information of study participants
| Characteristics | Number (%) |
|---|---|
| Age in months | |
| 1-30 | 242 (68.4) |
| 31-60 | 112 (31.6) |
| Gender | |
| Female | 170 (48) |
| Male | 184 (52) |
| Source of water | |
| Piped water | 351 (99.2) |
| Borehole | 3 (0.8) |
| Water treatment | 206 (58.2) |
| Travel history | 10 (2.8) |
| Occupation of guardian | |
| Health care practitioner | 2 (0.6) |
| Office/administrative/support | 17 (4.8) |
| Construction/installation/repair | 5 (1.4) |
| Education/ training | 14 (4.0) |
| Sales | 178 (50.3) |
| Unemployed | 138 (39.0) |
| Body temperature | |
| <38.0 | 54 (15.3) |
| ≥ 38.0 | 300 (84.7) |
| Duration of diarrhea | |
| 1-3 | 293 (82.8) |
| 4-6 | 32 (9.0) |
| ≥7 | 29 (8.2) |
| Diarrhea type | |
| Mucoid | 299 (84.5) |
| Bloody | 55 (15.5) |
| Symptoms | |
| Vomiting | 280 (79.1) |
| Fever | 293 (82.8) |
| Abdominal cramp | 235 (66.4) |
| Headache | 11 (3.1) |
| Nausea | 48 (13.6) |
| Appetite loss | 326 (92.1) |
| Sunken eyeball | 295 (83.3) |
| Dry tongue | 110 (31.1) |
| Reduced skin elasticity | 182 (51.4) |
Data are presented as number and proportions (%) of study participants. ≤, less than or equal to. <, less than. ≥, greater than or equal to. >, greater than. Health care practitioner (Nurse, Clinical officer). Office administrative support (secretary, clerical officer, social worker, driver, househelp, and caretaker). Construction/installation/repair (welder, carpenter, mason, tailor). Education/ training (teacher).Sales (saloonist, hawkers and small scale business, sales agents
Temperature of <38.0°C and ≥ 38.0°C was recorded in 54 (15.3%) and 300 (84.7%) children, respectively. In this study, 293 (82.8%), 32 (9.0%) and 29 (8.2%), respectively, reported having diarrhea for 1-3, 4-6 and ≥7 days. Of which, 299 (84.5%) were mucoid diarrhea and 55 (15.5%) were bloody diarrhea. Vomiting was evidenced in 280 (79.1%) patients, fever in 293 (82.8%), abdominal cramp in 235 (66.4%), headache in 11 (3.1%), nausea in 48 (13.6%), and appetite loss in 326 (92.1%) children. Clinical diagnosis of dehydration revealed that 295 (83.3%) had sunken eyeballs, 110 (31.1%) children had dry tongue and 182 (51.4%) had reduced skin elasticity.
Prevalence of diarrheagenic E. coli pathotypes and Shigella sero-groups isolated from study participants
The prevalence of diarrheagenic E. coli pathotypes and Shigella sero-groups in diarrheic children under five years of age in Nairobi City, Kenya, is presented in table 3. The prevalence of EAEC, ETEC, EPEC (eae), EIEC (ipaH), was 75 (21.2%), 37 (10.5%), 16 (4.5%), and 2 (0.6%) pure strains, respectively, while the prevalence of mixed infections was 2 (0.6) for ETEC/EAEC and 1 (0.3) for EAEC/EPEC/ETEC. No EHEC strain was isolated from the children. Among the EAEC pure isolate infections, 21 (5.9%) had aspU gene, 29 (8.2%) had both aspU and aggR genes and 25 (7.1%) had a combination of aspU, aggR and pcvd432 genes among the EAEC pure isolate infections. The pathogenetic profile of ETEC pure isolate infections was 8 (2.3%) for elt gene, 23 (6.5%) for both elt and est genes and 6 (1.7%) for est gene. The combination of aspU with aggR, elt and est, and pcvd432 with aggR, aspU and est was detected in 1(0.3%) case each in ETEC/EAEC mixed infection. The aspU gene co-existed with aggR, pcvd432, eae and elt in the 1 (0.3%) EAEC/EPEC/ETEC mixed infection case.
Table 3.
Prevalence of E. coli pathotype and Shigella sero-groups
| Isolate type | Strain | Number (%) |
|---|---|---|
| Diarrheagenic E. coli pathotypes | ||
| EAEC (all) | 75 (21.2) | |
| aspU | 21 (5.9) | |
| aspU/aggR | 29 (8.2) | |
| aspU/aggR/pcvd432 | 25 (7.1) | |
| EHEC | Stx | 0 (0.0) |
| EPEC | Eae | 17 (4.8) |
| ETEC (all) | 37(10.5) | |
| Elt | 8 (2.3) | |
| elt/est | 23 (6.5) | |
| Est | 6 (1.7) | |
| EIEC | ipaH | 5 (1.4) |
| ETEC/EAEC (all) | 2 (0.6) | |
| aspU/aggR/elt/est | 1 (0.3) | |
| aspU/aggR/pcvd432/est | 1 (0.3) | |
| EAEC/EPEC/ETEC | aspU/aggR/pcvd432/eae/elt | 1 (0.3) |
| Shigella sero-groups | ||
| S. boydii | ipaH | 3 (0.8) |
| S. dysenteriae | ipaH | 2 (0.6) |
| S. flexneri | ipaH | 6 (1.7) |
| S. sonnei | ipaH | 3 (0.8) |
Data are presented as number and proportions (%) of isolates. EPEC, enteropathogenic E. coli. ETEC, enterotoxigenic E. coli. EAEC, enteroaggregative E. coli. EIEC, enteroinvasive E. coli. EHEC.Shigella boydii.S. dysenteriae, Shigella dysenterae.S. flexneri,Shigella flexneri. S. sonnei, Shigella sonnei
A total of 14 (4.0%) Shigella isolates were observed in this study comprising of 3 (0.8%) S. boydii, 2 (0.6%) S. dysenteriae, 6 (1.7%) S. flexneri and 3 (0.8%) S. sonnei. All the Shigella isolates harbored ipaH gene. No shigella and pathogenic E. coli co-infection was detected in this study.
Antimicrobial susceptibility patterns of diarrheagenic E. coli and Shigella species
The antimicrobial susceptibility patterns of the diarrheagenic E. coli and Shigella species is presented in table 4. About 77.4%, 66.8%, 3.0%, 80.5%, 14.3%, 72.2%, 56.4%, 9.0%, 75.9%, 1.5%, 11.3%, 64.7% of DEC was resistant to ampicillin, trimethoprim/sulfamethoxazole, ceftriaxone, streptomycin, amoxicillin/clavulanic acid, gentamycin, kanamycin, ciprofloxacin, chloramphenicol, erythromycin, nalidixic acid, and tetracycline, respectively. Although none of the Shigella isolate was resistant to gentamycin, kanamycin and erythromycin, 85.8%, 57.1%, 14.3%, 92.9%, 7.1%, 14.3%, 57.1%, 14.3%, 85.8%, of the children were infected with Shigella species resistant to ampicillin, trimethoprim/ sulfamethoxazole, ceftriaxone, streptomycin, amoxicillin/ clavulanic acid, ciprofloxacin, chloramphenicol, nalidixic acid, and tetracycline, respectively.
Discussion
The prevalence diarrheagenic E. coli pathotypes and Shigella sero-groups as etiologic agents of diarrhea vary considerably from region to region and over time in a given region (17, 21, 37, 38). In addition, clinical manifestations of E. coli and shigella species are influenced by the type of virulence factor present which differ by pathotype and sero-group, respectively (8, 21). While antibiotics have proved successful in the treatment of E. coli and Shigella infections, emergence and spread of acquired and transmitted antimicrobial resistant strains is common (33). Thus, continuous epidemiological surveillance is key for planning antimicrobial treatment.
The findings of this study showing the presence of aspU, aggR, pcvd432, eae, elt, est, and ipaH, virulent factors in E. coli pathotypes, partly mirrors previous studies in Nairobi city, Kenya (14, 15). The EAEC predominates in this study with the remainder being EIEC, EPEC, ETEC and mixed diarrheagenic E. coli infections is partly consistent with previous studies in Kenya (14). This observation may be attributed to zinc deficiency that affects about 61% of children under five years in Kenya (39). Zinc inhibits diarrheagenic E. coli endothelial adherence, biofilm formation, virulence gene expression as well as promoting host immune clearance of diarrheagenic E. coli (40, 41). This and other studies in Kenya did not detect EHEC (15). Perhaps, EHEC is predominantly present in the environment and reservoirs, and it does not play role in infantile diarrhea (8, 42). However, the findings of this study are inconsistent with previous studies that identified EPEC as the most common DEC in children under five years in Nairobi city, Kenya (15). Probably, because of the proved EAEC genome heterogeneity (8, 18), using one virulence gene decreased the rate of isolation in that study (15). Taken together, EAEC, EIEC, EPEC and ETEC pathotypes harboring virulent factors are an important etiology of diarrhea in Kenyan children and require more attention from our public health services.
Several copies of ipaH gene being present on both plasmids and chromosomes may explain the gene being tested positive in all Shigella sero-groups (43) in this and previous studies (22, 23). The findings of this study reporting Shigella flexneri as the commonest strain and the remainder being S. dysenteriae, S. boydii and S. sonnei, although in low rates, is partly consistent with previous studies in Kenya (25-27). Shigella flexneri is less virulent than other Shigella sero-groups because it doesn’t kill its host immediately (44) explaining the dominancy of S. flexneri in Nairobi city, Kenya. However, the findings of this study are inconsistent with previous studies in industrialized countries reporting the dominancy of S. sonnei (45). Industrialized countries have greater levels of wealth and economic development positively influencing hospital care and treatment for diarrhea and dysentery (37, 46) which may drive the dominancy of S. sonnei due to its greater ability to develop resistance to antibiotic treatment (47). Therefore, heterogeneous distribution of Shigella species suggests that multivalent vaccine will be needed to prevent shigellosis in children in Kenya.
Many of the diarrheagenic Escherichia coli were resistant to ampicillin, chloramphenicol, tetracycline, trimethoprim/sulfamethoxazole, gentamycin, kanamycin, and streptomycin, which is partly consistent with the findings of previous studies in Kenya (24, 30, 31). On the other hand, increasing susceptibility of diarrheagenic E. coli to ampicillin, chloramphenicol, gentamycin, tetracycline, and trimethoprim/sulfamethoxazole has been reported among the Maasai community of Kenya (32). This is possibly due to the lower levels of exposure and usage of antimicrobials among the Maasai community of Kenya who mostly practice traditional medicine (48). The findings of this study showing Shigella species resistance to Ampicillin, trimethoprim/ sulfamethoxazole, streptomycin, chloramphenicol and tetracycline are in agreement with previous studies in Kenya (27) and Ethiopia (49). Likewise, the susceptible of Shigella species towards, ceftriaxone, amoxicillin/clavulanic acid, gentamycin, kanamycin, ciprofloxacin, erythromycin, and nalidixic acid is congruent to previous studies in Ethiopia (50). However, Shigella resistance to ceftriaxone, amoxicillin/clavulanic acid, gentamycin, kanamycin, ciprofloxacin, erythromycin, and nalidixic acid have been reported in China (51) and Iran (52) highlighting the importance of judicious use of these drugs to preserve their effectiveness for treatment in Kenya. Taken together, antimicrobial stewardship education-programs have to be developed to influence prescribing behavior hence optimizing the use of effective antimicrobials in Kenya.
It is important to outline the limitations of this study. The findings of this study must be interpreted with caution because molecular diagnosis of E. coli and Shigella species was performed on cultures but not stool samples. The presence of DAEC and AIEC pathotypes were not investigated in this study. In this study, the primers to detect the bundle-forming pilus (bfpA) gene which is present in typical EPEC and absent in some atypical EPEC were not included in diarrheagenic E. coli pathotyping since further analysis of eae-positive isolates for the presence of stx-gene is sufficient to distinguish EPEC from EHEC (6, 53). Although Shigella species expresses several virulent factors (22), singleplex PCR targeting ipaH gene was used in this study. We acknowledge the small sample size of Shigella isolates assayed for antimicrobial sensitivity.
We conclude that diarrheagenic E. coli pathotypes and Shigella sero-groups are an important etiology of diarrhea in children under five years in Kenya. These pathogens are of public health importance since they are highly heterogeneous and harbor very threatening virulent genes like aspU, aggR, pcvd342, est, elt, eae, stx, and ipaH for diarrheagenic E. coli pathotypes and ipaH for Shigella sero-groups. The heterogeneity of Shigella sero-groups is important in the implementation of vaccine prevention strategies. In addition, both diarrheagenic E. coli and Shigella species are resistant to ampicillin, trimethoprim/sulfamethoxazole, streptomycin, chloramphenicol and tetracycline while gentamycin and kanamycin resistance occur in diarrheagenic E. coli. This result is important in the treatment and prevention of the spread of antimicrobial resistant diarrheagenic E. coli pathotype and shigella species.
Acknowledgment
We thank the study participants for their participation in the study. We are grateful to the management and staff of Mbagathi Hospital, Nairobi city, Kenya,for their support during the study.
Conflict of interests
The authors declare that they have no conflict of interest.
References
- 1.Liu L, Oza S, Hogan D, Perin J, Rudan I, Lawn JE, et al. Global, regional, and national causes of child mortality in 2000-13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet. 2015;385:430–40. doi: 10.1016/S0140-6736(14)61698-6. [DOI] [PubMed] [Google Scholar]
- 2.Livio S, Strockbine NA, Panchalingam S, Tennant SM, Barry EM, Marohn ME, et al. Shigella isolates from the global enteric multicenter study inform vaccine development. Clin Infect Dis. 2014;59:933–41. doi: 10.1093/cid/ciu468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lanata CF, Fischer-Walker LC, Olascoaga AC, Torres CX, Lanata MCF, Fischer-Walker CL, et al. Global causes of diarrheal disease mortality in children <5 years of age: a systematic review. PLoS One. 2013;8:e72788. doi: 10.1371/journal.pone.0072788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, et al. World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PLoS Med. 2015;12:e1001921. doi: 10.1371/journal.pmed.1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ, et al. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ. 1999;77:651–66. [PMC free article] [PubMed] [Google Scholar]
- 6.Toma C, Lu Y, Higa N, Nakasone N, Chinen I, Baschkier A, et al. Multiplex PCR assay for identification of human diarrheagenic Escherichia coli. J Clin Microbiol. 2003;41:2669–71. doi: 10.1128/JCM.41.6.2669-2671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guyen TV, Le Van P, Le Huy C, Gia KN, Weintraub A. Weintraub, Detection and characterization of diarrheagenic Escherichia coli from young children in Hanoi, Vietnam. J Clin Microbiol. 2005;43:755–60. doi: 10.1128/JCM.43.2.755-760.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajendran P, S S Ajjampur, D Chidambaram, G Chandrabose, B Rajendran P, Ajjampur SS, Chidambaram D, Chandrabose G, Thangaraj B, Sarkar R, et al. Pathotypes of diarrheagenic Escherichia coli in children attending a tertiary care hospital in South India. Diagn Microbiol Infect Dis. 2010;68:117–22. doi: 10.1016/j.diagmicrobio.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke DJ, Chaudhuri RR, Martin HM, Campbell BJ, Rhodes JM, Constantinidou C, et al. Complete genome sequence of the Crohn's disease-associated adherent-invasive Escherichia coli strain HM605. J Bacteriol. 2011;193:4540. doi: 10.1128/JB.05374-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnich N, Darfeuille-Michaud A. Adherent-invasive Escherichia coli and Crohn's disease. Curr Opin Gastroenterol. 2007;23:16–20. doi: 10.1097/MOG.0b013e3280105a38. [DOI] [PubMed] [Google Scholar]
- 11.Sang WK, Boga HI, Waiyaki PG, Schnabel D, Wamae NC, Kariuki SM. Prevalence and genetic characteristics of Shigatoxigenic Escherichia coli from patients with diarrhoea in Maasailand, Kenya. J Infect Dev Ctries. 2012;6:102–8. doi: 10.3855/jidc.1750. [DOI] [PubMed] [Google Scholar]
- 12.Bii CC, Taguchi H, Ouko TT, Muita LW, Wamae N, Kamiya S. Detection of virulence-related genes by multiplex PCR in multidrug-resistant diarrhoeagenic Escherichia coli isolates from Kenya and Japan. Epidemiol Infect. 2005;133:627–33. doi: 10.1017/s0950268805003870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onyango AO, Kenya EU, Mbithi JJ, Ng'ayo MO. Pathogenic Escherichia coli and food handlers in luxury hotels in Nairobi, Kenya. Travel Med Infect Dis. 2009;7:359–66. doi: 10.1016/j.tmaid.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Iijima Y, Oundo JO, Hibino T, Saidi SM, Hinenoya A, Osawa K, et al. High prevalence of diarrheagenic Escherichia coli among children with diarrhea in Kenya. Jpn J Infect Dis. 2017;70:80–83. doi: 10.7883/yoken.JJID.2016.064. [DOI] [PubMed] [Google Scholar]
- 15.Makobe CK, Sang WK, Kikuvi G, Kariuki S. Molecular characterization of virulence factors in diarrhoeagenic Escherichia coli isolates from children in Nairobi, Kenya. J Infect Dev Ctries. 2012;6:598–604. doi: 10.3855/jidc.2082. [DOI] [PubMed] [Google Scholar]
- 16.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev. 2013;26:822–80. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gómez-Duarte OG, Arzuza O, Urbina D, Bai J, Guerra J, Montes O, et al. Detection of Escherichia coli enteropathogens by multiplex polymerase chain reaction from children's diarrheal stools in two Caribbean-Colombian cities. Foodborne Pathog Dis. 2010;7:199–206. doi: 10.1089/fpd.2009.0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Czeczulin JR, Whittam TS, Henderson IR, Navarro-Garcia F, Nataro JP. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect Immun. 1999;67:2692–9. doi: 10.1128/iai.67.6.2692-2699.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muthuirulandi Sethuvel DP, Devanga Ragupathi NK, Anandan S, Veeraraghavan B. Update on: Shigella new serogroups/serotypes and their antimicrobial resistance. Lett Appl Microbiol. 2017;64:8–18. doi: 10.1111/lam.12690. [DOI] [PubMed] [Google Scholar]
- 20.Mani S, Wierzba T, Walker RI. Status of vaccine research and development for Shigella. Vaccine. 2016;34:2887–94. doi: 10.1016/j.vaccine.2016.02.075. [DOI] [PubMed] [Google Scholar]
- 21.Von Seidlein L, Kim DR, Ali M, Lee H, Wang X, Thiem VD, et al. A multicentre study of Shigella diarrhoea in six Asian countries: disease burden, clinical manifestations, and microbiology. PLoS Med. 2006;3:e353. doi: 10.1371/journal.pmed.0030353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lluque A, Mosquito S, Gomes C, Riveros M, Durand D, Tilley DH, et al. Virulence factors and mechanisms of antimicrobial resistance in Shigella strains from periurban areas of Lima (Peru) Int J Med Microbiol. 2015;305:480–90. doi: 10.1016/j.ijmm.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sangeetha AV, Parija SC, Mandal I, Krishnamurthy S. Clinical and microbiological profiles of shigellosis in children. J Health Popul Nutr. 2014;32:580–6. [PMC free article] [PubMed] [Google Scholar]
- 24.Karambu S, Matiru V, Kiptoo M, Oundo J. Characterization and factors associated with diarrhoeal diseases caused by enteric bacterial pathogens among children aged five years and below attending Igembe District Hospital, Kenya. Pan Afr Med J. 2013;16:37. doi: 10.11604/pamj.2013.16.37.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brooks JT, Ochieng JB, Kumar L, Okoth G, Shapiro RL, Wells JG, et al. Surveillance for bacterial diarrhea and antimicrobial resistance in rural western Kenya, 1997-2003. Clin Infect Dis. 2006;43:393–401. doi: 10.1086/505866. [DOI] [PubMed] [Google Scholar]
- 26.Njuguna C, Njeru I, Mgamb E, Langat D, Makokha A, Ongore D, et al. Enteric pathogens and factors associated with acute bloody diarrhoea, Kenya. BMC Infect Dis. 2016;16:477. doi: 10.1186/s12879-016-1814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Njuguna HN, Cosmas L, Williamson J, Nyachieo D, Olack B, Ochieng JB, et al. Use of population-based surveillance to define the high incidence of shigellosis in an urban slum in Nairobi, Kenya. PLoS One. 2013.8:e58437. doi: 10.1371/journal.pone.0058437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.World Health Organization. Antimicrobial resistance: global report on surveillance. Geneva, Switzerland: WHO; 2014. [Google Scholar]
- 29.Cole ST. Who will develop new antibacterial agents? Philos Trans R Soc Lond B Biol Sci. 2014;369:20130430. doi: 10.1098/rstb.2013.0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sang WK, Oundo V, Schnabel D. Prevalence and antibiotic resistance of bacterial pathogens isolated from childhood diarrhoea in four provinces of Kenya. J Infect Dev Ctries. 2012;6:572–8. doi: 10.3855/jidc.2196. [DOI] [PubMed] [Google Scholar]
- 31.Oundo JO, Kariuki SM, Boga HI, Muli FW, Iijima Y. High incidence of enteroaggregative Escherichia coli among food handlers in three areas of Kenya: a possible transmission route of travelers' diarrhea. J Travel Med. 2008;15:31–8. doi: 10.1111/j.1708-8305.2007.00174.x. [DOI] [PubMed] [Google Scholar]
- 32.Sang WK, SM Kariuki, D Schnabel, HI Boga, PG Waiyaki, CN Wamae. Antibiotic susceptibility of Enteric pathogens from the Maasai community, Narok and Kajiado Districts, Kenya. Afr J Health Sci. 2011;19:74–79. [Google Scholar]
- 33.Pickering LK. Antimicrobial resistance among enteric pathogens. Semin Pediatr Infect Dis. 2004;15:71–7. doi: 10.1053/j.spid.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 34.World Health Organization. The treatment of diarrhoea: a manual for physicians and other senior health workers. 4th revision. Geneva, Switzerland: WHO; 2005. 50 pp. [Google Scholar]
- 35.Perilla , Mindy J, Bopp Cheryl, Elliott John, Facklam Richard, Popovic Tanja, Wells Joy. Manual for the Laboratory Identification and Antimicrobial Susceptibility Testing of Bacterial Pathogens of Public Health Importance in the Developing World. Geneva, Switzerland: World Health Organization; 2003. 359 pp. [Google Scholar]
- 36.National Committee for Clinical and Laboratory Standards. Performance Standards for Antimicrobial Disk Susceptibility Tests. Approved Standard-Twelfth Edition; 2015. [Google Scholar]
- 37.Chompook P, Samosornsuk S, von Seidlein L, Jitsanguansuk S, Sirima N, Sudjai S, et al. Estimating the burden of shigellosis in Thailand: 36-month population-based surveillance study. Bull World Health Organ. 2005;83:739–46. [PMC free article] [PubMed] [Google Scholar]
- 38.Mao Y, Cui E, Bao C, Liu Z, Chen S, Zhang J, et al. Changing trends and serotype distribution of Shigella species in Beijing from 1994 to 2010. Gut Pathog. 2013;5:21. doi: 10.1186/1757-4749-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.GOK UNICEF. Anaemia and the Status of Iron, Vitamin A and Zinc in Kenya. The 1999 Micronutrient Survey Report. Nairobi: 2002. [Google Scholar]
- 40.Bolick DT, Kolling GL, Moore JH 2nd, de Oliveira LA, Tung K, Philipson C, et al. Zinc deficiency alters host response and pathogen virulence in a mouse model of enteroaggregative Escherichia coli-induced diarrhea. Gut Microbes. 2014;5:618–27. doi: 10.4161/19490976.2014.969642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Medeiros P, Bolick DT, Roche JK, Noronha F, Pinheiro C, Kolling GL, et al. The micronutrient zinc inhibits EAEC strain 042 adherence, biofilm formation, virulence gene expression, and epithelial cytokine responses benefiting the infected host. Virulence. 2013;4:624–33. doi: 10.4161/viru.26120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan A, Das SC, Ramamurthy T, Sikdar A, Khanam J, Yamasaki S, et al. Antibiotic resistance, virulence gene, and molecular profiles of Shiga toxin-producing Escherichia coli isolates from diverse sources in Calcutta, India. J Clin Microbiol. 2002;40:2009–15. doi: 10.1128/JCM.40.6.2009-2015.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang S, Wang S, Yin W, Liang M, Li J, Zhang Q, et al. Epidemic characteristics of hemorrhagic fever with renal syndrome in China, 2006-2012. BMC Infect Dis. 2014;14:384. doi: 10.1186/1471-2334-14-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kahsay AG, Muthupandian S. A review on Sero diversity and antimicrobial resistance patterns of Shigella species in Africa, Asia and South America, 2001-2014. BMC Res Notes. 2016;9:422. doi: 10.1186/s13104-016-2236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta A, Polyak CS, Bishop RD, Sobel J, Mintz ED. Laboratory-confirmed shigellosis in the United States, 1989-2002: epidemiologic trends and patterns. Clin Infect Dis. 2004;38:1372–7. doi: 10.1086/386326. [DOI] [PubMed] [Google Scholar]
- 46.Samosornsuk S, Jitsanguansuk S, Sirima N, Sudjai S, Tapchaisri P, Chompook P, et al. Preferences for treatment of diarrhoea and dysentery in Kaengkhoi district, Saraburi province, Thailand. J Health Popul Nutr. 2004;22:113–8. [PubMed] [Google Scholar]
- 47.Thompson CN, Duy PT, Baker S. The rising dominance of shigella sonnei: an intercontinental shift in the etiology of bacillary dysentery. PLoS Negl Trop Dis. 2015;9:e0003708. doi: 10.1371/journal.pntd.0003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kimondo J, Miaron J, Mutai P, Njogu P. Ethnobotanical survey of food and medicinal plants of the Ilkisonko Maasai community in Kenya. J Ethnopharmacol. 2015;175:463–9. doi: 10.1016/j.jep.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 49.Gebrekidan A, Dejene TA, Kahsay G, Wasihun AG. Prevalence and antimicrobial susceptibility patterns of Shigella among acute diarrheal outpatients in Mekelle hospital, Northern Ethiopia. BMC Res Notes. 2015;8:611. doi: 10.1186/s13104-015-1606-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lamboro T, Ketema T, Bacha K. Prevalence and Antimicrobial Resistance in Salmonella and Shigella Species Isolated from Outpatients, Jimma University Specialized Hospital, Southwest Ethiopia. Can J Infect Dis Med Microbiol. 2016;2016:4210760. doi: 10.1155/2016/4210760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qu M, Lv B, Zhang X, Yan H, Huang Y, Qian H, et al. Prevalence and antibiotic resistance of bacterial pathogens isolated from childhood diarrhea in Beijing, China (2010-2014) Gut Pathog. 2016;8:31. doi: 10.1186/s13099-016-0116-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jomezadeh N, Babamoradi S, Kalantar E, Javaherizadeh H. Isolation and antibiotic susceptibility of Shigella species from stool samples among hospitalized children in Abadan, Iran. Gastroenterol Hepatol Bed Bench. 2014;7:218–23. [PMC free article] [PubMed] [Google Scholar]
- 53.Teixeira NB, Rojas TC, da Silveira WD, Matheus-Guimarães C, Silva NP, Scaletsky IC. Genetic analysis of enteropathogenic Escherichia coli (EPEC) adherence factor (EAF) plasmid reveals a new deletion within the EAF probe sequence among O119 typical EPEC strains. BMC Microbiol. 2015;15:200. doi: 10.1186/s12866-015-0539-9. [DOI] [PMC free article] [PubMed] [Google Scholar]