ABSTRACT
We previously characterized LPAATδ/AGPAT4 as a mitochondrial lysophosphatidic acid acyltransferase that regulates brain levels of phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylinositol (PI). Here, we report that Lpaatδ−/− mice display impaired spatial learning and memory compared to wild-type littermates in the Morris water maze and our investigation of potential mechanisms associated with brain phospholipid changes. Marker protein immunoblotting suggested that the relative brain content of neurons, glia, and oligodendrocytes was unchanged. Relative abundance of the important brain fatty acid docosahexaenoic acid was also unchanged in phosphatidylserine, phosphatidylglycerol, and cardiolipin, in agreement with prior data on PC, PE and PI. In phosphatidic acid, it was increased. Specific decreases in ethanolamine-containing phospholipids were detected in mitochondrial lipids, but the function of brain mitochondria in Lpaatδ−/− mice was unchanged. Importantly, we found that Lpaatδ−/− mice have a significantly and drastically lower brain content of the N-methyl-d-asparate (NMDA) receptor subunits NR1, NR2A, and NR2B, as well as the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor subunit GluR1, compared to wild-type mice. However, general dysregulation of PI-mediated signaling is not likely responsible, since phospho-AKT and phospho-mTOR pathway regulation was unaffected. Our findings indicate that Lpaatδ deficiency causes deficits in learning and memory associated with reduced NMDA and AMPA receptors.
KEYWORDS: AMPA receptors, brain lipid metabolism, enzymes, glycerophosphatidylinositols, learning, memory, mitochondrial metabolism, NMDA receptors, phospholipids
INTRODUCTION
The acylglycerophosphate acyltransferase (AGPAT) family of proteins is a group of homologous lysophosphatidic acid acyltransferases (LPAATs) that catalyze the de novo formation of phosphatidic acid (PA), a precursor to all glycerophospholipids, as well as to di- and triacylglycerols (1, 2). To date, 11 AGPATs have been identified in mice and humans, although only AGPAT1 to -5 preferentially utilize lysophosphatidic acid (LPA) as an acyl donor, while AGPAT6 to -11 have been reclassified based on alternate lysophospholipid substrate preferences, or preference for glycerol-3-phosphate. Thus, of the 11 known AGPAT family members, only AGPAT1 to -5 function as true LPAATs (3, 4).
We and others have studied the enzymatic and biochemical functions of LPAATδ/AGPAT4, which are present at ubiquitously high levels in whole mouse brain (5–7) and in multiple mouse brain subregions (6). In previous work, we reported that Lpaatδ deficiency in mouse brain is compensated for by induction of Lpaatα, -β, -γ, and -ε (Agpat1, -2, -3, and -5), which serves to maintain total brain levels of PA but, interestingly, does not functionally compensate for loss of the enzyme (6). Brain levels of specific glycerophospholipids synthesized downstream of PA, including phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylinositol (PI), were all significantly reduced, suggesting that LPAATδ provides a functionally distinct source of PA in this tissue.
Reductions in brain PC, PE, and PI in Lpaatδ−/− mice were significant in magnitude (∼30 to 50%), and we predicted that these changes would impact important brain processes involved in memory and learning. Phosphatidylinositol is enriched in the brain compared to most other tissues and acts as a source of polyphosphorylated phosphoinositides (PIPs) (8–10). PIPs function in a multitude of brain signaling pathways, including neurotransmission (11) and neuronal exocytosis (12), and have been implicated in regulating functional and structural plasticity (13–15). PC is a major structural component of neuronal and glial cell membranes (16). Dietary supplementation with sources of choline, an essential nutrient required for PC synthesis, improves the performance of mice on behavioral tasks (17) and has been implicated in fetal and neonatal brain development (18, 19), highlighting the importance of the lipid in the brain. Finally, PE is highly enriched in neuronal membranes (20, 21) and is commonly esterified with docosahexanoic acid (DHA), which promotes neurite outgrowth (22, 23), synaptogenesis (23), and neurogenesis (23, 24).
Therefore, using the Morris water maze (MWM), we investigated whether Lpaatδ−/− mice manifest a cognitive phenotype and found evidence of severe impairments in spatial learning and memory. To uncover the mechanisms underlying the poor performance of Lpaatδ−/− mice in the MWM, we examined differences in neuronal and glial content; brain phospholipid DHA (22:6n-3) content and fatty acyl profiles; mitochondrial phospholipid composition; mitochondrial function; downstream targets of PIP signaling, including AKT and mTOR pathways; and content of N-methyl-d-asparate (NMDA) and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors. To the best of our knowledge, this work constitutes the first report of a role for any LPAAT or AGPAT in development of a behavioral brain phenotype.
RESULTS
Lpaatδ−/− mice have significant impairments in spatial learning and memory compared to wild-type littermates.
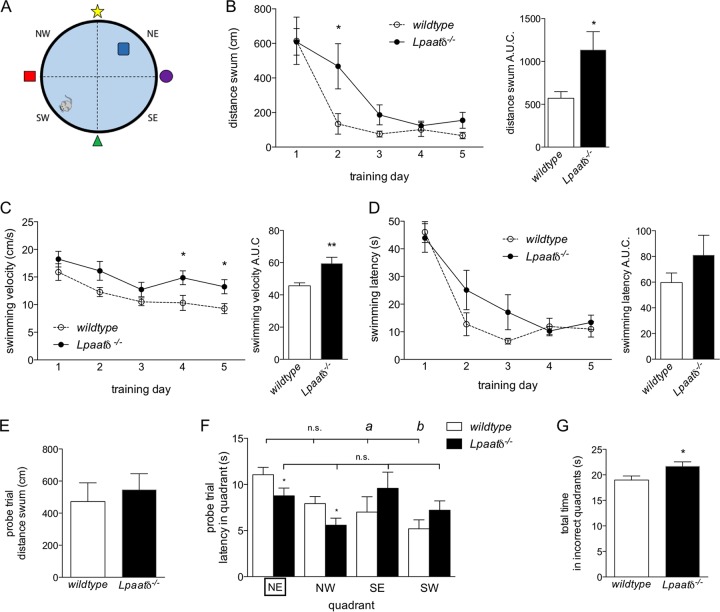
We have previously reported that Lpaatδ−/− mice have significantly lower brain levels of three abundant phospholipids, PC, PE, and PI (6). Therefore, in the current study, we first investigated whether a cognitive phenotype is associated with these decreases. We tested spatial learning and memory of wild-type and Lpaatδ−/− mice using the MWM paradigm (Fig. 1A). Researchers were blinded to the genotypes of the mice until after study completion to limit bias. Wild-type and Lpaatδ−/− mice swam similar distances on average during the first day of training (Fig. 1B, left). This was not unexpected, since swimming was a new experience for these mice, and neither group knew initially where the hidden platform was located. However, Lpaatδ−/− mice were significantly delayed in learning the MWM task compared to wild-type mice (Fig. 1B). On the second day of training, a significant difference was observed in the average distance swum between the two groups (Fig. 1B, left). While most wild-type mice remembered the location of the hidden platform and swam directly for it, Lpaatδ−/− littermates swam over three times the distance as they continued to search for it. By the third day, there was no longer a significant difference between Lpaatδ−/− mice and their wild-type littermates in the distance swum to reach the platform, indicating that both groups were eventually able to learn the task (Fig. 1B, left).
FIG 1.
Lpaatδ−/− mice display impaired spatial learning and memory in the MWM compared to wild-type littermates. (A) Schematic of the MWM, with the hidden platform placed in the NE quadrant during the five training days. (B) Average distances swum by wild-type and Lpaatδ−/− mice over 5 days of training (left) and area under the curve (AUC) analysis (right). (C) Average swimming velocities of mice during 5 days of training in the MWM (left) and area under the curve analysis (right). (D) Average swimming latencies (left) and area under the curve analysis (right) of mice during 5 days of training in the MWM. (E) Total distances swum by wild-type and Lpaatδ−/− mice during the 30-s probe trial performed on day 6, with the platform removed. (F) Latencies of mice in each quadrant during the day 6 probe trial (the correct NE quadrant is boxed). a, P < 0.05; b, P < 0.01; n.s., not significant versus NE platform quadrant for within-genotype comparison. (G) Total time spent in incorrect quadrants (i.e., NW, SE, and SW) during the probe trial by wild-type and Lpaatδ−/− mice. Data are means ± SEM; n = 7 or 8. *, P < 0.05; **, P < 0.01 versus wild type.
Examination of the swimming velocities of both groups during the training period indicated that Lpaatδ−/− mice were not impaired in their swimming ability and, indeed, swam faster than their wild-type littermates on average (Fig. 1C), as well as on training days 4 and 5, specifically (Fig. 1C, left). Because of this greater velocity, the mice were able to swim a farther distance in a shorter time, and the increased time required for Lpaatδ−/− mice to reach the platform (i.e., latency) was not statistically significantly different from the times required by wild-type littermates, who tended to take a more direct, remembered path (Fig. 1D).
To test memory after the MWM task was learned, we performed a probe trial in which the hidden platform was removed. The time spent by mice swimming and searching in each quadrant was then recorded to measure the ability to remember the learned task of finding the hidden platform. Although wild-type and Lpaatδ−/− mice swam the same distance during the probe trial (Fig. 1E), differences in swimming patterns were observed. During the probe trial, wild-type mice spent the majority of their time searching in the correct quadrant (northeast [NE]). They spent the least amount of their time in the quadrant from which they entered the pool (southwest [SW]) and significantly less time in the adjacent southeast (SE) quadrant, as well (Fig. 1F). In comparison, Lpaatδ−/− mice spent significantly less time than their wild-type littermates swimming in either the correct NE quadrant or the northwest (NW) quadrant (Fig. 1F). Indeed, Lpaatδ−/− mice showed no significant differences in time spent swimming in any region, indicating an inability to distinguish quadrants or even remember their entry point. In total, Lpaatδ−/− mice spent a significantly greater amount of time than wild-type mice swimming in incorrect quadrants (NW, SW, and SE) during the probe trial (Fig. 1G).
We next performed a series of studies to better understand the mechanisms underlying the changes in memory and learning observed in Lpaatδ−/− mice.
Lpaatδ deficiency does not alter brain levels of neuronal or glial marker proteins.
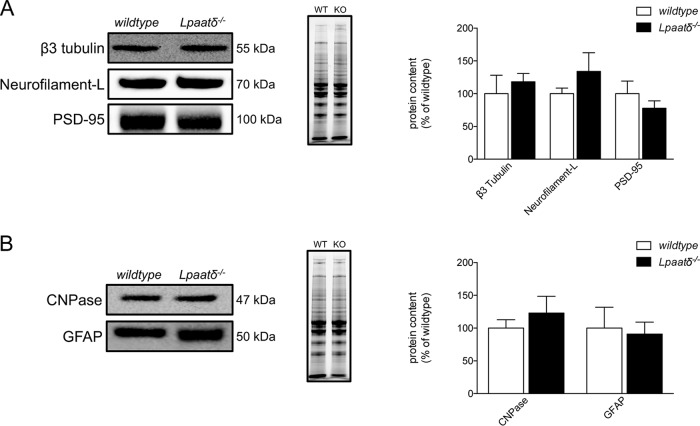
Changes in phospholipids could affect the content of major brain cell types. We first probed for markers of neuronal content, including β3 tubulin, neurofilament L, and the postsynaptic density scaffold protein PSD-95, but found no differences in the relative levels of any of these proteins in whole-brain homogenates from wild-type compared to Lpaatδ−/− mice (Fig. 2A). Likewise, we found no significant differences in the relative contents of CNPase, a marker of oligodendrocyte content, or the glial structural protein glial fibrillary acidic protein (GFAP) (Fig. 2B), indicating that impaired spatial learning in Lpaatδ−/− mice is not likely the result of altered brain cell composition.
FIG 2.
Lpaatδ deficiency does not alter brain levels of neuronal or glial marker proteins. Panels of neuronal cell (A) and glial cell (B) marker proteins were assessed by immunoblotting of whole-brain homogenates. Shown are representative immunoblots (left), images of total protein per lane in TGX stain-free gels as a loading control (middle), and quantification (right). The data are means ± SEM (n = 4 to 11). WT, wild type; KO, Lpaatδ−/−.
Lpaatδ deficiency does not reduce the relative content of docosahexaenoic acid in whole-brain phospholipids.
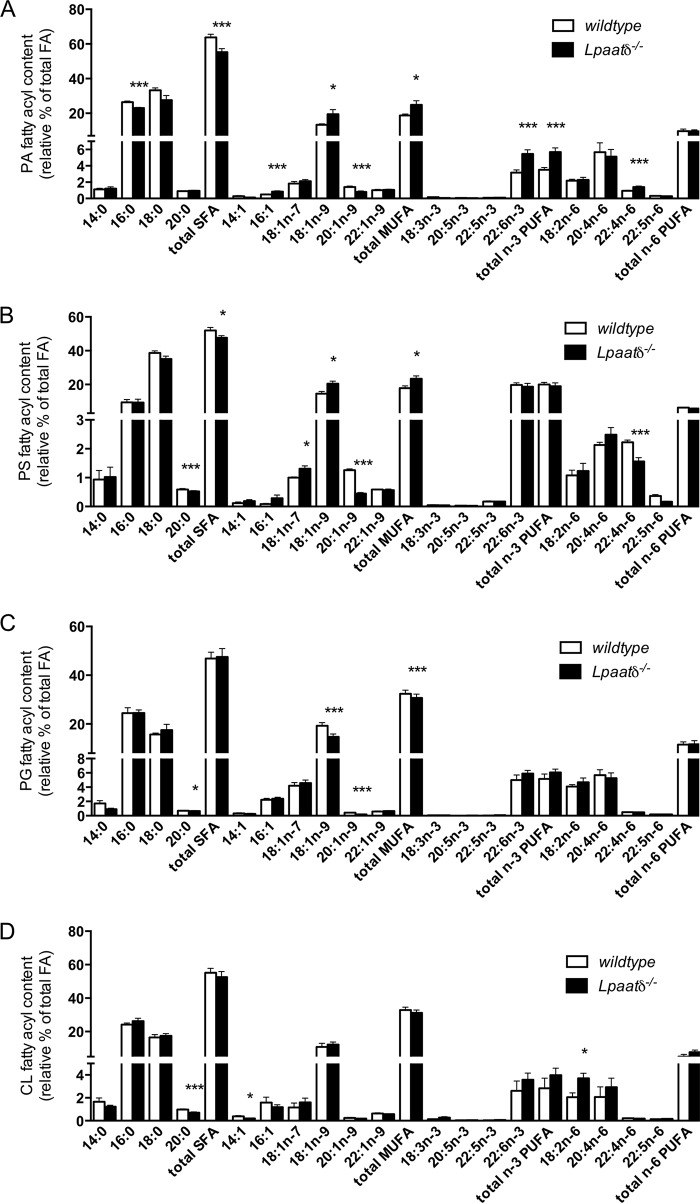
Deficiency of the n-3 polyunsaturated fatty acid (PUFA) DHA in neural phospholipids is associated with impairments in cognitive processes, such as learning and memory (25–27). To investigate if decreased DHA content was associated with the poor performance of Lpaatδ−/− mice in the MWM, we harvested whole brains and analyzed the fatty acyl composition of major phospholipid groups. Previously, we reported that the relative abundance of DHA was not significantly altered in PC, PE, or PI, and indeed, no other significant differences in total n-3 PUFA, or any other individual n-3 PUFA species, were evident within these phospholipid groups (6). In the present work, we investigated changes in the relative abundance of major fatty acyl species, including DHA and other n-3 PUFAs, within the following additional major brain phospholipid groups: PA (Fig. 3A), phosphatidylserine (PS) (Fig. 3B), phosphatidylglycerol (PG) (Fig. 3C), and cardiolipin (CL) (Fig. 3D). No significant decreases were observed in the DHA content of any of the additional phospholipid groups examined with Lpaatδ deficiency, and conversely, the relative abundance of DHA was increased in the PA fraction derived from brains of Lpaatδ−/− mice relative to wild-type brains (Fig. 3A).
FIG 3.
Lpaatδ deficiency does not reduce the relative content of docosahexaenoic acid in whole-brain phospholipids. Shown is the fatty acyl species composition of PA (A), PS (B), PG (C), and CL (D) from whole brains of wild-type or Lpaatδ−/− mice as a relative percentage of total fatty acid (FA) mass. The data are means ± SEM (n = 4). *, P < 0.05; ***, P < 0.001 versus the wild type.
Differences in the phospholipid fatty acyl compositions of individual phospholipid groups were evident. PA from brains of Lpaatδ−/− mice showed a reduction in total saturated fatty acids that was largely due to a significant reduction in palmitate (Fig. 3A). This was proportionately offset by increases in the PA content of 18:1n-9, DHA, and adrenic acid (22:4n-6), although a significant decrease in 20:1n-9 was also observed. In PS, there were decreases in arachidic acid (20:0), 20:1n-9, and adrenic acid but significant increases in the monounsaturated fatty acids (MUFAs) 18:1n-7 and 18:1n-9 (Fig. 3B). Arachidic acid was also significantly decreased in PG (Fig. 3C), but the MUFA 18:1n-9 was decreased, as was 20:1n9, in this phospholipid group. In cardiolipin, there were relatively few changes in the fatty acyl profile, and they included a decrease in arachidic acid and 14:1n-9 but an increase in linoleic acid.
Because AGPAT4 is a mitochondrial enzyme (6), we next examined whether there were specific changes in the phospholipid profile of mitochondria isolated from brains of Lpaatδ−/− mice and their wild-type littermates.
Lpaatδ deficiency reduces the abundance of specific ethanolamine-type phospholipids in brain mitochondria but does not affect the relative contents of most other major species.
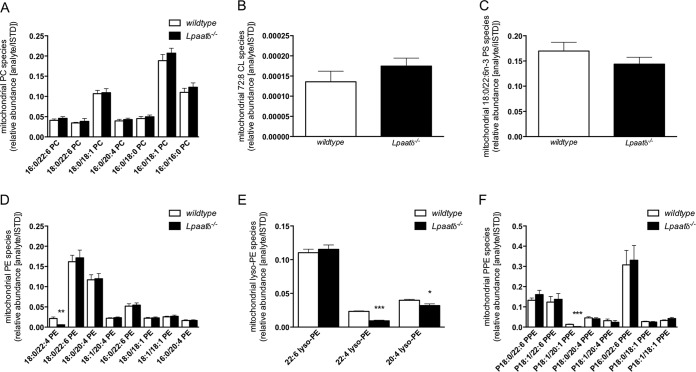
Isolated brain mitochondria were analyzed by tandem mass spectrometry (MS/MS). The identities and relative abundances of acyl species of lipids were manually determined and compared for the highly abundant species. While we noted no difference in any identifiable major PC species (Fig. 4A), tetralinoleoyl (72:8) CL (Fig. 4B), or 18:0/22:6n-3 PS (Fig. 4C), we observed a significant reduction in 18:0/22:4-PE (Fig. 4D), 22:4-lyso-PE, 20:4-lyso-PE (Fig. 4E), and P-18:1/20:1 plasmenyl-PE (Fig. 4F) in the brain mitochondrial fractions derived from Lpaatδ−/− mice compared to those from wild-type mice. These differences were also confirmed using principal-component analysis (PCA).
FIG 4.
Lpaatδ deficiency reduces the abundance of specific ethanolamine-type phospholipids in brain mitochondria. Shown are the specific fatty acyl composition of brain mitochondrial phosphatidylcholine (A), 72:8 CL (B), 18:0/22:6 PS (C), phosphatidylethanolamine (D), lysophosphatidylethanolamine (E), and plasmenyl phosphatidylethanolamine (PPE) (F) species from wild-type or Lpaatδ−/− mice as the abundance of the analyte relative to the internal standard. The data are means ± SEM (n = 4). *, P < 0.05; ***, P < 0.001 versus the wild type.
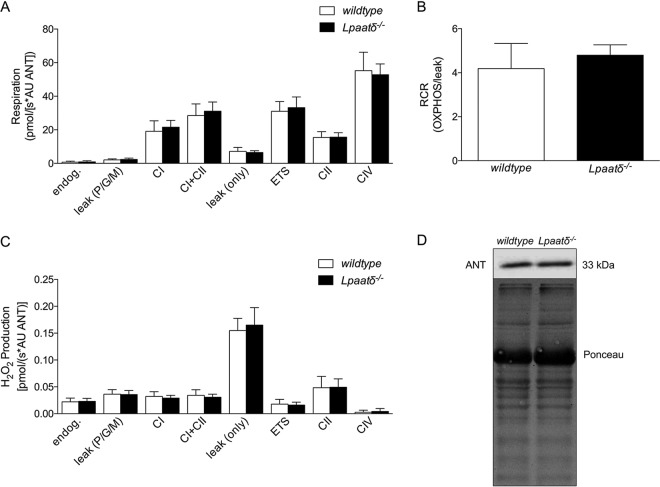
Mitochondrial function is similar in brains of Lpaatδ−/− and wild-type littermates.
Since LPAATδ localizes to the outer mitochondrial membrane (6), we investigated whether the function of isolated brain mitochondria was impaired in Lpaatδ−/− mice compared to their wild-type littermates. No statistically significant differences were seen in the activities of the electron transport chain (ETC) complexes tested, including complex I (CI), CI plus CII, CII, and CIV (Fig. 5A). Oxygen leak (leak only), which compensates for proton leak, proton slip, cation cycling, and electron leak, was not found to be different between wild-type and Lpaatδ−/− mice. In addition, the electron transport system (ETS) maximum oxygen flux (measured at optimum uncoupler concentration) was not different between wild-type and Lpaatδ−/− mice. Endogenous background mitochondrial respiration, as well as pyruvate, glutamate, and malate leak [leak (P/G/M)] was not significantly different between wild-type and Lpaatδ−/− mice. There was no significant difference between Lpaatδ−/− and wild-type littermate mice in the respiratory control ratio (RCR) (Fig. 5B). Examination of H2O2 production by brain mitochondria showed no statistically significant differences in any respiratory complexes or in leak (Fig. 5C). To estimate the brain mitochondrial content, immunoblots of the inner mitochondrial membrane protein adenine nucleotide translocator (ANT) were prepared from homogenates of isolated mitochondria (Fig. 5D). The protein content of ANT was not different between wild-type and Lpaatδ−/− protein lysates, suggesting no difference in the mitochondrial contents between wild-type and knockout mouse brains.
FIG 5.
Lpaatδ deficiency does not alter respiratory capacity or relative content of isolated brain mitochondria. (A) Respiratory activities (AU, arbitrary units) of individual mitochondrial electron transport chain complexes (CI, CI plus CII, CII, and CIV) were determined in isolated brain mitochondria, along with background activity (endog.), leak (P/G/M), oxygen leak (leak only), and electron transport system maximum oxygen flux at optimum uncoupler concentration (ETS). (B) Respiratory control ratio (RCR) indicating the relative oxidative phosphorylation (OXPHOS) activity compared to proton leak in isolated mitochondria, determined as a ratio between CI plus CII and the leak respiration following the addition of oligomycin [leak (only)]. (C) Hydrogen peroxide production by brain mitochondria isolated from wild-type and Lpaatδ−/− mice. (D) Representative immunoblot and Ponceau stain (loading control) demonstrating no significant difference in the endogenous content of ANT, a marker of mitochondrial content, from brains of wild-type and Lpaatδ−/− mice. The data are means ± SEM (n = 6 to 9).
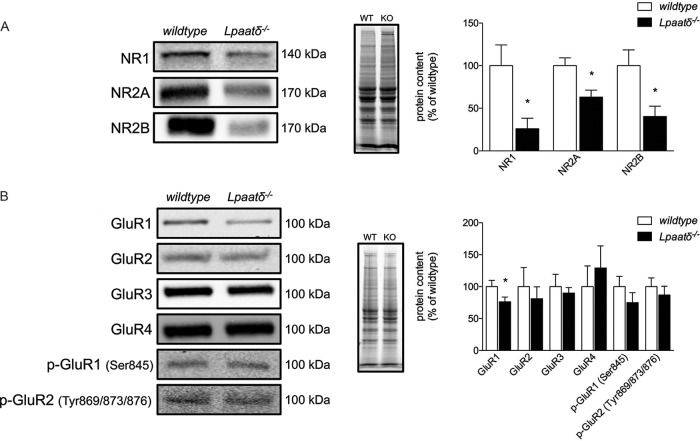
Specific NMDA and AMPA receptor subunit contents are decreased in brains of Lpaatδ−/− mice.
Lpaatδ−/− mice have a substantial decrease in total brain PI levels (6). NMDA and AMPA receptors are integral membrane proteins involved in synaptic plasticity and memory (28–31) that are regulated by changes in membrane phosphoinositide composition (32, 33). We investigated the brain content of the NMDA receptor subunits NR1, NR2A, and NR2B (Fig. 6A) and detected drastically lower levels of all three subunits in the brains of Lpaatδ−/− mice compared to those in wild-type mice (P = 0.023, P = 0.011, and P = 0.013, respectively). Since AMPA receptors are also critical for glutamate transfer between neurons, we probed for a number of AMPA receptor subunits, including GluR1, GluR2, GluR3, GluR4, GluR1 phosphorylated at serine 845 (Ser845), and GluR2 phosphorylated at Tyr869/873/876. We found that Lpaatδ−/− mouse brains had significantly lower contents of total GluR1 versus those in wild-type mice (P = 0.034) but no significant difference in the content of any other AMPA receptor subunits (Fig. 6B).
FIG 6.
Lpaatδ−/− mice have lower brain levels of NR1, NR2A, NR2B, and GluR1. Shown are representative immunoblots (left), images of total protein per lane in TGX stain-free gels as a loading control (middle), and quantification (right) of NMDA receptor subunits (A) and AMPA receptor subunits (B) from whole-brain homogenates of wild-type (WT) and Lpaatδ−/− (KO) mice. The data are means ± SEM. *, P < 0.05 versus the wild type (n = 4 to 12).
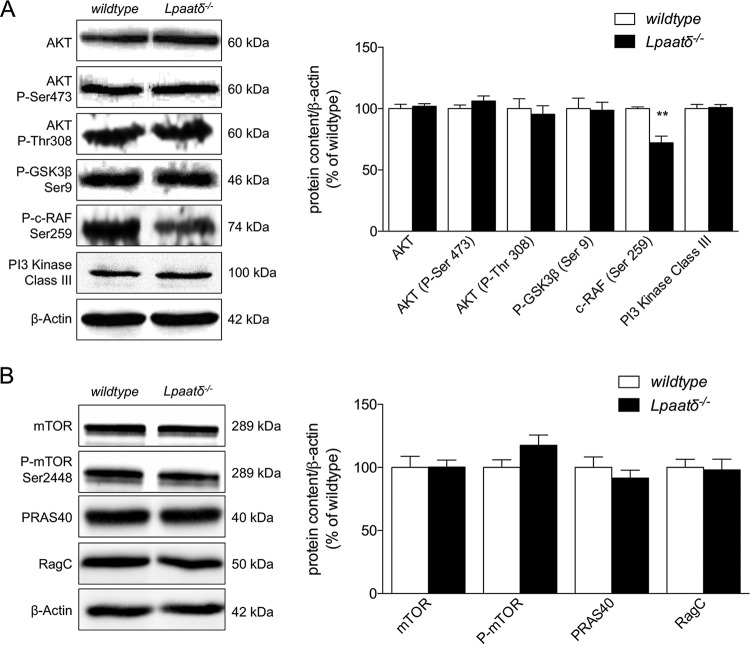
AKT and mTOR pathway proteins are largely unaffected by deficiency of Lpaatδ.
Since significant disruption of NMDA and AMPA receptors that are regulated by PIP-mediated signaling was observed, we investigated whether general changes were evident in AKT/protein kinase B (PKB) and mTOR pathways regulated downstream of PIP3 that have known roles in neural function (34–38).
Using specific antibodies, we measured immunodetectable levels of total AKT, as well as AKT activated by phosphorylation at Ser473 or threonine 308 (Thr308) in brains of wild-type and Lpaatδ−/− mice (Fig. 7A). We also immunoblotted with phospho-specific antibodies to detect levels of phosphorylation of two AKT target proteins, phospho-glycogen synthase kinase 3β (phospho-GSK3β) and phospho-c-RAF (Fig. 7A). There was no measurable difference in protein levels between wild-type and Lpaatδ−/− mice for total AKT or phospho-AKT phosphorylated at Ser473 or Thr308. Similarly, there was no difference in GSK3β phosphorylation at the Ser9 AKT target site. There was, however, a statistically significant (P < 0.01) decrease in phosphorylation of c-RAF at Ser259 in the brains of Lpaatδ−/− mice compared to wild-type mice (Fig. 7A). We then investigated the levels of total mTOR and mTOR phosphorylated at Ser2448 but did not see a significant difference in the content of either protein (Fig. 7B). We measured the content of two additional proteins in the mTOR pathway, proline-rich AKT substrate (PRAS40), which helps relay AKT signaling to the mTOR protein complex (39), and RagC, a GTPase that interacts with raptor in the mTORC1 complex (40, 41). However, we did not observe any significant changes in the content of either protein between wild-type and knockout brains (Fig. 7B).
FIG 7.
Brain levels of total and phosphorylated AKT and mTOR pathway proteins are largely unchanged in Lpaatδ−/− mice. Shown are representative immunoblots and quantification of proteins of the AKT signaling pathway (A) and the mTOR signaling pathway (B) expressed relative to β-actin in whole-brain homogenates of wild-type and Lpaatδ−/− mice. The data are means ± SEM. **, P < 0.01 versus the wild type (n = 4 to 10).
DISCUSSION
Together, PC and PE are two of the most abundant phospholipid species in mammalian membranes, jointly constituting approximately 71% of total brain phospholipids in humans and ∼79% in rodents (9). While PI makes up a much smaller percentage of total brain phospholipid (∼2.6% in humans and ∼1.6% in rodents) (9), it can be phosphorylated into highly bioactive PIP derivatives that are involved in a glut of signaling pathways. In work reported previously by our laboratory, we have shown that mice deficient in Lpaatδ have specific reductions in brain content of these phospholipids (6), which warranted the investigation of a possible cognitive phenotype and associated molecular changes.
In the current study, we report that Lpaatδ−/− mice have significant impairments in spatial learning and memory. On the initial training day, as expected, both wild-type and Lpaatδ−/− mice swam similar distances, consistent with the fact that neither group knew the location of the platform. On the second day of training, however, there were clear differences between the genotypes. While wild-type mice quickly learned the location of the hidden platform and displayed proficiency in reaching it, Lpaatδ−/− mice showed significant impairments and required a third day to learn the location. The ability to remember the location of the platform was also notably compromised, since Lpaatδ−/− mice spent significantly less time swimming in the correct quadrant after it was removed on a “test” day. Spatial learning and memory are highly integrated and dynamic processes. We therefore investigated key factors associated with altered phospholipid composition that could contribute to the poor performance of Lpaatδ−/− mice compared to their wild-type littermates.
Alterations in the content of major phospholipid species, such as PC and PE, have been shown to contribute to long-term potentiation (24, 42), an important process underlying the synaptic plasticity of neurons (43, 44). Glial cells also contribute to synaptic function, important in learning and memory (45). We hypothesized that a lower content of brain PC and PE, which are critical components of neuronal and glial membranes, may alter the brain content of these cell types in Lpaatδ−/− mice. However, immunoblotting for a panel of neuronal, glial, and oligodendrocyte markers demonstrated no significant difference in content between the brains of wild-type and Lpaatδ−/− mice, indicating that gross neuronal and glial losses were not likely major contributors to the observed impairments in spatial learning and memory observed in Lpaatδ−/− mice.
We next investigated differences in whole-brain phospholipid fatty acyl composition, since changes in the relative abundances of different acyl species within phospholipids can have significant effects on cellular function and health (46, 47). Previous in vitro studies by Eto et al. (7) have suggested that LPAATδ may play a role in enriching phospholipid DHA stores in neural cells. The n-3 polyunsaturated fatty acid DHA is abundant in the brain, particularly in PE, and deficiency of DHA in brain phospholipids is associated with impairments in cognitive processes, including learning and memory (20, 23, 26). However, examination of individual phospholipid groups (i.e., PA, PS, PG, and CL) in the current work, and in prior work (i.e., PC, PE, and PI) (6), demonstrated an increase of DHA in brain PA but no difference in DHA in any other fraction analyzed, strongly suggesting that relative DHA deficiency within brain phospholipids is not responsible for impaired memory and learning in Lpaatδ−/− mice. Overall, changes in fatty acyl species in the brain phospholipid groups examined were diverse, showing few consistent patterns of increases or decreases, which highlights the relative importance of Lands' pathway remodeling downstream of PA in the formation of mature cellular phospholipids.
Since we had previously identified LPAATδ as integral to the outer mitochondrial membrane, we next reasoned that loss of the enzyme may alter the mitochondrial phospholipid profile or reduce the function or brain content of the organelle. The most abundant PC, PS, and CL species, however, were present at similar levels in brain mitochondrial membranes from wild-type and Lpaatδ−/− mice, while the only differences observed were in specific ethanolamine-containing phospholipids. Of note, two of these contained adrenic acid, 18:0/22:4-PE, and 22:4-lyso-PE, which are among the brain species that have consistently been reported to undergo a progressive, age-related decline during normal aging (48, 49). In humans, ethanolamine phospholipids containing adrenic acid are decreased by approximately 20 to 25% in the hippocampus from age 20 to 80 years (48, 49). Adrenic acid content has also been found to be inversely correlated with cognitive performance (50) and also with the development of cognitive disorders, including attention deficit hyperactivity disorder (51) and autism spectrum disorders (52, 53). The significant loss of these PE species from brain mitochondrial phospholipids may, therefore, contribute to the poor performance of Lpaatδ−/− mice in the MWM.
Studies were next performed to determine whether the significant loss of these PE species from brain mitochondrial phospholipids was mechanistically associated with changes in the function or content of brain mitochondria. Overall, the respiratory capacity, the function of individual electron transport chain complexes, and other measures of mitochondrial oxidative phosphorylation and bioenergetic activity were not significantly different between brain mitochondria isolated from Lpaatδ−/− mice and their wild-type littermates. Similarly, mitochondrial marker analysis also indicated no difference in the relative content of this organelle between brains of wild-type and Lpaatδ−/− mice. Together, these data therefore indicate that changes in overall mitochondrial number, function, and composition are unlikely to explain the poor performance of Lpaatδ−/− mice in the MWM.
Lpaatδ−/− mice have a substantial decrease in total brain PI levels (6). Although PI is quantitatively a relatively minor phospholipid in most tissues (8, 9), it is enriched in the brain (10) and is important in myriad biological processes and cell signaling pathways as a precursor for PIPs (54, 55). In the brain, PI-mediated signaling events are involved in regulation of neuronal exocytosis (12) and neurotransmission (11), with phosphoinositides implicated in regulating functional and structural plasticity (13–15). We therefore reasoned that the brains of Lpaatδ−/− mice, which contain approximately half the PI content of wild-type brains, may have dysregulated signaling through pathways regulated by PIPs.
NMDA and AMPA receptors are glutamate-gated transmembrane proteins (56). Brain content and activation of NMDA receptors are associated with learning and memory (28–31, 57, 58), while AMPA receptors have been shown to contribute to hippocampal synaptic plasticity required for the formation of new memories (59, 60). Decreases in brain NMDA and AMPA receptor content are correlated with a reduced capacity for learning and memory in both mice (58) and humans (61). Changes in membrane PIP composition affect the conformation, localization, and function of both NMDA and AMPA receptors (32, 33). PIP2 mediates interaction of NMDA receptors with a cytoskeletal adaptor protein that is required for maintenance of the open state (32) and for synaptic localization (33). PIP3 is required for maintenance of AMPA receptor clustering at the postsynaptic membrane (15). We determined the levels of the NMDA receptor subunits NR1, NR2A, and NR2B, as well as a host of AMPA receptor subunits, in total brain lysates and found that Lpaatδ−/− mice have a significantly lower content of NR1, NR2A, NR2B, and GluR1 than wild-type mice. Since NMDA receptors are comprised of a heterotetramer formed from two NR1 subunits and either or both NR2 subunits (62), our finding of a global decrease in all components suggests that brain Lpaatδ deficiency leads to reduced total NMDA receptor levels, rather than a shift in the makeup of these receptors.
Most studies on the depletion/repletion of PIPs report redistribution of NMDA and AMPA receptors, rather than overt changes in total levels (15). Given the drastic reductions that were evident (i.e., ∼75% decrease in NR1), we did not further explore changes in subcellular localization, since global effects would likely predominate. The lack of difference in total receptor levels in PIP depletion/repletion experiments may be due to the short-term nature of those in vitro experiments in contrast to the Lpaatδ-deficient mouse, which represents a chronic model of altered PI. It is also possible that NMDA and AMPA receptor levels were not reduced as a result of changes in PI/PIPs per se. Brain PC and PE contents are also reduced in Lpaatδ-deficient mice. Moderate decreases in PC and PE are observed in the prefrontal cortex in rats with normal aging, and this is associated with subunit-specific decreases in AMPA and NMDA receptors (63). It is thus possible that the loss of PC and PE in brains of Lpaatδ−/− mice alters membrane composition in a manner that impairs assembly or enhances degradation of NMDA and AMPA receptors, causing the reductions in content observed. Therefore, we next tested whether there was evidence of generalized dysfunction in other well-characterized PIP-regulated signaling pathways.
PIPs in membranes form docking sites for proteins such as AKT that contain a pleckstrin homology domain (64). PIP3 is important for anchoring activated AKT/PKB to the membrane, where it acts as a kinase regulator of other proteins, including mTOR, which it phosphorylates directly (65) and also regulates indirectly (66). We performed an initial investigation examining AKT/PKB and mTOR signaling pathways, which are involved in a multitude of cellular processes in the brain, including cell survival and proliferation, glucose metabolism, and protein synthesis (67–72). However, we found no differences in brain levels of AKT or AKT activated by phosphorylation at Ser473 or Thr308. We also found no significant difference in levels of phosphorylation of GSK3β at Ser9, which is a target site for AKT phosphorylation, together suggesting that AKT-mediated signaling is not significantly different between wild-type and Lpaatδ−/− mice. Finally, we investigated the protein contents of mTOR and mTOR phosphorylated at the AKT consensus sites Ser2448, PRAS40, and RagC but found no significant differences between the brains of wild-type and Lpaatδ−/− mice. Although we did observe a reduction in phosphorylated cRAF, this protein is a target for kinases other than AKT (73–76). Taken together, our findings suggest that PI is present in the brain at a level that well exceeds cellular requirements for the production of phosphoinositides, such that a >50% decrease in total PI does not cause a generalized impairment of PI-mediated signaling.
Overall, the results from this study suggest a striking difference in learning and memory between wild-type and Lpaatδ−/− mice determined using the MWM. However, inferences should be interpreted with some caution. Although the MWM is generally regarded as a gold standard in assessing spatial learning and memory (77, 78), other confounding factors could affect the performance of Lpaatδ−/− mice in this testing paradigm, such as a greater level of anxiety than their wild-type littermates. In this regard, we tested Lpaatδ−/− mice using a computerized comprehensive laboratory animal modeling system and found no significant differences from wild-type littermates in total or directional activity when the mice were introduced to their new environment (79). This suggests that differences in anxiety or exploratory behavior are not responsible for impaired performance in the MWM. However, future studies using alternate testing paradigms may be of value for expanding the understanding of intricate relationships between neural lipid dysregulation and learning behavior.
In summary, our current study has demonstrated for the first time that deficiency of an Lpaat or Agpat gene causes significant impairments in spatial learning and memory as assessed by the MWM. These deficits are attributed, at least in part, to a significantly reduced neural content of the NMDA receptor subunits NR1, NR2A, and NR2B, as well as the AMPA receptor subunit GluR1. Significantly lower protein levels of these subunits are likely related to a significant decrease in the membrane glycerophospholipids PC, PE, and PI. However, the exact mechanism through which alterations in membrane phospholipid content and composition modulate glutamate receptor function in neurons is currently unknown. We speculate that significant loss of these phospholipids may alter the biophysical properties of neuronal membranes in a manner that is nonadvantageous to NMDA and AMPA receptor function, but further work will be required to fully understand these associations.
MATERIALS AND METHODS
Animals and genotyping.
Lpaatδ gene ablation was performed by Genentech/Lexicon and was achieved by homologous recombination that replaced exons 4 to 6 with a LacZ-neomycin selection cassette (80). Although exon 3 contains catalytic motif I, the HX4D catalytic dyad, exons 4 to 6 contain motifs III and IV, the highly conserved “EGTR” sequence, and a conserved proline that are critical for acyltransferase activity of AGPAT enzymes (81). We have previously reported confirmation of Lpaatδ disruption and PCR conditions for genotyping (6). Mice were produced by intercrossing heterozygous littermates to generate wild-type, heterozygous, and homozygous null mice, of which only the wild-type and homozygous null mice were used for studies. Twelve- to sixteen-week-old male wild-type and Lpaatδ−/− littermates were used. Animals were housed in a temperature- and humidity-controlled environment on a 12-h/12-h light/dark cycle, and standard rodent chow and water were provided ad libitum. All animal procedures were approved by the University of Waterloo Animal Care Committee.
Morris water maze.
Lpaatδ−/− and wild-type littermate mice were tested for spatial learning and memory using the MWM, as described previously (77, 82), with minor modifications for mice (78). The researchers were blinded to the genotypes of the mice while running the study. The pool was 26.5 cm in height and 69 cm in diameter and was divided into four quadrants. The hidden platform measured 5 cm in height by 9 cm in width by 14 cm in length. The water was maintained at 23°C ± 1°C to prevent hypothermia and stained white to obscure the platform and improve camera tracking of black and brown mice. The platform was hidden 1 cm below the water surface and placed in the northeast quadrant. Landmark cues were posted around the walls of the water maze room to help the mice orient themselves in the pool and find the platform. All MWM experiments took place between 10:00 a.m. and 12:00 p.m. The mice were placed into the pool facing away from the platform in the southwest quadrant and were allowed to swim for up to 30 s. Mice who found the platform in under 30 s were allowed to sit on the platform for 20 s. The researchers guided mice that failed to find the platform in 30 s to the platform, where they also sat for 20 s. The training period lasted for 5 days, and the mice were subjected to one block of three training repetitions each day. On the sixth day, the platform was removed, and the mice were allowed to swim for 30 s to assess spatial learning and procedural memory. The performance of the mice in the MWM was monitored by an overhead video camera connected to a computer with EthoVision XT video tracking software (Noldus Information Technology, Wageningen, The Netherlands).
High-resolution respirometry and hydrogen peroxide production.
Mitochondrial fractions were obtained as previously described (83, 84), with minor modifications. Briefly, 1/2 of the brain, split along the medial longitudinal fissure, was homogenized using a Polytron homogenizer (VWR, Radnor, PA) set at maximum speed in 2 ml of MiR05 buffer (0.5 mM EGTA, 3 mM MgCl2, 60 mM lactobionic acid, 20 mM taurine, 10 mM potassium dihydrogenphosphate, 20 mM HEPES, 110 mM sucrose, 1 g/liter essential-fatty-acid-free bovine serum albumin [BSA], and 10 μl/ml protease inhibitor cocktail), a mitochondrial respiration buffer developed by Oroboros Instruments (Innsbruck, Austria). All homogenization and centrifugation steps were performed at 4°C. To remove nuclei, brain homogenates were centrifuged at 1,400 × g for 7 min, and the supernatant was removed to a clean tube and spun again at 1,400 × g for an additional 3 min. The supernatant was removed to a fresh tube, which was spun at 10,000 × g for 5 min to pellet mitochondria. The mitochondrial pellet was gently resuspended in 1 ml of MiR05, and centrifuged at 1,400 × g for 3 min to pellet and remove any remaining nuclei, further purifying the mitochondrial fraction. The supernatant was transferred into a fresh tube and spun at 10,000 × g for 5 min to pellet the mitochondria. The supernatant was removed, and the mitochondrial pellet was then resuspended in 250 μl of MiR05. Forty-microliter aliquots of this uniform suspension were immediately injected into multiple chambers of an Oxygraph-2k respirometer. The remaining suspension was frozen at −80°C for subsequent protein analysis.
The rate of mitochondrial oxygen consumption was measured using the Oxygraph-2k respirometer (Oroboros, Innsbruck, Austria) equipped with the O2k-Fluorescence LED2 module for simultaneous measurement of H2O2 production using Amplex Red (85). All measurements were performed at 37°C. Prior to the addition of mitochondria, 2 ml of MiR05 was equilibrated to the chamber and oxygenated to approximately 300 nmol/ml of O2. Components of the Amplex Red detection system were added to each chamber (final concentrations, 10 μM Amplex Red, 1 U/ml horseradish peroxidase, and 5 U/ml superoxide dismutase), as previously described (85). After a baseline was recorded, H2O2 was injected into each chamber (0.1 μM final concentration) to allow initial calibration of the fluorescent signal. To examine components of the electron transport chain in detail, a substrate-uncoupler-inhibitor titration (SUIT) procedure adapted from Afshordel et al. (83) was performed. Forty microliters of the isolated brain mitochondria solution was added to each of 2 chambers and allowed to equilibrate (endogenous respiration [endog]). A solution of substrates for complex I, including glutamate (5 mM), pyruvate (5 mM), and malate (2 mM), was added to each chamber in the absence of ADP to determine mitochondrial leak [leak (P/G/M)]. ADP (2 mM) was then added to determine CI capacity, followed by addition of the complex II substrate succinate (10 mM) to determine the capacity of CI plus CII. Cytochrome c (10 μM) was subsequently added to test for mitochondrial integrity. The inhibition of ATP synthase with oligomycin (2.5 μM) was then tested to determine the substrate-limited leak state (leak [only]), which is a measure of intrinsic uncoupling due to proton and electron slip, as well at cation cycling. As a maximal measure of the ETS capacity, the uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP) (2.5 μM) was added. CII-supported noncoupled respiration was determined with the addition of rotenone (0.5 μM). The addition of antimycin A (2.5 μM), a complex III inhibitor, allowed the quantification of residual oxygen consumption (ROX). Finally, tetramethylphenylenediamine (TMPD) (0.5 mM) and ascorbate (2 mM) were added to each chamber to stimulate cytochrome c oxidase CIV activity. Autoxidation was determined with the subsequent addition of sodium azide (100 mM). All values were corrected for ROX, and CIV activity was also corrected for autoxidation. The total protein content was determined with a bicinchoninic acid (BCA) assay, and endogenous levels of ANT were determined by Western blotting as a marker of mitochondrial content per sample. All respirometry and Amplex Red fluorescence values were normalized to ANT content. The respiratory control ratio (RCR) of the isolated brain mitochondria was determined as a ratio between CI plus CII and the leak respiration following the addition of oligomycin (leak [only]).
Immunoblotting.
Immunoblot analysis was performed as previously described (6). Briefly, whole mouse brains were homogenized in 1 ml of buffer A (50 mM Tris-HCl, pH 6.8, 1 mM EDTA, and 0.5% Triton X-100) with 10 μl/ml of protease inhibitor cocktail, using a Polytron homogenizer set at the highest speed. Samples were incubated on ice for 15 min and then centrifuged at 7,600 × g for 10 min at 4°C. Supernatants were collected, transferred to a new tube, and then sonicated on ice with three 6-s bursts at 65% output. Samples that were not used immediately were stored at −80°C. For immunoblot analysis, samples were mixed with 2× Laemmli buffer (125 mM Tris-HCl, pH 6.8, 20% glycerol, 4% SDS, 10% 2-mercaptoethanol, and 0.05% bromophenol blue) and denatured by heating to 95°C for 5 min. Samples were electrophoresed on 12% SDS-PAGE gels at 120 V for 1 h and then transferred to nitrocellulose membranes at 350 mA for 90 min. Membranes were blocked for 1 h with 5% (wt/vol) skim milk blocker in TBST (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% Tween 20) or 3% (wt/vol) BSA blocker for phosphospecific proteins and then probed overnight in TBST with 1% (wt/vol) blocker using primary antibodies (1:1,000 dilution) directed against AKT, phospho-AKT (Ser 473), phospho-AKT (Thr308), phospho-GSKβ (Ser9), phospho-c-RAF (Ser259), phosphatidylinositol 3 (PI3)-kinase class III, mTOR, phospho-mTOR (Ser2448), PRAS40, RagC, GFAP, neurofilament L, GluR1, GluR2, GluR3, GluR4, phospho-GluR1 (Ser845), phospho-GluR2 (Tyr869/873/876), CNPase, β3 tubulin, β-actin, and NR1 (Cell Signaling, Beverly, MA) or PSD-95, NR2A, and NR2B (EMD Millipore, Billerica, MA). The membranes were washed 3 times with TBST and then probed with horseradish peroxidase (HRP)-conjugated secondary antibodies in TBST with 1% blocker. The blots were washed 3 times with TBST, and bands were detected by enhanced chemiluminescence (Luminata Forte reagent; EMD Millipore, Etobicoke, Ontario, Canada). Protein expression was normalized to one of three different loading controls: β-actin levels, total protein per lane following Ponceau staining of membranes, or total protein per lane following fluorescence-activated imaging of proteins in TGX stain-free gels (Bio-Rad, Mississauga, Ontario, Canada).
Lipid extraction and gas chromatography.
To assess the relative content of fatty acids within phospholipids from whole brains of wild-type and Lpaatδ−/− mice, brains were harvested and lipids were extracted by Folch's method as previously described (6, 86). Briefly, whole brains were homogenized in 2:1 (vol/vol) chloroform-methanol with butylated hydroxytoluene as an antioxidant, using a Polytron homogenizer set at the highest speed, followed by addition of sodium phosphate (86). The organic phase was collected, dried, applied to a silica gel H plate, and resolved by thin-layer chromatography (TLC) on a chloroform–methanol–2-propanol–0.25% KCl–triethylamine solvent front (30:9:25:6:18 [vol/vol/vol/vol/vol]). Bands corresponding to individual phospholipid species were identified using known standards, overlaid with 10 μg of 22:3n-3 ethyl ester internal standard (Nu-Check Prep, Elysian, MN), and scraped for determination of fatty acyl composition and content by gas chromatography with flame ionization detection as previously described (87). Briefly, fatty acyls in lipids were derivatized to fatty acid methyl esters by transesterification using 14% boron trifluoride in methanol (Thermo Scientific, Bellfonte, PA) with hexane on a 95°C heat block for 1 h. Fatty acid methyl esters were analyzed on a Varian 3900 gas chromatograph equipped with a DB-FFAP 15-m by 0.10-mm inside diameter by 0.10-μm film thickness nitroterephthalic acid-modified polyethylene glycol capillary column (J&W Scientific from Agilent Technologies, Mississauga, Ontario, Canada) with hydrogen as the carrier gas. Samples (2 μl) were introduced by a Varian CP-8400 autosampler into the injector heated to 250°C with a split ratio of 200:1. The initial temperature was 150°C with a 0.25-min hold followed by a 35°C/minute ramp to 200°C, an 8°C/minute ramp to 225°C with a 3.2-min hold, and then an 80°C/minute ramp up to 245°C with a 15-min hold at the end. The flame ionization detector temperature was 300°C with air and nitrogen makeup gas flow rates of 300 and 25 ml/min, respectively, and a sampling frequency of 50 Hz (87).
UHPLC-MS/MS.
To investigate differences in mitochondrial phospholipid content and acyl-specific lipid species, mitochondria were isolated by subfractionation, as we and others have previously described (6, 88), with minor modifications. Briefly, mouse brains were harvested and homogenized at 1,000 × g in buffer A (250 mM sucrose, 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2) and then centrifuged at 800 × g for 15 min to isolate the nuclear fraction. All centrifugation steps were performed at 4°C. The postnuclear supernatants were spun at 11,000 × g for 10 min to pellet mitochondria. The postmitochondrial supernatant was removed, and mitochondrial pellets were resuspended in 100 μl of phosphate-buffered saline (PBS), upon which lipids were extracted using a modified Folch protocol (86) containing 1,2-diheptadecanoyl-sn-glycero-3-phosphatidylcholine as the internal standard. A Thermo Q-Exactive Quadrupole-Orbitrap mass spectrometer (Thermo-Fisher Scientific, Waltham, MA) was used and coupled to a Dionex UltiMate 3000 UPLC System (Dionex Corporation, Bannockburn, IL). A reversed-phase, binary multistep, ultrahigh-performance liquid chromatography (UHPLC) protocol was used with a C18 Ascentis Express column with dimensions of 15 cm by 2.1 mm by 2.0 μm (Sigma-Aldrich, St. Louis, MO). The mobile phase consisted of 80:20 acetonitrile-water plus 2% glacial acetic acid plus 1% 14.8 M ammonium hydroxide, pH 6.7 (A) and 90:10 isopropanol-water plus 5% glacial acetic acid plus 2% 14.8 M ammonium hydroxide, pH 6.6 (B). The gradient protocol used was as follows: from 0 to 5 min, 40% B; from 5 to 10 min, 80% B; from 10 to 14 min, 100% B. From 14 to 15.2 min, B was decreased to 40% and allowed to equilibrate until the 20-min mark. The flow rate was 0.23 ml/min, column temperature was 55°C, and tray temperature was 4°C. The mass spectrometer was operated in negative electrospray ionization mode, 70,000 resolution, with a scan range of m/z 100 to 1,500, spray voltage of −2.5 kV, sheath gas flow rate of 35 units, and capillary temperature of 300°C. MS/MS experiments were done under data-dependent conditions with top 5 ions and 17,500 resolution, and the normalized collision energy was 17.5. Thermo Xcalibur QualBrowser software (version 2.1; Thermo-Fisher Scientific, Waltham, MA) was used for extracting ion chromatograms, integrating peak areas, and exporting MS-MS spectra. Chromeleon Xpress (version 7.2; Thermo-Fisher Scientific, Waltham, MA) was used to monitor and control the Dionex UHPLC settings. MS/MS spectra were analyzed using the NIST MS search program (version 2.0; National Institute of Standards and Technology, Gaithersburg, MD) and LipidBlast libraries. Principal-component analysis was also performed on compounds with MS/MS profiles using Progenesis QI (Durham, NC), and compounds with P values of <0.05 were identified manually. Extracted ion profiles were integrated for phosphatidylcholine species using the acetate adduct [M + acetate]−, phosphatidylethanolamine [M-H]−, phosphatidylserine [M-H]−, and cardiolipin [M-H]−; the data are expressed as the analyte/internal standard ratio.
Statistical analysis.
Results are expressed as means ± standard errors of the mean (SEM). Statistically significant differences between two groups were assessed by Student's t test, and differences across multiple groups were assessed by one-way analysis of variance (ANOVA) with Bonferroni's post hoc test. Significance was accepted at a P value of <0.05.
ACKNOWLEDGMENTS
We thank Erika Lui and Elham Satvat for technical assistance with MWM. We also thank Angela Wagler and Jean Flanagan for expert assistance in animal care. We acknowledge John Mielke for helpful discussions on learning and memory in mice.
This work was supported by grants to R.E.D. from the Canada Foundation for Innovation-Leader's Opportunity Fund and Ontario Research Fund (project 30259) and a Discovery Grant (418213-2012) and a Research Tools and Instruments Grant (EQPEQ-2015-472393) from the Natural Sciences and Engineering Research Council (NSERC) of Canada. E.B.M. is the recipient of an NSERC Master's Scholarship (PGS-M). R.M.B., D.B., J.J.A.H., A.S.M., and P.M.M. are the recipients of NSERC Doctoral Scholarships (PGS-D). D.B. was the recipient of an Ontario Graduate Scholarship (OGS).
REFERENCES
- 1.Kennedy EP, Weiss SB. 1956. The function of cytidine coenzymes in the biosynthesis of phospholipids. J Biol Chem 222:193–214. [PubMed] [Google Scholar]
- 2.Yamashita A, Hayashi Y, Matsumoto N, Nemoto-Sasaki Y, Oka S, Tanikawa T, Sugiura T. 2014. Glycerophosphate/acylglycerophosphate acyltransferases. Biology 3:801–830. doi: 10.3390/biology3040801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitson AP, Stark KD, Duncan RE. 2012. Enzymes in brain phospholipid docosahexaenoic acid accretion: a PL-ethora of potential PL-ayers. Prostaglandins Leukot Essent Fatty Acids 87:1–10. doi: 10.1016/j.plefa.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Bradley RM, Mardian EB, Moes KA, Duncan RE. 2017. Acute fasting induces expression of acylglycerophosphate acyltransferase (AGPAT) enzymes in murine liver, heart, and brain. Lipids 52:457–461. doi: 10.1007/s11745-017-4251-4. [DOI] [PubMed] [Google Scholar]
- 5.Bradley RM, Mardian EB, Marvyn PM, Vasefi MS, Beazely MA, Mielke JG, Duncan RE. 2016. Data on acylglycerophosphate acyltransferase 4 (AGPAT4) during murine embryogenesis and in embryo-derived cultured primary neurons and glia. Data Brief 6:28–32. doi: 10.1016/j.dib.2015.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradley RM, Marvyn PM, Aristizabal Henao JJ, Mardian EB, George S, Aucoin MG, Stark KD, Duncan RE. 2015. Acylglycerophosphate acyltransferase 4 (AGPAT4) is a mitochondrial lysophosphatidic acid acyltransferase that regulates brain phosphatidylcholine, phosphatidylethanolamine, and phosphatidylinositol levels. Biochim Biophys Acta 1851:1566–1576. doi: 10.1016/j.bbalip.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Eto M, Shindou H, Shimizu T. 2014. A novel lysophosphatidic acid acyltransferase enzyme (LPAAT4) with a possible role for incorporating docosahexaenoic acid into brain glycerophospholipids. Biochem Biophys Res Commun 443:718–724. doi: 10.1016/j.bbrc.2013.12.043. [DOI] [PubMed] [Google Scholar]
- 8.Daum G. 1985. Lipids of mitochondria. Biochim Biophys Acta 822:1–42. doi: 10.1016/0304-4157(85)90002-4. [DOI] [PubMed] [Google Scholar]
- 9.Diagne A, Fauvel J, Record M, Chap H, Douste-Blazy L. 1984. Studies on ether phospholipids. II. Comparative composition of various tissues from human, rat and guinea pig. Biochim Biophys Acta 793:221–231. [DOI] [PubMed] [Google Scholar]
- 10.Modi HR, Katyare SS, Patel MA. 2008. Ageing-induced alterations in lipid/phospholipid profiles of rat brain and liver mitochondria: implications for mitochondrial energy-linked functions. J Membr Biol 221:51–60. doi: 10.1007/s00232-007-9086-0. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, McCartney AJ, Zolov SN, Ferguson CJ, Meisler MH, Sutton MA, Weisman LS. 2012. Modulation of synaptic function by VAC14, a protein that regulates the phosphoinositides PI(3,5)P(2) and PI(5)P. EMBO J 31:3442–3456. doi: 10.1038/emboj.2012.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vicinanza M, D'Angelo G, Di Campli A, De Matteis MA. 2008. Function and dysfunction of the PI system in membrane trafficking. EMBO J 27:2457–2470. doi: 10.1038/emboj.2008.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ueno T, Falkenburger BH, Pohlmeyer C, Inoue T. 2011. Triggering actin comets versus membrane ruffles: distinctive effects of phosphoinositides on actin reorganization. Sci Signal 4:ra87. doi: 10.1126/scisignal.2002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanna PP, Cammalleri M, Berton F, Simpson C, Lutjens R, Bloom FE, Francesconi W. 2002. Phosphatidylinositol 3-kinase is required for the expression but not for the induction or the maintenance of long-term potentiation in the hippocampal CA1 region. J Neurosci 22:3359–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arendt KL, Royo M, Fernandez-Monreal M, Knafo S, Petrok CN, Martens JR, Esteban JA. 2010. PIP3 controls synaptic function by maintaining AMPA receptor clustering at the postsynaptic membrane. Nat Neurosci 13:36–44. doi: 10.1038/nn.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wells K, Farooqui AA, Liss L, Horrocks LA. 1995. Neural membrane phospholipids in Alzheimer disease. Neurochem Res 20:1329–1333. doi: 10.1007/BF00992508. [DOI] [PubMed] [Google Scholar]
- 17.Lim SY, Suzuki H. 2008. Dietary phosphatidylcholine improves maze-learning performance in adult mice. J Med Food 11:86–90. doi: 10.1089/jmf.2007.060. [DOI] [PubMed] [Google Scholar]
- 18.Zeisel SH. 2006. Choline: critical role during fetal development and dietary requirements in adults. Annu Rev Nutr 26:229–250. doi: 10.1146/annurev.nutr.26.061505.111156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeisel SH. 1992. Choline: an important nutrient in brain development, liver function and carcinogenesis. J Am Coll Nutr 11:473–481. doi: 10.1080/07315724.1992.10718251. [DOI] [PubMed] [Google Scholar]
- 20.Kubo K, Saito M, Tadokoro T, Maekawa A. 2000. Preferential incorporation of docosahexaenoic acid into nonphosphorus lipids and phosphatidylethanolamine protects rats from dietary DHA-stimulated lipid peroxidation. J Nutr 130:1749–1759. [DOI] [PubMed] [Google Scholar]
- 21.Sastry PS. 1985. Lipids of nervous tissue: composition and metabolism. Prog Lipid Res 24:69–176. doi: 10.1016/0163-7827(85)90011-6. [DOI] [PubMed] [Google Scholar]
- 22.Mita T, Mayanagi T, Ichijo H, Fukumoto K, Otsuka K, Sakai A, Sobue K. 2016. Docosahexaenoic acid promotes axon outgrowth by translational regulation of Tau and collapsin response mediator protein 2 expression. J Biol Chem 291:4955–4965. doi: 10.1074/jbc.M115.693499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao D, Kevala K, Kim J, Moon HS, Jun SB, Lovinger D, Kim HY. 2009. Docosahexaenoic acid promotes hippocampal neuronal development and synaptic function. J Neurochem 111:510–521. doi: 10.1111/j.1471-4159.2009.06335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su HM. 2010. Mechanisms of n-3 fatty acid-mediated development and maintenance of learning memory performance. J Nutr Biochem 21:364–373. doi: 10.1016/j.jnutbio.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Gamoh S, Hashimoto M, Sugioka K, Shahdat Hossain M, Hata N, Misawa Y, Masumura S. 1999. Chronic administration of docosahexaenoic acid improves reference memory-related learning ability in young rats. Neuroscience 93:237–241. doi: 10.1016/S0306-4522(99)00107-4. [DOI] [PubMed] [Google Scholar]
- 26.Petursdottir AL, Farr SA, Morley JE, Banks WA, Skuladottir GV. 2008. Effect of dietary n-3 polyunsaturated fatty acids on brain lipid fatty acid composition, learning ability, and memory of senescence-accelerated mouse. J Gerontol A Biol Sci Med Sci 63:1153–1160. doi: 10.1093/gerona/63.11.1153. [DOI] [PubMed] [Google Scholar]
- 27.Xiao Y, Wang L, Xu R, Chen Z. 2006. DHA depletion in rat brain is associated with impairment on spatial learning and memory. Biomed Environ Sci 19:474. [PubMed] [Google Scholar]
- 28.Cao X, Cui Z, Feng R, Tang YP, Qin Z, Mei B, Tsien JZ. 2007. Maintenance of superior learning and memory function in NR2B transgenic mice during ageing. Eur J Neurosci 25:1815–1822. doi: 10.1111/j.1460-9568.2007.05431.x. [DOI] [PubMed] [Google Scholar]
- 29.Ng D, Pitcher GM, Szilard RK, Sertie A, Kanisek M, Clapcote SJ, Lipina T, Kalia LV, Joo D, McKerlie C, Cortez M, Roder JC, Salter MW, McInnes RR. 2009. Neto1 is a novel CUB-domain NMDA receptor-interacting protein required for synaptic plasticity and learning. PLoS Biol 7:e41. doi: 10.1371/journal.pbio.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez J, Gamache K, Schneider R, Nader K. 2015. Memory retrieval requires ongoing protein synthesis and NMDA receptor activity-mediated AMPA receptor trafficking. J Neurosci 35:2465–2475. doi: 10.1523/JNEUROSCI.0735-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris RG, Steele RJ, Bell JE, Martin SJ. 2013. N-methyl-d-aspartate receptors, learning and memory: chronic intraventricular infusion of the NMDA receptor antagonist d-AP5 interacts directly with the neural mechanisms of spatial learning. Eur J Neurosci 37:700–717. doi: 10.1111/ejn.12086. [DOI] [PubMed] [Google Scholar]
- 32.Michailidis IE, Helton TD, Petrou VI, Mirshahi T, Ehlers MD, Logothetis DE. 2007. Phosphatidylinositol-4,5-bisphosphate regulates NMDA receptor activity through alpha-actinin. J Neurosci 27:5523–5532. doi: 10.1523/JNEUROSCI.4378-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandal M, Yan Z. 2009. Phosphatidylinositol (4,5)-bisphosphate regulation of N-methyl-d-aspartate receptor channels in cortical neurons. Mol Pharmacol 76:1349–1359. doi: 10.1124/mol.109.058701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heras-Sandoval D, Perez-Rojas JM, Hernandez-Damian J, Pedraza-Chaverri J. 2014. The role of PI3K/AKT/mTOR pathway in the modulation of autophagy and the clearance of protein aggregates in neurodegeneration. Cell Signal 26:2694–2701. doi: 10.1016/j.cellsig.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 35.Norrmen C, Suter U. 2013. Akt/mTOR signalling in myelination. Biochem Soc Trans 41:944–950. doi: 10.1042/BST20130046. [DOI] [PubMed] [Google Scholar]
- 36.Ahn JY. 2014. Neuroprotection signaling of nuclear akt in neuronal cells. Exp Neurobiol 23:200–206. doi: 10.5607/en.2014.23.3.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang PC, Yang CH, Huang CC, Hsu KS. 2008. Phosphatidylinositol 3-kinase activation is required for stress protocol-induced modification of hippocampal synaptic plasticity. J Biol Chem 283:2631–2643. doi: 10.1074/jbc.M706954200. [DOI] [PubMed] [Google Scholar]
- 38.Tao R, Gong J, Luo X, Zang M, Guo W, Wen R, Luo Z. 2010. AMPK exerts dual regulatory effects on the PI3K pathway. J Mol Signal 5:1. doi: 10.1186/1750-2187-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kovacina KS, Park GY, Bae SS, Guzzetta AW, Schaefer E, Birnbaum MJ, Roth RA. 2003. Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J Biol Chem 278:10189–10194. doi: 10.1074/jbc.M210837200. [DOI] [PubMed] [Google Scholar]
- 40.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. 2008. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones JP III, Meck HW, Williams CL, Wilson WA, Swartzwelder HS. 1999. Choline availability to the developing rat fetus alters adult hippocampal long-term potentiation. Brain Res Dev Brain Res 118:159–167. doi: 10.1016/S0165-3806(99)00103-0. [DOI] [PubMed] [Google Scholar]
- 43.Cooke SF, Bliss TV. 2006. Plasticity in the human central nervous system. Brain 129:1659–1673. doi: 10.1093/brain/awl082. [DOI] [PubMed] [Google Scholar]
- 44.Bliss TV, Collingridge GL. 1993. A synaptic model of memory: long-term potentiation in the hippocampus. Nature 361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 45.Witcher MR, Kirov SA, Harris KM. 2007. Plasticity of perisynaptic astroglia during synaptogenesis in the mature rat hippocampus. Glia 55:13–23. doi: 10.1002/glia.20415. [DOI] [PubMed] [Google Scholar]
- 46.Stubbs CD, Smith AD. 1984. The modification of mammalian membrane polyunsaturated fatty acid composition in relation to membrane fluidity and function. Biochim Biophys Acta 779:89–137. doi: 10.1016/0304-4157(84)90005-4. [DOI] [PubMed] [Google Scholar]
- 47.Bradley RM, Stark KD, Duncan RE. 2016. Influence of tissue, diet, and enzymatic remodeling on cardiolipin fatty acyl profile. Mol Nutr Food Res 60:1804–1818. doi: 10.1002/mnfr.201500966. [DOI] [PubMed] [Google Scholar]
- 48.Hancock SE, Friedrich MG, Mitchell TW, Truscott RJ, Else PL. 2015. Decreases in phospholipids containing adrenic and arachidonic acids occur in the human hippocampus over the adult lifespan. Lipids 50:861–872. doi: 10.1007/s11745-015-4030-z. [DOI] [PubMed] [Google Scholar]
- 49.Norris SE, Friedrich MG, Mitchell TW, Truscott RJ, Else PL. 2015. Human prefrontal cortex phospholipids containing docosahexaenoic acid increase during normal adult aging, whereas those containing arachidonic acid decrease. Neurobiol Aging 36:1659–1669. doi: 10.1016/j.neurobiolaging.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 50.de Groot RH, Hornstra G, Jolles J. 2007. Exploratory study into the relation between plasma phospholipid fatty acid status and cognitive performance. Prostaglandins Leukot Essent Fatty Acids 76:165–172. doi: 10.1016/j.plefa.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 51.Sumich AL, Matsudaira T, Heasman B, Gow RV, Ibrahimovic A, Ghebremeskel K, Crawford MA, Taylor E. 2013. Fatty acid correlates of temperament in adolescent boys with attention deficit hyperactivity disorder. Prostaglandins Leukot Essent Fatty Acids 88:431–436. doi: 10.1016/j.plefa.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 52.Yui K, Imataka G, Kawasaki Y, Yamada H. 2016. Increased omega-3 polyunsaturated fatty acid/arachidonic acid ratios and upregulation of signaling mediator in individuals with autism spectrum disorders. Life Sci 145:205–212. doi: 10.1016/j.lfs.2015.12.039. [DOI] [PubMed] [Google Scholar]
- 53.Yui K, Imataka G, Kawasaki Y, Yamada H. 2016. Down-regulation of a signaling mediator in association with lowered plasma arachidonic acid levels in individuals with autism spectrum disorders. Neurosci Lett 610:223–228. doi: 10.1016/j.neulet.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 54.Chen HC, Ziemba BP, Landgraf KE, Corbin JA, Falke JJ. 2012. Membrane docking geometry of GRP1 PH domain bound to a target lipid bilayer: an EPR site-directed spin-labeling and relaxation study. PLoS One 7:e33640. doi: 10.1371/journal.pone.0033640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rhee SG, Choi KD. 1992. Regulation of inositol phospholipid-specific phospholipase C isozymes. J Biol Chem 267:12393–12396. [PubMed] [Google Scholar]
- 56.Groc L, Choquet D. 2006. AMPA and NMDA glutamate receptor trafficking: multiple roads for reaching and leaving the synapse. Cell Tissue Res 326:423–438. doi: 10.1007/s00441-006-0254-9. [DOI] [PubMed] [Google Scholar]
- 57.Kim R, Moki R, Kida S. 2011. Molecular mechanisms for the destabilization and restabilization of reactivated spatial memory in the Morris water maze. Mol Brain 4:9. doi: 10.1186/1756-6606-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sakimura K, Kutsuwada T, Ito I, Manabe T, Takayama C, Kushiya E, Yagi T, Aizawa S, Inoue Y, Sugiyama H. 1995. Reduced hippocampal LTP and spatial learning in mice lacking NMDA receptor 1 subunit. Nature 373:151–154. doi: 10.1038/373151a0. [DOI] [PubMed] [Google Scholar]
- 59.Park P, Sanderson TM, Amici M, Choi SL, Bortolotto ZA, Zhuo M, Kaang BK, Collingridge GL. 2016. Calcium-permeable AMPA receptors mediate the induction of the protein kinase A-dependent component of long-term potentiation in the hippocampus. J Neurosci 36:622–631. doi: 10.1523/JNEUROSCI.3625-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zamanillo D, Sprengel R, Hvalby Ø, Jensen V, Burnashev N, Rozov A, Kaiser KMM, Köster HJ, Borchardt T, Worley P, Lübke J, Frotscher M, Kelly PH, Sommer B, Andersen P, Seeburg PH, Sakmann B. 1999. Importance of AMPA receptors for hippocampal synaptic plasticity but not for spatial learning. Science 284:1805–1811. doi: 10.1126/science.284.5421.1805. [DOI] [PubMed] [Google Scholar]
- 61.Malhotra AK, Pinals DA, Weingartner H, Sirocco K, David Missar C, Pickar D, Breier A. 1996. NMDA receptor function and human cognition: the effects of ketamine in healthy volunteers. Neuropsychopharmacology 14:301–307. doi: 10.1016/0893-133X(95)00137-3. [DOI] [PubMed] [Google Scholar]
- 62.Furukawa H, Singh SK, Mancusso R, Gouaux E. 2005. Subunit arrangement and function in NMDA receptors. Nature 438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- 63.Dyall SC, Michael GJ, Whelpton R, Scott AG, Michael-Titus AT. 2007. Dietary enrichment with omega-3 polyunsaturated fatty acids reverses age-related decreases in the GluR2 and NR2B glutamate receptor subunits in rat forebrain. Neurobiol Aging 28:424–439. doi: 10.1016/j.neurobiolaging.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 64.Miao B, Skidan I, Yang J, Lugovskoy A, Reibarkh M, Long K, Brazell T, Durugkar KA, Maki J, Ramana CV, Schaffhausen B, Wagner G, Torchilin V, Yuan J, Degterev A. 2010. Small molecule inhibition of phosphatidylinositol-3,4,5-triphosphate (PIP3) binding to pleckstrin homology domains. Proc Natl Acad Sci U S A 107:20126–20131. doi: 10.1073/pnas.1004522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mendoza MC, Er EE, Blenis J. 2011. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci 36:320–328. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sengupta S, Peterson TR, Sabatini DM. 2010. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell 40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim SY, Yoo SJ, Ronnett GV, Kim EK, Moon C. 2015. Odorant stimulation promotes survival of rodent olfactory receptor neurons via PI3K/Akt activation and Bcl-2 expression. Mol Cells 38:535–539. doi: 10.14348/molcells.2015.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Le Belle JE, Orozco NM, Paucar AA, Saxe JP, Mottahedeh J, Pyle AD, Wu H, Kornblum HI. 2011. Proliferative neural stem cells have high endogenous ROS levels that regulate self-renewal and neurogenesis in a PI3K/Akt-dependent manner. Cell Stem Cell 8:59–71. doi: 10.1016/j.stem.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rathmell JC, Fox CJ, Plas DR, Hammerman PS, Cinalli RM, Thompson CB. 2003. Akt-directed glucose metabolism can prevent Bax conformation change and promote growth factor-independent survival. Mol Cell Biol 23:7315–7328. doi: 10.1128/MCB.23.20.7315-7328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Akama KT, McEwen BS. 2003. Estrogen stimulates postsynaptic density-95 rapid protein synthesis via the Akt/protein kinase B pathway. J Neurosci 23:2333–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bergeron R, Russell RR, Young LH, Ren J-M, Marcucci M, Lee A, Shulman GI. 1999. Effect of AMPK activation on muscle glucose metabolism in conscious rats. Am J Physiol 276:E938–E944. [DOI] [PubMed] [Google Scholar]
- 72.Cota D, Proulx K, Smith KAB, Kozma SC, Thomas G, Woods SC, Seeley RJ. 2006. Hypothalamic mTOR signaling regulates food intake. Science 312:927–930. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
- 73.Chen J, Fujii K, Zhang L, Roberts T, Fu H. 2001. Raf-1 promotes cell survival by antagonizing apoptosis signal-regulating kinase 1 through a MEK–ERK independent mechanism. Proc Natl Acad Sci U S A 98:7783–7788. doi: 10.1073/pnas.141224398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yeung K, Janosch P, McFerran B, Rose DW, Mischak H, Sedivy JM, Kolch W. 2000. Mechanism of suppression of the Raf/MEK/extracellular signal-regulated kinase pathway by the raf kinase inhibitor protein. Mol Cell Biol 20:3079–3085. doi: 10.1128/MCB.20.9.3079-3085.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.English JM, Pearson G, Hockenberry T, Shivakumar L, White MA, Cobb MH. 1999. Contribution of the ERK5/MEK5 pathway to Ras/Raf signaling and growth control. J Biol Chem 274:31588–31592. doi: 10.1074/jbc.274.44.31588. [DOI] [PubMed] [Google Scholar]
- 76.Zang M, Hayne C, Luo Z. 2002. Interaction between active Pak1 and Raf-1 is necessary for phosphorylation and activation of Raf-1. J Biol Chem 277:4395–4405. doi: 10.1074/jbc.M110000200. [DOI] [PubMed] [Google Scholar]
- 77.Morris R. 1984. Developments of a water-maze procedure for studying spatial learning in the rat. J Neurosci Methods 11:47–60. doi: 10.1016/0165-0270(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 78.Vorhees CV, Williams MT. 2006. Morris water maze: procedures for assessing spatial and related forms of learning and memory. Nat Protoc 1:848–858. doi: 10.1038/nprot.2006.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mardian EB, Bradley RM, Aristizabal Henao JJ, Marvyn PM, Moes KA, Bombardier E, Tupling AR, Stark KD, Duncan RE. 16 August 2017. Agpat4/Lpaatδ deficiency highlights the molecular heterogeneity of epididymal and perirenal white adipose depots. J Lipid Res. doi: 10.1194/jlr.M079152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tang T, Li L, Tang J, Li Y, Lin WY, Martin F, Grant D, Solloway M, Parker L, Ye W, Forrest W, Ghilardi N, Oravecz T, Platt KA, Rice DS, Hansen GM, Abuin A, Eberhart DE, Godowski P, Holt KH, Peterson A, Zambrowicz BP, de Sauvage FJ. 2010. A mouse knockout library for secreted and transmembrane proteins. Nat Biotechnol 28:749–755. doi: 10.1038/nbt.1644. [DOI] [PubMed] [Google Scholar]
- 81.Lewin TM, Wang P, Coleman RA. 1999. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry 38:5764–5771. doi: 10.1021/bi982805d. [DOI] [PubMed] [Google Scholar]
- 82.Chen A, Bao C, Tang Y, Luo X, Guo L, Liu B, Lin C. 2015. Involvement of protein kinase zeta in the maintenance of hippocampal long-term potentiation in rats with chronic visceral hypersensitivity. J Neurophysiol 113:3047–3055. doi: 10.1152/jn.00929.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Afshordel S, Hagl S, Werner D, Rohner N, Kogel D, Bazan NG, Eckert GP. 2015. Omega-3 polyunsaturated fatty acids improve mitochondrial dysfunction in brain aging—impact of Bcl-2 and NPD-1 like metabolites. Prostaglandins Leukot Essent Fatty Acids 92:23–31. doi: 10.1016/j.plefa.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 84.Hagl S, Kocher A, Schiborr C, Eckert SH, Ciobanu I, Birringer M, El-Askary H, Helal A, Khayyal MT, Frank J, Muller WE, Eckert GP. 2013. Rice bran extract protects from mitochondrial dysfunction in guinea pig brains. Pharmacol Res 76:17–27. doi: 10.1016/j.phrs.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 85.Krumschnabel G, Fontana-Ayoub M, Sumbalova Z, Heidler J, Gauper K, Fasching M, Gnaiger E. 2015. Simultaneous high-resolution measurement of mitochondrial respiration and hydrogen peroxide production. Methods Mol Biol 1264:245–261. doi: 10.1007/978-1-4939-2257-4_22. [DOI] [PubMed] [Google Scholar]
- 86.Folch J, Lees M, Sloane Stanley GH. 1957. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem 226:497–509. [PubMed] [Google Scholar]
- 87.Metherel AH, Taha AY, Izadi H, Stark KD. 2009. The application of ultrasound energy to increase lipid extraction throughput of solid matrix samples (flaxseed). Prostaglandins Leukot Essent Fatty Acids 81:417–423. doi: 10.1016/j.plefa.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 88.Graham JM, Rickwood D (ed). 1997. Subcellular fractionation: a practical approach. Oxford University Press, New York, NY. [Google Scholar]