ABSTRACT
The long noncoding RNA (lncRNA) MEG3 is significantly downregulated in pancreatic neuroendocrine tumors (PNETs). MEG3 loss corresponds with aberrant upregulation of the oncogenic hepatocyte growth factor (HGF) receptor c-MET in PNETs. Meg3 overexpression in a mouse insulin-secreting PNET cell line, MIN6, downregulates c-Met expression. However, the molecular mechanism by which MEG3 regulates c-MET is not known. Using chromatin isolation by RNA purification and sequencing (ChIRP-Seq), we identified Meg3 binding to unique genomic regions in and around the c-Met gene. In the absence of Meg3, these c-Met regions displayed distinctive enhancer-signature histone modifications. Furthermore, Meg3 relied on functional enhancer of zeste homolog 2 (EZH2), a component of polycomb repressive complex 2 (PRC2), to inhibit c-Met expression. Another mechanism of lncRNA-mediated regulation of gene expression utilized triplex-forming GA-GT rich sequences. Transfection of such motifs from Meg3 RNA, termed triplex-forming oligonucleotides (TFOs), in MIN6 cells suppressed c-Met expression and enhanced cell proliferation, perhaps by modulating other targets. This study comprehensively establishes epigenetic mechanisms underlying Meg3 control of c-Met and the oncogenic consequences of Meg3 loss or c-Met gain. These findings have clinical relevance for targeting c-MET in PNETs. There is also the potential for pancreatic islet β-cell expansion through c-MET regulation to ameliorate β-cell loss in diabetes.
KEYWORDS: epigenetic, insulinoma, Meg3, pancreatic neuroendocrine tumors, triplex-forming oligonucleotides, c-MET
INTRODUCTION
The c-MET tyrosine kinase receptor, activated by the ligand hepatocyte growth factor (HGF), is associated with various biological processes for promoting embryonic development, wound repair, tissue regeneration, and cancer metastasis (1). HGF-mediated or autonomous activation of c-MET has been observed in a variety of aggressive, invasive cancers (2, 3). The aberrant upregulation of c-MET through gene amplification or increased expression correlates with metastatic disease and confers poor prognosis. Therefore, c-MET-targeted therapies that inhibit the activity of the receptor tyrosine kinase are under development or being evaluated in clinical trials for several cancers (4). Controlling the expression or activity of c-MET holds promise not only for inhibiting tumor progression but also for the development of novel tissue regeneration strategies for diabetes.
Several studies have collectively demonstrated significant c-MET upregulation in a subset of pancreatic neuroendocrine tumors (PNETs), ranging from 33% to 60% of primary nonmetastatic samples to 12% to 17% of samples from primary disease with concurrent metastases (5–8). PNETs are either hormone-secreting (functional) or hormone-nonsecreting (nonfunctional) tumors of the endocrine pancreas originating from the pancreatic islet cells. The increased incidence of PNETs over the past few decades, a lack of effective treatment options for advanced disease, and the poor prognostic survival outlook for metastatic disease warrant further investigation into the molecular pathways driving the disease and identification of suitable targets for drug treatment (9–11).
Seen mainly as sporadic tumors, PNETs can also occur as a manifestation of inherited endocrine tumor syndromes, such as multiple endocrine neoplasia type 1 (MEN1), von Hippel-Lindau (VHL) syndrome, von Recklinghausen's syndrome (neurofibromatosis 1 [NF1]), and the tuberous sclerosis complex (TSC) (12, 13). The most common sporadic functioning PNET is an insulinoma, an insulin-secreting tumor of the pancreatic islet β cell, with patients exhibiting symptoms of severe hypoglycemia (14). Interestingly, among the inherited syndromes, insulinomas are seen only in MEN1 patients (15).
The MEN1 syndrome is caused by inactivating germ line mutations in the MEN1 tumor suppressor gene, which encodes menin and is located on chromosome 11q13 (Online Mendelian Inheritance in Man number 131100) (16–18). Tissue-specific biallelic loss of the MEN1 gene results in tumors of mainly three endocrine tissues, namely, the pituitary gland, the parathyroid glands, and the endocrine pancreas (14, 19). While 40% of the sporadic nonfunctional PNETs display MEN1 mutations, sporadic insulinomas rarely exhibit MEN1 mutations (20–24). Also, PNETs in the mouse models of MEN1 loss are all insulinomas (25, 26). Therefore, the MEN1-encoded protein menin and/or its downstream targets are expected to play important roles in the pathogenesis of not only nonfunctional PNETs but also insulinomas.
We have recently demonstrated that maternally expressed gene 3 (MEG3), a long noncoding RNA, is a downstream target of menin (7, 27). MEG3 is ubiquitously expressed and is particularly abundant in endocrine tissues (28). While no mutations in MEG3 have been detected, downregulation of the MEG3 transcript has been observed in tumors of the pituitary gland, pancreas, liver, and brain (7, 29–33). PNETs with or without menin loss also showed downregulation of MEG3 RNA (7). These studies posit a putative tumor suppressor function for the lncRNA MEG3, prompting further investigation of downregulated MEG3 in the context of menin loss.
Menin is a subunit of the trithorax-like mixed-lineage leukemia (MLL) complex, which is responsible for depositing the epigenetic chromatin-activating histone H3 lysine 4 trimethyl mark (H3K4me3) (34, 35). A systematic analysis of genome-wide changes in H3K4me3 enrichment in menin-null mouse embryonic stem cells (mESCs) compared with wild-type mESCs revealed a conspicuous absence of H3K4me3 at the Meg3 promoter in the menin-null cells, along with an increase in DNA methylation at the promoter and an upstream region of the Meg3 locus (27). Meg3 downregulation in menin-null mESCs was recapitulated in murine and human PNETs, including insulinomas (7). Stable expression of Meg3 in a mouse insulinoma cell line showed a decreased expression of c-Met. Interestingly, the PNETs and insulinomas displaying downregulated MEG3 also showed a corresponding reciprocal elevation in c-MET (7). However, the precise molecular mechanisms pertaining to the regulatory events for MEG3–c-MET expression and downstream signaling are not known.
In order to investigate the therapeutic potential of modulating MEG3-mediated regulation of c-MET expression in PNETs, we investigated the mechanisms by which Meg3 could regulate c-Met. In this study, we show a direct correlation between Meg3 expression and the substantive abrogation of c-Met gene transcription. We have identified Meg3 binding to a few discrete regions in the c-Met DNA to regulate gene expression, wherein Meg3 also associates with enhancer of zeste homolog 2 (EZH2), a component of polycomb repressive complex 2 (PRC2). A multisubunit master regulator of the epigenetic machinery, PRC2 comprises the essential core components EZH2, embryonic ectoderm development (EED), suppressor of zeste 12 (SUZ12), and retinoblastoma-associated protein 46 (RBAP46). The catalytic EZH2 methyltransferase activity synergizes with the EED subunit to deposit, propagate, and maintain the repressive histone H3 lysine 27 trimethyl mark (H3K27me3) for gene silencing (36, 37). Triplex-forming oligonucleotides (TFOs) designed from GA-GT-rich tracts in lncRNA and their target genes have been shown to direct the formation of gene-regulatory lncRNA-genomic DNA triplexes (38, 39). We demonstrate that the GA-GT-rich TFO motifs in Meg3 can modulate c-Met gene transcription and insulinoma cell proliferation.
Collectively, these data posit a complex, multipronged model of Meg3-governed regulation of c-Met. Deciphering the molecular mechanisms of c-Met regulation by Meg3 is relevant to an improved understanding of PNET pathogenesis and guiding better clinical management practices and outcomes.
RESULTS
Identification and characterization of m-Meg3 genomic binding sites.
The tumor suppressor lncRNA MEG3 has been reported to exist as alternatively spliced isoforms (7, 31). There is an inverse correlation between the expression profiles of MEG3 and the oncogenic hepatocyte growth factor (HGF) receptor c-MET in human and mouse insulinoma cells (7). We sought to explore whether the various mouse Meg3 (m-Meg3) isoforms in mouse insulinoma cells could impact m-c-Met expression. In MIN6-4N cells, ectopic expression of the three predominant m-Meg3 isoforms found in pancreatic islet β cells (see Fig. S1A in the supplemental material) resulted in a statistically significant decrease in the m-c-Met mRNA mediated by two out of the three isoforms at 24 h or 48 h posttransfection (see Fig. S1B in the supplemental material). To determine whether the repressive effect of m-Meg3 affected the m-c-Met promoter activity, an m-c-Met promoter luciferase construct was transfected into three different m-Meg3-expressing stable cell lines (14M, M5, and Men1) and their respective vector control cell lines (9V, V3, and V6-1). In the m-Meg3-expressing cell lines, the m-c-Met promoter activity was slightly but significantly compromised (P ≤ 0.05) (see Fig. S1C in the supplemental material). These observations demonstrated a distinct repressive potential exhibited by m-Meg3 on the expression and transcription of the gene for the proto-oncogenic factor m-c-Met. Since the effect on the m-c-Met promoter was modest, this indicated a possibility that other m-c-Met genomic regions are responsive to m-Meg3-mediated repression. However, the molecular mechanisms underlying Meg3-governed regulation of c-Met in normal or tumor cells are not well understood.
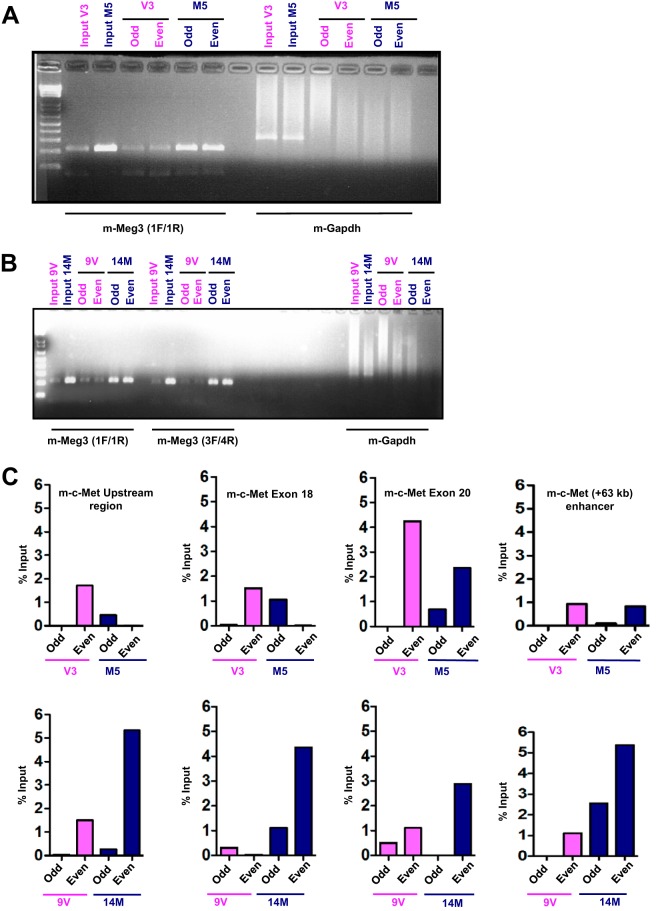
To uncover the putative regulatory genomic DNA binding locations of m-Meg3, we performed chromatin isolation by RNA purification and sequencing (ChIRP-Seq) using established protocols (40). For the m-Meg3 ChIRP-Seq, we used the m-Meg3 stable cell line M5 and its corresponding vector control cell line, V3. RNA isolated after m-Meg3 ChIRP employing odd and even probes produced an expected 205-bp product with m-Meg3-specific primers. The same RNA did not produce a specific PCR product with mouse glyceraldehyde-3-phosphate dehydrogenase gene (m-gapdh) primers (Fig. 1A). Thus, these data validate the ChIRP assay and the specificity of the probes used to retrieve m-Meg3. Similar specificity for m-Meg3 ChIRP was observed in another m-Meg3 stable cell line, 14M, stably expressing full-length m-Meg3-1 (Fig. 1B). These data provide evidence for the successful isolation of m-Meg3, presumably together with its DNA binding regions and protein targets, using ChIRP.
FIG 1.
ChIRP-Seq reveals m-Meg3 enrichment at multiple m-c-Met loci. (A) Representative agarose gel image showing the specificity of the m-Meg3 ChIRP probes. RNA was isolated after m-Meg3 ChIRP from the V3 (vector) and the M5 (stable MIN6-4N cells stably expressing the m-Meg3-3 isoform which lacks exon 4) cell lines. The RNA was then used for RT-PCR. Input corresponds to the RT-PCR product using RNA isolated before m-Meg3 ChIRP-Seq. Odd and even correspond to RT-PCR using RNA after m-Meg3 ChIRP with probes located at odd and even locations on the m-Meg3 RNA. The gel image represents products of the RT-PCR performed with primers 1F/1R (flanking exon 7 and exon 8) that recognize all m-Meg3 isoforms. gapdh served as the negative control. cDNAs from three replicates of V3 and M5 ChIRP-Seq were pooled due to low yields and sequenced. (B) m-Meg3 ChIRP-PCR in a stable cell line expressing full-length m-Meg3. RNA was isolated after m-Meg3 ChIRP from the 9V (vector) and the 14M (stable MIN6-4N cells with the m-Meg3-1 isoform, which encompasses all 10 exons) cell lines. The RNA was then used for RT-PCR. Input corresponds to RT-PCR from RNA isolated before m-Meg3 ChIRP. RT-PCR was performed with primers 1F/1R (flanking exons 7 and 8) that recognize all m-Meg3 isoforms and further confirmed with the ex3F/ex4R primer pair (flanking exons 3 and 4), specific for the m-Meg3-1 isoform. gapdh served as the negative control. (C) m-Meg3 enrichment patterns at discrete m-c-Met genomic regions in different m-Meg3 stable cell lines. DNA was isolated after m-Meg3 ChIRP from two different m-Meg-3 stable MIN6-4N cell lines and their respective vector controls. The DNA was then subjected to whole-genome amplification (WGA) and subsequent purification. The purified WGA DNA was then used to set up qPCRs in duplicate with primers specific for m-c-Met genomic regions identified by m-Meg3 ChIRP-Seq, namely, the m-c-Met upstream region, the m-c-Met exon 18 region, the m-c-Met exon 20 region, and also the previously identified kb +63 enhancer. The qPCR data are represented as percent input of DNA.
The DNA obtained after m-Meg3 ChIRP-Seq was subjected to next-generation sequencing (NGS). Sequence analyses of the m-Meg3 ChIRP-Seq data set were performed using both filtered and unfiltered bioinformatic approaches. After filtering the M5 versus V3 data set for m-Meg3-occupied genes (±5 kb from the transcriptional start site [TSS] and >5 ChIRP-Seq tags), a list of genes that showed no direct relevance to human insulinomas or other MEN1-related neuroendocrine tumors was obtained (see Fig. S2A in the supplemental material). Interestingly, unfiltered analyses showed Meg3 occupancy several kilobases from the TSS near Meg3-regulated genes in MIN6-4N cells (7) (see Table S1 in the supplemental material). Gene regulatory regions at a long distance from the TSS have been documented (41). Specifically, interrogating m-Meg3 occupancy sites in and near the c-Met gene yielded four potential m-c-Met candidate genomic loci: a kb −117 region upstream of the TSS, a region spanning m-c-Met exon 18, m-c-Met exon 20, and a fourth site at kb +153 from the TSS (see Fig. S2B in the supplemental material). A fifth site at kb +63 from the c-Met TSS was previously shown to act as an enhancer in human melanoma cells (42). The region at kb +153 from the TSS was discounted from further study due to a lack of genomic sequence conservation between mouse and human. The regions with high sequence conservation between mouse and human genomes, namely, the m-c-Met kb −117 upstream region and the exon 18, exon 20, and kb +63 enhancer regions, were assessed further for m-Meg3 enrichment. Quantitative PCR (qPCR) analyses of DNA isolated after m-Meg3 ChIRP showed m-Meg3 occupancy at the m-c-Met exon 18, exon 20, and kb +63 enhancer regions, and modest occupancy in the upstream region, in two different m-Meg3-expressing stable cell lines (Fig. 1C; see Fig. S2C in the supplemental material). These data identify m-Meg3 occupancy sites at different regions near the m-c-Met gene that could potentially serve to regulate m-c-Met gene transcription.
Defining a direct transcriptional regulatory effect of m-Meg3 on m-c-Met genomic loci.
In order to determine whether the multiple m-c-Met genomic loci identified by m-Meg3 ChIRP-Seq could affect m-c-Met expression, we used those regions in luciferase reporter assays. Luciferase reporter constructs consisting of a minimal promoter and each of the four m-c-Met loci, i.e., the upstream region, the kb +63 enhancer, exon 18, and exon 20 were generated. These constructs were transiently transfected into MIN6-4N cells along with a plasmid expressing m-Meg3-1. At 48 h posttransfection, none of the selected regions displayed any enhancer activity and instead suppressed the activity of the minimal promoter. m-Meg3-1 exerted a significant repression on the exon 20 region of m-c-Met and some minimal repression on exon 18, although no significant transcriptional repressive effects of m-Meg3 were observed, at either 24 h or 72 h posttransfection, at any of the other m-c-Met genomic locations (see Fig. S3A in the supplemental material). These results demonstrated that the four m-c-Met genomic regions did not possess enhancer activity in a heterologous promoter luciferase reporter assay and that there was a direct effect of m-Meg3 on one of the four m-c-Met loci examined. The lack of a direct effect on the other regions may reflect a combined inability of the heterologous promoter and the genomic region to respond to m-Meg3, when isolated from the chromatinized genomic context, or a requirement for the c-Met promoter. It may also be surmised that transient transfection could serve as a deterrent to any necessary m-Meg3 secondary structure formation and proper folding.
Hence, we explored the role of endogenous m-Meg3-governed repression of m-c-Met in the Men1 cell line, which expresses high endogenous m-Meg3. The MIN6-4N cell line is deficient in m-Meg3 but is m-c-Met proficient; on the other hand, the Men1 cell line stably expressing Men1 in a MIN6-4N background is m-Meg3 proficient and m-c-Met deficient. We performed luciferase reporter assays concurrently in both cell lines, using luciferase reporter constructs described above. As noted above, none of the regions analyzed acted as enhancers relative to the luciferase vector control in either cell line (see Fig. S3B in the supplemental material). In fact, they appeared to act as repressive regions, relative to the control, with the luciferase activity being similar in both the MIN6-4N and Men1 cell lines. It is possible that these genomic regions act as repressive regions for promoter sequences not related to c-Met. These data imply that m-Meg3 might not act alone but likely acts in concert with other repressive regulatory molecular machinery to suppress m-c-Met gene transcription.
m-Meg3 participates in the recruitment of epigenetic regulatory factors at the m-c-Met genomic regions.
m-Meg3-1 has been shown to recruit, and to interact with, components of the epigenetic regulator polycomb repressive complex 2 (PRC2) in mouse embryonic stem cells (mESCs) (43). This warranted an examination of the role of PRC2 in m-Meg3-mediated m-c-Met repression.
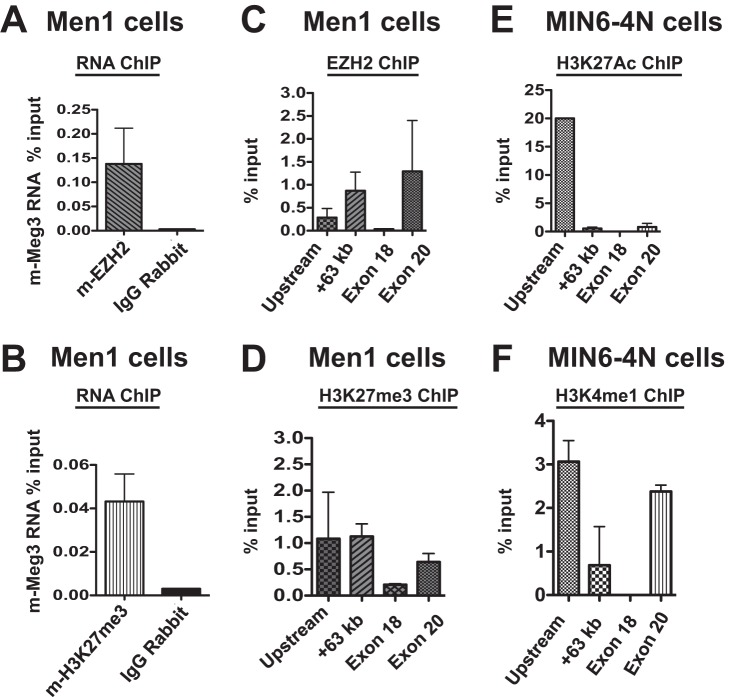
First, we sought to establish whether m-Meg3 interacted with important components of the PRC2 complex, such as EZH2. The catalytic methyltransferase activity of EZH2 has been shown to deposit the chromatin mark H3K27me3, which is associated with transcriptional repression (37). Both EZH2 and H3K27me3 were easily detectable by immunofluorescence assays in Men1 cells (see Fig. S4 in the supplemental material). These cells were further used in RNA chromatin immunoprecipitation (RNA ChIP) because they also have abundant endogenous m-Meg3 expression and low endogenous expression of m-c-Met. The RNA ChIP assays revealed that m-Meg3 specifically associated with both EZH2 and H3K27me3 relative to the IgG controls (Fig. 2A and B). Quality control was provided by RNA ChIP assays in HeLa cells showing association of the SFPQ long intergenic noncoding RNA (lincRNA) with SUZ12 (see Fig. S5A in the supplemental material). Under the same assay conditions, m-Nkx2-2 RNA, the negative control, was not detected in the EZH2 RNA ChIP assays (see Fig. S5B in the supplemental material). These data clearly establish the specificity of the m-Meg3 association with PRC2 components.
FIG 2.
m-Meg3 associates with PRC2 components in mouse insulinoma cell line models. (A and B) Association of m-Meg3 RNA with EZH2 and H3K27me3. RNA-ChIP in Men1 cells (Meg3 proficient) was performed using the antibodies directed toward EZH2 (A) and H3K27me3 (B). qPCR was performed on cDNA obtained from RNA isolated after RNA-ChIP. qPCR data were calculated as percent RNA input and are shown as the average from two independent biological replicates and multiple technical replicates (mean ± SD). IgG served as the negative control for the RNA-ChIP assay. (C and D) EZH2 and H3K27me3 enrichment at the m-c-Met regions identified by m-Meg3 ChIRP-Seq. ChIP assay was performed in Men1 cells using the indicated antibodies. DNA isolated after ChIP was used for qPCR, with primers specific for the m-c-Met upstream region, the +63-kb enhancer, the m-c-Met exon 18 region, and the m-c-Met exon 20 region. EZH2 (C) and H3K27me3 (D) enrichment was calculated as percent chromatin DNA input and constitutes the average from three independent biological replicates and multiple technical replicates (mean ± SD). (E and F) Enhancer-signature histone modifications at the m-c-Met loci identified by m-Meg3 ChIRP-Seq. MIN6-4N cells were subjected to ChIP assays and qPCR analyses to detect the enrichment of H3K27Ac (E) and H3K4me1 (F) at the m-c-Met upstream region, the kb +63 enhancer, the m-c-Met exon 18 region, and the m-c-Met exon 20 region. The data represent an average from two independent biological replicates and multiple technical replicates (mean ± SD).
Utilizing DNA ChIP assays in Meg3-proficient Men1 cells that express little or no m-c-Met, we next confirmed EZH2 occupancy and the presence of the repressive chromatin mark H3K27me3 at the m-c-Met genomic regions identified by m-Meg3 ChIRP-Seq. EZH2 was maximally enriched at the m-c-Met kb +63 and exon 20 locations, whereas H3K27me3 was detected mostly in the upstream and kb +63 enhancer regions, with some enrichment at exon 20 in Men1 cells (Fig. 2C and D). Both EZH2 and H3K27me3 displayed conspicuously low enrichment at the exon 18 locus, suggesting that there might be a locus-specific difference in PRC2-dependent m-c-Met regulation. In contrast, in the MIN6-4N cells, except for some marked H3K27me3 enrichment at the kb +63 enhancer locus, all other regions showed little to no enrichment of either EZH2 or H3K27me3 (see Fig. S5C and D in the supplemental material).
The presence of repressive marks at the m-c-Met genomic loci in an m-Meg3-expressing cell line indicated that the same regions in the MIN6-4N cells (which are deficient in endogenous m-Meg3), may have marks for active chromatin. Hence, we assessed enhancer-specific chromatin marks such as histone H3 lysine 27 acetylation (H3K27Ac) and histone H3 lysine 4 monomethylation (H3K4me1). In the Meg-3 deficient MIN6-4N cells that express c-Met abundantly, ChIP assays showed H3K27Ac enrichment predominantly at the m-c-Met upstream locus, whereas H3K4me1 enrichment was detected at the m-c-Met upstream and exon 20 regions, with little to no enrichment of either mark at the exon 18 locus (Fig. 2E and F). Concordantly, little to no enrichment of enhancer-specific chromatin signatures could be detected at the designated c-Met loci in Men1 cells (see Fig. S5E and F in the supplemental material). Taken collectively, these data offer compelling evidence of a functional interaction between m-Meg3 and PRC2 in Men1 cells to suppress the transcription of the m-c-Met gene, with the possibility of distinct m-c-Met loci acting as enhancers in the absence of m-Meg3.
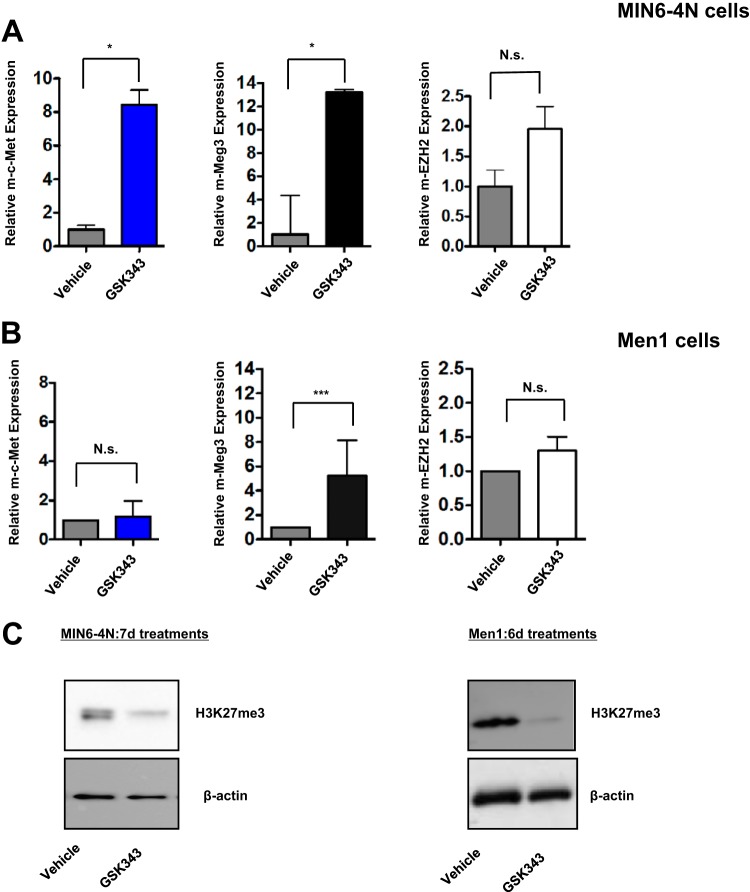
To further consolidate the evidence for PRC2 involvement in m-c-Met repression, RNA interference (RNAi) was used to inhibit EZH2, but the MIN6-4N cells were refractory to EZH2 knockdown (data not shown). Therefore, we used GSK343, a pharmacological small-molecule inhibitor of EZH2 catalytic methyltransferase activity (44). A 7-day GSK343 treatment of MIN6-4N cells evoked a dramatic increase in not only the m-c-Met transcript but also, surprisingly, the m-Meg3 transcript, with no effect on the m-EZH2 transcript (Fig. 3A). Similarly, a 6-day GSK343 regimen in Men1 cells elicited little or no change in m-c-Met and m-EZH2 transcripts. However, there was a significantly increased expression of the m-Meg3 transcript (Fig. 3B). Men1 cells have high endogenous Meg3 and very low endogenous c-Met. Therefore, any net effects of EZH2 inhibition on c-Met may not be observed as a significant decline in c-Met levels over the already low, basal c-Met transcript levels. We expected to see m-c-Met upregulation upon GSK343 treatment, due to GSK343 disrupting the Meg3-PRC2 interaction, but did not expect to see any significant upregulation in Meg3 expression. These results support the ideas that (i) m-Meg3 expression itself might be a target for PRC2-mediated regulation and (ii) m-Meg3 derives its tumor suppressor capabilities from cooperating with a functional EZH2 component of PRC2 to effectively inhibit the oncogenic effects of c-Met transcript upregulation. H3K27me3 levels were used to evaluate the effectiveness of the EZH2 inhibitor in both MIN6-4N and Men1 cells at the end of the treatment periods. Western blot analysis showed depletion of H3K27me3 upon GSK343 treatments, relative to the vehicle controls, in both cell lines, confirming drug efficacy (Fig. 3C). The presence of the H3K27me3 mark prior to drug treatments in Men1 cells may also be indicative of a bivalently poised region and not necessarily indicative of repression alone.
FIG 3.
m-Meg3 requires PRC2 components to repress the m-c-Met transcript. (A and B) Effect of the EZH2 inhibitor GSK343 on the expression of m-Meg3 and m-c-Met transcripts. RNA was isolated from MIN6-4N and Men1 cells after vehicle or GSK343 treatments at the indicated times. The RNA isolated was used in qPCR analyses using primers specific for m-c-Met, m-Meg3-1, and m-EZH2. The qPCR transcript data shown are from representative experiments where technical replicates were set up in triplicate. ***, P ≤ 0.001; *, P ≤ 0.05; N.s., nonsignificant. (C) Depletion of H3K27me3 is a confirmatory readout for the inhibition of EZH2 methyltransferase activity. Representative Western blots for H3K27me3, using whole-cell extracts (WCEs) from MIN6-4N and Men1 cells after 7-day and 6-day treatments, respectively, with 2.5 μM GSK343 or dimethyl sulfoxide (DMSO) vehicle controls are shown. β-Actin served as the loading control.
In summary, these data show that m-Meg3 acts in tandem with, and requires, functional PRC2 components to effectively silence m-c-Met gene transcription. These findings underscore one of several plausible molecular mechanisms of m-Meg3 tumor suppressor function in pancreatic islet β cells.
Structure-function analyses of m-Meg3.
Emerging investigations suggest that the lncRNA three-dimensional (3D) structure is a critical determinant of biological function (45). To elucidate the repressor domains of m-Meg3-1, we generated a series of m-Meg3-1 deletion constructs (see Fig. S6A in the supplemental material). No significant effect on the endogenous m-c-Met transcript in MIN6-4N cells was discernible by transient expression of the m-Meg3 N- and C-terminal halves corresponding, respectively, to the N- and C-terminal regions of m-Meg3 (see Fig. S6B, left panel, in the supplemental material). Further deletion of the N and C termini into one-fourth portions showed that only the C3–m-Meg3-1 construct could repress m-c-Met mRNA (see Fig. S6B, center panel, in the supplemental material). However, the repressive effects of the C3–m-Meg3-1 construct in MIN6-4N cells were not consistent (see Fig. S6B, right panel, in the supplemental material). Hence, the structure-function analyses of m-Meg3 confirmed that the m-c-Met repressive activity was not restricted to any single region and that m-Meg3 repressive action may require complete structural integrity of the Meg3 lncRNA. It is probable that the deletion constructs may have adversely perturbed structural conformations in m-Meg3, thereby impinging on functionality.
Triplex-forming motifs in m-Meg3 regulate m-c-Met.
Previous studies have attributed mechanistic significance to RNAs harboring GA-GT-rich sequences used to design triplex-forming oligonucleotides (TFOs); TFOs form triple helices with double-stranded DNA (dsDNA) and thereby regulate gene transcription (46, 47). Therefore, we investigated whether specific sequence motifs in m-Meg3 were responsible for targeted m-c-Met repression. Using the TFOs previously identified (46), we searched for TFO sequences that were conserved between mouse and human Meg3. Based on this filtering rationale, we utilized TFO-8 and a modified TFO-9 (see Materials and Methods for details) for our studies. TFO-1, which was not found in the mouse Meg3 sequence, served as a negative control. The TFO-9 construct was located in the extreme C-terminal portion of m-Meg3-1, depicted in the m-Meg3-1 schematic in Fig. 4A.
FIG 4.
m-Meg3 TFO-9 regulates the m-c-Met transcript. (A) Schematic of full-length m-Meg3-1 exon structure showing the TFO-9 coordinates. Full-length m-Meg3-1 is depicted, with exons 1 to 10 numbered (top) and the sequence length in base pairs (bottom). TFO-9, a GA- and GT-rich 16-mer sequence, is shown mapping to the C-terminal portion of exon 10 in m-Meg3-1. TFO-9, spanning bp 1822 to 1838, was predicted by Triplexator to form triplexes with double-stranded DNA. (B) Effect of TFO-9 on the expression of m-Meg3 and m-c-Met transcripts. RNA was isolated at 48 h and 96 h posttransfection from MIN6-4N cells transiently transfected with TFO RNA oligonucleotides. Purified RNA converted to cDNA was subjected to qPCR analyses with primers specific for m-Meg3-1 or m-c-Met. The data represent an average from three independent experiments and multiple technical replicates (mean ± SD) *, P ≤ 0.05.
Transfection experiments in MIN6-4N cells with TFO-8 (data not shown) and TFO-9 showed that at 48 h posttransfection, there was a significant decline in the already-low endogenous levels of the m-Meg3 transcript relative to the TFO-1 control. This was accompanied by a slight decrease in the abundantly expressed m-c-Met transcript, while all transcript changes were more modest at 96 h (Fig. 4B). Concurrent fluorescence detection of TFOs and immunostaining of m-c-Met revealed predominantly cytoplasmic staining for both TFOs and m-c-Met, with some nuclear TFO staining (see Fig. S7A in the supplemental material). This is suggestive of Meg3-TFO and m-c-Met DNA triplex interactions. At 96 h posttransfection, cells with TFO-9 exhibited increased c-Met staining, although there was a significant decline in transcript levels at the same time point. Moreover, with the principal TFO distribution being confined to the cytoplasm, rather than in the nucleus with DNA, the TFO localization might signal posttranscriptional control of m-c-Met.
To monitor the biological significance of TFO activity on MIN6-4N cells, we used bromodeoxyuridine (BrdU) incorporation as a marker of proliferation. TFO-9 transfection almost doubled the proliferation rate of MIN6-4N cells at 96 h posttransfection in comparison with either TFO-1 or TFO-8 (see the TFO schematic in Fig. S7B, top panel, in the supplemental material), whereas proliferation attained a plateau at 144 h posttransfection (see Fig. S7B, bottom panels, in the supplemental material). Since the Meg3 TFO-9 could downregulate both endogenous m-Meg3 and m-c-Met, it is difficult to infer whether there is a direct repressive effect on m-c-Met. We expected that downregulation of the oncogenic m-c-Met transcript expression would trigger a concomitant decrease in proliferation. Therefore, the unexpected increase in cell proliferation might entail TFO-9-mediated off-target effects on other genes or the activation of alternate survival signaling pathways.
DISCUSSION
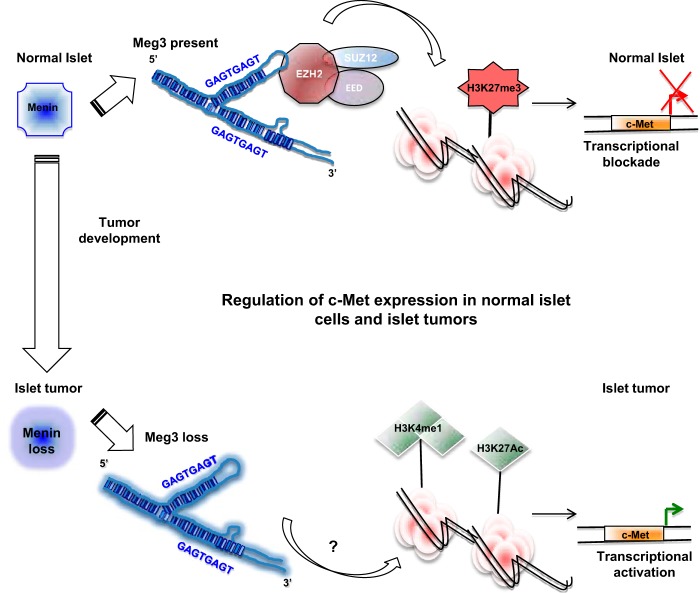
The mechanisms governing the tumor suppressor activity of the lncRNA Meg3 in the context of the c-Met signaling axis in normal pancreatic islets and the concomitant deregulation in MEN1-associated and sporadic PNETs remain poorly understood. We performed a comprehensive study examining multiple molecular processes by which the lncRNA Meg3 mediates the regulation of c-Met signaling. In summary, these studies identify Meg3 interaction with PRC2 and Meg3-TFO-dsDNA-mediated triplex formation as multiple regulatory mechanisms controlling m-c-Met gene transcription. In this proposed model of islet tumor development, Meg3 downregulation, driven by menin loss or by other mechanisms, triggers loss of the inhibitory Meg3-PRC2 interaction. Activating pathways and as-yet-unknown components that favor c-Met enhancer-specific activities are then able to promote the agenda of oncogenic c-Met gene transcription (Fig. 5).
FIG 5.
Model depicting the mechanisms of Meg3-mediated c-Met regulation in normal pancreatic islets and tumor cells. The model presented here suggests that in the context of intact menin in normal islet beta cells, the presence of Meg3 deters the proto-oncogenic activity of c-Met by multiple mechanisms. c-Met signaling can be repressed by genomic Meg3 binding, perhaps through contact with Meg3 TFO regions and Meg3 interaction with PRC2 components. Meg3 downregulation driven by menin loss and menin-independent Meg3 loss could both favor gene-activating epigenetic changes. These epigenetic alterations in turn potentiate aberrant c-Met signaling, leading to pancreatic islet β-cell tumor formation.
m-Meg3 ChIRP-Seq and ChIRP-PCR in two different cell line models stably expressing Meg3 mapped Meg3 binding sites with high specificity to discrete c-Met genomic regions, namely, the c-Met region 117 kb upstream of the TSS, the c-Met exon 18 region, the c-Met exon 20 region, and a kb +63 enhancer region previously identified in melanoma cells (42). Luciferase reporter assays using a heterologous promoter did not provide strong confirmatory evidence for c-Met enhancer activities in these regions. Concurrent comparative assessments of enhancer activity of the c-Met genomic regions on a minimal promoter failed to yield any distinct differences in Meg3-deficient and Meg3-proficient cell lines. We entertain the possibility that there are other missing regulatory elements from the c-Met promoter not used in the luciferase reporter assays. In the light of these findings, we speculate that Meg3 requires a complex chromatinized context in vivo, possibly in conjunction with other corepressors, in order to exert its c-Met repressive activities.
Chromatin-remodeling perturbations and concomitant transcriptional dysregulation as a consequence of MEN1 loss are now acknowledged as constituents of the genomic landscape of PNET pathogenesis and potential therapeutic targets (24, 48, 49). Whether histone acetyltransferases or the pioneering factors such as GATA2 and FOXA1 (50) are recruited to the c-Met genomic regions in the absence of menin and Meg3 to facilitate c-Met signaling and concomitant tumor development remains to be investigated. Also, Meg3 binding to components of PRC2 is critical for mediating gene silencing in pluripotent stem cells (51). To date, the role of the PRC2-Meg3 interaction in regulating m-c-Met gene transcription in normal cells and pancreatic islet β-cell tumors has not been studied. Therefore, we investigated the epigenetic mechanisms underlying Meg3 regulation of c-Met. We provide compelling new evidence for Meg3 acting in concert with PRC2 to epigenetically suppress c-Met expression. While the PRC2-Meg3 association was confirmed, the significance, of differential PRC2 enrichment (especially low at exon 18) at the different c-Met loci is unclear. Upon pharmacological inhibition of EZH2 by GSK343, we noticed a surprising lack of c-Met inhibition, despite marked Meg3 upregulation. These data suggest that Meg3 expression itself is a target for PRC2-mediated regulation and that Meg3 requires a functional EZH2 to mediate c-Met inhibition. These findings exemplify a scaffolding role for m-Meg3, perhaps in the assembly and recruitment of complex repressive machinery for m-c-Met regulation. In the absence of Meg3, PRC2 may be replaced by enhancer complexes and could have applications for clinical intervention.
Functional characterization in Meg3-deficient MIN6-4N cells revealed canonical enhancer-associated epigenetic signatures of H3K27Ac and H3K4me1 at distinct c-Met sites. Intriguingly, there was an overlap in the observed occupancy of H3K4me1 at the upstream region and exon 20 and H3K27me3 enrichment in MIN6-4N cells at the kb +63 enhancer locus, pointing to cell line specific contexts and the existence of poised bivalent domains.
lncRNAs are known for complex architecture and folding (45). Previous studies have identified a role for residues 345 to 348 of MEG3-1, a region conserved between human and mouse Meg3, in facilitating the interaction with EZH2 (46, 51). Given the complexity and length of the m-Meg3-1 structure, perhaps multiple repressive domains in addition to the PRC2-interacting N-terminal m-Meg3-1 domain are required to repress c-Met. Our deletion strategies using progressively smaller multiple N- and C-terminal portions of Meg3 did not recapitulate the c-Met repression exhibited by full-length Meg3. Hence, these data point to the potential for secondary and tertiary structural conformations that are likely required for m-Meg3 tumor-suppressive activity and are possibly lost upon deletion. Also, it is currently not known if the occurrence of spatial/temporal mislocalization of Meg3, the occurrence of alternatively spliced Meg3 isoforms, and the putative posttranscriptional processing of c-Met by lncRNA can serve as pro-oncogenic signaling triggers.
Recent studies have uncovered the importance of GA-GT-rich targeting sequences present in lncRNAs that help facilitate interactions with chromatin by forming RNA-DNA triplexes to mediate transcriptional regulation (46, 47). We provide evidence for the involvement of GA-GT-rich Meg3 TFO tracts in modulating the c-Met transcript and affecting the proliferation of MIN6-4N cells. Meg3 TFOs localized predominantly to the cytoplasm, and we observed increased c-Met protein and increased proliferation at 96 h with TFO-9. These observations likely point to additional c-Met regulatory processes downstream of Meg3 TFO-9 or to important off-target effects of Meg3 TFO-9.
The proliferation of MIN6-4N and the observed increase in c-Met protein mediated by Meg3 TFO-9 have important implications for the treatment of diabetes. Several studies collectively substantiate the necessity for c-Met induction and signaling in β-cell regeneration (52–55). Therefore, the events and signaling components downstream of Meg3 TFO action justify further investigation as potential activators of pathways controlling β-cell differentiation and proliferation.
In summary, our study exemplifies the complexity attendant upon the loss of lncRNA Meg3-mediated transcriptional and epigenetic mechanisms of c-Met regulation in the pathogenesis of PNETs. A better understanding of the pancreatic islet β-cell regulatory processes that govern cell proliferation could help elucidate potential therapeutic modulation strategies for c-Met expression and activity.
MATERIALS AND METHODS
Cell culture, stable cell lines, and inhibitor treatments.
Mouse MIN6-4N insulinoma cells (56–58) were maintained in low-glucose Dulbecco modified Eagle medium (DMEM) supplemented with 15% fetal bovine serum (FBS) and antibiotic/antimycotic (Thermo Fisher Scientific, Waltham, MA) at 37°C and 5% CO2. Vec-4N (here referred to as V6-1) and M27-4N (here referred to as Men1) are stable MIN6 lines containing the vector pcDNA3.1-Myc-His or pcDNA3.1-Myc-His-menin, respectively (7). Vec-3 (V3) and Meg-5 (M5) are stable cell lines of MIN6-4N containing vector or pcDNA3.1-mMeg3-3, respectively (7). Vec-9 (9V) and Meg-14 (14M) are MIN6-4N stable cell lines containing vector and pcDNA3.1-mMeg3-1, respectively (7). The various m-Meg3-1 isoforms are described in Fig. S1 in the supplemental material. m-Meg3-1 is full length with all 10 exons, m-Meg3-2 lacks exon 4, m-Meg3-3 lacks exon 2b and exon 4, and the m-Meg3-variant-1 has a unique first exon and a unique last exon that retains intron 9 sequences. Stable cell lines were maintained in DMEM supplemented as described above along with 300 μg/ml G418 (Life Technologies, Carlsbad, CA). MIN6 cell lines were authenticated by karyotype analyses, validated for pancreatic β-cell-specific gene expression signatures, and confirmed to secrete insulin (59).
For EZH2 inhibitor treatments, MIN6-4N cells (8 × 104 cells per well) or Men1 cells (1.2 × 106 cells) were seeded in a 12-well dish. At 24 h postseeding, cells were treated in fresh replacement medium every 24 h for a period of 6 or 7 days with vehicle or 2.5 μM GSK343 (Sigma-Aldrich, St. Louis, MO). On day 8 (MIN6-4N) or day 7 (Men1), the cells were processed for both RNA and protein whole-cell extract (WCE) preparations.
Transfection.
Plasmids were transfected into 3 × 105 MIN6-4N cells per well of 6-well dishes, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Cells were harvested using RLT buffer at 24 h and 48 h posttransfection for RNA isolation and protein WCE or were processed for luciferase reporter assays.
For transfection of triplex-forming oligonucleotides (TFOs), MIN6-4N cells (2 × 106) were subjected to nucleofection using a Nucleofector T kit (Lonza, Walkersville, MD), with 1 μM each TFO. Fresh medium was replenished at the end of 24 h and subsequently at 72 h following transfection. Cells were harvested in RLT buffer at 48 h, 96 h, and 144 h posttransfection for RNA isolation or were processed to detect the TFOs and c-Met protein.
Primers and antibodies.
All primers used for PCR and cloning are listed in Table S2 in the supplemental material. All antibodies used in the study are described in detail in Table S3 in the supplemental material.
ChIRP-Seq and ChIRP-qPCR.
Antisense oligonucleotide probes for m-Meg3 (n = 30) were designed using the online probe designer from LGC Biosearch Technologies, Petaluma, CA. Probes were designed such that 1 probe covered every 100 bp of RNA, with a target GC% of 45 and oligonucleotide length of 20. All repeat sequences and homologous regions were omitted during probe design. All probes were designed with 3′-biotinylated ends. The probes were numerically labeled according to the sequential position on the RNA as “even” (n = 14) and “odd” (n = 16). ChIRP was performed as per previously determined conditions and protocols (60). Stable vector or m-Meg3 cell lines (2 × 107 cells) were seeded and grown to confluence. Cells were cross-linked with 1% glutaraldehyde at room temperature for 15 min, lysed with lysis buffer, and sonicated 20 times in a Bioruptor (Diagenode, Denville, NJ). The lysate was sonicated to a fragment size of 100 to 500 bp and verified for chromatin smear size ranges by agarose gel electrophoresis. The chromatin so obtained was hybridized separately with the biotinylated antisense odd and even probes for 4 h at 37°C. Streptavidin-conjugated magnetic beads were used to capture the biotinylated Meg3-bound chromatin, followed by five stringent washes. From the ChIRP chromatin, RNA was purified from the beads using TRIzol (Thermo Fisher Scientific, Waltham, MA). DNA was purified after RNase A, RNase H, and proteinase K treatment. RNA was subjected to first-strand cDNA synthesis with random hexamers for reverse transcription-PCR (RT-PCR) to detect Meg3 enrichment; DNA was subjected to high-throughput sequencing at the NIDDK genomics core on the Illumina platform (ChIRP-Seq). Bioinformatic analyses of the ChIRP-Seq data were performed by Genomatix (Genomatix Software Inc., Ann Arbor, MI).
For validation of the ChIRP-Seq data, the DNA obtained from ChIRP was subjected to two rounds of whole-genome amplification (WGA) to amplify the ChIRP DNA using a WGA3 kit with WGA polymerase (Sigma-Aldrich, St. Louis, MO). The amplified DNA was then purified using the All prep DNA/RNA minikit (catalog number 80204; Qiagen, Germantown, MD). The purified DNA was analyzed by quantitative PCR (qPCR) reactions to validate m-Meg3 recruitment at c-Met regions identified by ChIRP-Seq. Enrichment of Meg3 occupancy was computed as a percentage of the DNA input.
ChIP assays and qPCR analyses.
MIN6-4N and Men1 cells were formaldehyde cross-linked, and processed for chromatin preparation and subsequent ChIP assays, using a ChIP kit (Millipore-EMD, Danvers, MA). Immunoprecipitation was directed against the PRC2 complex components EZH2 and the histone modifications H3K27-trimethylation mark (H3K27me3), H3K27-acetylation mark (H3K27Ac), and H3K4-monomethylation mark (H3K4me1). Rabbit or Mouse IgG served as the negative controls. ChIP DNA was purified using the All-Prep DNA/RNA minikit (Qiagen, Germantown, MD). Subsequent qPCR analyses of ChIP DNA were carried out with EZH2 and histone modification occupancy/enrichment computed as a percentage of input chromatin DNA.
RNA isolation and qPCR for gene expression analyses.
The RNeasy minikit (Qiagen, Germantown, MD) was used to isolate RNA. The total RNA was subjected to DNase I treatment (Ambion, Foster City, CA) and subsequent cleanup using the RNeasy minikit (Qiagen, Germantown, MD). DNase I-treated RNA was directly used in quantitative reverse transcription-PCRs (qRT-PCRs) using a Brilliant II SYBR green qRT-PCR kit (Agilent, Santa Clara, CA) and an Mx3000p thermal cycler (Stratagene, La Jolla, CA). RNA was also converted to cDNA using either the first-strand cDNA synthesis kit (Invitrogen) or the iScript kit (Bio-Rad, Hercules, CA), followed by qPCR for transcript analyses with the SYBR Q-PCR kit (Agilent, Santa Clara, CA). Mouse or human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used to normalize the threshold cycle (CT) values obtained. Relative gene expression changes were calculated by the 2−ΔCT method (61), and all fold changes computed relative to the appropriate vehicle or vector controls.
RNA-ChIP.
V6-1 and Men1 cells at 1 × 107 cells per 15-cm dish were cultured for 7 days, cross-linked with formaldehyde for 10 min, and processed for chromatin preparation. Chromatin was subjected to RNA chromatin immunoprecipitation (RNA-ChIP) using the RNA-CHIP-IT kit (catalog number 53024; Active Motif, Carlsbad, CA) with antibodies directed against EZH2 and H3K27me3 and IgG (rabbit) as a negative control. RNA was extracted from the eluted chromatin, using the RNeasy minikit (Qiagen, Germantown, MD). Following DNase I treatment, RNA was purified and used for first-strand cDNA synthesis. Subsequent qPCR analyses used cDNA to validate Meg3 enrichment as a percentage of chromatin RNA input. m-Nkx2-2 was used as a negative control for Meg3 enrichment. As a positive control for the RNA-ChIP assay, we used the RNA-CHIP-IT Control kit-Human (catalog number 53025; Active Motif, Carlsbad, CA), containing antibody directed toward Suz12, and primers for detecting the associated SFPQ noncoding long intergenic RNA (lincRNA).
TFOs.
MEG3 triplex-forming oligonucleotides (TFOs) designed from the human MEG3 cDNA sequence were previously described using the Triplexator algorithm (46, 62, 63). Originally designated TFO-8 and TFO-9 (46), we chose to synthesize only these two TFOs based on their conservation in both human and mouse Meg3-1. TFO-1, which lacked sequence conservation between mouse and human, served as a negative control for the TFO analysis. The TFOs used in our studies were TFO-1 (5′ to 3′, GGAGAGCAGAGAGGGAGCG) and TFO-8 (5′ to 3′, TAGGGTTGTTGTGAG). The originally published TFO-9 sequence (5′ to 3′, GGGCTGTTGTGAGGGG) (46) was modified in our studies, with the 3′-terminal GG sequence replaced by CA, to permit an exact match with the mouse sequence. The modified TFO-9 sequence used in our studies is (5′ to 3′) GGGCTGTTGTGAGGCA. All TFOs were custom synthesized (Sigma-Aldrich, St. Louis, MO) with psoralen modification at the 5′end for cross-linking studies and a d-biotin modification at the 3′end to enable detection as previously described (46).
TFO detection, c-Met staining, and BrdU assays.
MIN6-4N cells were transfected by nucleofection with TFO-1, -8, and -9 on chamber slides (5 × 105 cells per chamber well) for 48 h, 96 h, or 144 h. Untransfected cells and TFO-1-transfected cells served as controls. Posttransfection, cells were formaldehyde fixed for 30 min, washed twice with phosphate-buffered saline (PBS), and permeabilized with 0.5% Triton X-100 for 10 min. The washed slides were then blocked at room temperature for 2.5 h with immunofluorescence (IF) blocking buffer (1× Dulbecco PBS [DPBS] plus 0.1% Triton X-100 plus 2% bovine serum albumin [BSA]). Streptavidin-Alexa Fluor 488 (Molecular Probes, Eugene, OR) conjugate in IF blocking buffer was added and slides incubated overnight at 4°C. The next day, the slides were washed three times with IF washing buffer (1× DPBS plus 0.1% Triton X-100) and then mounted using 4′,6′-diamidino-2-phenylindole (DAPI) Prolong antifade mounting reagent (Invitrogen, Carlsbad, CA). The mounting reagent was left to set overnight in the dark at 4°C. Slides were imaged by fluorescence microscopy using the Keyence imaging system (Keyence Corporation, King of Prussia, PA) at a magnification of ×400.
To determine m-c-Met expression and TFO localization in transfected cells, fixing, permeabilization, and blocking procedures were performed as described above. After overnight streptavidin incubation and washing, the chamber slides were incubated for 1 h in the dark at room temperature with primary rabbit anti-c-Met antibody (Santa Cruz, Dallas, TX). No-primary-antibody controls were used, with additional controls of untransfected cells and TFO-1-transfected cells. After washing with IF wash buffer, slides were incubated with secondary anti-rabbit Alexa Fluor 594-conjugated antibody (Molecular Probes, Eugene, OR) for 1 h in the dark, at 37°C. Slides were then washed, mounted, and imaged as described in the preceding section. Brightness and contrast for all images were adjusted with Adobe Photoshop CS4 and applied to the entire image.
BrdU labeling was used to detect proliferation indices for TFO-transfected cells. MIN6-4N cells were transfected with TFOs on chamber slides as described above. Posttransfection, cells were treated at 36 h, 84 h, and 132 h with 1× BrdU labeling reagent (Gibco, Waltham, MA) for 16 h, and cells were fixed and stained for TFOs as noted above. The next day, the slides were washed three times with 1× DPBS and then incubated for 1 h in the dark at 37°C with primary rat anti-BrdU antibody (GeneTex, Irvine, CA). No-primary-antibody controls were used with additional controls of untransfected BrdU-labeled cells and TFO-1 transfected cells. After washing with IF wash buffer, slides were incubated with secondary anti-rat Alexa Fluor 594-conjugated antibody (Molecular Probes, Eugene, OR) for 1 h in the dark at 37°C. The slides were then washed twice with IF wash buffer, followed by two washes with 1× DPBS, and mounted with DAPI Prolong antifade mounting reagent. The mounting reagent was left to set overnight in the dark at 4°C. Slides were subject to fluorescence microscopy at a magnification of ×200, using an inverted fluorescence microscope (Axiovert 40CFL; Zeiss, Germany). Cell count fractions were determined for BrdU-positive cells over the total number of TFO-positive cells. The fractions were estimated as a percentage relative to the TFO-1 BrdU positive-control fraction to estimate the proliferation index.
Immunoblot analyses.
WCEs were prepared in 2× protein-loading SDS buffer or immunoprecipitation (IP) lysis buffer (1× Tris-buffered saline, 0.5% NP-40, Roche protease Inhibitor). Protein extracts were subjected to SDS-PAGE and Western blotting with antibodies directed against H3K27me3, with β-actin as a loading control. An enhanced chemiluminescence (ECL) kit (Millipore EMD, Danvers, MA) was used to detect the proteins, and the blots were imaged on a G-Box (Syngene, Frederick, MD).
Meg3 deletion constructs.
Full-length m-Meg3-1 cDNA was amplified from wild-type mouse islet total RNA and cloned into pcDNA3.1. The various deletion constructs of m-Meg3-1 were generated by PCR from the full-length m-Meg3-1 plasmid, using primers specifically designed to span the relevant regions into the pcDNA3.1 vector between BamHI and EcoRI cloning sites. All m-Meg3 deletion constructs were verified by sequencing.
Reporter assay plasmid constructs.
The c-Met promoter was amplified from mouse genomic DNA (−1307 to +180) and cloned into the XhoI and HindIII site of a promoterless pGL4.10 luciferase reporter vector (Promega, Madison, WI). m-c-Met genomic regions identified by Meg-3 ChIRP-Seq and a previously identified enhancer region in melanoma cells, the kb +63 region downstream of the transcription start site (42), were amplified from mouse genomic DNA and cloned into a pGL4.23 luciferase reporter vector containing a minimal promoter (Promega, Madison, WI). The sizes of the regions cloned were as follows: upstream region of c-Met, 355 bp; kb +63 region, 551 bp; exon 18 region, 447 bp; and exon 20 region, 449 bp.
Statistical analysis.
For statistical comparisons, data from at least three independent experiments were considered and computed using the Prism 5/6 software (GraphPad Software, San Diego, CA). Standard deviations (SD) were calculated. For EZH2 inhibitor treatments, two-tailed unpaired t tests were computed using GraphPad Quickcalcs. When comparing three or more experimental conditions or groups, one-way analysis of variance (ANOVA), using the Newman-Keuls test, was employed.
Accession number(s).
The Meg3 ChIRP-Seq data sets have been submitted to NCBI's Gene Expression Omnibus (GEO) public database and bear accession number GSE99798.
Supplementary Material
ACKNOWLEDGMENTS
We thank the NIDDK genomics core for processing the ChIRP-Seq samples and Susan Dombrowski of Genomatix Software, Inc., for bioinformatic analyses of ChIRP-Seq data.
This work was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases (Project ZIADK075035-07 and Project ZIADK075085-03, to S.K.A.).
We have no financial disclosures to make and declare no conflicts of interest.
S.I. and S.K.A. conceptualized, designed, and performed the experiments. S.D.M. and S.K.A. conceived of, designed, and performed the initial ChIRP-Seq experiments. S.I. and S.K.A. collected, analyzed, and interpreted the data. S.K.A. supervised the project and obtained project funding. S.I. and S.K.A. drafted and wrote the manuscript. All authors provided consent and approval for the final manuscript version submitted.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00278-17.
REFERENCES
- 1.Trusolino L, Bertotti A, Comoglio PM. 2010. MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol 11:834–848. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- 2.Cooper CS, Park M, Blair DG, Tainsky MA, Huebner K, Croce CM, Vande Woude GF. 1984. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature 311:29–33. doi: 10.1038/311029a0. [DOI] [PubMed] [Google Scholar]
- 3.Maroun CR, Rowlands T. 2014. The Met receptor tyrosine kinase: a key player in oncogenesis and drug resistance. Pharmacol Ther 142:316–338. doi: 10.1016/j.pharmthera.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 4.Cui JJ. 2014. A new challenging and promising era of tyrosine kinase inhibitors. ACS Med Chem Lett 5:272–274. doi: 10.1021/ml500091p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansel DE, Rahman A, House M, Ashfaq R, Berg K, Yeo CJ, Maitra A. 2004. Met proto-oncogene and insulin-like growth factor binding protein 3 overexpression correlates with metastatic ability in well-differentiated pancreatic endocrine neoplasms. Clin Cancer Res 10:6152–6158. doi: 10.1158/1078-0432.CCR-04-0285. [DOI] [PubMed] [Google Scholar]
- 6.Krampitz GW, George BM, Willingham SB, Volkmer JP, Weiskopf K, Jahchan N, Newman AM, Sahoo D, Zemek AJ, Yanovsky RL, Nguyen JK, Schnorr PJ, Mazur PK, Sage J, Longacre TA, Visser BC, Poultsides GA, Norton JA, Weissman IL. 2016. Identification of tumorigenic cells and therapeutic targets in pancreatic neuroendocrine tumors. Proc Natl Acad Sci U S A 113:4464–4469. doi: 10.1073/pnas.1600007113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Modali SD, Parekh VI, Kebebew E, Agarwal SK. 2015. Epigenetic regulation of the lncRNA MEG3 and its target c-MET in pancreatic neuroendocrine tumors. Mol Endocrinol 29:224–237. doi: 10.1210/me.2014-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murat Cde B, da Rosa PW, Fortes MA, Correa L, Machado MC, Novak EM, Siqueira SA, Pereira MA, Correa-Giannella ML, Giannella-Neto D, Giorgi RR. 2015. Differential expression of genes encoding proteins of the HGF/MET system in insulinomas. Diabetol Metab Syndr 7:84. doi: 10.1186/s13098-015-0079-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halfdanarson TR, Rabe KG, Rubin J, Petersen GM. 2008. Pancreatic neuroendocrine tumors (PNETs): incidence, prognosis and recent trend toward improved survival. Ann Oncol 19:1727–1733. doi: 10.1093/annonc/mdn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao JC, Eisner MP, Leary C, Dagohoy C, Phan A, Rashid A, Hassan M, Evans DB. 2007. Population-based study of islet cell carcinoma. Ann Surg Oncol 14:3492–3500. doi: 10.1245/s10434-007-9566-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Francois R, Iyer R, Seshadri M, Zajac-Kaye M, Hochwald SN. 2013. Current understanding of the molecular biology of pancreatic neuroendocrine tumors. J Natl Cancer Inst 105:1005–1017. doi: 10.1093/jnci/djt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norton JA, Krampitz G, Jensen RT. 2015. Multiple endocrine neoplasia: genetics and clinical management. Surg Oncol Clin N Am 24:795–832. doi: 10.1016/j.soc.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agarwal SK. 2017. Molecular genetics of MEN1-related neuroendocrine tumors, p 47–64. In Pacak K, Taïeb D (ed), Diagnostic and therapeutic nuclear medicine for neuroendocrine tumors. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 14.Thakker RV, Newey PJ, Walls GV, Bilezikian J, Dralle H, Ebeling PR, Melmed S, Sakurai A, Tonelli F, Brandi ML, Endocrine S. 2012. Clinical practice guidelines for multiple endocrine neoplasia type 1 (MEN1). J Clin Endocrinol Metab 97:2990–3011. doi: 10.1210/jc.2012-1230. [DOI] [PubMed] [Google Scholar]
- 15.Shin JJ, Gorden P, Libutti SK. 2010. Insulinoma: pathophysiology, localization and management. Future Oncol 6:229–237. doi: 10.2217/fon.09.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandrasekharappa SC, Guru SC, Manickam P, Olufemi SE, Collins FS, Emmert-Buck MR, Debelenko LV, Zhuang Z, Lubensky IA, Liotta LA, Crabtree JS, Wang Y, Roe BA, Weisemann J, Boguski MS, Agarwal SK, Kester MB, Kim YS, Heppner C, Dong Q, Spiegel AM, Burns AL, Marx SJ. 1997. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science 276:404–407. doi: 10.1126/science.276.5311.404. [DOI] [PubMed] [Google Scholar]
- 17.Lemmens I, Van de Ven WJ, Kas K, Zhang CX, Giraud S, Wautot V, Buisson N, De Witte K, Salandre J, Lenoir G, Pugeat M, Calender A, Parente F, Quincey D, Gaudray P, De Wit MJ, Lips CJ, Hoppener JW, Khodaei S, Grant AL, Weber G, Kytola S, Teh BT, Farnebo F, Thakker RV, et al. . 1997. Identification of the multiple endocrine neoplasia type 1 (MEN1) gene. The European Consortium on MEN1. Hum Mol Genet 6:1177–1183. doi: 10.1093/hmg/6.7.1177. [DOI] [PubMed] [Google Scholar]
- 18.Lemmens I, Merregaert J, Van de Ven WJ, Kas K, Zhang CX, Giraud S, Wautot V, Buisson N, De Witte K, Salandre J, Lenoir G, Calender A, Parente F, Quincey D, Courseaux A, Carle GF, Gaudray P, De Wit MJ, Lips CJ, Hoppener JW, Khodaei S, Grant AL, Weber G, Kytola S, Thakker RV, et al. . 1997. Construction of a 1.2-Mb sequence-ready contig of chromosome 11q13 encompassing the multiple endocrine neoplasia type 1 (MEN1) gene. The European Consortium on MEN1. Genomics 44:94–100. [DOI] [PubMed] [Google Scholar]
- 19.Agarwal SK. 2013. Multiple endocrine neoplasia type 1. Front Horm Res 41:1–15. doi: 10.1159/000345666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao Y, Gao Z, Li L, Jiang X, Shan A, Cai J, Peng Y, Li Y, Jiang X, Huang X, Wang J, Wei Q, Qin G, Zhao J, Jin X, Liu L, Li Y, Wang W, Wang J, Ning G. 2013. Whole exome sequencing of insulinoma reveals recurrent T372R mutations in YY1. Nat Commun 4:2810. doi: 10.1038/ncomms3810. [DOI] [PubMed] [Google Scholar]
- 21.Cromer MK, Choi M, Nelson-Williams C, Fonseca AL, Kunstman JW, Korah RM, Overton JD, Mane S, Kenney B, Malchoff CD, Stalberg P, Akerstrom G, Westin G, Hellman P, Carling T, Bjorklund P, Lifton RP. 2015. Neomorphic effects of recurrent somatic mutations in Yin Yang 1 in insulin-producing adenomas. Proc Natl Acad Sci U S A 112:4062–4067. doi: 10.1073/pnas.1503696112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, Velculescu VE, Diaz LA Jr, Vogelstein B, Kinzler KW, Hruban RH, Papadopoulos N. 2011. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lichtenauer UD, Di Dalmazi G, Slater EP, Wieland T, Kuebart A, Schmittfull A, Schwarzmayr T, Diener S, Wiese D, Thasler WE, Reincke M, Meitinger T, Schott M, Fassnacht M, Bartsch DK, Strom TM, Beuschlein F. 2015. Frequency and clinical correlates of somatic Ying Yang 1 mutations in sporadic insulinomas. J Clin Endocrinol Metab 100:E776–E782. doi: 10.1210/jc.2015-1100. [DOI] [PubMed] [Google Scholar]
- 24.Scarpa A, Chang DK, Nones K, Corbo V, Patch AM, Bailey P, Lawlor RT, Johns AL, Miller DK, Mafficini A, Rusev B, Scardoni M, Antonello D, Barbi S, Sikora KO, Cingarlini S, Vicentini C, McKay S, Quinn MC, Bruxner TJ, Christ AN, Harliwong I, Idrisoglu S, McLean S, Nourse C, Nourbakhsh E, Wilson PJ, Anderson MJ, Fink JL, Newell F, Waddell N, Holmes O, Kazakoff SH, Leonard C, Wood S, Xu Q, Nagaraj SH, Amato E, Dalai I, Bersani S, Cataldo I, Dei Tos AP, Capelli P, Davi MV, Landoni L, Malpaga A, Miotto M, Whitehall VL, Leggett BA, Harris JL, et al. . 2017. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature 543:65–71. doi: 10.1038/nature21063. [DOI] [PubMed] [Google Scholar]
- 25.Agarwal SK. 2014. Exploring the tumors of multiple endocrine neoplasia type 1 in mouse models for basic and preclinical studies. Int J Endocr Oncol 1:153–161. doi: 10.2217/ije.14.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piret SE, Thakker RV. 2011. Mouse models for inherited endocrine and metabolic disorders. J Endocrinol 211:211–230. doi: 10.1530/JOE-11-0193. [DOI] [PubMed] [Google Scholar]
- 27.Agarwal SK, Jothi R. 2012. Genome-wide characterization of menin-dependent H3K4me3 reveals a specific role for menin in the regulation of genes implicated in MEN1-like tumors. PLoS One 7:e37952. doi: 10.1371/journal.pone.0037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Assie G, Jouinot A, Bertherat J. 2014. The ‘omics’ of adrenocortical tumours for personalized medicine. Nat Rev Endocrinol 10:215–228. doi: 10.1038/nrendo.2013.272. [DOI] [PubMed] [Google Scholar]
- 29.Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM, Patel T. 2011. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 30:4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang P, Ren Z, Sun P. 2012. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem 113:1868–1874. doi: 10.1002/jcb.24055. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Rice K, Wang Y, Chen W, Zhong Y, Nakayama Y, Zhou Y, Klibanski A. 2010. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology 151:939–947. doi: 10.1210/en.2009-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Zhou Y, Klibanski A. 2010. Isolation and characterization of novel pituitary tumor related genes: a cDNA representational difference approach. Mol Cell Endocrinol 326:40–47. doi: 10.1016/j.mce.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR, Klibanski A. 2003. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab 88:5119–5126. doi: 10.1210/jc.2003-030222. [DOI] [PubMed] [Google Scholar]
- 34.Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, Kay GF, Hayward NK, Hess JL, Meyerson M. 2004. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell 13:587–597. doi: 10.1016/S1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 35.Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML. 2004. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol 24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen KH, Helin K. 2009. Epigenetic inheritance through self-recruitment of the polycomb repressive complex 2. Epigenetics 4:133–138. doi: 10.4161/epi.4.3.8483. [DOI] [PubMed] [Google Scholar]
- 37.Margueron R, Reinberg D. 2011. The Polycomb complex PRC2 and its mark in life. Nature 469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. 2011. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. 2007. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 40.Chu C, Chang HY. 2016. Understanding RNA-chromatin interactions using chromatin isolation by RNA purification (ChIRP). Methods Mol Biol 1480:115–123. doi: 10.1007/978-1-4939-6380-5_10. [DOI] [PubMed] [Google Scholar]
- 41.Gupta RM, Hadaya J, Trehan A, Zekavat SM, Roselli C, Klarin D, Emdin CA, Hilvering CRE, Bianchi V, Mueller C, Khera AV, Ryan RJH, Engreitz JM, Issner R, Shoresh N, Epstein CB, de Laat W, Brown JD, Schnabel RB, Bernstein BE, Kathiresan S. 2017. A genetic variant associated with five vascular diseases is a distal regulator of endothelin-1 gene expression. Cell 170:522–533. doi: 10.1016/j.cell.2017.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Webster DE, Barajas B, Bussat RT, Yan KJ, Neela PH, Flockhart RJ, Kovalski J, Zehnder A, Khavari PA. 2014. Enhancer-targeted genome editing selectively blocks innate resistance to oncokinase inhibition. Genome Res 24:751–760. doi: 10.1101/gr.166231.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. 2013. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol 20:1258–1264. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verma SK, Tian X, LaFrance LV, Duquenne C, Suarez DP, Newlander KA, Romeril SP, Burgess JL, Grant SW, Brackley JA, Graves AP, Scherzer DA, Shu A, Thompson C, Ott HM, Aller GS, Machutta CA, Diaz E, Jiang Y, Johnson NW, Knight SD, Kruger RG, McCabe MT, Dhanak D, Tummino PJ, Creasy CL, Miller WH. 2012. Identification of potent, selective, cell-active inhibitors of the histone lysine methyltransferase EZH2. ACS Med Chem Lett 3:1091–1096. doi: 10.1021/ml3003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu F, Somarowthu S, Pyle AM. 2017. Visualizing the secondary and tertiary architectural domains of lncRNA RepA. Nat Chem Biol 13:282–289. doi: 10.1038/nchembio.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, Mitra S, Mohammed A, James AR, Hoberg E, Moustakas A, Gyllensten U, Jones SJ, Gustafsson CM, Sims AH, Westerlund F, Gorab E, Kanduri C. 2015. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat Commun 6:7743. doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vance KW, Ponting CP. 2014. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet 30:348–355. doi: 10.1016/j.tig.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin W, Watanabe H, Peng S, Francis JM, Kaplan N, Pedamallu CS, Ramachandran A, Agoston A, Bass AJ, Meyerson M. 2015. Dynamic epigenetic regulation by menin during pancreatic islet tumor formation. Mol Cancer Res 13:689–698. doi: 10.1158/1541-7786.MCR-14-0457. [DOI] [PubMed] [Google Scholar]
- 49.Lines KE, Stevenson M, Filippakopoulos P, Muller S, Lockstone HE, Wright B, Grozinsky-Glasberg S, Grossman AB, Knapp S, Buck D, Bountra C, Thakker RV. 2017. Epigenetic pathway inhibitors represent potential drugs for treating pancreatic and bronchial neuroendocrine tumors. Oncogenesis 6:e332. doi: 10.1038/oncsis.2017.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dreijerink KM, Groner AC, Vos ES, Font-Tello A, Gu L, Chi D, Reyes J, Cook J, Lim E, Lin CY, de Laat W, Rao PK, Long HW, Brown M. 2017. Enhancer-mediated oncogenic function of the menin tumor suppressor in breast cancer. Cell Rep 18:2359–2372. doi: 10.1016/j.celrep.2017.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaneko S, Bonasio R, Saldana-Meyer R, Yoshida T, Son J, Nishino K, Umezawa A, Reinberg D. 2014. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell 53:290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alvarez-Perez JC, Ernst S, Demirci C, Casinelli GP, Mellado-Gil JM, Rausell-Palamos F, Vasavada RC, Garcia-Ocana A. 2014. Hepatocyte growth factor/c-Met signaling is required for beta-cell regeneration. Diabetes 63:216–223. doi: 10.2337/db13-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mellado-Gil J, Rosa TC, Demirci C, Gonzalez-Pertusa JA, Velazquez-Garcia S, Ernst S, Valle S, Vasavada RC, Stewart AF, Alonso LC, Garcia-Ocana A. 2011. Disruption of hepatocyte growth factor/c-Met signaling enhances pancreatic beta-cell death and accelerates the onset of diabetes. Diabetes 60:525–536. doi: 10.2337/db09-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roccisana J, Reddy V, Vasavada RC, Gonzalez-Pertusa JA, Magnuson MA, Garcia-Ocana A. 2005. Targeted inactivation of hepatocyte growth factor receptor c-met in beta-cells leads to defective insulin secretion and GLUT-2 downregulation without alteration of beta-cell mass. Diabetes 54:2090–2102. doi: 10.2337/diabetes.54.7.2090. [DOI] [PubMed] [Google Scholar]
- 55.Sun B, Liu R, Xiao ZD. 24 April 2015. Induction of insulin-producing cells from umbilical cord blood-derived stromal cells by activation of the c-Met/HGF axis. Dev Growth Differ doi: 10.1111/dgd.12214. [DOI] [PubMed] [Google Scholar]
- 56.Desai SS, Kharade SS, Parekh VI, Iyer S, Agarwal SK. 2015. Pro-oncogenic roles of HLXB9 protein in insulinoma cells through interaction with Nono protein and down-regulation of the c-Met inhibitor Cblb (Casitas B-lineage lymphoma b). J Biol Chem 290:25595–25608. doi: 10.1074/jbc.M115.661413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Desai SS, Modali SD, Parekh VI, Kebebew E, Agarwal SK. 2014. GSK-3beta protein phosphorylates and stabilizes HLXB9 protein in insulinoma cells to form a targetable mechanism of controlling insulinoma cell proliferation. J Biol Chem 289:5386–5398. doi: 10.1074/jbc.M113.533612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miyazaki J, Araki K, Yamato E, Ikegami H, Asano T, Shibasaki Y, Oka Y, Yamamura K. 1990. Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology 127:126–132. doi: 10.1210/endo-127-1-126. [DOI] [PubMed] [Google Scholar]
- 59.Shi K, Parekh VI, Roy S, Desai SS, Agarwal SK. 2013. The embryonic transcription factor Hlxb9 is a menin interacting partner that controls pancreatic beta-cell proliferation and the expression of insulin regulators. Endocr Relat Cancer 20:111–122. doi: 10.1530/ERC-12-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chu C, Quinn J, Chang HY. 25 March 2012. Chromatin isolation by RNA purification (ChIRP). J Vis Exp doi: 10.3791/3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buske FA, Bauer DC, Mattick JS, Bailey TL. 2012. Triplexator: detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res 22:1372–1381. doi: 10.1101/gr.130237.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buske FA, Bauer DC, Mattick JS, Bailey TL. 2013. Triplex-Inspector: an analysis tool for triplex-mediated targeting of genomic loci. Bioinformatics 29:1895–1897. doi: 10.1093/bioinformatics/btt315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.