ABSTRACT
DNA replication involves the inherent risk of genome instability, since replisomes invariably encounter DNA lesions or other structures that stall or collapse replication forks during the S phase. In the fission yeast Schizosaccharomyces pombe, the multi-BRCT domain protein Brc1, which is related to budding yeast Rtt107 and mammalian PTIP, plays an important role in maintaining genome integrity and cell viability when cells experience replication stress. The C-terminal pair of BRCT domains in Brc1 were previously shown to bind phosphohistone H2A (γH2A) formed by Rad3/ATR checkpoint kinase at DNA lesions; however, the putative scaffold interactions involving the N-terminal BRCT domains 1 to 4 of Brc1 have remained obscure. Here, we show that these domains bind Rhp18/Rad18, which is an E3 ubiquitin protein ligase that has crucial functions in postreplication repair. A missense allele in BRCT domain 4 of Brc1 disrupts binding to Rhp18 and causes sensitivity to replication stress. Brc1 binding to Rhp18 and γH2A are required for the Brc1 overexpression suppression of smc6-74, a mutation that impairs the Smc5/6 structural maintenance of chromosomes complex required for chromosome integrity and repair of collapsed replication forks. From these findings, we propose that Brc1 provides scaffolding functions linking γH2A, Rhp18, and Smc5/6 complex at damaged replication forks.
KEYWORDS: Brc1, Rhp18, Rad18, BRCT domain, DNA damage, DNA repair, DNA replication stress, genome integrity
INTRODUCTION
Genome stability is especially at risk during the DNA synthesis (S) phase of the cell cycle, when replisomes encounter DNA lesions or chromatin-bound proteins, or they collide with transcriptional machinery, potentially leading to replication fork collapse and ensuing deleterious genomic alterations. Faced with the critical requirement for replication fidelity to maintain cell viability and prevent disease (1–3), eukaryotic organisms have evolved a complex and highly regulated network of DNA damage response (DDR) pathways that work in conjunction with the replicative machinery to maintain genome integrity during the S phase (4–8). Thus, DDRs coordinate DNA replication, repair, and cell cycle progression to safeguard the genome.
In Schizosaccharomyces pombe, as in all eukaryotes, the replication stress response is initiated by the detection of replication protein A (RPA)-coated single-stranded DNA (ssDNA) that forms at stalled or damaged replication forks. This accumulation of RPA-bound ssDNA serves as a signal to activate the master checkpoint kinase Rad3/ATR, which phosphorylates key substrates, including an SQ motif on the C-terminal tail of histone H2A in chromatin flanking the stalled or collapsed replication fork (9, 10). Phosphohistone H2A, known as γH2A in yeast and equivalent to γH2AX in mammals, serves as a recruitment platform for key DDR proteins, including Brc1, Crb2, and Mdb1 in S. pombe (11–15).
Brc1 is an 878-amino-acid (aa) protein that contains four N-terminal and two C-terminal BRCT domains separated by an intervening linker region containing a nuclear localization signal (Fig. 1A) (16). This domain organization is shared with Saccharomyces cerevisiae Rtt107, which is an important genome protection protein (17–21). The mammalian genome protection protein PTIP also has six BRCT domains, although its linker region is located between domains 2 and 3 (18, 22). Brc1 was first identified in S. pombe as an allele-specific high-copy-number suppressor of the hypomorphic smc6-74 allele, which impairs the function of the essential Smc5/6 structural maintenance of chromosomes complex (23). Brc1 is not required for cell viability unless cells are challenged with DNA-damaging agents that collapse replication forks or they are defective in specific processes related to DNA replication. For example, Brc1 is essential for cell viability in mutants lacking Rqh1, which is a RecQ-like DNA helicase involved in DNA replication and repair (9, 24). Rqh1 is homologous to Sgs1 in S. cerevisiae and the BLM, WRN, and RTS/RECQ4 enzymes in humans that are associated with cancer predisposition and/or premature aging (25). Brc1 is also crucial when loss of deoxycytidylate deaminase (dCMP deaminase) creates imbalanced pools of deoxyribonucleoside triphosphates required for DNA synthesis and repair (26), or when there are defects in replication factor C, which loads proliferating cell nuclear antigen (PCNA) clamp onto duplex DNA (10). Perhaps most notably, Brc1 becomes essential in cells with compromised Smc5/6 complex function (23, 24). The reported function of the Smc5/6 complex at collapsed replication forks (27–29), combined with the observed sensitivity of Brc1-deficient cells to agents known to generate lesions and disrupt replication fork progression during S phase, suggests that Brc1 functions in the response to DNA damage during replication stress (24). Further supporting this idea, Brc1 was shown to localize at sites of replication stress through the interaction of its C-terminal BRCT domains with γH2A (12). Collectively, these data suggest that Brc1 stabilizes stalled replication forks and assists in the repair of collapsed replication forks (9, 10, 12, 16, 30).
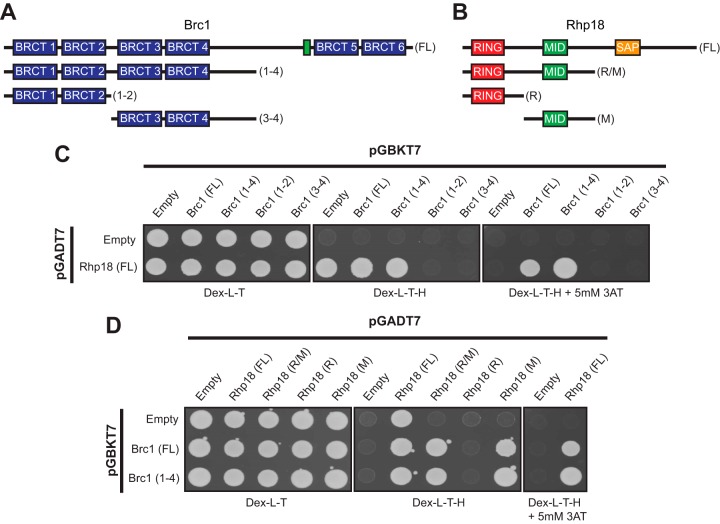
FIG 1.
The Brc1-Rhp18 interaction requires the N-terminal BRCT domains of Brc1 and the Mid domain of Rhp18. (A and B) Schematic representations of Brc1 (A) and Rhp18 (B) fragments tested for physical interactions by Y2H analysis (fragments sizes for both Brc1 and Rhp18 are listed in Materials and Methods). The location of Brc1's nuclear localization signal is depicted by the green box in panel A. (C) Y2H results showing full-length Brc1 and BRCT domains 1 to 4 interact with full-length Rhp18. Interactions were judged from the Dex-L-T-H plus 5 mM 3-AT due to observed one-hybrid activity of full-length Rhp18 in pGADT7 on Dex-L-T-H. (D) Y2H results indicating Rhp18's Mid domain is sufficient to support the interaction with Brc1. Interactions for full-length Rhp18 were evaluated from the Dex-L-T-H plus 5 mM 3-AT due to observed one-hybrid activity of full-length Rhp18 in pGADT7 on Dex-L-T-H. Removal of the SAP domain of Rhp18 alleviated the observed one-hybrid activity, allowing assessment of physical interactions on Dex-L-T-H for the R/M, R, and M fragments of Rhp18.
Exactly how Brc1 protects genomic stability during S phase has remained elusive. Extensive genetic interaction analysis has established that Brc1 is especially crucial for replication stress resistance and cell viability when DNA replication or other genome protection mechanisms are impaired (9, 10, 23, 24, 31–33); however, the lack of specific measureable enzymatic activity and limited protein interaction data have hindered progression in understanding Brc1's role in the maintenance of genomic stability under these circumstances. Brc1 binds γH2A, but this interaction probably serves to properly localize Brc1 at DNA lesions, where it engages with other proteins that are currently unknown. Moreover, mutations that disrupt Brc1 binding to γH2A only partially impair Brc1 function, indicating γH2A-independent roles for Brc1 that do not absolutely require formation of extensive domains of chromatin-bound Brc1 flanking stalled or collapsed replication forks (12, 16). The smc6-74 suppression by Brc1 overexpression was shown to require Rhp18, but whether this dependence reflects physical associations between Brc1, Rhp18, or the Smc5/6 holocomplex is unknown (24, 31). Rhp18, known as Rad18 in other organisms, is an E3 ubiquitin protein ligase, which binds RPA-coated ssDNA, where it functions with its cognate E2 enzyme, Rad6, to control the initial steps of post-replicative repair (PRR) via PCNA ubiquitination (34).
In this report, we identify Rhp18 as a binding partner for Brc1 and describe a mutation that disrupts this interaction. Binding studies and functional analysis suggest the interaction with Rhp18 is essential for Brc1 overexpression to suppress smc6-74. Moreover, Brc1 binding to γH2A is critical when the function of the Smc5/6 complex is impaired. These results suggest that Brc1's role in promoting genomic stability during S phase is mediated through coordinated binding with γH2A and Rhp18 at sites of replication stress.
RESULTS
Brc1 physically interacts with Rhp18.
BRCT domains often mediate physical interactions among proteins (35, 36). Brc1 contains six BRCT domains but no documented enzymatic activity; thus, it seemed likely that it functions as a scaffold for other DDR proteins at stalled or damaged replication forks. To date, the only interacting partner identified for Brc1 has been γH2A (12). Therefore, we sought to identify additional Brc1 interacting proteins through a yeast two-hybrid (Y2H) screen using full-length Brc1 as bait. The results from this preliminary screen returned multiple hits for Rhp18, which was notable given that Rhp18 is required for the rescue of smc6-74 by Brc1 overexpression (24, 31).
To confirm these preliminary results, we cloned full-length brc1 cDNA into pGBKT7 and full-length rhp18 cDNA into pGADT7 and assessed their two-hybrid interactions. Rhp18 displayed one-hybrid activity on standard −His selective medium (Dex-L-T-H), but the addition of 5 mM 3-amino-1,2,4 triazole (3-AT) to the medium suppressed this activity and confirmed the Brc1-Rhp18 interaction reported from our initial screen (Fig. 1C). These results were later validated through coimmunoprecipitation, as described below.
Identification of domains mediating the Brc1-Rhp18 interaction.
With Rhp18 identified as a binding partner of Brc1, we sought to narrow down the protein domains mediating this interaction. Fragments of Brc1 were evaluated for their ability to bind full-length Rhp18 using the Y2H method (Fig. 1A). This analysis revealed that a Brc1 fragment containing BRCT domains 1 to 4 plus part of the linker region was sufficient for the Y2H interaction with Rhp18. However, division of this fragment between the BRCT domains 2 and 3 eliminated the Y2H signal, suggesting that BRCT domains 1 to 4 likely function as a binding module for Rhp18 (Fig. 1C).
Rhp18 is a 387-aa protein that contains an N-terminal really interesting new gene (RING) domain, a central Mid zinc finger domain, and a C-terminal SAF-A/B, acinus, Pias (SAP) domain. The N-terminal RING domain coordinates the binding of Rad6, an E2 ubiquitin-conjugating enzyme, along with the C-terminal Rad6 binding interface. Aside from coordinating Rad6 binding, the RING domain is known to mediate Rhp18's E3 ubiquitin ligase activity to initiate PRR pathways (31, 37). The C-terminal SAP domain has been suggested to possess ssDNA-binding activity, which might recruit Rhp18 to sites of DNA lesions in concert with Rhp18's ability to bind RPA (38–40). However, the function of the central Mid zinc finger domain is uncertain, as some reports have claimed it mediates replication-independent DNA binding (38), while others have shown that this domain functions as a ubiquitin-binding zinc finger domain (37).
To identify the regions of Rhp18 required for its interaction with Brc1, we utilized our Y2H approach. Fragments of Rhp18 (Fig. 1B) were cloned into pGADT7 and tested for their ability to interact with full-length Brc1 or the Brc1(1-4) fragment expressed from pGBKT7 (Fig. 1A). Removal of the SAP domain alleviated the observed one-hybrid activity in pGADT7 and allowed scoring of interactions on restrictive plates lacking 3-AT. The Rhp18(R/M) fragment supported growth on the restrictive plates, suggesting that the SAP domain is not essential for the Brc1 interaction (Fig. 1D). The N-terminal RING domain failed to support growth on the restrictive plates when combined with either full-length Brc1 or the N-terminal BRCT domains (Fig. 1D), suggesting that the RING domain alone is insufficient to mediate the interaction with Brc1. The combined results above implied that the Mid domain alone may be sufficient to interact with Brc1, and the two-hybrid analysis expressing only the Mid domain in pGADT7 in conjunction with the Brc1 fragments in pGBKT7 corroborated that assumption (Fig. 1D). Therefore, our results suggest the Mid zinc finger domain of Rhp18 alone is sufficient to support the physical interaction with Brc1.
Mutation of BRCT domain 4 severely attenuates the Brc1-Rhp18 physical interaction.
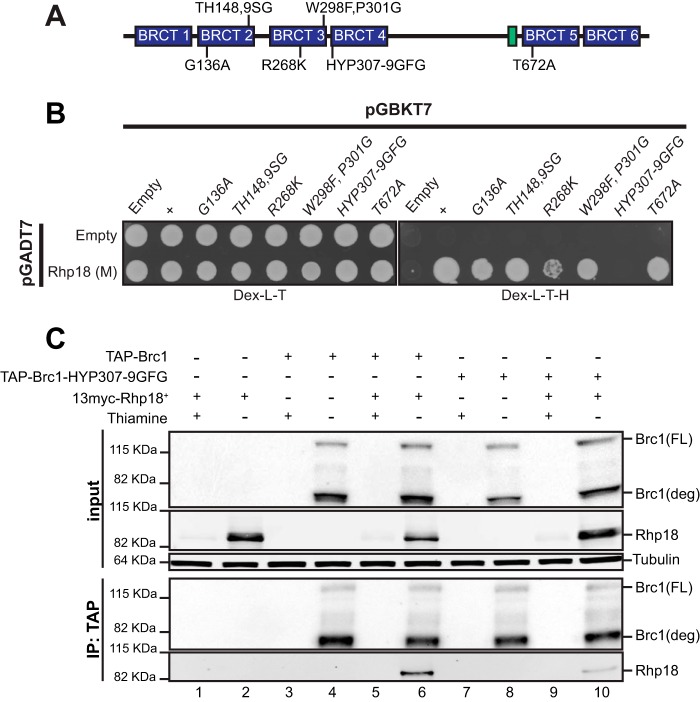
Having identified the regions of Brc1 and Rhp18 required for their physical interaction, we next turned our attention to analyzing the effects of previously characterized point mutations altering conserved residues in the BRCT domains of Brc1 (12, 16). As expected, the brc1-T672A mutation that disrupts binding to γH2A did not diminish binding to Rhp18 (Fig. 2B). Brc1 proteins with altered residues in BRCT domain 2 (G136A and TH148,149SG) or BRCT domain 3 (R268K and W298F,P301G) also maintained the Y2H interaction with Rhp18, although the R268K mutation appeared to significantly weaken the Y2H interaction. In contrast, the brc1-HYP307-9GFG allele, which alters three residues in BRCT domain 4 near the BRCT 3-4 linker, eliminated the Y2H interaction with Rhp18 (Fig. 2B). Importantly, previous work with mutations at the brc1 locus showed that brc1-HYP307-9GFG impaired Brc1 function in resistance to the genotoxic drugs hydroxyurea (HU) and camptothecin (CPT) without noticeably disrupting its ability form γH2A-dependent nuclear foci (16), suggesting that this allele disrupted critical scaffolding properties of Brc1. In the same assays the brc1-TH148, -149SG, and -R268K mutations also caused HU and CPT sensitivity, whereas the brc1-G136A and -W298F,P301G mutations had no effect (16).
FIG 2.
Mutation of BRCT domain 4 attenuates the Brc1-Rhp18 interaction. (A) A schematic representation of published Brc1 point mutations (16) tested for physical interaction with the Mid domain of Rhp18 by yeast two-hybrid analysis. (B) Yeast two-hybrid results indicating that mutation of BRCT4 inhibits the interaction with the Mid domain of Rhp18. (C) Results from coimmunoprecipitation experiments demonstrating the interaction between full-length Brc1 and Rhp18, as well as the reduction in Rhp18 binding observed with TAP-Brc1-HYP307-9GFG versus TAP-Brc1.
To confirm the yeast-two hybrid assays we performed coimmunoprecipitation experiments. Utilizing nmt41 promoter driven expression of N-terminally tagged Rhp18 and Brc1, we found that 13myc-tagged Rhp18 readily coprecipitated with TAP-tagged Brc1 (Fig. 2C, lane 6). In contrast, Rhp18 coimmunoprecipitation with TAP-tagged Brc1-HYP307-9GFG was strongly diminished (Fig. 2C, lane 10).
Efficient rescue of brc1Δ by expression of brc1-T672A but not brc1-HYP307-9GFG.
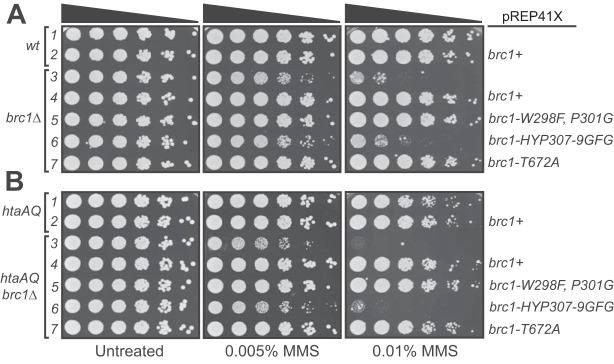
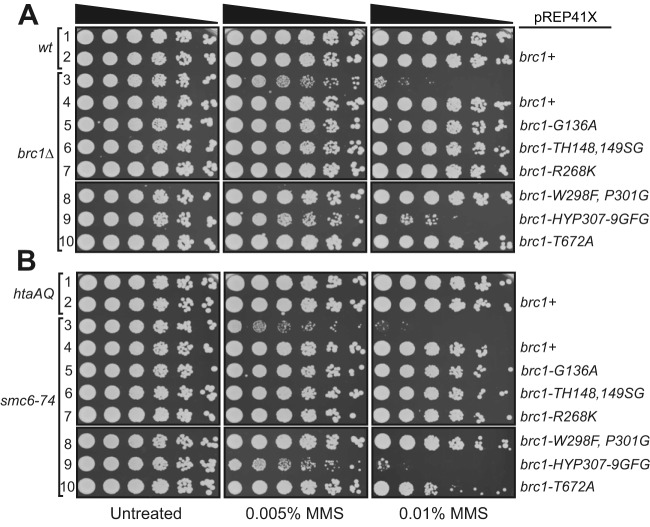
With the identification of point mutations of Brc1 that disrupt binding to Rhp18 or γH2A, we directly compared the effects of these mutations in determining cellular resistance to DNA damage. We expressed brc1+, the Rhp18-binding defective mutant brc1-HYP307-9GFG, or the previously described γH2A-binding defective mutant brc1-T672A (12), from the moderate strength nmt41 promoter in plasmid pREP41X. As an additional control, we also expressed brc1-W298F,P301G, containing mutations in BRCT domain 3, which did not disrupt the Brc1-Rhp18 Y2H interaction (Fig. 2B). We found that expression of brc1+, brc1-W298F:P301G, and brc1-T672A were all able to fully rescue brc1Δ MMS sensitivity (Fig. 3A, rows 4, 5, and 7). In contrast, expression of brc1-HYP307-9GFG resulted in an extremely weak rescue of the methyl methanesulfonate (MMS) phenotype compared to the vector only control (Fig. 3A, row 6). We obtained essentially the same results when we repeated the experiment in an htaAQ genetic background (hta1-S129A hta2-S128S) (Fig. 3B), which lacks the ability to form γH2A (11). Thus, defects in Brc1 binding to γH2A can be suppressed by Brc1 overexpression; however, the impacts of the Rhp18-binding defective brc1-HYP307-9GFG mutation on Brc1 function cannot be compensated for by merely increasing its cellular concentrations.
FIG 3.
Efficient rescue of brc1Δ by expression of brc1-T672A but not brc1-HYP307-9GFG. (A) Functional evaluation of four brc1 alleles in response to MMS treatment in a brc1Δ genetic background suggests that brc1-HYP307-9GFG (16) retains more activity than brc1Δ but significantly less than the previously published γH2A binding mutant brc1-T672A (12), which rescued brc1Δ, as well as brc1+ and brc1-W298F,P301G. (B) Functional evaluation of the brc1 alleles in response to MMS treatment in the htaAQ brc1Δ genetic background, demonstrating the Brc1-Rhp18 interaction is more essential for Brc1 function in an overexpression situation than its ability to bind γH2A. In each panel, rows 1 and 3 contain cells transformed with empty pREP41X.
Brc1 binding to Rhp18 and γH2A is important for suppression of smc6-74 by Brc1 overexpression.
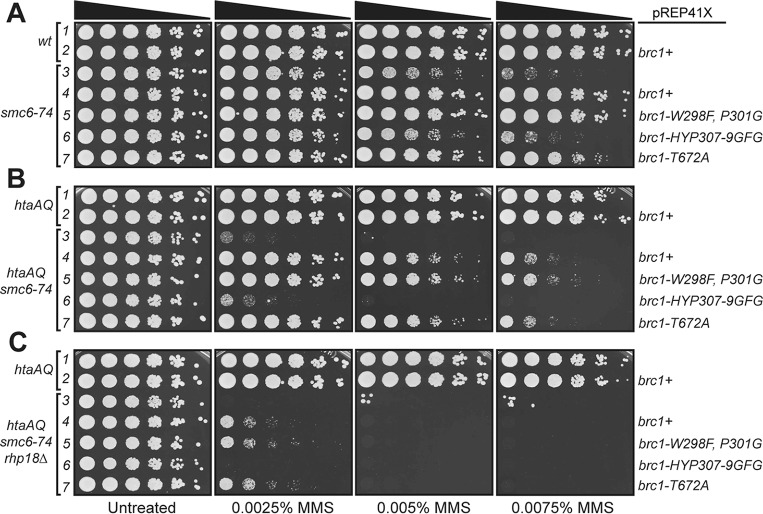
We next investigated the relationships between Brc1 binding to γH2A or Rhp18 and its ability to rescue smc6-74. Utilizing the same approach as described above, we expressed the brc1 alleles from pREP41X in the smc6-74 genetic background and tested MMS sensitivity. As seen for the brc1Δ rescue experiments, expression of brc1+ or brc1-W298F,P301G fully suppressed the smc6-74 MMS-sensitive phenotype in these assays (Fig. 4A, rows 4 and 5). In contrast, expression of brc1-HYP307-9GFG resulted in no suppression of smc6-74 (Fig. 4A, row 6). The correlation of attenuated Rhp18 binding and lack of smc6-74 suppression observed for brc1-HYP307-9GFG overexpression suggests that binding to Rhp18 is critical for Brc1 function. Interestingly, suppression smc6-74 by brc1-T672A overexpression was weakened in comparison to brc1+ overexpression (Fig. 4A, row 7), suggesting that γH2A-binding by Brc1 is important for suppression of smc6-74.
FIG 4.
Rhp18 and γH2A binding are required for efficient rescue of smc6-74 by Brc1 overexpression. (A) Results from smc6-74 suppression experiments comparing the rescue efficiency of the four brc1 alleles. The brc1-HYP307-9GFG allele that disrupts Brc1 binding to Rhp18 prevents Brc1 overexpression suppression of smc6-74. The brc1-T672A mutation that abrogates binding to γH2A impairs Brc1 overexpression suppression of smc6-74. (B) Results from smc6-74 htaAQ suppression experiments comparing the four brc1 mutations, supporting the MMS dose dependence for γH2A binding in mediating the scm6-74 rescue by Brc1. (C) Results from brc1 overexpression assays in the smc6-74 htaAQ rhp18Δ background, suggesting the failure of brc1-HYP307-9GFG to rescue smc6-74 is not completely explained by its inability to bind Rhp18. In each panel, rows 1 and 3 contain cells transformed with empty pREP41X.
To further investigate whether binding to γH2A is important for the Brc1 overexpression rescue of smc6-74, we assessed the ability of our brc1 alleles to suppress smc6-74 in an htaAQ background. Importantly, combining the smc6-74 and htaAQ mutations in the same strain caused an apparent synergistic increase in MMS sensitivity (compare Fig. 4A, row 3, to Fig. 4B, rows 1 and 3). Expression of brc1+ or brc1-W298F,P301G suppressed the smc6-74 MMS-sensitive phenotype, but MMS resistance was diminished in the htaAQ background (compare Fig. 4A and B, rows 4 and 5). As expected, an approximately equal level of suppression was observed for brc1-T672A overexpression in the htaAQ background (Fig. 4B, row 7), and no suppression was observed for brc1-HYP307-9GFG overexpression (Fig. 4B, row 6).
These results suggest that interactions of Brc1 with γH2A and Rhp18 are important for suppression of smc6-74 by Brc1 overexpression. To further test this hypothesis, we examined suppression in the htaAQ rhp18Δ smc6-74 genetic background. As expected, Brc1 overexpression only weakly suppressed MMS sensitivity of htaAQ rhp18Δ smc6-74 cells (Fig. 4C). This very weak suppression effect was observed for brc1+, brc1-W298F,P301G, and brc1-T672A overexpression. However, this weak suppression was eliminated when we overexpressed brc1-HYP307-9GFG (Fig. 4C, row 6). This result implies that the smc6-74 suppression defect of brc1-HYP307-9GFG is not fully explained by its inability to interact with Rhp18.
Analysis of additional brc1 alleles.
To extend these analyses we analyzed the other brc1 alleles mentioned above. The brc1-G136A, -TH148,149SG, and -R268K mutant proteins all rescued brc1Δ MMS sensitivity when they were expressed from the moderate strength nmt41 promoter in plasmid pREP41X (Fig. 5A). Similarly, these mutant proteins suppressed the MMS sensitivity of smc6-74 (Fig. 5B). These results are discussed below.
FIG 5.
Expression of brc1-G136A, -TH148,149SG, and -R268K rescues brc1Δ and smc6-74 MMS sensitivity. Constructs were expressed from the moderate strength nmt41 promoter in plasmid pREP41X and tested for rescue of brc1Δ (A) or suppression of smc6-74 (B). In each panel, rows 1 and 3 contain cells transformed with empty pREP41X.
DISCUSSION
Two-thirds of the mutations associated with human cancers arise from DNA replication errors, emphasizing the need to understand how cells protect genome integrity during S phase (1). Brc1 preserves genomic stability in response to replication stress, but the mechanism has remained elusive (23, 24, 31–33). One well-defined property of Brc1 is its ability to bind γH2A through its C-terminal BRCT domains. In this report, we identified Rhp18 as another binding partner of Brc1. This physical interaction was detected by coimmunoprecipitation and Y2H screening, which collectively indicate that the interaction is likely to be direct and independent of these proteins binding chromatin. This interaction is mediated through the N-terminal region of Brc1 containing BRCT domains 1 to 4, and it was disrupted by the brc1-HYP307-9GFG allele containing clustered mutations at the beginning of BRCT domain 4. As observed for wild-type Brc1, the Brc1 protein encoded by brc1-HYP307-9GFG properly localizes in the nucleus, where it forms foci in response to replication stress (16). Thus, the brc1-HYP307-9GFG mutation does not appear to grossly disrupt Brc1 protein stability or localization or its ability to bind γH2A-marked chromatin flanking stalled or damaged replication forks. We cannot exclude the possibility that the HYP307-9GFG mutation has minor effects on the ability to bind γH2A, but because complete ablation of Brc1 binding to γH2A only partially impairs Brc1 function (12), it seems unlikely that a subtle defect in binding to γH2A could explain the marked defects of the HYP307-9GFG mutant. From these results, we propose that the brc1-HYP307-9GFG mutation most likely disrupts a scaffolding function of Brc1 that involves binding Rhp18. This model is consistent with the requirements for Rhp18 to tolerate genotoxins that cause replication fork stalling and collapse and the requirement for Rhp18 in the suppression of smc6-74 by Brc1 overexpression (24, 31). Importantly, the brc1-HYP307-9GFG mutation abrogates smc6-74 suppression by Brc1 overexpression.
As a technical note, we used the Y2H method for our studies because we could not reliably precipitate full-length Brc1 in nondenaturing buffers. However, as shown in Fig. 2C, we have largely solved this problem using an N-terminal TAP tag, although a substantial amount of Brc1 appears to still be proteolytically cleaved. Importantly, we could confirm our two-hybrid findings with these coimmunoprecipitation studies. The ability to precipitate TAP-tagged Brc1 in nondenaturing buffers will make it possible to use proteomic methods in future experiments, which has been a very profitable strategy for analyzing the function of Rtt107 (21).
We note that the coimmunoprecipitation studies were performed using Brc1 and Rhp18 expressed from plasmid-borne constructs under the control of nmt41, which is an attenuated version of nmt1 promoter. Coimmunoprecipitation assays are ideally performed with proteins expressed from the endogenous locus, but we could not reliably detect full-length Brc1 using the endogenously tagged constructs that are currently available. We propose that in this case, coimmunoprecipitation studies involving overexpression are relevant because Brc1 was discovered as a multicopy suppressor of smc6-74, and this suppression requires Rhp18 (23, 31). It is formally possible that Brc1 overexpression drives the formation of a Brc1-Rhp18 subcomplex; thus, in future studies it will be important to determine whether Brc1 and Rhp18 coprecipitate when expressed at endogenous levels. It will also be interesting to determine whether phosphorylation of Rhp18 is required for its binding to Brc1.
We observed that suppression of smc6-74 MMS sensitivity by Brc1 overexpression is largely ablated when Brc1 cannot bind to γH2A. Moreover, elimination of γH2A strongly sensitizes smc6-74 cells to MMS. These data strengthen the evidence linking Brc1 to the proposed role for the Smc5/6 complex in homologous recombination (HR)-mediated repair of stalled replication forks (27–29). One possibility is that diminished Smc5/6 function creates a greater demand for Brc1 to act in fork stabilization and potential channeling of fork repair, or resolution, through alternate pathways that likely depend on Mus81-Eme1 or Slx1-Slx4 (24, 41, 42).
We analyzed additional brc1 mutations containing missense alterations in the N-terminal BRCT domains (16). The brc1-W298F, P301G, -G136A, -TH148,149SG, and -R268K mutants all maintained the Y2H interaction with Rhp18 (Fig. 2), albeit reduced in brc1-R268K, and they also retained the ability to rescue brc1Δ MMS sensitivity and suppress smc6-74 MMS sensitivity when expressed nmt41 promoter in plasmid pREP41X. Thus, in all assays these mutants appear to be functional. However, previous analyses indicated that the brc1-TH148,149SG and -R268K mutations conferred HU and CPT sensitivity when expressed from the endogenous locus (16), which is surprising in view of the current data. Experiments are planned to reconstruct these mutations in the endogenous loci to test their genotoxin sensitivities.
It has been reported that Rhp18 is recruited to ssDNA and RPA-bound ssDNA (39, 40). Given that RPA bound to ssDNA is sensed by Rad3/ATR, which phosphorylates H2A to form γH2A (5, 43), it is unlikely that Rhp18 localization at DNA lesions requires binding to Brc1. However, the potential presence of Rhp18 at the site of stalled replication forks, through its interaction with RPA, could provide a potential binding surface at the fork to explain the γH2A-independent function for Brc1 that has been suggested in previous publications (12, 16).
It was previously reported that the dependence of the smc6-74 rescue on Rhp18 was due to a requirement for the translesion synthesis (TLS) branch of PRR at higher MMS doses; however, the requirement at lower MMS concentrations could not be attributed to a known function of Rhp18 (24, 31). The results presented here combined with previously published data suggesting that RPA-coated ssDNA can negatively regulate Rad51 strand invasion (44–46), suggest the potential for the binding of Brc1 to RPA-bound Rhp18 to stabilize RPA on its ssDNA substrate, thus potentially inhibiting HR-mediated fork resolution. In the presence of a low level of DNA alkylation damage, this stabilization of RPA could serve to inhibit Rad51 strand invasion, therefore inhibiting the onset of HR-mediated fork resolution and allowing excision pathways to repair alkylated bases. This idea would be consistent with the requirement for Rhp18 in the smc6-74 rescue in response to low-level MMS treatment, accompanied by no known Rhp18 activity under those circumstances (31). Furthermore, the interaction between Brc1 and RPA bound Rhp18 would allow for the process of TLS to be mediated at MMS doses that can be tolerated by this method of PRR. Under this scenario, it is also possible that in response to high accumulation of alkylated bases that cannot be repaired via the previously mentioned mechanisms, the fork could be held in a stable confirmation, whereas Brc1 stimulates the licensing of surrounding dormant replication origins (47), allowing the resolution of the stalled fork via HR-mediated pathways late in the S phase or possibly in the G2 phase. If this hypothetical series of events is correct, it could potentially explain the complex epistatic relationships between Brc1 and factors involved in multiple DDR pathways (23, 24, 31–33).
Finally, we note that a recent proteomics study with human cells led to the discovery of a putative DNA repair factor, consisting of SLF1 and SLF2, which physically links Smc5/6 complex to Rad18 bound to RNF168-catalyzed ubiquitin chains at certain types of DNA lesions (48). SLF1, also known as BRCTx, is a multi-BRCT domain protein that uses its BRCT domains to bind Rad18 (49, 50). Indeed, this SLF1/BRCTx-Rad18 interaction was first discovered through a two-hybrid screen that used BRCTx as bait, just as we discovered the Brc1-Rhp18 interaction in fission yeast. The protein interaction network involving SLF1/SLF2, Rad18 and Smc5/6 complex does not include Rad6, suggesting that Rad18 functions structurally and not catalytically in this network (48). Strikingly, the suppression of smc6-74 by Brc1 overexpression on low-dose MMS does not require Rhp6, the fission yeast ortholog of Rad6 (31). Rtt107, the Brc1-like protein in budding yeast, was shown to mediate Smc5/6 recruitment to double-strand breaks (51). Thus, there appear to be several striking parallels of protein interactions involving Brc1/Rtt107/PTIP BRCT domain proteins, Rad18/Rhp18, and the Smc5/6 complex. We look forward to learning whether these similarities reflect evolutionary divergence of a conserved mechanism of localizing Smc5/6 complex to DNA lesions.
MATERIALS AND METHODS
S. pombe cultivation and general methods.
Standard S. pombe methods were conducted as previously described (52), and all S. pombe strains used in this study are listed in Table 1. The rhp18Δ strain was generated using a targeting construct that replaced the entire rhp18+ open reading frame with a hygromycin B (hphMX6) cassette, and Rhp18 was N-terminally tagged at its endogenous locus using pFA6-natMX6-p41nmt-13myc (described below) using described methods (53). Both the deletion of Rhp18 and the epitope tag were subsequently verified by PCR and sequencing before use in any experiments. Double and triple mutants were generated using random spore analysis, the resulting genotypes were validated by growth on appropriate selective media and then subsequently verified by PCR.
TABLE 1.
Strains generated for and used in this study
| Strain | Genotype (all strains are leu1-32 and ura4-D18) |
|---|---|
| MR5456 | h– pREP41X |
| MR5457 | h– pREP41Xbrc1+ |
| MR5458 | h– brc1::kanMX6 pREP41X |
| MR5459 | h– brc1::kanMX6 pREP41Xbrc1+ |
| MR5546 | h– brc1::kanMX6 pREP41Xbrc1-G136A |
| MR5547 | h– brc1::kanMX6 pREP41Xbrc1-TH148,149SG |
| MR5548 | h– brc1::kanMX6 pREP41Xbrc1-R268K |
| MR5460 | h– brc1::kanMX6 pREP41Xbrc1-W298F, P01G |
| MR5461 | h– brc1::kanMX6 pREP41Xbrc1-HYP307-9GFG |
| MR5462 | h– brc1::kanMX6 pREP41Xbrc1-T672A |
| MR5464 | h– smc6-74 pREP41X |
| MR5465 | h– smc6-74 pREP41Xbrc1+ |
| MR5549 | h– smc6-74 pREP41Xbrc1-G136A |
| MR5550 | h– smc6-74 pREP41Xbrc1-TH148,149SG |
| MR5551 | h– smc6-74 pREP41Xbrc1-R268K |
| MR5466 | h– smc6-74 pREP41Xbrc1-W298F,P01G |
| MR5467 | h– smc6-74 pREP41Xbrc1-HYP307-9GFG |
| MR5468 | h– smc6-74 pREP41Xbrc1-T672A |
| MR5469 | h– hta1-S129A:ura4+ hta2-S128A:his3+ his3-D1 pREP41X |
| MR5470 | h– hta1-S129A:ura4+ hta2-S128A:his3+ his3-D1 pREP41Xbrc1+ |
| MR5471 | h– hta1-S129A:ura4+ hta2-S128A:his3+ brc1::kanMX6 his3-D1 pREP41X |
| MR5472 | h– hta1-S129A:ura4+ hta2-S128A:his3+ brc1::kanMX6 his3-D1 pREP41Xbrc1+ |
| MR5473 | h– hta1-S129A:ura4+ hta2-S128A:his3+ brc1::kanMX6 his3-D1 pREP41Xbrc1-W298F,P301G |
| MR5474 | h– hta1-S129A:ura4+ hta2-S128A:his3+ brc1::kanMX6 his3-D1 pREP41Xbrc1-HYP307-9GFG |
| MR5475 | h– hta1-S129A:ura4+ hta2-S128A:his3+ brc1::kanMX6 his3-D1 pREP41Xbrc1-T672A |
| MR5477 | h– hta1-S129A:ura4+ hta2-S128A:his3+ smc6-74 his3-D1 pREP41X |
| MR5478 | h– hta1-S129A:ura4+ hta2-S128A:his3+ smc6-74 his3-D1 pREP41Xbrc1+ |
| MR5479 | h– hta1-S129A:ura4+ hta2-S128A:his3+ smc6-74 his3-D1 pREP41Xbrc1-W298F,P301G |
| MR5480 | h– hta1-S129A:ura4+ hta2-S128A:his3+ smc6-74 his3-D1 pREP41Xbrc1-HYP307-9GFG |
| MR5481 | h– hta1-S129A:ura4+ hta2-S128A:his3+ smc6-74 his3-D1 pREP41Xbrc1-T672A |
| MR5485 | h– hta1-S129A:ura4+ hta2-S128A:his3+ smc6-74 rhp18::hphMX6 his3-D1 pREP41X |
| MR5486 | h– hta1-S129A:ura4+ hta2-S128A:his3+ smc6-74 rhp18::hphMX6 his3-D1 pREP41Xbrc1+ |
| MR5487 | h– hta1-S129A:ura4+ hta2-S128A:his3+ smc6-74 rhp18::hphMX6 his3-D1 pREP41Xbrc1-W298F,P301G |
For immunoprecipitation experiments, exponentially growing cultures of indicated strains were cultivated in appropriately supplemented Edinburgh minimal medium 2 (EMM2) in the presence or absence of 5 μg/ml thiamine for 25 h at 30°C to actively regulate the expression from the nmt41 promoter. For MMS survival assays, the indicated strains were cultivated in appropriately supplemented EMM2 media in the presence of 5 μg/ml thiamine for 25 h; log-phase cultures were then suspended to an optical density at 600 nm (OD600) of 0.4 and serially diluted 5-fold onto yeast extract, glucose, and supplements (YES) agar plates containing the designated concentration of MMS. Cell growth was evaluated after 4 days at 30°C as previously described (24, 31).
Plasmid construction.
For the Y2H analysis constructs, all brc1 fragments (FL, aa 1 to 878; 1-4, aa 1 to 553; 1-2, aa 1 to 202; and 3-4, aa 195 to 553) and point mutants were isolated using standard PCR methods with NdeI linkers on upstream primers and BamHI linkers on downstream primers. All rhp18 fragments (FL, aa 1 to 387; R/M, aa 1 to 202; R, aa 1 to 117; and M, aa 111 to 202) were generated using a similar PCR-mediated strategy except for using XmaI linkers on the downstream primers. The resulting brc1 inserts were then ligated into NdeI- and BamHI-digested pGBKT7, and the rhp18 fragments were ligated into NdeI and XmaI digested pGADT7. Expression of TAP-Brc1 was achieved by cloning full-length brc1+ or brc1-HYP307-9GFG cDNA into NdeI- and BamHI-digested pREP41-NTAP as previously described (54). For MMS survival assays, pREP41Xbrc1+ was used to rescue the MMS phenotypes as previously described (24, 31), and all evaluated brc1 point mutants were generated by site-directed mutagenesis (Agilent Technologies) using pREP41Xbrc1+ as the template. To N-terminally tag Rhp18 at its endogenous locus, pFA6a-natMX6-p41nmt-13myc was constructed by cleaving the 3×FLAG from pFA6a-natMX6-p41nmt-3×FLAG (55) and replacing it with the 13myc tag, without its stop codon, isolated from pFA6a-13myc-natMX6 (53). All plasmids generated for use in this study were sequence verified before use.
Yeast two-hybrid analysis.
All Brc1 and Rhp18 fusion constructs were generated as described above. The resulting fusion protein constructs were transformed into the S. cerevisiae AH109 reporter strain (Clontech Matchmaker Gold system), and cotransformants were selected for by plating on Dex-L-T media. Y2H analysis was carried out by diluting the indicated log-phase cultures to an OD600 of 0.4 and then spotting them onto restrictive and permissive plates. Control growth was evaluated on Dex-L-T, and protein interactions were scored either Dex-L-T-H with or without 3-AT based on the presence of one-hybrid activity. All Y2H growth was scored after 3 days of growth at 32°C.
Immunoblotting and immunoprecipitation.
Whole-cell extracts were generated from 15-ml cultures of the indicated strains cultivated as described above. Cell pellets were lysed in lysis buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 5 mM EDTA, 10% glycerol, 0.1% Nonidet P-40, 1 mM NaF, 1 mM phenylmethylsulfonyl fluoride, and Complete protease inhibitors) using a FastPrep-24 (MP Biomedicals) according to the manufacturer's protocol. For each lysate, 1.5 mg of total protein was incubated with rabbit IgG (Sigma)-conjugated tosylactivated Dynabeads M-280 (Thermo Fisher Scientific) for 3 h at 4°C with rotation. The beads were collected and washed three times in lysis buffer before eluting the proteins from the beads by boiling in 1× SDS-PAGE loading buffer (100 mM Tris [pH 6.8], 4% SDS, 20% glycerol, 0.2% bromophenol blue). Proteins were resolved on Novex WedgeWell 4 to 20% Tris-glycine gels (Thermo Fisher Scientific), transferred via iBlot2 (Thermo Fisher Scientific) to nitrocellulose membranes, and blocked and probed using standard techniques and manufacturer-recommended protocols. TAP-Brc1 was detected using peroxidase-antiperoxidase-soluble complex antibody produced in rabbit (catalog no. P12291; Sigma-Aldrich) diluted 1:2,000, 13myc-Rhp18 was detected using anti-myc antibody (9E10; Covance) diluted 1;1,000, and tubulin was detected using monoclonal anti-α-tubulin antibody produced in mice (T5168; Sigma-Aldrich) diluted 1:10,000.
ACKNOWLEDGMENTS
We thank Matthew O'Connell for plasmids and technical insights, Oliver Limbo for technical support, and members of the Russell laboratory and Nick Boddy for helpful discussions.
This research was supported by NIH grants GM059347, CA077325, and CA117638 to P.R.
We declare no conflicts of interest.
REFERENCES
- 1.Tomasetti C, Li L, Vogelstein B. 2017. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 355:1330. doi: 10.1126/science.aaf9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tubbs A, Nussenzweig A. 2017. Endogenous DNA damage as a source of genomic instability in cancer. Cell 168:644–656. doi: 10.1016/j.cell.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilera A, Garcia-Muse T. 2013. Causes of genome instability. Annu Rev Genet 47:1–32. doi: 10.1146/annurev-genet-111212-133232. [DOI] [PubMed] [Google Scholar]
- 4.Gadaleta MC, Noguchi E. 2017. Regulation of DNA replication through natural impediments in the eukaryotic genome. Genes 8:98. doi: 10.3390/genes8030098. [DOI] [Google Scholar]
- 5.Branzei D, Foiani M. 2009. The checkpoint response to replication stress. DNA Repair 8:1038–1046. doi: 10.1016/j.dnarep.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Cortez D. 2015. Preventing replication fork collapse to maintain genome integrity. DNA Repair 32:149–157. doi: 10.1016/j.dnarep.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer DR, Rhind N. 2017. The intra-S checkpoint responses to DNA damage. Genes 8:74. doi: 10.3390/genes8020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Branzei D, Foiani M. 2010. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 9.Rozenzhak S, Mejía-Ramírez E, Williams JS, Schaffer L, Hammond JA, Head SR, Russell P. 2010. Rad3ATR decorates critical chromosomal domains with γH2A to protect genome integrity during S-phase in fission yeast. PLoS Genet 6:e1001032. doi: 10.1371/journal.pgen.1001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mejia-Ramirez E, Limbo O, Langerak P, Russell P. 2015. Critical function of γH2A in S-phase. PLoS Genet 11:e1005517. doi: 10.1371/journal.pgen.1005517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura TM, Du L-L, Redon C, Russell P. 2004. Histone H2A Phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol Cell Biol 24:6215–6230. doi: 10.1128/MCB.24.14.6215-6230.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams JS, Williams RS, Dovey CL, Guenther G, Tainer JA, Russell P. 2010. γH2A binds Brc1 to maintain genome integrity during S-phase. EMBO J 29:1136–1148. doi: 10.1038/emboj.2009.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du LL, Nakamura TM, Russell P. 2006. Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev 20:1583–1596. doi: 10.1101/gad.1422606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sofueva S, Du LL, Limbo O, Williams JS, Russell P. 2010. BRCT domain interactions with phospho-histone H2A target Crb2 to chromatin at double-strand breaks and maintain the DNA damage checkpoint. Mol Cell Biol 30:4732–4743. doi: 10.1128/MCB.00413-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei Y, Wang HT, Zhai Y, Russell P, Du LL. 2014. Mdb1, a fission yeast homolog of human MDC1, modulates DNA damage response and mitotic spindle function. PLoS One 9:e97028. doi: 10.1371/journal.pone.0097028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SY, Russell P. 2013. Brc1 links replication stress response and centromere function. Cell Cycle 12:1665–1671. doi: 10.4161/cc.24900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hang LE, Peng J, Tan W, Szakal B, Menolfi D, Sheng Z, Lobachev K, Branzei D, Feng W, Zhao X. 2015. Rtt107 is a multifunctional scaffold supporting replication progression with partner SUMO and ubiquitin ligases. Mol Cell 60:268–279. doi: 10.1016/j.molcel.2015.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan B, Hang LE, Zhao X. 2016. Multi-BRCT scaffolds use distinct strategies to support genome maintenance. Cell Cycle 15:2561–2570. doi: 10.1080/15384101.2016.1218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leung GP, Brown JA, Glover JN, Kobor MS. 2016. Rtt107 BRCT domains act as a targeting module in the DNA damage response. DNA Repair 37:22–32. doi: 10.1016/j.dnarep.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Balint A, Kim T, Gallo D, Cussiol JR, Bastos de Oliveira FM, Yimit A, Ou J, Nakato R, Gurevich A, Shirahige K, Smolka MB, Zhang Z, Brown GW. 2015. Assembly of Slx4 signaling complexes behind DNA replication forks. EMBO J 34:2182–2197. doi: 10.15252/embj.201591190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohouo PY, Bastos de Oliveira FM, Almeida BS, Smolka MB. 2010. DNA damage signaling recruits the Rtt107-Slx4 scaffolds via Dpb11 to mediate replication stress response. Mol Cell 39:300–306. doi: 10.1016/j.molcel.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 22.Ray Chaudhuri A, Callen E, Ding X, Gogola E, Duarte AA, Lee JE, Wong N, Lafarga V, Calvo JA, Panzarino NJ, John S, Day A, Crespo AV, Shen B, Starnes LM, de Ruiter JR, Daniel JA, Konstantinopoulos PA, Cortez D, Cantor SB, Fernandez-Capetillo O, Ge K, Jonkers J, Rottenberg S, Sharan SK, Nussenzweig A. 2016. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 535:382–387. doi: 10.1038/nature18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verkade HM, Bugg SJ, Lindsay HD, Carr AM, O'Connell MJ. 1999. Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol Biol Cell 10:2905–2918. doi: 10.1091/mbc.10.9.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheedy DM, Dimitrova D, Rankin JK, Bass KL, Lee KM, Tapia-Alveal C, Harvey SH, Murray JM, O'Connell MJ. 2005. Brc1-mediated DNA repair and damage tolerance. Genetics 171:457–468. doi: 10.1534/genetics.105.044966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernstein KA, Gangloff S, Rothstein R. 2010. The RecQ DNA helicases in DNA repair. Annu Rev Genet 44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez A, Sharma S, Rozenzhak S, Roguev A, Krogan NJ, Chabes A, Russell P. 2012. Replication fork collapse and genome instability in a deoxycytidylate deaminase mutant. Mol Cell Biol 32:4445–4454. doi: 10.1128/MCB.01062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pebernard S, McDonald WH, Pavlova Y, Yates JR, Boddy MN. 2004. Nse1, Nse2, and a novel subunit of the Smc5-Smc6 complex, Nse3, play a crucial role in meiosis. Mol Biol Cell 15:4866–4876. doi: 10.1091/mbc.E04-05-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morikawa H, Morishita T, Kawane S, Iwasaki H, Carr AM, Shinagawa H. 2004. Rad62 protein functionally and physically associates with the Smc5/Smc6 protein complex and is required for chromosome integrity and recombination repair in fission yeast. Mol Cell Biol 24:9401–9413. doi: 10.1128/MCB.24.21.9401-9413.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pebernard S, Wohlschlegel J, McDonald WH, Yates JR, Boddy MN. 2006. The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol Cell Biol 26:1617–1630. doi: 10.1128/MCB.26.5.1617-1630.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee SY, Rozenzhak S, Russell P. 2013. γH2A-binding protein Brc1 affects centromere function in fission yeast. Mol Cell Biol 33:1410–1416. doi: 10.1128/MCB.01654-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee KM, Nizza S, Hayes T, Bass KL, Irmisch A, Murray JM, O'Connell MJ. 2007. Brc1-mediated rescue of Smc5/6 deficiency: requirement for multiple nucleases and a novel Rad18 function. Genetics 175:1585–1595. doi: 10.1534/genetics.106.067801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sánchez A, Roguev A, Krogan NJ, Russell P. 2015. Genetic interaction landscape reveals critical requirements for Schizosaccharomyces pombe Brc1 in DNA damage response mutants. G3 (Bethesda) 5:953–962. doi: 10.1534/g3.115.017251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sánchez A, Russell P. 2015. Ku stabilizes replication rorks in the absence of Brc1. PLoS One 10:e0126598. doi: 10.1371/journal.pone.0126598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao Y, Mutter-Rottmayer E, Zlatanou A, Vaziri C, Yang Y. 2017. Mechanisms of postreplication DNA repair. Genes 8:64. doi: 10.3390/genes8020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung CCY, Glover JNM. 2011. BRCT domains: Easy as one, two, three. Cell Cycle 10:2461–2470. doi: 10.4161/cc.10.15.16312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reinhardt HC, Yaffe MB. 2013. Phospho-Ser/Thr-binding domains: navigating the cell cycle and DNA damage response. Nat Rev Mol Cell Biol 14:563–580. doi: 10.1038/nrm3640. [DOI] [PubMed] [Google Scholar]
- 37.Notenboom V, Hibbert RG, van Rossum-Fikkert SE, Olsen JV, Mann M, Sixma TK. 2007. Functional characterization of Rad18 domains for Rad6, ubiquitin, DNA binding and PCNA modification. Nucleic Acids Res 35:5819–5830. doi: 10.1093/nar/gkm615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakajima S, Lan L, Kanno S-i, Usami N, Kobayashi K, Mori M, Shiomi T, Yasui A. 2006. Replication-dependent and -independent responses of RAD18 to DNA damage in human cells. J Biol Chem 281:34687–34695. doi: 10.1074/jbc.M605545200. [DOI] [PubMed] [Google Scholar]
- 39.Huttner D, Ulrich HD. 2008. Cooperation of replication protein A with the ubiquitin ligase Rad18 in DNA damage bypass. Cell Cycle 7:3629–3633. doi: 10.4161/cc.7.23.7166. [DOI] [PubMed] [Google Scholar]
- 40.Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. 2008. Activation of ubiquitin-dependent DNA damage bypass Is mediated by replication protein A. Mol Cell 29:625–636. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Froget B, Blaisonneau J, Lambert S, Baldacci G. 2008. Cleavage of stalled forks by fission yeast Mus81/Eme1 in absence of DNA replication checkpoint. Mol Biol Cell 19:445–456. doi: 10.1091/mbc.E07-07-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roseaulin L, Yamada Y, Tsutsui Y, Russell P, Iwasaki H, Arcangioli B. 2008. Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J 27:1378–1387. doi: 10.1038/emboj.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Branzei D, Foiani M. 2008. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol 9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 44.Dou H, Huang C, Singh M, Carpenter PB, Yeh ETH. 2010. Regulation of DNA repair through de-SUMOylation and SUMOylation of replication protein A complex. Mol Cell 39:333–345. doi: 10.1016/j.molcel.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heyer W-D, Ehmsen KT, Liu J. 2010. Regulation of homologous recombination in eukaryotes. Annu Rev Genet 44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X, Heyer W-D. 2008. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res 18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bass KL, Murray JM, O'Connell MJ. 2012. Brc1-dependent recovery from replication stress. J Cell Sci 125:2753–2764. doi: 10.1242/jcs.103119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raschle M, Smeenk G, Hansen RK, Temu T, Oka Y, Hein MY, Nagaraj N, Long DT, Walter JC, Hofmann K, Storchova Z, Cox J, Bekker-Jensen S, Mailand N, Mann M. 2015. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science 348:1253671. doi: 10.1126/science.1253671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams DJ, van der Weyden L, Gergely FV, Arends MJ, Ng BL, Tannahill D, Kanaar R, Markus A, Morris BJ, Bradley A. 2005. BRCTx is a novel, highly conserved RAD18-interacting protein. Mol Cell Biol 25:779–788. doi: 10.1128/MCB.25.2.779-788.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu T, Chen H, Kim H, Huen MS, Chen J, Huang J. 2012. RAD18-BRCTx interaction is required for efficient repair of UV-induced DNA damage. DNA Repair 11:131–138. doi: 10.1016/j.dnarep.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leung GP, Lee L, Schmidt TI, Shirahige K, Kobor MS. 2011. Rtt107 is required for recruitment of the SMC5/6 complex to DNA double-strand breaks. J Biol Chem 286:26250–26257. doi: 10.1074/jbc.M111.235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Forsburg SL, Rhind N. 2006. Basic methods for fission yeast. Yeast 23:173–183. doi: 10.1002/yea.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bahler J, Wu JQ, Longtine MS, Shah NG, AMcKenzie 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943–951. doi:. [DOI] [PubMed] [Google Scholar]
- 54.Tasto JJ, Carnahan RH, Hayes McDonald W, Gould KL. 2001. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast 18:657–662. doi: 10.1002/yea.713. [DOI] [PubMed] [Google Scholar]
- 55.Noguchi C, Garabedian MV, Malik M, Noguchi E. 2008. A vector system for genomic FLAG epitope-tagging in Schizosaccharomyces pombe. Biotechnol J 3:1280–1285. doi: 10.1002/biot.200800140. [DOI] [PubMed] [Google Scholar]