ABSTRACT
Human respiratory syncytial virus (RSV) is the most prevalent worldwide cause of severe respiratory tract infection in infants and young children. Human parainfluenza virus type 1 (HPIV1) also causes severe pediatric respiratory illness, especially croup. Both viruses lack vaccines. Here, we describe the preclinical development of a bivalent RSV/HPIV1 vaccine based on a recombinant HPIV1 vector, attenuated by a stabilized mutation, that expresses RSV F protein modified for increased stability in the prefusion (pre-F) conformation by previously described disulfide bond (DS) and hydrophobic cavity-filling (Cav1) mutations. RSV F was expressed from the first or second gene position as the full-length protein or as a chimeric protein with its transmembrane and cytoplasmic tail (TMCT) domains substituted with those of HPIV1 F in an effort to direct packaging in the vector particles. All constructs were recovered by reverse genetics. The TMCT versions of RSV F were packaged in the rHPIV1 particles much more efficiently than their full-length counterparts. In hamsters, the presence of the RSV F gene, and in particular the TMCT versions, was attenuating and resulted in reduced immunogenicity. However, the vector expressing full-length RSV F from the pre-N position was immunogenic for RSV and HPIV1. It conferred complement-independent high-quality RSV-neutralizing antibodies at titers similar to those of wild-type RSV and provided protection against RSV challenge. The vectors exhibited stable RSV F expression in vitro and in vivo. In conclusion, an attenuated rHPIV1 vector expressing a pre-F-stabilized form of RSV F demonstrated promising immunogenicity and should be further developed as an intranasal pediatric vaccine.
IMPORTANCE RSV and HPIV1 are major viral causes of acute pediatric respiratory illness for which no vaccines or suitable antiviral drugs are available. The RSV F glycoprotein is the major RSV neutralization antigen. We used a rHPIV1 vector, bearing a stabilized attenuating mutation, to express the RSV F glycoprotein bearing amino acid substitutions that increase its stability in the pre-F form, the most immunogenic form that elicits highly functional virus-neutralizing antibodies. RSV F was expressed from the pre-N or N-P gene position of the rHPIV1 vector as a full-length protein or as a chimeric form with its TMCT domain derived from HPIV1 F. TMCT modification greatly increased packaging of RSV F into the vector particles but also increased vector attenuation in vivo, resulting in reduced immunogenicity. In contrast, full-length RSV F expressed from the pre-N position was immunogenic, eliciting complement-independent RSV-neutralizing antibodies and providing protection against RSV challenge.
KEYWORDS: human parainfluenza virus type 1, intranasal vaccine, live attenuated vaccine, mucosal vaccines, pediatric vaccine, prefusion F, respiratory syncytial virus, vaccine
INTRODUCTION
Human respiratory syncytial virus (RSV) is an enveloped virus with a nonsegmented, negative-sense RNA genome of approximately 15.2 kb, which is classified in the family Pneumoviridae (1). RSV is the most common viral cause of hospitalization associated with bronchiolitis and pneumonia in infants and young children worldwide. It is associated with an estimated 34 million lower-respiratory-tract infections, 4 million hospitalizations, and 66,000 to 199,000 annual deaths in children younger than 5 years of age (2). The annual RSV related death burden in all age groups is 234,000 to 520,000 worldwide (3). As a single agent, RSV is second only to malaria in the burden of mortality in the postneonatal (age 28 days to 1 year) population worldwide (3). Although mortality due to RSV is low in the United States, morbidity is substantial and includes an estimated 132,000 to 172,000 RSV-associated hospitalizations annually in children <5 years of age (4).
The human parainfluenza viruses (HPIVs) are second to RSV as a leading viral cause of severe pediatric lower respiratory tract disease. The HPIVs are enveloped viruses with nonsegmented, negative-sense RNA genomes of approximately 15.4 to 17 kb and are classified in the family Paramyxoviridae. They comprise four serotypes, 1 to 4. Of these, HPIV1 is the most common cause of croup. The aggregate burden of morbidity and mortality of the HPIVs is somewhat less than that of RSV.
Despite their impact, RSV and the HPIVs lack licensed vaccines and suitable antiviral drugs. Experimental vaccines based on inactivated preparations of RSV or HPIV3 have been shown to prime for enhanced RSV or HPIV3 disease in virus-naive humans (RSV) (5) and experimental animals (RSV and HPIV3) (6), and similar observations have been made for purified RSV subunit vaccines in rodents (7). In contrast, replication-competent vaccines based on live attenuated RSV or a chimeric live PIV3 vector have been shown to be free of enhanced disease in pediatric clinical trials (8–11). We have been developing live-attenuated PIV vectors expressing the RSV F protein, which is the major RSV neutralization and protective antigen. This type of construct provides a bivalent vaccine against RSV and the HPIV serotype represented by the vector.
The HPIV genome contains six genes arranged in the order: N (nucleoprotein), P (phosphoprotein), M (internal matrix protein), F (fusion glycoprotein), HN (hemagglutinin-neuraminidase glycoprotein), and L (large polymerase protein). Depending on the HPIV species, the P open reading frame (ORF) can have additional overlapping open reading frames encoding various proteins, including ones that antagonize the host interferon and apoptosis responses (12). Foreign genes can readily be inserted into the HPIV genome: when modified to be flanked by HPIV gene-start and gene-end transcription signals and inserted between HPIV genes, foreign genes are transcribed by the HPIV polymerase into separate mRNAs.
The RSV F protein is a type I integral membrane glycoprotein that mediates fusion of the viral envelope with cellular membranes during infection. F protein is synthesized as a precursor F0 that is proteolytically cleaved into F1 and F2 subunits that remain linked by disulfide bonds. During fusion, F undergoes a dramatic, irreversible conformational rearrangement from a metastable prefusion (pre-F) form to a highly stable postfusion (post-F) form. RSV F is sufficiently metastable that it can also readily undergo spontaneous triggering, with the result that RSV virions and infected cell surfaces can contain a large proportion of F present in the post-F configuration (13). Although both pre- and post-F possess neutralizing epitopes (14–17), pre-F-specific antibodies represent most of the RSV-neutralizing activity in human sera (18). This suggests that pre-F is the antigen of choice for an RSV vaccine. RSV F can be modified to have increased stability in the pre-F conformation by the introduction of mutations described below.
Most of the previous work with HPIV vectors employed a recombinant chimeric bovine/human PIV3 called rB/HPIV3 to express the RSV F protein. A number of modifications were evaluated to increase RSV-specific immunogenicity, and the greatest increases were observed with modifications that (i) increase pre-F stability and (ii) increase the incorporation of RSV F into the vector particle (19–21). McLellan and coworkers previously described increasing RSV pre-F stability by the introduction of disulfide bond (DS), generating mutations (S155C and S290C), as well as the further introduction of hydrophobic cavity-filling (Cav1) substitutions (S190F and V207L) into the pre-F head (14, 15). In addition, while there is little packaging of unmodified RSV F protein into the rB/HPIV3 virus particle, packaging can be made efficient by the replacement of the transmembrane (TM) and cytoplasmic tail (CT) domains of RSV F with their counterparts from the vector HPIV3 F protein (20). These two factors—pre-F stabilization and RSV F packaging—individually and additively increased the immunogenicity of the RSV F insert and, in particular, increased the induction of serum antibodies that efficiently neutralized RSV in vitro in the absence of added complement. In contrast, vectors expressing native RSV F induced little or no detectable antibodies capable of neutralizing RSV in the absence of added complement.
Recently, we also initiated evaluation of HPIV1 as a vector to express the RSV F protein. Evaluating a second HPIV serotype is of value because it is unknown a priori how the different HPIV serotypes will perform and compare as vectors for RSV F. In addition, the availability of different HPIV serotypes expressing RSV antigen would allow for the sequential use of different serotype vectors to boost immunity. Also, a particular interest of HPIV1 as a vector is that natural infection with HPIV1 typically occurs between 1 and 4 years of age (22), whereas HPIV3 resembles RSV in its ability to infect and cause disease during the first months of life. Therefore, an HPIV1-based vector expressing RSV F could be optimally used following 1 year of age as a primary immunization against HPIV1 and to boost immune responses to an RSV vaccine that had been administered previously at several months of age.
We previously evaluated two different attenuated HPIV1 backbones as vectors for expressing unmodified RSV F (23). One backbone was attenuated by the amino acid substitution Y942A in the L protein, which proved to be overattenuated. The other backbone contained a 6-nucleotide deletion in the P gene (called CΔ170) that, in the P protein, deletes amino acids 172G and 173F and, in the C protein, deletes amino acids 168R and 169D and changes 170F to 170S (24). The HPIV1-CΔ170 backbone was moderately attenuated and stable for RSV F expression during in vitro and in vivo replication and was immunogenic and therefore appeared to be promising. In the present study, engineered RSV F stabilized in the pre-F conformation by DS-Cav1 mutations was expressed from this backbone from the first (pre-N) or second (N-P) gene position, either as a full-length protein or a chimera in which its transmembrane and cytoplasmic tail (TMCT) domain was derived from HPIV1 F. The four vaccine candidates recovered by reverse genetics were characterized in vitro and evaluated in hamsters for replication, immunogenicity, and protection against wild-type (wt) RSV challenge.
RESULTS
Construction of HPIV1 vectors expressing RSV pre-F protein.
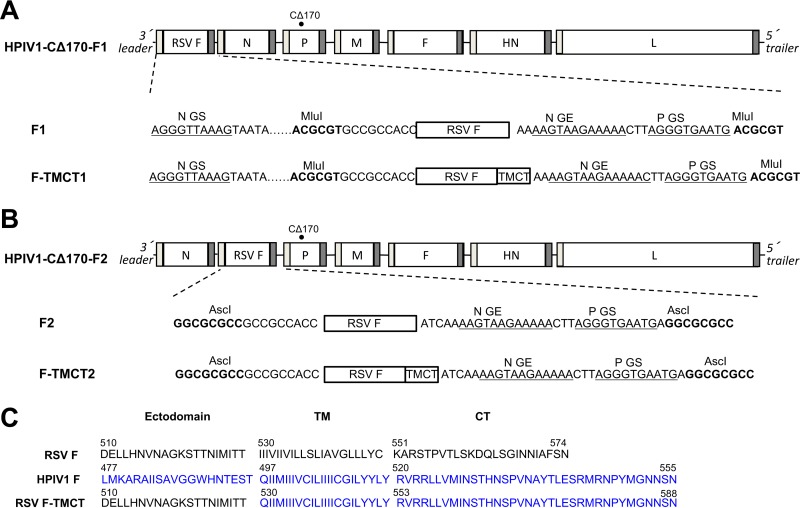
The RSV F ORF was modified so that the encoded protein contained an added disulfide bond (DS; S155C and S290C) and two cavity-filling mutations in the pre-F head (Cav1; S190F and V207L). The DS and Cav1 mutations confer increased stability to the pre-F conformation, including unique pre-F-specific protective epitopes such as antigenic site Ø (15). The RSV F ORF also was codon optimized for expression in human cells. The RSV F ORF was inserted into the rHPIV1 backbone at the pre-N (position 1; Fig. 1A) or N-P (position 2; Fig. 1B) gene position under the control of added HPIV1 transcription signals. The ORF was designed to encode either the full-length RSV F protein or a chimeric version containing the RSV F ectodomain fused with the TMCT domain of the HPIV1 F protein. The TMCT domain of HPIV1 F was identified based on the predicted hydrophobic TMCT region (Fig. 1C). The TMCT modification was an effort to facilitate RSV F packaging in the vector particle (20). The rHPIV1 backbone in each construct contained the stabilized attenuating mutation CΔ170 (25). Thus, four vector constructs were made: two had the insert at position 1 and were called F1 and F-TMCT1, and two had the insert at position 2 and were called F2 and F-TMCT2.
FIG 1.
Attenuated HPIV1-CΔ170 expressing RSV pre-F protein, full length or containing the TMCT domains of vector F protein, inserted at the first (pre-N) (A) or second (N-P) (B) gene position. The RSV F ORF was codon optimized for human expression and modified so that the encoded protein had increased stability in the pre-F conformation by the introduction of a disulfide bond (DS) mutations (S155C and S290C) and two cavity-filling (Cav1) mutations (S190F and V207L). (A and B) The rHPIV1 backbone contained the stabilized attenuating CΔ170 mutation in the P/C ORF (filled circle). The RSV F insert was flanked by the HPIV1 gene-end (GE) and gene-start (GS) transcription signals and the intergenic CTT trinucleotide, so that it would be expressed as a separate mRNA. (C) Amino acid sequences of the C-terminal end of the ectodomain, transmembrane (TM), and cytoplasmic tail (CT) domains of the RSV F, HPIV1 F (predicted), and the chimeric RSV F-TMCT proteins.
Stability and replication of rHPIV1-CΔ170-RSV F vectors in Vero cells.
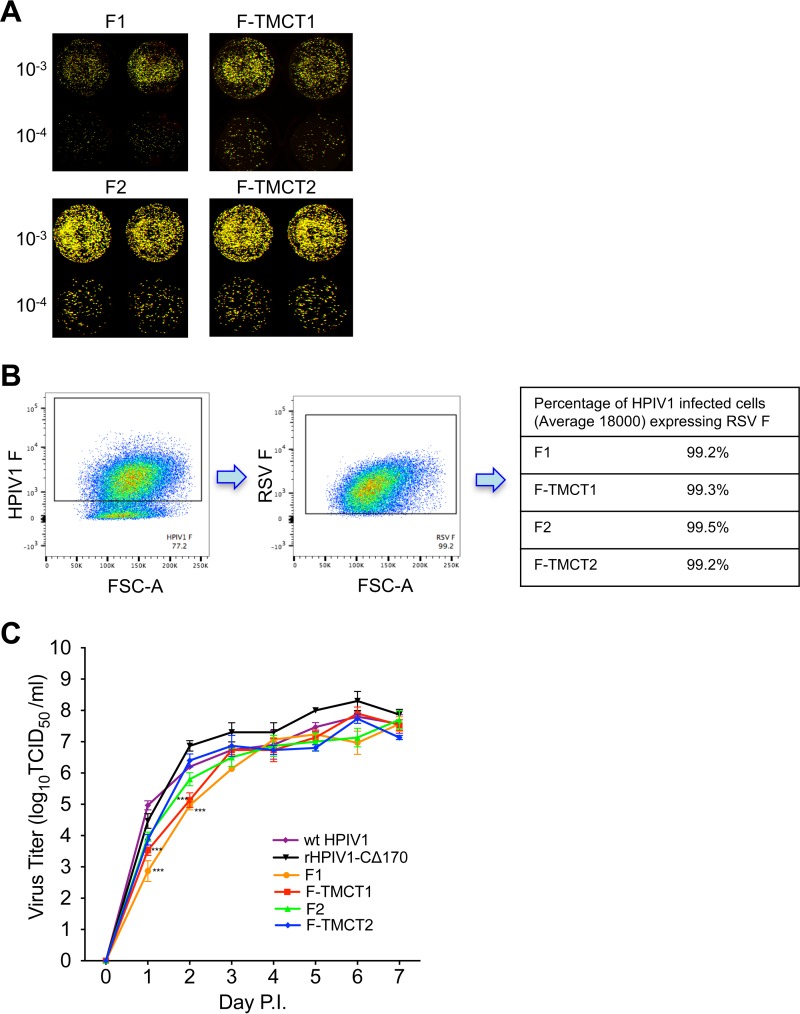
The four vectors were readily recovered. The stability of expression of the RSV F protein was analyzed by a dual-staining fluorescent plaque assay that detects the coexpression of rHPIV1 proteins and RSV F protein. The PFU of the working stocks of the F1, F-TMCT1, and F-TMCT2 vectors exhibited nearly 100% expression of RSV F, whereas the percentage of F2 vector PFU that expressed RSV F was substantially less (∼50%). Examination of a total of 16 independent recoveries for the F2 vector provided identification of a single stock with a high level of stability of F expression (Fig. 2A). The F1 and F-TMCT1 vectors containing RSV F at the pre-N position formed very small plaques (Fig. 2A), making it difficult to assess the stability of expression. Thus, we also examined the expression of RSV F by flow cytometry performed by staining infected Vero cells for HPIV1 F and RSV F expression. Live, single, HPIV1 F+ cells were gated and analyzed for RSV F coexpression, more than 98% of which expressed RSV F for all viruses (Fig. 2B). The consensus genome sequences of all four vectors were determined and were confirmed to be free of adventitious mutations.
FIG 2.
Virus replication and stability in vitro. (A) Vero cells were infected with 10-fold serially diluted viruses and incubated under a methylcellulose overlay at 32°C for 6 days. The cells were stained with RSV F MAbs and a polyvalent HPIV1-specific antiserum, which were pseudocolored red and green, respectively. Images were acquired using an Odyssey infrared imaging system, and plaques coexpressing RSV F and HPIV1 proteins were detected as yellow when merged. The virus fold dilution is indicated to the left of the images. (B) Vero cells were infected with viruses at an MOI of 1 TCID50 per cell and incubated for 48 h. Cells were stained with anti-HPIV1 F and anti-RSV F MAbs and subjected to flow cytometry analysis. Single live cells gated on HPIV1 F+ cells were analyzed for RSV F expression, and the percentages of cells expressing both HPIV1 F and RSV F are shown. (C) Multicycle replication in Vero cells. Cell monolayers in three replicate wells per virus in a six-well plate were infected at an MOI of 0.01 TCID50 per cell with rHPIV-CΔ170 empty vector or the four rHPIV-CΔ170 vectors F1, F-TMCT1, F2, and F-TMCT2, or wt HPIV1. Aliquots of cell culture medium were collected at 24-h intervals, and virus titers were determined by serial dilution on LLC-MK2 cells and hemadsorption assay. Mean titers ± the standard errors of the mean (SEM) are shown. The statistical significance of the differences among viruses was determined by one-way analysis of variance (ANOVA) with Dunnett's multiple comparisons posttest and is indicated by asterisks (***, P < 0.001).
Multicycle growth curves were determined on Vero cells, the substrate for vaccine manufacture (Fig. 2C). During the exponential phase of replication (days 0 to 3), the F1 and F-TMCT1 vectors replicated significantly slower than the F2 and F-TMCT2 vectors, particularly on days 1 and 2 postinfection (p.i.). However, the peak titers on days 6 to 7 were comparable for all vectors and exceeded 107 log10 50% tissue culture infective doses (TCID50) per ml. This would permit efficient vaccine manufacture in Vero cells.
Expression of RSV F and HPIV1 proteins in vitro.
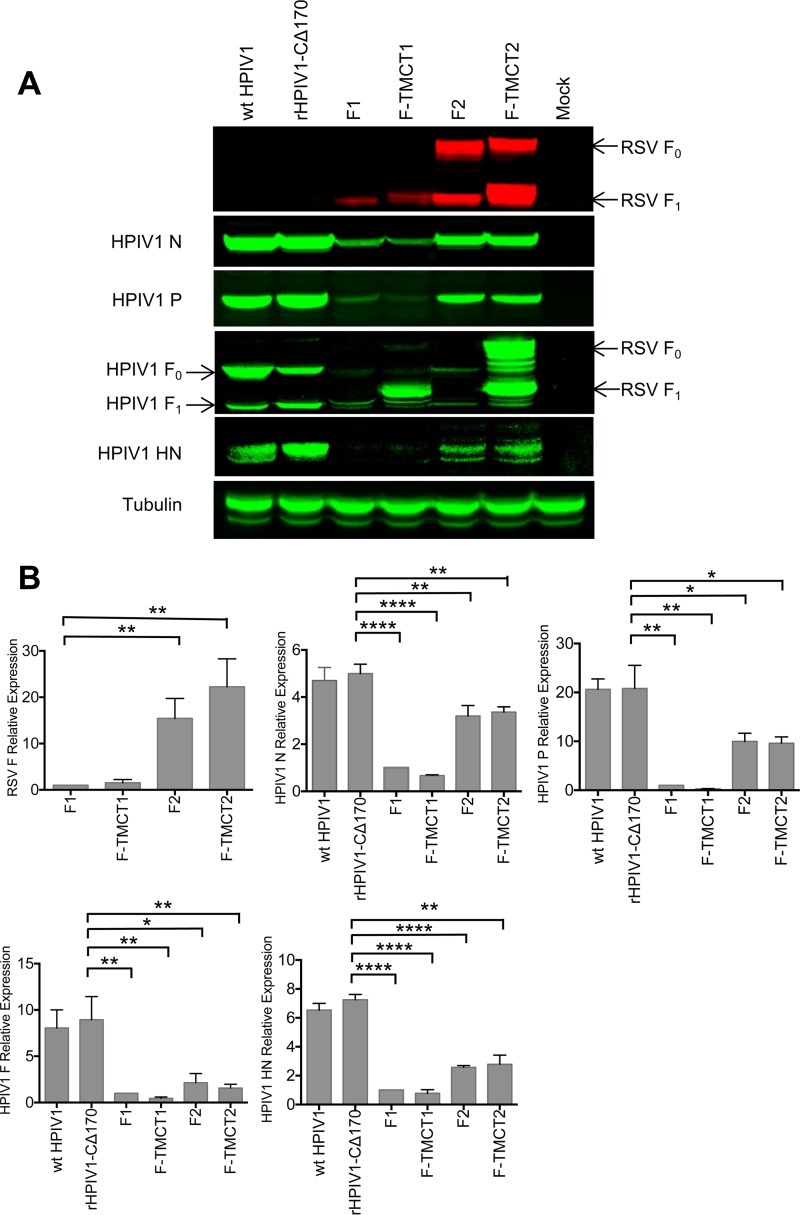
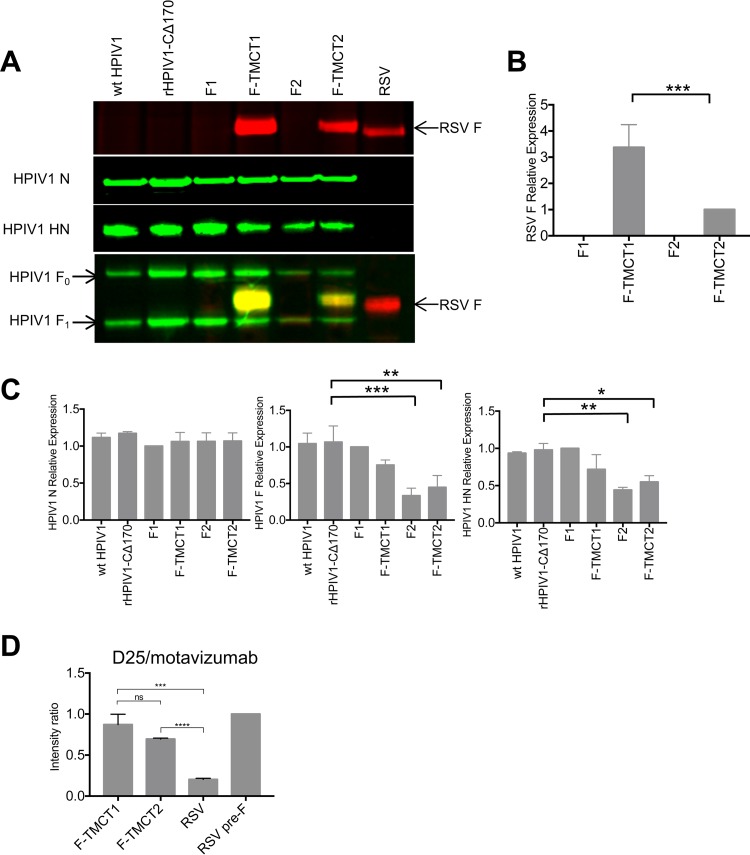
The vectors were evaluated for the expression of RSV F protein and vector proteins in vector-infected Vero cells at 48 h p.i., as analyzed by Western blotting. This showed that F-TMCT2 had the greatest expression (Fig. 3A and B), followed by F2, whereas expression by F1 and F-TMCT1 was substantially reduced. This was contrary to the expectation that, due to the viral transcriptional gradient, a gene in position 1 would be expressed more efficiently than one in position 2.
FIG 3.
Western blot analysis of the expression of RSV F and HPIV1 proteins by the rHPIV1-CΔ170-RSV F vectors. (A) Vero cells were infected with the indicated viruses at an MOI of 5 TCID50 per cell. Cellular lysates were harvested at 48 h p.i. and subjected to gel electrophoresis under reducing and denaturing conditions, followed by Western blotting with a MAb specific for RSV F (top panel, red) and individual anti-peptide sera specific for vector proteins (N, P, F, and HN: second, third, fourth, and fifth panels from the top, respectively, green). Note that the antiserum specific to HPIV1 F protein was raised against a synthetic peptide representing the CT domain and therefore also reacted with the forms of RSV F bearing the HPIV1 TMCT. (B) The intensities of RSV F, and HPIV1 (N, P, F, and HN) protein bands from three independent experiments were quantified, and expression is shown as means ± the SEM relative to the F1 virus (set at a value of 1.0). The statistical significance of the differences between the indicated viruses was analyzed by one-way ANOVA with Dunnett's multiple-comparison test using a 95% confidence interval and is indicated by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
The expression levels of the HPIV1 N, P, F and HN proteins in the same samples were investigated using monospecific sera raised separately against a synthetic peptide from each vector protein. This showed that expression of these vector proteins by the F1, F-TMCT1, F2, and F-TMCT2 vectors was significantly reduced compared to wt HPIV1 and the empty vector. Expression was particularly reduced for the F1 and F-TMCT1 vectors (Fig. 3A and B). Note that the peptide used to generate HPIV1 F-specific antibodies was from the CT domain; hence, this antibody reacted with both the vector HPIV1 F protein and the RSV F-TMCT chimeric proteins (Fig. 3A, fourth panel from the top). Curiously, the use of this C-terminal-specific antibody appeared to detect considerably more RSV F protein from the F-TMCT1 vector (Fig. 3A, fourth panel from the top) compared to the RSV F-specific monoclonal antibody (MAb; top panel). This was also observed in two additional independent repetitions of this experiment (data not shown), but the basis for this is unclear. In the case of the F2 and F-TMCT2 vectors, we also observed reduction in the expression of the N protein, even though its gene is upstream of the inserted RSV F gene. Since transcription of this upstream gene should not be directly affected by a downstream insertion, this reduction in N expression suggests that there also was a reduction in RNA replication, resulting in fewer genome templates (Fig. 3A and B).
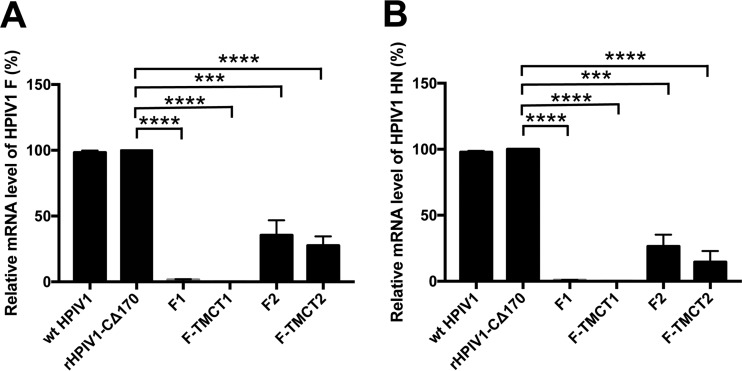
We also investigated the reduced expression of vector genes by quantifying the levels of intracellular HPIV1 F and HN RNA by quantitative real-time PCR (qRT-PCR). This assay was not strand specific or mRNA specific and thus would quantify the aggregate sum of genomic, antigenomic, and mRNA, but in a wt HPIV1-infected cell the amount of mRNA typically is at least 10-fold greater than that of genomic and antigenomic RNA. This analysis showed that the level of intracellular RNA specific to HPIV1 F and HN was substantially reduced (∼5-fold) for the F2 and F-TMCT2 vectors compared to wt HPIV1 and empty vector and was drastically reduced for the F1 and F-TMCT1 vectors (Fig. 4). This confirmed that insertion of the RSV F gene at the pre-N or N-P position indeed reduced downstream gene transcription, with the pre-N insertion having the greater suppressive effect.
FIG 4.
qRT-PCR analyses of HPIV1 F and HN RNA. Vero cells were infected with the indicated viruses at an MOI of 5 TCID50 per cell at 32°C for 48 h. Total RNA was extracted from infected cells and reverse transcribed into cDNA. The transcription levels of HPIV1 F (A) and HN (B) were quantified using Power SYBR green PCR master mix according to the manufacturer's instruction. GAPDH was used as an endogenous control. All samples were analyzed in triplicate and the results are shown as mean level of transcripts ± the SEM relative to rHPIV1-CΔ170 empty vector, whose expression is set at 100%.
Incorporation of RSV F in the rHPIV1 virion particles.
To examine the effects of TMCT modification on the incorporation of RSV F protein into the vector particle, we analyzed sucrose gradient-purified virions to measure the relative levels of RSV F protein and HPIV1 vector proteins. Consistent with the observation that the various vectors had similar yields in the multistep replication kinetics experiment (Fig. 2A), the relative yields (in μg protein) of purified F1, F-TMCT1, F2, and F-TMCT2 vector particles were similar and were ∼2-fold less than for the empty vector (the respective values, normalized to F1, were 1.0, 1.2, 1.0, 1.1, and 2.4). The purified vector particles were lysed, and 0.5 μg of total protein per sample was subjected to Western blotting. As shown in Fig. 5A and C, the amounts of HPIV1 N protein were similar for the vectors, as well as for wt HPIV1 and the empty vector, and were used to normalize the values of the other proteins.
FIG 5.
Western blot analysis of RSV F and HPIV1 proteins in sucrose purified rHPIV1-CΔ170-RSV F vector particles. (A) LLC-MK2 or Vero cells were infected with HPIV1 or wt RSV at an MOI of 0.1 TCID50 or 0.01 PFU per cell, respectively, and incubated at 32°C. The supernatant was collected on day 7 p.i. and centrifuged on discontinuous sucrose gradients to obtain partially purified virus. The virus was lysed in sodium dodecyl sulfate sample buffer, and 0.5 μg of protein per sample was subjected to gel electrophoresis under reducing and denaturing conditions, followed by Western blotting with an MAb specific for RSV F (top panel, red), anti-peptide sera specific for individual vector proteins (N and HN: second and third panels from the top, respectively, green), or a mixture of the RSV F-specific MAb (red) and anti-peptide sera for vector F protein (green) that merge to yellow when they both bind to the chimeric TMCT proteins (fourth panel from the top). (B and C) The intensities of protein bands from three separate Western blots were quantified, and expression is shown relative to the F-TMCT2 virus (B) and F1 virus (C), respectively, as 1.0. The statistical analysis was performed as described for Fig. 3. (D) Relative amounts of RSV F protein in the pre-F conformation in vector and RSV virions. Dot blot immunostaining was performed for the indicated virus stocks, together with a sample of purified recombinant stabilized pre-F DS-Cav1 protein, in triplicate on nitrocellulose membranes with MAbs D25 (specific to pre-F) or motavizumab (specific to both pre- and post-F). The mean ratios ± the SEM of the binding intensities of D25 versus motavizumab are shown, normalized to a value of 1.0 for the purified pre-F protein control. The statistical significance of the differences between the indicated viruses was analyzed by one-way ANOVA with Dunnett's multiple-comparison test using a 95% confidence interval and is indicated by asterisks (***, P < 0.001; ****, P < 0.0001; ns, not significantly different [P > 0.05]).
There was no detectable packaging of full-length RSV F protein into the purified F1 and F2 particles (Fig. 5A and B). In contrast, both the F-TMCT1 and F-TMCT2 particles exhibited significant incorporation that was three times greater for F-TMCT1 than for F-TMCT2 (Fig. 5A and B). In both cases, the amount of packaged RSV F protein per 0.5 μg of virion protein was greater than for wt RSV (Fig. 5A).
The packaging of HPIV1 F and HN was significantly reduced for the F2 and F-TMCT2 vectors, whereas there was little or no reduction for F1 and F-TMCT1, compared to the empty vector or wt HPIV1. As already noted (Fig. 3), the antiserum to the HPIV1 F protein had been raised against a synthetic peptide representing the CT domain and thus also reacted with the chimeric RSV F protein expressed by the F-TMCT1 and F-TMCT2 vectors. The identity of the RSV F band that reacted with the HPIV1 F-specific antiserum was confirmed by costaining with the anti-RSV F MAb (RSV F bands appeared yellow due to merging). This confirmed the identity and nature of the chimeric RSV F protein expressed by the TMCT constructs and packaged in the vector particles.
We also evaluated the relative amount of pre-F protein present in vector and RSV virions. Virus stocks comprised of clarified cell culture medium supernatants from infected LLC-MK2 cells were analyzed by dot blot immunostaining. Virus stocks were dot blotted on nitrocellulose membranes and immunostained with either the MAb motavizumab, which measured total RSV F by binding to site II conserved on both pre-F and post-F, or by MAb D25, which binds site Ø unique to the pre-F conformation (26). Wild-type RSV and HPIV1-CΔ170, as well as an aliquot of previously described purified stabilized RSV pre-F DS-Cav1 protein (15, 18), were included as controls. The results were expressed as a ratio of binding intensity for D25 versus motavizumab and were normalized to the ratio for purified pre-F protein as 1.0 (Fig. 5D). This showed that the relative proportion of packaged RSV F protein present in the pre-F conformation was somewhat higher for F-TMCT1 than for F-TMCT2. The values of D25/motavizumab binding for the vectors were almost as high as for the purified stabilized pre-F protein, suggesting that the vector virions had a high content of pre-F protein. Both vectors had a much higher relative content of pre-F protein than RSV virions.
Cell surface expression of the pre-F form of RSV F.
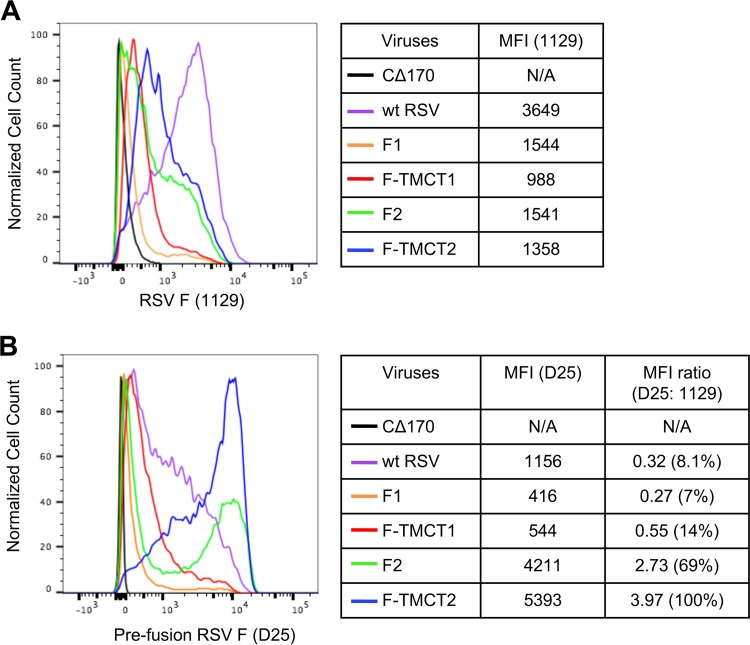
To analyze the expression of RSV F on the cell surface, vector-infected Vero cells were stained with MAb 1129 (the murine precursor of palivizumab and motavizumab) and analyzed by flow cytometry (Fig. 6A). The MAb 1129 recognizes antigenic site II present in both the pre-F and post-F conformations and thus reflects the total amount of RSV F. As indicated by the median fluorescence intensity (MFI), the surface expression of total RSV F protein was greatest for wt RSV (Fig. 6A), whereas that of HPIV1 vectors was relatively lower. Consistent with the Western blots (Fig. 3A), F-TMCT2 viruses demonstrated relatively higher surface expression compared to F-TMCT1. F1 and F2 showed similar intensity of surface expression of RSV F, which was modestly higher than the TMCT versions.
FIG 6.
Cell surface expression of the pre-F RSV F protein. Vero cells were infected with the indicated vectors at an MOI of 5 TCID50/cell or with wt RSV at an MOI of 5 PFU/cell and incubated at 32°C for 48 h. Nonfixed, nonpermeabilized cells were stained with an RSV F mouse MAb 1129 (A), which binds site II present in both pre- and post-F, or with human MAb D25 (B), which is specific for site Ø present only in pre-F. The x axis shows the intensity of RSV F expression, and the y axis is the percentage of cell count normalized to the maximal count in a distribution. The MFI values for MAb 1129 and D25 staining are shown for all viruses, along with their ratios.
Infected cells were also analyzed for pre-F surface expression. This was measured with human MAb D25 that specifically recognizes antigenic site Ø uniquely present in pre-F (14, 27) (Fig. 6B). This showed that the F2 vector expressed substantial cell surface pre-F protein, and this was further enhanced by the TMCT modification in F-TMCT2, while cell surface expression of RSV F was substantially reduced for F-TMCT1 and F1 (Fig. 6B), consistent with their Western blot profile (Fig. 3A). The MFI ratio for wt RSV was lower than that of F-TMCT2 and F2 (Fig. 6B), suggesting that, although wt RSV-infected cells expressed a relatively larger amount of total cell surface RSV F (MAb 1129, Fig. 6A), a much lower proportion of that was present in the pre-F conformation (MAb D25, Fig. 6B).
Replication of vectors in the respiratory tract of hamsters.
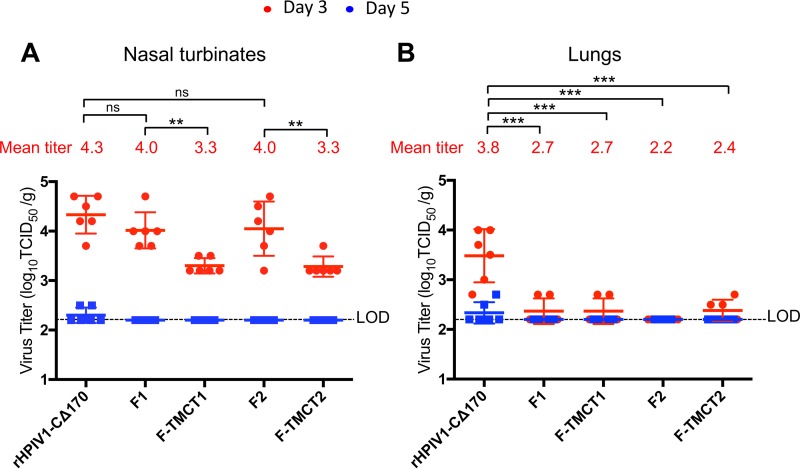
Hamsters were inoculated intranasally (i.n.) with 106 TCID50 of each vector. To evaluate virus replication, nasal turbinates and lungs were collected on days 3 and 5 p.i., and tissues were homogenized and titrated by serial dilution on LLC-MK2 cells (Fig. 7). In the nasal turbinates on day 3 p.i., the F1 and F2 vectors were not attenuated compared to the empty vector, whereas the F-TMCT1 and F-TMCT2 vectors were significantly attenuated (Fig. 7A). In the lungs on day 3, all four vectors showed significant attenuation compared to the empty vector, with titers that were either undetectable or slightly above the detection limit (Fig. 7B). This indicates that the presence of the RSV insert is attenuating in the lower but not in the upper respiratory tract, and the introduction of the additional TMCT modification to the F protein is further attenuating in the upper respiratory tract. The attenuating effect of TMCT modification was not discernible in the lung due to the overall poor replication of all vectors at this site. All vectors expressing RSV F were undetectable on day 5 p.i. in both tissues, whereas trace amounts of the empty vector were detected in both tissues. Analysis of hamster tissue homogenates by fluorescent dual-stained plaque assay showed that for all four vectors, the percentage of PFU that expressed the RSV F protein remained at a level close to 100% after in vivo replication (not shown).
FIG 7.
Replication of the rHPIV1-CΔ170-RSV F vectors in the nasal turbinates (A) and lungs (B) of hamsters. Golden Syrian hamsters (n = 6 per group per day) were inoculated i.n. with 106 TCID50 of the indicated vectors in 100 μl of inoculum. Hamsters were euthanized on day 3 or 5 p.i., and nasal turbinates (A) and lungs (B) were collected and homogenized; the viral titers were determined by limiting dilution on LLC-MK2 cells at 32°C and hemadsorption. The red and blue dots indicate the titers for individual animals euthanized on days 3 and 5, respectively. Mean titers of each group per day are indicated by a horizontal line and for day 3 are also shown numerically as red numbers. The shorter horizontal lines indicate the SEM. The limit of detection (LOD) was 2.2 log10 TCID50/g of tissue, indicated with a dotted line. The statistical significance of the differences between viruses was determined as described for Fig. 3 (**, P ≤ 0.01; ***, P ≤ 0.001; ns, not significantly different [P > 0.05]). Replication of wt HPIV1 in hamsters was determined in previous studies and this control was not included here to minimize the number of animals used. When administered at 106 TCID50 i.n., wt HPIV1 replicates to peak titers of 5 and 4.5 log10 TCID50/g in the nasal turbinates and lungs, respectively (40).
Induction of RSV- and HPIV1-specific serum NAbs.
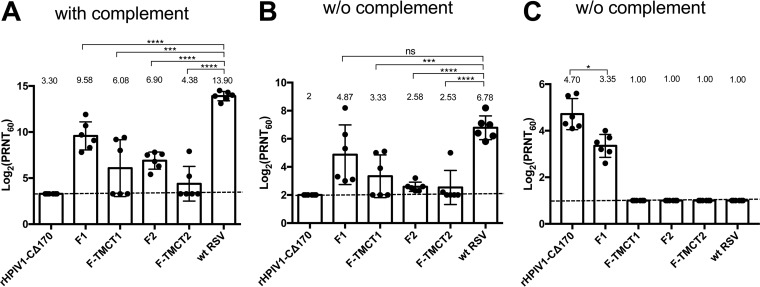
Additional hamsters were inoculated i.n. with 106 TCID50 of the various vectors, and sera were collected on day 28 p.i. The RSV-specific neutralizing antibody (NAb) titers were determined by using a 60% plaque reduction neutralization test (PRNT60) with (Fig. 8A) or without (Fig. 8B) complement. The presence of added complement provides for more sensitive detection of virus-specific antibodies because the presence of complement can confer neutralizing activity by viral lysis and steric hindrance effects (28). In contrast, the complement-independent assay would detect only those antibodies that can directly neutralize the virus, which we dubbed “high-quality” antibodies.
FIG 8.
RSV- and HPIV1-neutralizing serum antibody responses induced by rHPIV1-CΔ170-RSV F vectors. Additional hamsters (n = 6 per group) were immunized as described in Fig. 7, and serum samples were collected on day 28 postimmunization. RSV-neutralizing antibody titers were determined by a PRNT60 performed on Vero cells at 37°C with (A) or without (B) added guinea pig complement. The height of each bar represents the log2 mean titer ± the SEM with mean values shown numerically above each bar. The values of individual animals are shown as dots. The LOD is indicated with a dotted line. The statistical significance of the differences between individual groups and wt RSV was determined as described for Fig. 3 (***, P < 0.001; ****, P < 0.0001; ns, not significantly different [P > 0.05]). (C) HPIV1-neutralizing serum antibody titers were determined by PRNT60 performed on Vero cells at 32°C without guinea pig complement. The values represent means ± the SEM.
In the presence of complement (Fig. 8A), the F1 vector induced the highest mean titer of RSV-specific NAbs, followed, in order of decreasing magnitude, by F2, F-TMCT1, and F-TMCT2. In the absence of complement (Fig. 8B), the mean NAb titer of F1 was the highest, followed by F-TMCT1. Both F2 and F-TMCT2 induced weak complement-independent NAb titers. Overall, the vectors expressing RSV F with TMCT modification induced relatively lower mean NAb titers compared to their full-length RSV F counterparts, which is likely due to their significantly reduced replication in the respiratory tract (Fig. 7A and B). In the presence of complement, the NAb titers of all constructs were significantly lower than that of wt RSV. However, in the complement free assay, the mean titer of NAb induced by F1 was similar to that of wt RSV, suggesting that, despite significant attenuation in the respiratory tract, F1 induced high-quality RSV NAbs comparable in titer to wt RSV.
The HPIV1-specific NAb titers were determined by PRNT60 performed without added complement because the commercial guinea pig complement that we use directly neutralized HPIV1. Only the F1 construct and rHPIV1-CΔ170 empty vector induced detectable HPIV1-specific NAbs. F-TMCT1, F2, and F-TMCT2 failed to induce detectable HPIV1 NAbs. This indicates a high level of restriction of vector replication.
Protection against wt RSV challenge.
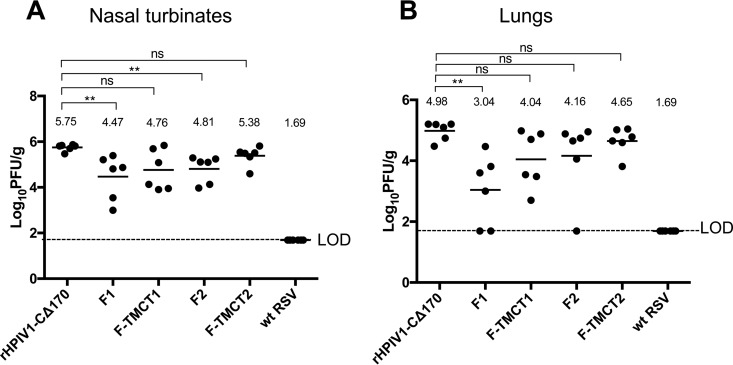
The hamsters immunized in the experiment shown in Fig. 8 were challenged i.n. on day 30 postimmunization with 106 PFU of wt RSV. The nasal turbinates and lungs were collected on day 3 postchallenge, and tissue RSV loads were determined by plaque titration on Vero cells. In the nasal turbinates, the F1 and F2 vectors provided significant restriction of RSV replication (P < 0.01) compared to the empty vector (Fig. 9A). In the lungs, only F1 significantly reduced RSV replication (Fig. 9B), with two of six animals showing no detectable RSV. Constructs with the TMCT modification failed to restrict RSV in either the nasal turbinates or lungs (Fig. 9), which was consistent with their poor replication and weak immunogenicity (Fig. 7 and 8).
FIG 9.
Protection against wt RSV challenge. The immunized hamsters (n = 6 per group) from the experiment described in Fig. 8 were challenged i.n. on day 30 postimmunization with 106 PFU of wt RSV in a 100-μl inoculum. On day 3 postchallenge, the hamsters were euthanized, and nasal turbinates (A) and lungs (B) were collected. The virus load in tissue homogenates was determined by RSV plaque titration on Vero cells at 37°C. Each dot represents the RSV titer for an individual hamster. The mean titer of each group is shown as a horizontal line and also is indicated numerically on the top. The LOD was 1.7 log10 PFU/g of tissue, indicated by a dotted line. The statistical significance of the difference between each group, and the rHPIV1-CΔ170 empty vector was determined as described for Fig. 3 (**, P < 0.01; ns, not significantly different [P > 0.05]).
DISCUSSION
The RSV disease enhancement caused by a formalin-inactivated RSV vaccine tested in the 1960s in RSV-naive infants, together with other factors such as the immunosuppressive effects of maternal antibodies, the immunosuppressive nature of RSV itself, and immaturity of the neonatal immune system, have delayed development of a pediatric RSV vaccine. However, the recent discovery of the molecular structure of the pre-F conformation of RSV F protein and the possibility of increasing the stability of this optimal antigenic form by introducing structure-based mutations provide a new opportunity of developing improved pediatric RSV vaccines based on vectors expressing the RSV pre-F protein (29, 30).
We have been evaluating HPIV1 as a vector system in addition to our previous studies with B/HPIV3 in order to enlarge our armamentarium of vectors, because (i) the properties of HPIV1 as a vector were largely unknown, (ii) different serotype vectors likely can be used one after another with minimal cross-neutralization, and (iii) the optimal use of an HPIV1-based vector would be after 1 year of age, based on the epidemiology of HPIV1, providing a strategy for a boost to RSV immunity in the second year of life. The use of the RSV pre-F protein in this boost would specifically boost the response to the most effective RSV-neutralization epitopes, and the HPIV1 backbone would provide immunization at an appropriate age against HPIV1. The goal of present study was to improve the immunogenicity of RSV F in the attenuated rHPIV1-CΔ170 vector by (i) increasing the stability of the pre-F conformation by introducing the structure-based mutations DS and Cav1 and (ii) introducing the TMCT domain of the HPIV1 F protein in an effort to increase packaging of RSV F in the vector particle.
All four vector constructs expressing full-length or TMCT versions of RSV pre-F from the first or second gene position were readily rescued and replicated efficiently in vitro. However, the F2 vector alone exhibited a high level of instability of expression of RSV pre-F protein: only 1 of 16 independent recoveries had stable expression of RSV pre-F protein. This was unexpected because, in our earlier study (23), instability was not observed for unmodified RSV F expressed from any of the tested sites (F1, F2, and F3) in a nearly identical CΔ170 backbone. However, in that study, instability at the F2 position was observed in the backbone bearing the Y942A attenuating mutation in the L protein. Therefore, in some circumstances, the RSV F insert (unmodified or pre-F) was unstable in the N-P gene junction, but this effect was unpredictable, and its mechanism is unknown. It also was surprising that, once a stable preparation of the F2 vector was identified (and confirmed to have the correct sequence), it remained stable.
Western blot analysis of the expression of RSV pre-F protein in cells infected with the various vectors showed that expression was very low for the F1 and F-TMCT1 vectors compared to F2 and F-TMCT2. This observation was in contrast to the expectation that an insert in the first gene position should have a higher level of expression than from a downstream position, due to the 3′-to-5′ transcription gradient. This very low level of expression by F1/F-TMCT1 also contrasted with our previous study, in which unmodified RSV F was expressed with similar efficiencies from the first, second, and third positions of the HPIV1-CΔ170 vector (23). The HPIV1-CΔ170 vectors in the present study were identical to those used previously by Mackow et al. (23) with one exception: the vectors differed by a small number of nucleotides in the nontranslated region immediately upstream and downstream of the RSV F ORF (Fig. 1A and B) (23), due mainly to differences in restriction sites. Therefore, the possibility exists that these few sequence differences may be responsible for the observed expression differences, although inspection of the sequences did not suggest any deviations from efficient contexts for translation.
In addition to the difference in the magnitude of expression of RSV F protein between the two studies, we note that expression of the vector proteins by the F1/F-TMCT1 vectors in particular and, to a lesser extent, the F2/F-TMCT2 vectors also was substantially reduced in the present study compared to the previous one (23). This suggested another possibility; namely, that the differences in expression are due to differences in vector spread in the monolayer cultures caused by difference in the functionality of the RSV F protein. Specifically, the unmodified RSV F protein expressed in the previous study was fully functional and highly fusogenic, as was evident from the efficient formation of syncytia involving the entire monolayer, whereas the HPIV1 vector alone did not form observable syncytia (23). In contrast, in the present study, RSV F protein stabilized in the pre-F conformation should be nonfunctional, and indeed no syncytia were observed (data not shown). Note that, even though we used an input multiplicity of infection (MOI) of 5 and should have infected every cell, a substantial fraction of cells in cultures infected with the vectors bearing RSV pre-F (nonfunctional) protein expressed RSV F poorly even at 48 h p.i. (e.g., as evident by flow cytometry; Fig. 6A and B), which is suggestive of uneven infection in the cultures. Thus, we speculate that expression of functional RSV F, resulting in fusion between highly expressing cells and their neighbors, would augment the spread and increase the total expression of RSV F and vector proteins. In contrast, the lack of cell-to-cell fusion observed in the present study could be a factor in the reduced vector expression. We suggest that the present study provides the more accurate view of expression from the first gene position: namely, that it results in substantially reduced expression of both the RSV F insert and the vector proteins, particularly early in infection. The present study likely also provides a more realistic view of the effects of insertion into the second gene position, which also resulted in the reduced expression of vector proteins (compared to empty vector and wt HPIV1), although not as reduced as with the first position.
Packaging of full-length RSV pre-F into the HPIV1 vector particles was below the level of detection and evidently very inefficient. In contrast, the F-TMCT chimeric molecules were packaged with an efficiency that was greater, per 0.5 μg of purified virion protein, than for RSV virions. Unexpectedly, the relative amounts of RSV F protein packaged for the F-TMCT1 and F-TMCT2 vectors did not reflect the relative levels of intracellular expression. Specifically, whereas the intracellular expression of RSV F from the F-TMCT1 vector was much lower than for F-TMCT2 (Fig. 3), packaging of RSV F protein by the F-TMCT1 vector was very efficient, and indeed appeared to be somewhat greater than for F-TMCT2 (Fig. 5A, B, and D). The incongruity between the intracellular and packaged levels of RSV F protein was not associated with differences in the yields of progeny vectors. We speculate that this incongruity reflects differences in the timing of the experiments. Specifically, the Western blots showed the total amount of RSV F in the infected cells at 48 h p.i. In this situation, the initial slower replication of F-TMCT1 (Fig. 2A) might result in substantially less accumulation of RSV F and vector proteins compared to F-TMCT2 and empty vector. In contrast, the vector particles were harvested 7 days p.i., a time when the efficiencies of the infections became similar (Fig. 2A). Thus, the two experiments together illustrate the slower initial replication and expression of the F1/F-TMCT1 vectors, which later catch up (at least in vitro) and indeed might have a higher final level of RSV F protein per cell than the F2/F-TMCT2 vectors. Dot blot immunostaining with D25 and motavizumab MAbs to assess the integrity of the pre-F conformation on the virion surface also indicated a modestly higher proportion of pre-F protein in F-TMCT1 than F-TMCT2. The observation that the values of D25/motavizumab for the vectors were almost as high as for the purified stabilized pre-F protein suggested that the vector virions had a high content of pre-F protein. In contrast, the relative proportion of pre-F protein in wt RSV was much lower than for either vector.
When inoculated into the respiratory tract of hamsters, the F1 and F2 vectors were not significantly attenuated in the nasal turbinates compared to the empty vector but showed a significant (P < 0.001) 10-fold reduction in replication in the lungs on day 3. Thus, the presence of the RSV F insert was attenuating in this vector. This likely reflects the added burden of a longer genome and an additional gene, as well as reductions in vector protein expression. It is common for such effects to be more evident in vivo than in vitro. The F-TMCT1 and F-TMCT2 vectors were more attenuated (P < 0.01) than F1 and F2, respectively, in the nasal turbinates on day 3, showing that the TMCT modification was further attenuating. This likely reflects the burden of packaging of an additional surface glycoprotein into the vector particle. All of the vectors bearing RSV F were nearly completely restricted in the lungs on day 3 and were completely restricted in both the nasal turbinates and the lungs on day 5, indicating a high level of attenuation. We previously evaluated the empty vector and versions of F1 and F2 expressing unmodified RSV F protein in hamsters and observed titers that were similar or somewhat greater in the nasal turbinates and essentially the same in the lungs (23).
In hamsters, the F1 vector induced the highest titers of RSV-neutralizing serum antibodies, as analyzed in vitro in the presence or absence of complement. The other three vectors induced low levels of RSV-neutralizing antibodies when analyzed with or without complement. Consistent with their immunogenicity, the F1 and F2 vectors provided significant protection against RSV infection in the nasal turbinates, but only F1 was also protective in the lungs. Also, consistent with their low immunogenicity, F-TMCT1 and F-TMCT2 did not provide significant protection in the nasal turbinates or the lungs. F-TMCT1 and F-TMCT2 showed efficient RSV F virion packaging, most of which had maintained its pre-F conformation, a feature we had previously shown to significantly improve immunogenicity (20). However, in this case these constructs were poorly immunogenic due to their overattenuation in vivo.
Whereas all of the vectors induced detectable RSV-neutralizing serum antibodies in hamsters, only the F1 vector (and empty vector) induced detectable serum neutralizing antibodies against the HPIV1 vector. Thus, the RSV F insert was more immunogenic than the HPIV1 vector. We also note that F1 expressed less RSV F protein than F2 but was comparatively more immunogenic. It could be that F1 was more fit in vivo, since it was also the only vector conferring neutralizing antibodies against the HPIV1 vector. The failure of most of the vectors bearing RSV F to induce detectable vector-specific responses is a clear indication of strongly restricted vector replication, due to (i) the presence of the CΔ170 attenuating mutation, (ii) the attenuating effects of the RSV F insert, (iii) the further attenuating effects of the TMCT modification, when present, and (iv) the semipermissive nature of HPIV1 replication in this rodent model. A more realistic view of the immunogenicity of HPIV1 vectors might be obtained by evaluation in African Green monkeys, which offer a better infection model for HPIV1 (31) compared to hamsters, albeit still semipermissive. Ultimately, however, the characteristics of replication, safety, and immunogenicity of an HPIV1-based vector can only be determined in seronegative children. We recently evaluated, in seronegative children, a live-attenuated HPIV1 vaccine candidate that contained three attenuating elements: (i) the CΔ170 mutation, (ii) missense mutations at amino acids 84 and 533 in the C and HN proteins, respectively, that are attenuating when present together, and (iii) the previously mentioned stabilized missense mutation Y942A in the L protein (10). This candidate was insufficiently infectious and immunogenic in HPIV1-seronegative children, indicating that the three attenuating elements were too restrictive. In the present study, the F1 and F2 vectors contained two attenuating elements (CΔ170 and the F insert), and the F-TMCT1/F-TMCT2 vectors contained in addition a third attenuating element (the TMCT mutation). Therefore, although these vectors may appear to be substantially overattenuated in the available animal models, they provide a range of attenuated candidates that may include one or more that might be suitable in seronegative children. This could be investigated starting with clinical evaluation of the F-TMCT1 vector, which is one of the more attenuated vectors, and proceed by stepwise evaluation in adults, seropositive children, and seronegative children, as described previously (10).
In summary, we demonstrated that the attenuated rHPIV1-CΔ170 vector stably expressed RSV pre-F protein as the full-length form or as a chimera with the ectodomain fused to the TMCT domain of the vector F. The TMCT modification resulted in very efficient incorporation of RSV F in the vector particle but overattenuated the vectors for the semipermissive hamster model, resulting in poor immunogenicity and lack of protection. We expect the attenuated HPIV1 vectors and RSV pre-F inserts to be substantially more immunogenic in a permissive host. Building on experience with a live-attenuated HPIV1 vaccine candidate previously evaluated in seronegative children, the present vectors are promising candidates for clinical evaluation.
MATERIALS AND METHODS
Cells and viruses.
African green monkey Vero cells and rhesus monkey LLC-MK2 cells were maintained in Opti-MEMI medium (Life Technologies, Carlsbad CA) supplemented with 5% fetal bovine serum (FBS; HyClone, Logan, UT) and 1 mM l-glutamine (Life Technologies). BHK BSR T7/5 hamster kidney cells, which constitutively express T7 RNA polymerase, were maintained as described previously (32). HPIV1 sequences were derived from strain Washington/20993/1964 (GenBank accession number AF457102). The RSV F sequence was derived from strain A2 (GenBank accession number KT992094). The wild-type (wt) RSV used in this study was recombinantly derived strain A2 (GenBank accession number KT992094). The wt HPIV1 virus was a biologically derived virus that was identical in sequence to the recombinant derivatives except for the introduced mutations. rHPIV1 viruses were propagated at 32°C in LLC-MK2 cells in serum-free Opti-MEMI medium containing 1.2% trypsin (TrypLE Select; Life Technologies) and 1 mM l-glutamine. RSV was propagated in Vero cells in the presence of Opti-MEMI medium containing 2.5% FBS and 1 mM l-glutamine.
Design of rHPIV1-CΔ170 viruses expressing pre-F form of RSV F protein.
The RSV F ORF was codon optimized (GenScript, Piscataway, NJ) for human expression and was modified by two amino acid substitutions (K66E and Q101P) to be identical in amino acid sequence to an early passage of strain A2 that originally had been propagated in human embryonic kidney cells (HEK F protein) (33). RSV F was stabilized in pre-F conformation by introducing the DS and Cav1 substitutions (15).
Using a previously described reverse genetics system (34), rHPIV1 bearing the CΔ170 attenuating mutation (24) was modified to express RSV F from the pre-N (first position, 3′ proximal to HPIV1 N; Fig. 1A) or N-P (second position, between N and P ORFs; Fig. 1B) gene positions, where it was inserted using the MluI or AscI restriction sites, respectively, as described previously (23). At each site, RSV F was inserted either as a full-length or as a chimeric ORF in which the RSV F TMCT coding sequence was replaced with that of the HPIV1 F protein (Fig. 1C). Four constructs were thus made: full-length or TMCT F protein in the first position (F1 and F-TMCT1) or second position (F2 and F-TMCT2) of the HPIV1 genome.
The genome nucleotide lengths of all engineered viruses conformed to the “rule of six” (35). The RSV F ORF was flanked by the HPIV1 transcription signals to enable transcription as a separate mRNA. Vector genes maintained their wt hexamer phasing except N for the pre-N constructs that had the P gene phasing; the inserts at pre-N and N-P sites had the hexamer phasing of the N and P genes, respectively. All inserts were synthetically derived and cloned into the rHPIV1-CΔ170 antigenome plasmid, and their sequences were confirmed by automated sequencing.
Rescue of rHPIV1-CΔ170 viruses expressing the RSV F protein.
Recombinant viruses were recovered in BHK BSR T7/5 cells as previously described (23) and passaged twice on LLC-MK2 cells. Their sequences were confirmed by automated consensus sequencing of uncloned RT-PCR fragments, as previously described (23).
Fluorescent double-staining plaque assay.
The fluorescent double-staining plaque assay was performed as previously described (23) to determine the stability of RSV F expression by vectors. Briefly, Vero cell monolayers were infected with 10-fold serially diluted virus stocks and then overlaid with culture medium containing 0.8% methylcellulose and 4% trypsin, followed by incubation at 32°C for 6 days. Immunostaining was performed with a mixture of the three RSV F-specific mouse MAbs 1129, 1243, and 1269 and HPIV1-specific goat polyclonal serum, followed by infrared dye-conjugated secondary antibodies as described previously (23). Images were acquired using an Odyssey infrared imaging system (LI-COR, Inc., Lincoln, NE). The plaques showing costaining of HPIV1 proteins and RSV F were pseudocolored to appear green (800 nm) and red (680 nm), respectively, that appeared yellow when merged.
Analysis of RSV F and HPIV1 protein expression by Western blotting.
Vero cells were infected with the viruses at an MOI of 5 TCID50 per cell, incubated at 32°C, and harvested at 48 h p.i. by lysis with 200 μl of LDS sample buffer (Life Technologies). Cell lysates were reduced and denatured at 95°C for 10 min. Total proteins were separated on 4 to 12% Bis-Tris NuPAGE gels (Life Technologies) and transferred onto polyvinylidene difluoride membranes using an iBlot protein transfer system (Life Technologies). Membranes were blocked for 1 h in blocking buffer (LI-COR) and probed with an RSV F-specific murine MAb (Abcam, Cambridge, MA) and the previously described (23) rabbit polyclonal HPIV1 N-specific anti-peptide serum HPIV1-N-485, at a 1:1,000 dilution in blocking buffer. Replicate blots were probed with previously described anti-peptide antisera for HPIV1 P, F, or HN at a 1:200 dilution. Replicate blot probed with anti-tubulin antibody (Cell Signaling Technology) was used as a loading control. After overnight incubation with indicated antibodies, the membranes were washed four times for 5 min each time, followed by incubation for 1 h with secondary antibodies (goat anti-mouse IRDye 680LT and goat anti-rabbit IRDye 800CW [LI-COR]) diluted in LI-COR blocking buffer.
Packaging of RSV F protein in the rHPIV1 particles.
LLC-MK2 cells were inoculated with rHPIV1 at an MOI of 0.1 TCID50 per cell. Vero cells were inoculated with wt RSV at 0.01 PFU per cell. After an incubation for up to 7 days at 32°C, supernatant media containing virus particles was harvested and clarified at 1,000 rpm for 5 min. Viruses were layered onto the top of a discontinuous (30 to 60%) sucrose gradient and centrifuged for 90 min at 26,000 × g. Virion bands were collected and centrifuged again at 13,000 rpm for 1 h. Virion pellets were resuspended in 50 μl of radioimmunoprecipitation assay buffer and protein concentration was determined with a Pierce BCA protein assay kit (Thermo Fisher Scientific). A total 0.5 μg of protein per sample was subjected to polyacrylamide gel electrophoresis under denaturing and reducing conditions, and Western blotting was performed as described above.
The relative amounts of F protein present in the pre-F conformation in vector and RSV virions was assessed by dot blot immunostaining with the site Ø-specific MAb D25, which binds only to pre-F protein, versus the site II-specific MAb motavizumab, which binds to both pre- and post-F. Virus stocks were prepared by infecting cells with rHPIV1 or wt RSV at MOIs of 0.1 TCID50 and 0.01 PFU per cell, respectively. After an incubation for up to 7 days at 32°C, supernatant medium containing virus particles was harvested and clarified at 1,000 rpm for 5 min. To determine an appropriate virus dilution, each virus stock was serially diluted in phosphate-buffered saline (PBS), blotted at equal volumes onto a nitrocellulose membrane, and dried. The membranes were blocked in 5% nonfat dry milk for 1 h at room temperature on a shaker. After three washes in 1× PBS (pH 7.4) containing 0.2% Tween for 10 min each, the membranes were incubated on a shaker for 1 h with 2 μg/ml motavizumab in 5% nonfat dry milk. After washing steps as described above, the membranes were incubated with anti-human horseradish peroxidase-conjugated antibodies diluted at 1:3,000 in 5% nonfat dry milk for 1 h. After washing as described above, the membranes were treated with 1.5 ml of ECL Prime Western blotting detection reagent (GE Healthcare) for 5 min. Images were captured with the GeneSys system. This method identified a dilution for each virus that produced dot signal intensity comparable to that of other viruses and the purified recombinant DS-Cav1 pre-F protein (15, 18) on the same blot. F-TMCT1 and wt RSV were diluted at 1:4 and 1:8, respectively, whereas F-TMCT2 and HPIV1-CΔ170 were used undiluted. Next, 3-μl portions of these viruses and purified recombinant stabilized pre-F DS-Cav1 protein (5 μg/ml) control were blotted in triplicates on duplicate nitrocellulose membranes and assessed for binding either by MAb motavizumab or the MAb D25. Blots were developed as described above. Densitometry readings for each virus were used to determine the ratio of binding of D25 versus motavizumab. The calculated ratios of three replicates were further normalized to those of the purified stabilized pre-F protein control (set at 1.0) and were used to calculate the mean ratio of D25/motavizumab. All reported values per virus represent an average of triplicates.
Analysis of RSV F expression by flow cytometry.
To analyze the stability of RSV F expression by rHPIV1 constructs, Vero cells were infected with rHPIV1 at an MOI of 1 TCID50 per cell or with wt RSV at an MOI of 1 PFU per cell. After incubation for 48 h at 32°C (rHPIV1 constructs) or 37°C (RSV), the infected cells were harvested by incubation in 1 mM EDTA in PBS at 37°C and then washed twice with fluorescence-activated cell sorting buffer (2% FBS in PBS). The cells were stained using a Fixable Aqua dead cell staining kit (Life Technologies) for 30 min and then fixed using a fixation/permeabilization kit (BD Biosciences, San Jose CA). Fixed and permeabilized cells were stained for HPIV1 F protein by incubation with the HPIV1 F-specific MAb HPIV1 F7.1 diluted at 1:500 for 1 h, followed by Alexa Fluor 488-conjugated goat anti-mouse IgG (Life Technologies) diluted at 1: 2,000 for 30 min. The stained cells were washed twice and incubated with Alexa Fluor 647-conjugated RSV F MAb 1129 diluted at 1:500 for 1 h. The stained cells were analyzed by using a BD FACSCanto II flow cytometer.
To determine the expression of pre-F form of RSV F on the infected cell surface, Vero cells were infected with rHPIV1 at an MOI of 5 TCID50 per cell or with wt RSV at an MOI of 5 PFU per cell. Cells were stained using a Fixable Aqua dead cell staining kit for 30 min, washed, and then incubated with an optimized dilution of unconjugated D25, a pre-F site Ø-specific human MAb, or Alexa Fluor 647-conjugated RSV F site II-specific MAb 1129. After a washing step, the cells stained with D25 were incubated with Alexa Fluor 488-conjugated goat anti-human IgG (Life Technologies) for 30 min. The Alexa Fluor 647-conjugated RSV F MAb was prepared using an Alexa Fluor 647 antibody labeling kit (Life Technologies). The stained cells were washed twice and analyzed using a BD FACSCanto II flow cytometer, and data analysis was performed using FlowJo software version 10.1 (Ashland OR).
qRT-PCR.
Vero cells were infected with the viruses using the same conditions as described for Western blot analysis. Total RNA was extracted using TRIzol Reagent (Life Technologies), and cDNA was reverse transcribed using SuperScript III first-strand synthesis system (Life Technologies). The relative transcription levels of HPIV1 F and HN were quantitated by real-time PCR using Power SYBR green PCR master mix (Life Technologies) according to manufacturer's manual. The oligonucleotide primers used in this assay were as follows: HPIV1 F, forward (5′-CATCCTTAGGAGGTGCAGATGT-3′) and reverse (5′-GATGCAATGCAATTAGCCACTA-3′); HPIV1 HN, forward (5′-TGCCGAAGGTAGGCTACTTAAA-3′) and reverse (5′-TATGCATTCTCTCGGACATCTG-3′); and GAPDH, forward (5′-ATGAGAAGTATGACAACAGCCTCAAG-3′) and reverse (5′-ATGAGTCCTTCCACGATACCAAAGTT-3′).
All PCRs were run in triplicate on 7900HT fast real-time PCR system (Applied Biosystems), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as an endogenous control. The F and HN threshold cycle (CT) values obtained for the rHPIV1-CΔ170 empty vector were used as the calibrator for calculating the relative fold change in F and HN expression for the rest of the viruses. Fold change values were converted to relative percent expression keeping rHPIV1-CΔ170 at 100%.
Evaluation of vaccine candidates in hamsters: replication, immunogenicity, and protective efficacy against wt RSV challenge.
All animal studies were approved by the NIH Institutional Animal Care and Use Committee (IACUC). Six-week-old Golden Syrian hamsters (Envigo Laboratories, Frederick, MD) were confirmed to be seronegative for HPIV1 and RSV by hemagglutination inhibition assay and RSV neutralization assay, respectively (36, 37). To evaluate virus replication, hamsters were anesthetized and inoculated i.n. with 0.1 ml of L15 medium (Life Technologies) containing 106 TCID50 of virus per animal. Six hamsters were euthanized on days 3 and 5 postinoculation, and nasal turbinates and lungs were collected. To assess virus replication, nasal turbinates and lungs were homogenized, and clarified homogenates were titrated by serial dilution on LLC-MK2 cells and hemadsorption with guinea pig erythrocytes. Virus titers are reported as the log10 TCID50/g of tissue. The stability of RSV F expression after in vivo replication was determined for all vectors by performing dual-stained fluorescent plaque assay with tissue homogenates. To assess vector immunogenicity, six hamsters per group were immunized i.n. as described above with 106 TCID50 of the various vectors or 106 PFU of wt RSV, and sera were collected on day 28 postinoculation. RSV- and HPIV1-specific virus serum NAb titers were determined by PRNT60 on Vero cells as previously described (23, 37) using recombinant RSV and HPIV1 expressing green fluorescent protein, respectively, and are reported as the log2 PRNT60 (23, 38). RSV NAb assay was performed in the presence or absence of guinea pig complement (Lonza, Walkersville, MD), while that for HPIV1 was performed only without complement. The protective efficacy of the vaccine candidates against RSV infection was determined by challenge infection of hamsters on day 30 postimmunization: hamsters were inoculated i.n. with 0.1 ml L15 medium containing 106 PFU of wt RSV strain A2. Hamsters were euthanized, and nasal turbinates and lungs were collected on day 3 postchallenge. The RSV load in the respiratory tract was determined by titrating RSV in the nasal turbinate and lung homogenates by plaque assay on Vero cells (39).
ACKNOWLEDGMENTS
This study was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID) and by the National Institutes of Health (NIH) and was supported in part by a Cooperative Research and Development Agreement between NIAID, NIH, and Sanofi Pasteur, Inc.
We thank the Comparative Medicine Branch, NIAID, NIH (Bethesda, MD), for technical assistance in the hamster experiments and for the care and management of the animals. We thank Ursula Buchholz for providing BHK BSR T7/5 cells for virus rescues. We also thank Mario Skiadopoulos and Brian R. Murphy for providing the anti-peptide antisera against the HPIV1 N, P, F, and HN proteins.
REFERENCES
- 1.Afonso CL, Amarasinghe GK, Banyai K, Bao Y, Basler CF, Bavari S, Bejerman N, Blasdell KR, Briand FX, Briese T, Bukreyev A, Calisher CH, Chandran K, Cheng J, Clawson AN, Collins PL, Dietzgen RG, Dolnik O, Domier LL, Durrwald R, Dye JM, Easton AJ, Ebihara H, Farkas SL, Freitas-Astua J, et al. 2016. Taxonomy of the order Mononegavirales: update 2016. Arch Virol 161:2351–2360. doi: 10.1007/s00705-016-2880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, et al. 2012. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stockman LJ, Curns AT, Anderson LJ, Fischer-Langley G. 2012. Respiratory syncytial virus-associated hospitalizations among infants and young children in the United States, 1997-2006. Pediatr Infect Dis J 31:5–9. doi: 10.1097/INF.0b013e31822e68e6. [DOI] [PubMed] [Google Scholar]
- 5.Kapikian AZ, Mitchell RH, Chanock RM, Shvedoff RA, Stewart CE. 1969. An epidemiologic study of altered clinical reactivity to respiratory syncytial (RS) virus infection in children previously vaccinated with an inactivated RS virus vaccine. Am J Epidemiol 89:405–421. doi: 10.1093/oxfordjournals.aje.a120954. [DOI] [PubMed] [Google Scholar]
- 6.Ottolini MG, Porter DD, Hemming VG, Prince GA. 2000. Enhanced pulmonary pathology in cotton rats upon challenge after immunization with inactivated parainfluenza virus 3 vaccines. Viral Immunol 13:231–236. doi: 10.1089/vim.2000.13.231. [DOI] [PubMed] [Google Scholar]
- 7.Murphy BR, Sotnikov AV, Lawrence LA, Banks SM, Prince GA. 1990. Enhanced pulmonary histopathology is observed in cotton rats immunized with formalin-inactivated respiratory syncytial virus (RSV) or purified F glycoprotein and challenged with RSV 3 to 6 months after immunization. Vaccine 8:497–502. doi: 10.1016/0264-410X(90)90253-I. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein DI, Malkin E, Abughali N, Falloon J, Yi T, Dubovsky F, Investigators M-C. 2012. Phase 1 study of the safety and immunogenicity of a live, attenuated respiratory syncytial virus and parainfluenza virus type 3 vaccine in seronegative children. Pediatr Infect Dis J 31:109–114. doi: 10.1097/INF.0b013e31823386f1. [DOI] [PubMed] [Google Scholar]
- 9.Karron RA, Luongo C, Thumar B, Loehr KM, Englund JA, Collins PL, Buchholz UJ. 2015. A gene deletion that upregulates viral gene expression yields an attenuated RSV vaccine with improved antibody responses in children. Sci Transl Med 7:312ra175. doi: 10.1126/scitranslmed.aac8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karron RA, Mateo JC, Thumar B, Schaap-Nutt A, Buchholz UJ, Schmidt AC, Bartlett EJ, Murphy BR, Collins PL. 2014. Evaluation of a live-attenuated human parainfluenza type 1 vaccine in adults and children. J Pediatric Infect Dis Soc 4:e143–e146. doi: 10.1093/jpids/piu104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright PF, Karron RA, Belshe RB, Shi JR, Randolph VB, Collins PL, O'Shea AF, Gruber WC, Murphy BR. 2007. The absence of enhanced disease with wild-type respiratory syncytial virus infection occurring after receipt of live, attenuated, respiratory syncytial virus vaccines. Vaccine 25:7372–7378. doi: 10.1016/j.vaccine.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartlett EJ, Cruz AM, Esker J, Castano A, Schomacker H, Surman SR, Hennessey M, Boonyaratanakornkit J, Pickles RJ, Collins PL, Murphy BR, Schmidt AC. 2008. Human parainfluenza virus type 1 C proteins are nonessential proteins that inhibit the host interferon and apoptotic responses and are required for efficient replication in nonhuman primates. J Virol 82:8965–8977. doi: 10.1128/JVI.00853-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liljeroos L, Krzyzaniak MA, Helenius A, Butcher SJ. 2013. Architecture of respiratory syncytial virus revealed by electron cryotomography. Proc Natl Acad Sci U S A 110:11133–11138. doi: 10.1073/pnas.1309070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, Kumar A, Modjarrad K, Zheng Z, Zhao M, Xia N, Kwong PD, Graham BS. 2013. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLellan JS, Chen M, Joyce MG, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Chen L, Srivatsan S, Zheng A, Zhou T, Graepel KW, Kumar A, Moin S, Boyington JC, Chuang GY, Soto C, Baxa U, Bakker AQ, Spits H, Beaumont T, Zheng Z, Xia N, Ko SY, Todd JP, Rao S, Graham BS, Kwong PD. 2013. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swanson KA, Settembre EC, Shaw CA, Dey AK, Rappuoli R, Mandl CW, Dormitzer PR, Carfi A. 2011. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc Natl Acad Sci U S A 108:9619–9624. doi: 10.1073/pnas.1106536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLellan JS, Yang Y, Graham BS, Kwong PD. 2011. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol 85:7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ngwuta JO, Chen M, Modjarrad K, Joyce MG, Kanekiyo M, Kumar A, Yassine HM, Moin SM, Killikelly AM, Chuang GY, Druz A, Georgiev IS, Rundlet EJ, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Nason MC, Capella C, Peeples ME, Ledgerwood JE, McLellan JS, Kwong PD, Graham BS. 2015. Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci Transl Med 7:309ra162. doi: 10.1126/scitranslmed.aac4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang B, Munir S, Amaro-Carambot E, Surman S, Mackow N, Yang L, Buchholz UJ, Collins PL, Schaap-Nutt A. 2014. Chimeric bovine/human parainfluenza virus type 3 expressing respiratory syncytial virus (RSV) F glycoprotein: effect of insert position on expression, replication, immunogenicity, stability, and protection against RSV infection. J Virol 88:4237–4250. doi: 10.1128/JVI.03481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang B, Ngwuta JO, Herbert R, Swerczek J, Dorward DW, Amaro-Carambot E, Mackow N, Kabatova B, Lingemann M, Surman S, Yang L, Chen M, Moin SM, Kumar A, McLellan JS, Kwong PD, Graham BS, Schaap-Nutt A, Collins PL, Munir S. 2016. Packaging and prefusion stabilization separately and additively increase the quantity and quality of RSV-neutralizing antibodies induced by respiratory syncytial virus (RSV) fusion protein expressed by a parainfluenza vector. J Virol 90:10022–10038. doi: 10.1128/JVI.01196-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang B, Surman S, Amaro-Carambot E, Kabatova B, Mackow N, Lingemann M, Yang L, McLellan JS, Graham BS, Kwong PD, Schaap-Nutt A, Collins PL, Munir S. 2015. Enhanced neutralizing antibody response induced by respiratory syncytial virus pre-fusion F protein expressed by a vaccine candidate. J Virol 89:9499–9510. doi: 10.1128/JVI.01373-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reed G, Jewett PH, Thompson J, Tollefson S, Wright PF. 1997. Epidemiology and clinical impact of parainfluenza virus infections in otherwise healthy infants and young children <5 years old. J Infect Dis 175:807–813. doi: 10.1086/513975. [DOI] [PubMed] [Google Scholar]
- 23.Mackow N, Amaro-Carambot E, Liang B, Surman S, Lingemann M, Yang L, Collins PL, Munir S. 2015. Attenuated human parainfluenza virus type 1 (HPIV1) expressing the fusion glycoprotein of human respiratory syncytial virus (RSV) as a bivalent HPIV1/RSV vaccine. J Virol 89:10319–10332. doi: 10.1128/JVI.01380-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartlett EJ, Castano A, Surman SR, Collins PL, Skiadopoulos MH, Murphy BR. 2007. Attenuation and efficacy of human parainfluenza virus type 1 (HPIV1) vaccine candidates containing stabilized mutations in the P/C and L genes. Virol J 4:67. doi: 10.1186/1743-422X-4-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartlett EJ, Amaro-Carambot E, Surman SR, Collins PL, Murphy BR, Skiadopoulos MH. 2006. Introducing point and deletion mutations into the P/C gene of human parainfluenza virus type 1 (HPIV1) by reverse genetics generates attenuated and efficacious vaccine candidates. Vaccine 24:2674–2684. doi: 10.1016/j.vaccine.2005.10.047. [DOI] [PubMed] [Google Scholar]
- 26.McLellan JS. 2015. Neutralizing epitopes on the respiratory syncytial virus fusion glycoprotein. Curr Opin Virol 11:70–75. doi: 10.1016/j.coviro.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwakkenbos MJ, Diehl SA, Yasuda E, Bakker AQ, van Geelen CM, Lukens MV, van Bleek GM, Widjojoatmodjo MN, Bogers WM, Mei H, Radbruch A, Scheeren FA, Spits H, Beaumont T. 2010. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat Med 16:123–128. doi: 10.1038/nm.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoder SM, Zhu Y, Ikizler MR, Wright PF. 2004. Role of complement in neutralization of respiratory syncytial virus. J Med Virol 72:688–694. doi: 10.1002/jmv.20046. [DOI] [PubMed] [Google Scholar]
- 29.Giersing BK, Modjarrad K, Kaslow DC, Moorthy VS, WHO Product Development for Vaccines Advisory Committee. 2016. Report from the World Health Organization's Product Development for Vaccines Advisory Committee (PDVAC) meeting, Geneva, 7–9th Sep 2015. Vaccine 34:2865–2869. doi: 10.1016/j.vaccine.2016.02.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Modjarrad K, Giersing B, Kaslow DC, Smith PG, Moorthy VS, WHO RSV Vaccine Consultation Expert Group. 2016. WHO consultation on Respiratory Syncytial Virus Vaccine Development Report from a World Health Organization Meeting held on 23–24 March 2015. Vaccine 34:190–197. doi: 10.1016/j.vaccine.2015.05.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durbin AP, Elkins WR, Murphy BR. 2000. African green monkeys provide a useful nonhuman primate model for the study of human parainfluenza virus types-1, -2, and -3 infection. Vaccine 18:2462–2469. doi: 10.1016/S0264-410X(99)00575-7. [DOI] [PubMed] [Google Scholar]
- 32.Buchholz UJ, Finke S, Conzelmann KK. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol 73:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitehead SS, Juhasz K, Firestone CY, Collins PL, Murphy BR. 1998. Recombinant respiratory syncytial virus (RSV) bearing a set of mutations from cold-passaged RSV is attenuated in chimpanzees. J Virol 72:4467–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newman JT, Surman SR, Riggs JM, Hansen CT, Collins PL, Murphy BR, Skiadopoulos MH. 2002. Sequence analysis of the Washington/1964 strain of human parainfluenza virus type 1 (HPIV1) and recovery and characterization of wild-type recombinant HPIV1 produced by reverse genetics. Virus Genes 24:77–92. doi: 10.1023/A:1014042221888. [DOI] [PubMed] [Google Scholar]
- 35.Kolakofsky D, Pelet T, Garcin D, Hausmann S, Curran J, Roux L. 1998. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol 72:891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clements ML, Belshe RB, King J, Newman F, Westblom TU, Tierney EL, London WT, Murphy BR. 1991. Evaluation of bovine, cold-adapted human, and wild-type human parainfluenza type 3 viruses in adult volunteers and in chimpanzees. J Clin Microbiol 29:1175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coates HV, Alling DW, Chanock RM. 1966. An antigenic analysis of respiratory syncytial virus isolates by a plaque reduction neutralization test. Am J Epidemiol 83:299–313. doi: 10.1093/oxfordjournals.aje.a120586. [DOI] [PubMed] [Google Scholar]
- 38.Munir S, Le Nouen C, Luongo C, Buchholz UJ, Collins PL, Bukreyev A. 2008. Nonstructural proteins 1 and 2 of respiratory syncytial virus suppress maturation of human dendritic cells. J Virol 82:8780–8796. doi: 10.1128/JVI.00630-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luongo C, Winter CC, Collins PL, Buchholz UJ. 2013. Respiratory syncytial virus modified by deletions of the NS2 gene and amino acid S1313 of the L polymerase protein is a temperature-sensitive, live-attenuated vaccine candidate that is phenotypically stable at physiological temperature. J Virol 87:1985–1996. doi: 10.1128/JVI.02769-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartlett EJ, Amaro-Carambot E, Surman SR, Newman JT, Collins PL, Murphy BR, Skiadopoulos MH. 2005. Human parainfluenza virus type I (HPIV1) vaccine candidates designed by reverse genetics are attenuated and efficacious in African green monkeys. Vaccine 23:4631–4646. doi: 10.1016/j.vaccine.2005.04.035. [DOI] [PubMed] [Google Scholar]