ABSTRACT
Exposure to dengue virus (DENV) is thought to elicit lifelong immunity, mediated by DENV-neutralizing antibodies (nAbs). However, Abs generated by primary infections confer serotype-specific protection, and immunity against other serotypes develops only after subsequent infections. Accordingly, the induction of these nAb responses acquired after serial DENV infections has been a long-sought-after goal for vaccination. Nonetheless, it is still unclear if tetravalent vaccines can elicit or recall nAbs. In this study, we have characterized the responses from a volunteer who had been previously exposed to DENV and was immunized with the live attenuated tetravalent vaccine Butantan-DV, developed by the NIH and Butantan Institute. Eleven days after vaccination, we observed an ∼70-fold expansion of the plasmablast population. We generated 21 monoclonal Abs (MAbs) from singly sorted plasmablasts. These MAbs were the result of clonal expansions and had significant levels of somatic hypermutation (SHM). Nineteen MAbs (90.5%) neutralized at least one DENV serotype at concentrations of 1 μg/ml or less; 6 of the 21 MAbs neutralized three or more serotypes. Despite the tetravalent composition of the vaccine, we observed a neutralization bias in the induced repertoire: DENV3 was targeted by 18 of the 19 neutralizing MAbs (nMAbs). Furthermore, the P3D05 nMAb neutralized DENV3 with extraordinary potency (concentration to achieve half-maximal neutralization [Neut50] = 0.03 μg/ml). Thus, the Butantan-DV vaccine engendered a mature, antigen-selected B cell repertoire. Our results suggest that preexisting responses elicited by a previous DENV3 infection were recalled by immunization.
IMPORTANCE The dengue epidemic presents a global public health challenge that causes widespread economic burden and remains largely unchecked by existing control strategies. Successful control of the dengue epidemic will require effective prophylactic and therapeutic interventions. Several vaccine clinical efficacy trials are approaching completion, and the chances that one or more live attenuated tetravalent vaccines (LATVs) will be introduced worldwide is higher than ever. While it is widely accepted that dengue virus (DENV)-neutralizing antibody (nAb) titers are associated with protection, the Ab repertoire induced by LATVs remain uncharacterized. Here, we describe the isolation of potent (Neut50 < 0.1 μg/ml) nAbs from a DENV-seropositive volunteer immunized with the tetravalent vaccine Butantan-DV, which is currently in phase III trials.
KEYWORDS: B cell, Butantan-DV, TV003, dengue, monoclonal antibodies, plasmablast, vaccines
INTRODUCTION
Infection with dengue virus (DENV) remains a major cause of morbidity and mortality worldwide. The four serotypes of DENV infect approximately 400 million individuals every year (1). With more than 100 countries affected and over 3 billion people at risk of DENV infection, the development of a safe and efficacious DENV vaccine is an international priority. Several vaccines have advanced through clinical development, with the live attenuated tetravalent vaccines (LATVs) by Sanofi Pasteur (2), NIH (3), and Takeda (4) showing some promise.
The goal of most DENV vaccine candidates is to induce the natural immunity observed after multiple exposures to different serotypes of DENV. The prevailing thought is that individuals exposed to DENV serotypes in a serial manner will have lifelong protection against infection with any DENV. In contrast, exposure to a single DENV serotype protects only against reinfections with the same serotype (homotypic) and not against different DENV serotypes (heterotypic). Secondary infections with a heterotypic serotype are, however, associated with a higher risk of severe disease. This pathogenic process, likely mediated by DENV-specific antibodies, is referred to as antibody (Ab)-dependent enhancement (ADE) (5, 6). Thus, the primary challenge in creating a safe vaccine against DENV is engendering immunity against all four unique serotypes similar to that in serially exposed individuals. Several vaccine approaches are attempting to simultaneously induce immunity against all four DENV serotypes (DENV1 to DENV4).
In order to induce a tetravalent response, the Sanofi Pasteur (2), NIH (3), and Takeda (4) LATVs are composed of four different constructs, given at the same time. Unfortunately, it has been challenging to achieve balanced immunity against all four serotypes concurrently (7–10). Results from the Sanofi Pasteur, NIH, and Takeda preclinical and clinical trials show that each of the four attenuated viruses in the tetravalent formulations has different viremia levels, kinetics, and response magnitudes. Furthermore, these LATVs will be delivered to a population with various levels preexisting immunity from previous encounters with diverse DENV serotypes. This creates additional complexities for developing an effective vaccine. Thus, to engender anti-DENV immunity by vaccination, one needs to consider the interactions of the four vaccine viruses, the host's serological status, and the likely challenge virus serotype.
The Sanofi Pasteur LATV has shown some degree of protection in efficacy studies. Nonetheless, this vaccine-induced protection depended on the infecting DENV serotype, serological status at vaccination, and age (11). In contrast, the NIH LATV was 100% efficacious in preventing DENV2 infections in a human challenge study (12). Phase III clinical studies for the NIH and Takeda LATVs are ongoing, and their performances in the population remain to be evaluated. A number of questions persist from the Sanofi Pasteur LATV clinical trials. Why was the incidence of hospitalization higher in vaccinees younger than 9 years? Why was vaccine efficacy greater in seropositive individuals? Why was the protection against DENV2 reduced compared to that against other DENV serotypes? These questions are also likely to be relevant to the NIH and Takeda vaccines. While there are no clear answers to these questions yet, an analysis and characterization of the antibody repertoire elicited after vaccination may give some insights (13).
Recent advances in monoclonal Ab (MAb) isolation now allow us to characterize B cell repertoires in more detail at the single-cell level (14). Analyses of single memory B cells suggest that DENV neutralization activity in serum is likely due to a few neutralizing Abs (nAbs) instead of a pool of weakly reactive nAbs (15, 16). Additionally, studies of the primary B cell repertoire against DENV revealed both DENV serotype-specific nAbs and cross-serotype-reactive Abs, conferring different levels of cross-serotype-neutralization (15, 16). Higher titers cross-serotype-neutralizing Ab in serum have been associated with a reduced risk of severe disease in longitudinal studies (17). In contrast, low neutralization titers have been associated with enhanced secondary disease (5, 6). Nonetheless, the majority of these cross-reactive Abs elicited in primary DENV infection lack neutralization function (15, 16, 18–20). These weakly cross-neutralizing Abs elicited by primary infection may, however, prime the B cell repertoire that is engaged during a subsequent exposure. Analysis of secondary repertoires shows a broadening of neutralization activity, increase in somatic hypermutation (SHM) levels, and neutralization potency. In fact, several potent (concentration to achieve half-maximal neutralization [Neut50] < 0.1 μg/ml) and broadly DENV-neutralizing Abs have been isolated from secondary DENV infections (18, 20–22). Although immune correlates of infection- or vaccine-induced protection have not been definitely established, DENV nAb titers in serum have been largely associated with protection (23). Therefore, vaccination should, ideally, induce potent nAbs similar to those observed after secondary infection (13). Unfortunately, the only study describing the B cell repertoire induced by vaccination (using only the DENV1 component of the NIH LATV) showed a high frequency of B cells encoding weakly neutralizing nAbs, and no potent nAbs were isolated (24). Critically, the B cell repertoire induced by tetravalent vaccine mixtures, as opposed to exposure to a single serotype immunogen, remains unexplored.
The focus in this study was whether nAb-producing B cells are expanded after immunization with the LATV Butantan-DV. This vaccine was produced by the Butantan Institute, with the same vaccine vectors as used in the NIH LATV (3). It is possible to envision multiple possible scenarios of memory B cell activation following vaccination of seropositive individuals; for example, responses against a particular serotype may dominate the humoral responses. Alternatively, it is possible that simultaneous exposure to all four constructs would favor the activation of broadly DENV-reactive B cells.
To investigate the Ab repertoire induced by Butantan-DV, we collected plasmablasts from a DENV-seropositive volunteer (2028) following vaccination. The volunteer had measurable prevaccination serum neutralizing activity against DENV3 and, to a lesser extent, the other serotypes. A robust expansion (∼70-fold) of the plasmablast population was observed postvaccination. The serum neutralization titer against DENV3 quickly increased, a hallmark of anamnestic responses. At day 91 postvaccination, neutralizing activity against all four serotypes was detected in serum. From sorted single plasmablasts (day 15 postvaccination), we generated 21 MAbs and have characterized their abilities to neutralize DENV serotypes 1 to 4. Nineteen of the 21 MAbs (90.5%) neutralized DENV (Neut50 < 1 μg/ml). Eighteen of the 19 nAbs neutralized the DENV3 serotype; 11 nAbs neutralized two or more DENV serotypes. Finally, we isolated a few nAbs exhibiting exceptional potency (Neut50 < 0.1 μg/ml). Of the 21 MAbs, we identified P3D05, a serotype-specific nAb that potently neutralizes DENV3 (Neut50 = 0.03 μg/ml). In summary, here we show that vaccinating a DENV-exposed volunteer with the NIH LATV activated nAb-producing B cells. This is the first report of the isolation of potent nAbs after LATV immunization.
RESULTS
Vaccination of DENV-exposed volunteer 2028 induces broadly reactive DENV-neutralizing serum activity.
To study the Butantan-DV-induced nAb repertoire, we collected B cells from volunteers participating in a double-blinded phase II trial to evaluate the safety and immunogenicity of the NIH LATV vaccine produced by the Butantan Institute (Clinical Trials.gov identifier NCT01696422). The Butantan-DV vaccine contains attenuated DENV1, DENV2, DENV3, and DENV4 components given in a single mixture subcutaneously (s.c.) at a dose of 1,000 PFU of each virus. This phase of the study was designed with a ratio of 3:1 individuals in the vaccine to placebo groups. After vaccination, some of the volunteers presented a maculopapular rash (data not shown). This mild, known adverse event of dengue immunization was previously reported as a correlate of seroconversion to all four DENV serotypes (3, 12, 25). Since the rash was associated with vaccine take, we focused our study on individuals that presented this symptom postimmunization. Thus, without a priori knowledge of the vaccination statuses, volunteer 2028 (Table 1) was selected based on the development of a rash presented at day 11 postimmunization. Following vaccination, we observed a robust increase of DENV-neutralizing activity in serum to all four serotypes (Table 1). At day 91, the titers of vaccine-induced nAbs against the DENV3 serotype were 20-fold higher than titers of nAbs against other serotypes. This suggests a classic anamnestic response, with the recall of preexisting B cells against DENV3 and B cells targeting DENV1 and -2 to a lesser extent. In summary, these results show that Butantan-DV vaccination of previously DENV-exposed volunteer 2028 was immunogenic and induced nAb responses against all four DENV serotypes.
TABLE 1.
Details for volunteer 2028
| Status at trial enrollment | Vaccinea | Serotype | Serum neutralization titer (Neut50) |
|
|---|---|---|---|---|
| Prevaccination | Day 91 | |||
| DENV ELISA reactive | Butantan-DV (DENV1–4, 1,000 PFU, s.c.) | D1 | 56 | 1,102 |
| D2 | 83 | 865 | ||
| D3 | 417 | 20,192 | ||
| D4 | 79 | 114 | ||
Vaccine lots were produced by the Butantan Institute.
Robust vaccine-induced plasmablast expansion.
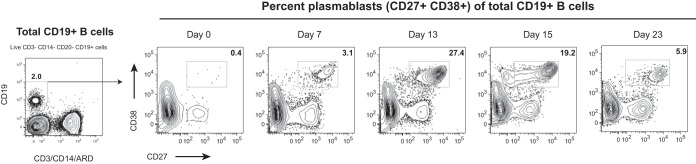
DENV infection typically leads to massive expansion of antigen-specific B cells (22, 26, 27). These B cell responses can be used to study the effects of vaccination, as acute plasmablast frequency and specificity are thought to be associated with the development of long-term nAb titers. We have thus monitored the expansion of plasmablasts (live CD3− CD14− CD19+ CD27high CD38high lymphocytes) postvaccination. While the plasmablast responses have been well characterized for dengue virus infections, less is known for the generation of responses postvaccination with attenuated viruses. To our surprise, we detected a very robust expansion of plasmablasts, starting as early as day 7 and peaking at day 13 postimmunization (Fig. 1). At peak, the expansion of this population was approximately 70-fold over the baseline level.
FIG 1.
Plasmablasts expand after Butantan-DV vaccination. Blood samples were collected longitudinally from volunteer 2028, and the frequency of plasmablasts postimmunization was analyzed by flow cytometry. The numbers of plasmablasts in fresh whole blood are expressed as the percent CD27high CD38high lymphocytes among total live CD19+ CD3− CD14− B cells.
Butantan-DV-induced plasmablast repertoire.
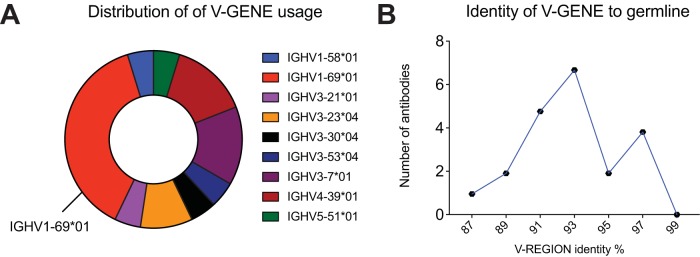
To study the characteristics of the Ab repertoire induced by Butantan-DV, we sorted single plasmablasts at day 15 postvaccination and amplified reverse-transcribed DNA encoding Abs using primer mixes (see Table S1 in the supplemental material). IgG Abs were amplified using a reverse primer that aligns to a CH1 region conserved on IgG1, IgG2, IgG3, and IgG4. Using this strategy, we obtained 356 PCR amplifications. If a positive heavy amplification was obtained, we attempted amplifying the cognate light chain from the same cell. We obtained a positive lambda or kappa PCR from 160 wells. The Ab sequences were cloned into rhesus pFUSE (InvivoGen) expression plasmids, by either direct restriction cloning from PCR products or the synthesis of codon optimized inserts. The PCR and plasmids were confirmed to be authentic Abs by Sanger sequencing. Antibodies without paired cloned heavy (H) and light chains and plasmids with stop codons and fragmented variable chains were discarded. Additionally, we discontinued testing if plasmid transfection resulted in inconsistent, poor, or no Ab expression. Consequently, from the initial PCR amplifications, we selected 21 MAbs with consistent expression, from paired heavy and light chains cloned into expression vectors. Examination of the cloned sequences showed that a few of the MAbs were related (Table 2; Fig. 2). The most frequent clone was represented by 3 of the 21 MAb sequences (P1A10, P7F09, and P7H10; depicted in blue in Table 2). Interestingly, 8 of the 21 MAbs possessed the same variable heavy chain allele (IGHV1-69*01) but were not clonally related, as shown by different rearrangements of the IGVD and IGVJ genes, CDR3 lengths, and light chain sequences (depicted in red in Table 2). Finally, analysis of levels of SHM—defined as the percent divergence from germ line genes at the nucleotide level—revealed a bimodal distribution, suggesting the presence of two distinct Ab populations (Fig. 2).
TABLE 2.
Genetic characteristics of isolated MAbsa
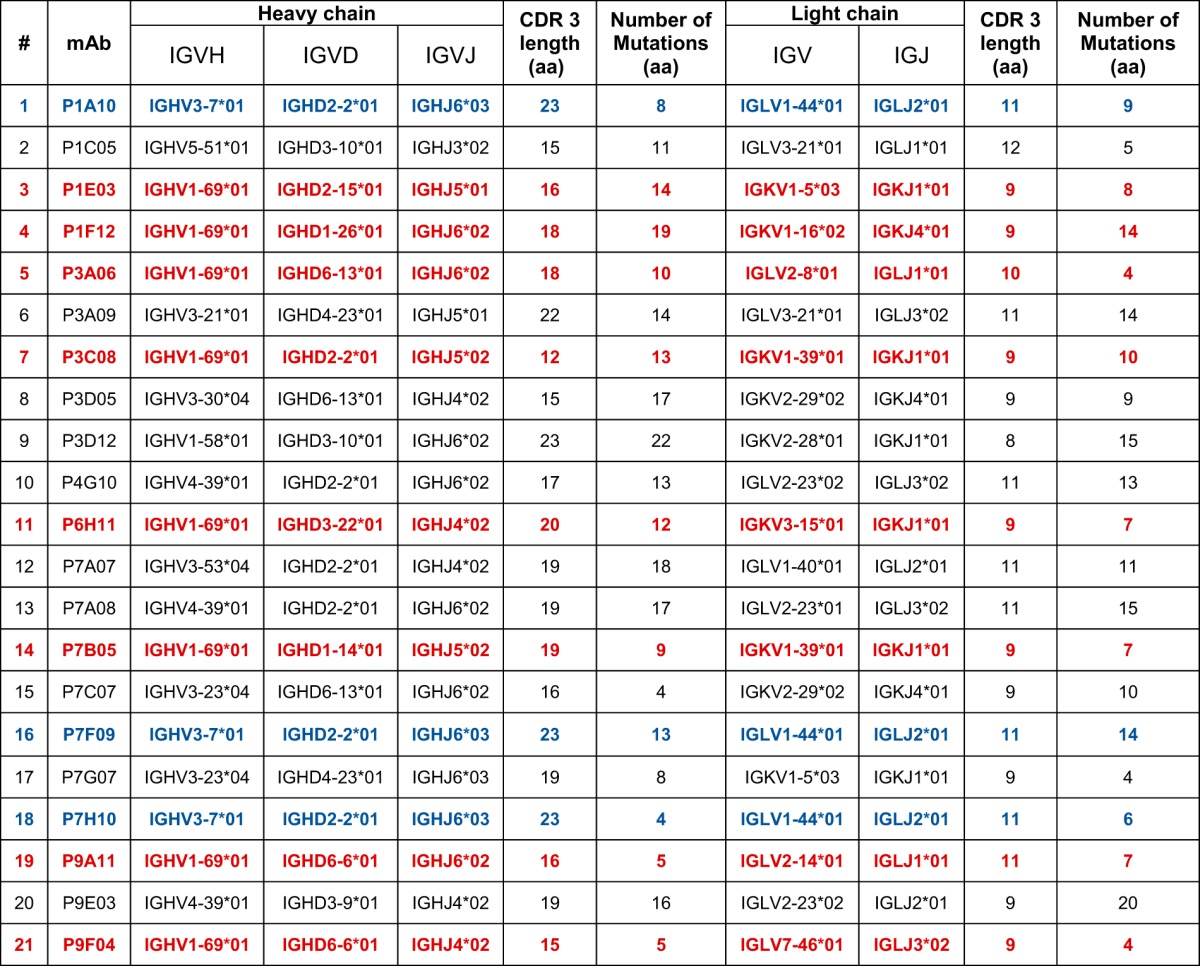
In blue are clonally related MAbs. In red are MAbs with same V gene but which are not clonally related. aa, amino acids.
FIG 2.
Plasmablast repertoire characteristics. Shown are the distributions of V-gene usage (A) and identity (nucleotide) to germ line sequences (B) among the sequences of 21 MAbs isolated from single plasmablasts after vaccination.
Functional characteristics of Butantan-DV-induced, plasmablast-derived MAbs.
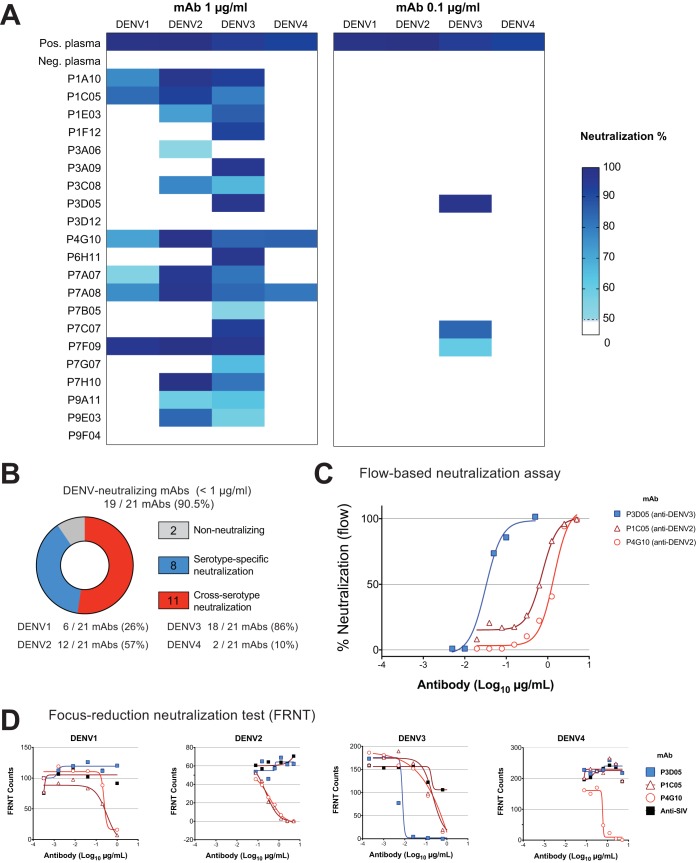
To determine the ability of our MAbs to neutralize DENV1, DENV2, DENV3, and DENV4, we used a flow-based Vero cell neutralization assay (28). We first incubated virus with the transfection supernatant containing MAbs at 1 μg and 0.1 μg (Fig. 3A). Strikingly, 19 of 21 (90.5%) of our isolated MAbs neutralized at least one DENV strain at 1 μg/ml. Of these, 8 nAbs were serotype specific and 11 nAbs neutralized two or more DENV serotypes. Furthermore, most of the nAbs neutralized DENV3 (18 of 19 nAbs [Fig. 3B]). We then proceeded to define the neutralization potency of our best nAbs (Fig. 3C). Remarkably, the human monoclonal nAb P3D05 exhibited an extraordinary DENV3-neutralizing capability (Neut50 = 0.03 μg/ml). In contrast, P1C05 and P4G10 neutralized DENV2 best (Neut50 = 1.41 μg/ml and 0.73 μg/ml, respectively). These selected nAbs were further characterized by the focus reduction neutralization test (FRNT), which yielded comparable results (Fig. 3A, B, and D). The P3D05 DENV3 Neut50 was determined as ∼0.03 μg/ml by flow assay and ∼0.01 μg/ml by FRNT (Fig. 3D). In contrast to the cross-neutralizer P1C05 and P4G10 nAbs, P3D05 selectively neutralized DENV3 but did not neutralize DENV1, DENV2, or DENV4 (Fig. 3D). This novel, vaccine-derived nAb is among the most potent described to date (21, 29, 30). Importantly, we also isolated three Abs with cross-serotype neutralizing activity.
FIG 3.
DENV neutralization by LATV-derived MAbs. A flow-based DENV neutralization assay was used to determine the neutralization potential of the isolated MAbs. (A) Each MAb-containing transfection supernatant (with 1 μg and 0.1 μg of the MAb) was initially screened for neutralization against the four DENV serotypes. The neutralization activity of supernatants was then validated in a follow-up assay, using purified MAbs. Values represent the percent reduction of DENV infectivity (>50%). (B) Summary of the DENV serotype neutralization pattern of the 21 MAbs studied. (C) Representative virus neutralization curves of three isolated nAbs. The neutralization potencies (Neut50) of the selected MAbs were determined by assaying Vero cell infectivity in the presence of serial dilutions of purified MAbs. (D) The neutralization of DENV serotypes by P1C05, P3D05, P4G10 nAbs was validated by the focus reduction neutralization test (FRNT). The purified P3D05 MAb selectively neutralized DENV3. A control anti-SIV antibody was used as a negative control.
DISCUSSION
The dengue epidemic presents a global public health challenge that causes widespread economic burden and remains largely unchecked by existing control strategies. Successful control of the dengue epidemic will require effective prophylactic and therapeutic interventions. Several vaccine clinical efficacy trials are approaching completion, and the chance that one or more LATVs will be introduced worldwide is higher than ever. The safety and effectiveness of vaccines against dengue will likely be determined by the nature of the elicited Abs (13). Inducing protective responses is no trivial task: vaccines must generate serum responses that control but do not enhance disease in a population with preexisting responses to diverse DENV serotypes. Given the increased hospitalization in younger individuals who received the Sanofi Pasteur LATV (11, 31), the possibility of vaccine-induced disease is now more than theoretical. While it is widely accepted that DENV nAb titers are associated with protection, the Ab repertoire induced by LATVs remains uncharacterized. Here we describe the isolation of potent (Neut50 < 0.1 μg/ml) nAbs from a DENV-seropositive volunteer immunized with the tetravalent vaccine Butantan-DV, which is currently in phase III trials.
To our knowledge, this is the first characterization of a LATV-derived B cell repertoire against DENV. We isolated 21 MAbs from volunteer 2028, a DENV-exposed individual that received the Butantan-DV during a phase II clinical trial. Among the plasmablast-derived MAbs, we isolated a potent vaccine-induced DENV3-specific nAb, P3D05. Vaccination of volunteer 2028 allowed us to monitor the magnitude and kinetics of vaccine-induced B cell responses in detail. We observed a massive plasmablast expansion postvaccination (∼70-fold from prevaccination levels), comparable to previous reports of patients with secondary wild-type DENV infections (18, 22, 27, 32, 33). The accumulation of plasmablasts in circulation peaked at day 13 postimmunization. Similarly, others have reported infection-induced peak plasmablast responses at days 3 to 8 after onset of dengue symptoms (22, 27, 32, 33), which is thought to occur 4 to 6 days after mosquito transmission (34). Interestingly, the plasmablast response against DENV occurs later than the responses in several other viral infections and vaccines, which usually peak around 7 days postexposure (35, 36). The reasons for these differences are not immediately clear, but it is possible that the viral incubation period and replication kinetics could account for the different timing.
Rapid accumulation of plasmablasts in the circulation after immunization against influenza has been previously associated with the activation and expansion of a few responding B cell clones (35). Similarly, a recent study of plasmablast activation during secondary DENV infections describes expansion of only a limited number of B cell clones (22). While we isolated clonally related MAbs (e.g., P1A10, P7F09, and P7H10, depicted in blue in Table 2), most MAbs appeared to be independently generated. However, our study was not designed to identify clonal relationships and is not sufficiently powered to detect less frequent but related MAbs. Another noticeable feature of our LATV-derived repertoire was the high frequency of IGHV1-69*01 allele usage. However, this came from distinct clones with diverse D and J gene arrangements (depicted in red in Table 2). IGHV1-69 and other VH1 genes were also found to be enriched in naturally infection-induced repertoires against DENV (37). Curiously, Ab repertoires enriched for the IGHV1-69 gene have also been reported for responses against other pathogens, such as influenza (38) and hepatitis C (39) viruses. The bias toward VH1 usage suggests that these germ line genes are common precursors of DENV Abs and might recognize similar epitopes. In conclusion, from our limited data set, we report an IGHV1-69-rich repertoire but did not find support for a pauci-clonal response against the DENV LATV.
In addition to the recombination events that generate the Ab diversity, SHM facilitates the development of clones with increasing affinities (40). Interestingly, we observed a bimodal distribution of SHM levels, suggesting that two discrete Ab populations were elicited by vaccination. While we could not determine the origin of these cells, one possible explanation is that the most mutated Abs were recalled from preexisting memory cells generated in the original DENV infection, hence the higher levels of SHM. In this scenario, the less mutated counterparts were de novo B cell responses induced by the LATV. The median plasmablast-derived MAb had 22 nucleotide mutations and 13 amino acid changes in the heavy chain. These levels of SHM are comparable to responses induced by natural secondary wild-type DENV infections (22, 32).
The Butantan-DV-expanded plasmablast population was also highly enriched for cells responding to DENV. Ninety percent of the derived MAbs neutralized at least one serotype at low concentrations (Neut50 < 1 μg/ml). Several groups have isolated infection-derived DENV nAbs with various neutralization potencies in recent years (15, 16, 18–22, 30, 41, 42). Dejnirattisai and colleagues have described a novel class of DENV nAbs that bind to conformational epitopes on the viral particle and are capable of neutralizing all four strains with exceptional potency (21). Because of the simultaneous stimulation with the four vaccine constructs, we hypothesized that immunization with the NIH LATV would favor the development of this type of potent, broadly reactive nAb. Accordingly, we opted to isolate the MAbs without a binding selection step so as not to exclude potential nAbs that might bind to conformational epitopes. Contrary to our initial expectations, we did not find that LATV elicited potent and broadly reactive nAbs. Only two antibodies had neutralizing activity against all four serotypes. Instead, we found that 8 nAbs were serotype specific and 11 of 21 nAbs had Neut50 potencies below 1 μg/ml for two or more DENV serotypes. Furthermore, the most potent MAbs neutralized a preferred serotype (e.g., P3D05 for DENV3 and P4G10 for DENV2). Remarkably, these vaccine-elicited MAbs are in the neutralization range of the most potent nAbs described to date (15, 16, 18–22, 24, 30, 32, 41, 42).
A previous report of the memory B cell repertoire induced by the NIH DENV1 live attenuated vaccine did not identify nAbs with neutralization potencies below 1 μg/ml (24). It is still unclear why our isolated nAbs had such a superior neutralization capability. Likely, the preexisting DENV-specific responses of volunteer 2028 were recalled by vaccination. Supporting evidence from a recent study shows that several plasmablast-derived MAbs isolated during a secondary infection targeted a serotype different than that of the infecting virus (22), presumably the virus responsible for the primary infection. Additionally, 90 to 100% of plasmablast-derived MAbs from secondary infections were DENV specific, with virus-neutralizing activity (21, 32). These results strongly suggest that plasmablasts in secondary DENV infections are already antigen selected and consequently originate from memory cells. The high number of isolated nAbs and SHM levels identified after Butantan-DV vaccination is compatible with cells originating from a memory pool. Intriguingly, however, MAbs isolated from memory B cells do not appear to be as potent as our vaccine-induced plasmablast-derived MAbs (24). In fact, most of the memory B cell-derived repertoires have extensive cross-reactive, weakly DENV-neutralizing MAbs (16, 30, 43). Also, Abs that do not contribute to the serum neutralizing activity, such as the ones against precursor membrane (prM), are often detected (16). Interestingly, the fact that we did not observe a high frequency of nonneutralizing MAbs also implies that prM and NS1 MAbs were not vastly represented in our repertoire, despite our lack of selection steps. The picture emerging from these MAb studies is that plasmablast-derived repertoires are fundamentally distinct from circulating memory B cell-derived repertoires.
Vaccination of seropositive individuals with the Sanofi Pasteur LATV had a higher degree of efficacy than vaccination of seronegative individuals in clinical trials (11, 44). This strongly suggests that their vaccine can build on the patient's preexisting immunity. Similarly, it is tempting to speculate that the primary infection of volunteer 2028 was a DENV3 exposure, given the high frequency of isolated nAbs targeting this serotype. Importantly, however, there are alternative explanations for the serotype bias observed in 2028's LATV-derived repertoire. For instance, the DENV3 component of Butantan-DV is known to replicate well in the host, and its viremia is detected more frequently than others (recovery of immunizing virus in vaccines is 63% for DENV3 but only 21% for DENV1, 17% for DENV4, and 4% for DENV2) (12). Thus, the unbalanced replication of individual components of the Butantan-DV vaccine could favor the generation of potent nAbs against the DENV3 serotype. Volunteer 2028's serum neutralization activity matched this pattern, showing a much lower titer against DENV4 and DENV2 than other DENV serotypes, in agreement with the reported NIH LATV replication levels. Lastly, previously reported Ab repertoires had considerable donor variation (22). Thus, it is possible that volunteer 2028's genetic composition or immune history and/or our B cell isolation techniques largely influenced the generation of the particular kinds of MAbs obtained after Butantan-DV immunization.
The size and quality of the plasmablast response have been suggested as an early indicator of serological responses (45). The detection of the P3D05 nAb—and other nAbs against DENV3—from a sort on day 15 postvaccination was indeed consistent with volunteer 2028's serum responses, which showed a higher neutralizing activity against DENV3 at day 91 postvaccination. However, we cannot presently infer vaccine efficacy based on the isolation of these nAbs. While nAbs are widely believed to be essential components of DENV vaccines; the immune correlates of vaccine protection against DENV are not yet known. Future studies that associate vaccine-induced nAb frequency, potency, and perhaps other functional properties will be critical for establishing the requirements for protective efficacy.
MATERIALS AND METHODS
Vaccination.
We collected samples from volunteer 2028 as a part of the phase II clinical trial to evaluate the safety and immunogenicity of an attenuated tetravalent lyophilized dengue vaccine manufactured by the Butantan Institute (ClinicalTrials.gov identifier NCT01696422). This volunteer had been previously exposed to DENV and received a single subcutaneous dose of the lyophilized vaccine consisting of 1,000 PFU of each attenuated virus [rDEN1Δ30, rDEN2/4Δ30(ME), rDEN3Δ30/31, and rDEN4Δ30]. Blood samples were collected prevaccination and at days 0, 7, 13, 15, 23, and 91 postvaccination.
Flow cytometry.
We determined the frequency of plasmablasts in circulation by cytometric analysis of whole blood collected in tubes containing acid-citrate-dextrose (ACD). We stained 100 μl of blood (room temperature, in the dark) with 100 μl of a cocktail containing the following fluorophore-antibody conjugates: phycoerythrin (PE)-CF594 anti-human CD3 (clone UCHT1; Becton Dickinson [BD]), PE-CF594 anti-human CD14 (clone MφP9; BD), allophycocyanin (APC)-cyanine 7 (Cy7) anti-human CD19 (clone SJ25C1; BD), peridinin chlorophyll protein complex (PerCP) anti-human CD20 (clone L27; BD), APC anti-human CD27 antibody (clone O323; Biolegend), fluorescein isothiocyanate (FITC) anti-human CD38 (clone HB7; BD), and PE anti-human CD138 (clone MI15; BD). In addition, a viability dye LIVE/DEAD fixable red dead cell stain kit (Life Technologies) was included in the staining mix to discriminate between live and dead cells. Following a 30-min incubation, red blood cells we lysed by hypotonic shock with a 1:10 dilution (200 μl into 2 ml) in water for 10 min. Next, the cells were washed twice with FACS buffer (phosphate-buffered saline [PBS], 0.5% fetal bovine serum [FBS], 2 mM EDTA), fixed with a 1% paraformaldehyde (PFA) solution, and stored at 4°C until acquisition on the same day. Samples were acquired using a BD FACSCanto II flow cytometer and analyzed using FlowJo 9 (FlowJo). The frequency of the plasmablast population in circulation was expressed as a fraction of the total live CD19+ cells (see gating strategy in Fig. S1).
Single plasmablast sorting.
We sorted single plasmablasts (CD3− CD14− CD19+ CD20−/low CD27high CD38high) at day 15 postvaccination, using the plasmablast staining panel described above. Before staining, peripheral blood mononuclear cells (PBMCs) were extracted from blood collected in ACD using a Ficoll-Paque (GE Lifesciences) gradient. The fresh PBMC samples were sorted on a BD FACSAria II flow cytometer. Single plasmablasts were sorted into 96-well plates containing a lysis buffer designed to extract and preserve the RNA (250 mM Tris-HCl [pH 8.3], 375 mM KCl, 15 mM MgCl2, 6.25 mM dithiothreitol [DTT], and 250 ng/well of yeast tRNA [Life Technologies], 20 U of RNase inhibitor [New England BioLabs], and 0.0625 μl/well of IGEPAL CA-630 [Sigma]). After sorting, the RNA plates were immediately frozen in dry ice for subsequent cloning of the reverse-transcribed RNA encoding the variable (V) region of the heavy (H), lambda (L), and kappa (K) Ab chains.
Ab repertoire analysis.
We carried out a reverse transcription-PCR (RT-PCR) and a nested PCR to amplify the V region of the immunoglobulin chains using previously described protocols with minor modifications (14). First, cDNA was synthesized in a 25-μl reaction mixture using the original sort plates. Each reaction had 1 μl of 150-ng random hexamers (IDT), 2 μl of 10 mM deoxynucleoside triphosphate (dNTP; Life Technologies), 1 μl of SuperScript III reverse transcriptase (Life Technologies), 1 μl of molecular-biology-grade water, and 20 μl of single-sorted cell sample in lysis buffer (described above). The reverse transcription reaction was performed at 42°C for 10 min, 25°C for 10 min, 50°C for 60 min, and 94°C for 10 min. Upon completion of the reaction, cDNA was stored at −20°C. DNAs encoding the H, L, and K chains were amplified in three different nested PCRs, using a mix of 5′ V-specific primers with matching 3′ primers designed to bind DNA encoding the constant regions of the IgGH, IgL, and IgK chains (see Table S1). PCRs were prepared using HotStarTaq Plus DNA polymerase (Qiagen). The second PCRs were carried out with primers adapted from those of Tiller et al. (14). These new oligonucleotides incorporated restriction sites compatible with subcloning into rhesus IgG1 expression vectors (pFUSEss-CHIg-rhG1 and pFUSE2ss-CLIg-rhK pFuse expression vectors, which were purchased from InvivoGen), and a similar expression construct encoding rhIgLC1*01 (created by gene synthesis). Amplified and cloned products were sequenced using primers complementary to the pFuse plasmids (HTLV-5UTR [5′-TGCTTGCTCAACTCTACGTC-3′]) (Genscript). Sequences were then analyzed using IMGT/V-QUEST (46) to identify V(D)J gene rearrangements, as well as SHM levels.
Ab expression and purification.
We expressed MAbs in human embryonic kidney (HEK) 293 cell lines. The plasmids encoding heavy and light chains were cotransfected using the jetPRIME transfection reagent (Polyplus-transfection SA). After 4 to 6 days, we harvested the secreted MAb in the supernatant. Before proceeding with the functional assays, Ig concentration in the supernatant was determined by an anti-rhesus IgG enzyme-linked immunosorbent assay (ELISA). For the experiments to determine the Neut50 potencies, we used Protein A Plus (Pierce)-containing columns to purify the MAbs. The concentration of purified protein was determined by measuring absorbance at 280 nm (NanoDrop; Thermo Scientific).
Flow-based neutralization assays.
The neutralizing potency of the MAbs was measured using a flow cytometry-based assay (28, 47). Recombinant MAbs (transfection supernatant or purified) were diluted and preincubated with the reference DENV serotypes in a final volume of 220 μl for 1 h at 37°C. The virus and MAb mixture were then added to duplicate Vero cell layers in 24-well cell culture plates. After infection for 1 h at 37°C, the DENV-containing supernatants were washed, and the plates were incubated for a total of 24 h. Cells were then permeabilized, and the DENV envelope was detected using Ab clone 4G2, followed by staining with an anti-mouse IgG2a APC fluorophore-conjugated secondary reagent (Biolegend). The concentration to achieve half-maximal neutralization (Neut50) was calculated by nonlinear regression analysis with Prism 7.0 software (GraphPad Software, Inc.). The following strains were used in our neutralization assays (GenBank accession numbers are in parentheses): DENV1-West Pac (U88535.1), DENV2-NGC (AF038403.1), DENV3-Sleman/78 (AY648961), and DENV4-Dominica (AF326573.1).
FRNT.
The neutralization potency and specificity of selected antibodies were validated by the focus reduction neutralization test (FRNT), as previously described (48). Briefly, purified antibody was serially diluted in minimal essential medium (Corning Cellgro, Manassas, VA) containing 5% heat-inactivated fetal bovine serum (Gibco-Invitrogen, Gaithersburg, MD) and incubated 1 h at 37°C with virus. After incubation, the antibody-virus mixture was added in triplicate to 96-well plates containing monolayers of Vero cells. Plates were incubated for 1.5 h at 37°C. Following the incubation, wells were overlaid with 1% methylcellulose in supplemented MEM with 2% heat-inactivated fetal bovine serum (Gibco-Invitrogen, Gaithersburg, MD) and 1:100 HEPES. Plates were incubated at 37°C and 5% CO2 for 40 h, after which cells were fixed and permeabilized with Perm/Wash buffer (BD Biosceinces, San Jose, CA) for 5 min. After permeabilization, cells were incubated with a 1:2,000 dilution of antiflavivirus antibody (MAB10216; EMD Millipore, Darmstadt, Germany) for 2 h and then washed with PBS. Following washing, cells were incubated with anti-mouse horseradish peroxidase (HRP)-conjugated secondary Ab (115035146; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) in Perm/Wash buffer for 2 h. Following washing of cells with PBS, plates were developed with peroxidase substrate (KPL, Milford, MA). Control wells with no virus, virus only, simian immunodeficiency virus (SIV)-specific purified MAbs, and positive and negative sera were run concurrently with wells with the test MAb. The DENV strains used for FRNT were the same as for the flow-based neutralization assay.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ana Carolina Soares, Carla Sant'Anna Corrêa, Ovini Senelka, and Meagan Read for help provided in different stages of this study. We also thank Beatriz Thome, Ricardo Palacios, and Alexander Precioso for support at the Butantan Institute.
This work was supported by funding from the Wallace H. Coulter Center for Translational Research, Miami, FL, the Brazilian Development Bank (BNDES), and the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID, NIH).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00867-17.
REFERENCES
- 1.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guy B, Briand O, Lang J, Saville M, Jackson N. 2015. Development of the Sanofi Pasteur tetravalent dengue vaccine: one more step forward. Vaccine 33:7100–7111. doi: 10.1016/j.vaccine.2015.09.108. [DOI] [PubMed] [Google Scholar]
- 3.Whitehead SS. 2016. Development of TV003/TV005, a single dose, highly immunogenic live attenuated dengue vaccine; what makes this vaccine different from the Sanofi-Pasteur CYD vaccine? Expert Rev Vaccines 15:509–517. doi: 10.1586/14760584.2016.1115727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osorio JE, Wallace D, Stinchcomb DT. 2016. A recombinant, chimeric tetravalent dengue vaccine candidate based on a dengue virus serotype 2 backbone. Expert Rev Vaccines 15:497–508. doi: 10.1586/14760584.2016.1128328. [DOI] [PubMed] [Google Scholar]
- 5.Halstead SB. 2003. Neutralization and antibody-dependent enhancement of dengue viruses. Adv Virus Res 60:421–467. doi: 10.1016/S0065-3527(03)60011-4. [DOI] [PubMed] [Google Scholar]
- 6.Halstead SB, Nimmannitya S, Cohen SN. 1970. Observations related to pathogenesis of dengue hemorrhagic fever. IV. Relation of disease severity to antibody response and virus recovered. Yale J Biol Med 42:311–328. [PMC free article] [PubMed] [Google Scholar]
- 7.Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengsaa K, Wartel TA, Moureau A, Saville M, Bouckenooghe A, Viviani S, Tornieporth NG, Lang J. 2012. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet 380:1559–1567. doi: 10.1016/S0140-6736(12)61428-7. [DOI] [PubMed] [Google Scholar]
- 8.Halstead SB. 2013. Identifying protective dengue vaccines: guide to mastering an empirical process. Vaccine 31:4501–4507. doi: 10.1016/j.vaccine.2013.06.079. [DOI] [PubMed] [Google Scholar]
- 9.Halstead SB. 2012. Dengue vaccine development: a 75% solution? Lancet 380:1535–1536. doi: 10.1016/S0140-6736(12)61510-4. [DOI] [PubMed] [Google Scholar]
- 10.Swaminathan S, Khanna N, Herring B, Mahalingam S. 2013. Dengue vaccine efficacy trial: does interference cause failure? Lancet Infect Dis 13:191–192. doi: 10.1016/S1473-3099(13)70028-8. [DOI] [PubMed] [Google Scholar]
- 11.Wilder-Smith A, Massad E. 2016. Age specific differences in efficacy and safety for the CYD-tetravalent dengue vaccine. Expert Rev Vaccines 15:437–441. doi: 10.1586/14760584.2016.1143366. [DOI] [PubMed] [Google Scholar]
- 12.Kirkpatrick BD, Whitehead SS, Pierce KK, Tibery CM, Grier PL, Hynes NA, Larsson CJ, Sabundayo BP, Talaat KR, Janiak A, Carmolli MP, Luke CJ, Diehl SA, Durbin AP. 2016. The live attenuated dengue vaccine TV003 elicits complete protection against dengue in a human challenge model. Sci Transl Med 8:330ra336. doi: 10.1126/scitranslmed.aaf1517. [DOI] [PubMed] [Google Scholar]
- 13.Acosta EG, Bartenschlager R. 2016. Paradoxical role of antibodies in dengue virus infections: considerations for prophylactic vaccine development. Expert Rev Vaccines 15:467–482. doi: 10.1586/14760584.2016.1121814. [DOI] [PubMed] [Google Scholar]
- 14.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. 2008. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods 329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dejnirattisai W, Jumnainsong A, Onsirisakul N, Fitton P, Vasanawathana S, Limpitikul W, Puttikhunt C, Edwards C, Duangchinda T, Supasa S, Chawansuntati K, Malasit P, Mongkolsapaya J, Screaton G. 2010. Cross-reacting antibodies enhance dengue virus infection in humans. Science 328:745–748. doi: 10.1126/science.1185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NT, Sukupolvi-Petty S, Navarro-Sanchez E, Young PR, de Silva AM, Rey FA, Varani L, Whitehead SS, Diamond MS, Harris E, Lanzavecchia A, Sallusto F. 2010. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 8:271–283. doi: 10.1016/j.chom.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katzelnick LC, Montoya M, Gresh L, Balmaseda A, Harris E. 2016. Neutralizing antibody titers against dengue virus correlate with protection from symptomatic infection in a longitudinal cohort. Proc Natl Acad Sci U S A 113:728–733. doi: 10.1073/pnas.1522136113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai W-Y, Lai C-Y, Wu Y-C, Lin H-E, Edwards C, Jumnainsong A, Kliks S, Halstead S, Mongkolsapaya J, Screaton GR, Wang W-K. 2013. High-avidity and potently neutralizing cross-reactive human monoclonal antibodies derived from secondary dengue virus infection. J Virol 87:12562–12575. doi: 10.1128/JVI.00871-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith SA, Zhou Y, Olivarez NP, Broadwater AH, de Silva AM, Crowe JE Jr. 2012. Persistence of circulating memory B cell clones with potential for dengue virus disease enhancement for decades following infection. J Virol 86:2665–2675. doi: 10.1128/JVI.06335-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith SA, de Alwis AR, Kose N, Harris E, Ibarra KD, Kahle KM, Pfaff JM, Xiang X, Doranz BJ, de Silva AM, Austin SK, Sukupolvi-Petty S, Diamond MS, Crowe JE Jr. 2013. The potent and broadly neutralizing human dengue virus-specific monoclonal antibody 1C19 reveals a unique cross-reactive epitope on the bc loop of domain II of the envelope protein. mBio 4:e00873-13. doi: 10.1128/mBio.00873-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinski A, Jumnainsong A, Edwards C, Quyen NT, Duangchinda T, Grimes JM, Tsai WY, Lai CY, Wang WK, Malasit P, Farrar J, Simmons CP, Zhou ZH, Rey FA, Mongkolsapaya J, Screaton GR. 2015. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat Immunol 16:170–177. doi: 10.1038/ni.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Priyamvada L, Cho A, Onlamoon N, Zheng NY, Huang M, Kovalenkov Y, Chokephaibulkit K, Angkasekwinai N, Pattanapanyasat K, Ahmed R, Wilson PC, Wrammert J. 2016. B cell responses during secondary dengue virus infection are dominated by highly cross-reactive, memory-derived plasmablasts. J Virol 90:5574–5585. doi: 10.1128/JVI.03203-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srikiatkhachorn A, Yoon IK. 2016. Immune correlates for dengue vaccine development. Expert Rev Vaccines 15:455–465. doi: 10.1586/14760584.2016.1116949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith SA, de Alwis R, Kose N, Durbin AP, Whitehead SS, de Silva AM, Crowe JE Jr. 2013. Human monoclonal antibodies derived from memory B cells following live attenuated dengue virus vaccination or natural infection exhibit similar characteristics. J Infect Dis 207:1898–1908. doi: 10.1093/infdis/jit119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durbin AP, Kirkpatrick BD, Pierce KK, Elwood D, Larsson CJ, Lindow JC, Tibery C, Sabundayo BP, Shaffer D, Talaat KR, Hynes NA, Wanionek K, Carmolli MP, Luke CJ, Murphy BR, Subbarao K, Whitehead SS. 2013. A single dose of any of four different live attenuated tetravalent dengue vaccines is safe and immunogenic in flavivirus-naive adults: a randomized, double-blind clinical trial. J Infect Dis 207:957–965. doi: 10.1093/infdis/jis936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwissa M, Nakaya HI, Onlamoon N, Wrammert J, Villinger F, Perng GC, Yoksan S, Pattanapanyasat K, Chokephaibulkit K, Ahmed R, Pulendran B. 2014. Dengue virus infection induces expansion of a CD14(+)CD16(+) monocyte population that stimulates plasmablast differentiation. Cell Host Microbe 16:115–127. doi: 10.1016/j.chom.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wrammert J, Onlamoon N, Akondy RS, Perng GC, Polsrila K, Chandele A, Kwissa M, Pulendran B, Wilson PC, Wittawatmongkol O, Yoksan S, Angkasekwinai N, Pattanapanyasat K, Chokephaibulkit K, Ahmed R. 2012. Rapid and massive virus-specific plasmablast responses during acute dengue virus infection in humans. J Virol 86:2911–2918. doi: 10.1128/JVI.06075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Alwis R, de Silva AM. 2014. Measuring antibody neutralization of dengue virus (DENV) using a flow cytometry-based technique. Methods Mol Biol 1138:27–39. doi: 10.1007/978-1-4939-0348-1_3. [DOI] [PubMed] [Google Scholar]
- 29.Fibriansah G, Tan JL, Smith SA, de Alwis R, Ng TS, Kostyuchenko VA, Jadi RS, Kukkaro P, de Silva AM, Crowe JE, Lok SM. 2015. A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins. Nat Commun 6:6341. doi: 10.1038/ncomms7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Alwis R, Smith SA, Olivarez NP, Messer WB, Huynh JP, Wahala WM, White LJ, Diamond MS, Baric RS, Crowe JE Jr, de Silva AM. 2012. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc Natl Acad Sci U S A 109:7439–7444. doi: 10.1073/pnas.1200566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halstead SB, Russell PK. 2016. Protective and immunological behavior of chimeric yellow fever dengue vaccine. Vaccine 34:1643–1647. doi: 10.1016/j.vaccine.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Xu M, Hadinoto V, Appanna R, Joensson K, Toh YX, Balakrishnan T, Ong SH, Warter L, Leo YS, Wang CI, Fink K. 2012. Plasmablasts generated during repeated dengue infection are virus glycoprotein-specific and bind to multiple virus serotypes. J Immunol 189:5877–5885. doi: 10.4049/jimmunol.1201688. [DOI] [PubMed] [Google Scholar]
- 33.Garcia-Bates TM, Cordeiro MT, Nascimento EJ, Smith AP, Soares de Melo KM, McBurney SP, Evans JD, Marques ET Jr, Barratt-Boyes SM. 2013. Association between magnitude of the virus-specific plasmablast response and disease severity in dengue patients. J Immunol 190:80–87. doi: 10.4049/jimmunol.1103350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishiura H, Halstead SB. 2007. Natural history of dengue virus (DENV)-1 and DENV-4 infections: reanalysis of classic studies. J Infect Dis 195:1007–1013. doi: 10.1086/511825. [DOI] [PubMed] [Google Scholar]
- 35.Li GM, Chiu C, Wrammert J, McCausland M, Andrews SF, Zheng NY, Lee JH, Huang M, Qu X, Edupuganti S, Mulligan M, Das SR, Yewdell JW, Mehta AK, Wilson PC, Ahmed R. 2012. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci U S A 109:9047–9052. doi: 10.1073/pnas.1118979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halliley JL, Kyu S, Kobie JJ, Walsh EE, Falsey AR, Randall TD, Treanor J, Feng C, Sanz I, Lee FE. 2010. Peak frequencies of circulating human influenza-specific antibody secreting cells correlate with serum antibody response after immunization. Vaccine 28:3582–3587. doi: 10.1016/j.vaccine.2010.02.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Godoy-Lozano EE, Tellez-Sosa J, Sanchez-Gonzalez G, Samano-Sanchez H, Aguilar-Salgado A, Salinas-Rodriguez A, Cortina-Ceballos B, Vivanco-Cid H, Hernandez-Flores K, Pfaff JM, Kahle KM, Doranz BJ, Gomez-Barreto RE, Valdovinos-Torres H, Lopez-Martinez I, Rodriguez MH, Martinez-Barnetche J. 2016. Lower IgG somatic hypermutation rates during acute dengue virus infection is compatible with a germinal center-independent B cell response. Genome Med 8:23. doi: 10.1186/s13073-016-0276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Avnir Y, Tallarico AS, Zhu Q, Bennett AS, Connelly G, Sheehan J, Sui J, Fahmy A, Huang CY, Cadwell G, Bankston LA, McGuire AT, Stamatatos L, Wagner G, Liddington RC, Marasco WA. 2014. Molecular signatures of hemagglutinin stem-directed heterosubtypic human neutralizing antibodies against influenza A viruses. PLoS Pathog 10:e1004103. doi: 10.1371/journal.ppat.1004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan CH, Hadlock KG, Foung SK, Levy S. 2001. V(H)1-69 gene is preferentially used by hepatitis C virus-associated B cell lymphomas and by normal B cells responding to the E2 viral antigen. Blood 97:1023–1026. [DOI] [PubMed] [Google Scholar]
- 40.Victora GD, Nussenzweig MC. 2012. Germinal centers. Annu Rev Immunol 30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 41.Deng YQ, Dai JX, Ji GH, Jiang T, Wang HJ, Yang HO, Tan WL, Liu R, Yu M, Ge BX, Zhu QY, Qin ED, Guo YJ, Qin CF. 2011. A broadly flavivirus cross-neutralizing monoclonal antibody that recognizes a novel epitope within the fusion loop of E protein. PLoS One 6:e16059. doi: 10.1371/journal.pone.0016059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Alwis R, Beltramello M, Messer WB, Sukupolvi-Petty S, Wahala WM, Kraus A, Olivarez NP, Pham Q, Brien JD, Tsai WY, Wang WK, Halstead S, Kliks S, Diamond MS, Baric R, Lanzavecchia A, Sallusto F, de Silva AM. 2011. In-depth analysis of the antibody response of individuals exposed to primary dengue virus infection. PLoS Negl Trop Dis 5:e1188. doi: 10.1371/journal.pntd.0001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friberg H, Jaiswal S, West K, O'Ketch M, Rothman AL, Mathew A. 2012. Analysis of human monoclonal antibodies generated by dengue virus-specific memory B cells. Viral Immunol 25:348–359. doi: 10.1089/vim.2012.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hadinegoro SR, Arredondo-Garcia JL, Capeding MR, Deseda C, Chotpitayasunondh T, Dietze R, Muhammad Ismail HI, Reynales H, Limkittikul K, Rivera-Medina DM, Tran HN, Bouckenooghe A, Chansinghakul D, Cortes M, Fanouillere K, Forrat R, Frago C, Gailhardou S, Jackson N, Noriega F, Plennevaux E, Wartel TA, Zambrano B, Saville M, CYD-TDV Dengue Vaccine Working Group. 2015. Efficacy and long-term safety of a dengue vaccine in regions of endemic disease. N Engl J Med 373:1195–1206. [DOI] [PubMed] [Google Scholar]
- 45.Fink K. 2012. Origin and function of circulating plasmablasts during acute viral infections. Front Immunol 3:78. doi: 10.3389/fimmu.2012.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brochet X, Lefranc MP, Giudicelli V. 2008. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res 36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kraus AA, Messer W, Haymore LB, de Silva AM. 2007. Comparison of plaque- and flow cytometry-based methods for measuring dengue virus neutralization. J Clin Microbiol 45:3777–3780. doi: 10.1128/JCM.00827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brien JD, Lazear HM, Diamond MS. 2013. Propagation, quantification, detection, and storage of West Nile virus. Curr Protoc Microbiol 31:15D.3.1–15D.3.18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.