Abstract
The life cycle of a primary cilium begins in quiescence and ends prior to mitosis. In quiescent cells, primary cilium insulates itself from contiguous dynamic membrane processes on the cell surface to function as a stable signaling apparatus. Here, we demonstrate that basal restriction of ciliary structure dynamics is established by cilia-enriched phosphoinositide 5-phosphatase, Inpp5e. Growth induction displaces ciliary Inpp5e and accumulates phosphatidylinositol 4,5-bisphosphate to distal cilia. This triggers otherwise forbidden actin polymerization in primary cilia, which excises cilia tips in a process we call cilia decapitation. Whilst cilia disassembly is traditionally thought to occur solely through resorption, we show that an acute loss of IFT-B through cilia decapitation precedes resorption. Finally, we propose that cilia decapitation induces mitogenic signaling and constitutes a molecular link between the cilia life cycle and cell-division cycle. This newly defined ciliary mechanism may find significance in cell proliferation control during normal development and cancer.
Introduction
Primary cilia function as specialized sensory antennae for cells to detect signals critical to cell proliferation and differentiation (Christensen et al., 2012; Ezratty et al., 2011; Lancaster et al., 2011; Nauli et al., 2003; Phua et al., 2015; Rohatgi et al., 2007; Singla and Reiter, 2006). Activity of cilia-dependent Gli transcription factors controls expression of cell cycle regulators (Hui and Angers, 2011; Regl et al., 2004; Yoon et al., 2002), and mis-activation of ciliary signaling leads to cancers including basal cell carcinoma and medulloblastoma (Han et al., 2009; Wong et al., 2009). Hence there is growing interest in understanding how primary cilia affect cell cycle decisions (Basten and Giles, 2013; Kim et al., 2011; Plotnikova et al., 2008, 2009; Yeh et al., 2013). Indeed, the cilia life cycle is tightly coupled with the cell cycle; cilia assemble in non-dividing cells of G0/G1 phases, and disassemble with cell cycle entry (Plotnikova et al., 2009). Yet, current literature does not explain how the ciliary structure is insulated from contiguous dynamic membrane processes occurring on the cell surface, such as endo/exocytosis and membrane ruffling, to function as a stable signal transduction apparatus.
Almost all membrane processes on the cell surface depend on phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2), a key molecular identifier of the plasma membrane. PI(4,5)P2 recruits PI(4,5)P2-binding actin regulators to modulate actin polymerization at the cell cortex required to drive membrane activities on the cell surface (Saarikangas et al., 2010). Despite contiguity with plasma membrane, ciliary membrane contains PI(4,5)P2 only proximally, and is enriched with phosphatidylinositol 4-phosphate (PI(4)P) instead, creating a unique molecular identifier for ciliary membrane that distinguishes it from surrounding PI(4,5)P2-enriched plasma membrane (Chávez et al., 2015; Garcia-Gonzalo et al., 2015). Such strict PI(4)P/PI(4,5)P2 compartmentalization is established by Inpp5e, a cilia-enriched phosphoinositide 5-phosphatase associated with Joubert syndrome and MORM syndrome (Chávez et al., 2015; Garcia-Gonzalo et al., 2015). Loss-of-function of Inpp5e replenishes ciliary PI(4,5)P2 and accelerates cilia disassembly depending on phosphoinositide 3-kinase, platelet-derived growth factor receptor alpha and Aurora A kinase (AurA), implicating phosphoinositides in primary cilia stability (Bielas et al., 2009; Jacoby et al., 2009; Plotnikova et al., 2014). Moreover, primary cilia are supported by a microtubule-based axoneme and devoid of F-actin (Francis et al., 2011), unlike other F-actin-based membrane protrusions such as filopodia (Mattila and Lappalainen, 2008).
Primary cilia disassemble via gradual resorption of ciliary material into the cell body (Plotnikova et al., 2009). AurA, a master regulator of cilia disassembly, induces cilia resorption partly via histone deacetylase 6 (HDAC6)-dependent deacetylation of ciliary microtubules, which destabilizes the axoneme (Pugacheva et al., 2007). Interestingly, Chlamydomonas flagella also disassemble via excision and release into the extracellular environment, in response to environmental stress such as high acidity (Pan et al., 2004). Recent reports suggest that vertebrate primary cilia could possess similar capacity in releasing vesicles into the extracellular environment (Dubreuil et al., 2007; Wood and Rosenbaum, 2015). While monitoring primary cilia of cycling kidney fibroblasts, Paridaen et al. occasionally observed release of vesicular structures from distal cilia (Paridaen et al., 2013). Active release of ciliary contents was also observed in retinal pigment epithelial cells over-expressing a CEP162 mutant (Wang et al., 2013). Furthermore, vesicular structures were closely apposed with tip-dilated primary cilia in cystic kidneys of Inpp5e mutant mice (Jacoby et al., 2009), suggesting a connection between phosphoinositides and extracellular vesicle formation. Together, these evidence supports the notion that primary cilia undergo vesicle release under specific conditions, and this process could be involved in cilia disassembly during cell cycle entry.
Results
Ciliary vesicle release is promoted by Inpp5e loss and growth stimulation
We first determined factors promoting primary cilia vesicle release. Fluorescent protein (FP)-tagged ciliary membrane markers (5HT6 or Arl13b) enabled live-cell visualization of ciliary vesicle release, and primary cilia were induced by culturing cells in 0–0.1% fetal bovine serum (FBS) for 24–48 hours to attain quiescence. In NIH/3T3, hTERT RPE-1 and mIMCD-3, we occasionally observed release of ciliary vesicles from distal cilia (Figures 1A, 1B and S1A; Movie S1). Since Inpp5e is implicated in cilia stability and extracellular vesicles (Bielas et al., 2009; Jacoby et al., 2009), we established Inpp5e+/− and Inpp5e−/− mouse embryonic fibroblasts (MEF) and compared ciliary vesicle release (Garcia-Gonzalo et al., 2015). In quiescent cells, the length of Inpp5e−/− cilia was shorter than Inpp5e+/− cilia and displayed a higher tendency of bulging at ciliary tips (Figures S1B, S1D–S1F). The percentage of ciliation was also lower in Inpp5e−/− MEF, which displayed an accelerated cilia disassembly rate than Inpp5e+/− MEF (Figures S1B, S1C and S1G). Ciliary vesicle release was observed in quiescent Inpp5e+/− MEF with a low three-hour occurrence of 23.8 ± 1.5% (mean ± SEM), while a complete loss of Inpp5e resulted in a higher three-hour occurrence of 68.3 ± 6.3% (mean ± SEM) (Figures 1C and 1D; Movie S2). Released ciliary vesicles frequently associate with the cell surface and could be traced in fixed cell samples as Arl13b-positive and acetylated tubulin (Ac tub)-negative particles which were harbored by a higher proportion of Inpp5e−/− MEF than Inpp5e+/− MEF (Figures 1E–1G, S1H). These data demonstrate a role of Inpp5e in modulating ciliary vesicle release.
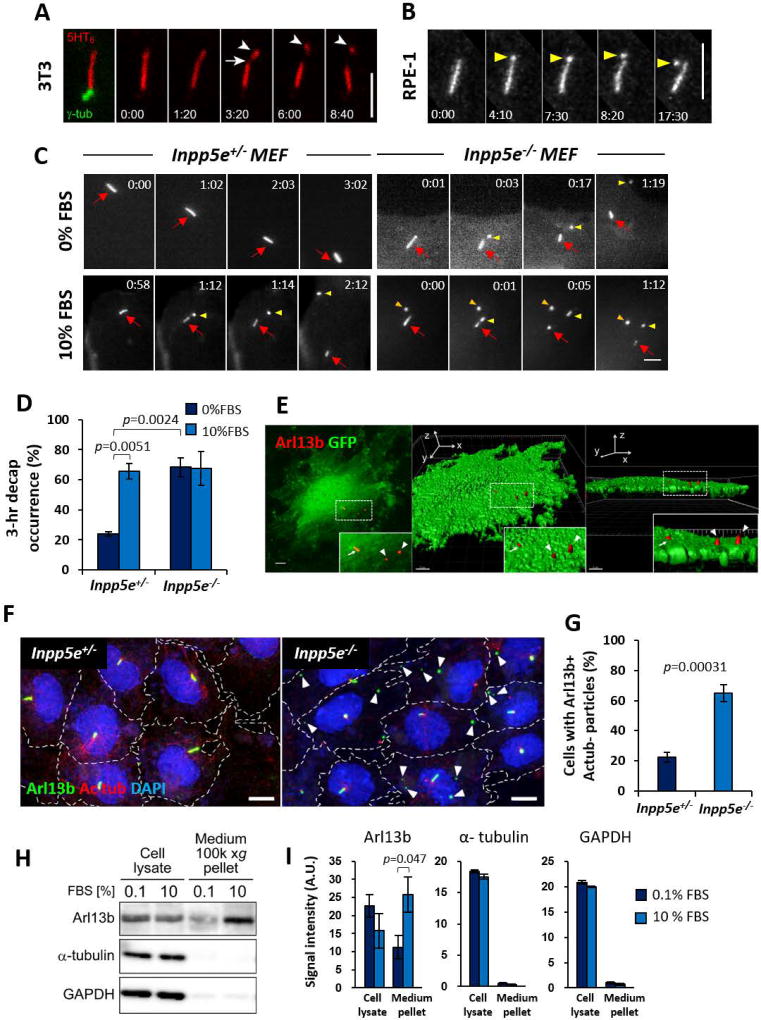
Figure 1. Growth stimulation induces cilia decapitation regulated by Inpp5e.
(A) Time-lapse images of a NIH/3T3 primary cilium expressing 5HT6-mCherry and γ-tub-GFP at quiescent state. Arrow: ciliary membrane thinning prior to excision. Arrowheads: excised cilium tip.
(B) Time-lapse images of an hTERT RPE-1 primary cilium expressing Arl13b-GFP at quiescent state. Note ciliary tip bulging prior to excision (yellow arrowheads).
(C) Time-lapse images of Inpp5e+/− and Inpp5e−/− MEF primary cilia expressing 5HT6-YFP at quiescent or growth-stimulated states. Red arrows: cilia. Yellow arrows: excised cilia tips. Orange arrow: YFP+ particles present from beginning.
(D) Scoring % of cells displaying cilia decapitation over 3 hours, as in (C). (n= 51, 49, 28, 34 cells from left to right; 3–5 experiments)
(E) Maximum intensity projection of Arl13b immunofluorescence and GFP in Inpp5e−/− MEF. Middle and right panels are 3D reconstructions of confocal image on left panel. Insets are magnifications of dotted regions. Arrows: cilia. Arrowheads: Arl13b+ particles residing on cell surface.
(F) Arl13b/Ac tub immunofluorescence and nuclear staining (DAPI) of quiescent Inpp5e+/− and Inpp5e+/− MEF. Arrowheads: Arl13b+Ac tub- particles.
(G) Scoring % of cells with associated Arl13b+Actub- particles, as in (F). (n= 344, 376 cells from left to right; 3 experiments)
(H) Representative Western blots in cell lysates and conditioned culture media pellets from mIMCD-3 cells treated with 0.1% or 10% FBS for 24 hours.
(I) Quantification of band signal intensities in (H). (n = 4 experiments for Arl13b, n = 3 experiments for α-tubulin and GAPDH)
Data are shown as mean ± SEM; Student’s T-tests were performed with p values indicated. Time in min:sec for (A) and (B), and hr:min for (C). Scale bars: 5µm in (A), (B), (C) and (E), and 10µm in (F). See also Figure S1 and Movies S1 and S2.
We next assessed the effect of growth stimulation on ciliary vesicle release. A three-hour 10% FBS treatment on Inpp5e+/− MEF stimulated ciliary vesicle release in 65.5 ± 5.3% of total cells (mean ± SEM), but the same treatment on Inpp5e−/− MEF did not additionally induce a higher occurrence of ciliary vesicle release (Figures 1C and 1D; Movie S2). To further demonstrate the effect of growth signals on extracellular ciliary vesicle release, culture media conditioned for twenty-four hours with quiescent (0–0.1% FBS) or growth-stimulated (10% FBS) mIMCD-3 cells were analyzed for ciliary markers (Figures S5D). Growth stimulation of mIMCD-3 resulted in significantly larger amounts of Arl13b detected in conditioned culture media, whereas extracellular levels of α-tubulin and GADPH were low and unaffected by growth stimulation (Figures 1H, 1I, S1I and S1J). Ciliary vesicle release is thus promoted by Inpp5e loss and growth stimulation. Since this process occurs through an excision of primary cilia tips, we hereon refer to this process as cilia decapitation.
Growth-stimulated cilia decapitation entails ciliary Inpp5e depletion and PI(4,5)P2 re-distribution
We next investigated growth stimulation-Inpp5e interplay. A four-hour 10% FBS stimulation to quiescent Inpp5e+/− MEF depleted endogenous Inpp5e in primary cilia (Figures 2A and 2B). To probe the functional role of Inpp5e on cilia decapitation, we targeted exogenous Inpp5e to primary cilia through fusion with a ciliary membrane marker and assessed the effect on cilia decapitation. As compared with 5HT6-YFP, 5HT6-YFP-Inpp5e(WT) reduced the proportion of Inpp5e+/− MEF harboring extracellular YFP-labeled ciliary vesicles over a four-hour 10% FBS stimulation, while 5HT6-YFP-Inpp5e(D477N; phosphatase-dead(PD)) did not exert similar effect (Figures S2A and S2B). In quiescent Inpp5e−/− MEF, 5HT6-YFP-Inpp5e(WT) also exerted a stronger repressive effect on ciliary vesicle formation than 5HT6-YFP-Inpp5e(PD) (Figures S2A and S2B). Overall, these results show that Inpp5e functions as a decapitation repressor in primary cilia.
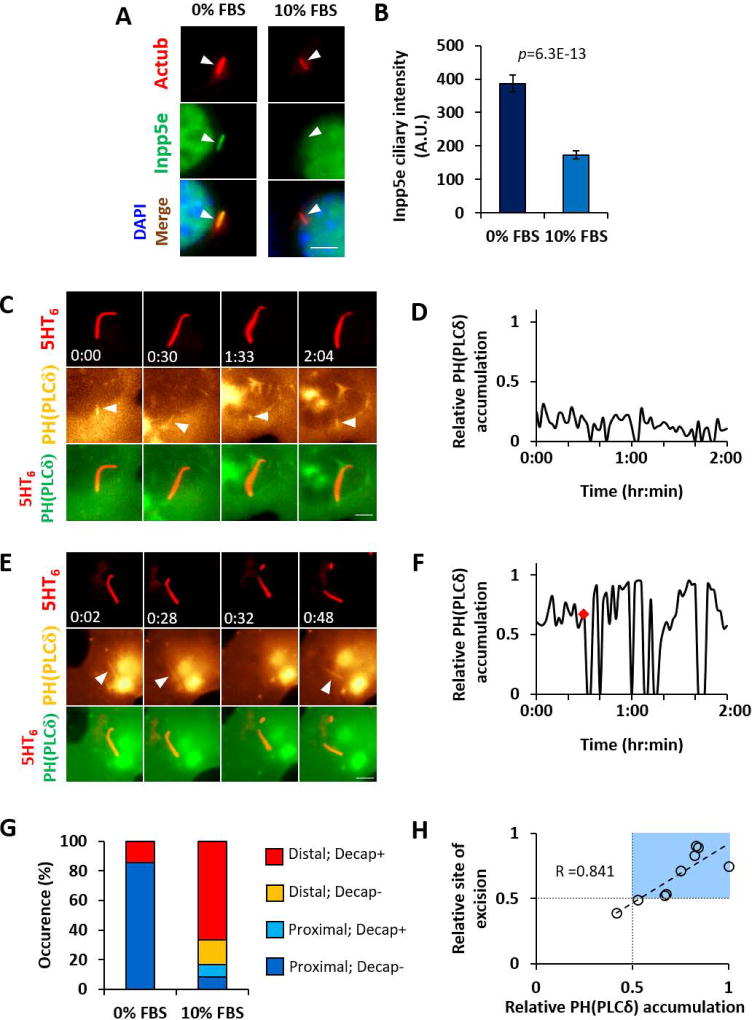
Figure 2. Growth stimulation regulates ciliary Inpp5e and PI(4,5)P2 localization.
(A) Ac tub/Inpp5e immunofluorescence and nuclear staining (DAPI) of Inpp5e+/− MEF treated with 0% FBS or 10% FBS for 4 hours. Images of each wavelength are scaled to same intensity range. Arrowheads mark cilia positions.
(B) Inpp5e immunofluorescence signal intensity measurements in primary cilia, as in (A). Data shown as mean ± SEM. Student’s T-test was performed with p values indicated. (n= 131, 148 cells from left to right; 2 experiments)
(C) Time-lapse images of Inpp5e+/− MEF primary cilium expressing 5HT6-mCeru3 and YFP-PH(PLCδ) (a PI(4,5)P2 sensor) at quiescent state. Arrows marks PH(PLCδ) at proximal cilia.
(D) Time-lapse measurements of PH(PLCδ) proximal-distal accumulation in cilium in (C), given as relative ratio of cilium length.
(E) Time-lapse images of Inpp5e+/− MEF primary cilium expressing 5HT6-mCeru3 and YFP-PH(PLCδ) at 4–6 hours of 10% FBS stimulation. Arrows marks ciliary PH(PLCδ).
(F) Time-lapse measurements of PH(PLCδ) accumulation in cilium in (E), given as relative ratio of cilium length. Red diamond marks cilia decapitation time point.
(G) Classifying ciliary PH(PLCδ) accumulation with cilia decapitation. “Distal” or “proximal” indicates PH(PLCδ) accumulating to distal or proximal half of ciliary length. “Decap+” or “Decap-” indicates presence or absence of cilia decapitation over 2-hour imaging periods. (n= 7,12 cells from left to right; 2–3 experiments)
(H) Correlation plot of ciliary PH(PLCδ) accumulation with site of excision, each given as a relative ratio of ciliary length. In each case, the maximal ciliary PH(PLCδ) accumulation value within ten minutes prior to excision is shown. Light blue box highlights distal location of ciliary PH(PLCδ) accumulation and site of cilia excision. Linear regression is drawn in dashed line, with Pearson correlation coefficient R value indicated. (n=9 cells from 10% FBS data in (G))
Time in hr:min for (C) and (E). Scale bars: 5µm. See also Figure S2 and Movie S3.
Since AurA is essential for cilia disassembly (Pugacheva et al., 2007), we examined if inhibition of AurA and its downstream effector, HDAC6, could affect ciliary Inpp5e. AurA inhibition using alisertib (Ast) countered growth-stimulated ciliary Inpp5e depletion, whereas HDAC6 inhibition using tubacin (Tub) did not (Figures S2E and S2F). In addition, AurA was responsible for cilia decapitation, with alisertib treatment suppressing growth-stimulated ciliary vesicle release in Inpp5e+/− MEF (Figure S2G). Although HDAC6 does not regulate Inpp5e localization, tubacin treatment did exert a mild effect on ciliary vesicle release in Inpp5e+/− MEF (Figure S2G), implying that HDAC6-mediated deacetylation of ciliary microtubules could indirectly affect cilia decapitation. Notably, neither alisertib nor tubacin inhibited ciliary vesicle release in Inpp5e−/− MEF (Figure S2G). The concomitant Inpp5e absence and AurA-independent cilia decapitation in Inpp5e−/− MEF supports that cilia tip excision requires ciliary Inpp5e depletion. Thus, growth-stimulated AurA activity drives Inpp5e depletion in primary cilia as well as cilia decapitation.
A net Inpp5e reduction in primary cilia could result in a lower capacity to deplete PI(4,5)P2. Whilst PI(4,5)P2 was confined within the proximal half of cilia in quiescent Inpp5e+/− MEF (Figures 2C, 2D and S2H; Movie S3 (Garcia-Gonzalo et al., 2015)), a 10% FBS stimulation for two hours or more led to PI(4,5)P2 accumulation in distal half of Inpp5e+/− cilia, concomitant with cilia decapitation (Figures 2E, 2F and S2I–K; Movie S3). Decapitation primarily occurred in cilia with PI(4,5)P2 accumulating to the distal half of ciliary length, and the position of cilia excision was correlated with the maximal ciliary PI(4,5)P2 accumulation prior to decapitation (Figures 2G and 2H). Interestingly, cilia decapitation was often followed by wide oscillations in ciliary PI(4,5)P2 accumulation, which suggests bi-directional regulation between PI(4,5)P2 and cilia decapitation (Figures 2E, 2F, S2J and S2K; Movie S3). Thus, growth-stimulated PI(4,5)P2 redistribution in primary cilia could organize signalling events leading to cilia decapitation.
Ciliary PI(4,5)P2 induces F-actin in ciliary lumen
Since PI(4,5)P2 re-organization could affect ciliary localization of actin regulators, we inspected F-actin formation in primary cilia of Inpp5e+/− and Inpp5e−/− MEF using mCerulean3(mCeru3)-Lifeact biosensor. Of note, an elevated steady state of ciliary PI(4,5)P2 in Inpp5e−/− MEF was associated with a tenfold increase in the probability of detecting Lifeact-positive structures in primary cilia of quiescent cells (Figures 3A and 3B). We also performed three-dimensional super-resolution structured illumination microscopy on cells stained with phalloidin, and detected similar phalloidin-labelled structures within ciliary lumen that was juxtaposed with the microtubule axoneme (Figure S3A). Direct PI(4,5)P2 amplification by targeting constitutively active PI(4)P 5-kinase Iγ (PIPK) to cilia (5HT6-FP-PIPK (Garcia-Gonzalo et al., 2015)) was similarly associated with a higher frequency of intraciliary F-actin than cilia-targeted kinase-dead PIPK (5HT6-FP-PIPK (D253A; KD)) (Figures 3A and 3B, S3B and S3C). Therefore, ciliary PI(4,5)P2 exerts direct control on intraciliary F-actin assembly. A screen for actin regulators further revealed PI(4,5)P2-dependent ciliary localization of cofilin-1, fascin and Kras small GTPase that could work together to induce actin polymerization in primary cilia (Figures S3B–G).
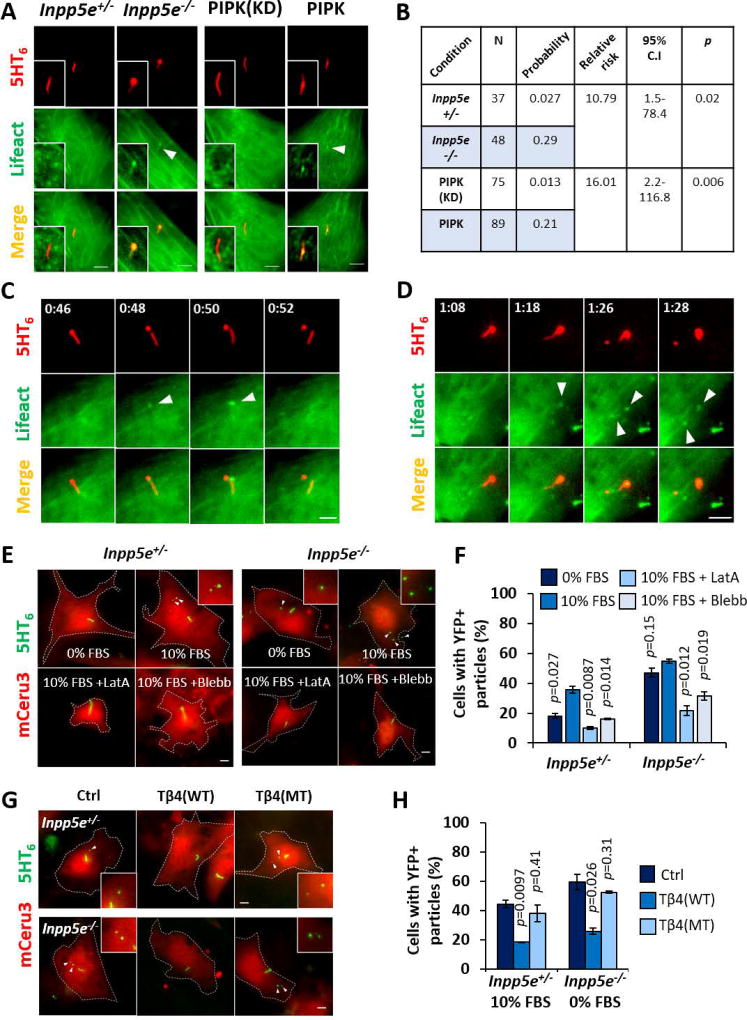
Figure 3. Ciliary PI(4,5)P2 induces intraciliary F-actin assembly which executes cilia decapitation.
(A) Live fluorescence images of (two leftmost columns) Inpp5e+/− and Inpp5e−/− MEF primary cilia expressing 5HT6-YFP and mCeru3-Lifeact, and (two rightmost columns) NIH/3T3 primary cilia expressing 5HT6-YFP-PIPK or 5HT6-YFP-PIPK(KD) and mCeru3-Lifeact. Insets are magnifications of respective cilia. Arrowheads mark ciliary Lifeact signals.
(B) Relative risk ratio analyses on the effect of ciliary PI(4,5)P2 in influencing intraciliary F-actin incidence, as in (A).
(C) Time-lapse images of a primary cilium from Inpp5e+/− MEF expressing 5HT6-YFP and mCeru3-Lifeact at 4–6 hours of 10% FBS stimulation. Arrowheads mark F-actin at site of cilia excision.
(D) Time-lapse images of primary cilium from Inpp5e−/− MEF expressing 5HT6-YFP and mCeru3-Lifeact at 0–2 hours of 10% FBS stimulation. Arrowheads mark F-actin at site of cilia excision. Note that F-actin first appeared in bulged cilia tip before expanding to proximal cilia region. F-actin was also detected in excised cilia tip.
(E) Live fluorescence images of Inpp5e+/− and Inpp5e−/− MEF expressing 5HT6-YFP with mCeru3 after 3 hours in 0% FBS, 10% FBS, 10% FBS + 200nM latrunculin A (LatA), or 10% FBS + 50µM blebbistatin (Blebb). Arrowheads indicate cell-associated YFP+ particles that were likely vesicles released from primary cilia, and insets are respective magnified images.
(F) Quantification of % cells with associated extracellular YFP+ particles, as in (E). (n=99, 82, 70, 62, 75, 69, 73, 67 cells from left to right; 2 experiments)
(G) Live fluorescence images of Inpp5e+/− MEF and Inpp5e−/− MEF expressing 5HT6-YFP (control), 5HT6-YFP-Tβ4(WT) or 5HT6-YFP-Tβ4(MT) with mCeru3 respectively after 3 hours of 10% FBS stimulation or at quiescence (0% FBS). Arrowheads indicate cell-associated YFP+ particles, and insets are respective magnified images.
(H) Quantification of % cells with associated extracellular YFP+ particles, as in (G). (n=83, 88, 96, 69, 95, 76 cells from left to right; 2 experiments)
Data shown as mean ± SEM. Student’s T-tests were performed with respect to (E) each 10% FBS condition and (F) each control condition, p values indicated. Time in hr:min for (A) and (B). Scale bars: 5µm. See also Figures S3 and S4 and Movie S4.
Intraciliary actin polymerization is required for cilia decapitation
We investigated F-actin involvement in cilia decapitation. By monitoring Inpp5e+/− and Inpp5e−/− MEF over two-hour periods during quiescence, or between zero and six hours of growth stimulation, we detected F-actin assembly at the site of cilia excision (Figures 3C and 3D, S4A–I; Movie S4). These intraciliary actin polymerization events were acute and transient, occurring a few minutes prior to each decapitation event (Figures 3C and 3D, S4H and S4I; Movie S4). Furthermore, growth-stimulated Inpp5e−/− MEF tend to assemble larger intraciliary F-actin foci than quiescent Inpp5e−/− MEF and growth-stimulated Inpp5e+/− MEF, suggesting stronger actin polymerization that was associated with cilia excision occurring at more proximal positions (Figures S4B and S4C).
Next, we determined the essentiality of F-actin in cilia decapitation. Latrunculin A restricted growth-stimulated ciliary vesicle release in Inpp5e+/− and Inpp5e−/− MEF, indicating that whole-cell F-actin turnover affects membrane excision in primary cilia (Figures 3E and 3F). Additionally, we devised a genetic strategy to suppress intraciliary F-actin by targeting thymosin β4 (Tβ4) to primary cilia, which sequesters G-actin from incorporation into actin filaments (Van Troys et al., 1996). As compared with 5HT6-YFP, 5HT6-YFP-Tβ4(WT) expression robustly suppressed ciliary vesicle formation in quiescent Inpp5e−/− MEF, as well as in Inpp5e+/− MEF over a four-hour growth stimulation period (Figures 3G and 3H). In contrast, 5HT6-YFP-Tβ4(KK18,19EE; actin-binding mutant MT (Van Troys et al., 1996)) expression did not exert similar effect. Thus, intraciliary actin polymerization is essential for cilia decapitation, and suppression could be achieved via a genetically-encoded ciliary actin inhibitor.
We then attempted to determine factors that regulate F-actin to promote ciliary membrane excision. Myosin II Inhibition with blebbistatin suppressed growth-stimulated ciliary vesicle release in Inpp5e+/− and Inpp5e−/− MEF (Figures 3E and 3F). We also explored sorting nexin 9 (SNX9), as the molecule was detected in the proteomic profile of ciliary vesicles described in the following section. Previous reports demonstrate SNX9 as a PI(4,5)P2 and PI(3,4)P2 binder which couples actin assembly with membrane remodelling at endocytic sites (Posor et al., 2013; Yarar et al., 2007). Accordingly, we observed accumulation of YFP-SNX9 in the vicinity of cilia excision site, shortly before cilia decapitation occurred in Inpp5e+/− MEF (23/23; 100% of decapitation events) (Figures S4J and S4K). These results suggest a role of actomyosin contractility and SNX9-mediated actin nucleation in cilia decapitation.
Released ciliary vesicles contain IFT-B and related cargoes
We dissected the contents of ciliary vesicles released into the extracellular milieu. Global proteomic profiling and comparative analyses were performed on culture media conditioned for twenty-four hours with quiescent (0.1% FBS) and growth-stimulated (10% FBS) wild-type (WT) or cilia-deficient Ift88-knockout (Ift88-KO) mIMCD-3 (Figures 4 and S5A–E), and we identified 1376 proteins that were detected two times or more in conditioned media of at least one experiment condition (Figure 4A; Table S1). Within this list, 477 represent proteins that were extracellularly released with growth stimulation, and 71 are indicative of proteins that were extracellularly released depending on functional cilia (Figure 4A; see legend for details). A 57-protein overlap between these two categories revealed putative ciliary vesicle components, which consist of a large majority of intraflagellar transport-related proteins, a subset of ciliary proteins, and Hedgehog signaling effectors (Figure 4B; Table S1). On average, identified IFT-B components were ranked higher than IFT-A components in terms of the relative abundance between growth-stimulated WT and Ift88-KO mIMCD-3 culture media pellets (Figures 4C and 4D; Table S2). At the protein level, larger amounts of Arl13b, Ift88 and Ift81 (IFT-B components) were indeed detected in conditioned media pellets from growth-stimulated WT mIMCD-3 than from quiescent WT mIMCD-3 or growth-stimulated Ift88-KO mIMCD-3, whereas Ift122 and Ift140 (IFT-A components) were scarcely detected in conditioned media pellets from both conditions (Figures 4E and 4F). This confirms that growth-stimulated ciliary vesicle release removes IFT-B, rather than IFT-A, from primary cilia. We also validated extracellular accumulation of Sufu and full-length Gli3 (Gli3FL) in conditioned media pellets of growth-stimulated WT mIMCD-3, while Gli2 and a truncated form of Gli3 (Gli3R) were hardly detected (Figures 4E and 4F).
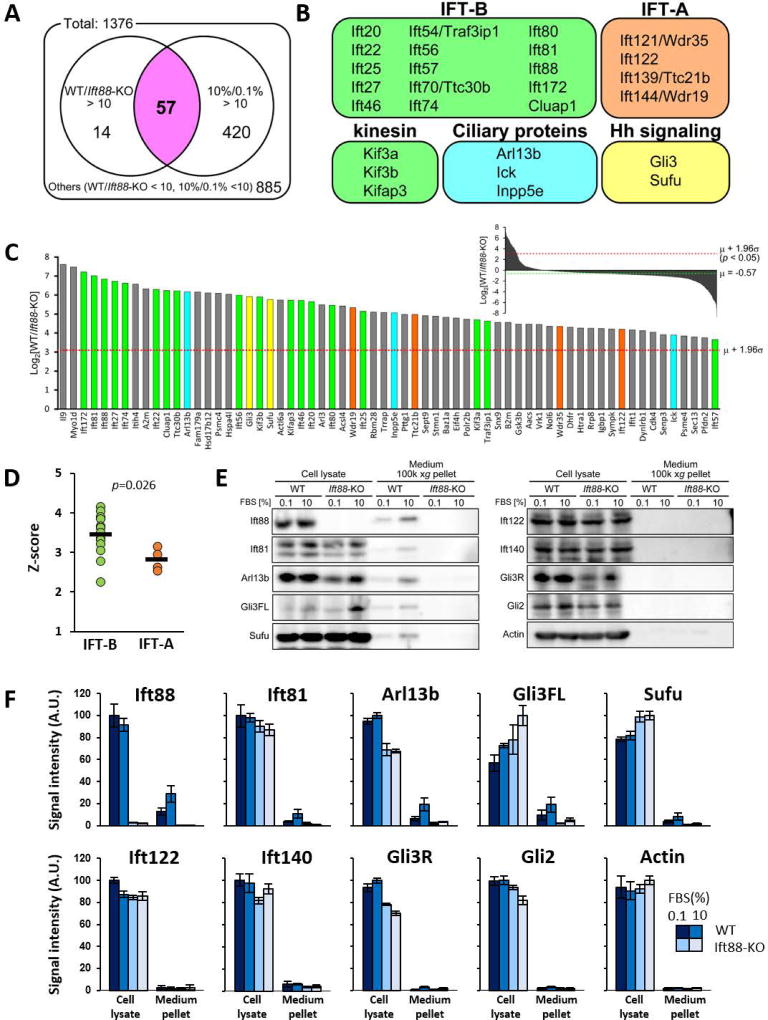
Figure 4. Global proteomic profiling of conditioned culture media reveals growth-stimulated extracellular release of IFT-B dependent on primary cilia.
(A) Venn diagram classification of proteins detected in conditioned culture media under indicated conditions, with a threshold of false discovery rate=0.01. “10%/0.1% >10”: 477 proteins with mass spectrometry signal areas ≥10 fold higher in 10% FBS condition than in 0.1% FBS condition, in WT mIMCD-3 (growth stimulation-dependent). “WT/Ift88KO >10”: 71 proteins with mass spectrometry signal areas ≥10 fold higher in WT mIMCD-3 than Ift88-KO mIMCD-3, in 10% FBS condition (cilia-dependent). There is a 57-protein (purple) overlap, i.e. both cilia- and growth stimulation- dependent.
(B) Classification of ciliary proteins amongst the 57 proteins highlighted in (A).
(C) Comparative analyses of proteins extracellularly released in cilia-dependent manner (WT/Ift88KO). Mass spectrometry signal area ratios for respective proteins are represented as logarithm to the base 2 (log2), and ranked in descending order of Z-scores. Only top 1.2% of proteins with Z-scores higher than 2.25 are shown. The mean (µ) log2 ratios of area values is -0.57, and standard deviation (σ) is 1.87. Color coding as in (B).
(D) Z-score comparison between IFT-B and IFT-A components identified from proteomic analysis, as in (C). Ift88 is excluded from IFT-B group, since a high WT/Ift88KO ratio would occur with Ift88-KO. Bars indicate mean of Z-scores. A Mann-Whitney U-test was performed with p values indicated.
(E) Representative Western blot analyses in total cell lysates and conditioned culture media pellets.
(F) Quantification of respective band signal intensities in (E). Data shown as mean ± SEM. (n=3 experiments)
We proceeded to understand the mechanism for relative IFT-B enrichment in released ciliary vesicles. In quiescent cells, Ift140(IFT-A) and Ift81(IFT-B) were detected along ciliary shafts of Inpp5e+/− and Inpp5e−/− MEF, with Inpp5e−/− primary cilia harboring high enrichment of Ift140 as previously reported (Figures S5F and S5G) (Garcia-Gonzalo et al., 2015). Interestingly, a four-hour 10% FBS stimulation led to increased ciliary Ift81 in Inpp5e+/− and Inpp5e−/− MEF, while ciliary Ift140 underwent reduction in both cell types (Figures S5F and S5G). In a mIMCD-3 cell line with endogenous Ift81 tagged with yellow Nano-lantern (YNL) (Figures S5H–J), Ift81-YNL signals were detected throughout the ciliary shaft at quiescence, with stronger accumulation in proximal cilia (Figure S5K). A four-hour growth stimulation not only elevated the overall Ift81-YNL in primary cilia, but also slightly shifted Ift81-YNL distribution towards the distal cilia (Figures S5K–O). Thus, an IFT-B increase with a complementary IFT-A decrease could potentially account for the selective release of IFT-B from primary cilia upon ciliary vesicle release.
Cilia decapitation is required for cilia disassembly
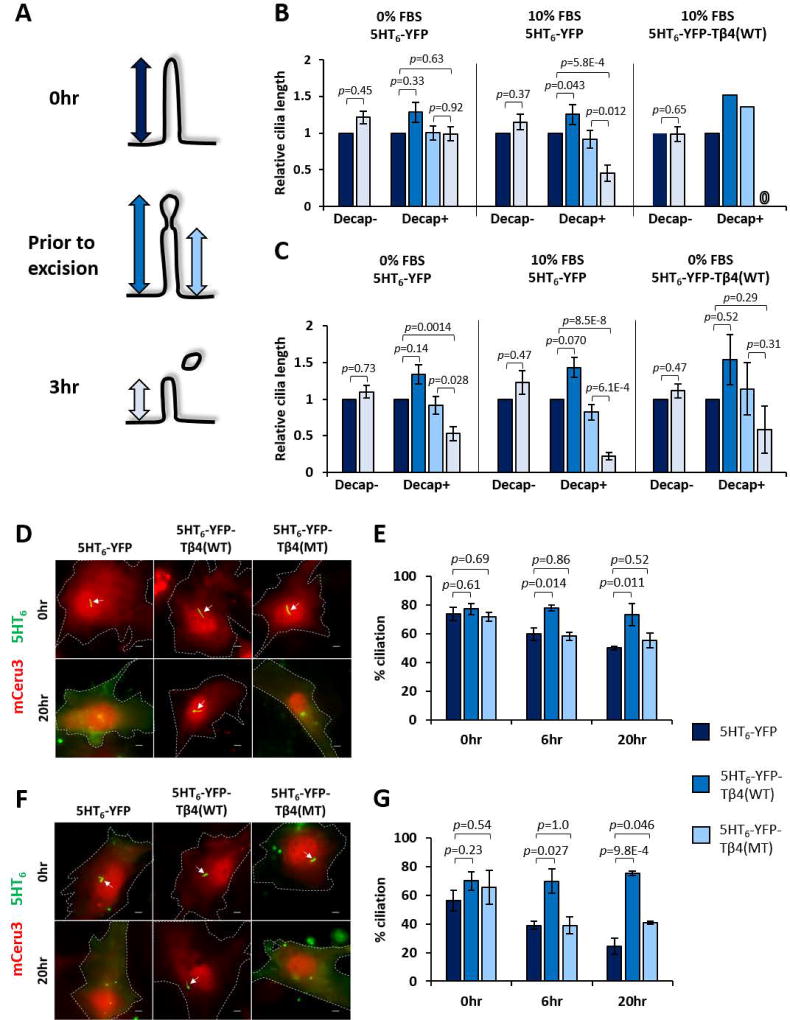
Cilia decapitation culminates in an acute loss of ciliary proteins that may compromise cilia stability. Likewise, Inpp5e loss triggers accelerated cilia instability upon growth stimulation (Figure S1G) (Bielas et al., 2009; Jacoby et al., 2009). We therefore investigated the functional consequences of decapitation in Inpp5e+/− and Inpp5e−/− MEF ciliary resorption. In growth-stimulated Inpp5e+/− MEF expressing 5HT6-YFP, the lengths of intact primary cilia were maintained (Decap- in 10% FBS; Figures 5A and 5B), while decapitated cilia underwent significant resorption within three hours post-decapitation (Decap+ in 10% FBS; Figures 5A and 5B). Consistently, growth-stimulated Inpp5e+/− primary cilia inhibited for decapitation via 5HT6-YFP-Tβ4(WT) expression were also suppressed for resorption (Decap+ in 10% FBS Tβ4(WT); Figures 5A and 5B). Thus, decapitation is necessary for cilia resorption induced by growth stimulation. In Inpp5e absence, growth-stimulated cilia decapitation in Inpp5e−/− MEF was accompanied with a greater extent of cilia resorption as compared with Inpp5e+/− counterpart (Decap+ in 10% FBS; Figures 5A and 5C). Cilia decapitation in quiescent Inpp5e−/− MEF was also sufficient to activate cilia resorption within three hours post-decapitation, albeit to a smaller extent than in growth stimulated events (Decap+ in 0% FBS; Figures 5A and 5C). Thus, cilia resorption post-decapitation was amplified in the absence of Inpp5e.
Figure 5. Inhibition of cilia decapitation suppresses growth-stimulated cilia disassembly.
(A) Schematic illustrating the time points when respective cilia lengths, indicated by double-headed arrows, were measured over a 3-hour live imaging period. Color coding for arrows applies to (B) and (C).
(B) Quantification of Inpp5e+/− MEF cilia lengths over 3-hour imaging period. Cilia are classified according to occurrence of cilia decapitation, and cilia lengths are given as a ratio to cilia length at T=0. Only a single cell was available for analysis under the category of “Decap+; 10% FBS Tβ4(WT)” due to high efficacy of 5HT6-YFP-Tβ4(WT) in inhibiting cilia decapitation. “0” indicates complete disassembly. Data in left and middle panels were derived from data in Figure 1D. (n=26,11,19,22,9,1 cells; 1–5 experiments)
(C) Quantification of Inpp5e−/− MEF cilia lengths over 3-hour imaging period, with similar data representation as in (B). Data in left and middle panels were derived from data in Figure 1D. (n=8,16,8,20,11,3 cells; 2–5 experiments)
(D) Live fluorescence images of Inpp5e+/− MEF under indicated conditions at 0 and 20 hours of 10% FBS stimulation. Arrows mark cilia.
(E) Quantification of % cells possessing primary cilia, as in (D). (n=167, 182, 162, 157, 153, 164, 176, 131, 139 cells from left to right; 3 experiments)
(F) Live fluorescence images of Inpp5e−/− MEF under indicated conditions at 0 and 20 hours of 10% FBS stimulation. Arrows mark cilia.
(G) Quantification of % cells possessing primary cilia, as in (F). (n=165, 155, 112, 156, 107, 123, 14, 136, 144 cells from left to right; 3 experiments)
Data shown as mean ± SEM; Student’s T-tests were performed with p values indicated. In (B–C), Student’s T-tests were performed on absolute cilia lengths. Scale bars: 5µm.
We further probed the role of decapitation on the extent of ciliation in Inpp5e+/− and Inpp5e−/− MEF expressing 5HT6-YFP, 5HT6-YFP-Tβ4(WT) and 5HT6-YFP-Tβ4(MT) at zero, six and twenty hours of 10% FBS stimulation. While a similar proportion of cells were ciliated at zero hour across all three conditions, a larger proportion of 5HT6-YFP-Tβ4(WT)-expressing cells harbored primary cilia at six and twenty hours of growth stimulation (Figures 5D–G). Thus, ciliary actin inhibition suppressed cilia decapitation that was required for cilia disassembly. Moreover, the observed role of 5HT6-YFP-Inpp5e(WT) in suppressing growth-stimulated cilia disassembly underscores how Inpp5e controls the disassembly process (Figures S2C and S2D).
Cilia decapitation ensures timely quiescence exit
Since growth stimulation of quiescent cells would induce cell cycle entry, we determined the timing of cilia decapitation in relation to cell cycle. Co-expression of Venus-p27K− (a G0 marker) and mCherry-hCdt1(30/120) (a G0/G1 marker) enables cells in G0, G1 or S phase to be respectively labelled with Venus/mCherry, mCherry only, or null nuclear fluorescence (Oki et al., 2014). G0-G1 transit will be manifested as a steep decrease in Venus-p27K−, while G1-S transit will be marked by a subsequent abrupt decrease in mCherry-hCdt1(30/120). By considering the relative expression ratio of Venus-p27K− and mCherry-hCdt1(30/120), we arbitrarily defined Venus-p27K−/mCherry-hCdt1(30/120) = 0.5 as the mid-point of G0-G1 transit.
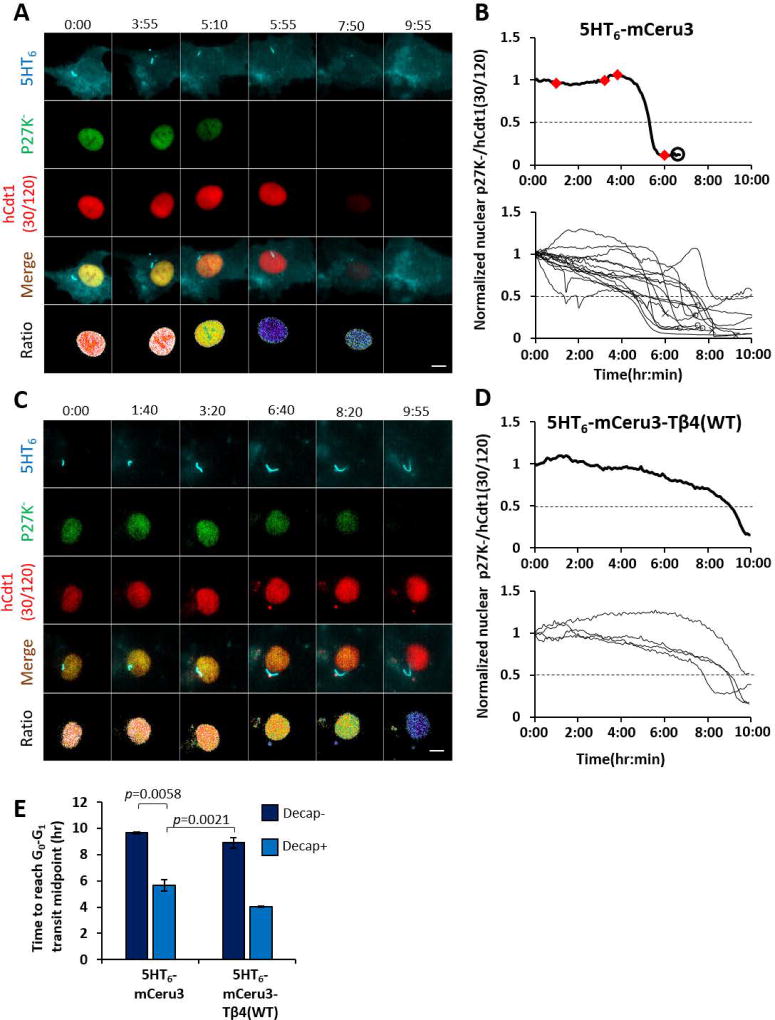
We expressed these cell cycle probes in Inpp5e+/− MEF with either 5HT6-mCeru3 or 5HT6-mCeru3-Tβ4(WT), and subjected quiescent cells to a ten-hour growth stimulation. In 5HT6-mCeru3-expressing cells, 20/25 (80%) cells underwent at least a single cilia decapitation event (Figures 6A, 6B, S6A and S6B). Out of these 20 cells, 19 (95%) of them exhibited the first cilia decapitation event prior to G0-G1 transit, while one cell underwent cilia decapitation during the transit (Figures 6A, 6B and S6A). Thus, growth stimulation at quiescence induces cilia decapitation in G0 phase. We further compared the time taken by cells with intact or decapitated primary cilia to reach the G0-G1 transit midpoint upon growth stimulation. Within the cell population which exhibited at least one cilia decapitation event at quiescence, 15/20 (75%) cells were determined to exit quiescence within ten hours of growth stimulation (i.e. these cells demonstrated a sharp decrease in Venus-p27K− at some point), and they took an average time of 5.7 ± 0.4 hours to reach the G0-G1 transit mid-point (Figures 6A, 6B, 6E and S6A; Movie S5). In contrast, 7/10 (70%) of 5HT6-mCeru3-Tβ4(WT)-expressing cells suppressed for cilia decapitation were either not determined for quiescence exit (43% (3/7)), or took a prolonged time of 8.9 ± 0.4 hours to reach G0-G1 transit mid-point (57% (4/7)) (Figures 6C–E and S6D; Movie S5). Thus, cilia decapitation is required by quiescent cells for timely G1 entry.
Figure 6. Cilia decapitation occurs in G0 and regulates G1 phase entry.
(A) Representative time-lapse images of prompt G1 entry that occurs with cilia decapitation in Inpp5e+/− MEF expressing 5HT6-mCeru3. Venus-p27K− was abruptly degraded at approximately 5 hours post-FBS stimulation, and rapid mCherry-hCdt1(30/120) depletion ensued by approximately 8 hours post-growth stimulation, indicating transit into G1 and S phase respectively. Images of each panel are scaled to same intensity range.
(B) Quantification of basal-normalized nuclear Venus-p27K-/mCherry-hCdt1 ratio (pseudo-colored in (A)) over 10 hours. Upper panel: plot for cell in (A). Red diamonds indicate cilia decapitation time points. Open circle marks beginning of mCherry-hCdt1(30/120) degradation. Lower panel: plot for all 15 cells that were determined for quiescence exit within the 10-hour period (i.e. these cells demonstrated a sharp decrease in Venus-p27K− at some point). Cilia decapitation events not denoted here; refer to Figure S6A for individual plots. Open circle marks beginning of mCherry-hCdt1(30/120) degradation. Crosses mark end of imaging period.
(C) Representative time-lapse images of prolonged G1 entry that occurs with suppressed cilia decapitation in Inpp5e+/− MEF expressing 5HT6-mceru3-Tβ4(WT). Note gradual Venus-p27K− degradation which indicates delayed G1 entry. Images of each panel are scaled to same intensity range.
(D) Quantification of basal-normalized nuclear Venus-p27K−/mCherry-hCdt1 ratio (pseudo-colored in (C)) over 10 hours. Upper panel: plot for cell in (C). No cilia decapitation was observed over the 10-hour imaging period. Lower panel: plot for all 4 cells that were determined to exit quiescence; refer to Figure S6D for individual plots.
(E) Quantification of time duration to reach G0-G1 transit mid-point, derived from time points when basal-normalized p27K−/hCdt1=0.5. Only cells that reached ratio value ≤0.5 by 10 hours were considered here. Data shown as mean ± SEM. Student’s T-tests were performed with p values indicated. (n=2, 15, 4, 2 cells; 3–7 experiments)
Time in hr:min. Scale bars: 10µm. See also Figure S6 and Movie S5.
Cilia decapitation modulates Gli transcription factor activity
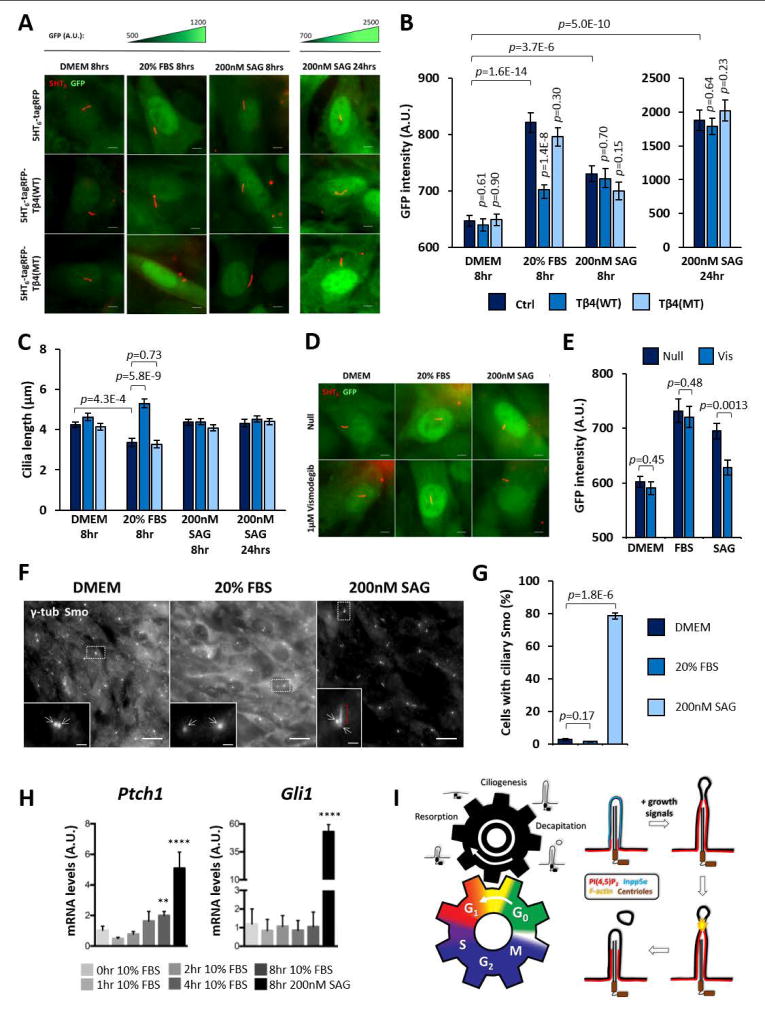
We speculated that cilia decapitation might promote quiescence exit through mitogenic Hedgehog (Hh) signaling (Roy and Ingham, 2002). We established an NIH/3T3 cell line stably expressing a GFP reporter driven by a minimal promoter and 8xGli-Binding-Site (GBS) sequence to measure Gli1/2 (but not Gli3) transcription activity in single cells (Sasaki et al., 1997; Stamataki et al., 2005). This reporter cell line was transfected with either 5HT6-tagRFP, 5HT6-tagRFP-Tβ4(WT) or 5HT6-tagRFP-Tβ4(MT) prior to quiescence induction, and Gli1/2 activity in these cells was measured in response to growth stimulation (20% FBS) or Smoothened (Smo)-dependent Hh activation (SAG; Smo agonist). All three groups of cells similarly expressed a low basal level of GFP at quiescence (Figures 7A and 7B). In SAG positive controls, a modest increase in GFP fluorescence was detected at eight hours post-stimulation across these cells, which robustly increased by twenty-four hours (Figures 7A and 7B). Remarkably, an eight-hour growth stimulation induced an intermediate GFP fluorescence increase in 5HT6-tagRFP-expressing cells that was accompanied by an average decrease in cilia length (Figures 7A–C). Similar observations were made for 5HT6-tagRFP-Tβ4(MT)-expressing cells, whereas 5HT6-tagRFP-Tβ4(WT)-expressing cells, which were suppressed for cilia decapitation, exhibited neither comparable changes in GFP fluorescence nor cilia length (Figures 7A–C). Moreover, Smo inhibitor, Vismodegib was able to abolish the GFP fluorescence increase in SAG-induced conditions but not in FBS-treated conditions (Figures 7D and 7E). We also verified that an eight-hour treatment with 20% FBS did not promote Smo translocation into primary cilia (Figures 7F and 7G). Overall, these results implied a Smo-independent role of cilia decapitation in growth-induced Gli1/2 activation. To further verify this, we assessed how the expression of general Hh targets Gli1 and Ptch1 were affected by growth stimulation. Control treatment with SAG induced a robust increase in Gli1 and Ptch1 gene expression in NIH/3T3 at eight hours. Interestingly, an approximately two-fold increase in Ptch1 gene expression was observed in NIH/3T3 cells at eight hours post-stimulation with 10% FBS, while no change was observed for Gli1 gene (Figure 7H). Growth-induced Ptch1 expression supports an increase in Gli1/2 activity. Lastly, a lack of Gli1 gene up-regulation suggests that Gli1 activation upon growth stimulation would likely occur at the post-transcriptional level.
Figure 7. Growth-induced Gli activation is dependent on cilia decapitation.
(A) Representative fluorescence images of NIH/3T3: 8xGBS-GFP reporter line under indicated conditions. GFP fluorescence intensities of the same column group are scaled to the same intensity ranges indicated above each group.
(B) GFP fluorescence intensity measurements, as in (A). (n= 70, 89, 72, 64, 73, 70, 61, 77, 61, 39, 56 and 59 cells from left to right; 4 experiments)
(C) Cilia length measurements in response to indicated stimuli and cilia-targeted probes; data derived from same experiments in (B).
(D) Representative fluorescence images of NIH/3T3: 8xGBS-GFP reporter line at 8 hours under indicated conditions. GFP fluorescence intensities are scaled to the same intensity range.
(E) GFP fluorescence intensity measurements, as in (D). (n= 39, 54, 48, 49, 43 and 44 cells from left to right; 3 experiments)
(F) γ-tub and Smoothened (Smo) immunofluorescence on NIH/3T3: 8xGBS-GFP reporter line upon 8 hours with indicated conditions. Insets are magnifications of dotted regions. Arrows indicate centrioles. Red bracket indicates ciliary Smo signals.
(G) Scoring % cells with ciliary Smo signals, as in (F). (n=249, 206, 239 cells from left to right; 3 experiments)
(H) Quantitative real-time PCR assay for Ptch1 and Gli1 gene expression performed on NIH/3T3 post-stimulation with 10% FBS. An 8-hour 200nM SAG positive control for Smo-dependent Hedgehog signaling activation was included. Ubc was used to normalize Ptch1 and Gli1 transcript levels. One-way ANOVA was performed to compare 0-hour 10% FBS samples with all other samples. ** p< 0.01; **** p< 0.0001 (n= 8 experiments)
(I) Summary model of cilia decapitation during quiescence exit. (Left panel) Two meshing gears represent the mutual dependency between cell division cycle and primary cilium life cycle. As known, growth induction (counterclockwise rotation of the bottom gear) promotes cilia disassembly (clockwise rotation of the top gear). The present study characterizes cilia decapitation as one of the key gear teeth enmeshing the two biological cycles. Cilia decapitation stimulates disassembly of cilia, and also modulates cell proliferation by inducing G1 entry. (Right panel) Growth-stimulated cilia decapitation occurs through four major steps: (1) Inpp5e re-localization (2) PI(4,5)P2 elevation (3) Actin polymerization (4) Cilia tip excision.
In (B), (C), (E), (G), data are shown as mean ± SEM; Student’s T-tests were performed between indicated sample pairs (horizontal p values), or with respect to each 5HT6-tagRFP condition (vertical p values). Scale bars indicate 5µm in (A) and (D), 20µm and 2µm in (F) and (F) insets respectively.
Discussion
A primary cilium is traditionally defined by the presence of a microtubular axoneme and an absence of actin filaments. Here, we demonstrate that primary cilia possess capacity for actin polymerization under specific conditions. F-actin-powered cilia decapitation not only triggers resorption of the ciliary structure, but also regulates proliferative signaling which connects the cilia life cycle and cell division cycle (Figure 7I).
The molecular players of cilia decapitation
Inpp5e and HDAC6 act in complementary pathways downstream of AurA to disassemble the ciliary structure. Earlier work determined that AurA phosphorylates Inpp5e to modulate its 5-phosphatase activity (Plotnikova et al., 2014), and we discovered that AurA also dictates Inpp5e re-localization. It is therefore tempting to speculate that phosphorylated Inpp5e may have altered affinity against binding partners which coordinate its ciliary localization (Humbert et al., 2012). Within the primary cilia, Inpp5e functions as a rheostat which tunes PI(4,5)P2 required for actin polymerization. The resultant assembly of F-actin in distal cilia could be attributed to the presence of a bulky microtubular axoneme which physically obstructs stable F-actin foci organization in the proximal ciliary lumen. Upon growth stimulation, concomitant HDAC6-dependent axonemal resorption and Inpp5e-dependent PI(4,5)P2 re-distribution could culminate in a position in distal cilia that is devoid of microtubules while harboring PI(4,5)P2, permitting stable actin nucleation at that point. Moreover, the tip region of a primary cilium has been revealed as a sub-compartment harboring specific signaling molecules(He et al., 2014). The distal cilia may similarly possess properties promoting actin polymerization, and this is supported by our observed cofilin-1 enrichment in the tips of PI(4,5)P2-elevated cilia (Figures S3B and S3D).
The functional role of IFT-B elimination in cilia disassembly
Cilia decapitation could function as a ciliary outlet for IFT-B during cilia disassembly. Many components of IFT-B are associated with cilia growth; Ift81 and Ift74 constitute a tubulin-binding module which captures and delivers αβ-tubulin dimers to the plus end of ciliary axoneme for elongation (Bhogaraju et al., 2013). During the process of cilia resorption, the large pore size (7.9–9nm) of a diffusion barrier positioned at the ciliary base would allow rapid diffusion of resorbed IFT-B constituents from the cytosol back into the ciliary lumen to result in inappropriate cilia re-growth (Breslow et al., 2013; Lin et al., 2013). Acute removal of IFT-B from primary cilia through decapitation could limit cilia re-growth, thereby playing an active, facilitating role in cilia disassembly. Up-regulation of the ciliary IFT-B/IFT-A ratio by growth stimulation could enhance IFT-B removal from ciliary tips, and could involve the serine/threonine protein kinase ICK, which is a candidate ciliary vesicle component involved in regulating the ciliary anterograde motor, Kif3a (Figure 4C; Table S2) (Chaya et al., 2014).
Cilia decapitation as a driver of quiescence exit
In differentiating neural progenitors, primary cilia dismantling as a result of apical abscission terminates mitogenic Hh signals for quiescence entry (Das and Storey, 2014). Conversely, we propose that cilia decapitation enhances Hh signaling to drive quiescence exit. Growth induction likely activates the mTOR signaling pathway in which S6K1 phosphorylates Gli1 to release it from Sufu inhibition (Wang et al., 2012), and active Gli1 could promote downstream cyclin D2 expression which functions in quiescence exit (Susaki et al., 2007; Yoon et al., 2002). Exploring cilia decapitation-mTOR signaling crosstalk thus holds exciting potential for understanding ciliary control of cell cycle, and is motivated by: (i) a role of primary cilia in mTOR signaling suppression via Lkb1 (Boehlke et al., 2010; Orhon et al., 2016) (ii) detection of an mTOR inhibitor, glycogen synthase kinase-3 beta (GSK3B) in ciliary vesicle proteomics (Figure 4C; Table S2) (Inoki et al., 2006). The Smo-independent role of growth stimulation on Gli1 activity is distinct from the Hh signaling repression caused by ciliary accumulation of Smo antagonists with complete Inpp5e loss (Chávez et al., 2015; Garcia-Gonzalo et al., 2015). Future work would entail understanding how Inpp5e loss affects Smo-independent Gli1 regulation, and how growth-induced changes in ciliary PI(4,5)P2 distribution could affect Smo antagonists in primary cilia.
Conclusion
Taken together, our present work describes a cell-autonomous role of cilia decapitation in cell cycle regulation. Interestingly, earlier proposals characterized ciliary vesicles as signaling devices in algae and worms (Bergman et al., 1975; Cao et al., 2015; Wang et al., 2014, 2015; Wood and Rosenbaum, 2015; Wood et al., 2013). In line with these reports, mammalian primary cilia also associate with extracellular vesicles in vivo (Dubreuil et al., 2007; Hogan et al., 2009; Jacoby et al., 2009; Wood and Rosenbaum, 2015). Thus, the vesicles generated from cilia decapitation may engage in signal transmission in a non-cell-autonomous manner, and exhibit potential as diagnostic readouts in development and cancer.
STAR Methods Text
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the lead author Takanari Inoue (jctinoue@jhmi.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
We derived MEF from littermate E19.5 Inpp5e+/− and Inpp5e−/− embryonic tails using mouse protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of California, San Francisco. Primary MEF were maintained in DMEM, 15% FBS, and PenStrep, and immortalized by infection with a lentivirus expressing SV40 large T antigen.
Immortalized MEF and NIH/3T3 were cultured in DMEM medium containing 10% FBS. mIMCD-3 and hTERT RPE-1 cells were cultured in DMEM/F-12 (1:1; Invitrogen) medium supplemented with 10% FBS. NIH/3T3-Flp-in: 8xGBS-GFP line was cultured in DMEM medium containing 10% FBS and 2mM GlutaMAX (Gibco). In all experiments, cells were first induced for ciliogenesis by attaining quiescent state with following conditions: NIH/3T3 and MEF were cultured in Opti-MEM® I reduced serum medium (Thermofisher Scientific) with 0% FBS for 24 hours, NIH/3T3-Flp-in: 8xGBS-GFP line was cultured in DMEM containing 2mM GlutaMAX for 48 hours, and mIMCD-3 and hTERT-RPE1 were cultured in DMEM/F-12 (1:1; Invitrogen) medium containing 0–0.1% FBS for 24 hours. To stimulate growth in quiescent cells, 10–20% FBS were added to respective media depending on cell type. In some experiments, cells were treated with 200nM Alisertib (MedChem Express) and 2µM Tubacin (Cayman Chemical Company). For live cell imaging, cells were plated on poly(d-lysine)-coated borosilicate glass Lab-Tek 8-well chambers (Thermo Scientific).
METHOD DETAILS
Transient and stable transfection
For transient transfection, cells were transfected with the respective DNA constructs by plating them directly in a transfection solution containing DNA plasmids and Xtremegene 9 (Roche). For mIMCD-3 and hTERT-RPE1 Arl13b-GFP stable lines, cells were transfected with Arl13b-pEGFP (Clontech) using FuGENE HD transfection reagents (Promega), and stable clones were selected in culture media containing 750 mg/ml G418. The plasmids were transfected
DNA plasmid construction
To construct 5HT6-YFP/mCeru3/tagRFP expression plasmids, we amplified DNA encoding 5HT6 with 5′ and 3′ AgeI cleavage sites by PCR from 5HT6-EGFP and subcloned it into pEYFP/mCerulean3/tagRFP-C1 vectors (Clontech). To construct 5HT6-YFP/mCeru3-PIPK and 5HT6-YFP/mCeru3-PIPK(KD) expression plasmids, we digested DNAs encoding the wild-type or mutant forms of PIPK from CFP-FKBP-PIPK (Suh et al., 2006) with 5′ EcoRI and 3′ BamHI cleavage sites and subcloned them into 5HT6-YFP/mCeru3 expression plasmids. 5HT6-mCherry was constructed by inserting stop codon-lacking mouse 5HT6 cDNA into pmCherry-N1 using 5’ EcoRI and 3’ BamHI sites. To construct 5HT6-YFP/mCeru3/tagRFP-Tβ4(WT), full-length thymosinβ4 cDNA was amplified from NIH/3T3 total cDNA using fwd: 5’-TTGG GAATTCGATGTCTGACAAACCCGAT-3’ and rev: 5’-CCAAGGATCCCGATTCGCCAGCTTGCTT-3’ primers, and inserted into 5HT6-YFP/mCeru3/tagRFP using 5’ EcoRI and 3’ BamHI sites. Thymosinβ4 KK18,19EE mutant (Tβ4(MT)) was subsequently constructed via site-directed mutagenesis using fwd: 5’-TCGATAAGTCGAAGTTGGAGGAAACAGAAACGCAAGAG-3’ and rev: 5’-CTCTTGCGTTTCTGTTTC-CTCCAACTTCGACTTATCGA-3’ primers. For 5HT6-YFP-Inpp5e(WT), the human Inpp5e CaaX domain was first abolished using a C641A mutation to make Inpp5e non-membrane bound, and the resultant catalytic domain was amplified using fwd: 5’-TTGGGAATTCGCGGATCTTGCAGACTACAAGCTC-3’ and rev: 5’-AACCGGATCCTCAAGAAACGGAGGCGATGGTGC-3’ primers and inserted into 5HT6-YFP using 5’ EcoRI and 3’ BamHI sites. Inpp5e D477N mutant was constructed via site-directed mutagenesis using fwd: 5’-AGGTGTTCTGGTTTGGAAACTTCAACTTCCGCCTGAG-3’ and rev: 5’- CTCAGGCGGAAGTTGAAGTTTCCAAACCAGAACACCT-3’ primers. γ-tubulin-EGFP was constructed through cloning stop codon-lacking human TUBG2 cDNA into pEGFP-N2 plasmid using 5’ BglII and 3’ EcoRI sites. PMXs-IP-mVenus-p27K− and pCSII-EF-mCherry-hCdt1(30/120) plasmids were kind gifts from Toshio Kitamura and Atsushi Miyawaki respectively.
Generation of Ift88-KO mIMCD-3
Ift88-KO mIMCD-3 was generated with a CRISPR/CAS9-based genome editing technique. mIMCD-3 cells were transfected transiently by an all-in-one plasmid U6-gRNA(Ift88)/CMV-Cas9-2A-GFP harboring CAS9, GFP, and guide RNA, available from Sigma-Aldrich. The target sequence of guide RNA was 5’-GGAGGTCTTCTGCCATGAC-3’. Cells expressing CAS9 was sorted next day with a fluorescence-activated cell sorter, ARIA (BD). The sorted cells were cloned in 96-well plate. Screening was performed with western blot analyses. Cells absent for Ift88 protein signals were further verified by analyzing target site DNA sequence in the genome.
Generation of Ift81:YNL knock-in mIMCD-3
Ift81:YNL knock-in mIMCD-3 cell line was generated using CRISPR/CAS9-based genome editing technique. mIMCD-3 cells were co-transfected with an all-in-one plasmid U6-gRNA(Ift81)/CMV-Cas9-2A-GFP harboring CAS9, GFP, and guide RNA, and a plasmid vector pCR-LA(Ift81)-YNL-RA(Ift81) harboring the left arm (834 bp), YNL (1614bp), and right arm (715 bp). The target sequence of gRNA was selected by using an algorism provided by Broad Institute (http://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design). The target sequence of guide RNA was 5’-ACAGGGCTCAGAGAACCAGC-3’. Cells expressing CAS9 were sorted next day with a fluorescence-activated cell sorter, ARIA (BD). The sorted cells were cloned in 96-well plate. Initial screening was performed by PCR with primers: 5’-CAGTTGGCAGTTAAGAAACGGAG-3’; 5’-GTAGTCCACATGGAACAGAGGC-3’. Expression of Ift81-YNL was verified using Western blot analyses with anti-GFP antibody (Medical & Biological Laboratories, Nagoya, Japan).
Generation of NIH/3T3-Flp-In: 8xGBS-GFP and reporter assay
A minimal promoter and 8xGLI-binding-site (GBS) sequences were inserted upstream of GFP within a pRRL.sin-18.PPT.GFP.pre lentiviral plasmid vector (Monje et al., 2011), and prepared lentiviruses were used to infect NIH/3T3-Flp-In cells (Life Technologies) in the presence of 4 µg/ml polybrene (Sigma). Cells were grown to confluence prior to serum starvation in the presence of 200nM SAG (Enzo Life Sciences) for 24 hours. GFP-expressing cells were single cell sorted into a 96-well plate using FACSAria II (BD Biosciences), and multiple single cell-derived clones were analyzed for SAG-induced GFP expression. For all reporter assay experiments, a specific clone with low basal GFP expression and high SAG-induced GFP expression was used. 40000 cells were seeded into one well of Lab-Tek 8-well chamber (Thermo Scientific), and cells were induced for quiescent state by culturing in serum starvation media (DMEM containing 2mM GlutaMAX) for 48 hours. Afterwards, cells were treated with serum starvation media containing 20% FBS or 200nM SAG (Enzo Life Sciences), in the presence or absence of 1µM Vismodegib (LC labs) for 8 hours or 24 hours. Cells were then fixed with 4% paraformaldehyde at room temperature for 10 mins prior to imaging of reporter GFP expression.
Immunofluorescence
For most experiments, cells were fixed with 4% (v/v) paraformaldehyde at room temperature for 10 min, permeabilized with 0.1% (v/v) Triton-X100 for 5 min, blocked with 2% bovine serum albumin for 1 hour. Antibodies used are as follows: rabbit anti-Inpp5e (1:500; a kind gift from Stéphane Schurmans), rabbit anti-Smo (1:500; as reported in (Rohatgi et al., 2007)) rabbit anti-Arl13b (1:500; Proteintech 17711-1-AP), mouse anti-acetylated tubulin (1:1000; Sigma, T7451), mouse anti-gamma tubulin (1:1000; Sigma T6557). For immunofluorescence with rabbit anti-Ift81 (1:250; Proteintech,11744-1-AP), cells were fixed with 4% (v/v) paraformaldehyde for 10 min, prior to −20°C methanol fixation for 3 min. For immunofluorescence with rabbit anti-Ift140 (1:100; Proteintech, 17460-1-AP), cells were fixed with −20°C methanol for 3 min. F-actin probe, Alexa Fluor 488 Phalloidin (ThermoFisher Scientific) was also used.
Epi-fluorescence imaging
Live-cell imaging experiments were mostly performed using an IX-71 (Olympus) microscope with a 63x oil objective (Olympus) (with additional 1.6x optical zoom) and a Cool-SNAP HQ charge-coupled device camera (Photometrics) or an ORCA-Flash4.0 LT Digital CMOS camera (Hamamatsu). Time-lapse imaging experiments were performed with either 2-min or 5-min intervals, and between 3 and 11 0.5-µm z-stacks were taken at each time point. Images shown were mostly maximum intensity projection. Micrographs were taken and analyzed using MetaMorph 7.5 imaging software (Molecular Devices).
Confocal imaging
Confocal live cell imaging of NIH/3T3 cilia decapitation was performed on FV1000 (Olympus, Japan) equipped with a stage top incubator (Tokai Hit, Japan). Confocal imaging of Arl13b+Actub- particles and live cell imaging of mIMCD-3 and hTERT RPE-1 cilia decapitation were performed using a LSM710 confocal microscope (Carl Zeiss microscopy) equipped with a Plan Aprochromat 63X oil immersion objective lens (NA 1.4) and processed using Zeiss Zen software. mIMCD-3 ciliary Ift81-YNL signals were visualized via direct 488-nm laser excitation of Venus on SP8 confocal microscope (Leica), and 3D image reconstruction were performed through volume rendering in 3D View software equipped in LAS X (Leica).
Structured illumination microscopy imaging
3D-SIM imaging was performed on an ELYRA S.1 microscope (Carl Zeiss microscopy) equipped with an Andor iXon 885 EMCCD camera, a 100X/1.40 NA oil-immersion objective and four laser beams (405, 488, 561, 642 nm). Serial z-stack sectioning was carried out at 101 nm intervals. Z-stacks were recorded with 3 phase-changes and 5 grating rotations for each section. The microscope was routinely calibrated with 100 nm fluorescent beads to calculate both lateral and axial limits of image resolution. Images were reconstituted with Zeiss Zen software. 3D rendering was produced by using Imaris 7.4.2 (Bitplane). Images were extracted from Imaris and then used to obtain the final images and movies used in the figures.
Quantitative image analyses
Inpp5e, Ift81 and Ift140 ciliary signals were quantified via subtraction of ciliary fluorescence signals from vicinity background signals; the boundary of ciliary axoneme was defined by Actub immunofluorescence, while γtub immunofluorescence enables distinguishing between ciliary base and tip. For ciliary Ift81-YNL measurements, each cilium was divided into 10 equal compartments along cilia length. YNL fluorescence intensities along cilia length was obtained using a line scan spanning from base to tip of each primary cilium, and mean YNL fluorescence intensity in each compartment was obtained by averaging across entire sample. Total YNL fluorescence intensity in cilium was calculated by integrating fluorescence intensities across all 10 compartments. Mean YNL fluorescence intensity distribution along cilia length was obtained by expressing mean YNL fluorescence intensity in each compartment as a percentage of Total YNL fluorescence intensity in cilium.
Relative ciliary PI(4,5)P2 and small GTPase accumulation measurements were as previously reported (Garcia-Gonzalo et al., 2015). Probes were scored as cilia-localized if probe fluorescence signals were co-localized with 5HT6 or Arl13b fluorescence signals in x-, y- and z- planes. Cell-associated ciliary vesicles were either defined as Arl13b+Actub- particles (on fixed cells) or 5HT6/Arl13b-YFP-positive particles that were associated with cell surface. To determine mid-point of G0-G1 transit, 10-hour time series of nuclear Venus-p27K− and mCherry-hCdt1(30/120) fluorescence signals were subtracted with background signals, divided to obtain Venus-p27K−/mCherry-hCdt1(30/120) signal ratio values, and further normalized with basal ratio values at T=0. For 8xGBS-GFP Gli activity reporter measurements, average nuclear GFP signal in best-focused z-plane was measured for each cell under specified conditions, since: (1) expressed GFP show nuclear enrichment in NIH/3T3 cells (2) high cell densities preclude ability to measure whole-cell signals.
It is of note that we occasionally observed primary cilia of fibroblasts extending from the ventral cell surface and adhering to underlying substratum. Subsequent cellular movements could result in mechanical pulling and breaking of primary cilia (Figure S1K). Care was taken to exclude instances of passive cilia breaking throughout all experiments.
Ciliary vesicle collection
FBS used for vesicle collection was pre-depleted via overnight centrifugation at 100,000 xg to remove FBS-intrinsic tiny vesicles, exosomes and ectosomes. Wild-type or Ift88-KO mIMCD-3 cells were subjected to serum starvation by reducing FBS concentration to 0–0.1% for 12–24 hours after reaching confluence. Culture media were then replaced by fresh media containing 0–0.1% or 10% pre-depleted FBS. Conditioned culture media were collected 24 to 30 hours later. Small vesicles released from cultured cells into media were collected through a three-step centrifugation. Culture media were first centrifuged at 1,500 xg for 20 min to remove large cell debris. The supernatant of the first centrifuge was further centrifuged at 10,000 xg for 20 min. The supernatant of the second centrifuge was centrifuged at 100,000 xg for 3 hr with MLA-55 angle rotor (Beckman). Pellets were rinsed once with PBS, and then subjected to western blotting or proteomics.
Western blotting
Collected small vesicles were resolved in 1.5x SDS sample buffer. Cultured cells were also lysed with the same buffer. Extracted proteins were denatured at 95 °C for 5 min. Proteins were separated with SDS-PAGE, and transferred to PVDF membrane (Millipore). The membrane was blocked with 10% goat serum or 5% bovine serum albumin. The blocked membrane was incubated with primary antibodies in the blocking solution at 4 °C overnight. Primary antibodies used were as follows: rabbit anti-Ift88 (1:2000; ProteinTech, 13967-1-AP); mouse anti-Kif3A (1:2000; BD Transduction, 611508); goat anti-Gli3 (1:200; R&D Systems, AF3690); rabbit anti-Gli2 (1:1000; Abcam, ab26056); rabbit anti-Sufu (1:2500; as reported in (Humke et al., 2010)); rabbit anti-Arl13b (1:1000; ProteinTech, 17711-1-AP); rabbit anti-actin (1:2000; Sigma, A2066); rabbit anti-Ift122 (1:2000; ProteinTech, 19304-1-AP); rabbit anti-Ift140 (1:2000; ProteinTech, 17460-1-AP); mouse anti-α-tubulin (1:10000; Sigma, T9026); mouse anti-GAPDH (1:2000; Millipore, MAB374). The primary antibodies were labeled with horseradish peroxidase-conjugated secondary antibodies (Jackson Immuno Lab). Signals were developed with enhanced chemiluminescent substrate (GE) and detected with a cooled CCD camera system, LAS-3000 mini (Fuji film). Band intensities were quantified with free software, ImageJ.
LC-MS/MS of proteins in pelleted conditioned culture media
Collected vesicles were lysed in 50 mM Tris, pH 8.0, containing 1.5% Triton X-100 with vigorous vortexing. Detergent and salt were removed with 2-D Clean-Up kit (GE Healthcare), and with cold acetone if further removal of detergent was required. Protein pellets were resolved in 50 mM NH4HCO3 with vigorous vortexing. The proteins were subjected to reductive alkylation of sulfur group with iodoacetamide (ThermoFisher), and then digested with trypsin (ThermoFisher) at 37 °C overnight. Digested peptides were desalted by means of C-18 column (ThermoFisher) with 5% acetonitrile/0.5% trifluoroacetic acid used as wash buffer, and eluted with 70% acetonitrile. The acetonitrile was evaporated with a Speed-Vac concentrator (Tomy), and the eluted peptides were resolved in 0.1% formic acid.
The peptides were analyzed with a Q Exactive-Orbitrap mass spectrometer (ThermoFisher) connected to EASY-nLC liquid chromatography (ThermoFisher). Peptides were loaded into a micro capillary column (NTCC-360; Nikkyo Technos, Japan) of 75 µm inner diameter, and separated with a linear gradient of acetonitrile from 0 to 35% in 0.1% formic acid at a flow rate of 325 nL/min. Separated peptides were ionized with Nanospray Flex Ion Source, NSI (ThermoFisher) at spray voltage of 2.0 kV. Full mass spectra were acquired with a resolution of 70,000, a maximum injection time of 60 msec, and a scan range between m/z 350 to 1800, at positive ion mode. dd-MS2 was acquired with a resolution of 17,500, a maximum injection time of 60 msec, and an isolation window of 2.0 m/z. All processes were operated with software XcaliburTM (ThermoFisher).
Proteomic analyses
Data acquired with LC-MS/MS were analyzed with Proteome Discoverer 1.4 software (ThermoFisher). All parameters for Spectrum Selector were set to default values. Identification of peptide/protein was carried out by means of MASCOT (MatrixScience) with Swiss-Prot used as database. Mass tolerances were 10 ppm for precursor mass and 0.02 Da for fragment mass. A miscleavage was allowed. Obtained peptide lists were validated by Percolator (Käll et al., 2007) based on q-values with a stringent threshold of target false discovery rate (FDR) of 1% using reverse decoy database (Elias and Gygi, 2007). The label-free semi-quantification was performed by using the precursor ions area detector (PIAD) node mounted in Proteome Discoverer 1.4.
Proteins that were detected at least two times in 9 independent experiments for serum-stimulated wild-type or Ift88-KO mIMCD-3 cells or 6 independent experiments for serum-starved wild-type mIMCD-3 cells were selected. The protein list and area values were exported as a table for further analyses in Microsoft Excel. Zero area values were replaced with one tenth of minimal area value of each sample for calculating fold changes. Ratio of mean area values were calculated for serum-stimulated wild type versus Ift88-KO cells (WT/Ift88-KO) and for serum-stimulated versus serum-starved wild-type cells (10%/0.1%). Ratios were converted to logarithm to the base 2 (log2), and Z-scores were calculated.
Quantitative real-time PCR
NIH/3T3 cells were seeded in OptiMEM (Thermo Fisher Scientific) for 24 hours to induce ciliogenesis at quiescent state. Cells were subsequently treated with DMEM containing 10% FBS for 0, 1, 2, 4, and 8 hr prior to sample collection. As a positive control, cells were also treated with 200 nM SAG (Cayman Chemicals) for 8hr. Cells were lysed with Buffer RLT, and RNA samples were isolated with the RNeasy Mini Kit with DNase I digestion (Qiagen). Samples were reverse transcribed using the iScript cDNA synthesis kit (BioRad). Gli1 (F: 5’-GGATGAAGAAGCAGTTGGGA-3’; R: 5’-ATTGGATTGAACATGGCGTC-3’) and Ptch1 (F: 5’-CTCTGGAGCAGATTTCCAAGG-3’; R: 5’-TGCCGCAGTTCTTTTGAATG-3’) primers were used to test for Hedgehog signaling activation. Ubc primers (F: 5’-TCCAGAAAGAGTCCACCCTG-3’; R: 5’-GACGTCCAAGGTGATGGTCT-3’) were used to normalize Gli1 and Ptch1 transcript levels. Transcripts were quantified using SYBR-GreenER (Thermo Fisher Scientific) on a 7900HT Fast Real-Time PCR system (Thermo Fisher Scientific).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical parameters including the definition and exact values of n (number of cells and experiments), distribution and deviation are reported in figures and corresponding legends. Most data are represented as mean ± SEM using two-tailed Student’s T tests. Probability data in Figures 3 and S3 were analyzed using statistical risk ratio analyses; probability was determined by dividing number of positives by total number of cells analyzed, and relative risk was determined by dividing probability value in each test condition by that of control condition. Mass spectrometry signal area ratios were presented with Z-scores in Table S2. Z-scores in Figure 4D and data in Figure S5N were analyzed using Mann-Whitney U-test. Quantitative real-time PCR assays in Figure 7H were analyzed using one-way ANOVA. Statistical analyses were performed in Microsoft Excel, MedCalc or GraphPad Prism.
DATA AND SOFTWARE AVAILABILITY
ADDITIONAL RESOURCES
Chávez, M., Ena, S., Van Sande, J., de Kerchove d’Exaerde, A., Schurmans, S., and Schiffmann, Serge N. (2015). Modulation of Ciliary Phosphoinositide Content Regulates Trafficking and Sonic Hedgehog Signaling Output. Developmental Cell 34, 338–350.
Garcia-Gonzalo, Francesc R., Phua, Siew C., Roberson, Elle C., Garcia Iii, G., Abedin, M., Schurmans, S., Inoue, T., and Reiter, Jeremy F. (2015). Phosphoinositides Regulate Ciliary Protein Trafficking to Modulate Hedgehog Signaling. Developmental Cell 34, 400–409.
Jacoby, M., Cox, J., Gayral, S., Hampshire, D., Ayub, M., Blockmans, M., Pernot, E., Kisseleva, M., Compere, P., Schiffmann, S., et al. (2009). INPP5E mutations cause primary cilium-signaling defects, ciliary instability and ciliopathies in human and mouse. Nat Genet 41, 1027 – 1031.
Pugacheva, E., Jablonski, S., Hartman, T., Henske, E., and Golemis, E. (2007). HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 129, 1351 – 1363.
Supplementary Material
Cell was expressed with 5HT6-mCherry (red) and γ-tub-GFP (green). Ciliary tip is positioned at the top of image, as in Figure 1A. Time in min:sec. Bar indicates 5µm.
(I) Example of a primary cilium in quiescent Inpp5e+/− MEF remaining intact over a 3-hour live imaging period, as in Figure 1C top left panel. (II) Example of a primary cilium in quiescent Inpp5e−/− MEF undergoing a single decapitation event, as in Figure 1C top right panel; two cell-associated YFP+ particles were observed from beginning. (III) Example of a primary cilium in growth-stimulated Inpp5e+/− MEF undergoing two decapitation events, as in Figure 1C bottom left panel. (IV) Example of a primary cilium in growth-stimulated Inpp5e−/− MEF undergoing multiple decapitation events prior to resorption, as in Figure 1C bottom right panel; one cell-associated YFP+ particle was observed from beginning. In all cases, cells were expressed with 5HT6-YFP. Time in hr:min. Bars indicate 5µm.
(I) In quiescent Inpp5e+/− MEF over two hours in 0% FBS, as in Figure 2C. (II) In Inpp5e+/− MEF between 4 hours and 6 hours of 10% FBS stimulation, as in Figure 2E; bright YFP particles were cell debris. (III) In Inpp5e+/− MEF between 0 hour and 2 hours of 10% FBS stimulation, as in Figure S2J; note the robust ciliary PI(4,5)P2 oscillation post-decapitation. In all cases, cells were expressed with 5HT6-mCeru3 (red) and YFP-PH(PLCδ) (gold/green). Time in hr:min. Bars indicate 5µm.
(I) In growth-stimulated Inpp5e+/− MEF between 4 hours and 5 hours of 10% FBS stimulation, as in Figure 3C. (II) In growth-stimulated Inpp5e−/− MEF between 1 hour and 2 hours of 10% FBS stimulation, as in Figure 3D. In both cases, cells were expressed with 5HT6-YFP (red) and mCeru3-lifeact (green). Time in hr:min. Bars indicate 5µm.
(I) An example of timely G1 entry that occurs with cilia decapitation, as in Figure 6A. Inpp5e+/− MEF was expressed with Venus-p27K− (green), mCherry-hCdt1(30/120) (red) and 5HT6-mCeru3 (cyan), and imaged for ten hours post-stimulation with 10% FBS. Four decapitation events were observed. Venus-p27K− was abruptly degraded at approximately 5 hours, while mCherry-hCdt1 began degradation from approximately 7 hours onwards, indicating transit into G1 phase and S phase respectively. Imaging position was adjusted between 03:20 and 03:26 to accommodate for cell movements. (II) An example of prolonged G1 entry that occurs with suppressed cilia decapitation, as in Figure 6C. Inpp5e+/− MEF was expressed with Venus-p27K− (green), mCherry-hCdt1(30/120) (red) and 5HT6-mCeru3-Tβ4(WT) (cyan), and imaged for ten hours post-stimulation with 10% FBS. Venus-p27K− underwent slowly degradation over 10 hours, indicating delayed G1 entry. Note that bright mCeru3+Venus+mCherry+ particle that appeared from 03:35 onwards was cell debris. Time in hr:min. Bars indicate 10µm.
Green, IFT-B components including related motor proteins; orange, IFT-A components; yellow, hedgehog signaling proteins; cyan, known ciliary proteins. Grey-shaded cells in signal intensity columns indicate data points where no signal was detected, and null values in these cells were replaced with one tenth of minimum peak area in each sample condition to enable calculation of fold changes.
Proteins were ranked according to Log2[WT/Ift88KO] and corresponding Z-scores. Color coding as in Table S1.
Acknowledgments
We thank Stéphane Schurmans for rabbit anti-Inpp5e antibody, Toshio Kitamura for PMXs-IP-mVenus-p27K− plasmid, Atsushi Miyawaki for pCSII-EF-mCherry-hCdt1(30/120) plasmid, Akiko Yuba-Kubo for γ-Tubulin-EGFP plasmid, Kiyoshi Shibata, Kousuke Fukui, and Hang Thi Vu for technical assistance on FACS and Western blot analyses. We also appreciate Erica Golemis for helpful discussions and critical reading of the manuscript, as well as Robert DeRose for writing suggestions. This work was supported in part by the US National Institutes of Health (NIH) grant to T.I. (DK102910), J.F.R. (AR054396 and GM095941) and R.R. (DP2 GM105448), Japan Grant-in-Aid for Scientific Research on Innovative Areas (15H01207) to K.I., the Japan Agency for Medical Research and Development (AMED) grant (921910520) to M. Setou, and Japan Grant-in-Aid for Young Scientists (A) to S.C. G.V.P. is supported by American Heart Association fellowship. S.C.P. is supported by the Agency for Science, Technology and Research (Singapore).
Footnotes
Author Contributions
S.C.P., S.C., K.I. and T.I. devised experiments. S.C.P. performed and analyzed most live-imaging experiments with assistance from E.S. S.C. performed most confocal and super-resolution imaging experiments. K.I. established Ift88-KO and Ift81:YNL knock-in mIMCD-3. M. Suzuki performed proteomics experiments under supervision of K.I. M. Setou advised K.I. and provided infrastructure. E.R. performed qPCR assays under supervision of J.F.R. G.V.P. established NIH/3T3: 8xGBS-GFP reporter line under supervision of R.R. Inpp5e+/− and Inpp5e−/− MEF were provided by J.F.R. S.C.P., K.I. and T.I. wrote the manuscript. K.I. and T.I. supervised the work.
References
- Basten SG, Giles RH. Functional aspects of primary cilia in signaling, cell cycle and tumorigenesis. Cilia. 2013;2:6. doi: 10.1186/2046-2530-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman K, Goodenough UW, Goodenough DA, Jawitz J, Martin H. Gametic differentiation in Chlamydomonas reinhardtii. II. Flagellar membranes and the agglutination reaction. J. Cell Biol. 1975;67:606–622. doi: 10.1083/jcb.67.3.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S, Cajanek L, Fort C, Blisnick T, Weber K, Taschner M, Mizuno N, Lamla S, Bastin P, Nigg EA, et al. Molecular Basis of Tubulin Transport Within the Cilium by IFT74 and IFT81. Science. 2013;341:1009–1012. doi: 10.1126/science.1240985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielas SL, Silhavy JL, Brancati F, Kisseleva MV, Al-Gazali L, Sztriha L, Bayoumi Ra, Zaki MS, Abdel-Aleem A, Rosti RO, et al. Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nat. Genet. 2009;41:1032–1036. doi: 10.1038/ng.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehlke C, Kotsis F, Patel V, Braeg S, Voelker H, Bredt S, Beyer T, Janusch H, Hamann C, Gödel M, et al. Primary cilia regulate mTORC1 activity and cell size through Lkb1. Nat. Cell Biol. 2010;12:1115–1122. doi: 10.1038/ncb2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Koslover EF, Seydel F, Spakowitz AJ, Nachury MV. An in vitro assay for entry into cilia reveals unique properties of the soluble diffusion barrier. J. Cell Biol. 2013;203:129–147. doi: 10.1083/jcb.201212024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Ning J, Hernandez-Lara CI, Belzile O, Wang Q, Dutcher SK, Liu Y, Snell WJ. Uni-directional ciliary membrane protein trafficking by a cytoplasmic retrograde IFT motor and ciliary ectosome shedding. Elife. 2015;2015 doi: 10.7554/eLife.05242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez M, Ena S, Van Sande J, de Kerchove d’Exaerde A, Schurmans S, Schiffmann SN. Modulation of Ciliary Phosphoinositide Content Regulates Trafficking and Sonic Hedgehog Signaling Output. Dev. Cell. 2015;34:338–350. doi: 10.1016/j.devcel.2015.06.016. [DOI] [PubMed] [Google Scholar]
- Chaya T, Omori Y, Kuwahara R, Furukawa T. ICK is essential for cell type-specific ciliogenesis and the regulation of ciliary transport. EMBO J. 2014;33:1227–1242. doi: 10.1002/embj.201488175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen ST, Clement CA, Satir P, Pedersen LB. Primary cilia and coordination of receptor tyrosine kinase (RTK) signalling. J. Pathol. 2012;226:172–184. doi: 10.1002/path.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das RM, Storey KG. Apical abscission alters cell polarity and dismantles the primary cilium during neurogenesis. Science. 2014;343:200–204. doi: 10.1126/science.1247521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil V, Marzesco AM, Corbeil D, Huttner WB, Wilsch-Bräuninger M. Midbody and primary cilium of neural progenitors release extracellular membrane particles enriched in the stem cell marker prominin-1. J. Cell Biol. 2007;176:483–495. doi: 10.1083/jcb.200608137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Ezratty EJ, Stokes N, Chai S, Shah AS, Williams SE, Fuchs E. A role for the primary cilium in notch signaling and epidermal differentiation during skin development. Cell. 2011;145:1129–1141. doi: 10.1016/j.cell.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis SS, Sfakianos J, Lo B, Mellman I. A hierarchy of signals regulates entry of membrane proteins into the ciliary membrane domain in epithelial cells. J. Cell Biol. 2011;193:219–233. doi: 10.1083/jcb.201009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Phua SC, Roberson EC, Garcia G, Abedin M, Schurmans S, Inoue T, Reiter JF. Phosphoinositides Regulate Ciliary Protein Trafficking to Modulate Hedgehog Signaling. Dev. Cell. 2015;34:400–409. doi: 10.1016/j.devcel.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y-G, Kim HJ, Dlugosz AA, Ellison DW, Gilbertson RJ, Alvarez-Buylla A. Dual and opposing roles of primary cilia in medulloblastoma development. Nat. Med. 2009;15:1062–1065. doi: 10.1038/nm.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Subramanian R, Bangs F, Omelchenko T, Liem KF, Kapoor TM, Anderson KV. The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat. Cell Biol. 2014;16:663–672. doi: 10.1038/ncb2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan MC, Manganelli L, Woollard JR, Masyuk AI, Masyuk TV, Tammachote R, Huang BQ, Leontovich AA, Beito TG, Madden BJ, et al. Characterization of PKD Protein-Positive Exosome-Like Vesicles. J. Am Soc Nephrol. 2009;20:278–288. doi: 10.1681/ASN.2008060564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui C, Angers S. Gli Proteins in Development and Disease. Annu. Rev. Cell Dev. Biol. 2011;27:513–537. doi: 10.1146/annurev-cellbio-092910-154048. [DOI] [PubMed] [Google Scholar]
- Humbert MC, Weihbrecht K, Searby CC, Li Y, Pope RM, Sheffield VC, Seo S. ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc. Natl. Acad. Sci. U.S.A. 2012;109:19691–19696. doi: 10.1073/pnas.1210916109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 2010;24:670–682. doi: 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, et al. TSC2 Integrates Wnt and Energy Signals via a Coordinated Phosphorylation by AMPK and GSK3 to Regulate Cell Growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- Jacoby M, Cox JJ, Gayral S, Hampshire DJ, Ayub M, Blockmans M, Pernot E, Kisseleva MV, Compère P, Schiffmann SN, et al. INPP5E mutations cause primary cilium signaling defects, ciliary instability and ciliopathies in human and mouse. Nat. Genet. 2009;41:1027–1031. doi: 10.1038/ng.427. [DOI] [PubMed] [Google Scholar]
- Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- Kim S, Zaghloul Na, Bubenshchikova E, Oh EC, Rankin S, Katsanis N, Obara T, Tsiokas L. Nde1-mediated inhibition of ciliogenesis affects cell cycle re-entry. Nat. Cell Biol. 2011;13:351–360. doi: 10.1038/ncb2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster Ma, Schroth J, Gleeson JG. Subcellular spatial regulation of canonical Wnt signalling at the primary cilium. Nat. Cell Biol. 2011;13:700–707. doi: 10.1038/ncb2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-C, Niewiadomski P, Lin B, Nakamura H, Phua SC, Jiao J, Levchenko A, Inoue TT, Rohatgi R. Chemically inducible diffusion trap at cilia reveals molecular sieve-like barrier. Nat. Chem. Biol. 2013;9:437–443. doi: 10.1038/nchembio.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- Monje M, Mitra SS, Freret ME, Raveh TB, Kim J, Masek M, Attema JL, Li G, Haddix T, Edwards MSB, et al. Hedgehog-responsive candidate cell of origin for diffuse intrinsic pontine glioma. Proc. Natl. Acad. Sci. U.S.A. 2011;108:4453–4458. doi: 10.1073/pnas.1101657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauli SM, Alenghat FJ, Luo Y, Williams E, Vassilev P, Li X, Elia AEH, Lu W, Brown EM, Quinn SJ, et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat. Genet. 2003;33:129–137. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- Oki T, Nishimura K, Kitaura J, Togami K, Maehara A, Izawa K, Sakaue-Sawano A, Niida A, Miyano S, Aburatani H, et al. A novel cell-cycle-indicator, mVenus-p27K-, identifies quiescent cells and visualizes G0-G1 transition. Sci. Rep. 2014;4:4012. doi: 10.1038/srep04012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orhon I, Dupont N, Zaidan M, Boitez V, Burtin M, Schmitt A, Capiod T, Viau A, Beau I, Wolfgang Kuehn E, et al. Primary-cilium-dependent autophagy controls epithelial cell volume in response to fluid flow. Nat. Cell Biol. 2016;i:657–667. doi: 10.1038/ncb3360. [DOI] [PubMed] [Google Scholar]
- Pan J, Wang Q, Snell WJ. An aurora kinase is essential for flagellar disassembly in Chlamydomonas. Dev. Cell. 2004;6:445–451. doi: 10.1016/s1534-5807(04)00064-4. [DOI] [PubMed] [Google Scholar]
- Paridaen JTML, Wilsch-Bräuninger M, Huttner WB. Asymmetric inheritance of centrosome-associated primary cilium membrane directs ciliogenesis after cell division. Cell. 2013;155 doi: 10.1016/j.cell.2013.08.060. [DOI] [PubMed] [Google Scholar]
- Phua SC, Lin YC, Inoue T. An intelligent nano-antenna: Primary cilium harnesses TRP channels to decode polymodal stimuli. Cell Calcium. 2015;58:415–422. doi: 10.1016/j.ceca.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikova OV, Seo S, Cottle DL, Conduit S, Hakim S, Dyson JM, Mitchell CA, Smyth IM. INPP5E interacts with AURKA, linking phosphoinositide signalling to primary cilium stability. J. Cell Sci. 2014:364–372. doi: 10.1242/jcs.161323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikova OV, Golemis EA, Pugacheva EN. Cell cycle-dependent ciliogenesis and cancer. Cancer Res. 2008;68:2058–2061. doi: 10.1158/0008-5472.CAN-07-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikova OV, Pugacheva EN, Golemis EA. Primary cilia and the cell cycle. Methods Cell Biol. 2009;94:137–160. doi: 10.1016/S0091-679X(08)94007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posor Y, Eichhorn-Gruenig M, Puchkov D, Schöneberg J, Ullrich A, Lampe A, Müller R, Zarbakhsh S, Gulluni F, Hirsch E, et al. Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature. 2013;499:233–237. doi: 10.1038/nature12360. [DOI] [PubMed] [Google Scholar]
- Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regl G, Kasper M, Schnidar H, Eichberger T, Neill GW, Ikram MS, Quinn AG, Philpott MP, Frischauf A-M, Aberger F. The zinc-finger transcription factor GLI2 antagonizes contact inhibition and differentiation of human epidermal cells. Oncogene. 2004;23:1263–1274. doi: 10.1038/sj.onc.1207240. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Roy S, Ingham PW. Hedgehogs tryst with the cell cycle. J. Cell Sci. 2002;115:4393–4397. doi: 10.1242/jcs.00158. [DOI] [PubMed] [Google Scholar]
- Saarikangas J, Zhao H, Lappalainen P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol. Rev. 2010;90:259–289. doi: 10.1152/physrev.00036.2009. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- Singla V, Reiter JF. The primary cilium as the cell’s antenna: signaling at a sensory organelle. Science. 2006;313:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- Stamataki D, Ulloa F, Tsoni SV, Mynett A, Briscoe J. A gradient of Gli activity mediates graded Sonic Hedgehog signaling in the neural tube. Genes Dev. 2005;19:626–641. doi: 10.1101/gad.325905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh BC, Inoue T, Meyer T, Hille B. Rapid chemically induced changes of PtdIns (4,5) P2 gate KCNQ ion channels. Science. 2006;314:1454–1457. doi: 10.1126/science.1131163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susaki E, Nakayama K, Nakayama KI. Cyclin D2 translocates p27 out of the nucleus and promotes its degradation at the G0-G1 transition. Mol. Cell. Biol. 2007;27:4626–4640. doi: 10.1128/MCB.00862-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Troys M, Dewitte D, Goethals M, Carlier MF, Vandekerckhove J, Ampe C. The actin binding site of thymosin beta 4 mapped by mutational analysis. EMBO J. 1996;15:201–210. [PMC free article] [PubMed] [Google Scholar]
- Wang J, Silva M, Haas LA, Morsci NS, Nguyen KCQ, Hall DH, Barr MM. C. elegans ciliated sensory neurons release extracellular vesicles that function in animal communication. Curr. Biol. 2014;24:519–525. doi: 10.1016/j.cub.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Kaletsky R, Silva M, Williams A, Haas LA, Androwski RJ, Landis JN, Patrick C, Rashid A, Santiago-Martinez D, et al. Cell-Specific Transcriptional Profiling of Ciliated Sensory Neurons Reveals Regulators of Behavior and Extracellular Vesicle Biogenesis. Curr. Biol. 2015;25:3232–3238. doi: 10.1016/j.cub.2015.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W-J, Tay HG, Soni R, Perumal GS, Goll MG, Macaluso FP, Asara JM, Amack JD, Tsou M-FB. CEP162 is an axoneme-recognition protein promoting ciliary transition zone assembly at the cilia base. Nat. Cell Biol. 2013;15:591–601. doi: 10.1038/ncb2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ding Q, Yen CJ, Xia W, Izzo JG, Lang JY, Li CW, Hsu JL, Miller SA, Wang X, et al. The Crosstalk of mTOR/S6K1 and Hedgehog Pathways. Cancer Cell. 2012;21:374–387. doi: 10.1016/j.ccr.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SY, Seol AD, So P-L, Ermilov AN, Bichakjian CK, Epstein EH, Dlugosz Aa, Reiter JF. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat. Med. 2009;15:1055–1061. doi: 10.1038/nm.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CR, Rosenbaum JL. Ciliary ectosomes: Transmissions from the cell’s antenna. Trends Cell Biol. 2015;25:276–285. doi: 10.1016/j.tcb.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CR, Huang K, Diener DR, Rosenbaum JL. The cilium secretes bioactive ectosomes. Curr. Biol. 2013;23:906–911. doi: 10.1016/j.cub.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D, Waterman-Storer CM, Schmid SL. SNX9 Couples Actin Assembly to Phosphoinositide Signals and Is Required for Membrane Remodeling during Endocytosis. Dev. Cell. 2007;13:43–56. doi: 10.1016/j.devcel.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Yeh C, Li A, Chuang JZ, Saito M, Cáceres A, Sung CH. IGF-1 Activates a cilium-localized noncanonical gβγ signaling pathway that regulates cell-cycle progression. Dev. Cell. 2013;26:358–368. doi: 10.1016/j.devcel.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JW, Kita Y, Frank DJ, Majewski RR, Konicek BA, Nobrega MA, Jacob H, Walterhouse D, Iannaccone P. Gene expression profiling leads to identification of GLI1-binding elements in target genes and a role for multiple downstream pathways in GLI1-induced cell transformation. J. Biol. Chem. 2002;277:5548–5555. doi: 10.1074/jbc.M105708200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cell was expressed with 5HT6-mCherry (red) and γ-tub-GFP (green). Ciliary tip is positioned at the top of image, as in Figure 1A. Time in min:sec. Bar indicates 5µm.
(I) Example of a primary cilium in quiescent Inpp5e+/− MEF remaining intact over a 3-hour live imaging period, as in Figure 1C top left panel. (II) Example of a primary cilium in quiescent Inpp5e−/− MEF undergoing a single decapitation event, as in Figure 1C top right panel; two cell-associated YFP+ particles were observed from beginning. (III) Example of a primary cilium in growth-stimulated Inpp5e+/− MEF undergoing two decapitation events, as in Figure 1C bottom left panel. (IV) Example of a primary cilium in growth-stimulated Inpp5e−/− MEF undergoing multiple decapitation events prior to resorption, as in Figure 1C bottom right panel; one cell-associated YFP+ particle was observed from beginning. In all cases, cells were expressed with 5HT6-YFP. Time in hr:min. Bars indicate 5µm.
(I) In quiescent Inpp5e+/− MEF over two hours in 0% FBS, as in Figure 2C. (II) In Inpp5e+/− MEF between 4 hours and 6 hours of 10% FBS stimulation, as in Figure 2E; bright YFP particles were cell debris. (III) In Inpp5e+/− MEF between 0 hour and 2 hours of 10% FBS stimulation, as in Figure S2J; note the robust ciliary PI(4,5)P2 oscillation post-decapitation. In all cases, cells were expressed with 5HT6-mCeru3 (red) and YFP-PH(PLCδ) (gold/green). Time in hr:min. Bars indicate 5µm.
(I) In growth-stimulated Inpp5e+/− MEF between 4 hours and 5 hours of 10% FBS stimulation, as in Figure 3C. (II) In growth-stimulated Inpp5e−/− MEF between 1 hour and 2 hours of 10% FBS stimulation, as in Figure 3D. In both cases, cells were expressed with 5HT6-YFP (red) and mCeru3-lifeact (green). Time in hr:min. Bars indicate 5µm.
(I) An example of timely G1 entry that occurs with cilia decapitation, as in Figure 6A. Inpp5e+/− MEF was expressed with Venus-p27K− (green), mCherry-hCdt1(30/120) (red) and 5HT6-mCeru3 (cyan), and imaged for ten hours post-stimulation with 10% FBS. Four decapitation events were observed. Venus-p27K− was abruptly degraded at approximately 5 hours, while mCherry-hCdt1 began degradation from approximately 7 hours onwards, indicating transit into G1 phase and S phase respectively. Imaging position was adjusted between 03:20 and 03:26 to accommodate for cell movements. (II) An example of prolonged G1 entry that occurs with suppressed cilia decapitation, as in Figure 6C. Inpp5e+/− MEF was expressed with Venus-p27K− (green), mCherry-hCdt1(30/120) (red) and 5HT6-mCeru3-Tβ4(WT) (cyan), and imaged for ten hours post-stimulation with 10% FBS. Venus-p27K− underwent slowly degradation over 10 hours, indicating delayed G1 entry. Note that bright mCeru3+Venus+mCherry+ particle that appeared from 03:35 onwards was cell debris. Time in hr:min. Bars indicate 10µm.
Green, IFT-B components including related motor proteins; orange, IFT-A components; yellow, hedgehog signaling proteins; cyan, known ciliary proteins. Grey-shaded cells in signal intensity columns indicate data points where no signal was detected, and null values in these cells were replaced with one tenth of minimum peak area in each sample condition to enable calculation of fold changes.
Proteins were ranked according to Log2[WT/Ift88KO] and corresponding Z-scores. Color coding as in Table S1.