Abstract
The primate-specific serum protein apolipoprotein L1 (APOL1) is the only secreted member of a family of cell death promoting proteins1–4. APOL1 kills the bloodstream parasite Trypanosoma brucei brucei, but not the human sleeping sickness agents T.b. rhodesiense and T.b. gambiense3. We considered the possibility that intracellular members of the APOL1 family, against which extracellular trypanosomes could not have evolved resistance, could kill pathogenic T. brucei subspecies. Here we show that recombinant APOL3 (rAPOL3) kills all African trypanosomes including T.b. rhodesiense, T.b. gambiense and the animal pathogens T. evansi, T. congolense and T. vivax. In contrast, rAPOL3 did not kill more distant trypanosomes such as T. theileri or T. cruzi. This trypanolytic potential was partially shared by rAPOL1 from Papio papio (rPpAPOL1). The differential killing ability between rAPOL3 and rAPOL1 was associated with distinct dependence on acidic pH for activity. Due to both instability and toxicity when injected into mice rAPOL3 cannot be used for treatment of infection, but an experimental rPpAPOL1 mutant inspired by APOL3 exhibited enhanced trypanolytic activity in vitro and ability to completely inhibit T.b. gambiense infection in mice. We conclude that pH dependence influences the trypanolytic potential of rAPOLs.
African trypanosomes (prototype T.b. brucei) are fly-borne protozoan flagellates able to infect both animals and humans. T.b. brucei, T. congolense and T. vivax cause a severe bovine disease termed nagana, which impedes cattle raising over one third of the African continent. The closely related T. evansi is common in South America and Asia, where this parasite causes acute disease in camels and horses, and chronic disease in cattle and buffaloes. Humans and some primates exhibit specific innate immunity towards T.b. brucei and T. evansi owing to the activity of the serum protein APOL1, which triggers apoptotic-like death of the parasite following the formation of pores, initially in endosomal membranes and subsequently in the mitochondrial membrane5–7, possibly also the plasma membrane8. A first association with endosomal membranes is mandatory for APOL1 activity, because this protein requires acidic conditions for membrane insertion3,5,7,9–11. If not inserted in a membrane, APOL1 is probably quickly degraded3,12. APOL1 cannot kill T.b. gambiense and T.b. rhodesiense because these subspecies express different APOL1 resistance proteins in the endocytic pathway3,5,13,14. While the T.b. rhodesiense endolysosomal Serum Resistance-Associated (SRA) protein neutralizes APOL1 through direct interaction with the C-terminal amphipathic helix of APOL13,5,13, the T.b. gambiense-specific glycoprotein TgsGP, characteristic of type I but not type II T.b. gambiense, inactivates APOL1 through its ability to stiffen membranes, presumably hampering APOL1 insertion in endosomal membranes3,14,15. Both resistance proteins evolved from the main surface component of the parasite, termed VSG for Variant Surface Glycoprotein, whose continuous sequence changes underlie antigenic variation3. Experimental sequence variation within the C-terminal helix can allow rAPOL1 to escape neutralization by SRA and therefore, to kill T.b. rhodesiense16. Interestingly, natural amino acid substitutions in the C-terminal region of APOL1 have been strongly selected in Africa presumably because they confer human resistance to T.b. rhodesiense, despite the fact that these substitutions are also associated with renal disease17. Similarly, the variant C-terminal sequence of Papio papio APOL1 (PpAPOL1) allows this primate APOL1 to escape neutralization by SRA and to kill T.b. rhodesiense18.
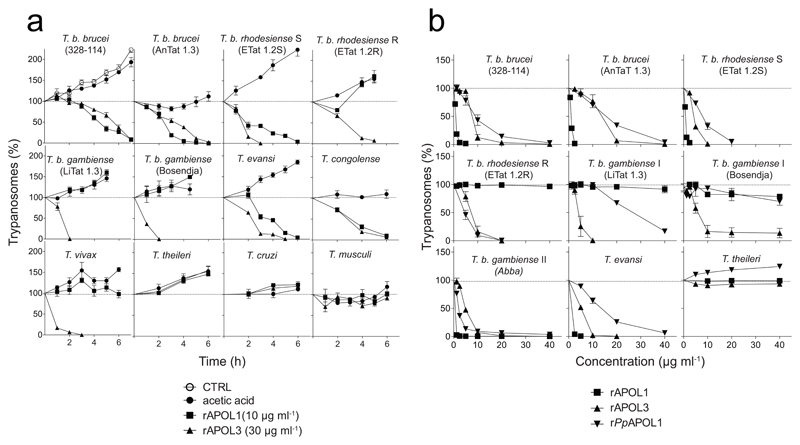
From sequence comparison, the six members of the human APOL family appear to be highly similar19. Thus, we speculated that these proteins could share the membrane pore-forming activity of APOL1. If so, intracellular APOLs could also be trypanolytic, and we envisaged that African trypanosomes, which are exclusively extracellular, could not have evolved natural resistance against these APOLs. Therefore, we incubated various trypanosome species and subspecies with different recombinant APOLs (rAPOLs). Among these proteins, rAPOL3 exhibited potent trypanolytic activity on T.b. brucei, in keeping with the relative conservation of the helices defined for APOL16 (Suppl. Figs. 1, 2). Moreover, close to the concentration range of APOL1 in plasma, 0.4 – 15 µg ml-1 (ref. 20), rAPOL3 also efficiently killed different trypanosomes closely related to T. brucei, including T.b. rhodesiense, T.b. gambiense types I and II, T. evansi, T. congolense and T. vivax (Fig. 1). In contrast, rAPOL3 did not kill more distantly related trypanosomes such as T. theileri, T. lewisi or T. musculi (Fig. 1). Under the same conditions and concentration, rPpAPOL1 efficiently killed T.b. rhodesiense, but exhibited only weak activity on T.b. gambiense type I (Fig. 1b).
Fig. 1.
In vitro trypanolytic potential of various rAPOLs (error bars: s.e.m.; 3 technical replicates; n=3 independent experiments). The effect of different rAPOLs or acetic acid solvent on growth of various trypanosomes was evaluated in kinetics (a) and dose-effect experiments after 24 h incubation (b) (CTRL in first panel = control growth without addition of rAPOL or solvent). The volume of APOL samples was identical in all assays.
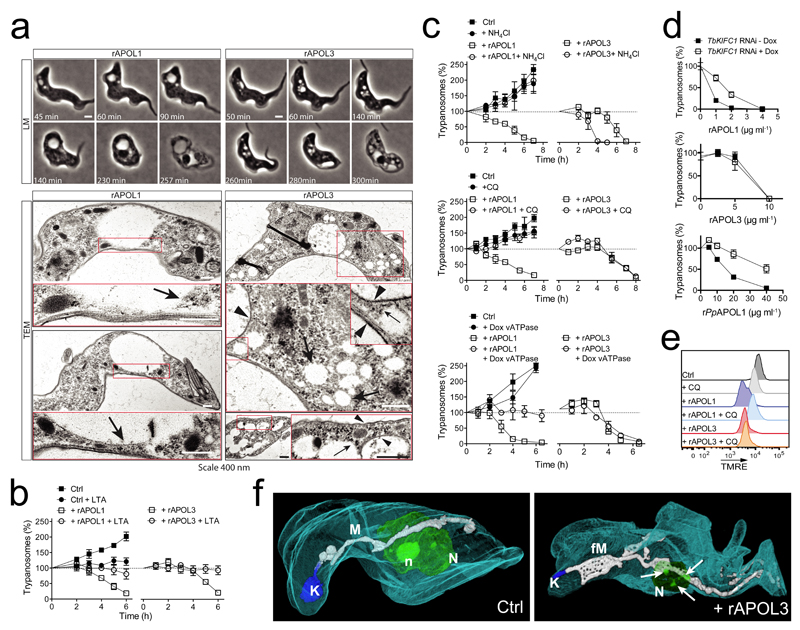
The phenotype of rAPOL3-mediated lysis of T.b. brucei differed from that induced by rAPOL1. rAPOL3 triggered the formation of multiple intracellular vesicles instead of the unique large vesicle typically observed with APOL16,7,21,22 (Fig. 2a). Moreover, some of the rAPOL3-induced vesicles exhibited a dense coat comparable with that of the plasma membrane, in clear contrast with rAPOL1-induced vesicles that never showed such characteristics3,5–7,21,22 (Fig. 2a). As the swollen vesicle induced by rAPOL1 is the lysosome3,5,7, these findings suggested that rAPOL3 could trigger endosome swelling earlier than rAPOL1. The actin inhibitor latrunculin A inhibited lysis by both rAPOL1 and rAPOL3, suggesting active protein uptake in both cases (Fig. 2b). Like rAPOL1 (ref. 5,7,9,12), rAPOL3 was taken up through the endocytic compartment between the kinetoplast and the nucleus, and was sensitive to cathepsin digestion (Suppl. Fig. 3a). However, contrary to rAPOL1, rAPOL3 efficiently induced trypanolysis at low temperatures, suggesting less dependence on endocytosis for membrane insertion (Suppl. Fig.3b). While as expected3,6,7,9–11 rAPOL1 activity on membranes was inhibited by preventing vacuolar acidification as occurs after incubation with the endosome basifying agents NH4Cl or chloroquine, or by RNAi-mediated knock-down of the vacuolar ATPase, rAPOL3-mediated lysis resisted all these treatments (Fig. 2c). Altogether, these results suggested that contrary to APOL1, rAPOL3 does not require acidic pH for activity, and could insert into membranes before rAPOL1. In support of this conclusion, rAPOL3-mediated lysis did not involve membrane transport by the kinesin TbKIFC1, which appears to recruit acidic vesicles23 and participates in both rAPOL1 (ref. 7) and rPpAPOL1 activity as demonstrated by the reduction of APOL1-mediated trypanolysis in parasites with RNAi-mediated TbKIFC1 knock-down (Fig. 2d; Suppl. Fig. 4). Further downstream in the process of lysis no difference could be evidenced between APOLs, since typical features linked to APOL1 death-promoting activity like mitochondrial membrane depolarization and fenestration or nuclear heterochromatinization7, were also observed with rAPOL3 (Fig. 2e,f; Suppl. Fig. 5).
Fig. 2.
Phenotype of rAPOL3-mediated trypanolysis (error bars: s.e.m.; 3 technical replicates; n=3 independent experiments). (a) LLM and TEM imaging of T.b. brucei during lysis by either 10 µg ml-1 rAPOL1 or 30 µg ml-1 rAPOL3 (see Methods regarding the effective APOL3 concentration). Arrowheads and arrows respectively point to coated and non-coated membranes. (b) Effect of 2.5 µg ml-1 latrunculin A on T.b. brucei trypanolysis by 10 µg ml-1 rAPOL1 or 30 µg ml-1 rAPOL3 (c) Effect of 20 mM NH4Cl, 10 µM chloroquine or V-ATPase subunit F gene knock-down following RNAi induction by 1 µg ml-1 doxycycline10, on T.b. brucei trypanolysis by 10 µg ml-1 rAPOL1 or 30 µg ml-1 rAPOL3 (d) Effect of TbKIFC1 RNAi induction by 1 µg ml-1 doxycycline7 for 24h on T.b. brucei overnight trypanolysis by rAPOL1, rAPOL3 or rPpAPOL1 (e) Effect of T.b. brucei incubation with 10 µg ml-1 rAPOL1 or 30 µg ml-1 rAPOL3, in the presence or not of 10 µM chloroquine, on mitochondrial membrane polarity as determined by TMRE fluorescence and FACS analysis. (n = 3 independent experiments) (f) FIB-SEM tomography of T.b. brucei after 2 h incubation with 30 µg ml-1 rAPOL3 (K=kinetoplast; M=mitochondrion; fM= fenestrated mitochondrion; N=nucleus; n=nucleolus; arrows: heterochromatin patches).
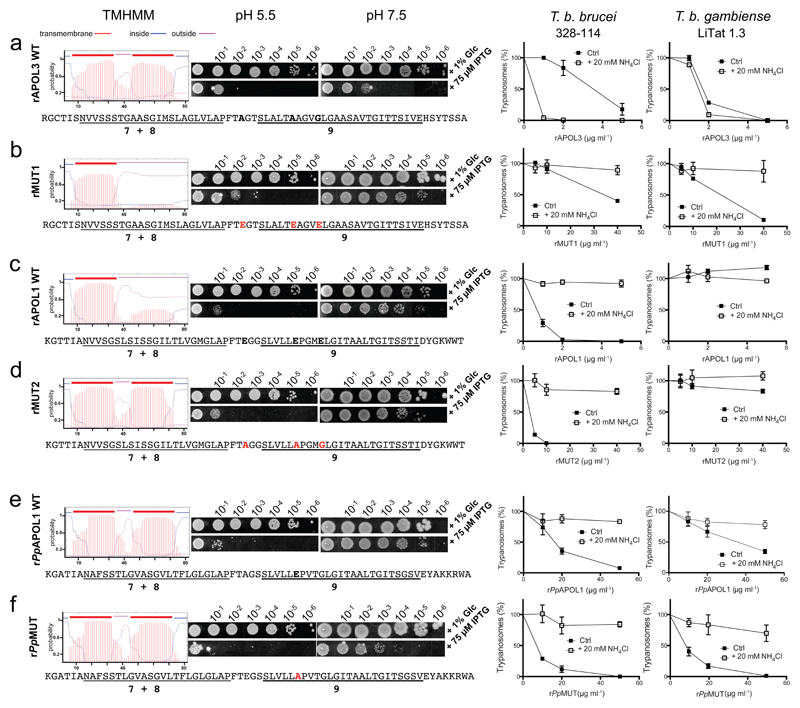
The reason for the differential dependence on acidic conditions for activity could be the clear distinction between the two proteins with respect to their theoretical ability to insert a double-stranded hairpin structure in membranes at neutral pH, this hairpin being the core of the colicin-like pore-forming domain of APOL16,7. While APOL1 only exhibited a single transmembrane span, APOL3 contained two (Fig. 3a,c). As expected from these theoretical predictions, when assayed in Escherichia coli16 rAPOL3 was fully active under both neutral and acidic conditions, in sharp contrast with rAPOL1 which was only active at acidic pH (Fig. 3a,c). This difference could be attributed to the presence in APOL1, but not in APOL3, of three glutamic acid residues within or close to helix 9 (highlighted in bold in Fig. 3c). Indeed, negatively charged side chains hinder membrane insertion at neutral pH, and their protonation at low pH is required to trigger transmembrane insertion24. We evaluated this possibility by swapping residues at these positions between APOL1 and APOL3, both in silico and experimentally. As shown in Fig. 3b (left), inserting in APOL3 the three APOL1-specific glutamic acid residues resulted in the theoretical inability of helix 9 to span a membrane at neutral pH. Accordingly, in E. coli this variant version of rAPOL3 (rMUT1) exhibited almost complete inactivation at neutral pH (Fig. 3b, center). Similarly, the trypanolytic activity of rMUT1 on both T.b. brucei and T.b. gambiense was fully inhibited after vacuolar basification with NH4Cl (Fig. 3b, right), which is remarkable in the former case given the particularly high trypanolytic activity of rAPOL3 in the presence of NH4Cl (Fig. 3a, right). Introducing only some of the three glutamic acids did not result in reversion of rAPOL3 independence to acidic pH (Suppl. Fig. 6). Thus, for APOL3 the absence of these three glutamic acids was necessary and sufficient to explain the protein pore-forming activity at neutral pH. For APOL1 this conclusion could not be confirmed, as in this case no change of activity was observed following amino acid swapping with APOL3 (rMUT2), despite the theoretical prediction that the transmembrane potential of helix 9 can be affected (Fig. 3d). Thus, even with two potential transmembrane spans at neutral pH such as in rMUT2, rAPOL1 activity remained dependent on acidic conditions. This observation was also valid for rPpAPOL1, which only contains a single glutamic acid in helix 9 and exhibits two possible transmembrane segments at neutral pH (Fig. 3e). We attempted to define which APOL1 segment would be responsible for the pH dependence, by swapping domains between rAPOL1 and rAPOL3 and evaluating the ability of the APOL chimaeras to kill T.b. gambiense. Among five such chimaeras, only two resisted inactivation, and from their relative activity on T.b. gambiense it could be concluded that the APOL sensitivity to pH resides in the pore-forming domain (Suppl. Fig. 7a,c). Attempts to define this character more precisely by site-directed mutagenesis within this domain were unsuccessful (Suppl. Fig. 7b).
Fig. 3.
Differential pH-dependence between rAPOL3, rAPOL1 and rPpAPOL1. In each row, the left panels show the predicted patterns of transmembrane spans (TMHMM program, Expasy) in the APOL sequences defined underneath (numbers refer to key helices in the pore-forming domain6), followed by measurements of the colicin-like activity of the different rAPOLs in E. coli as determined by scoring the plating efficiency after overnight incubation at 37°C, comparing induction of protein expression following IPTG addition with control following glucose (Glc) addition16, and comparing the effect of pH. The right panels show the trypanolytic potential of the different rAPOLs, as determined on T.b. brucei 328-114 and T.b. gambiense LiTat 1.3 after 24 h incubation in vitro with or without 20 mM NH4Cl (error bars: s.e.m.; 3 technical replicates; n=3 independent experiments). (a) wild-type rAPOL3, (b) rMUT1, a rAPOL3 mutant (residues in bold red), (c) wild-type rAPOL1, (d) rMUT2, a rAPOL1 mutant (residues in bold red), (e) wild-type Papio papio APOL1 (rPpAPOL1), (f) rPpMUT, a rPpAPOL1 mutant (residues in bold red).
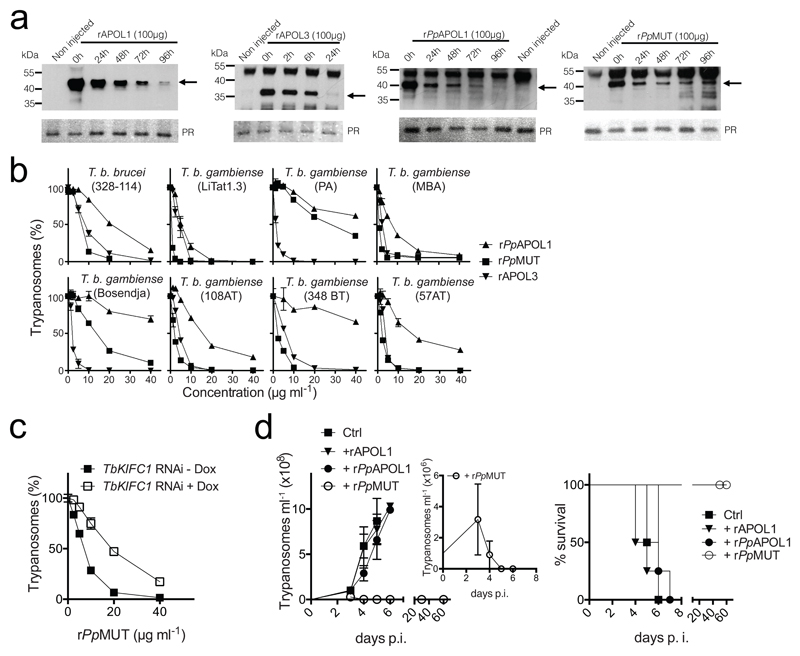
In order to evaluate the usefulness of rAPOL3 to treat sleeping sickness we tested the effect of intravenous injection of rAPOL3 or rAPOL1 into mice. As expected for a protein normally not present in serum, rAPOL3 disappeared from the bloodstream more rapidly than rAPOL1, with a half-life of approximately 13 h and 50 h respectively (Fig. 4a; Suppl. Fig. 8). Moreover, injection of rAPOL3 was clearly toxic, as depending on the dose it induced pathology or death of the mice within one day whereas all mice safely survived injection of either rAPOL1, rPpAPOL1or an inactivated mutant of rAPOL3 (rMUT-null) (Suppl. Table; Suppl. Fig. 9). Being dependent on protein activity, rAPOL3 toxicity could hypothetically result from pore formation into host membranes. Thus, due to both instability and toxicity rAPOL3 cannot be considered for treatment of infection in vivo.
Fig. 4.
In vivo trypanolytic potential of various rAPOLs. (a) Western blot detection in the blood of 6 weeks-old female BALB/c mice, of different rAPOLs (100 µg) intravenously injected at t=0. The arrows point to the rAPOLs. PR=Ponceau Red staining, used as loading control (the band represents APOA1). (b) Trypanolytic activity of different rAPOLs on T.b. brucei 328-114 and different T.b. gambiense strains after 24 h incubation in vitro (error bars: s.e.m.; 3 technical replicates; n=3 independent experiments; error bars are often smaller than the symbol size). (c) Effect of TbKIFC1 RNAi induction by 1 µg ml-1 doxycycline7 on T.b. brucei trypanolysis by rPpMUT (error bars: s.e.m.; 3 technical replicates; n=3 independent experiments). (d) Mouse infection and survival after intravenous inoculation of 106 T.b. gambiense LiTat 1.3 parasites followed after 24 h by intravenous injection of 100 µg of different rAPOLs (Ctrl=inactive rAPOL3: see Suppl. Fig. 9; p.i.=post-inoculation; error bars: s.e.m.; 4 technical replicates; n=3 independent experiments).
So far, PpAPOL1 was presented as the best trypanolytic protein for use in the field25, because PpAPOL1 cannot be inactivated by the T.b. rhodesiense resistance protein SRA, and therefore kills this human pathogen18. Moreover, it was suggested that PpAPOL1 also efficiently kills T.b. gambiense, although this issue remains controversial because it has only been documented in vitro, and there is evidence for natural tolerance of both Papio papio and Papio hamadryas to infection by T.b. gambiense26–28. While rAPOL3 and rPpAPOL1 resistance to SRA is expected to result from sequence difference in the C-terminal region16,18, rAPOL3 ability to kill T.b. gambiense, thus, rAPOL3 resistance to TgsGP, could hypothetically be linked to independence of acidic conditions for membrane insertion. TgsGP protects a restricted fraction (7%) of endosomal membranes against APOL1, in a defined region close to the flagellar pocket14. Given the results presented in Fig. 2a-d and Suppl. Fig. 3, we hypothesize that contrary to rAPOL1, rAPOL3 could be able to insert into membranes before reaching the TgsGP-containing endosomal compartment. Along this speculative view, the ability of rPpAPOL1 to partially resist TgsGP could result from some membrane insertion before the acidic compartment, owing to the absence of two of the three APOL1-specific glutamic acid residues in helix 9. We tested this hypothesis by replacing the last glutamic acid of PpAPOL1 helix 9 (E195) with the corresponding amino acid in APOL3 (A221) (Fig. 3f). Strikingly, this single substitution increased by 100-fold the ability of PpAPOL1 to kill E. coli at neutral pH (Fig. 3f, center), and it allowed more efficient killing activity of PpAPOL1 on all different strains of T.b. gambiense that we tested (Fig. 3f, right; Fig. 4b; Suppl. Fig. 10). In contrast to what is observed with rAPOL3, the activity of rPpAPOL1 E195A (rPpMUT) was still inhibited by NH4Cl (Fig. 3f, right) and was still increased by TbKIFC1 (Fig. 4c), suggesting that as occurs for rAPOL1, rPpMUT still contains a pH-sensitive element outside helix 9. Despite this residual pH dependence, when injected into mice 24 h after intravenous inoculation of 106 LiTat 1.3 T.b. gambiense parasites, 100 µg rPpMUT was necessary and sufficient to strongly reduce the first parasitaemia peak and then completely clear infection, in sharp contrast to observations made with similar amounts of either rAPOL1 or rPpAPOL1 (Fig. 4d).
In all trypanolysis experiments conducted in this work, the killing efficiency of the various APOLs was variable between different trypanosome populations. Regarding T.b. gambiense this variability could possibly depend on the relative level of TgsGP, but it could also be influenced by the type of VSG coat or the relative adaptation of the parasites to in vitro conditions. As shown in Suppl. Fig. 10, the rate of T.b. gambiense trypanolysis by rPpMUT was inversely correlated with the relative growth of trypanosomes (panel a), and not with the VSG type (panel b), or TgsGP level (panel c). Differential in vitro adaptation could explain the discrepancy between the reported efficient T.b. gambiense killing activity of rPpAPOL1 in vitro28 and both our observations of poor in vitro activity of this protein and the known natural tolerance of different Papio species to in vivo infection by T.b. gambiense27.
In summary, we have identified two rAPOL variants able to lyse efficiently both T.b. rhodesiense and T.b. gambiense in vitro, associated with reduced dependence on acidic pH for activity. The high trypanolytic activity of one of these variants in mice offers promising prospects for treatment of sleeping sickness.
Methods
Ethics statement
This research was approved by the ethics committee of the Institute for Molecular Biology and Medicine (IBMM). All mice were housed in our pathogen-free facility and the experiments were performed in compliance with the relevant laws and institutional guidelines (license LA2500482).
Parasites
Trypanosoma brucei brucei 328-114 were grown in HMI9 supplemented with 10% foetal bovine serum, 10% Serum Plus at 37°C in 5% CO2.The RNAi were induced for 24h prior to experiments by 1 µg ml-1 doxycycline7. T.b. rhodesiense ETat 1.2 APOL1-resistant (R) and APOL1-sensitive (S) clones12 were grown in IMDM medium supplemented with 20% foetal bovine serum. The T.b. gambiense LiTat 1.3 clone and the T.b. gambiense strains Bosendja, MBA and 348BT were grown in IMDM medium supplemented with 15% foetal bovine serum + 5% human serum.
The T.b. gambiense type I (see ref. 29 for the distinction between T.b. gambiense types) were ELIANE (LiTAR 1; MHOM/CI/ITMAP 2188, man, Côte d'Ivoire, 1952), MBA (AnTAR 11; KINKOLE/ITMAP, man, Bandundu Province, DRC, 1974), Bosendja (AnTAR 6; ZR/KIN001, man, Equateur Province, DRC, 1972), PA (AnTAR 22; MHOM/CG/ITMAP1843, man, Congo, 1975) and 108AT, 348 BT and 57AT (respectively MHOM/CD/INRB/2007/25A, MHOM/CD/INRB/2006/23A and MHOM/CD/INRB/2007/28, man, East Kasai Province, DRC (ref. 30)). The T.b. gambiense type II was ABBA (MHOM/CI/DAL 626, man, Côte d'Ivoire, 1983). The T.b. rhodesiense parasites were from the ETAR repertoire (TREU 164; Glossina pallipides, Busoga, Uganda, 1960). Other parasites were respectively the T. evansi RoTat 1.2 clone (buffalo, Indonesia, 1982), the T. congolense Tc13 clone (IL 3000, Trans Mara, bovine, Kenya, 1966), the T. vivax IL 1392 clone (Y486, bovine, Nigeria, 1972), a T. theileri isolate from a primary bovine reticulocyte culture, the T. musculi Partinico II strain and the T. cruzi Dm28c clone.
Production of rAPOLs
The various recombinant proteins were all prepared according to the procedure already described5,6. Briefly, C-terminal His6-tagged rAPOLs were purified after expression from pET21d vector in Escherichia coli following 4 h-induction at 37°C with 1 mM isopropyl β-D-thiogalactoside. After washing, inclusion bodies were dissolved in 6 M guanidium–hydrochloric acid and 50 mM phosphate buffer (pH 8.0) and incubated with Ni-NTA beads for 16 h at 4°C. All washing steps occurred at pH 8.0. After elution with 250 mM imidazole, the proteins were extensively dialysed against 20 mM acetic acid. APOL3 isoform 2 was chosen, as this isoform appears to be the most expressed one in different human cell lines. The purity and concentration were verified by SDS-PAGE and Coomassie blue staining. At least three independent preparations were analysed, with similar results. Regarding rAPOL3, a fraction of the protein systematically precipitated in the trypanosome incubation medium. This fraction represented approximately 60% of the protein (Suppl. Fig. 11). Therefore, the effective concentration of rAPOL3 in our in vitro trypanolysis assays was probably lower than indicated. For rPpAPOL1 the synthetic gene based on the available coding sequence (EMBL-EBI: AGM15879.1) was ordered from Life technologies. For rPpMUT one codon was synthetically replaced resulting in E195A sequence modification.
Antibodies
Rabbit anti-APOL1 antibodies were from Sigma-Aldrich (HPA018885). To detect rAPOL3 and rPpAPOL1 we used rat antibodies raised against rAPOL3 and rAPOL1 respectively. To detect TbKIFC1, we used the mouse monoclonal anti-TbKIFC1 antibody (H3)23.
Western blotting
The protein extracts were separated on SDS-PAGE gel. Protein transfer was realized onto Hybond-P membrane (Amersham) by electrotransfer in 25 mM Tris, 192 mM glycine, 20% methanol (pH 8.3) for 1h at 100V. Membranes were blocked by incubation with 5% skim milk powder in 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 (pH 7.4) and were then incubated with antibodies in 150 mM NaCl, 50 mM Tris (pH 7.5) and 3.5% milk powder overnight at 4°C. Secondary antibodies were diluted in 150 mM NaCl, 50 mM Tris (pH 7.5) and the bound antibodies were detected by chemiluminescence (Perkin Elmer).
In vitro trypanolysis and growth assays
Lysis assays were performed as described5,6. Briefly, trypanosomes isolated from mice were incubated at 105 ml-1 in HMI-9 supplemented medium at 37°C in a CO2-equilibrated incubator. At the indicated times, living trypanosomes were counted in triplicate under the microscope. In these assays the volume of the APOL samples, in 20 mM acetic acid (pH 5), was kept constant so that there was no difference of acetic acid content between samples. In vitro growth assays were performed by daily dilutions of trypanosomes at 105 ml-1 in HMI-9 supplemented medium. Normalizations were performed to untreated controls. For the comparative measurement of the temperature effect on rAPOL1 and rAPOL3 uptake in T.b. gambiense, TgsGP KO LiTat 1.3 parasites were diluted in IMDM complete medium containing 25 mM Hepes (pH 7.5) and cooled down at 4°C. rAPOL1 or rAPOL3 (or the acetic acid solvent alone for control growth) were added to the parasites and the mixture was divided in 0.5ml tubes. Parasites were further incubated for 18 h at 15°C, 20°C, 26°C, 30°C and 37°C before counting.
In vivo growth assays
106 trypanosomes in 100 μl PSGS medium were injected intravenously in 6 weeks-old female BALB/c mice. Daily, 2 μl tail blood samples were diluted in erythrocyte lysis buffer (0.85% (w/v) NH4Cl, 10 mM Tris pH 7.4) and counted with a haemocytometer. Sample size was empirically estimated and validated by the stability of the standard deviation of the growth curves. Within each experiment, animals used were morphologically undistinguishable (same strain, age and weight) and allocated randomly to each group. No blinding was done.
Pore-forming activity in bacteria
Cultures of SE1 (Staby®) strains transfected with pStaby1.2 (Delphi Genetics) harboring the different APOL constructs were grown at 37°C in LB containing 1% glucose and 50 μg ml-1 ampicillin from freshly plated colonies until optical density at 600nm (OD600) reached 0.7 to 0.8. Ten-times serial dilutions were plated (5µl spots) onto squared LB petri dish containing agar in pH-buffered LB of 5.5 or 7.5 (1% Tryptone, 0.5% yeast extract, 0.5% NaCl, 1.5% agar, 50 mM MES, 50 mM HEPES, pH adjusted), 50 μg ml-1 ampicillin and either 1% glucose (non-induced control) or 75 μM IPTG (induction of recombinant proteins). Spots strips were compared after overnight incubation at 37°C. These results reflect the pore-forming activity of APOLs6,16.
Live light microscopy (LLM) and flow cytometry
For live microscopy, trypanosomes were mounted on a 1% low melting point agarose pad sealed with rubber glue. Cells were imaged in single plane with Axioimager M2 widefield fluorescence microscope with a 100x Plan-APOCHROMAT 1.4 objective. For the mitochondrial membrane polarity evaluation, trypanosomes were incubated for at least 15 min with 25 pM Tetramethylrhodamine Ethyl ester perchlorate (TMRE) (Life technology) before treatment. For flow cytometry, cells were analysed with a FACS canto II. 20,000 live cells were analyzed based on the gating. One morphological FSC/SSC gate followed by one FSC-H/FSC-A gate for effective singlet isolation was performed. Mean fluorescence intensity (mfi) of the gated cells was measured for the TMRE staining (PE-filter). The FACS data was further analysed in FlowJo.
Transmission Electron Microscopy (TEM)
Cells were fixed for 1h at room temperature in 2.5% glutaraldehyde in culture medium, and postfixed in 2% OsO4 in the same buffer. After serial dehydration in increasing ethanol concentrations, samples were embedded in agar 100 (Agar Scientific Ltd., United Kingdom) and left to polymerize for 2 days at 60°C. Ultrathin sections (50 to 70 nm thick) were collected in Formvar-carbon-coated copper grids by using a Leica EM UC6 ultramicrotome and stained with uranyl acetate and lead citrate. Observations were made on a Tecnai10 electron microscope (FEI), and images were captured with an Olympus VELETA camera and processed with AnalySIS software.
Focused Ion Beam-Scanning Electron Microscopy (FIB-SEM)
Samples were incubated in fixative (2% paraformaldehyde (PFA, Applichem), 2.5% gluteraldehyde (GA, EMS) in 0.15M Sodium Cacodylate (Sigma-Aldrich) buffer (pH7.4) at room temperature (RT) for 30 min. Fixative was removed by washing 5 x 3 min in 0.15M cacodylate buffer and samples were incubated in 1% osmium (OsO4, EMS), 1.5% potassium ferrocyanide (Sigma-Aldrich) in 0.15M cacodylate buffer for 40 min at RT. This was immediately followed by a second incubation in OsO4 (1% Osmium in double distilled H2O (ddH2O) for 40 min at RT. After washing in ddH2O for 5 x 3 min, samples were incubated overnight at 4°C in 1% Uranyl Acetate (UA, EMS). Uranyl acetate was removed by washing in ddH2O for 5 x 3 min and subsequently dehydrated and embedded as indicated above. Embedded samples were then mounted on aluminium SEM stubs (diameter 12 mm) and coated with ~8nm of Platinum (Quorum Q150T ES). FIB-SEM imaging was performed using a Zeiss Auriga Crossbeam system with Atlas3D software. The Focused Ion Beam (FIB) was set to remove 5 nm-sections by propelling Gallium ions at the surface. Imaging was done at 1.5 kV using an ESB (back-scattered electron) detector. 3D reconstruction and segmentation were generated from the images stacks using Fiji ImageJ (NIH, USA) and ilastik softwares.
Fluorescence imaging of rAPOL3 uptake
Parasites were preincubated in culture medium supplemented with 20 µM FMK-064 for 15 min at 37°C before addition of rAPOL3 for 15 min. Detection of rAPOL3 was performed with anti-rAPOL3 rat antibodies and Alexa488 anti-rat antibodies after Triton X100 permeabilization.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files.
Supplementary Material
Acknowledgments
We thank Dr. Annecke Kremer (Ghent) for the FIB-SEM acquisitions. This work was supported by the European Research Council (ERC 669007-APOLs), the Interuniversity Attraction Poles Programme – Belgian Science Policy (PAI P7-41) and the PDR-FNRS (PDR T.0159.13). The Center for Microscopy and Molecular Imaging is supported by the European Regional Development Fund and Wallonia.
Footnotes
Author contributions
E.P., F.F. and D.P.M. designed the research; F.F., L.L., G.V., P.U., M.S. and P. T. performed the research; B.V. supervised some experiments; N.V.R. and P.B. adapted the trypanosome strains to in vitro growth; E.P. and F.F. wrote the paper.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Vanhollebeke B, Pays E. The function of apolipoproteins L. Cell Mol Life Sci. 2006;63:1937–1944. doi: 10.1007/s00018-006-6091-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu CA, Klopfer EI, Ray PE. Human apolipoprotein L1 (ApoL1) in cancer and chronic kidney disease. FEBS Lett. 2012;586:947–955. doi: 10.1016/j.febslet.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pays E, Vanhollebeke B, Uzureau P, Lecordier L, Pérez-Morga D. The molecular arms race between African trypanosomes and humans. Nat Rev Microbiol. 2014;12:575–584. doi: 10.1038/nrmicro3298. [DOI] [PubMed] [Google Scholar]
- 4.Uzureau S, et al. Apolipoproteins L control cell death triggered by TLR3/TRIF signaling in dendritic cells. Eur J Immunol. 2016;46:1854–1866. doi: 10.1002/eji.201546252. [DOI] [PubMed] [Google Scholar]
- 5.Vanhamme L, et al. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature. 2003;422:83–87. doi: 10.1038/nature01461. [DOI] [PubMed] [Google Scholar]
- 6.Pérez-Morga D, et al. Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science. 2005;309:469–472. doi: 10.1126/science.1114566. [DOI] [PubMed] [Google Scholar]
- 7.Vanwalleghem G, et al. Coupling of lysosomal and mitochondrial membrane permeabilization in trypanolysis by APOL1. Nat Commun. 2015;6:8078. doi: 10.1038/ncomms9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greene AS, Hajduk SL. Trypanosome lytic factor-1 initiates oxidation-stimulated osmotic lysis of Trypanosoma brucei brucei. J Biol Chem. 2016;291:3063–3075. doi: 10.1074/jbc.M115.680371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimamura M, Hager KM, Hajduk SL. The lysosomal targeting and intracellular metabolism of trypanosome lytic factor by Trypanosoma brucei brucei. Mol Biochem Parasitol. 2001;115:227–237. doi: 10.1016/s0166-6851(01)00292-4. [DOI] [PubMed] [Google Scholar]
- 10.Lecordier L, et al. Identification of Trypanosoma brucei components involved in trypanolysis by normal human serum. Mol Microbiol. 2014;94:625–636. doi: 10.1111/mmi.12783. [DOI] [PubMed] [Google Scholar]
- 11.Thomson R, Finkelstein A. Human trypanolytic factor APOL1 forms pH-gated cation-selective channels in planar lipid bilayers: Relevance to trypanosome lysis. Proc Natl Acad Sci USA. 2015;112:2894–2899. doi: 10.1073/pnas.1421953112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alsford S, Currier RB, Guerra-Assunção JA, Clark TG, Horn D. Cathepsin-L can resist lysis by human serum in Trypanosoma brucei brucei. PLoS Pathog. 2014;10:e1004130. doi: 10.1371/journal.ppat.1004130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xong HV, et al. A VSG expression site-associated gene confers resistance to human serum in Trypanosoma rhodesiense. Cell. 1998;95:839–846. doi: 10.1016/s0092-8674(00)81706-7. [DOI] [PubMed] [Google Scholar]
- 14.Uzureau P, et al. Mechanism of Trypanosoma gambiense resistance to human serum. Nature. 2013;501:430–434. doi: 10.1038/nature12516. [DOI] [PubMed] [Google Scholar]
- 15.Capewell P, et al. The TgsGP gene is essential for resistance to human serum in Trypanosoma brucei gambiense. PLoS Pathog. 2013;9:e1003686. doi: 10.1371/journal.ppat.1003686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lecordier L, et al. C-terminal mutants of apolipoprotein L-I efficiently kill both Trypanosoma brucei brucei and Trypanosoma brucei rhodesiense. PLoS Pathog. 2009;5:e1000685. doi: 10.1371/journal.ppat.1000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genovese G, et al. Association of trypanolytic apoL1 variants with kidney disease in African-Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomson R, Molina-Portela P, Mott H, Carrington M, Raper J. Hydrodynamic gene delivery of baboon trypanosome lytic factor eliminates both animal and human-infective African trypanosomes. Proc Natl Acad Sci U S A. 2009;106:19509–19514. doi: 10.1073/pnas.0905669106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Page NM, Butlin DJ, Lomthaisong K, Lowry PJ. The human apolipoprotein L gene cluster: identification, classification, and sites of distribution. Genomics. 2001;74:71–78. doi: 10.1006/geno.2001.6534. [DOI] [PubMed] [Google Scholar]
- 20.Bruggeman LA, et al. Plasma apolipoprotein L1 levels do not correlate with CKD. J Am Soc Nephrol. 2014;25:634–644. doi: 10.1681/ASN.2013070700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanhollebeke B, Lecordier L, Perez-Morga D, Amiguet-Vercher A, Pays E. Human serum lyses Trypanosoma brucei by triggering uncontrolled swelling of the parasite lysosome. J Eukaryot Microbiol. 2007;54:448–451. doi: 10.1111/j.1550-7408.2007.00285.x. [DOI] [PubMed] [Google Scholar]
- 22.Pays E, et al. The trypanolytic factor of human serum. Nat Rev Microbiol. 2006;4:477–486. doi: 10.1038/nrmicro1428. [DOI] [PubMed] [Google Scholar]
- 23.Dutoya S, et al. A novel C-terminal kinesin is essential for maintaining functional acidocalcisomes in Trypanosoma brucei. J Biol Chem. 2001;276:49117–49124. doi: 10.1074/jbc.M105962200. [DOI] [PubMed] [Google Scholar]
- 24.Barrera FN, Weerakkody D, Anderson M, Andreev OA, Reshetnyak YK, Engelman DM. Roles of carboxyl groups in the transmembrane insertion of peptides. J Mol Biol. 2011;413:359–371. doi: 10.1016/j.jmb.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willyard C. Putting sleeping sickness to bed. Nat Medicine. 2011;17:14–17. doi: 10.1038/nm0111-14. [DOI] [PubMed] [Google Scholar]
- 26.Raper J, Friedman DJ. Parasitology: Molecular one-upmanship. Nature. 2013;501:322–323. doi: 10.1038/501322a. [DOI] [PubMed] [Google Scholar]
- 27.Kageruka P, et al. Infectivity of Trypanosoma (Trypanozoon) brucei gambiense for baboons (Papio hamadryas, Papio papio) Ann Soc Belg Med Trop. 1991;71:39–46. [PubMed] [Google Scholar]
- 28.Cooper A, et al. A primate APOL1 variant that kills Trypanosoma brucei gambiense. PLoS Negl Trop Dis. 2016;10:e0004903. doi: 10.1371/journal.pntd.0004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radwanska M, et al. Novel primer sequences for a polymerase chain reaction-based detection of Trypanosoma brucei gambiense. Am J Trop Med Hyg. 2002;67:289–295. doi: 10.4269/ajtmh.2002.67.289. [DOI] [PubMed] [Google Scholar]
- 30.Pyana Pati P, et al. Melarsoprol sensitivity profile of Trypanosoma brucei gambiense isolates from cured and relapsed sleeping sickness patients from the Democratic Republic of the Congo. PLoS Negl Trop Dis. 2014;8:e3212. doi: 10.1371/journal.pntd.0003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files.