Abstract
In this study, we report that the tetraspanin CD81 enhances HIV-1 reverse transcription in HIV-1-infected cells. This is enabled by the direct interaction of CD81 with the deoxynucleoside triphosphate phosphohydrolase SAMHD1. This interaction prevents the endosomal accumulation and favours the proteasome-dependent degradation of SAMHD1. Consequently, CD81 depletion results in SAMHD1 increased expression, decreasing the availability of deoxynucleoside triphosphates (dNTP) and thus HIV-1 reverse transcription. Conversely, CD81 overexpression, but not the expression of a CD81 C-term deletion mutant, increases cellular dNTP content and HIV-1 reverse transcription. Our results demonstrate that the interaction of CD81 with SAMHD1 controls the metabolic rate of HIV-1 replication by tuning the availability of building blocks for reverse transcription, i.e. dNTPs. Together with its role in HIV-1 entry and budding into host cells, the data herein indicate that HIV-1 uses CD81 as a rheostat that controls different stages of the infection.
Keywords: CD81, SAMHD1, HIV-1, Reverse transcription, dNTP content, Tetraspanin-enriched microdomains
Introduction
Human immunodeficiency virus 1 (HIV-1) infects mainly CD4+ T lymphocytes, monocytes and dendritic cells. After attachment to the CD4 receptor, viral envelope glycoproteins interact with a coreceptor protein (CXCR4 or CCR5), and trigger conformational changes that allows the fusion between the viral and cellular membranes1. HIV-1 replication requires uncoating, reverse transcription (RT), and viral DNA insertion into the cell genome. Uncoating may occur at the plasma membrane just after fusion, triggering RT and then the pre-integration complex (PIC) would be actively transported into the nucleus2, 3. Alternatively, the viral core could be transported to the perinuclear region, with gradual uncoating and RT2, 3. A third model proposes that, after fusion, the viral capsid would remain intact, and RT would occur during transport, being completed at the nuclear pore before PIC transfer into the nucleus2, 3.
Numerous cellular molecules, including tetraspanins, regulate HIV-1 infection4, 5. Tetraspanins interact with other tetraspanins, as well as with transmembrane receptors, lipids and intracellular proteins, organizing tetraspanin-enriched microdomains (TEMs)6. Through TEMs, tetraspanins modulate the function of their associated partners, playing important roles in physiological and pathological processes7–9. In infected cells, HIV-1 assembly preferentially occurs at TEMs10, 11, although tetraspanin role as budding co-factors is not yet well established12. Tetraspanins incorporation into HIV-1 particles reduces virus-to-cell and viral-induced cell-cell fusion13–15. In target cells, CD63 controls CXCR4 surface expression, regulating viral entry16, and CD9 and CD81inhibit HIV-1-induced membrane fusion17. Inhibition of membrane fusion by tetraspanins would prevent syncytia formation and support efficient viral cell-cell transfer4. Indeed, HIV-1 vpu protein downregulates CD81 in HIV-1 infected cells, promoting virion infectivity18. On the contrary, CD63 is important for viral RT19.
SAMHD1 is a ubiquitous deoxyribonucleoside triphosphate triphosphohydrolase (dNTPase) that regulates intracellular dNTP levels20. SAMHD1 activity limits HIV-1 RT in resting monocytes, macrophages, dendritic cells and CD4+ T cells21. Low dNTP levels restrain viral DNA synthesis, blocking later stages of viral replication22, 23. In activated CD4+ T cells, SAMHD1 expression is reduced24, and the high levels of dNTPs contribute to the viral infection22. SAMHD1 knock-out mice also display increased RT and higher intracellular dNTP concentrations25, 26. The specific mechanisms regulating SAMHD1 enzymatic function remain undefined. In cycling cells, which display no HIV-1 restriction, SAMHD1 is phosphorylated at threonine 592 (T592) by cyclin-dependent kinases27–29. This phosphorylation is induced by interleukin-2 and -7 in CD4+ T lymphocytes30. SAMHD1 also shows in vitro exonuclease activity against single-stranded DNAs and RNAs, displaying specificity for DNA-RNA duplexes which occur during RT31, and this activity is also involved in HIV-1 restriction32, 33. However, the control of phosphohydrolase and nuclease activities by T592 phosphorylation has been questioned34, 35, suggesting that additional regulatory mechanisms may be involved.
In this study, we show that SAMHD1 is a molecular partner of tetraspanin CD81, and that CD81 regulates RT by controlling SAMHD1 expression, thereby modulating the intracellular dNTP content.
Results
CD81 associates with SAMHD1
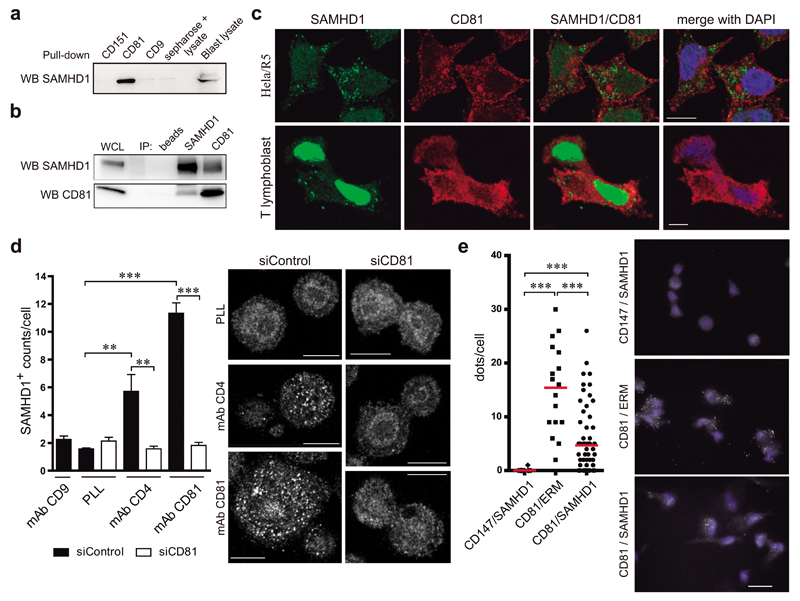
A putative interaction between CD81 and SAMHD1 was suggested by high-throughput mass spectrometry studies in primary human T lymphoblasts; not being validated at that time36. The association between CD81 and SAMHD1was first probed by pull-down experiments using biotinylated tetraspanin C-terminal peptides. SAMHD1 specifically bound to CD81 C-terminal peptides, whereas no association was observed with tetraspanins CD9 and CD151 (Figure 1a). Then, the association between endogenous molecules was assessed by co-immunoprecipitation, and SAMHD1 was detected in CD81 immunoprecipitates and vice versa (Figure 1b). These results indicate that SAMHD1 and CD81 directly associate through the CD81 C-terminal domain.
Figure 1. The C-terminal domain of CD81 mediates its association with SAMHD1.
a) SAMHD1 immunoblot of primary T lymphoblast lysates pulled-down with biotinylated peptides of tetraspanins CD81, CD9 and CD151 C-terminal domains. Sepharose-negative control and whole cell lysate are shown. b) Primary T lymphoblast lysates were immunoprecipitated and immunoblotted with SAMHD1 or CD81 antibodies. Control beads incubated with cell lysate, and whole cell lysate are shown. c) Hela/R5 cells or primary T lymphoblasts were plated onto PLL, fixed, permeabilized in PBS 0.5% Triton X-100 for 5 min, stained for CD81 (red) and SAMHD1 (green), and analysed by confocal microscopy. One single confocal plane is shown, nuclei are in blue. Bar = 10μm. d) Hela/R5 cells transfected with control siRNA (siControl) or CD81 siRNA (siCD81) were plated onto PLL (10μg/ml), anti-CD9 (VJ1/20, 10μg/ml), anti-CD4 (HP2/6, 10μg/ml) or anti-CD81 (5A6, 10μg/ml) monoclonal antibodies for 2h, fixed, permeabilized in 0.5% Triton X-100 for 5min, and stained for SAMHD1 (polyclonal antibody). Images show a single confocal plane at a ventral position, bar = 10μm. Graph shows means ± SEM of the number (counts/cell) of SAMHD1+ clusters (n=50 cells, 3 independent experiments), analysed by one-way ANOVA with Tukey’s post-test. e) Duo-link immunoassay of primary T lymphoblasts plated onto PLL, permeabilized in PBS 0.5% Triton X-100 for 5min, and stained for SAMHD1 and CD81. SAMHD1/CD147 and CD81/ERM were used as negative and positive controls respectively, bar = 10μm. Graph shows the number of dots per cell; each dot represents a single cell, bars denote the mean of scatter plots, and data was analysed by one-way ANOVA with Dunns post-test.
Since SAMHD1 is not expressed in T cell lines, like Jurkat or CEM (Supplementary Figure 1), we have used primary T lymphoblasts and Hela cells to investigate its cellular distribution. Different permeabilizing conditions were used to co-stain CD81, which is a transmembrane protein expressed at the plasma membrane and in intracellular vesicular compartments, and SAMHD1, which has been mainly characterized as a nuclear protein, although also described at the cytoplasm of T lymphocytes22. Increasing concentrations of the permeabilizing agent allowed the detection of nuclear SAMHD1, but diminished the tetraspanin signal (Supplementary Figure 2a-d). Cytoplasmic localization of the enzyme could be observed with these permeabilizing conditions, as well as in Jurkat cells transfected with SAMHD1-GFP (Supplementary Figure 2a-e). Using mild permeabilizing conditions (0.5% Triton X-100 for 5min), intracellular CD81 was co-stained with cytoplasmic and nuclear SAMHD1 in primary T lymphoblasts and Hela/R5 cells (Figure 1c and Supplementary Figure 2c). However, the absence of clear co-localization in resting conditions indicates a transient interaction.
Therefore, we assessed SAMHD1 localization upon tetraspanin crosslinking in Hela/R5 cells stably expressing CD4 and CCR5. Cells plated onto poly-L-lysine (PLL), as a control, or onto anti-CD9 antibody showed weak SAMHD1 staining (Figure 1a). Interestingly, the hydrolase accumulated in patches at the cellular basal layer upon CD81 or CD4 (a CD81 partner7) crosslinking (Figure 1d). This clustering was abrogated by siRNA depletion of CD81 (siCD81) (Figure 1d). Moreover, in situ proximity ligation assays revealed SAMHD1-CD81 interactions in a significant number of primary T lymphoblasts (Figure 1e). We used SAMHD1/CD147 as negative control, since CD147 is a membrane receptor with higher expression than CD81; and CD81/ERM as positive control, corroborating that proximity ligation signal could be attained between a membrane-bound molecule and an intracellular connector that were previously shown to interact at the uropod of T-lymphoblasts37.
CD81 positively regulates HIV-1 reverse transcription
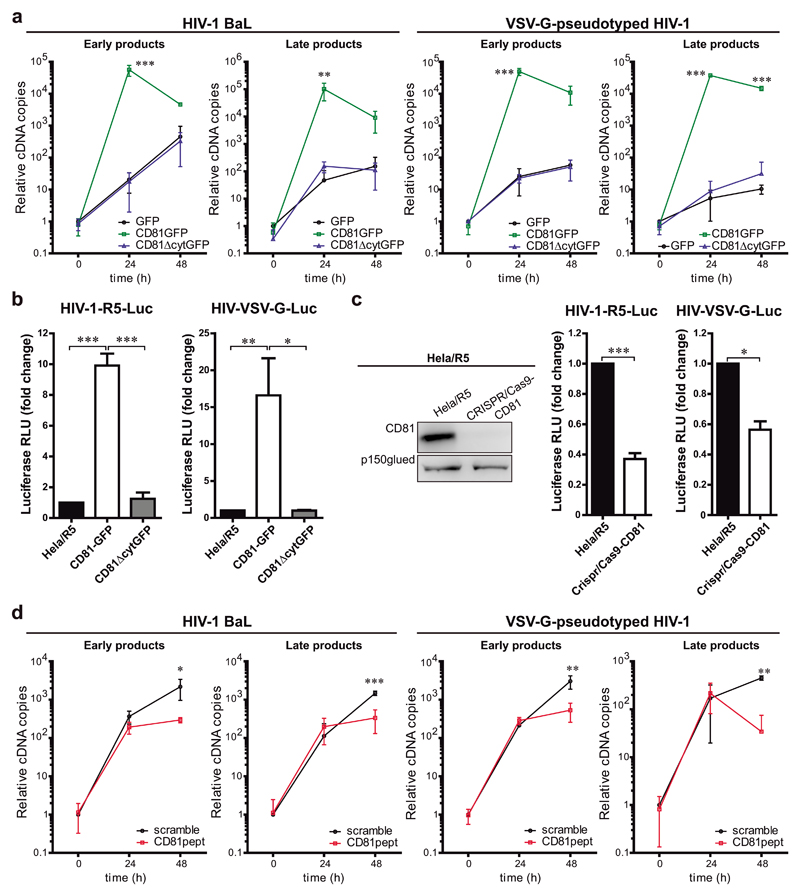
Since SAMHD1 regulates HIV-1 RT, we assessed the role of CD81 in this step of the viral cycle. Hela/R5 cells were transfected with GFP, GFP-tagged CD81 (CD81GFP) or a CD81 mutant lacking the C-terminal cytoplasmic region (CD81ΔcytGFP)38, and infected with wild-type R5-tropic HIV-1 (BaL strain). Early and late viral RT products were measured by qPCR. CD81GFP expression increased HIV-1 RT, and this effect was dependent on the CD81 C-terminal domain (Figure 2a, HIV-1 BaL). CD81GFP overexpression also highly increased RT of VSV-G-pseudotyped HIV-1 (HIV-VSV-G) (Figure 2a, HIV-VSV-G), which enters the cells via attachment of the VSV glycoprotein G, ruling out any effect of CD81 in HIV-1 entry, assembly or release10, 17. Moreover, CD81 positively regulated HIV-1 replication of single-cycle luciferase (Luc) reporter virus (pseudotyped with HIV-1 or VSV-G envelopes), through a process mediated by its C-terminal domain (Figure 2b).
Figure 2. CD81 expression supports R5-tropic HIV-1 RT.
a) Time course of HIV-1 RT measured by qPCR of early or late RT products at 24 or 48h post-infection. Hela/R5 expressing GFP, CD81GFP or CD81∆cytGFP were infected with HIV-1 (BaL) or HIV-VSV-G. Data are mean fold change ± SEM from 4 independent experiments performed in triplicate, and analysed by two-way ANOVA with Bonferroni’s post-test. b) Hela/R5 cells expressing GFP, CD81GFP or CD81∆cytGFP were infected with single-cycle HIV-1-R5-Luc or HIV-VSV-G-Luc. Data are mean fold-induction ± SEM of luciferase activity from 3 independent experiments performed in triplicate, and analysed by one-way ANOVA with Tukey’s post-test. c) Hela/R5 cells transfected with CRISPR/Cas9-CD81 or left untreated were infected and analysed as in b. Data are mean fold change ± SEM from 3 independent experiments performed in triplicate, and analysed by paired Student t-test, *** p<0.0001 (left) and * p = 0.0157 (right). Left immunoblots show whole cell lysates probed for CD81, and p150 as loading control. d) Hela/R5 cells pre-incubated for 5 days with 2μM cytopermeable peptides containing the CD81 C-terminal sequence (CD81pept) or a scrambled version were infected and analysed as in a.
Conversely, CD81 knock-down using siRNA (Supplementary Figure 3a), or full knock-out using the CRISPR/Cas9 technology (CRISPR/Cas9-CD81; Figure 2c) in Hela/R5 cells reduced RT of both HIV-1 wild-type and HIV-VSV-G (Supplementary Figure 3a-b). CD81 deficiency also diminished the luciferase activity after infection with single-cycle HIV-1-R5-Luc or HIV-VSV-G-Luc reporter viruses (Figure 2c). The expression levels of CD4, CCR5 or tetraspanins CD82, CD9, CD151 or CD63 were not affected by CD81 knock-down or knock-out (Supplementary Figure 3c-d). In addition, pre-treatment of Hela/R5 cells with fluorescently labelled cytopermeable peptides corresponding to the sequence of the CD81 C-terminal region (CD81pept), which functionally mimics the effects of CD81 knockdown in different models38, 39 significantly reduced RT of wild-type HIV-1 and HIV-VSV-G (Figure 2d), further corroborating the involvement of CD81 C-terminal domain in the regulation of viral RT.
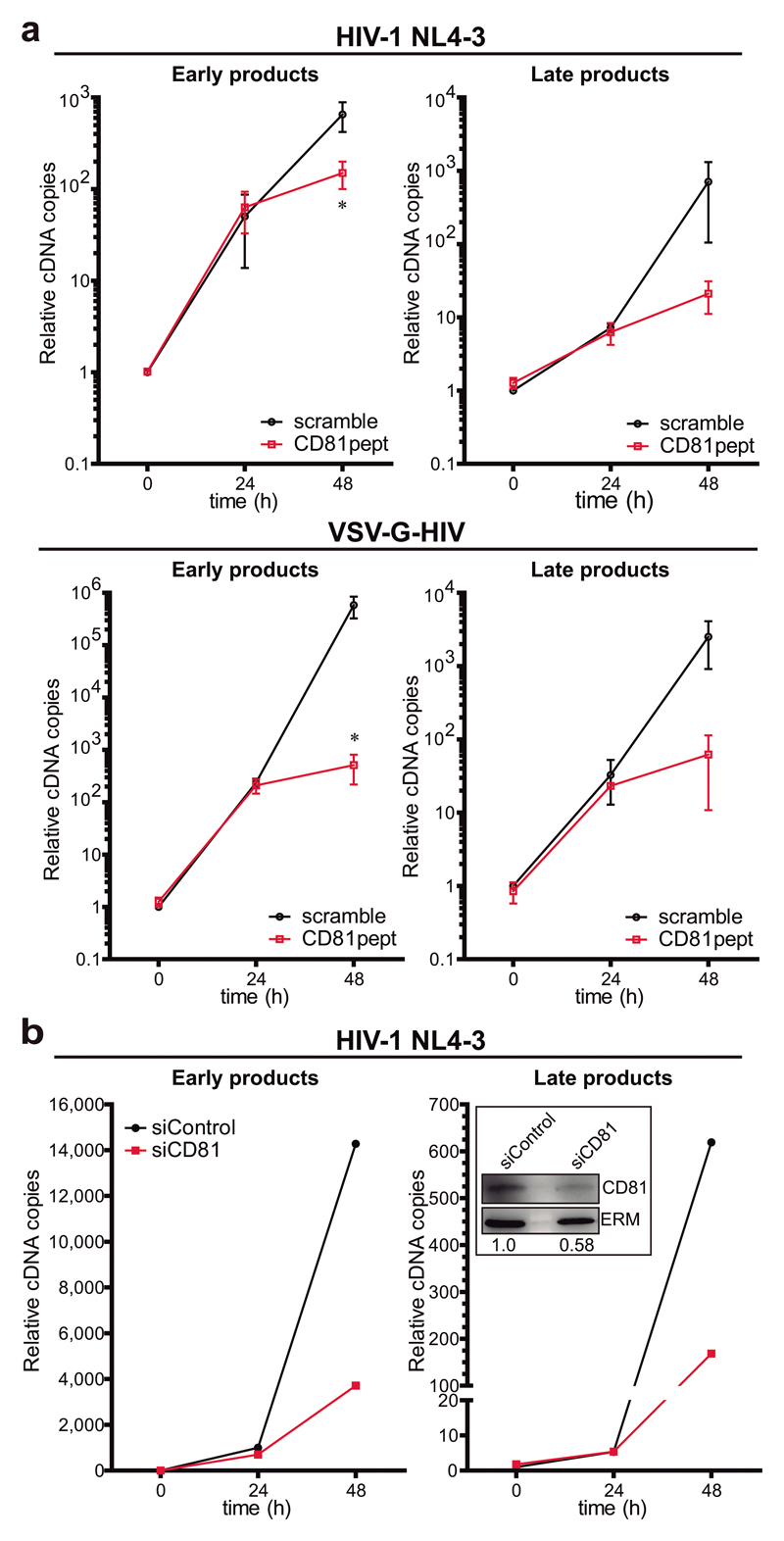
In a more physiological setting, pre-treatment of primary T lymphoblasts with CD81pept clearly impaired RT of wild-type HIV-1 (NL4-3 strain) or HIV-VSV-G (Figure 3a). Importantly, CD81 knock-down with siRNA in primary T lymphoblasts also specifically decreased RT (Figure 3b). Together, our results suggest that CD81 positively regulates HIV-1 RT of both R5- and X4-tropic viruses.
Figure 3. CD81 regulates X4-tropic HIV-1 RT.
a) Primary T lymphoblasts pre-treated with 2μM of scramble or CD81pept for 5 days, were infected with HIV-1 NL4-3 strain or HIV-VSV-G. Early or late RT products were measured as in Figure 2a. Data are mean fold change ± SEM of 2 independent experiments performed in triplicate, and analysed by two-way ANOVA with Bonferroni’s post-test. b) Primary T lymphoblasts transfected with control or CD81 siRNA were infected with NL4-3 strain, and RT was measured as in Figure 2a. Data are from a representative experiment out of two. In-box shows immunoblots of whole cell lysates from the represented experiment probed for CD81, and ERM as loading control. The CD81/ERM signal ratio is indicated.
CD81 regulates intracellular dNTP levels via SAMHD1
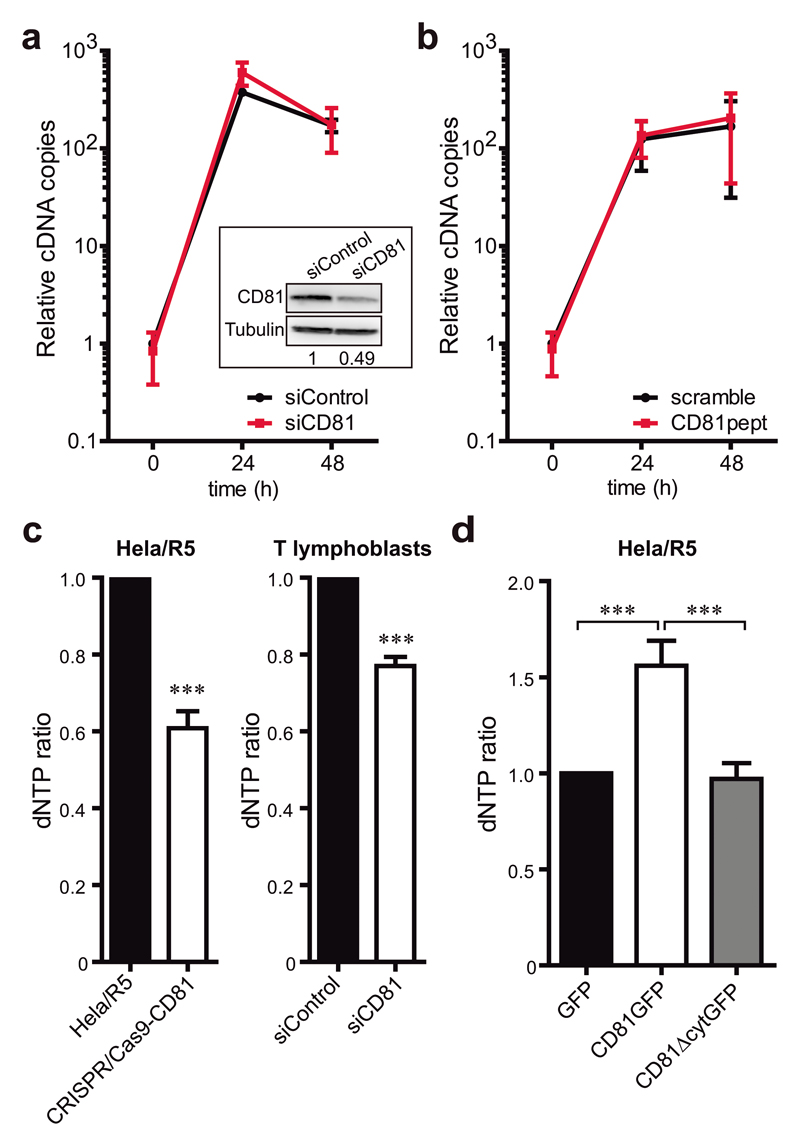
To determine whether the function of CD81 in RT relied on the regulation of SAMHD1, we analysed the effect of CD81 knock-down in Jurkat cells, which do not express SAMHD1 (Supplementary Figure 1). In these cells, CD81 knock-down or treatment with CD81pept did not alter the levels of RT products in comparison to control cells (Figure 4a-b), indicating that CD81 does not affect HIV-1 RT in the absence of SAMHD1.
Figure 4. CD81 negatively regulates cellular dNTP content through SAMHD1.
a) Jurkat T cells transfected with control or CD81 siRNA were infected with HIV-1 NL4-3 strain, and early RT products were measured by qPCR at the indicated times. Data are mean fold change ± SEM of 2 independent experiments performed in triplicate. In-box shows representative immunoblots of CD81 and tubulin as loading control, and the CD81/Tubulin signal ratio is indicated. b) Jurkat T cells pre-treated with 2μM of scramble or CD81pept for 5 days were infected with NL4-3 strain, and RT was analysed as in a. c) Mean fold change ± SEM of the dNTP content of Hela/R5 or Hela/R5 CRISPR/Cas9-CD81 cells (left graph, 4 independent experiments), and primary T lymphoblasts transfected with control or CD81 siRNA (right graph, 2 independent experiments) measured by a HIV RT-based dNTP assay. Analysis was performed by paired Student t-test, *** p<0.0001 (left) and *** p<0.0001 (right). d) dNTP content of Hela/R5 cells overexpressing GFP, CD81GFP or CD81∆cytGFP measured as in c. Data are mean fold change ± SEM from 3 independent experiments analysed by one-way ANOVA with Tukey’s post-test.
SAMHD1 dNTPase activity was then directly analysed by quantification of the intracellular dNTP content in cell lysates after CD81 overexpression or depletion. In parallel to the effects observed on viral RT, the dNTP pool was significantly reduced in both Hela/R5 cells and primary T lymphoblasts depleted for CD81 (Figure 4c) or treated with CD81pept (Supplementary Figure 3e). Conversely, CD81GFP overexpression in Hela/R5 cells increased the dNTP content, which remained unaffected by CD81ΔcytGFP expression (Figure 4d). These results indicate that CD81 regulates the intracellular dNTP content by associating with SAMHD1 through its C-terminal domain.
CD81 controls SAMHD1 degradation by the proteasome and its subcellular localization
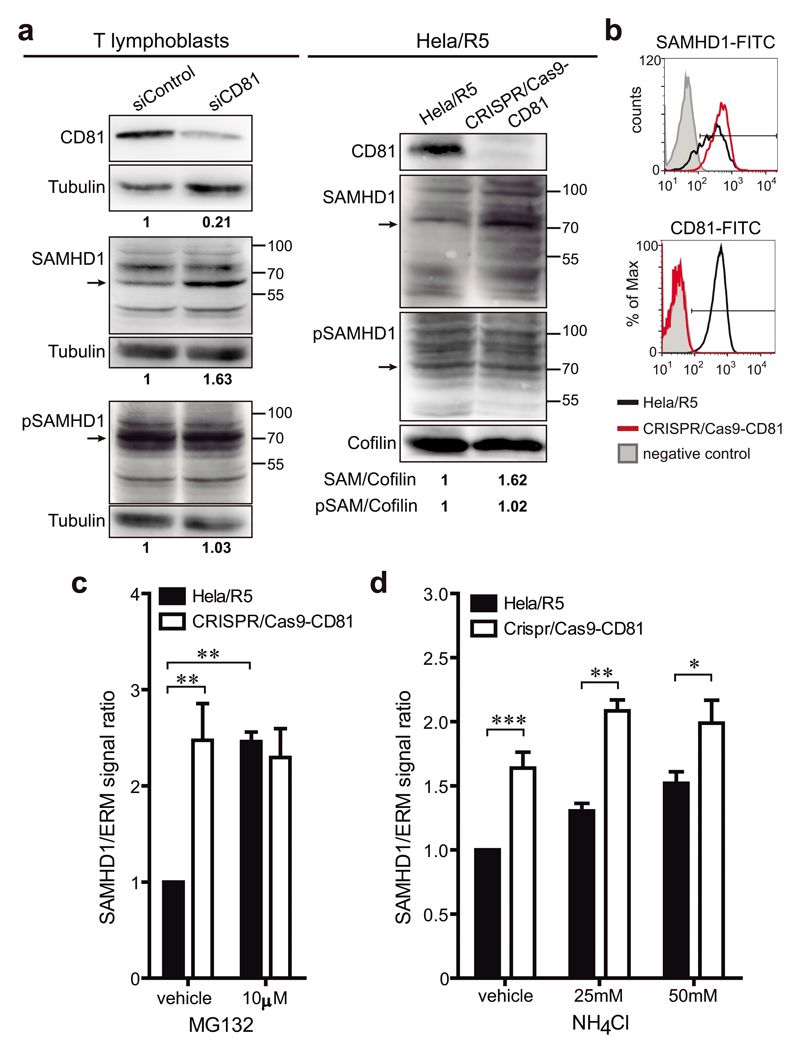
We next investigated the mechanism by which CD81 could regulate SAMHD1. Interestingly, CD81 deficiency increased the expression of SAMHD1, both in primary T lymphoblasts transfected with CD81 siRNA and in Hela/R5 CRISPR/Cas9-CD81 cells (Figure 5a-b). However, no differences were observed in the levels of SAMHD1 phosphorylation (Figure 5a). Although SAMHD1 has a predicted molecular weight of ~70kDa, additional bands with different molecular weights could be detected, suggesting cleavage, splicing variants40 or post-translational modifications of the enzyme27, 28, 41. Specificity of the antibody was confirmed using whole cell lysates from cells expressing SAMHD1-GFP or transfected with SAMHD1 siRNA (Supplementary Figure 4). To assess whether the increase in SAMHD1 expression was related to altered protein degradation, cells were treated with MG132, which prevents proteasome function, or with ammonium chloride (NH4Cl), which blocks the acidification of lysosomes. SAMHD1 expression was two-fold higher in Hela/R5 control cells upon treatment with MG132 (Figure 5c), while it was only slightly affected by increasing concentrations of NH4Cl (Figure 5d), indicating that the proteasome is the main route for SAMHD1 turnover. In CRISPR/Cas9-CD81 cells, despite the expected higher SAMHD1 basal levels in cells treated with the vehicle, the increase in the expression of the enzyme after MG132 treatment was completely abolished (Figure 5c), suggesting that CD81 is essential for SAMHD1 degradation by the proteasome. Treatment of CRISPR/Cas9-CD81 cells with NH4Cl slightly increased SAMHD1 expression, suggesting that lysosomal degradation was unaffected or slightly favoured upon proteasomal blockade by CD81 (Figure 5d).
Figure 5. CD81 regulates SAMHD1 expression.
a) Primary T lymphoblasts transfected with control or CD81 siRNA, and Hela/R5 cells transfected or not with CRISPR/Cas9-CD81 were lysed and immunoblotted for CD81, SAMHD1 and phosphorylated SAMHD1. Tubulin or cofilin were used as loading controls. Blots are from representative experiments out of 2 (lymphoblasts) and 3 (Hela/R5). Signal ratios in relation to the loading controls are depicted. Arrows indicate the SAMHD1 band that has the predicted molecular weight (~70kDa). b) Hela/R5 cells transfected or not with CRISPR/Cas9-CD81 were fixed, permeabilized, immunolabelled for SAMHD1 and CD81, and analysed by flow cytometry. Histograms show a representative experiment out of 3. The negative control corresponds to cells stained only with secondary antibody. c-d) CRISPR/Cas9-CD81 or control Hela/R5 cells were treated for 6h with the vehicle or with the depicted concentrations of (c) MG132 or (d) NH4Cl. Cells were lysed and immunoblotted for SAMHD1, and ERM as loading control. Graph shows the mean fold change ± SEM of SAMHD1/ERM signal ratio from (c) 3 or (d) 2 independent experiments analysed by one-way ANOVA with Bonferroni’s post-test.
Since CD81 deletion increases SAMHD1 expression by protecting the enzyme from proteasomal degradation (Figure 5); and CD81 crosslinking triggers SAMHD1 accumulation beneath the plasma membrane (Figure 1d), we investigated whether CD81 could regulate the subcellular distribution of SAMHD1. In the absence of CD81, SAMHD1 accumulated in cytoplasmic speckles, which displayed increased number and size with respect to control cells (Figure 6a and Supplementary Figure 5a,b). Similar results were obtained when cells were treated with CD81pept (Figure 6b and Supplementary Figure 5a,b). Accordingly, the presence and area of these SAMHD1 circular cytoplasmic structures was conversely reduced in CD81GFP-expressing cells, while no differences were observed between cells expressing CD81ΔcytGFP or GFP (Figure 6c and Supplementary Figure 5c).
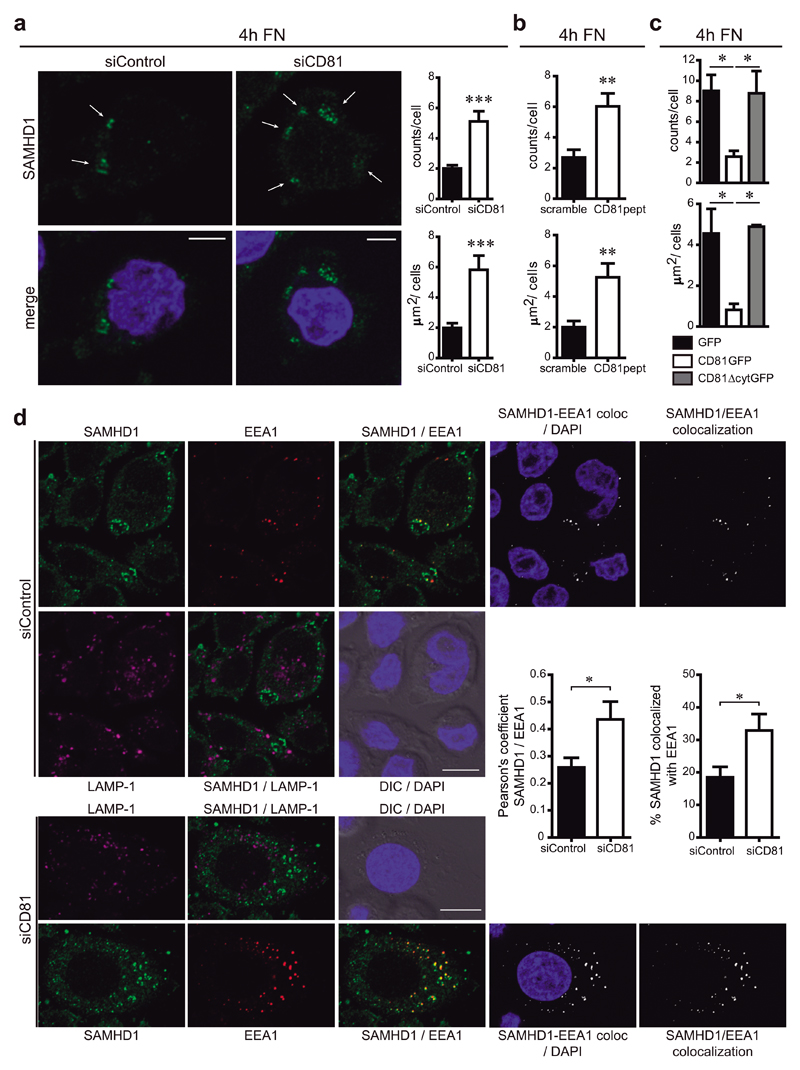
Figure 6. SAMHD1 is partially enriched at early endosomes.
a) Hela/R5 cells were transfected with control or CD81 siRNA, adhered for 4h onto fibronectin (FN), fixed, permeabilized in PBS 0.1% Triton X-100 for 5min, and immunolabelled for SAMHD1. Images show one single confocal plane, nuclei are in blue. Arrows indicate SAMHD1 accumulation in circular-shaped intracellular structures, bars = 10μm. Graphs show means ± SEM of the number (counts/cell) and area (μm2/cell) of the cytoplasmic structures observed (n=230 cells, 4 independent experiments analysed by Student t-test, *** p = 0.0005 (upper) and *** p = 0.0006 (bottom)). b) Hela/R5 cells treated with 2μM of scramble or CD81pept were analysed as in a (n=400 cells, 4 independent experiments analysed by Student t-test, ** p = 0.0063 (upper) and ** p = 0.0054 (bottom)). c) Hela/R5 cells transfected with GFP, CD81GFP or CD81∆cytGFP were analysed as in a (n=20 cells, 2 independent experiments analysed by one-way ANOVA with Tukey’s post-test). d) Hela/R5 transfected with control or CD81 siRNA were treated as in a. Images show SAMHD1 (green), EEA1 (red), LAMP-1 (magenta), nuclei (blue), DIC, SAMHD1/EEA1 co-localization channel (white), and merged images. One single confocal plane is shown, bars = 10μm. Graphs represent the quantification of SAMHD1-EEA1 co-localization performed in 3D stack confocal microscopy images, showing means ± SEM of the Pearson’s coefficient; and of the % of SAMHD1 signal co-localized with EEA1 signal with respect to the total SAMHD1 signal in the cell (n = 200 cells, 3 independent experiments analysed by Student t-test, * p = 0.0262 (left) and * p = 0.0479 (right)).
To characterize these intracellular structures, Hela/R5 cells were co-stained with antibodies against SAMHD1 and markers of different intracellular compartments. We could not observe any co-localization between SAMHD1 cytoplasmic speckles and markers of late endosomes (HGS/HRS), multi-vesicular bodies (CD63) or lysosomes (LAMP-1) (Figure 6d and Supplementary Figure 6). Interestingly, the circular-shaped intracellular structures that accumulated SAMHD1 partially co-localized with EEA1, a marker of early endosomes. SAMHD1/EEA1 co-localization was increased in CD81 knocked-down cells, as quantified by the Pearson’s coefficient and the frequency of SAMHD1 co-localizing with EEA1 when compared to the total SAMHD1 signal (Figure 6d).
Altogether, our results suggest that CD81 deletion controls SAMHD1 expression by protecting the enzyme from proteasomal degradation via its subcellular compartmentalization in early endosomes.
Discussion
In this study, we show that tetraspanin CD81 regulates HIV-1 RT through its molecular association with SAMHD1, and the control of the expression and subcellular localization of the hydrolase. We provide strong evidence of CD81 association with SAMHD1 in primary T lymphoblasts by: i) pull-down with synthetic peptides containing the CD81 C-terminal sequence but not of other tetraspanins; ii) co-immunoprecipitation of endogenous CD81 and SAMHD1 using detergent lysis conditions (0.5% NP-40) that mostly disrupt TEMs, thus impairing indirect connections; and iii) in situ proximity ligation assay, which provides strong evidence that the CD81-SAMHD1 molecular association occurs in vivo. Moreover, crosslinking with monoclonal antibodies against CD81 and CD4 triggers SAMHD1 juxta-membrane clustering in a CD81-dependent manner.
SAMHD1 is a cellular inhibitor of HIV-1 RT in resting cells. Although widely studied in the past years, the mechanisms that control this enzyme are not yet fully understood21, 42. SAMHD1 phosphorylation at T592 was shown to be important for the control of SAMHD1 RNase33 and dNTPase29, 43 activities. However, it was recently suggested that this phosphorylation cannot fully explain the restriction effect35. The SAMHD1 exonuclease activity is also controversial, with recent studies indicating that viral restriction is not related to this enzymatic activity34, 44, which could even derive from contaminants in the sample45. In the absence of CD81, SAMHD1 expression is induced and its dNTPase activity is increased, without noticeable differences in its phosphorylation levels.
Our results clearly indicate that CD81 is an important player in the regulation of SAMHD1-dependent restriction of HIV-1 replication, through a mechanism dependent on its C-terminal domain. When CD81 is depleted, SAMHD1 expression is increased and the subsequent reduction in the intracellular pool of dNTPs impairs viral RT and replication. The effects of CD81 depletion on RT are observed only 48h post-infection, probably because during the first 24h, despite increased SAMHD1 activity, the remaining intracellular pool of dNTPs would be sufficient to allow initial viral replication until exhaustion observed at 48h. Accordingly, HIV-1 reverse transcriptase can efficiently synthesize viral DNA in the presence of low dNTP concentrations46. On the contrary, when CD81 is overexpressed, SAMHD1 expression is reduced and the cellular dNTP content is higher, allowing a huge increase in HIV-1 RT even at 24h. The observed increase in dNTP levels in these cells was not as impressive as the increase in RT, further indicating that the viral reverse transcriptase is very sensitive to slight changes in the intracellular availability of dNTPs. Thus, even a small increase in the levels of these nucleotides is sufficient for the huge increase in viral genome replication.
The precise subcellular site where viral RT takes place remains to be determined2, 3. Although SAMHD1 has been mainly described as a nuclear protein, we and others22 could detect this enzyme in the cytoplasm. In this regard, it has been recently shown that oxidized SAMHD1 is specifically located at the cytoplasm41. Tetraspanins, which are transmembrane proteins present in both the plasma membrane and intracellular compartments, can either promote or inhibit HIV-1 transmission, playing negative or positive roles in different steps of the viral cycle4. The involvement of tetraspanins in RT would imply that the virus highjacks intracellular membranes to support its early replication. Interestingly, CD81 crosslinking triggers SAMHD1 enrichment beneath the plasma membrane, and CD81 knock-down increases SAMHD1 permanence in an early endosomal compartment, with partial SAMHD1 and EEA1 co-localization. Our results indicate that degradation of the hydrolase can partially occur through lysosomes but it is mostly dependent on the proteasome. While CD81 does not affect SAMHD1 degradation by acidic compartments, it clearly controls its degradation by the proteasome. Therefore, CD81 regulation of SAMHD1 sub-cellular localization seems to be important for the turnover of the enzyme. Other cellular proteins have been described to regulate SAMHD1 degradation by the proteasome. Cyclin L2 interacts with SAMHD1 at the nucleus, driving its proteasomal degradation through a mechanism dependent on the ubiquitin ligase adaptor DCAF147. Interaction with the eukaryotic elongation factor 1A1 (eEF1A1) at the cytoplasm also targets SAMHD1 for proteasomal turnover, through the association with Cullin4A and Rbx148. Interestingly, we also detected eEF1A1 as a putative molecular partner of CD81 in our previous proteomic study36.
In summary, CD81 regulates HIV-1 early replication via the direct association with SAMHD1, modulating the intracellular dNTP content through the control of SAMHD1 expression and subcellular localization. The evidence that SAMHD1 is included in TEMs highlights the importance of these membrane microdomains during HIV-1 replication, not only in the entry and assembly phases of the viral cycle, but also in RT. A more detailed knowledge on how SAMHD1 blocks HIV-1 infection will provide insights to reinforce its restriction activity and prevent HIV-1 progression and AIDS. Moreover, the regulation of cellular dNTP levels by CD81 might be important in other physiological and pathological processes, like the autoimmune disorder Aicardi-Goutières syndrome or even in tumour progression.
Methods
Cells
The Hela P4 MAGI CCR5+ cells (Hela/R5)17 were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: P4R5 MAGI from Dr. N. Landau, and were cultured in DMEM (Sigma) supplemented with 10% FCS (Invitrogen) and 1µg/ml of puromycin (Sigma). Vβ8 Jurkat T cell (J77 cl20) and human kidney cell line HEK293T (ATCC) were cultured in RPMI 1640 or DMEM (Sigma) supplemented with 10% FCS. Cell lines from ATCC were authenticated by ATCC and were not further validated in our laboratory. Cells from the NIH were not validated further in our laboratory. Cell lines were routinely tested for mycoplasma contamination and were mycoplasma free. Peripheral blood lymphocytes were isolated from Buffy coats of healthy donors, and maintained as described39. Buffy coats were received from the Blood Transfusion Center of Comunidad de Madrid, and all donors signed their consent for the use of samples for research purposes. All the procedures using primary human cells were approved by the Ethics Committee of the Hospital Universitario de la Princesa.
Hela/R5 cells (8x106), human T lymphoblasts (2x107), or Jurkat cells (2x107) were washed twice with HBSS (Lonza), and transiently transfected by electroporation with siRNA (1µM) or plasmids (20μg) in OPTIMEM (Gibco, Invitrogen) at 240V and 34ms (Gene Pulser II, Bio-Rad). Overexpression, knockdown or gene deletion were confirmed by flow cytometry or western-blot.
Reagents and constructs
Tetramethylrhodamine (TAMRA) N-terminal–labelled peptides with the sequences RRRRRRRCCGIRNSSVY (CD81) or RRRRRRRYSVNICRGCSS (Scrambled) were purchased from LifeTein. N-terminally biotinylated peptides containing a SGSG linker sequence connected to the cytoplasmic C-terminal domains of the proteins of interest were purchased from Ray Biotech, and have been previously described36, 38. The constructs CD81GFP and CD81ΔcytGFP were previously described38; and SAMHD1-GFP was kindly provided by Dr. Nathaniel Landau (New York, US). Control siRNA and siRNA for CD81 CAATTTGTGTCCCTCGGGC (siCD81) were purchased from Eurogentec. We have validated three different sequences to knock-down CD81 obtaining similar phenotypes in other systems38, 39. siRNA for SAMHD1 was purchased from Dharmacon (SMARTpool ON-TARGETplus containing four different siRNA sequences).
Pull-down, immunoprecipitation and immunoblot assays
For immunoprecipitation, primary T lymphoblasts (2x107) were lysed in PBS 0.5 % NP-40 containing protease and phosphastase inhibitors. Lysates were precleared for 2h at 4ºC with protein G-Sepharose (Amersham Biosciences), and incubated for 2h at 4ºC with anti-CD81 5A6 monoclonal (Dr. S. Levy, Stanford, USA) or mouse polyclonal anti-SAMHD1 (Sigma) antibodies immobilized on protein G-Sepharose beads. After rinsing with lysis buffer, complexes were eluted in Laemmli buffer, and resolved by SDS-PAGE.
N-terminally biotinylated peptides (30 nmol) were conjugated to 40μl of streptavidin sepharose (GE Healthcare). Pull-down assays were carried out as previously described38. Briefly, cells were washed once with ice-cold PBS and lysed in PBS 1% NP-40 containing protease and phosphatase inhibitors (Complete, PhosSTOP; Roche). Lysates were precleared for 2h at 4ºC with streptavidin sepharose (GE Healthcare) and incubated for 2h at 4ºC with biotinylated peptides immobilized on streptavidin sepharose beads.
For immunoblotting, untreated cells or cells incubated with different concentrations of ammonium chloride (NH4Cl, Sigma) or MG132 (Sigma) for 6h at 37ºC were lysed in PBS 1% Triton X-100 containing protease and phosphatase inhibitors. All blots were revealed with FUJIFILM LAS-4000 after membrane incubation with specific antibodies and peroxidase-conjugated secondary antibodies (Pierce). Primary antibodies were against: SAMHD1 (polyclonal antibody; Sigma), CD81 (5A6), α-tubulin (clone DM1A, Sigma), p150glued (BD Biosciences), cofilin (Abcam), and ERM (90.3; kindly provided by Dr. H Furthmayr, Stanford, USA). Band intensities were quantified using ImageGauge (FUJIFILM) and results normalized with respect to the intensity signal of the loading controls.
Flow Cytometry
Cells were fixed in 2% paraformaldehyde (PFA; Electron Microscopy Sciences), permeabilized using the BD Cytofix/Cytoperm kit when observing intracellular proteins, and stained with primary antibodies followed by species-matching secondary antibodies (Invitrogen). Primary antibodies were: anti-CCR5 (Santa Cruz), anti-SAMHD1 (polyclonal antibody; Sigma), anti-CD82 (TS82b, kindly provided by Dr. E. Rubinstein, Villejuif, France), and antibodies produced in our laboratory (anti-CD9 (VJ1/20), anti-CD151 (LIA1/1), anti-CD63 (Tea3/18), and anti-CD4 (HP2/6))17, 49, 50. Data was acquired with a FACSCantoII flow cytometer (BD), and analysed with BD FACSDIVA (BD) or FlowJo (FlowJo LLC) softwares.
Fluorescence Confocal Microscopy
Cells were adhered onto poly-L-lysine (PLL; Sigma), CD4, CD81 or CD9 monoclonal antibodies (10μg/ml) for 2h; or onto fibronectin (FN)-coated coverslips (20μg/ml, Sigma) for 2, 4 or 18h at 37ºC, fixed in 4% PFA, permeabilized with PBS 0.1%, 0.5% or 1% Triton X-100 for 5min or 30min, stained with primary antibodies followed by species-matching secondary antibodies coupled to Alexa Fluor fluorochromes (Invitrogen), and mounted in Prolong antifading medium (Invitrogen). Primary antibodies were: anti-CD81 (5A6), anti-SAMHD1 (monoclonal or polyclonal antibodies; Sigma), anti-EEA1, CD63 and LaminA (Santa Cruz), anti-HGS/HRS (Abcam), and Alexa-647-labeled anti-LAMP-1 (BioLegend). Duolink in situ proximity ligation assay (Sigma) was performed following the instructions provided with the kit. Primary antibodies were: SAMHD1 (polyclonal antibody), CD81 (5A6), ERM (90.3) or CD147 (VJ1/917; produced in our laboratory).
Confocal images were obtained with a Zeiss LSM700 confocal scanning laser unit attached to an inverted epifluorescence microscope (Observer.Z1) fitted with a Pan APO Chromat 63X/1.4 oil immersion objective, using ZEN 2009 acquisition software (Carl Zeiss Microscopy GmbH). Images were analysed with ImageJ (NIH) software, using the Particle Analysis plugin. Measurement of Pearson’s coefficient and % of co-localized SAMHD1 and EEA1 was performed using Imaris (Bitplane), analysing three-dimensional stack confocal microscopy images. The SAMHD1 nuclear signal was excluded from the analysis.
HIV-1 viral preparation and infection
Preparation of wild-type HIV-1 NL4-3 (X4-tropic) or BaL (R5-tropic), and VSV-G-pseudotyped HIV recombinant virus (HIV-VSV-G), which allows the analysis of replication independently of HIV entry, was performed as described50. Pseudotyped viruses with a luciferase reporter gene were produced by co-transfection of HEK293T cells with an equal mixture of the pNL4-3.LucR-E- reporter plasmid (Dr. N. Landau, NIH AIDS Reagent Program), and HIV-1 envelope expression plasmid (NIH AIDS Reagent Program) or pcDNA3-VSV plasmid (encoding the vesicular stomatitis virus G-protein) using calcium phosphate (Sigma). The pNL4-3.LucR-E- has frame shift mutations in Env (E-) and Vpr (R-), and a luciferase reporter gene inserted into the Nef gene. After 24h of transfection, the medium was replaced, and 2h later pseudotyped viruses were harvested, filtered (0.45μm), aliquoted, and frozen at -80°C until further use. Viral titre was quantified by using a Gag p24 ELISA kit (INNOTEST HIV-1 Antigen mAb; Innogenetic).
For analysis of HIV-1 RT, Hela/R5 cells were infected with 100ng per 24 well of HIV-1 BaL or with 50ng per 24well of HIV-VSV-G, and primary T lymphoblasts or Jurkat cells were infected with 100ng per 106 cells with HIV-1 NL4-3 or 200ng per 106 cells with HIV-VSV-G. After 2h of infection, cells were extensively washed and incubated at 37ºC for 24 or 48h, lysed in 0.2% NP-40, and total genomic DNA was extracted using QiAmp DNA mini kit (Qiagen). Quantitative PCR analysis for the measurement of HIV reverse transcription was performed by amplifying genomic DNA using Power SYBR Green PCR master mixture (Applied Biosystems): forward primer 5´-CAGGATTCTTGCCTGGAGCTG-3´ and reverse primer 5´-GGAGCAGCAGGAAGCACTATG-3´for early reverse transcription products, and forward primer 5´-TGTGTGCCCGTCTGTTGTGT-3´ and reverse primer 5´-CGAGTCCTGCGTCGAGAGAT-3´ for late reverse transcription products. The β-actin gene was amplified to measure DNA concentration and used for normalization. Each reaction was performed in triplicates.
For single-cycle infection assay, Hela/R5 cells were plated on a 24-well plate and infected with 50ng HIV-1 p24Gag/106 cells. After 2h of infection, cells were extensively washed and incubated at 37ºC for 48h. Then, cells were washed with PBS, and lysed with a Steady Glo luciferase assay buffer (Promega Corporation, WI, USA). The light intensity of each sample was measured on a luminometer 1450 Microbeta Luminescence Counter.
Intracellular dNTP measurement
For dNTP analysis and quantification, cells were harvested, lysed in ice cold 65% methanol, and vigorously vortexed for 2min. Extracts were incubated at 95ºC for 3min, then supernatants were collected and dried in a speed vacuum. Samples were processed in a blinded manner for the single nucleotide incorporation assay as described46. Each dNTP (dATP, dCTP, dGTP and dTTP) was detected separately, and for the analysis their levels in cells overexpressing or depleted for CD81 were normalized to the levels in control cells.
Statistical analysis
All statistical analyses were performed with GraphPad Prism (GraphPad Software Inc.). p values were calculated using two-tailed Student’s t-test, one-way ANOVA with Tukey’s or two-way ANOVA with Bonferroni’s post-hoc multiple comparison tests. When indicated, one-way ANOVA with Dunn’s post-test was used. Statistical significance was assigned at * p<0.05, ** p<0.01, and *** p<0.001.
Data availability
The data that support the findings of this study are available from the corresponding author upon request. Complete blots for all the figures and supplementary figures are shown in Supplementary Figures 7 and 8.
Supplementary Material
Acknowledgements
The authors thank M. Vicente-Manzanares (Hospital de la Princesa, UAM, Spain) for critical reading of the manuscript. Microscopy was performed at CNIC Microscopy & Dynamic Imaging Unit. This work was supported by grants to SL (Translational Cancer Award from Stanford Cancer Institute, SPARK at Stanford, and the Breast Cancer Research program from the Department of Defense grant W81XWH-14-1-0397); to BK (R01 GM104198; R01 AI049784); to MAM-F (RD16/0025/0019; PI16/01863; CYTED 214RT0482); to FS-M (SAF2011-25834; SAF2014-55579-R; INDISNET-S2011/BMD-2332; RD12-0042-0056; ERC-2011-AdG 294340-GENTRIS) and to MY-M (BFU2014-55478-R; Fundación Ramón Areces; RYC-2012-11025); and was co-funded by Fondo Europeo de Desarrollo Regional (FEDER). The CNIC is supported by the Spanish Ministry of Economy and Competitiveness (MINECO) and the Pro CNIC Foundation. FV-C was supported by The American Association of Immunologist through a Careers in Immunology Fellowship; and HS by a FPI-UAM Fellowship.
Footnotes
Author Contributions
V.R.P., F.S.M., M.Y.M. conceived and designed research. V.R.P., H.S., S.A., S.L.M., G.L., M.Y.M. performed experimental work. S.A., F.V.C., S.L., B.K, M.A.M.F., F.S.M., M.Y.M. provided reagents. V.R.P, M.Y.M. analysed the data. V.R.P. wrote the paper.
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.Blumenthal R, Durell S, Viard M. HIV entry and envelope glycoprotein-mediated fusion. J Biol Chem. 2012;287:40841–40849. doi: 10.1074/jbc.R112.406272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arhel N. Revisiting HIV-1 uncoating. Retrovirology. 2010;7:96. doi: 10.1186/1742-4690-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambrose Z, Aiken C. HIV-1 uncoating: connection to nuclear entry and regulation by host proteins. Virology. 2014;454–455:371–379. doi: 10.1016/j.virol.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thali M. The roles of tetraspanins in HIV-1 replication. Curr Top Microbiol Immunol. 2009;339:85–102. doi: 10.1007/978-3-642-02175-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rocha-Perugini V, Gordon-Alonso M, Sanchez-Madrid F. PIP: choreographer of actin-adaptor proteins in the HIV-1 dance. Trends Microbiol. 2014;22:379–388. doi: 10.1016/j.tim.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yanez-Mo M, et al. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 2009;19:434–446. doi: 10.1016/j.tcb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Levy S, Shoham T. The tetraspanin web modulates immune-signalling complexes. Nat Rev Immunol. 2005;5:136–148. doi: 10.1038/nri1548. [DOI] [PubMed] [Google Scholar]
- 8.van Spriel AB, Figdor CG. The role of tetraspanins in the pathogenesis of infectious diseases. Microbes Infect. 2010;12:106–112. doi: 10.1016/j.micinf.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Rocha-Perugini V, Sanchez-Madrid F, Martinez Del Hoyo G. Function and Dynamics of Tetraspanins during Antigen Recognition and Immunological Synapse Formation. Front Immunol. 2015;6:653. doi: 10.3389/fimmu.2015.00653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grigorov B, et al. A role for CD81 on the late steps of HIV-1 replication in a chronically infected T cell line. Retrovirology. 2009;6:28. doi: 10.1186/1742-4690-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ono A. Relationships between plasma membrane microdomains and HIV-1 assembly. Biol Cell. 2010;102:335–350. doi: 10.1042/BC20090165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thali M. Tetraspanin functions during HIV-1 and influenza virus replication. Biochem Soc Trans. 2011;39:529–531. doi: 10.1042/BST0390529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krementsov DN, et al. Tetraspanins regulate cell-to-cell transmission of HIV-1. Retrovirology. 2009;6:64. doi: 10.1186/1742-4690-6-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato K, et al. Modulation of human immunodeficiency virus type 1 infectivity through incorporation of tetraspanin proteins. J Virol. 2008;82:1021–1033. doi: 10.1128/JVI.01044-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weng J, et al. Formation of syncytia is repressed by tetraspanins in human immunodeficiency virus type 1-producing cells. J Virol. 2009;83:7467–7474. doi: 10.1128/JVI.00163-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida T, et al. A CD63 mutant inhibits T-cell tropic human immunodeficiency virus type 1 entry by disrupting CXCR4 trafficking to the plasma membrane. Traffic. 2008;9:540–558. doi: 10.1111/j.1600-0854.2007.00700.x. [DOI] [PubMed] [Google Scholar]
- 17.Gordon-Alonso M, et al. Tetraspanins CD9 and CD81 modulate HIV-1-induced membrane fusion. J Immunol. 2006;177:5129–5137. doi: 10.4049/jimmunol.177.8.5129. [DOI] [PubMed] [Google Scholar]
- 18.Lambele M, et al. Vpu is the main determinant for tetraspanin downregulation in HIV-1-infected cells. J Virol. 2015;89:3247–3255. doi: 10.1128/JVI.03719-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li G, et al. A post-entry role for CD63 in early HIV-1 replication. Virology. 2011;412:315–324. doi: 10.1016/j.virol.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franzolin E, et al. The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proc Natl Acad Sci U S A. 2013;110:14272–14277. doi: 10.1073/pnas.1312033110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ballana E, Este JA. SAMHD1: at the crossroads of cell proliferation, immune responses, and virus restriction. Trends Microbiol. 2015;23:680–692. doi: 10.1016/j.tim.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Baldauf HM, et al. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med. 2012;18:1682–1687. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lahouassa H, et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol. 2012;13:223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruffin N, et al. Low SAMHD1 expression following T-cell activation and proliferation renders CD4+ T cells susceptible to HIV-1. Aids. 2015;29:519–530. doi: 10.1097/QAD.0000000000000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behrendt R, et al. Mouse SAMHD1 has antiretroviral activity and suppresses a spontaneous cell-intrinsic antiviral response. Cell Rep. 2013;4:689–696. doi: 10.1016/j.celrep.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rehwinkel J, et al. SAMHD1-dependent retroviral control and escape in mice. Embo J. 2013;32:2454–2462. doi: 10.1038/emboj.2013.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cribier A, et al. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 2013;3:1036–1043. doi: 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 28.White TE, et al. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe. 2013;13:441–451. doi: 10.1016/j.chom.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan J, et al. CyclinA2-Cyclin-dependent Kinase Regulates SAMHD1 Protein Phosphohydrolase Domain. J Biol Chem. 2015;290:13279–13292. doi: 10.1074/jbc.M115.646588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coiras M, et al. IL-7 Induces SAMHD1 Phosphorylation in CD4+ T Lymphocytes, Improving Early Steps of HIV-1 Life Cycle. Cell Rep. 2016;14:2100–2107. doi: 10.1016/j.celrep.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beloglazova N, et al. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J Biol Chem. 2013;288:8101–8110. doi: 10.1074/jbc.M112.431148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi J, et al. SAMHD1 specifically restricts retroviruses through its RNase activity. Retrovirology. 2015;12:46. doi: 10.1186/s12977-015-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryoo J, et al. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat Med. 2014;20:936–941. doi: 10.1038/nm.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antonucci JM, et al. SAMHD1-mediated HIV-1 restriction in cells does not involve ribonuclease activity. Nat Med. 2016;22:1072–1074. doi: 10.1038/nm.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharya A, et al. Effects of T592 phosphomimetic mutations on tetramer stability and dNTPase activity of SAMHD1 can not explain the retroviral restriction defect. Scientific reports. 2016;6:31353. doi: 10.1038/srep31353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez-Hernandez D, et al. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J Biol Chem. 2013;288:11649–11661. doi: 10.1074/jbc.M112.445304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sala-Valdes M, et al. EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin-radixin-moesin proteins. J Biol Chem. 2006;281:19665–19675. doi: 10.1074/jbc.M602116200. [DOI] [PubMed] [Google Scholar]
- 38.Tejera E, et al. CD81 regulates cell migration through its association with Rac GTPase. Mol Biol Cell. 2013;24:261–273. doi: 10.1091/mbc.E12-09-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rocha-Perugini V, et al. CD81 controls sustained T cell activation signaling and defines the maturation stages of cognate immunological synapses. Mol Cell Biol. 2013;33:3644–3658. doi: 10.1128/MCB.00302-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welbourn S, et al. Identification and characterization of naturally occurring splice variants of SAMHD1. Retrovirology. 2012;9:86. doi: 10.1186/1742-4690-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mauney CH, et al. The SAMHD1 dNTP Triphosphohydrolase Is Controlled by a Redox Switch. Antioxid Redox Signal. 2017 doi: 10.1089/ars.2016.6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahn J. Functional organization of human SAMHD1 and mechanisms of HIV-1 restriction. Biol Chem. 2016;397:373–379. doi: 10.1515/hsz-2015-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pauls E, et al. Cell cycle control and HIV-1 susceptibility are linked by CDK6-dependent CDK2 phosphorylation of SAMHD1 in myeloid and lymphoid cells. J Immunol. 2014;193:1988–1997. doi: 10.4049/jimmunol.1400873. [DOI] [PubMed] [Google Scholar]
- 44.Welbourn S, Strebel K. Low dNTP levels are necessary but may not be sufficient for lentiviral restriction by SAMHD1. Virology. 2016;488:271–277. doi: 10.1016/j.virol.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seamon KJ, et al. SAMHD1 is a single-stranded nucleic acid binding protein with no active site-associated nuclease activity. Nucleic Acids Res. 2015;43:6486–6499. doi: 10.1093/nar/gkv633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diamond TL, et al. Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J Biol Chem. 2004;279:51545–51553. doi: 10.1074/jbc.M408573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kyei GB, Cheng X, Ramani R, Ratner L. Cyclin L2 is a critical HIV dependency factor in macrophages that controls SAMHD1 abundance. Cell Host Microbe. 2015;17:98–106. doi: 10.1016/j.chom.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morrissey C, et al. The eukaryotic elongation factor eEF1A1 interacts with SAMHD1. Biochem J. 2015;466:69–76. doi: 10.1042/BJ20140203. [DOI] [PubMed] [Google Scholar]
- 49.Yanez-Mo M, et al. Regulation of endothelial cell motility by complexes of tetraspan molecules CD81/TAPA-1 and CD151/PETA-3 with alpha3 beta1 integrin localized at endothelial lateral junctions. J Cell Biol. 1998;141:791–804. doi: 10.1083/jcb.141.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gordon-Alonso M, et al. Actin-binding protein drebrin regulates HIV-1-triggered actin polymerization and viral infection. J Biol Chem. 2013;288:28382–28397. doi: 10.1074/jbc.M113.494906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request. Complete blots for all the figures and supplementary figures are shown in Supplementary Figures 7 and 8.