Abstract
Over the last decade, microRNAs have emerged as critical regulators in the expression and function of animal genomes. This review article discusses the relationship between microRNA-mediated regulation and the biology of the fruit fly Drosophila melanogaster. We focus on the roles that microRNAs play in tissue growth, germ cell development, hormone action, and the development and activity of the central nervous system. We also discuss the ways in which microRNAs affect robustness. Many gene regulatory networks are robust; they are relatively insensitive to the precise values of reaction constants and concentrations of molecules acting within the networks. MicroRNAs involved in robustness appear to be nonessential under uniform conditions used in conventional laboratory experiments. However, the robust functions of microRNAs can be revealed when environmental or genetic variation otherwise has an impact on developmental outcomes.
Keywords: Drosophila, microRNAs, miRNAs
1. Introduction
This review explores some of the major advances in miRNA biology of Drosophila melanogaster. We focus primarily on the past five years of research, and on certain areas of Drosophila biology that are impacted by miRNAs. Due to the sheer size of the Drosophila miRNA literature, we made choices about which areas to review, in hopes that they would be of greatest interest to the reader. We apologize to our colleagues whose papers we had to omit because of the page limits of this review.
2.1 Germline Development
In the gonads, germline stem cells (GSCs) receive a signal from their niche to maintain their stem cell identity. With each GSC division, one daughter cell becomes displaced from the niche and is forced to differentiate, while the other, closer to the niche, retains its GSC fate [1]. miRNAs have been shown to play a role in regulating the balance between maintenance of stem cell fate and differentiation. The miRNAs miR-7 and miR-278 promote division of female GSCs by repressing the cyclin-dependent kinase inhibitor Dacapo [2]. The miRNA miR-184, works in the opposite direction, tipping the balance toward GSC differentiation in females [3]. It represses the Decapentaplegic (Dpp) receptor, which is found on GSCs and differentiating cells. The niche secretes Dpp, which signals to nearby GSCs to maintain their stemness. Daughter cells more distant from the Dpp source do not respond to the Dpp signal because miR-184 tunes down their responsiveness, and they accordingly differentiate.
In male GSCs, miR-7 has a different impact on GSC biology by maintaining the stemness of these cells [4]. It does so by directly repressing expression of Bag of Marbles (Bam), a protein required for germ cell differentiation [4]. The nuclear factor Maelstrom represses mir-7 transcription in daughter cells, leading to their expression of Bam and consequent development into spermatogonia. Later, when these cells switch from spermatogonia to spermocytes, Bam is down-regulated by the action of a different set of miRNAs, miR-275 and miR-306 [5].
Aging is associated with a decrease in numbers of GSCs. An age-dependent increase in let-7 miRNAs has been shown to promote this process in male flies [6]. The male niche is made up of hub cells that express Unpaired, a cytokine that activates the Jak/STAT pathway in nearby cells to promote male GSC fate. Unpaired protein levels decrease with age, resulting in age-related decline of GSC number. In hub cells, Unpaired mRNA is bound by the IGF-II mRNA-binding protein (Imp), and this interaction blocks degradation by of Unpaired mRNA by endogenous siRNAs. Imp levels decrease with age due to its direct repression by let-7 miRNAs, which show an age-dependent increase in their expression. Thus, Unpaired mRNA becomes more sensitive to degradation as hub cells age.
An important characteristic of GSCs is their resistance to genotoxic stress. In response to toxic stimuli such as irradiation, GSCs initiate a response pathway that promotes their survival. The bantam miRNA is a vital component of this process [7]. When gonadal cells are irradiated, they initiate apoptosis but not before secreting PDGF- and VEGF-related factor 1 (Pvf1). Pvf1 diffuses to nearby GSCs and activates the Tie-like receptor tyrosine kinase, resulting in GSC expression of bantam. Bantam then promotes GSC survival by inhibiting the proapoptotic gene head involution defective (hid) and preventing GSCs from undergoing apoptosis.
Somatic and germ cells also interact with one another under ordinary non-stressful conditions. Female egg chambers contain germ cells that are enveloped by a monolayer of somatic follicle cells. A pair of follicle cells secrete the cytokine Unpaired, which acts through the Jak-STAT pathway to the specify border cell fate in certain follicle cells [8]. Although Jak activity is graded across twelve follicle cells, only a small number of these differentiate into border cells. This is due to repression of STAT protein expression that is mediated by Apontic protein. It was shown that Apontic-mediated repression of STAT is dependent on Apontic activation of miR-279 expression [8]. In cells destined to become follicle cells, Apontic induces the expression of miR-279, which targets STAT for miRNA-mediated repression.
2.2 Maternal to Zygotic Transition
Genetic control of embryonic development is initially managed by maternal RNAs and proteins that are deposited into the egg. There is a time in which this control shifts to the embryo’s own genome, called the maternal to zygotic transition (MZT) [9]. During the MZT, thousands of maternal RNAs are degraded and thousands of newly synthesized RNAs appear. The miR-309 cluster of eight miRNAs are among these early zygotic RNAs that appear, and these miRNAs target the clearance of many maternally loaded RNAs [10]. Transcription of the miR-309 cluster is induced by the pioneer transcription factor Zelda [11], which plays a key role in zygotic genome activation. Zelda also induces early zygotic transcription of ten other miRNA clusters [12]. While there is no evidence that the other miRNA clusters also clear maternal RNAs, they do program many of the early patterning events of the fly embryo.
Although some miRNAs are synthesized during the MZT, maternally deposited miRNAs are degraded. A key trigger for their degradation is 3′ end modification of these miRNAs, which occurs in newly fertilized eggs [13]. The poly(A) polymerase Wispy associates with Argonaute-1 (Ago1) and adds short poly-Adenine tails to mature miRNAs, causing them to become unstable and turnover. Wispy activity dissipates before the MZT, ensuring that zygotic miRNAs are not destabilized.
While the functions of many maternal miRNAs are not known, the miR-2 family plays an important role in anterior-posterior pattern formation. Maternal mRNAs encoded by the caudal gene are uniformly distributed in early embryos, but translational repression creates a gradient of Caudal protein from low (anterior) to high (posterior) [14]. Although the morphogen protein Bicoid was known to generate the Caudal gradient [15], what was unappreciated was the key role played by miR-2 miRNAs [16]. Their binding site in the caudal 3′UTR overlaps with the Bicoid binding site, and is essential for Bicoid-induced repression. Although the precise mechanism is not known, miR-2 might synergize Bicoid binding or mediate the repression of translation induced by Bicoid binding.
2.3 Tissue Growth
A relationship between miRNAs and tissue growth has been known since the discovery of the bantam miRNA in 2003 [17]. This miRNA promotes the growth of imaginal tissues by both stimulating cell proliferation and antagonizing cell apoptosis. In cells that are being developmentally programmed to undergo either cell-cycle arrest or apoptosis, bantam miRNA levels are down-regulated [17, 18]. The mechanisms behind this regulation have been elucidated by focusing on key signal transduction pathways (Figure 1).
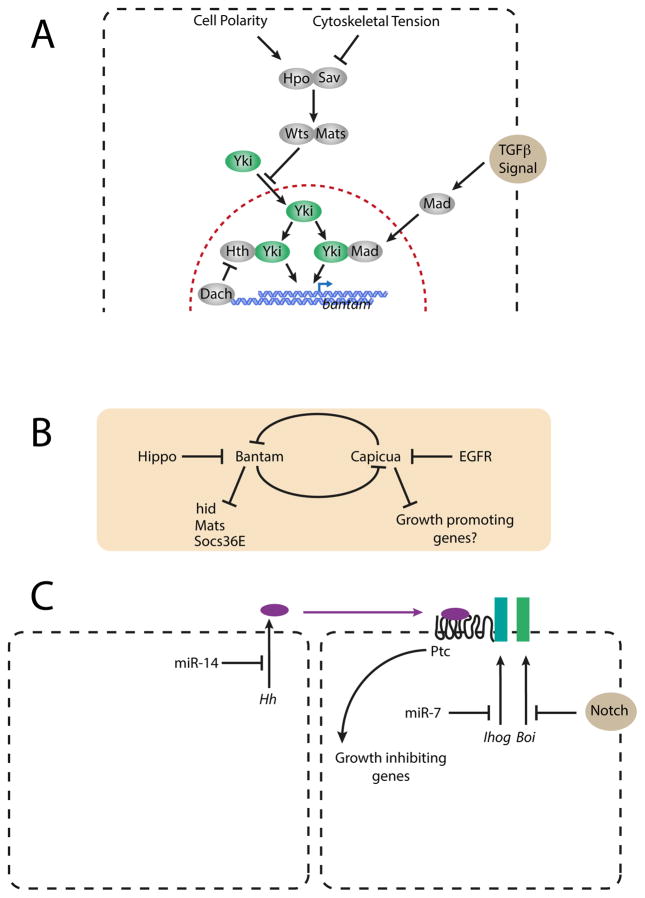
Figure 1. Control of tissue growth by miRNAs.
(A) Regulated bantam expression by the Hippo and TGF pathways. Signals are transduced via the Hippo (Hpo) and Wts kinases, aided by cofactors Sav and Mats, respectively. This effectively blocks Yorkie (Yki) from entering the nucleus and activating bantam transcription. Transcription also requires either Homothorax (Hth) or Mad. Dachshund (Dach) blocks this in the eye. (B) Cross-talk between the Hippo and EGFR pathways mediated by bantam and Capicua. (C) Regulation of growth through the Hedgehog (Hh) pathway is mediated by several miRNAs. Ihog and Boi enable productive interaction between Hh and its receptor Patched (Ptc).
The Hippo signaling pathway senses mechanical stress, among other cues, and it transduces these signals via a kinase cascade to repress a transcriptional co-activator called Yorkie. In the absence of signaling, Yorkie activates the transcription of genes that promote tissue growth including the bantam gene [18, 19]. Bantam is essential for Yorkie-induced overproliferation and is sufficient to rescue survival and proliferation of yorkie mutant cells. Yorkie interacts with the transcription factors Mad and Scalloped to promote bantam transcription in the wing imaginal disc [20, 21], and it interacts with the Homothorax transcription factor to activate bantam transcription in proliferating cells of the eye [21, 22].
Bantam transcription by Yorkie can be modulated in at least three ways (Figure 1A). Hippo signaling can cause retention of Yorkie in the cytoplasm, blocking Yorkie’s ability to activate bantam [18]. Second, TGFβ signaling can cause the nuclear translocation of Mad, which activates bantam so long as Yorkie is present [20]. Third, programmed expression of transcription factors can either enable or block Yorkie activity. In the lateral wing, the Bifid transcription factor is essential for Yorkie/Mad to up-regulate bantam [23]. In the eye, the Dachshund transcription factor prevents Yorkie/Homothorax from activating bantam transcription [24]. This occurs when eye cells undergo a cell-cycle arrest before they differentiate.
Other signaling pathways also regulate bantam expression (Figure 1B). Signals mediated by the epidermal growth factor receptor (EGFR) are transduced within wing disc cells to activate bantam transcription [25]. This is executed via regulation of the transcription factor Capicua, which represses bantam. When EGFR signaling occurs, Capicua protein is rendered unstable, providing a de-repressive effect on bantam.
Bantam executes its effects on tissue growth via multiple gene targets. One regulatory target is the pro-apoptotic gene hid, which provides a rationale for how bantam inhibits apoptosis [17]. Another direct target is the anti-proliferation gene Suppressor of cytokine signaling at 36E (Socs36E) [26]. Bantam represses Socs36E expression, and this effect is part of EGFR’s pro-growth role in imaginal discs. Bantam also affects the local organization of cells within the wing imaginal disc. It directly represses the expression of the Enabled protein, which is a regulator of cortical actin filament elongation [27]. Cells at the boundary between dorsal and ventral compartments of the wing experience lower levels of bantam due to local Notch signals. Hence, Enabled protein abundance is higher in these cells, and this correlates with their abundant actin-myosin cables. It is thought that the increased membrane tension of boundary cells is important for delimitation of each compartment.
Certain bantam targets feedback to regulate bantam expression (Figure 1B). One of these targets is the capicua gene, which makes a repressor of bantam transcription [25]. This double-negative feedback loop provides a regulatory link between the Hippo and EGFR pathways. Hippo signaling down-regulates bantam, which leads to greater Capicua and consequently a greater barrier for EGFR signaling to overcome [25]. This ensures that the impact of EGFR signaling is dampened when Hippo signaling is present. Conversely, bantam indirectly stimulates the level of Mob as tumor suppressor (Mats) protein, a component of the Hippo pathway [28]. This mechanism might provide negative feedback to bantam when Hippo signaling is operational.
Bantam is not the only growth regulatory miRNA. miR-7 is required for normal wing growth, and its loss results in smaller cells and a disrupted G1-S transition [29]. Conversely, overexpressing miR-7 in the eye causes overproliferation when Notch signaling is also hyperactive [30]. The mechanism of this effect converges on Hedgehog signal transduction [30]. The Interference hedgehog (Ihog) Hedgehog co-receptor is targeted by miR-7 while the paralogous Brother of hog (Boi) Hedgehog co-receptor is repressed by Notch (Figure 1C). Knockdown of both co-receptors leads to diminished Hedgehog signaling and strong tissue overgrowth. Interestingly, miR-932 also represses Boi protein expression and Hedgehog signaling [31], though it is unclear whether it interacts with miR-7 in promoting growth. Yet another growth-promoting miRNA is miR-14 (Figure 1C). miR-14 targets production of the Hedgehog ligand in wing cells, leading to normal tissue growth [32]. The miRNA miR-8 has more complex effects on growth. Overexpressing miR-8 potentiates the growth-promoting effects of EGFR in the wing [33]. It does so by repressing expression of the Peanut septin, which causes cells to become polyploid and induce apoptosis and engulfment of their neighbors. Conversely, miR-8 is a growth-inhibiting miRNA in the context of Notch signaling. Overexpressing miR-8 in the eye suppresses the overproliferation effects of Notch signaling [34]. miR-8 inhibits growth by blocking cell proliferation via the Notch ligand Serrate. Serrate is a direct target of miR-8 regulation.
miRNAs also regulate cell apoptosis during tissue growth. In the salivary gland, miR-14 targets production of Inositol 1,4,5-triphosphate kinase 1 (IP3K1), which enables cell autophagy and apoptosis of cells during the pupal process of gland shrinkage [35]. In the wing disc, miR-9a represses expression of the beadex (Bx) gene, which encodes a transcription factor [36, 37]. Maintained tuning of Bx protein levels suppresses apoptosis and allows for balanced growth of both dorsal and ventral wing compartments. The miR-11/miR-998 cluster is also involved in blocking apoptosis [38–40]. Both miRNAs are embedded within an intron of the E2F transcription factor 1 gene, and both miRNAs limit E2f1-dependent apoptosis. While miR-11 represses components of the core apoptotic machinery (Hid and Reaper), miR-998 elevates prosurvival signaling downstream of EGFR through inhibition of Cbl, a negative regulator of EGFR signaling.
2.4 Endocrinology
The neuroendocrine control center of Drosophila is located in the brain, in a region known as the pars intercerebralis. Here, insulin-like peptides (ILPs) that regulate body growth and metabolism are produced [41]. ILPs are produced and secreted from a cluster of fourteen cells known as the insulin producing cells (IPCs), which are regulated by various inputs such as nutrient availability and certain neuropeptides. One such neuropeptide, short Neuropeptide F (sNPF) modulates IPC function by binding its receptor, sNPFR on the IPC cell surface. This results in activation of ERK-mediated signaling and stimulation of ILP production to promote body growth.
Regulation of ILP production by sNPF is mediated by the miRNA miR-9a [42]. miR-9a modulates body size through its repression of sNPFR1 levels in IPCs. miR-9a-mediated sNPFR1 repression results in a decrease in ILP production and a concomitant decrease in body size (Fig. 2). Interestingly, this interaction is conserved in mammalian insulin endocrinology. The sNPF ortholog NPY modulates insulin production in the β-islet cells of mammals. The miR-9 ortholog represses NPY receptor NPY2R expression in a rat insulinoma cell line [42].
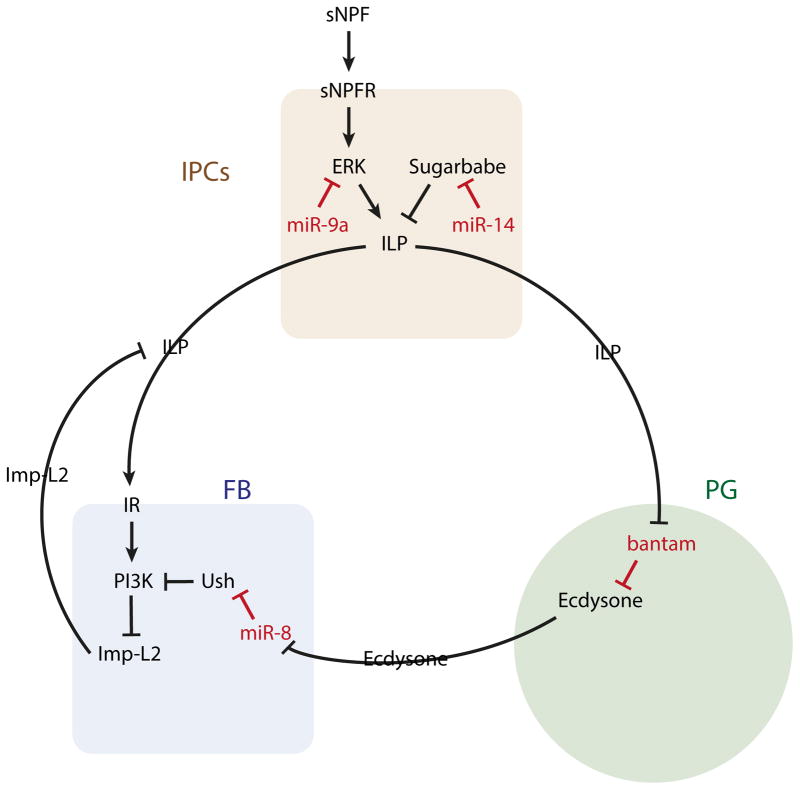
Figure 2. Cross-talk via miRNAs between various growth hormone pathways.
Feedback occurs between insulin producing cells (IPCs), the prothoracic gand (PG), and the fat body (FB). ILPs, ecdysone, and Imp-L2 mediate these interactions. Within each secretory gland, miRNAs directly target various genes including Sugarbabe and U-shaped (Ush). The identities of other direct targets is less understood.
ILP production is also under the control of miR-14 [43]. miR-14 mutants have decreased levels of ILPs, and mutants show elevated triglyceride stores as a consequence. Additionally, miR-14 mutants have increased sensitivity to starvation as a result of defective mobilization of energy stores, since a certain level of ILP production may be necessary for fat mobilization. The transcription factor Sugarbabe is directly targeted by miR-14, and Sugarbabe otherwise inhibits ILP transcription in IPCs (Fig. 2).
Another important hormone in Drosophila is the molting hormone ecdysone, which negatively regulates body growth. ILPs stimulate production of ecdysone in the prothoracic gland (PG) of the brain. They do so by inhibiting a miRNA in the PG (Fig. 2). The bantam miRNA promotes systemic growth through its inhibition of ecdysone production [44]. However, ILPs inhibit bantam expression in the PG, thus bantam mediates ILP-dependent expression of ecdysone.
Ecdysone not only triggers molting, but it also inhibits body growth. It does so by its action on the fat body [45]. The fat body is the insect liver, and in addition to metabolism, the fat body also regulates body growth. Fat body miR-8 has been shown to serve as a key link between ecdysone and body growth [46]. Without miR-8 expression in the fat body, ecdysone is unable to repress body growth. miR-8 directly represses expression of U-shaped, which is an inhibitor of PI3K [47]. Since PI3K is a key transducer of ILP signals in fat body cells, indirect upregulation of PI3K by miR-8 potentiates the response of the fat body to ILPs (Fig. 2). However, ecdysone represses the expression of miR-8 in the fat body, thereby antagonizing ILP-induced signal transduction [46].
The fat body is itself a site of hormone production. Production of several of these factors is down-regulated by miR-8 [48]. Imp-L2 is one of these factors, and it is indirectly down-regulated by miR-8 through miR-8’s action on U-shaped (Fig. 2). Imp-L2 protein is secreted by the fat body under starvation conditions, and it binds to and inhibits humoral ILPs [49]. Since Imp-L2 expression is induced by ecdysone [50], it is likely that ecdysone regulation of miR-8 in the fat body is one means by which ecdysone stimulates Imp-L2. In turn, this would antagonize ILP-induced body growth. However, this mechanism is not sufficient to account for the effect of miR-8 and ecdysone on body growth [48].
Ecdysone also antagonizes juvenile hormone (JH). Pulses of ecdysone trigger metamorphosis, whereas JH acts in the opposing direction to repress metamorphosis. The miR-2 family of miRNAs are involved in this process, with loss of miR-2 resulting in impaired induction of metamorphosis [51]. miR-2 acts by repressing the transcription factor Kruppel homolog-1, which functions downstream of JH. By rapidly clearing Kruppel homolog-1 mRNA in the last larval instar, miR-2 miRNAs ensure that the transition to metamorphosis is able to progress.
2.5 Growth of the Central Nervous System (CNS)
The adult brain attains its correct size and morphology as a result of precisely tuned formation of neural stem cells (neuroblasts), and their survival, proliferation and differentiation. MiRNAs tune key molecular players involved in all these processes by directly acting in neuroblast pools. In central brain neuroblasts, the bantam miRNA targets the pro-differentiation gene prospero to block premature cell cycle exit and differentiation [52]. Anachronism is a secreted growth inhibitor, which is expressed in neuroblasts. miR-124 synthesized in central brain neuroblasts, represses anachronism expression to aid cell proliferation [53]. Similarly, miR-92a/b expression in the optic lobe correlates cell autonomously with the inhibition of ectopic differentiation [54].
MiRNAs can also act as dynamic remodelers of stem cell niches by controlling glial cells. Glia are support cells, which carry out basic metabolic clearing functions. They also serve as anatomical niches by providing developmental cues to surrounding neuroblasts. This has been demonstrated by multiple lines of evidence in the optic lobes of the brain. Each optic lobe is derived from proliferating neuroepithelial cells, which undergo a transition to neuroblasts. This proliferation and transition is controlled by spatio-temporally regulated glial cell cues. The EGFR ligand Spitz is secreted by a subpopulation of glial cells that co-express miR-8. miR-8 directly targets the Spitz 3′UTR thereby regulating the rate of proliferation and the rate of transit of cells to a neuroblast lineage [55]. Moreover, miR-8 remodels glial cell architecture, possibly by regulating endo-replication. Glial loss of miR-8 leads to reduced cell size and reduced sprouting towards the neuroepithlium [55]. These glia also express bantam, which downregulates the gene Bifid. Bantam activation also upregulates myc expression, a known cell growth promoter [56]. Loss of bantam in glia affects glial cell numbers and distribution, and therefore negatively affects photoreceptor axon projection patterns which depend on glial cues [57]. Thus, miRNAs promote glial cell survival, architecture and physiology to dynamically regulate neural patterning.
2.6 Cell Differentiation in the CNS
The remarkable complexity of the brain is in part due to the diverse array of specialized neural subtypes that arise during the course of differentiation. A specific neural progenitor cell might transition into many different terminal cell fates. Maintaining the correct ratios of these fates is therefore crucial for function. One strategy for enforcing specific cell fate ratios is to carefully time the ordered production of neuronal subtypes. The let-7 miRNA transduces developmental timing to control these cell-type transitions [58]. It couples the systemic hormone ecdysone to neuronal differentiation in the brain’s mushroom bodies (MBs). The MB is the center for olfactory learning and memory. Multi-potent progenitor cells give rise to three different classes of MB neurons at different times during development – γ neurons in the larval stage, α′/β′ neurons in the prepupal stage, and α/β in the pupal stage. The larval to pupal transition corresponds to an ecdysone-mediated activation of the let-7-C gene [58]. The let-7-C gene cotranscribes two miRNAs: let-7 and miR-125. Both miRNAs target the cell fate regulator chinmo, causing its diminished abundance over time. However, a loss of let-7-C results in extended chinmo expression and a pre-ponderance of early-born fates (γ, α′/β′). In contrast, overexpression of let-7-C miRNAs leads to greater number of late-born fates (α/β) [58]. Incorrect subtype ratios are not a consequence of excessive growth of one population or apoptosis of another, but bona fide events of subtype fate switching.
2.7 Morphogenesis of the CNS
In the MB, let-7 also represses expression of the transcription factor Abrupt [59]. In turn, Abrupt controls the expression of a cell adhesion molecule Fasciclin 2, which regulates axon pathfinding and helps to cluster self-similar cells during morphogenesis. Predictably, loss of let-7 impairs proper morphogenesis of the MB, leading to defective olfactory processing [59]. miR-iab4/iab8 is involved in adult ovary innervation, and mutants display defective innervation and morphology of oviduct motorneurons [60]. This is presumably due to deprepression of target Hox genes in the posterior ventral nerve cord, resulting in female sterility.
Another aspect of neuronal morphogenesis is the extension of dendrites toward a specific receptor field and their arborization to cover the field. Dendrites find their cognate receptor fields by recognizing spatial cues in the form of special extra-cellular matrix (ECM) components and cell adhesion molecules (CAMs) generated by epithelial cells. MiRNAs act to fine tune receptor field properties, which are recognized by surrounding neurons. Loss of the bantam miRNA in epithelial cells leads to upregulated Akt1 kinase in adjacent neurons and hyperactive dendritic growth [61]. Activating Akt1 also stimulates dendritic regeneration of the same class of neurons [62]. Bantam likely exerts this affect on arborization by regulating the production of CAMs and ECM components generated by the epithelial cells. Bantam promotes epithelial endo-replication, which in turn leads to progressive changes in the ECM by promoting the expression of Myospheroid, a β-integrin [63]. Thus, bantam expression marks the boundaries of the permissive substrate onto which dendrite-epithelium contacts can be made.
After covering a receptor field, neurons must compensate for organismal growth or morphogenetic changes by adapting to their changing receptor fields over time. This phenomenon is called scaling growth of dendrites. Scaling growth requires bantam in epithelial body wall cells. Scaling also depends on miR-9a, which remodels cellular contacts in neurons by down-regulating the CAM protein Starry night [64]. Like bantam, miR-9a expression is restricted to epithelial cells and therefore attenuates dendritic scaling non-autonomously.
2.8 Synaptogenesis
The complexity of the brain arises not only from the multiple cell types it harbors but even more so from the connectivity between them. Thus, control of neuronal wiring specificity must be carefully regulated. The larval neuro-muscular junction (NMJ) affords an experimentally amenable system to study synapse formation. Here too, we find similar themes of miRNA controlled ECM and CAM modulation. miR-8 functions in post-synaptic muscle cells to repress the actin regulatory protein Enabled, which modifies the sub-synaptic reticulum during late stages of neuro-muscular synapse development [65, 66]. This maintains the expansion of a synapse as the larval muscles grow. Apart from a late post-synaptic role, miR-8 also has an early role in muscle innervation by motor neurons. miR-8 is required for the correct expression of synaptic CAMs Fasciclin 3 and Neuroglian. Both the pre-synaptic and post-synaptic termini express Fasciclin 3 and Neuroglian under miR-8 regulation to mediate neuron-muscle contact [67].
2.9 Behavior
Apart from spatio-temporal regulation of neuronal development and morphogenesis, miRNAs have also been shown to regulate CNS activity. This is apparent in the regulation of the circadian patterns of rest and activity that are set by clock or pacemaker neurons in the CNS. Pacemaker neurons transduce external environmental cues such as light and temperature into internal molecular oscillations of circadian gene products. These signals are then transmitted to other parts of the brain to produce pertinent animal behavior patterns. In clock neurons, ecdysone controls let-7-C miRNAs to repress the circadian gene clockwork orange [68]. Thus, gain of let-7C activity in pacemaker neurons lengthens the circadian period whereas loss of let-7C attenuates molecular oscillations. On the other hand, miR-279 acts downstream of the central clock to transduce the signal from pacemaker neurons, possibly via the Jak/STAT pathway. The Jak/STAT ligand Unpaired is a miR-279 target in the brain and it mediates the attenuation of circadian rhythms when miR-279 is mis-regulated [69].
MiRNAs have recently been implicated in modulating memory and behavior. The miR-iab4/iab8 locus, a known repressor of the Hox gene Ultrabithorax, controls self-righting behavior in larvae [70]. iab4/iab8 mutant animals take longer to correct their orientation when turned upside down. This was traced to derepression of Ultrabithorax in the two neurons composing the self righting node and abberrant neural activity patterns. More elusive behavioral phenotypes such as responses to naïve or conditioned stimuli invoke the role of memory formation and recall. There is a specific requirement for miRNAs miR-31a and miR-974 in cholinergic neurons and olfactory receptor neurons/MB-V2 neurons, respectively [71]. Other evidence comes from the study of miR-276 in Ellipsoid Body (EB) and MB neurons. In both cases, miR-276 is required to tune the levels of Dopamine receptor (DopR) to mediate an appropriate response to external stimuli [72]. The miR-276::DopR pairing in two distinct behavioural circuits involve dopaminergic neurons – naïve responses to odors in the EB, and conditioned memory responses in the MB.
MiRNAs also serve neuroprotective roles in the brain. For instance, miR-1000 controls post-synaptic glutamate excitotoxicity by down regulating vesicular glutamate transporter (VGlut) in the pre-synaptic terminal [73]. Loss of miR-1000 leads to elevated cell death as a result of excitotoxicity. miR-1000 tunes the repression of Vglut in an activity dependent manner, providing a dynamic mechanism to allow neuronal activity to be coupled to synaptic strength [73]. miR-34 promotes long-term brain integrity by preventing age associated neurodegeneration [74]. Strikingly, over-expression of miR-34 increases lifespan and counteracts polyglutamine mediated neurodegeneration. The developmental gene Ecdysone-induced protein 74EF is a key target of miR-34 in this process [74].
3. Robustness
Biological processes frequently exhibit a property known as robustness [75]. Robust processes occur reproducibly and uniformly even in the face of variability induced by the environment, genetic variation, or random chance. Certain biological processes are quite variable, and hence, do not require much robustness. However, other processes, particularly irreversible ones such as differentiation, are strongly robust to ensure a minimal impact of error [76].
A feature of many evolutionarily conserved miRNAs is their lack of strong phenotypic consequences when individually mutated. This has been attributed to the weak repression of target gene expression elicited by most miRNAs. Weak and tunable repression has led to the proposition that miRNAs generally elicit two distinct effects on their targets [77]. In the first type of effect, a miRNA reduces the level of target below a threshold that might act like a switch; in the second type of effect, a miRNA buffers fluctuations in the target, limiting undesired signal propagation. Each of these effects can potentially be harnessed to provide robustness to target gene regulation. Is there evidence that miRNAs potentiate robustness?
At a phenotypic level, loss of miRNAs can diminish the uniformity of developmental outcomes. Successful embryogenesis is resistant to temperature variation between 18° and 29°C, but when miR-9a is lost, embryogenesis fails at elevated temperature, concommitant with a disruption in myotendon formation [78]. In a wildtype embryo, there are 34 primordial germ cells (PGCs) on average, and there is low variation between embryos (coefficient of variation approximately 12%). If either miR-9c or miR-969 are absent, PGC variation increases 2- to 3-fold while the average number decreases slightly [79]. Other mutants that decrease average PGC number have no effect on variation.
A deeper analysis of the question has come from study of Drosophila sensory organ precursors (SOPs). SOPs are specified at precise locations throughout the developing adult body, where each SOP individually forms an independent sensory organ. Because of the discrete location of sensory organs, their number is highly reproducible between adult individuals. This uniformity breaks down when certain miRNAs are missing. mir-9a mutant adults bear a slightly excessive number of sensory organs [80]. However, variation in sensory organ numbers between mir-9a mutant individuals increases several fold compared to wildtype [81]. Upregulation of the miR-9a target gene senseless, a proneural factor, is primarily responsible for this enhanced variability. It suggests that a function for miR-9a is to ensure developmental robustness during SOP specification. Evidence for this interpretation has come from studies estimating how well mir-9a mutants are able to mitigate the effects of environmental and genetic variation on sensory organ numbers (Figure 2). Temperature variation has a greater effect on sensory organ numbers when either mir-9a is mutated or the miR-9a binding sites in the senseless mRNA are mutated [81]. Likewise, the natural genetic variation that favors greater numbers of sensory organs is also unleashed when either miR-9a or its binding to senseless mRNA are impaired [81]. However, natural genetic variation that favors smaller numbers of sensory organs is unaffected by miR-9a [82]. Thus, developmental robustness provided by miR-9a is differential towards some but not all types of genetic perturbation.
Another example of differential robustness is found with miR-7. Although miR-7 is not required under normal conditions for SOP development, when mir-7 mutants are subjected to modest temperature fluctuation, they exhibit impaired SOP patterning [83]. Several antineural E(spl) complex genes are direct targets of miR-7 [83, 84], accounting for the apparent proneural function of miR-7, but seen only under certain environmentally challenging conditions.
miR-7 and miR-9a appear to generate thresholds for the switch-like behavior of SOP development. SOP specification is guided by lateral inhibition between cells via Notch signaling, where a cell exerts positive auto-feedback by inhibiting its neighbors [85]. Positive feedback allows for a sharp switch-like response, but it makes the system very sensitive to fluctuations in signal processes. miR-9a repression is coupled to the positive auto-feedback loop by its effect on senseless, thereby making the system more stable. This is because lateral inhibition must first overcome the action of miR-9a before it can trigger the positive feedback loop. In contrast, miR-7 ensures that feedback occurs robustly when it needs to. There is negative feedback between the proneural and antineural transcription factors, which helps stabilize their levels but it comes at a cost; it is harder to activate the positive feedback loop. However, miR-7 neutralizes the negative feedback above a threshold of sustained proneural transcription factor activity, allowing auto-feedback to occur.
Another example of a miRNA regulating switch-like feedback is found in abdominal histoblasts. These cells are dormant until pupation, when ecdysone hormone triggers their proliferation and morphogenesis into the adult abdomen. In dormant histoblasts, miR-965 represses the expression of string, which encodes a cell-cycle phosphatase necessary for the G2/M transition [86]. miR-965 also represses the expression of the ecdysone receptor (EcR). At pupariation, ecdysone reduces the expression of miR-965, which leads to de-repression of EcR and even stronger ecdysone signal transduction. This mutual repression circuit can therefore contribute positive auto-feedback to EcR and a switch-like behavior. However, the delay between transcription and decay of miR-965 RNA means that only sustained changes in EcR activity will be sufficient to trigger the feedback loop.
A different kind of robustness is demonstrated by miR-263a/b in the eye [87]. Progenitor cells are overproduced in the eye, and excess cells are eliminated by apoptosis after all differentiated cells are specified. Differentiated cells are protected during the stochastic pruning process. However, in the absence of miR-263a/b, sensory organs are lost in a stochastic manner due to these cells up-regulating the miR-263a/b target gene hid.
4. Conclusions
Clearly, miRNAs play large and diverse roles in the biology of Drosophila melanogaster, ranging from development and physiology to behavior. Future challenges will be to integrate these effects into the larger biochemical networks operating to control such phenomena, and to understand what, if any, special roles that miRNAs might play in largescale networks.
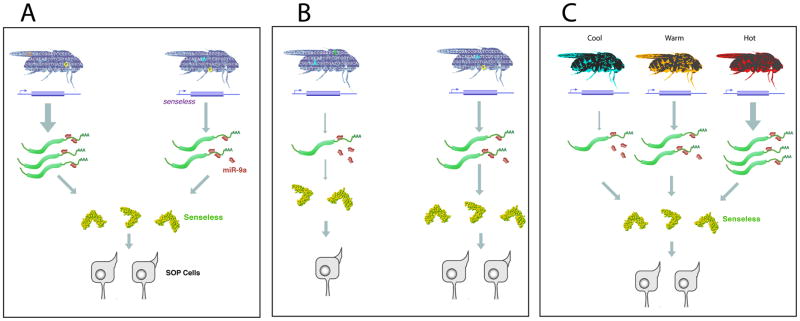
Figure 3. Control of developmental variation by miRNA miR-9a.
(A) Natural genetic variation in a population of flies can lead to above-average transcription of the senseless gene. Nevertheless, Senseless protein output is rendered uniform by miR-9a repression. This results in less variation in bristle number. (B) Natural genetic variation can also lead to below-average senseless transcription. miR-9a is unable to render Senseless protein output to a normal level, and flies can exhibit a below-average bristle number. (C) Raising flies at different temperatures has little effect on bristle number phenotypes because miR-9a represses Senseless protein output.
Acknowledgments
The following funding sources are acknowledged: Malkin Scholarship Fund (R.G.), the Cell and Molecular Basis of Disease NIGMS Training Grant (P.A.), and R01GM077581 (R.W.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gilboa L, Lehmann R. How different is Venus from Mars? The genetics of germ-line stem cells in Drosophila females and males. Development. 2004;131:4895–905. doi: 10.1242/dev.01373. [DOI] [PubMed] [Google Scholar]
- 2.Yu J-Y, Reynolds SH, Hatfield SD, Shcherbata HR, Fischer KA, Ward EJ, et al. Dicer-1-dependent Dacapo suppression acts downstream of Insulin receptor in regulating cell division of Drosophila germline stem cells. Development (Cambridge, England) 2009;136:1497–507. doi: 10.1242/dev.025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iovino N, Pane A, Gaul U. miR-184 has multiple roles in Drosophila female germline development. Developmental cell. 2009;17:123–33. doi: 10.1016/j.devcel.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 4.Pek JW, Lim AK, Kai T. Drosophila maelstrom ensures proper germline stem cell lineage differentiation by repressing microRNA-7. Developmental cell. 2009;17:417–24. doi: 10.1016/j.devcel.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 5.Eun SH, Stoiber PM, Wright HJ, McMurdie KE, Choi CH, Gan Q, et al. MicroRNAs downregulate Bag of marbles to ensure proper terminal differentiation in the Drosophila male germline. Development (Cambridge, England) 2013;140:23–30. doi: 10.1242/dev.086397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toledano H, D’Alterio C, Czech B, Levine E, Jones DL. The let-7-Imp axis regulates ageing of the Drosophila testis stem-cell niche. Nature. 2012;485:605–10. doi: 10.1038/nature11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xing Y, Su TT, Ruohola-Baker H. Tie-mediated signal from apoptotic cells protects stem cells in Drosophila melanogaster. Nature communications. 2015;6:7058. doi: 10.1038/ncomms8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon WH, Meinhardt H, Montell DJ. miRNA-mediated feedback inhibition of JAK/STAT morphogen signalling establishes a cell fate threshold. Nature cell biology. 2011;13:1062–9. doi: 10.1038/ncb2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–42. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- 10.Bushati N, Stark A, Brennecke J, Cohen SM. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol. 2008;18:501–6. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- 11.Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–3. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu S, Nien C-Y, Liang H-L, Rushlow C. Co-activation of microRNAs by Zelda is essential for early Drosophila development. Development (Cambridge, England) 2014;141:2108–18. doi: 10.1242/dev.108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee M, Choi Y, Kim K, Jin H, Lim J, Nguyen TA, et al. Adenylation of maternally inherited microRNAs by Wispy. Molecular cell. 2014;56:696–707. doi: 10.1016/j.molcel.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubnau J, Struhl G. RNA recognition and translational regulation by a homeodomain protein. Nature. 1996;379:694–9. doi: 10.1038/379694a0. [DOI] [PubMed] [Google Scholar]
- 15.Cho PF, Poulin F, Cho-Park YA, Cho-Park IB, Chicoine JD, Lasko P, et al. A new paradigm for translational control: inhibition via 5′-3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell. 2005;121:411–23. doi: 10.1016/j.cell.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 16.Rodel CJ, Gilles AF, Averof M. MicroRNAs act as cofactors in bicoid-mediated translational repression. Current biology: CB. 2013;23:1579–84. doi: 10.1016/j.cub.2013.06.041. [DOI] [PubMed] [Google Scholar]
- 17.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 18.Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–74. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 19.Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol. 2006;16:1895–904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- 20.Oh H, Irvine KD. Cooperative regulation of growth by Yorkie and Mad through bantam. Dev Cell. 2011;20:109–22. doi: 10.1016/j.devcel.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slattery M, Voutev R, Ma L, Negre N, White KP, Mann RS. Divergent transcriptional regulatory logic at the intersection of tissue growth and developmental patterning. PLoS Genet. 2013;9:e1003753. doi: 10.1371/journal.pgen.1003753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng HW, Slattery M, Mann RS. Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 2009;23:2307–19. doi: 10.1101/gad.1820009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Luo D, Pflugfelder GO, Shen J. Dpp signaling inhibits proliferation in the Drosophila wing by Omb-dependent regional control of bantam. Development (Cambridge, England) 2013;140:2917–22. doi: 10.1242/dev.094300. [DOI] [PubMed] [Google Scholar]
- 24.Bras-Pereira C, Casares F, Janody F. The retinal determination gene Dachshund restricts cell proliferation by limiting the activity of the Homothorax-Yorkie complex. Development. 2015;142:1470–9. doi: 10.1242/dev.113340. [DOI] [PubMed] [Google Scholar]
- 25.Herranz H, Hong X, Cohen SM. Mutual repression by bantam miRNA and Capicua links the EGFR/MAPK and Hippo pathways in growth control. Current biology: CB. 2012;22:651–7. doi: 10.1016/j.cub.2012.02.050. [DOI] [PubMed] [Google Scholar]
- 26.Herranz H, Hong X, Hung NT, Voorhoeve PM, Cohen SM. Oncogenic cooperation between SOCS family proteins and EGFR identified using a Drosophila epithelial transformation model. Genes & development. 2012;26:1602–11. doi: 10.1101/gad.192021.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becam I, Rafel N, Hong X, Cohen SM, Milan M. Notch-mediated repression of bantam miRNA contributes to boundary formation in the Drosophila wing. Development (Cambridge, England) 2011;138:3781–9. doi: 10.1242/dev.064774. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Lai Z-C. Mob as tumor suppressor is regulated by bantam microRNA through a feedback loop for tissue growth control. Biochemical and biophysical research communications. 2013;439:438–42. doi: 10.1016/j.bbrc.2013.08.095. [DOI] [PubMed] [Google Scholar]
- 29.Aparicio R, Simoes Da Silva CJ, Busturia A. MicroRNA miR-7 contributes to the control of Drosophila wing growth. Developmental dynamics: an official publication of the American Association of Anatomists. 2015;244:21–30. doi: 10.1002/dvdy.24214. [DOI] [PubMed] [Google Scholar]
- 30.Da Ros VG, Gutierrez-Perez I, Ferres-Marco D, Dominguez M. Dampening the signals transduced through hedgehog via microRNA miR-7 facilitates notch-induced tumourigenesis. PLOS Biology. 2013;11:e1001554. doi: 10.1371/journal.pbio.1001554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao L, Wu L, Hou X, Zhang Q, Zhang F, Ye X, et al. Drosophila miR-932 modulates hedgehog signaling by targeting its co-receptor Brother of ihog. Developmental biology. 2013;377:166–76. doi: 10.1016/j.ydbio.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Kim K, Vinayagam A, Perrimon N. A rapid genome-wide microRNA screen identifies miR-14 as a modulator of Hedgehog signaling. Cell reports. 2014;7:2066–77. doi: 10.1016/j.celrep.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eichenlaub T, Cohen SM, Herranz H. Cell Competition Drives the Formation of Metastatic Tumors in a Drosophila Model of Epithelial Tumor Formation. Curr Biol. 2016;26:419–27. doi: 10.1016/j.cub.2015.12.042. [DOI] [PubMed] [Google Scholar]
- 34.Vallejo DM, Caparros E, Dominguez M. Targeting Notch signalling by the conserved miR-8/200 microRNA family in development and cancer cells. The EMBO journal. 2011;30:756–69. doi: 10.1038/emboj.2010.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson C, Ambros V, Baehrecke EH. miR-14 regulates autophagy during developmental cell death by targeting ip3-kinase 2. Molecular cell. 2014;56:376–88. doi: 10.1016/j.molcel.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biryukova I, Asmar J, Abdesselem H, Heitzler P. Drosophila mir-9a regulates wing development via fine-tuning expression of the LIM only factor, dLMO. Developmental biology. 2009;327:487–96. doi: 10.1016/j.ydbio.2008.12.036. [DOI] [PubMed] [Google Scholar]
- 37.Bejarano F, Smibert P, Lai EC. miR-9a prevents apoptosis during wing development by repressing Drosophila LIM-only. Developmental biology. 2010;338:63–73. doi: 10.1016/j.ydbio.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Truscott M, Islam ABMMK, Lopez-Bigas N, Frolov MV. mir-11 limits the proapoptotic function of its host gene, dE2f1. Genes & development. 2011;25:1820–34. doi: 10.1101/gad.16947411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ge W, Chen Y-W, Weng R, Lim SF, Buescher M, Zhang R, et al. Overlapping functions of microRNAs in control of apoptosis during Drosophila embryogenesis. Cell death and differentiation. 2012;19:839–46. doi: 10.1038/cdd.2011.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Truscott M, Islam ABMMK, Lightfoot J, Lopez-Bigas N, Frolov MV. An intronic microRNA links Rb/E2F and EGFR signaling. PLoS genetics. 2014;10:e1004493. doi: 10.1371/journal.pgen.1004493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Q, Brown MR. Signaling and function of insulin-like peptides in insects. Annu Rev Entomol. 2006;51:1–24. doi: 10.1146/annurev.ento.51.110104.151011. [DOI] [PubMed] [Google Scholar]
- 42.Suh YS, Bhat S, Hong SH, Shin M, Bahk S, Cho KS, et al. Genome-wide microRNA screening reveals that the evolutionary conserved miR-9a regulates body growth by targeting sNPFR1/NPYR. Nat Commun. 2015;6:7693. doi: 10.1038/ncomms8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varghese J, Lim SF, Cohen SM. Drosophila miR-14 regulates insulin production and metabolism through its target, sugarbabe. Genes & development. 2010;24:2748–53. doi: 10.1101/gad.1995910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boulan L, Martin D, Milan M. bantam miRNA promotes systemic growth by connecting insulin signaling and ecdysone production. Current biology: CB. 2013;23:473–8. doi: 10.1016/j.cub.2013.01.072. [DOI] [PubMed] [Google Scholar]
- 45.Colombani J, Bianchini L, Layalle S, Pondeville E, Dauphin-Villemant C, Antoniewski C, et al. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science. 2005;310:667–70. doi: 10.1126/science.1119432. [DOI] [PubMed] [Google Scholar]
- 46.Jin H, Kim VN, Hyun S. Conserved microRNA miR-8 controls body size in response to steroid signaling in Drosophila. Genes & development. 2012;26:1427–32. doi: 10.1101/gad.192872.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G, et al. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell. 2009;139:1096–108. doi: 10.1016/j.cell.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 48.Lee GJ, Jun JW, Hyun S. MicroRNA miR-8 regulates multiple growth factor hormones produced from Drosophila fat cells. Insect molecular biology. 2015;24:311–8. doi: 10.1111/imb.12156. [DOI] [PubMed] [Google Scholar]
- 49.Honegger B, Galic M, Kohler K, Wittwer F, Brogiolo W, Hafen E, et al. Imp-L2, a putative homolog of vertebrate IGF-binding protein 7, counteracts insulin signaling in Drosophila and is essential for starvation resistance. J Biol. 2008;7:10. doi: 10.1186/jbiol72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Osterbur DL, Fristrom DK, Natzle JE, Tojo SJ, Fristrom JW. Genes expressed during imaginal discs morphogenesis: IMP-L2, a gene expressed during imaginal disc and imaginal histoblast morphogenesis. Dev Biol. 1988;129:439–48. doi: 10.1016/0012-1606(88)90391-0. [DOI] [PubMed] [Google Scholar]
- 51.Lozano J, Montanez R, Belles X. MiR-2 family regulates insect metamorphosis by controlling the juvenile hormone signaling pathway. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:3740–5. doi: 10.1073/pnas.1418522112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weng R, Cohen SM. Control of Drosophila Type I and Type II central brain neuroblast proliferation by bantam microRNA. Development (Cambridge, England) 2015;142:3713–20. doi: 10.1242/dev.127209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weng R, Cohen SM. Drosophila miR-124 regulates neuroblast proliferation through its target anachronism. Development (Cambridge, England) 2012;139:1427–34. doi: 10.1242/dev.075143. [DOI] [PubMed] [Google Scholar]
- 54.Yuva-Aydemir Y, Xu X-L, Aydemir O, Gascon E, Sayin S, Zhou W, et al. Downregulation of the Host Gene jigr1 by miR-92 Is Essential for Neuroblast Self-Renewal in Drosophila. PLoS genetics. 2015;11:e1005264. doi: 10.1371/journal.pgen.1005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morante J, Vallejo DM, Desplan C, Dominguez M. Conserved miR-8/miR-200 defines a glial niche that controls neuroepithelial expansion and neuroblast transition. Developmental cell. 2013;27:174–87. doi: 10.1016/j.devcel.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reddy BVVG, Irvine KD. Regulation of Drosophila glial cell proliferation by Merlin-Hippo signaling. Development (Cambridge, England) 2011;138:5201–12. doi: 10.1242/dev.069385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y, Padgett RW. bantam is required for optic lobe development and glial cell proliferation. PloS one. 2012;7:e32910. doi: 10.1371/journal.pone.0032910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu Y-C, Chen C-H, Mercer A, Sokol NS. Let-7-complex microRNAs regulate the temporal identity of Drosophila mushroom body neurons via chinmo. Developmental cell. 2012;23:202–9. doi: 10.1016/j.devcel.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kucherenko MM, Barth J, Fiala A, Shcherbata HR. Steroid-induced microRNA let-7 acts as a spatio-temporal code for neuronal cell fate in the developing Drosophila brain. EMBO J. 2012;31:4511–23. doi: 10.1038/emboj.2012.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garaulet DL, Castellanos MC, Bejarano F, Sanfilippo P, Tyler DM, Allan DW, et al. Homeotic function of Drosophila Bithorax-complex miRNAs mediates fertility by restricting multiple Hox genes and TALE cofactors in the CNS. Developmental cell. 2014;29:635–48. doi: 10.1016/j.devcel.2014.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parrish JZ, Xu P, Kim CC, Jan LY, Jan YN. The microRNA bantam functions in epithelial cells to regulate scaling growth of dendrite arbors in drosophila sensory neurons. Neuron. 2009;63:788–802. doi: 10.1016/j.neuron.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song Y, Ori-McKenney KM, Zheng Y, Han C, Jan LY, Jan YN. Regeneration of Drosophila sensory neuron axons and dendrites is regulated by the Akt pathway involving Pten and microRNA bantam. Genes & development. 2012;26:1612–25. doi: 10.1101/gad.193243.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang N, Soba P, Parker E, Kim CC, Parrish JZ. The microRNA bantam regulates a developmental transition in epithelial cells that restricts sensory dendrite growth. Development (Cambridge, England) 2014;141:2657–68. doi: 10.1242/dev.107573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y, Wang H, Li X, Li Y. Epithelial microRNA-9a regulates dendrite growth through Fmi-Gq signaling in Drosophila sensory neurons. Developmental neurobiology. 2015 doi: 10.1002/dneu.22309. [DOI] [PubMed] [Google Scholar]
- 65.Nesler KR, Sand RI, Symmes BA, Pradhan SJ, Boin NG, Laun AE, et al. The miRNA pathway controls rapid changes in activity-dependent synaptic structure at the Drosophila melanogaster neuromuscular junction. PLoS One. 2013;8:e68385. doi: 10.1371/journal.pone.0068385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loya CM, McNeill EM, Bao H, Zhang B, Van Vactor D. miR-8 controls synapse structure by repression of the actin regulator enabled. Development (Cambridge, England) 2014;141:1864–74. doi: 10.1242/dev.105791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu CS, Zhai B, Mauss A, Landgraf M, Gygi S, Van Vactor D. MicroRNA-8 promotes robust motor axon targeting by coordinate regulation of cell adhesion molecules during synapse development. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2014:369. doi: 10.1098/rstb.2013.0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen W, Liu Z, Li T, Zhang R, Xue Y, Zhong Y, et al. Regulation of Drosophila circadian rhythms by miRNA let-7 is mediated by a regulatory cycle. Nature communications. 2014;5:5549. doi: 10.1038/ncomms6549. [DOI] [PubMed] [Google Scholar]
- 69.Luo W, Sehgal A. Regulation of circadian behavioral output via a MicroRNA-JAK/STAT circuit. Cell. 2012;148:765–79. doi: 10.1016/j.cell.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Picao-Osorio J, Johnston J, Landgraf M, Berni J, Alonso CR. MicroRNA-encoded behavior in Drosophila. Science (New York, NY) 2015;350:815–20. doi: 10.1126/science.aad0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Busto GU, Guven-Ozkan T, Fulga TA, Van Vactor D, Davis RL. microRNAs That Promote or Inhibit Memory Formation in Drosophila melanogaster. Genetics. 2015;200:569–80. doi: 10.1534/genetics.114.169623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li W, Cressy M, Qin H, Fulga T, Van Vactor D, Dubnau J. MicroRNA-276a functions in ellipsoid body and mushroom body neurons for naive and conditioned olfactory avoidance in Drosophila. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2013;33:5821–33. doi: 10.1523/JNEUROSCI.4004-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Verma P, Augustine GJ, Ammar M-R, Tashiro A, Cohen SM. A neuroprotective role for microRNA miR-1000 mediated by limiting glutamate excitotoxicity. Nature neuroscience. 2015;18:379–85. doi: 10.1038/nn.3935. [DOI] [PubMed] [Google Scholar]
- 74.Liu N, Landreh M, Cao K, Abe M, Hendriks G-J, Kennerdell JR, et al. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature. 2012;482:519–23. doi: 10.1038/nature10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kitano H. Biological robustness. Nat Rev Genet. 2004;5:826–37. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- 76.Pelaez N, Carthew RW. Biological robustness and the role of microRNAs: a network perspective. Curr Top Dev Biol. 2012;99:237–55. doi: 10.1016/B978-0-12-387038-4.00009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–24. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yatsenko AS, Shcherbata HR. Drosophila miR-9a targets the ECM receptor Dystroglycan to canalize myotendinous junction formation. Dev Cell. 2014;28:335–48. doi: 10.1016/j.devcel.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 79.Kugler J-M, Chen Y-W, Weng R, Cohen SM. Maternal loss of miRNAs leads to increased variance in primordial germ cell numbers in Drosophila melanogaster. G3 (Bethesda, Md) 2013;3:1573–6. doi: 10.1534/g3.113.007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cassidy JJ, Jha AR, Posadas DM, Giri R, Venken KJT, Ji J, et al. miR-9a minimizes the phenotypic impact of genomic diversity by buffering a transcription factor. Cell. 2013;155:1556–67. doi: 10.1016/j.cell.2013.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cassidy JJ, Straughan AJ, Carthew RW. Differential Masking of Natural Genetic Variation by miR-9a in Drosophila. Genetics. 2016;202:675–87. doi: 10.1534/genetics.115.183822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–82. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–80. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Quan XJ, Hassan BA. From skin to nerve: flies, vertebrates and the first helix. Cell Mol Life Sci. 2005;62:2036–49. doi: 10.1007/s00018-005-5124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Verma P, Cohen SM. miR-965 controls cell proliferation and migration during tissue morphogenesis in the Drosophila abdomen. eLife. 2015:4. doi: 10.7554/eLife.07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hilgers V, Bushati N, Cohen SM. Drosophila microRNAs 263a/b confer robustness during development by protecting nascent sense organs from apoptosis. PLoS Biol. 2010;8:e1000396. doi: 10.1371/journal.pbio.1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]