Abstract
Predictive testing of anticancer drugs remains a challenge. Bioengineered systems, designed to mimic key aspects of the human tumor microenvironment, are now improving our understanding of cancer biology and facilitating clinical translation. We show that mechanical signals have major effects on cancer drug sensitivity, using a bioengineered model of human bone sarcoma. Ewing sarcoma (ES) cells were studied within a three-dimensional (3D) matrix in a bioreactor providing mechanical loadings. Mimicking bone-like mechanical signals within the 3D model, we rescued the ERK1/2-RUNX2 signaling pathways leading to drug resistance. By culturing patient-derived tumor cells in the model, we confirmed the effects of mechanical signals on cancer cell survival and drug sensitivity. Analyzing human microarray datasets, we showed that RUNX2 expression is linked to poor survival in ES patients. Mechanical loadings that activated signal transduction pathways promoted drug resistance, stressing the importance of introducing mechanobiological cues into preclinical tumor models for drug screening.
Keywords: Mechanical Stimulation, Tumor Microenvironment, RUNX2, Ewing Sarcoma, Drug Sensitivity
1. Introduction
Developing anti-cancer drugs is a long, costly and inefficient process (1). Although many drug candidates show promising preclinical results, less than 7% are approved for clinical use (2, 3). Drug safety and efficacy are currently studied in vitro (in cell monolayers and aggregates) and in vivo (in rodent models). When cultured in vitro, cancer cells are deprived of their native microenvironment and tend to lose the tumor phenotype due to undesired adaptation (4). Animal models, which are considered essential for cancer research, also fail to predict the clinical outcomes (5). To overcome these limitations, tumor features can be tailored in vitro using bioengineering techniques (6). Existing, 3D models replicate some properties of bone but have not fully reproduced the structural and cellular composition of the bone microenvironment. For instance, we recently developed a bioengineered model of human bone tumor that recapitulates three-dimensional (3D) tissue context, extracellular matrix and tumor-stroma interactions (7). In this model, cancer cells recovered their original hypoxic tumor phenotype and expression of important oncogenes. Among other factors, flow strongly affects tumor behavior and drug response, as shown using an Ewing Sarcoma 3D model cultured in a perfusion bioreactor (8). The use of patient-derived tumor xenografts (PDXs) is also becoming a viable alternative to cultures of cancer cell lines, as they better preserve the parental tumor heterogeneity and drug responses (9). Recent findings suggest that a PDX 3D model of prostate cancer recapitulates essential pathological properties of bone metastasis, enabling interrogation of complex tumor-stromal interactions (10).
However, critical microenvironmental cues such as mechanical signals remain elusive to study in vivo and are challenging to model in vitro. In fact, nearly every tissue in our body is subjected to mechanical forces. These forces, sensed by the cells, are transduced into biochemical signals activating intracellular pathways (11). As a result, mechanical stimuli play a major role in tissue development and diseases such as cancer (12). For instance, Ewing sarcoma (ES) – the second most frequent bone tumor in adolescents – thrives in a mechanically active microenvironment. Despite multi-modal therapy, survival rates in ES remain poor (13). Hence, novel therapeutic strategies and translational investment are needed to increase the life expectancy of young ES patients (14).
One promising approach targets a family of cell-surface receptors called receptor tyrosine kinases (RTKs). Ligand binding to these receptors activates downstream signaling pathways mediated by the extracellular-signal regulated kinase (ERK1/2). In a similar fashion, ERK1/2 is part of the mechanoregulatory circuit linking physical cues to molecular pathways in cancer cells (15). Therefore, blocking ERK1/2 leads to reduced cell proliferation and survival in many tumors. However, despite encouraging results in ES preclinical models, the use of RTK inhibitors showed little or no effects in ES patients (16). Recent studies have shown that mesenchymal stem cells exposure to mechanical loading stimulated ERK1/2-dependent activation of RUNX2, a transcription factor and master regulator of bone differentiation (17). In addition to its role in osteogenesis, RUNX2 promotes cancer cell survival, invasion and drug resistance (18, 19). Given Ewing sarcoma mesenchymal features and oncogenic potential of RUNX2 in the bone, it is surprising that there is little evidence linking RUNX2 to ES.
Our objective was to develop a bioengineered model of Ewing sarcoma that incorporates the application of mechanical loadings to investigate the role of RUNX2 in ES cells drug sensitivity. We hypothesized that the exposure of ES cells to mechanical forces, stimulates ERK1/2-dependent expression of RUNX2, altering RTK inhibitors efficacy. To test this hypothesis, we analyzed RUNX2 expression in ES tumor samples and ES cell lines. ES cell lines or patient-derived ES xenografts were grown in a previously validated biomimetic 3D matrix (20). The 3D tissue models were cultured in the bioreactor and exposed to external forces of physiologically relevant types and magnitudes, with static controls. The ERK1/2-RUNX2 transduction mechanism was studied by measuring gene and protein expression. Drug sensitivity to RTK inhibitors was assessed by analyzing cell phenotype, apoptosis and proliferation, with emphasis on the effects of mechanical forces on the ERK1/2-RUNX2 signaling pathway.
2. Materials and Methods
2.1 Drugs and chemicals
Sorafenib was purchased from Santa Cruz Biotechnology. Doxorubicin, sunitinib, and imatinib were purchased from Sigma Aldrich. U0126 was purchased from Cell Signaling Technology.
2.2 Cell lines
Ewing sarcoma cell lines SK-N-MC (HTB-10) and RD-ES (HTB-166) were purchased from the American Type Culture Collection (ATCC) and cultured according to the manufacturer’s specifications using ATCC-formulated EMEM or RPMI-1640 medium respectively, supplemented with 10% (v/v) Hyclone fetal bovine serum (FBS) and 1% penicillin/streptomycin (Gibco).
2.3 Patient-derived cancer cells
Processing of the patients’ samples, expansion, and isolation of the patient-derived xenografts were conducted as in our previous studies (21). Briefly, de-identified samples of the patients’ tumor tissue were collected under a protocol approved by the Memorial Sloan Kettering Cancer Center (MSKCC) Institutional Review Board (IRB). The patient-derived xenograft was established by engrafting and expanding the patient’s tumor tissue in NSG mice (second passage). Single cell suspensions were achieved by physical disruption and digestion of the explanted xenografts using collagenase type IV (Gibco). Early cell cultures (PS3 cells, p<3) were characterized, validated, and tested for mycoplasma contamination at MSKCC core facility. The cells were cultured in Dulbecco’s Modified Eagle’s Medium and supplemented with 10% FBS, L-glutamate, and antibiotics penicillin/streptomycin.
2.4 3D matrix preparation
3D porous matrices were prepared from collagen 1 and hyaluronic acid solutions using a freeze-drying technique as in our previous studies (20). A low molecular weight (10–20 kDa) Sodium Hyaluronate (Lifecore) was dissolved in distilled water to obtain a 1% (wt/v) solution. Four parts of Collagen 1 (8–11 mg/ml in 0.02 N acetic acid, Corning) were mixed with one part of HA solution (4:1). 75 μl of the solution was dispensed into a 3 mm high × 4 mm diameter well serving as a mold for scaffold formation, frozen at −40 °C for 4 hours, and lyophilized under vacuum of < 100 mTorr at −40°C for 12 hours. Sublimation of ice crystals formed in the frozen mixture results in the formation of interconnected pores within the 3D matrix. The collagen 1 – HA 3D matrices were cross-linked with a water-soluble carbodiimide and soaked in 95% ethanol containing 33 mM 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (Sigma Aldrich Co. Ltd.) and 6 mM N-hydroxysuccinimide (Sigma Aldrich) for 4 h at 25 °C. After crosslinking, the porous scaffolds were washed in distilled water (5 min × 10 times) and freeze-dried overnight.
2.5 3D cell culture
Each matrix was seeded with 4 million cells. To this end, 12 scaffolds and 24 ml of cell suspension containing 2 ×106 cells/ml were placed into a 50 ml Falcon tube on an orbital shaker (3h at 37°C and 5% CO 2) Cell-seeded matrices were then transferred to non-treated 24-multiwell plates (Nunc) and cultured in 1.5 ml of medium at 37°C and 5% CO 2 for 48 hrs to allow the cells to attach to matrix. These cultures were established using ES cell lines (SK-N-MC and RD-ES) and patient-derived xenografts (PS3 cells).
2.6 Bioreactor
A bioreactor developed in our previous studies (22, 23) was used to subject 3D cell cultures to dynamic compressive loading, by the vertical motion of plungers that were in contact with the tissues placed into standard 24 well-plates. A linear actuator and a stepper motor were used to control the displacement magnitude, and the stimulation frequency and waveform. Live monitoring of the motion was achieved using a linear variable differential transformer. To maintain the viability, the culture chamber was filled with culture medium, and all experiments were conducted within an incubator at 37°C and 5% C O2.
Porous scaffolds (3 mm high × 4 mm diameter cylinders), were prepared from collagen I and hyaluronic acid solutions using a freeze-drying technique. Cells were seeded into porous scaffolds (2 ×106 cells/ml, 2 ml/scaffold) and allowed to attach. The resulting tissue constructs were placed in the bioreactor and subjected to cycles of dynamic mechanical stimulation.
2.7 Mechanical stimulation protocol
3D tumor tissues were placed into the bioreactor and subjected to unconfined, dynamic compressive loading, applied periodically. Specifically, the compressive strains of 1, or 10% were applied using a sinusoidal waveform at a 0.25 Hz frequency. Each day, 1800 loading cycles were applied over 2 hours of stimulation. The protocol included 2 hours of bioreactor culture each day, where the 3D cultures (static and stimulated group), placed in regular tissue culture dishes, had top and bottom surfaces in contact with bioreactor parts. The stimulated group was exposed to dynamic loading, whereas controls were cultured in static conditions. After 2 hours, the stimulated samples and the static controls were removed from the bioreactor allowing for free diffusion of drugs and nutrients across all the construct’s surfaces.
2.8 Quantitative real-time PCR (qRT-PCR)
qRT-PCR was performed as previously described (7). Briefly, total RNA was extracted using Trizol (Thermo-Fisher) following the manufacturer’s instructions. RNA preparations (2 μg) were treated with “Ready-to-go you-prime first-strandbeads” (GE Healthcare) to generate cDNA. Quantitative real-time PCR was performed using DNA Master SYBR Green I mix (Applied Biosystems). mRNA expression levels were quantified applying the ΔCt method, ΔCt = (Ct of gene of interest - Ct of GAPDH). qRT-PCR primer sequences were obtained from the PrimerBank database (http://pga.mgh.harvard.edu/primerbank/) (Table S3).
2.9 Histology and immunohistochemistry (IHC)
Cell seeded 3D matrices were fixed in paraformaldehyde for 12 h at 25 °C and dehydrated with graded ethanol washes, embedded in paraffin and serial sections, 5 μm thick, were prepared for histology. Frozen sections of the Ewing sarcoma tumor samples were fixed in pre-cooled acetone (−20 °C) for 20 min immediately before immunostaining procedure. Sections were washed with Tris Buffered Saline (TBS) and treated with 0.3% H2O2 solution in TBS at room temperature for 10 min to block endogenous peroxidase activity and incubated with Vectastain Elite ABC Kit (Vector Labs), according to the manufacturer’s protocol. The sections were incubated with the following antibodies: CD99 (dilution 1:500; Signet antibodies, SIG-3620), OPN (1:200, Abcam, ab8448), BSP (1:500, Abcam, ab33022), RUNX2 (1:200, Abcam, ab76956), Ki67 (1:500, Abcam, ab15580) overnight at 4°C. After washing with TBS, samples were incubated with secondary antibodies for 1h at 25°C and developed (Vector Laboratories). Negative controls were prepared by omitting the primary antibody step. Slides were counterstained with Hematoxylin QS (Vector Labs). Apoptosis was evaluated using the terminal deoxynucleotidyl transferase-dUTP nick end labeling (TUNEL) assay following the manufacturer’s protocol (ThermoFisher).
2.10 Western blots
Cells were lysed in high-salt radioimmunoprecipitation assay (RIPA) lysis buffer (Cell Signaling) containing protease/phosphatase inhibitors (Cell Signaling). Cell lysates were centrifugated at 12000 g for 15 min at 4°C and supernatants were col lected. Total protein concentration was measured using Quick Start™Bradford (Bio-Rad Laboratories) and 15–30μg of proteins were loaded on a Bis-Tris gels (BioRad), transferred to a nitrocellulose membrane and incubated with antibodies against RUNX2 (1:500; Santa Cruz Biotech, M-70, sc-10758), c-KIT (1:100, Santa Cruz Biotech, C-19, sc-168), p-ERK1/2 (1:200, Cell Signaling, 4370), t-ERK1/2 (1:1000, Cell Signaling, 4695), CD99 (dilution 1:500; Signet antibodies, SIG-3620) at 4 degrees overnight and GAPDH (1:5000, Invitrogen 437000) or Actin (1:5000, Sigma Aldrich, A2066) at room temperature for one hour. Membranes were incubated with anti-rabbit or anti-mouse secondary antibody conjugated with Alexa Fluor 680 (1:5000; Thermo Fisher Scientific) at room temperature for one hour and imaged on Licor Odyssey scanner. Densitometric analysis of specific bands was performed with NIH Image J software.
2.11 Drug studies
Cells were incubated with sorafenib, sunitinib, imatinib, U0126 or doxorubicin at specific concentrations for 1 h before the start of mechanical stimulation. Samples were stimulated once a day for 2 h (1800 cycles at 0.25 Hz). For drug studies in 2D cell cultures that were used as additional controls, 5 × 10^3 cells were plated in a 96-well plate in 200 μL of medium. After 24 h, a drug was added (sorafenib, sunitinib, imatinib or doxorubicin) to each well at a specified concentration, and cell proliferation was measured after 48h of incubation. Drug concentrations were chosen based on pharmacokinetics data reported in clinical trials for Ewing sarcoma (imatinib, sorafenib) or pediatric patients (doxorubicin).
Imatinib (24): concentrations were obtained on day 8 of therapy from 36 patients. The median concentration was 6.1 μM, range 0.5–21.4 μM. The concentration used in this study was 6 μM.
Sorafenib (25): plasma pharmacokinetic parameters measured after dosing with sorafenib (400 mg twice daily) showed that the average maximum concentration at day 1 and 28 was 4.9 and 13.33 μM. Concentrations used in this study were 1 and 5 μM.
Doxorubicin (26): for a dose of 40 mg/m2, the median plasma concentration of doxorubicin was 0.115 μM the 10th percentile was 0.0677 μM and the 90th percentile was 0.211 μM. The concentration used in this study was 0.2 μM.
For sunitinib, we used the half maximum inhibitor concentration (IC50), determined on ES cells cultured in monolayer.
2.12 Cell proliferation and viability
DNA content was measured to monitor cell proliferation, using the Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies) according to the manufacturer’s protocol. Bioengineered tumor samples were digested at 60 °C over-night, the supernatant was collected and diluted as needed to a concentration within the linear range of the assay. A standard curve was used to convert fluorescence to total DNA. Samples were read using a fluorescent plate reader at an excitation wavelength of 480nm and an emission wavelength of 528 nm. CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS) (Promega) was used to evaluate anti-proliferative activity of the drugs in cell lines cultured in 2D. At the end of the incubation period medium was replaced with 20 μL of stock solution of the MTS assay kit and 100 μL of fresh medium. The absorbance at 490nm was recorded using a 96-well plate reader. Cell proliferation in drug-treated cells was normalized to their respective controls.
2.13 Computational study
A multiphysics computational finite element (FE) model was developed to predict both flow velocity and pressure in the 3D porous matrix following mechanical loading. The experimental set-up was designed using the software Comsol Multiphysics® (Comsol 2015). The geometry and boundary conditions for the computational study are shown in Supplementary Fig.2a. Briefly, A cylindrical sample with thickness t and diameter h placed between two rigid impervious platens in a chamber, where the specimen is immersed in a fluid (culture medium). The surfaces between the specimen and the platens are taken to be frictionless so that the specimen can expand freely in the lateral (radial) direction while the cyclic compressive load is applied in the axial direction using the motion waveform resulting from the function described in Supplementary Table 4. The fluid can flow out of the cylindrical matrix via the free surface. The specimen is modelled as an isotropic solid matrix containing fluid-saturated pores entrapped by a porous network (27). The matrix with pore fluid is considered as poroelastic, where the fluid, in this case the culture medium is assumed to be incompressible.
The material properties required to define the biphasic component are listed in Supplementary Table 4 and include the Young’s modulus E and Poisson’s ratio ν of the matrix. The Young’s modulus was determined as previously described (20). Briefly, samples were exposed to unconfined compression in phosphate-buffered saline (PBS) at room temperature. An initial load of 0.2 N was applied, and followed by a series of stress-relaxation steps, where specimens were compressed at a velocity of 1% per second up to 10% strain, and maintained at each position for 1,800 seconds. The Young’s modulus was calculated from the equilibrium force measured at the 10% strain. To determine the Poisson’s ratio, the same unconfined compression setup was used to control strain in the y direction and the resulting radial strain was measured optically using ImageJ software.
Modeling poroelasticity requires the coupling of two laws. The first is Darcy’s law, which describes the relation between fluid motion and pressure within a porous medium. The second law is the structural displacement of the porous matrix. Biot poroelasticity describes this coupled physics. The fluid equation comes from:
which in the pressure formulation translates to:
where ∂εvol/∂t is the rate of change in volumetric strain from the equations for solid displacements, ρf is the fluid density and αB is the Biot-Willis coefficient. The storage term S is calculated from:
The drained bulk modulus Kd is always smaller than the solid bulk modulus Ks, therefore Biot-Willis coefficient is always bounded to:
αB does not depend on the properties of the fluid, but on the properties of the porous matrix. A soft porous matrix has a Biot-Willis coefficient close to 1 (since Kd ≪ Ks). The software Comsol Multiphysics® (Comsol 2015) was used to set up, discretize and solve the equations. The final mesh was composed of 4×106 tetrahedral elements. The numerical analyses were run on a Linux workstation with 32 Gb RAM.
2.14 Collection of patients’ tumor samples
Fully de-identified Ewing sarcoma tumor samples were obtained from the Columbia University Tissue Bank, on an IRB-approved protocol. Human tumor sections were analyzed in a blinded fashion by a designated pathologist at the Columbia University Tissue Bank. Estimation of viable tumor cells relative to non-tumor cells, as well as tumor necrosis was made for all samples using hematoxylin and eosin staining.
2.15 Datasets for genomic analysis
The web-based genomics analysis and visualization application r2.amc (http://r2.amc.nl.) used to generate the graph allows the user to compare average mRNA expression of a gene of interest across microarray datasets. For consistency, this is possible only if the same microarray chips and normalization methods were used across studies. In the graph we studied the average mRNA expression of a RUNX2 probeset in the Affymetrix u133p2 chips with MAS5.0 normalization method (20).
Tumor Ewing’s Sarcom-Savola (73 samples)
Source: GEO ID: gse17679 Dataset Date: 2000-01-01. Inflammatory gene profiling of Ewing sarcoma family of tumors.
Tumor Ewing’s Sarcoma-Francesconi (37 samples)
Source: GEO ID: gse12102 Dataset Date: 2000-01-01. A genome-wide association study of at least 401 French ES patients compared to either 684 French or 3668 US self-described Caucasian controls consistently revealed candidate loci at chromosomes 1 and 10 (p<10–6).
Tumor Ewing’s Sarcoma-Delattre (117 samples)
Source: GEO ID: gse34620 Dataset Date: 2008-06-15. Expression profiling of Ewing sarcoma samples in the frame of the CIT program from the french Ligue Nationale Contre le Cancer.
Tumor Ewing’s Sarcoma-Volchenboum (47 samples)
Source: GEO ID: gse63157 Dataset Date: 2014-11-11 Gene Expression Profiling of Ewing Sarcoma Tumors Reveals the Prognostic Importance of Tumor-Stromal Interactions: A Report from the Childrens Oncology Group
Normal Various–Roth (353 samples)
Source: GEO ID: GSE3526 Dataset Date: 2006-03-30. Normal human tissue samples from ten post-mortem donors were processed to generate total RNA, which was analyzed for gene expression using Affymetrix U133 plus 2.0 arrays.
Normal Various–Roth (503 samples)
Source: GEO ID: GSE7307 Dataset Date: 2007-04-09. Affymetrix human U133 plus 2.0 array was used to transcriptionally profile both normal and diseased human tissues representing over 90 distinct tissue types.
2.16. Kaplan Meier curves
Kaplan scanning was performed within R2 (http://r2.amc.nl) as previously described (28). The Kaplan scanner in the genomic analysis tool r2.amc separates the samples of a dataset into 2 groups based on the gene expression of one gene (RUNX2). In the order of expression, it will use every increasing expression value as a cutoff to create 2 groups and test the p-value in a log-rank test (minimum group size was set to 8). The highest value is reported, accompanied by a Kaplan-Meier picture. It will find the most significant expression cutoff for survival analysis separating sample group in high and low expression values. The best possible Kaplan-Meier curve is based on the log-rank test. Such analysis is only possible for datasets where survival data is present (in our study Savola dataset and Volchenboum dataset). In the top plot (Savola dataset (29)) patients were enrolled in the Italian Cooperative Study (SE 91-CNR Protocol; started November 1991; ended November 1997) organized by the Italian Association for Pediatric Hematology-Oncology and the National Council of Research (CNR). In the bottom plot (Volchenboum dataset (30)) the survival data was obtained from the Children’s Oncology Group (COG) study (Protocol: AEWS0031; started May 2001; ended August 2006). The study is registered at ClinicalTrials.gov (identifier: NCT00006734).
2.17. Statistical analysis
Quantitative real-time PCR, image quantification, viability assay, DNA content and western blot densitometry results were presented as fold change and analyzed using a Student’s t-test (two-tailed; average + standard deviation; n≥4; p<0.05). All experiments were repeated at least twice with the number of samples for each experiment reported in the figures captions. The Kaplan-Meier curves were represented as the overall survival probability, expressed as a percentage over time (years), in ES patients as a function of RUNX2 mRNA expression. A log-rank test that gave the lowest P-value was calculated to separate tumor samples expressing high and low RUNX2 mRNA levels. Statistical significance was assessed using a two-way ANOVA with Bonferroni post hoc test (n≥50; p< 0.05). In the genomic analysis, data was represented as an average expression of a mRNA probeset + standard deviation and statistical significance was assessed using a two-way ANOVA with Bonferroni post hoc test (n≥ 10; p<0.05).
3. Results
3.1 Mechanical stimulation promotes RUNX2 expression
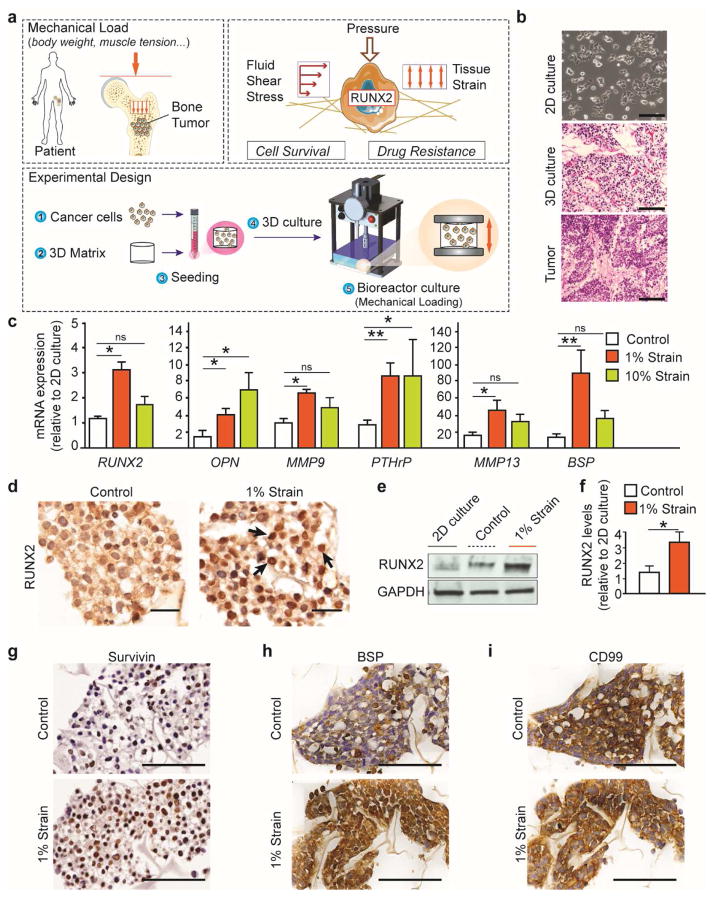
Strong evidence suggests that the inhibition of EWS-FLI1, Ewing sarcoma’s main oncogene, allows ES cells to recover the phenotype of their mesenchymal stem cells progenitors (31). Mesenchymal stem cells, exposed to mechanical loads, express RUNX2 (17) - a transcription factor master regulator of bone development also involved in cancer cell survival and drug resistance in the bone. We generated a bioengineered model to investigate whether ES cells responded to mechanical loads expressing higher RUNX2 levels. To generate the model, cancer cells were seeded onto a 3D collagen 1 -hyaluronic acid porous scaffold and cultured in free swelling conditions for 48 hours to promote cell attachment. After 48 hours, tumor models were placed in a bioreactor used to mimic stresses arising from mechanical loads in the bone. The stimulated group was exposed to different magnitudes of deformation measured in % strain, whereas controls were cultured in a similar configuration but unstimulated. After 2 hours of stimulation, the samples were cultured in free swelling conditions (Fig. 1a; Supplementary Fig. 1a, b; Supplementary Video 1). While cells cultured on tissue culture plastic (2D culture) formed flat sheets, histological staining showed that cells adhered to the porous matrix and formed a tissue (3D culture) (Fig. 1b).
Fig. 1. Mechanical stimulation promotes RUNX2 expression in the engineered tumor model.
(a) The bones are exposed to cyclic tissue deformation resulting from the mechanical loads generated mainly by body weight and muscle tension. Cancer cell growing in the bone are exposed to a mechano-environment where extracellular forces, resulting from pressurization of interstitial fluids, flow-induced shear and matrix strain, are dynamically superimposed and alter cell survival and drug response through the activation of transcription factors (RUNX2). To generate the bone tumor models, ES cells were seeded onto a 3D porous matrix, placed in a tissue culture dish and grown in free-swelling conditions for 48 hours. Plates were lodged in the bioreactor for 2 hours of stimulation followed by 24 hours of culture in free-swelling conditions. For longer experiments stimulation was repeated every 24 hours. (b) Representative images of ES cells cultured on a 2D tissue culture dish (top) or on a 3D porous scaffold (middle) compared to ES tumor sections obtained from patients (bottom). Scale bars: phase contrast 50 μm; H&E 100 μm. (c) Gene expression analysis of engineered ES models exposed to bone-like (1% strain) or cartilage-like (10% strain) mechanical loadings. mRNA levels of RUNX2 and several target genes (OPN, MMP9, PTHrP, MMP13 and BSP) was normalized to GAPDH and to the 2D culture. (average ± sd; n= 9; * p<0.05; ** p<0.01; ns not statistically significant). P values are determined by Student’s t-test (two-tailed). (d) Immunostaining revealed RUNX2 nuclear localization (black arrows) in cells exposed to stimulation. Scale bar: 20μm. (e,f) Western blot analysis of RUNX2 protein levels in 2D culture or in the ES models (1% strain or control). Quantified RUNX2 protein levels (f) are represented as relative changes in band density normalized to GAPDH and to RUNX2 levels in 2D cultures. (Average ± sd; n= 9; * p<0.05; ** p<0.01; ns not statistically significant). P values are determined by Student’s t-test (two-tailed). (g–i) Immunostaining analysis of RUNX2 transcriptional targets Survivin (g), BSP (h) and ES diagnostic marker CD99 (i). Scale bars: 50μm.
Our computational study shows that the 3D cultures cyclically deformed and were exposed to a combination of extracellular forces resulting from matrix strain, pressure, and hydrodynamic shear stress derived from interstitial flow (Supplementary Fig. 2a–f, Supplementary Video 2). Therefore we investigated the effect of bone-like (1% strain; (32)) or cartilage-like (10% strain (33)) mechanical loadings on gene expression in the tumor models (Fig. 1c; Supplementary Fig. 3a). Bone-like loads increased the mRNA levels of RUNX2 and its transcription targets (OPN, MMP9, PTHrP, MMP13 and BSP) associated with cancer progression in the bone (Fig. 1c). We also found that RUNX2 protein levels were higher in cancer cells exposed to 1% strain compared to the unstimulated controls and 2D cultures (Fig. 1d–f). Immunostaining revealed higher expression of Survivin and BSP in mechanically stimulated cells (Fig. 1g, h). Mechanical stimulation did not affect the mRNA and protein levels of the ES clinical marker CD99 and ES oncogene EWS-FLI (Fig. 1i; Supplementary Figure. 3a). Overall, RUNX2 transcriptional activity was rescued in the bioengineered ES model by mimicking the cyclic tissue deformation resulting from the mechanical loads acting on the bones.
3.2. Mechanotransduction pathway mediates RUNX2 transcriptional activity
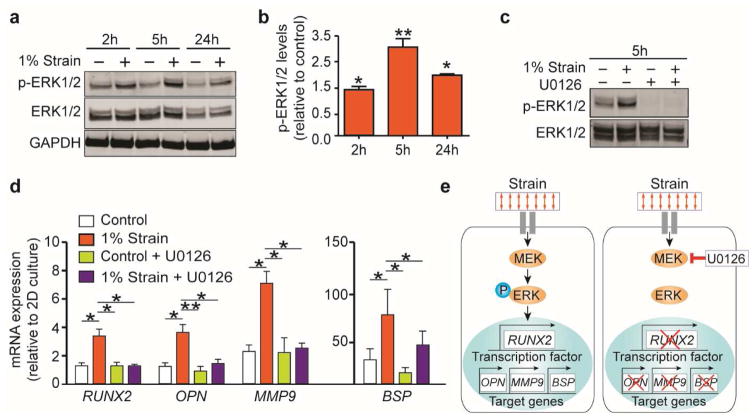
Extracellular signal-regulated kinase 1/2 (ERK1/2) phosphorylation mediates the activation of RUNX2 as a critical step in the responses to mechanical stimulation by mesenchymal stem cells (17). Given the phenotypical features shared between mesenchymal stem cells and Ewing cells, we investigated whether a mechanobiological mechanism could mediate RUNX2 activation in ES cells. Western blot analysis showed that ERK1/2 phosphorylation increased over time and peaked 5 hours after the onset of the stimulation in the bioreactor (Fig. 2a, b). ERK1/2 phosphorylation depends on the upstream activity of the kinase enzyme MEK which could be blocked with the highly selective inhibitor U0126. Exposing ES cells to 10 μM of U0126, inhibited ERK1/2 phosphorylation both in the stimulated and the unstimulated groups (Fig. 2c). ERK1/2 blockade led to a drop in RUNX2 mRNA levels to baseline levels (Fig. 2d). Reduced levels of RUNX2 transcription targets OPN, MMP9 and BSP suggested a decrease in RUNX2 activity (Fig. 2d, e). MEK-ERK1/2 inhibition did not alter the expression of ES oncogene EWS-FLI and the ES marker NKX2.2 (Supplementary Fig. 3b).
Fig. 2. Mechanotransduction pathway mediates RUNX2 expression and activation.
(a,b) Western blot analysis (a) showing ERK1/2 phosphorylation (p-ERK1/2) in ES models mechanically stimulated (+) or unstimulated (−). Quantified p-ERK1/2 protein levels (b) are represented as relative changes in band density normalized to total ERK1/2 and to relative p-ERK1/2 levels in the control. (average ± sd; n≥ 3; * p<0.05; ** p<0.01). P values are determined by Student’s t-test (two-tailed). (c) Upstream regulation of ERK1/2 phosphorylation by MEK1/2. Western blot analysis confirmed ERK1/2 phosphorylation blockade when cells were cultured with the MEK1/2 inhibitor U0126 (the + and the - indicate 10 and 0 μM respectively). (d) qRT-PCR analysis of RUNX2, OPN, MMP9, and BSP mRNA levels in ES models treated with U0126 at 10 μM. (average ± SD; n=5; * p<0.05; ** p<0.01). P values are determined by Student’s t-test (two-tailed). (e) Proposed mechanism for the mechano-regulation of RUNX2 expression in ES cells. Cancer cells sensing the bone-like tissue deformation (strain), transduce the acting external forces into biochemical signals. When ERK1/2 is phosphorylated by MEK1/2 it leads to the expression of RUNX2 and several target genes related to cancer progression in the bone.
3.3. Mechanical stimulation modulates drug sensitivity
Receptor tyrosine kinase inhibitors (RTKIs) are small molecules commonly used as therapy for cancer. Receptor tyrosine kinase inhibitors can bind to the active site of a RTK thus preventing phosphorylation and by doing so inhibit the downstream signaling often converging to ERK1/2. Despite RTKIs, such as sorafenib, imatinib and sunitinib have been successfully tested in preclinical ES models (mice and 2D culture) they failed in ES patients.
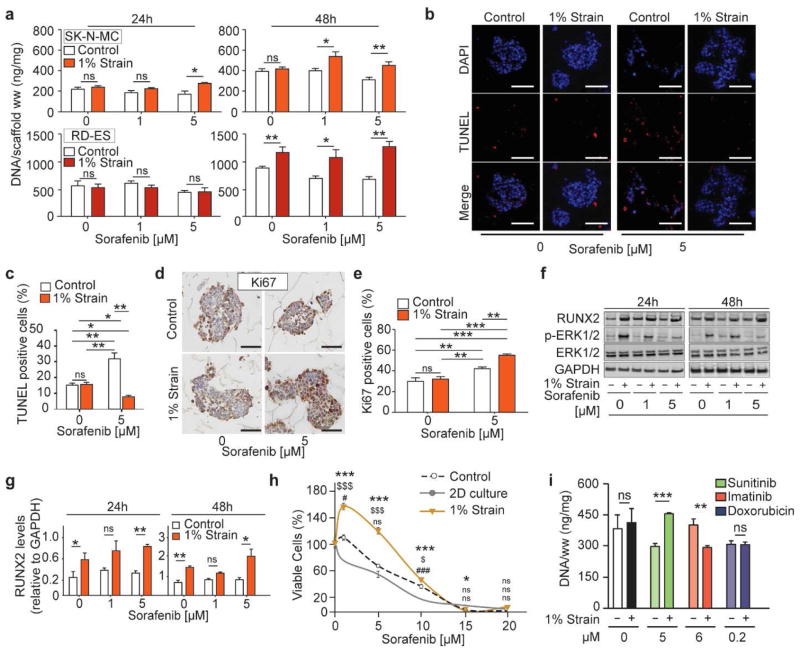
We tested whether the mechanobiological activation of ERK1/2 altered RTKI efficacy in the engineered ES model. Although titration studies were performed to measure the half maximal inhibitory concentration for each drug in 2D cultures, drug concentrations used in the bioreactor experiments were chosen based on pharmacokinetics data reported in clinical trials (Supplementary Fig. 4; Table S1). Bioengineered tumors populated with SK-N-MC or RD-ES cells were exposed to mechanical stimulation (1% strain) and treated with 0 (drug vehicle),1 or 5 μM of sorafenib for 24 or 48 hours (Fig. 3a; Supplementary Fig. 4). Unstimulated ES models treated with sorafenib for 48 hours had a modest decrease in DNA content (Fig. 3a) resulting from increased apoptosis (~40%) and drop in proliferation (~30%) (Fig. 3b–e). By contrast, tumor models exposed to mechanical stimulation during sorafenib treatment show, after 48 hours, a significant increase in DNA content (Fig. 3a) resulting from a reduction in apoptosis (~8%) and increase in proliferation (~50%) (Fig. 3b–e). Western blot analysis revealed that phosphorylated ERK1/2 levels decreased in a dose-dependent manner, confirming inhibitory effects of sorafenib (Fig. 3f). However, sorafenib failed to inhibit the ERK1/2 phosphorylation levels induced by mechanical stimulation. Notably, along with ERK1/2 activated state, RUNX2 protein levels increased with increasing drug concentrations (Fig. 3f–g).
Fig. 3. Mechanical stimulation modulates drug sensitivity.
(a) Cells growth (DNA content) in the tumor models following sorafenib treatment. DNA levels were normalized to the tissue wet weight (ww). (Average ± sd; n=6; * p<0.05; ** p<0.01; *** p< 0.001; ns not statistically significant). P values are determined by Student’s t-test (two-tailed). (b,c) TUNEL staining (b) revealed reduced apoptosis following sorafenib treatment, in ES models exposed to mechanical stimulation. Scale bar: 50 μm. Quantification of apoptotic cells (c) represented as the percentage of TUNEL-positive cells relative to the total number of cells. (Average ± sd (n=5; * p<0.05; ** p<0.01; *** p<0.001; ns not statistically significant). P values are determined by Student’s t-test (two-tailed). (d,e) Ki67 staining (d) showing increased proliferation, following sorafenib treatment, in ES cells exposed to mechanical stimulation. Scale bar: 50μm. Quantifications of proliferating cells (e) represented as the percentage number of Ki67-positive cells relative to the total number of cells. (Average ± sd (n=5; * p<0.05; ** p<0.01; *** p<0.001; ns not statistically significant). P values are determined by Student’s t-test (two-tailed). (f) Western blot analysis showing RUNX2, p-ERK1/2 and ERK1/2 protein levels in ES cells exposed to mechanical stimulation (+) or in the controls (−), treated with sorafenib. (g) Quantified RUNX2 protein levels represented as relative changes in band density normalized to GAPDH. (Average ± sd (n= 3; * p<0.05; ** p<0.01). P values are determined by Student’s t-test (two-tailed). (h) Cell viability of ES models dissociated into a single-cell suspension and re-plated in a tissue culture dish for drug studies. Re-plated cells treated with sorafenib evaluated using MTS assay (percent of control treated with drug vehicle). Symbols: * 1% strain and 2D culture; $ 1% strain and Control; # Control and 2D culture (average ± sd; n=6; * p<0.05; ** p<0.01; *** p< 0.001; ns not statistically significant). P values: one-way analysis of variance (ANOVA) with post hoc Bonferroni tests). (i) Cells growth (DNA content) in the engineered models following drug treatments. DNA levels were normalized to the tissue wet weight (ww). (Average ± sd; n=4; * p<0.05; ** p<0.01; *** p< 0.001; ns not statistically significant). P values: Student’s t-test (two-tailed).
To determine if mechanical stimulation induced a resistant phenotype in ES cells, we first dissociated the tumors models after the bioreactor culture and then re-plated the cells on standard 96-well plates (Fig. 3h). Thus, we could assess a resistant phenotype by comparing sorafenib efficacy in ES cells cultured in 2D, in 3D stimulated and unstimulated cells. Dissociated cells were initially seeded at 5×10^3 cells/well in a 96 well plate and treated with sorafenib or with drug vehicle for 48 hours. Re-plated cells previously exposed to mechanical stimulation showed higher IC50 values compared to the unstimulated controls and 2D cultures (10, 7.8, and 5 μM respectively) (Fig. 3h; Supplementary Table 2). Finally, we studied the efficacy of clinically relevant concentrations (24, 34) of RTKIs sunitinib and imatinib, using the anthracycline doxorubicin for comparison (Fig. 3i). Tumor models exposed to stimulation had increased DNA content when treated with sunitinib. By contrast imatinib treatment during mechanical stimulation led to a drop in DNA content in tumor models. No change in DNA content was measured in tumor models treated with doxorubicin. Overall, mechanical stimulation-mediated activation of the ERK1/2-RUNX2 axis led to reduced drug efficacy through increased cell proliferation and decreased apoptosis. Mechanically stimulated ES cells retained their drug resistance when re-plated in 2D culture. Finally, we showed that mechanical stimulation could reduce or potentiate the efficacy of RTKIs with different targets. Overall, our data hints to RUNX2 involvement in mechanotransduction-mediated drug resistance in the bone tumor models exposed to bone-like mechanical loads.
3.4. Patient-derived bone sarcoma model incorporating mechanical loadings
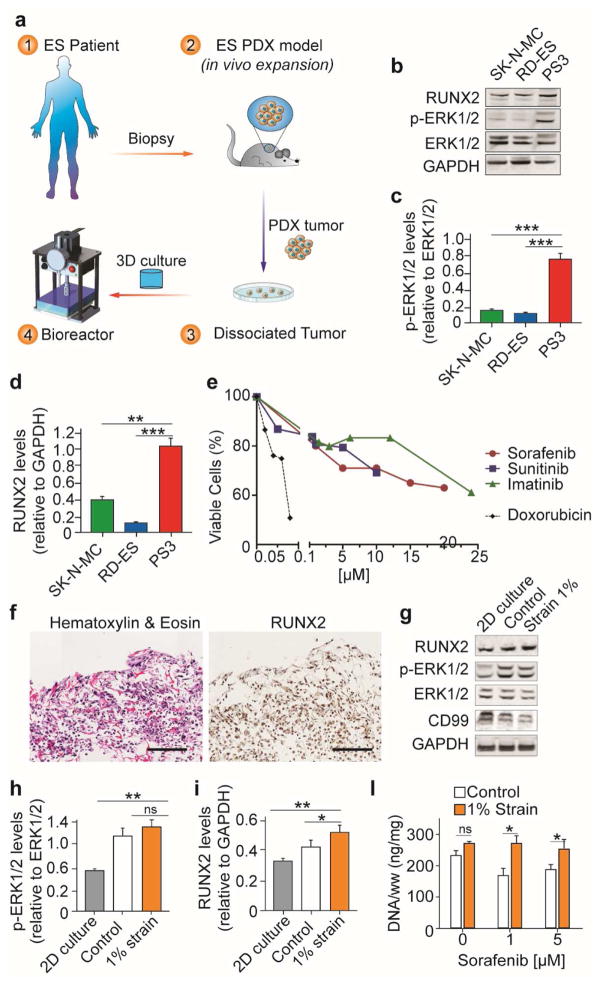
The patient-derived (PD) tumor biopsy was implanted in vivo to promote cancer cell growth while retaining tumor heterogeneity. Tumor xenografts were dissociated in vitro, seeded on the biomimetic 3D matrix and cultured in the bioreactor mimicking bone-like mechanical loads (Fig. 4a). Protein analysis revealed higher ERK1/2 phosphorylation and RUNX2 protein levels in PD cells (PS3) compared to ES cell line cultured in 2D (Fig. 4b–d). Following RTKI treatment in 2D culture, primary cancer cells had higher IC50 values compared to ES cell lines (Fig. 4e; Supplementary Table 1). Immunostaining analysis showed that PD cells, seeded onto the 3D matrix and exposed to mechanical stimulation in the bioreactor, expressed RUNX2 (Fig. 4f). Western blot analysis revealed higher ERK1/2 phosphorylation and RUNX2 protein levels in PD tumor models compared to 2D cell culture and unstimulated controls (Fig. 4g–i). PD tumor models exposed to mechanical loadings showed increased DNA content compared to unstimulated controls, following 48 hours of sorafenib treatment (Fig. 4l). Overall, these data suggest that patient-derived cells cultured in the bioreactor retain at least in part the native tumor phenotype, including RUNX2 expression and resistance to RTK inhibitors.
Fig. 4. Patient-derived ES model incorporating bone-like mechanical loadings.
(a) Workflow to generate the patient-specific ES model. A biopsy of the patients’ tumor was engrafted and expanded in vivo. The patient derived xenograft harvested from the animal, was digested and plated in vitro. The dissociated tumor cells were collected, seeded onto the 3D porous matrix and cultured in the bioreactor. (b–d) Western blot analysis of ES cell lines (SK-N-MC, RD-ES) and patient-derived cells (PS3) cultured in 2D. Quantified p-ERK1/2 (c) and RUNX2 (d) protein levels represented as relative changes in band density normalized to total ERK1/2 (c) or GAPDH (d). (Average ± sd (n≥ 3; ** p<0.01; *** p< 0.001). P values are determined by Student’s t-test (two-tailed). (e) Cell viability (MTS assay represented as percent of non-treated control) of patient-derived cells in 2D culture treated with different drugs for 48 hours. (f) Immunostaining (H&E left, RUNX2 right) of patient-derived ES models exposed to mechanical loading in the bioreactor. Scale bar: 200 μm. (g–i) Western blot analysis of patient-derived cells in 2D or bioreactor culture. Quantified p-ERK1/2 (h) and RUNX2 (i) protein levels represented as relative changes in band density normalized to total ERK1/2 (h) or GAPDH (i). Average ± sd (n= 5; * p<0.05; ** p<0.01; *** p< 0.001). P values are determined by Student’s t-test (two-tailed). (l) Cells growth (DNA content) following sorafenib treatment in the patient-derived models cultured in the bioreactor. DNA levels were normalized to tissue’s wet-weight (ww). (Average ± sd; n=3; * p<0.05; ns not statistically significant). P values are determined by Student’s t-test (two-tailed).
3.5. RUNX2 expression in Ewing sarcoma patients
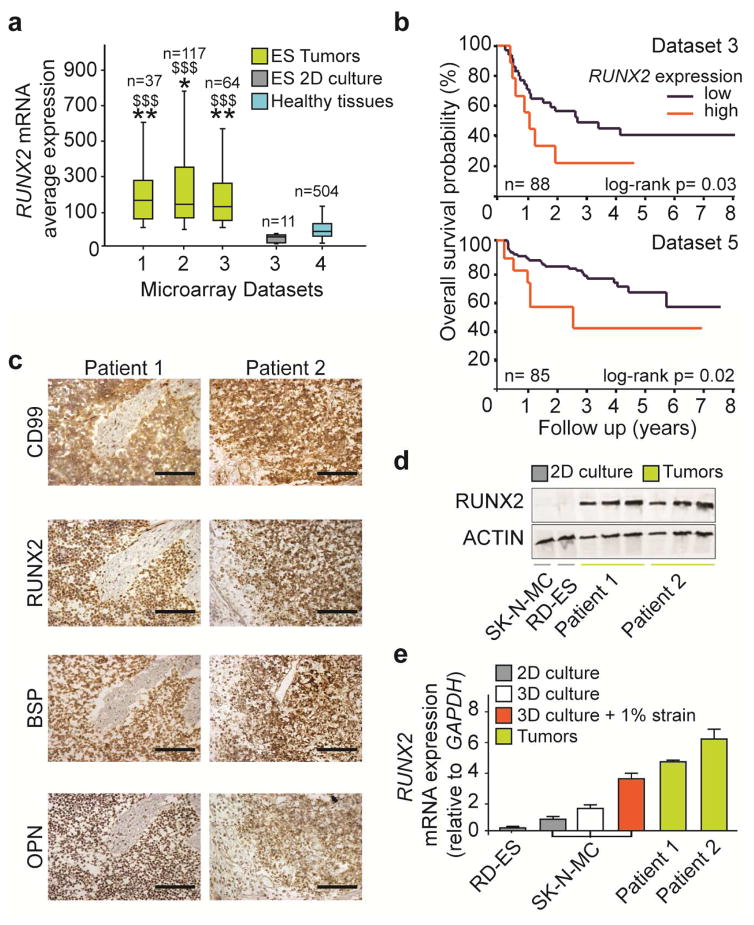
Although RUNX2 mediates proliferation, survival and drug resistance in many types of cancer thriving the bone, including breast and prostate, its expression in ES has not been well documented. To confirm our in vitro results, we performed a genomic analysis of ES microarray datasets that revealed higher RUNX2 mRNA levels in ES patients compared to ES cell lines cultured in 2D and healthy tissues (Fig. 5a). Datasets with survival entries were analysed to estimate the overall survival probability of ES patients as a function of RUNX2 mRNA levels (Fig. 5b). In both datasets studied, patients with high RUNX2 levels were associated with poor five-year survival probability (20–40%), compared to patients with low RUNX2 levels (60–80%).
Fig. 5. RUNX2 expression in Ewing sarcoma patients.
(a) RUNX2 gene expression levels in ES tumors, ES cell lines (2D culture) and healthy tissues. Raw data of gene expression microarrays was obtained from open access datasets (ds1: (35); ds2 (36); ds3 (29); ds4 (37)) and analysed for RUNX2 expression. Symbols: * ES tumors vs ES 2D culture; $ ES tumors vs Healthy tissues. * p<0.05; ** or $$ p<0.01; $$$ p<0.01. P values determined by one-way analysis of variance (ANOVA) with post hoc Bonferroni tests). (b) Poor survival probability of ES patients expressing high RUNX2 levels. The Kaplan-Meier plot shows the overall survival probability, expressed as a percentage over time (years), in ES patients as a function of RUNX2 mRNA expression. Two separate datasets were analysed: (29) (top); (30) (bottom. A log-rank test that gave the lowest P-value was calculated to separate tumor samples expressing high and low RUNX2 mRNA levels. P-values were corrected for multiple testing (one-way ANOVA). (c) Histological analysis of CD99, RUNX2, BSP and OPN in ES tumors. Scale bar: 500 μm. (d) Western blot analysis of RUNX2 protein levels in ES tumors and ES cell lines in 2D culture. Protein from tumors was loaded with increasing amounts (5, 10, 20 μg/lane), while cell lines protein was kept constant at the highest concentration (20 μg/lane). (e) RUNX2 gene expression analyzed by qRT-PCR in ES cell lines in 2D culture, 3D culture unstimulated, 3D culture stimulated (1% strain) and ES tumors. (Average ± sd; mRNA levels were normalized to GAPDH).
The survival data were collected from two separate clinical trials where, despite intrinsic differences in trial modalities, RUNX2 remains associated to poor overall survival. To validate these results, we performed gene and protein analysis on ES tumor biopsies. Because stromal contamination in the tumor sections could influence the measured RUNX2 levels, we estimated the percentage of ES cells relative to stromal cells and extracellular matrix, using immunostaining analysis of the ES diagnostic marker CD99. We found that the tumor samples had 70% of CD99 positive cells, with the remaining 30% of the tissue consisting largely of extracellular matrix, necrotic areas and some stromal cells (Supplementary Fig. 5a–c). Immunostainings also showed that CD99 positive cells express RUNX2 and its transcriptional targets OPN and BSP (Fig. 5c). Western blot assay confirmed that protein levels were higher in ES patients compared to ES cell lines in 2D culture (Fig. 5d). Finally, mechanically- stimulated ES models and ES tumors had higher RUNX2 mRNA levels than ES cell lines in 2D and simple 3D culture (Fig. 5e). Overall, we found that RUNX2 expression - highly expressed in ES patients but not in cell lines - is associated with poor survival probability and could be rescued in the tumor model exposed to bone-like mechanical loads.
4. Discussion
Although intravenous models of Ewing sarcoma show patterns of disease spread similar to that found in patients, only 23% of the experimental mice in these studies developed bone tumors (38). Currently, most preclinical models of bone cancer, including those of Ewing sarcoma, rely on subcutaneous or intramuscular xenografts (39). In these models, cancer cells lack the bone microenvironment where they naturally thrive. Daily activities, such as walking and muscle contractions, exert loads on the skeleton, giving rise to a variety of deformations within the bone tissue that are sensed by cells. Typically, these stresses maintain bone homeostasis by regulating cellular activities, directly through strains in the bone matrix and indirectly through fluid flow derived shear stresses. Cancer cells cross-talk with the bone microenvironment, including stromal cells and extracellular matrix, has been explored and its importance in disease progression widely recognized. However, how the cyclic mechanical signals arising from tissue loading affect cancer cells proliferation, survival and drug resistance has not been well studied and could be relevant to build more predictive preclinical models.
In our engineered tumor, cancer cell lines and patient-derived tumor cells were cultured on a 3D porous matrix. Previously, we showed that microenvironmental signals arising from this biomimetic matrix (including the 3-dimensionality, composition and stiffness) are necessary for recapitulating properties found in native tumors. By using a bioreactor, we exposed the 3D ES models to physiologically-relevant mechanical loads. The amplitude and frequency of the dynamic loads were chosen based on previously reported values in vivo and in vitro (32, 40).
Our computational modeling shows that in the mechanically loaded tumor models, extracellular forces, resulting from pressurization of interstitial fluids, flow-induced shear and matrix strain, are dynamically superimposed. Our model differs from others previously reported, in that it allows to evaluate drug efficacy in a milieu where mechanobiological components are combined in a synergistic way. In our bioengineered ES model, mechanical loads reactivated signaling pathways targeted by commonly used anticancer drugs and altered cancer cells drug sensitivity. We showed the involvement of the ERK1/2-RUNX2 axis. These results mirror the previous observation that ERK-mediated RUNX2 activation is a critical step in the response of mesenchymal cells to biomechanical forces (17). Given the mesenchymal features of ES tumors, it is not surprising that ES cells transduce mechanical forces activating similar signaling pathways. Although, it is not clear whether RUNX2 is a druggable candidate, targeting the ERK1/2-RUNX2 axis could emerge as a promising therapeutic strategy in ES.
Receptor tyrosine kinase inhibitors, that showed promising results in ES preclinical models, failed to show benefit in patients (16, 41). In the mechanically-competent tumor models, we found reduced apoptosis and increased proliferation following drug treatments. These findings tie in with a growing body of evidence linking mechanobiological factors to cancer resistance to therapy. Previous studies have shown that intracellular forces caused by cells response to matrix stiffness modulate proliferation and drug response (42, 43). In addition to the cytoskeletal tension related to cell adhesion, also extracellular forces can activate signaling pathways in cells (44). In fact, several in vitro studies have shown that shear stresses arising from fluid flow modulate drug response in many types of cancer including Ewing sarcoma (8).
Next generation preclinical models rely on the use of patient-derived cells as a viable alternative to cancer cell lines (45). We showed that ES patient-derived cells cultured in monolayer have elevated RUNX2 expression, ERK1/2 phosphorylation levels, and RTK inhibitor resistance compared to ES cell lines. Nonetheless, extensive monolayer culture would inevitably alter their heterogeneity and drug sensitivity. Currently the alternative is to expand the patient-derived xenografts in mice (9), but cost and ethical issues are potential limitations. A promising strategy to overcome these issues is the use of 3D models combined with bioreactor cultures that recapitualte key aspects of the tumor microenvironment in vitro. Here, we showed the feasibility of this approach by culturing patient-derived cells on the bioengineered tumor model in the presence of mechanical loads. In this biomimetic microenvironment, ES cells showed increased resistance to RTK inhibitors and high RUNX2 expression.
Despite RUNX2 is mediator of invasion and survival in primary and metastatic bone tumors, its expression in ES is not clear (19, 46). A recent study reported that the ES-specific fusion protein EWS-FLI binds and modulates RUNX2, inhibiting its expression in 2D cell cultures (47), consistent with our findings. RUNX2 is ectopically expressed in prostate and breast cancer metastasis into bone, suggesting microenvironmental regulation of its functions (46, 48, 49). By analyzing human tissue microarray datasets, we found that ES tumors have higher RUNX2 levels compared to a broad range of healthy tissues. Notably, RUNX2 levels drop sharply in ES cell lines cultured on flat (2D) tissue culture dishes. In addition, our genomic data showed that ES patients with high RUNX2 levels had a poor overall survival probability. These results support the clinical significance of RUNX2 in ES patients and highlight the need to retain its native expression in preclinical tumor models.
In summary, we demonstrate that mechanical stimulation regulates drug sensitivity of cancer cells through the activation of important mechanotransduction signaling pathways. Bioengineered models of human tumors – incorporating mechanical loading regimens – could thus find utility in basic research of solid tumors and studies of induced drug resistance.
Advances toward creating tissue-engineered bone for regenerative medicine applications have led to the development of in vitro models for studying bone cancer, disease progression, and drug screening in the bone microenvironment. However, existing 3D bone cancer models, including the one presented in this work, only incorporate some features of the complex microenvironment of a tumor. In the future, models will combine physical properties (rigidity, fluid flow, compression, pore size) and different cell types (osteoblasts, osteoclasts, endothelial cells, fibroblasts, immune cells), allowing scientist to better understand the interplay between cancer cells and the surrounding bone microenvironment.
We believe that combining tissue-engineering methods with organ-on-a-chip technologies will lead to new pharmacodynamics and pharmacokinetics insights, and facilitate drug development. Ultimately, advanced in vitro and in vivo tumor models are expected to have a major impact on both discovery and clinical translation in cancer, leading to significant patient benefit.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the NIH funding of this work (EB002520 and EB 17103).
Footnotes
Author contributions
A.M.-K., A.V., and G.V.-N designed research; A.M.-K., A.V., A.C., K.Y., S.R.A. performed research; A.M.-K., A.C. and G.V.-N analyzed data; and A.M.-K, M.R.T., and G.V.-N wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Begley CG, Ellis LM. Raise standards for preclinical cancer research. Nature. 2012;483(7391):531–3. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- 2.Waring MJ, Arrowsmith J, Leach AR, Leeson PD, Mandrell S, Owen RM, et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat Rev Drug Discov. 2015;14(7):475–86. doi: 10.1038/nrd4609. [DOI] [PubMed] [Google Scholar]
- 3.Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. Clinical development success rates for investigational drugs. Nat Biotechnol. 2014;32(1):40–51. doi: 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- 4.Jacks T, Weinberg RA. Taking the study of cancer cell survival to a new dimension. Cell. 2002;111(7):923–5. doi: 10.1016/s0092-8674(02)01229-1. [DOI] [PubMed] [Google Scholar]
- 5.Mak IW, Evaniew N, Ghert M. Lost in translation: animal models and clinical trials in cancer treatment. Am J Transl Res. 2014;6(2):114–8. [PMC free article] [PubMed] [Google Scholar]
- 6.Zervantonakis IK, Hughes-Alford SK, Charest JL, Condeelis JS, Gertler FB, Kamm RD. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc Natl Acad Sci U S A. 2012;109(34):13515–20. doi: 10.1073/pnas.1210182109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villasante A, Marturano-Kruik A, Vunjak-Novakovic G. Bioengineered human tumor within a bone niche. Biomaterials. 2014;35(22):5785–94. doi: 10.1016/j.biomaterials.2014.03.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santoro M, Lamhamedi-Cherradi SE, Menegaz BA, Ludwig JA, Mikos AG. Flow perfusion effects on three-dimensional culture and drug sensitivity of Ewing sarcoma. Proc Natl Acad Sci U S A. 2015;112(33):10304–9. doi: 10.1073/pnas.1506684112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, et al. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9(6):338–50. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fong EL, Wan X, Yang J, Morgado M, Mikos AG, Harrington DA, et al. A 3D in vitro model of patient-derived prostate cancer xenograft for controlled interrogation of in vivo tumor-stromal interactions. Biomaterials. 2016;77:164–72. doi: 10.1016/j.biomaterials.2015.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol. 2009;10(1):75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 12.Huang S, Ingber DE. Cell tension, matrix mechanics, and cancer development. Cancer Cell. 2005;8(3):175–6. doi: 10.1016/j.ccr.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Linch M, Miah AB, Thway K, Judson IR, Benson C. Systemic treatment of soft-tissue sarcoma-gold standard and novel therapies. Nat Rev Clin Oncol. 2014;11(4):187–202. doi: 10.1038/nrclinonc.2014.26. [DOI] [PubMed] [Google Scholar]
- 14.van Maldegem AM, Bhosale A, Gelderblom HJ, Hogendoorn PC, Hassan AB. Comprehensive analysis of published phase I/II clinical trials between 1990–2010 in osteosarcoma and Ewing sarcoma confirms limited outcomes and need for translational investment. Clin Sarcoma Res. 2012;2(1):5. doi: 10.1186/2045-3329-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8(3):241–54. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 16.Fleuren ED, Versleijen-Jonkers YM, Boerman OC, van der Graaf WT. Targeting receptor tyrosine kinases in osteosarcoma and Ewing sarcoma: current hurdles and future perspectives. Biochim Biophys Acta. 2014;1845(2):266–76. doi: 10.1016/j.bbcan.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Ge C, Long JP, Begun DL, Rodriguez JA, Goldstein SA, et al. Biomechanical stimulation of osteoblast gene expression requires phosphorylation of the RUNX2 transcription factor. J Bone Miner Res. 2012;27(6):1263–74. doi: 10.1002/jbmr.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito Y, Bae SC, Chuang LS. The RUNX family: developmental regulators in cancer. Nat Rev Cancer. 2015;15(2):81–95. doi: 10.1038/nrc3877. [DOI] [PubMed] [Google Scholar]
- 19.Sadikovic B, Thorner P, Chilton-Macneill S, Martin JW, Cervigne NK, Squire J, et al. Expression analysis of genes associated with human osteosarcoma tumors shows correlation of RUNX2 overexpression with poor response to chemotherapy. BMC Cancer. 2010;10:202. doi: 10.1186/1471-2407-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villasante A, Marturano-Kruik A, Ambati SR, Liu Z, Godier-Furnemont A, Parsa H, et al. Recapitulating the Size and Cargo of Tumor Exosomes in a Tissue-Engineered Model. Theranostics. 2016;6(8):1119–30. doi: 10.7150/thno.13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ambati SR, Shieh JH, Pera B, Lopes EC, Chaudhry A, Wong EW, et al. BO-1055, a novel DNA cross-linking agent with remarkable low myelotoxicity shows potent activity in sarcoma models. Oncotarget. 2016;7(28):43062–75. doi: 10.18632/oncotarget.9657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marturano-Kruik A, Villasante A, Vunjak-Novakovic G. Bioengineered Models of Solid Human Tumors for Cancer Research. Methods Mol Biol. 2016;1502:203–11. doi: 10.1007/7651_2016_353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marturano-Kruik A, Yeager K, Bach D, Villasante A, Cimetta E, Vunjak-Novakovic G. Mimicking biophysical stimuli within bone tumor microenvironment. Conference proceedings: Annual International Conference of the IEEE Engineering in Medicine and Biology Society IEEE Engineering in Medicine and Biology Society Annual Conference; 2015; pp. 3561–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bond M, Bernstein ML, Pappo A, Schultz KR, Krailo M, Blaney SM, et al. A phase II study of imatinib mesylate in children with refractory or relapsed solid tumors: a Children’s Oncology Group study. Pediatr Blood Cancer. 2008;50(2):254–8. doi: 10.1002/pbc.21132. [DOI] [PubMed] [Google Scholar]
- 25.Strumberg D, Awada A, Hirte H, Clark JW, Seeber S, Piccart P, et al. Pooled safety analysis of BAY 43–9006 (sorafenib) monotherapy in patients with advanced solid tumours: Is rash associated with treatment outcome? Eur J Cancer. 2006;42(4):548–56. doi: 10.1016/j.ejca.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 26.Frost BM, Eksborg S, Bjork O, Abrahamsson J, Behrendtz M, Castor A, et al. Pharmacokinetics of doxorubicin in children with acute lymphoblastic leukemia: multi-institutional collaborative study. Med Pediatr Oncol. 2002;38(5):329–37. doi: 10.1002/mpo.10052. [DOI] [PubMed] [Google Scholar]
- 27.Harley BA, Leung JH, Silva EC, Gibson LJ. Mechanical characterization of collagen-glycosaminoglycan scaffolds. Acta Biomater. 2007;3(4):463–74. doi: 10.1016/j.actbio.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 28.Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483(7391):589–93. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 29.Savola S, Klami A, Tripathi A, Niini T, Serra M, Picci P, et al. Combined use of expression and CGH arrays pinpoints novel candidate genes in Ewing sarcoma family of tumors. BMC Cancer. 2009;9:17. doi: 10.1186/1471-2407-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volchenboum SL, Andrade J, Huang L, Barkauskas DA, Krailo M, Womer RB, et al. Gene Expression Profiling of Ewing Sarcoma Tumors Reveals the Prognostic Importance of Tumor-Stromal Interactions: A Report from the Children’s Oncology Group. J Pathol Clin Res. 2015;1(2):83–94. doi: 10.1002/cjp2.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11(5):421–9. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 32.Burr DB, Milgrom C, Fyhrie D, Forwood M, Nyska M, Finestone A, et al. In vivo measurement of human tibial strains during vigorous activity. Bone. 1996;18(5):405–10. doi: 10.1016/8756-3282(96)00028-2. [DOI] [PubMed] [Google Scholar]
- 33.Guilak F, Ratcliffe A, Mow VC. Chondrocyte deformation and local tissue strain in articular cartilage: a confocal microscopy study. J Orthop Res. 1995;13(3):410–21. doi: 10.1002/jor.1100130315. [DOI] [PubMed] [Google Scholar]
- 34.Dubois SG, Shusterman S, Ingle AM, Ahern CH, Reid JM, Wu B, et al. Phase I and pharmacokinetic study of sunitinib in pediatric patients with refractory solid tumors: a children’s oncology group study. Clin Cancer Res. 2011;17(15):5113–22. doi: 10.1158/1078-0432.CCR-11-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scotlandi K, Remondini D, Castellani G, Manara MC, Nardi F, Cantiani L, et al. Overcoming resistance to conventional drugs in Ewing sarcoma and identification of molecular predictors of outcome. J Clin Oncol. 2009;27(13):2209–16. doi: 10.1200/JCO.2008.19.2542. [DOI] [PubMed] [Google Scholar]
- 36.Postel-Vinay S, Veron AS, Tirode F, Pierron G, Reynaud S, Kovar H, et al. Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nat Genet. 2012;44(3):323–7. doi: 10.1038/ng.1085. [DOI] [PubMed] [Google Scholar]
- 37.Roth RB, Hevezi P, Lee J, Willhite D, Lechner SM, Foster AC, et al. Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics. 2006;7(2):67–80. doi: 10.1007/s10048-006-0032-6. [DOI] [PubMed] [Google Scholar]
- 38.Vormoor B, Knizia HK, Batey MA, Almeida GS, Wilson I, Dildey P, et al. Development of a preclinical orthotopic xenograft model of ewing sarcoma and other human malignant bone disease using advanced in vivo imaging. PLoS One. 2014;9(1):e85128. doi: 10.1371/journal.pone.0085128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keir ST, Maris JM, Lock R, Kolb EA, Gorlick R, Carol H, et al. Initial testing (stage 1) of the multi-targeted kinase inhibitor sorafenib by the pediatric preclinical testing program. Pediatr Blood Cancer. 2010;55(6):1126–33. doi: 10.1002/pbc.22712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simmons CA, Matlis S, Thornton AJ, Chen S, Wang CY, Mooney DJ. Cyclic strain enhances matrix mineralization by adult human mesenchymal stem cells via the extracellular signal-regulated kinase (ERK1/2) signaling pathway. J Biomech. 2003;36(8):1087–96. doi: 10.1016/s0021-9290(03)00110-6. [DOI] [PubMed] [Google Scholar]
- 41.Maruwge W, D’Arcy P, Folin A, Brnjic S, Wejde J, Davis A, et al. Sorafenib inhibits tumor growth and vascularization of rhabdomyosarcoma cells by blocking IGF-1R-mediated signaling. OncoTargets and therapy. 2008;1:67–78. doi: 10.2147/ott.s3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schrader J, Gordon-Walker TT, Aucott RL, van Deemter M, Quaas A, Walsh S, et al. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology. 2011;53(4):1192–205. doi: 10.1002/hep.24108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin JW, Mooney DJ. Extracellular matrix stiffness causes systematic variations in proliferation and chemosensitivity in myeloid leukemias. Proc Natl Acad Sci U S A. 2016;113(43):12126–31. doi: 10.1073/pnas.1611338113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ozcivici E, Luu YK, Adler B, Qin YX, Rubin J, Judex S, et al. Mechanical signals as anabolic agents in bone. Nat Rev Rheumatol. 2010;6(1):50–9. doi: 10.1038/nrrheum.2009.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crystal AS, Shaw AT, Sequist LV, Friboulet L, Niederst MJ, Lockerman EL, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346(6216):1480–6. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Javed A, Barnes GL, Pratap J, Antkowiak T, Gerstenfeld LC, van Wijnen AJ, et al. Impaired intranuclear trafficking of Runx2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo. Proc Natl Acad Sci U S A. 2005;102(5):1454–9. doi: 10.1073/pnas.0409121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X, McGee-Lawrence ME, Decker M, Westendorf JJ. The Ewing’s sarcoma fusion protein, EWS-FLI, binds Runx2 and blocks osteoblast differentiation. J Cell Biochem. 2010;111(4):933–43. doi: 10.1002/jcb.22782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zaidi SK, Pande S, Pratap J, Gaur T, Grigoriu S, Ali SA, et al. Runx2 deficiency and defective subnuclear targeting bypass senescence to promote immortalization and tumorigenic potential. Proc Natl Acad Sci U S A. 2007;104(50):19861–6. doi: 10.1073/pnas.0709650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akech J, Wixted JJ, Bedard K, van der Deen M, Hussain S, Guise TA, et al. Runx2 association with progression of prostate cancer in patients: mechanisms mediating bone osteolysis and osteoblastic metastatic lesions. Oncogene. 2010;29(6):811–21. doi: 10.1038/onc.2009.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.