Abstract
Retroviral DNA integration takes place within the context of the intasome nucleoprotein complex. X-ray crystal structures of functional spumaviral intasomes were previously revealed to harbor a homotetramer of integrase, and it was generally believed that integrase tetramers catalyzed the integration of other retroviruses. The elucidation of new structures from four different retroviruses over the past year has however revealed this is not the case. The number of integrase molecules required to construct the conserved intasome core structure differs between viral species. While four subunits suffice for spumaviruses, α- and β-retroviruses require eight and the lentiviruses use up to sixteen. Herein we described these alternative architectures, highlighting both evolutionary and structural constraints that result in the different integrase-DNA stoichiometries across Retroviridae.
Introduction
Integration of the linear viral DNA that is made by reverse transcription into host cellular chromatin is a critical step in the retroviral life cycle. To accomplish integration, the viral enzyme integrase (IN) catalyzes two consecutive reactions, 3′ processing and strand transfer. During 3′ processing, IN hydrolyzes the nascent viral DNA adjacent to invariant CA dinucleotides to yield recessed CAOH-3′ ends. In the nucleus IN uses the 3′-hydroxyl groups to cleave chromosomal target DNA, which at the same joins the viral DNA ends to host DNA 5′-phosphates. Both reactions occur via SN2 transesterification, assisted by a pair of divalent metal cations in the IN active site [1,2].
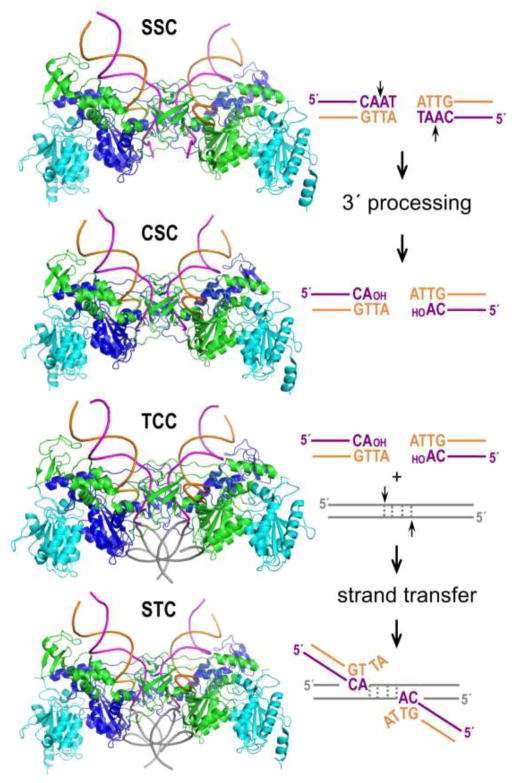
Retroviral integration occurs within the context of intasome nucleoprotein complexes that consist of an IN multimer engaging the two viral DNA ends. The initial assembly is most usually referred to as the stable synaptic complex (SSC), which becomes the cleaved synaptic complex (CSC) after 3′ processing. The target capture complex (TCC) forms when the CSC engages chromatin, and 3′ end joining yields the post-catalytic strand transfer complex (STC, Figure 1). Disassembly of the STC and DNA repair of the hemi-integrant, both of which are carried out by host enzymes, results in the formation of a stable provirus. The process of integration is highly similar to that of DNA transposition, and the retroviral INs belong to an extended superfamily that includes prokaryotic and eukaryotic DD(E/D) transposases (reviewed in [3]).
Figure 1.
Retroviral integration and intasome nucleoprotein complexes. Shown are representative PFV intasome structures (SSC, pdb accession code 4e7h; CSC, pdb code 3oy9; TCC, pdb code 3os2; STC, pdb code 3os0). The intasomes consist of purified recombinant IN protein and synthetic oligonucleotides that model the U5 ends of viral DNA. Two DNA binding IN protomers are painted blue and green, whereas supportive IN molecules are cyan. Adjacent DNA end sequences are color-coded to match the transferred magenta and non-transferred orange viral DNA strands in the structures. Short vertical arrows, scissile phosphodiester bonds. Target DNA is shown in grey; during strand transfer, the DNA is cleaved with a 4 bp staggered cut.
Prototype foamy virus intasome structures
The first retroviral intasome to be characterized structurally was the prototype foamy virus (PFV, from the spumavirus genus) CSC in 2010. Determined using X-ray crystallography, this structure was transformative in that it provided the initial glimpse of a functional IN multimer and also elucidated the mechanism of action of the clinical HIV IN strand transfer inhibitors [4]. Additional structures, which visualized the remaining PFV complexes along the integration pathway, were similarly reported over the ensuing 2 years (Figure 1) [2,5,6]. Today, the PFV intasome can be considered a minimalist assembly with the lowest order IN-to-viral DNA stoichiometry among retroviral intasomes characterized thus far [7]. Accordingly, we will overview its salient features before discussing the more complex assemblies that were elucidated only very recently.
Retroviral IN proteins are composed of three or four conserved domains joined via highly divergent flexible linkers. The N-terminal domain (NTD, a three-helical bundle stabilized by coordination of a Zn2+ ion), catalytic core domain (CCD, harboring the DDE catalytic triad within an RNase H fold), and C-terminal domain (CTD, a beta-barrel highly similar to SH3 and Tudor domains) are common to all of the proteins. The canonical domains do not have independent functions; each of them participates in essential protein-protein and protein-DNA interactions within the intasome structure. The INs from the spuma-, ε-, and γ-retroviruses additionally carry the NTD extension domain (NED), which is involved in interactions with viral DNA.
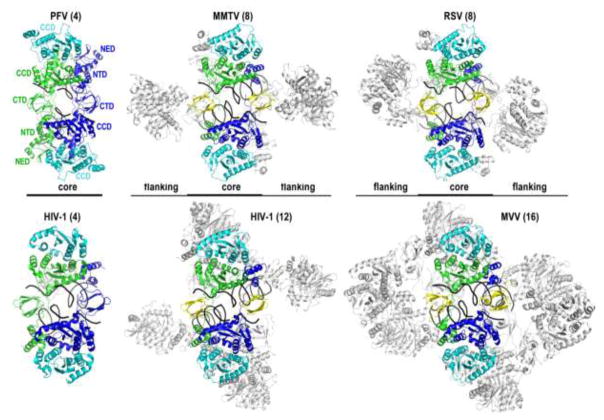
The PFV intasome is a 2-fold symmetric complex harboring four IN subunits, with viral DNA ends synapsed between a pair of IN dimers [4]. The inner IN molecules from each dimer (green and blue in Figures 1 and 2) are responsible for all interactions with viral DNA and accordingly provide their active sites to catalyze the 3′ processing and strand transfer reactions. The inner subunits interact with each other across the synaptic interface, and an underlying theme of the intasome is the swapping of the inner NTDs to function in trans with the opposing CCDs (Figure 2). The associated CTDs, which are referred to as synaptic CTDs, engage target DNA during integration [5] and help to rigidly bridge the halves of the intasome together (Figure 2). The outer IN subunits (cyan) fasten to the catalytic protomers via the CCD-CCD dimerization interface [4,8].
Figure 2.
Retroviral intasome structures and the CIC. The PFV structure is an underside view of the CSC relative to Figure 1, labeled for individual IN domains. The other intasome structures (MMTV CSC, pdb code 3jca; RSV STC, pdb code 5ejk; tetrameric HIV-1 STC, pdb code 5u1c; MVV CSC; pdb code 5m0q) are similarly oriented, size matched for common CIC components (colored). Synaptic CTDs that are donated from flanking IN dimers are shown in yellow. The portions of these structures that do not form part of the CIC are deemphasized by light gray. Viral DNA strands are black, and the target DNA strands from the RSV and HIV-1 STC structures, as well as the LEDGF/p75 IN-binding domain from the HIV-1 dodecamer, were omitted for clarity. Numbers indicate the number of IN molecules within each structure.
Characterization of a series of PFV intasomes revealed that the viral nucleoprotein architecture persists throughout the integration pathway without global perturbations. Hydrolysis of the 3′ dinucleotide(s) from viral DNA ends opens the intasomal active sites for the engagement of host DNA. The latter binds across the intasome structure, making contacts with both synaptic CTDs in a sharply bent conformation to provide the IN active sites with access to target phosphodiester bonds for strand transfer (Figure 1). Furthermore, the deformation may allow accumulation of conformational strain in target DNA, thought to be responsible for the ejection of the nascent virus-target DNA bond from the active site [2,5]. This subtle reconfiguration prevents the reversal of transesterification, which, by itself, is an energy-neutral process. More detailed reviews of the PFV intasome structure and the mechanism of retroviral DNA integration can be found elsewhere [9,10].
Surprising diversity of retroviral intasome architectures
Solution-based measures indicated that tetrameric IN forms could be the functionally relevant multimers for α-retroviral [11,12] and lentiviral [13–16] integration. Although arguments and some experimental evidence for larger species were also presented [15,17,18], the homo-tetrameric architecture observed in the PFV intasome was expected to be a universal feature of the retroviral integration machinery. However, recent intasome structures from four orthoretroviruses revealed both striking complexity and diversity. Reported from February 2016 to January 2017, the new structures elucidated intasomes from the β-retrovirus mouse mammary tumor virus (MMTV) [19], α-retrovirus Rous sarcoma virus (RSV) [20], lentivirus maedi-visna virus (MVV) [7], and lentivirus human immunodeficiency virus 1 (HIV-1) [21]. As three of the studies utilized single particle cryo-electron microscopy [7,19,21], much of the work benefited from the ongoing revolution in this structural biology platform (reviewed in [22]).
Whereas the PFV intasome is constructed from four IN molecules, eight were found in the MMTV and RSV structures (Figure 2). The IN assemblies within the latter two structures can be viewed as tetramers of dimers (Figure 2). Four core IN molecules situate in positions analogous to those in the PFV IN tetramer, forming the conserved catalytic IN core dimer (blue and green chains in Figure 2) with a pair of outer subunits (cyan). As in the PFV intasome, the catalytic subunits exchange their NTDs across the synaptic interface. The intasome is completed by insertion of CTDs, in these cases from the flanking IN dimers. An immediately obvious feature of the MMTV and RSV assemblies is the presence of a PFV intasome-like core, which we term the conserved intasome core (CIC, displayed in color in Figure 2). The factor necessitating the flanking subunits is the length of the linker that connects the CCD and CTD in α- and β-retroviral INs. While the CCD-CTD linker in PFV IN comprises 49 residues, which is sufficiently long to position the inner IN CTDs at the critical synaptic positions (Figure 2), the analogous regions in α-retroviral and β-retroviral INs harbor only 8 amino acids [19]. Accordingly, the CTDs of the catalytic IN protomers cannot assume the synaptic positions to complete the CIC structure. Instead, the α- and β-retroviruses evolved to utilize CTDs from additional IN molecules to fulfill the critical synaptic roles (Figure 2, yellow). As RSV and MMTV INs form stable dimers in solution [19,23,24], the resulting intasomes require two extra dimers, accounting for the observed octameric assembly. Results of biochemical complementation experiments indicate that the flanking IN subunits are required for MMTV IN strand transfer activity in vitro [19].
Analysis of representative IN sequences across Retroviridae indicated that the lengths of ε- and γ-retroviral IN CCD-CTD linkers, which are 55 and 61 residues, respectively, may support CIC formation from just four IN molecules, akin to the spumaviruses. The lengths of lentiviral and δ-retroviral IN CCD-CTD linkers, at ~19–22 residues, are intermediary to the comparatively short and long lengths noted for all other retroviruses [19]. The CCD-CTD linker of HIV-1 IN can be modeled sufficiently stretched to allow formation of the minimalist PFV-like tetrameric intasome [25,26]. However, the required fully extended conformation is incompatible with IN crystallography studies that observed this region in a compact α-helical configuration [27,28]. Moreover, the CCD-CTD linker is predicted to be α-helical across divergent members of the lentiviral genus [7].
HIV-1 intasome preparations revealed extensive heterogeneity, which we suspect resulted from the use of a mutant IN protein [21]. Wild-type HIV-1 IN displays limited solubility in isotonic salt-containing buffers (100–150 mM NaCl) and is minimally active with oligonucleotide viral DNA substrates in the absence of added cofactors such as the common lentiviral host factor LEDGF/p75 [18,29,30]. Given that PFV IN is much more amenable to in vitro studies [4,31,32], Craigie and colleagues assessed over 30 small DNA binding domains fused to the N-terminus of HIV-1 IN in an attempt to mimic the NED of the spumaviral protein. The screen identified Sso7d, an archaeal histone-like protein, which greatly improved the solubility and activity of the fusion construct [30]. While the mechanism of this effect is not fully understood, the mutation reduced the propensity of the protein to form multimers in solution [21]. Importantly, however, Sso7d-IN could support integration in the context of HIV-1 infection [30]. Perhaps owing to the approach of mimicking PFV IN, the Sso7d-IN construct formed a tetrameric IN intasome, although higher-order species were also observed in the assembly reactions [21]. The CIC in the tetrameric structure is made from the same components that formulate the PFV intasome (Figure 2), with the important caveat that the critical CCD-CTD linker could not be visualized in the electron density. The addition of the α helical IN-binding domain of LEDGF/p75 stabilized the higher-order species, including a dodecameric assembly that could be refined to high resolution [21]. In this complex, the CIC is formed between two tetramers, with the synaptic interface completed by insertion of CTDs from flanking IN subunits (Figure 2).
MVV intasomes assembled using wild-type protein harbor sixteen IN subunits that assume a tetramer-of-tetramers architecture. Although LEDGF/p75 was essential for the formation of the MVV intasome, the host factor dissociated during purification of the nucleoprotein complex. Just as in the case of the other characterized intasomes, the CIC is found at the center of the structure (Figure 2), and four IN tetramers participate in its formation. As in the dodecameric Sso7d-IN intasome, two tetramers provide a catalytic IN subunit each, and the synaptic CTDs are provided by flanking subunits. However, in contrast to the Sso7d-IN structure, the flanking subunits are each part of a complete tetramer. Overall, the dodecameric HIV-1 Sso7d-IN and the hexadecameric MVV intasomes are remarkably similar, although the former is missing an IN dimer from each flank. All of the CCD-CTD linkers in both structures assume nearly straight α-helical configurations, in agreement with crystallographic studies [7,27,28]. The CTD-CTD dimerization interface originally discovered via nuclear magnetic resonance spectroscopy is another previously underappreciated HIV-1 IN structure that is observed in the higher-order lentiviral intasome structures, but absent in the tetrameric Sso7d-IN intasome [7,21,33,34]. The remarkable conservation of the quaternary features in face of limited primary structure conservation between HIV-1 and MVV INs, which share less than 30% amino acid sequence identity [16], strongly argues for the relevance of the higher-order lentiviral assemblies. Although more work is clearly required, the results of mutagenesis experiments are consistent with the importance of the higher-order HIV-1 intasome during virus infection [21]. Nevertheless, the barebones tetrameric Sso7d-IN assembly, refined to a near-atomic resolution, provides the essential details of the CIC structure and the enzyme active sites much needed for IN strand transfer inhibitor development. Modifications that include an intact active site (an E152Q IN mutant was used in the recent work) and processed viral DNA end substrate (without target DNA) will optimally serve this purpose.
The compact α helical configuration of the CCD-CTD linkers in the higher-order lentiviral intasomes precludes the positioning of inner core protomer CTDs at the critical synaptic positions. Thus, the physical distance that can be spanned by inner core CCD-CTD linkers, prohibitively short for α- and β-retroviruses due to limited residue content and likewise short for the lentiviruses due to α helix formation, underlies the requirement for additional flanking IN protomers to complete the respective CIC structures. The propensity of lentiviral INs to form large multimers, such as tetramers, in the absence of DNA explains the resulting higher-order architectures, in particular for the MVV hexadecamer.
Collectively, the recent structural studies highlight a remarkable evolutionary plasticity in the construction of the intasome. While, in principle, one could envisage a functional intasome made of only two catalytic IN subunits, the smallest complex observed to-date is that of PFV, a bare-bones tetrameric CIC. Interestingly, the NED, NTD, and CTD of the outer IN subunits, which are disordered in all PFV intasome crystal structures [2,4–6], are dispensable for catalytic function in vitro [35]. However, the outer IN subunits were implicated in interactions with nucleosomal DNA during integration into chromatinized targets [36]. Therefore, the tetrameric CIC may represent the minimally functionally competent integration machine, although representative structures from three more retroviral genera are yet to be resolved.
As has been noted some time ago [37], DNA transposases tend to form functional multimers only on their DNA substrates, which is explained by the key roles that protein-DNA contacts play in the formation of transpososomes [38–40]. Applied to integration, the functional intasome would be constructed from available species in solution. Indeed, the available structures contain a consistent pattern, whereby intasomes assemble via the tetramerization of the predominant solution form of IN [7]. Thus, PFV IN is monomeric [31,32], whereas RSV and MMTV are predominantly dimers [19,23,24]. Although HIV-1 IN monomers can be detected, especially with Zn2+-free apo protein [41–43], tetramers were the prominent species reported by most studies [14,16,21,44–46], while MVV IN exists in a tetramer-octamer equilibrium in solution [7]. This model will benefit from additional IN biophysical characterizations and structural elucidation of representative intasomes from the remaining three retroviral genera.
Conclusions and perspectives
Recent work has significantly expanded our appreciation of retroviral intasome structure and function, as we now know that the CIC can form from a total of four, eight, or sixteen IN protomers. Based on intensive, decades-long approaches from several laboratories, elucidation of the lentiviral intasome structures represent a substantial leap forward. The elucidation of further intasome structures from wild-type primate lentiviral IN proteins should help to further clarify the structural basis of HIV DNA integration and the generation of clinical resistance to IN inhibitors. We also hope that current advancements will inspire characterization of the intasomes from the remaining retroviral genera, in particular from human T-lymphotropic virus 1, a δ-retrovirus that causes severe pathology.
One particular puzzling aspect of retroviral integration that has not been addressed is the mechanism of intasome assembly. Although we interpret the Sso7d-IN tetramer and dodecamer multimers as likely side effects of the heterologous fusion protein, the work raises other intriguing possibilities. It seems possible that the Sso7d IN tetramer could represent an intermediate in intasome assembly or disassembly. If so, the IN CCD-CTD linker would need to adopt two radically different forms, extended versus α-helical, during transition to (or from) the higher-order hexadecamer.
While the structural explanation for the seemingly excessive architectures of orthoretroviral intasomes is relatively straightforward, their functional consequences remain a mystery. One hypothesis, which could merit rigorous testing, is that higher-order stoichiometry facilitates high valency interactions with host chromatin, which could help the viral integration machinery to sense local enrichment of cues associated with transcriptional competency [7].
Highlights.
Retroviruses use 4, 8, or 16 integrase molecules for a common intasome core structure
A divergent linker region that connects two domains influences higher-order assembly
Intasomes seemingly represent the tetramerization of predominant solution IN species
Lentiviral structures will facilitate inhibitor design and drug resistance studies
Acknowledgments
Work in the authors laboratories is funded by US National Institutes of Health grant AI070042 (A.N.E.) and the Francis Crick Institute (P.C.), which receives its core funding from Cancer Research UK (FC001061), the UK Medical Research Council (FC001061), and the Wellcome Trust (FC001061). We thank Dmitry Lyumkis for providing coordinates of the higher-order Sso7d-IN STC intasome structure.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 2.Hare S, Maertens GN, Cherepanov P. 3′-processing and strand transfer catalysed by retroviral integrase in crystallo. EMBO J. 2012;31:3020–3028. doi: 10.1038/emboj.2012.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nowotny M. Retroviral integrase superfamily: the structural perspective. EMBO Rep. 2009;10:144–151. doi: 10.1038/embor.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4••.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232–236. doi: 10.1038/nature08784. Though now several years old, this breakthrough paper must nevertheless be highlighted, as it first revealed the functional architecture of the retroviral intasome. The structure, which was entirely unpredictable, forms the heart of all retroviral intasome structures since determined. This study also elucidated the mechanism of action of the clinical integrase inhibitors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maertens GN, Hare S, Cherepanov P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature. 2010;468:326–329. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin Z, Lapkouski M, Yang W, Craigie R. Assembly of prototype foamy virus strand transfer complexes on product DNA bypassing catalysis of integration. Protein Sci. 2012;21:1849–1857. doi: 10.1002/pro.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7••.Ballandras-Colas A, Maskell DP, Serrao E, Locke J, Swuec P, Jonsson SR, Kotecha A, Cook NJ, Pye VE, Taylor IA, et al. A supramolecular assembly mediates lentiviral DNA integration. Science. 2017;355:93–95. doi: 10.1126/science.aah7002. CryoEM structure of the functional maedi-virus virus cleaved synaptic complex reveals that a tetramer-of-tetramers (hexadecamer) integrase architecture mediates lentiviral DNA integration. The authors coin the “conserved intasome core” term to highlight a universal feature among all retroviral intasome structures elucidated to date. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 9.Cherepanov P, Maertens GN, Hare S. Structural insights into the retroviral DNA integration apparatus. Curr Opin Struct Biol. 2011;21:249–256. doi: 10.1016/j.sbi.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Lesbats P, Engelman AN, Cherepanov P. Retroviral DNA Integration. Chem Rev. 2016;116:12730–12757. doi: 10.1021/acs.chemrev.6b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bao KK, Wang H, Miller JK, Erie DA, Skalka AM, Wong I. Functional oligomeric state of avian sarcoma virus integrase. J Biol Chem. 2003;278:1323–1327. doi: 10.1074/jbc.C200550200. [DOI] [PubMed] [Google Scholar]
- 12.Bosserman MA, O’Quinn DF, Wong I. Loop202-208 in avian sarcoma virus integrase mediates tetramer assembly and processing activity. Biochemistry. 2007;46:11231–11239. doi: 10.1021/bi700197a. [DOI] [PubMed] [Google Scholar]
- 13.Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, Tarrago-Litvak L, Litvak S, Parissi V. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res. 2005;33:977–986. doi: 10.1093/nar/gki241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li M, Mizuuchi M, Burke TR, Jr, Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bera S, Pandey KK, Vora AC, Grandgenett DP. Molecular interactions between HIV-1 integrase and the two viral DNA ends within the synaptic complex that mediates concerted integration. J Mol Biol. 2009;389:183–198. doi: 10.1016/j.jmb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hare S, Di Nunzio F, Labeja A, Wang J, Engelman A, Cherepanov P. Structural basis for functional tetramerization of lentiviral integrase. PLoS Pathog. 2009;5:e1000515. doi: 10.1371/journal.ppat.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heuer TS, Brown PO. Photo-cross-linking studies suggest a model for the architecture of an active human immunodeficiency virus type 1 integrase-DNA complex. Biochemistry. 1998;37:6667–6678. doi: 10.1021/bi972949c. [DOI] [PubMed] [Google Scholar]
- 18.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 19••.Ballandras-Colas A, Brown M, Cook NJ, Dewdney TG, Demeler B, Cherepanov P, Lyumkis D, Engelman AN. Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function. Nature. 2016;530:358–361. doi: 10.1038/nature16955. The cryoEM structure of the mouse mammary tumor virus cleaved synaptic complex reveals that betaretroviral integration is mediated by a tetramer-of-dimers (octameric) integrase architecture. Phylogenetic analysis of integrase sequences across retroviruses indicates that the length of the linker that connects the catalytic core and carboxy terminal domains is an underlying feature of functional integrase-to-DNA stoichiometry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20••.Yin Z, Shi K, Banerjee S, Pandey KK, Bera S, Grandgenett DP, Aihara H. Crystal structure of the Rous sarcoma virus intasome. Nature. 2016;530:362–366. doi: 10.1038/nature16950. X-ray crystal structure of the Rous sarcoma virus strand transfer complex reveals an octameric tetramer-of-dimers integrase architecture mediates alpharetroviral integration. The target DNA portion of the nucleoprotein complex indicates integrase-host DNA contacts that are important for integration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21••.Passos DO, Li M, Yang R, Regensburg S, Ghirlando R, Jeon Y, Kvaratskhelia M, Craigie R, Lyumkis D. Cryo-EM structures and atomic model of the HIV-1 strand transfer complex intasome. Science. 2017;355:89–92. doi: 10.1126/science.aah5163. CryoEM analysis of the strand transfer complex constructed using a hyperactive mutant of HIV-1 integrase (Sso7d-IN) yields high resolution structures for the long-awaited HIV-1 intasome. The structures in particular should stimulate IN inhibitor development, and provide critical platforms for understanding the structural basis for drug resistance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bai X-c, McMullan G, Scheres SHW. How cryo-EM is revolutionizing structural biology. Trends Biochem Sci. 2015;40:49–57. doi: 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Andrake MD, Skalka AM. Multimerization determinants reside in both the catalytic core and C terminus of avian sarcoma virus integrase. J Biol Chem. 1995;270:29299–29306. doi: 10.1074/jbc.270.49.29299. [DOI] [PubMed] [Google Scholar]
- 24.Coleman J, Eaton S, Merkel G, Skalka AM, Laue T. Characterization of the self association of avian sarcoma virus integrase by analytical ultracentrifugation. J Biol Chem. 1999;274:32842–32846. doi: 10.1074/jbc.274.46.32842. [DOI] [PubMed] [Google Scholar]
- 25.Krishnan L, Li X, Naraharisetty HL, Hare S, Cherepanov P, Engelman A. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc Natl Acad Sci U S A. 2010;107:15910–15915. doi: 10.1073/pnas.1002346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson BC, Métifiot M, Ferris A, Pommier Y, Hughes SH. A homology model of HIV-1 integrase and analysis of mutations designed to test the model. J Mol Biol. 2013;425:2133–2146. doi: 10.1016/j.jmb.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen JC, Krucinski J, Miercke LJ, Finer-Moore JS, Tang AH, Leavitt AD, Stroud RM. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc Natl Acad Sci U S A. 2000;97:8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta K, Turkki V, Sherrill-Mix S, Hwang Y, Eilers G, Taylor L, McDanal C, Wang P, Temelkoff D, Nolte RT, et al. Structural basis for inhibitor-induced aggregation of HIV integrase. PLoS Biol. 2016;14:e1002584. doi: 10.1371/journal.pbio.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hare S, Shun MC, Gupta SS, Valkov E, Engelman A, Cherepanov P. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog. 2009;5:e1000259. doi: 10.1371/journal.ppat.1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, Jurado KA, Lin S, Engelman A, Craigie R. Engineered hyperactive integrase for concerted HIV-1 DNA integration. PLoS One. 2014;9:e105078. doi: 10.1371/journal.pone.0105078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delelis O, Carayon K, Guiot E, Leh H, Tauc P, Brochon J-C, Mouscadet J-F, Deprez E. Insight into the integrase-DNA recognition mechanism: A SPECIFIC DNA-BINDING MODE REVEALED BY AN ENZYMATICALLY LABELED INTEGRASE. J Biol Chem. 2008;283:27838–27849. doi: 10.1074/jbc.M803257200. [DOI] [PubMed] [Google Scholar]
- 32.Valkov E, Gupta SS, Hare S, Helander A, Roversi P, McClure M, Cherepanov P. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 2009;37:243–255. doi: 10.1093/nar/gkn938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eijkelenboom AP, Lutzke RA, Boelens R, Plasterk RH, Kaptein R, Hard K. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat Struct Biol. 1995;2:807–810. doi: 10.1038/nsb0995-807. [DOI] [PubMed] [Google Scholar]
- 34.Lodi PJ, Ernst JA, Kuszewski J, Hickman AB, Engelman A, Craigie R, Clore GM, Gronenborn AM. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry. 1995;34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 35.Li M, Lin S, Craigie R. Outer domains of integrase within retroviral intasomes are dispensible for catalysis of DNA integration. Protein Sci. 2016;25:472–478. doi: 10.1002/pro.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maskell DP, Renault L, Serrao E, Lesbats P, Matadeen R, Hare S, Lindemann D, Engelman AN, Costa A, Cherepanov P. Structural basis for retroviral integration into nucleosomes. Nature. 2015;523:366–369. doi: 10.1038/nature14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rice PA, Baker TA. Comparative architecture of transposase and integrase complexes. Nat Struct Biol. 2001;8:302–307. doi: 10.1038/86166. [DOI] [PubMed] [Google Scholar]
- 38.Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 39.Richardson JM, Colloms SD, Finnegan DJ, Walkinshaw MD. Molecular architecture of the Mos1 paired-end complex: the structural basis of DNA transposition in a eukaryote. Cell. 2009;138:1096–1108. doi: 10.1016/j.cell.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montano SP, Pigli YZ, Rice PA. The mu transpososome structure sheds light on DDE recombinase evolution. Nature. 2012;491:413–417. doi: 10.1038/nature11602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Gent DC, Elgersma Y, Bolk MW, Vink C, Plasterk RH. DNA binding properties of the integrase proteins of human immunodeficiency viruses types 1 and 2. Nucleic Acids Res. 1991;19:3821–3827. doi: 10.1093/nar/19.14.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taddeo B, Carlini F, Verani P, Engelman A. Reversion of a human immunodeficiency virus type 1 integrase mutant at a second site restores enzyme function and virus infectivity. J Virol. 1996;70:8277–8284. doi: 10.1128/jvi.70.12.8277-8284.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandey KK, Bera S, Grandgenett DP. The HIV-1 integrase monomer induces a specific interaction with LTR DNA for concerted integration. Biochemistry. 2011;50:9788–9796. doi: 10.1021/bi201247f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayouka Z, Rosenbluh J, Levin A, Loya S, Lebendiker M, Veprintsev D, Kotler M, Hizi A, Loyter A, Friedler A. Inhibiting HIV-1 integrase by shifting its oligomerization equilibrium. Proc Natl Acad Sci USA. 2007;104:8316–8321. doi: 10.1073/pnas.0700781104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKee CJ, Kessl JJ, Shkriabai N, Dar MJ, Engelman A, Kvaratskhelia M. Dynamic modulation of HIV-1 integrase structure and function by cellular lens epithelium-derived growth factor (LEDGF) protein. J Biol Chem. 2008;283:31802–31812. doi: 10.1074/jbc.M805843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jurado KA, Wang H, Slaughter A, Feng L, Kessl JJ, Koh Y, Wang W, Ballandras-Colas A, Patel PA, Fuchs JR, et al. Allosteric integrase inhibitor potency is determined through the inhibition of HIV-1 particle maturation. Proc Natl Acad Sci USA. 2013;110:8690–8695. doi: 10.1073/pnas.1300703110. [DOI] [PMC free article] [PubMed] [Google Scholar]