Abstract
Auxin regulates nearly all aspects of plant growth and development including cell division, cell elongation and cell differentiation, which are achieved largely by rapid regulation of auxin response genes. However, the functions of a large number of auxin response genes remain uncharacterized. Paclobutrazol Resistance (PRE) proteins are non-DNA binding basic helix-loop-helix transcription factors that have been shown to be involved in gibberellin and brassinosteroid signaling, and light responses in Arabidopsis. Here, we provide molecular and genetic evidence that PRE6, one of the six PRE genes in Arabidopsis, is an auxin response gene, and that PRE6 is involved in the regulation of auxin signaling. By using quantitative RT-PCR, we showed that the expression level of PRE6 was increased in response to exogenously applied IAA. GUS staining results also showed that the expression of GUS reporter gene in the PRE6p:GUS transgenic seedlings was elevated in response to auxin. Phenotypic analysis showed that overexpression of PRE6 in Arabidopsis resulted in auxin-related phenotypes including elongated hypocotyl and primary roots, and reduced number of lateral roots when compared with the Col wild type seedlings, whereas opposite phenotypes were observed in the pre6 mutants. Further analysis showed that PRE6 overexpression plants were hyposensitive, whereas pre6 mutants were hypersensitive to auxin in root and hypocotyl elongation and lateral root formation assays. By using protoplasts transfection, we showed that PRE6 functions as a transcriptional repressor. Consistent with this, the expression of the auxin response reporter DR5:GUS was decreased in PRE6 overexpression lines, but increased in pre6 mutants. When co-transfected into protoplasts, ARF5 and ARF8 activated the expression of the PRE6p:GUS reporter. Chromatin immunoprecipitation assays showed that ARF5 and ARF8 can be recruited to the promoter regions of PRE6. Taken together, these results suggest that PRE6 is an auxin response gene whose expression is directly regulated by ARF5 and ARF8, and that PRE6 is a transcriptional repressor that negatively regulates auxin responses in Arabidopsis.
Keywords: ARF5, ARF8, Arabidopsis, auxin, PRE6, transcription factor
Introduction
Auxin regulates many aspects of plant growth and development such as cell division, cell elongation and cell differentiation, mainly via rapid regulation of auxin response genes (Teale et al., 2006). The components involved in auxin signaling and the mechanisms of auxin perception and signal transduction have been largely elucidated during the last several decades (Chandler, 2016; Weijers and Wagner, 2016; Mironova et al., 2017). The core components involved in auxin signaling pathway include the TIR1/AFB (TRANSPORT INHIBITOR RESISTANT 1/AUXIN SIGNALING F-BOX) proteins, the ARF (AUXIN RESPONSE FACTOR) transcription factors, and the Aux/IAA (AUXIN/INDOLE-3-ACETIC ACID) proteins (Weijers and Wagner, 2016). The TIR1/AFB proteins are auxin receptors that can be activated by auxin molecules (Dharmasiri et al., 2005a,b; Kepinski and Leyser, 2005; Parry et al., 2009), the ARF transcription factors can bind to the TGTCTC auxin response elements in the promoter regions of the auxin response genes (Ulmasov et al., 1997a, 1999; Tiwari et al., 2003; Wang et al., 2005; Guilfoyle and Hagen, 2007; Chandler, 2016), whereas the Aux/IAA proteins are transcriptional repressors that can interact with ARF transcription factors (Tiwari et al., 2003, 2004).
In Arabidopsis, five ARFs including ARF5, ARF6, ARF7, ARF8, and ARF19 have been shown to be transcriptional activators (Wang et al., 2005; Guilfoyle and Hagen, 2007). When auxin levels in the cells are low, the Aux/IAA proteins are stable, and they can interact with the ARF activators that bound to the auxin response elements, thus repressing the activities of the ARF activators, resulting in repression of the auxin response genes (Li et al., 2011; Vernoux et al., 2011; Farcot et al., 2015). Elevated auxin levels in the cells will result in the activation of the TIR1 auxin receptor, leading to the ubiquitylation of the Aux/IAA proteins. Ubiquitinated Aux/IAA proteins will be degraded, allowing the activation of the auxin response genes by the ARF activators (Dharmasiri et al., 2005a,b; Kepinski and Leyser, 2005; Guilfoyle and Hagen, 2007; Mockaitis and Estelle, 2008). Although many auxin response genes have been reported, many others remain unidentified.
The bHLH (basic helix-loop-helix) transcription factor family is one of the largest transcription factor families in Arabidopsis, which regulates multiple aspects of plant growth and development (Bailey et al., 2003; Zhao et al., 2012). Among the bHLH genes in Arabidopsis, a total of six genes encode Paclobutrazol Resistances (PREs), which are atypical bHLH transcription factors that lack the basic domain required for DNA binding (Lee et al., 2006; Mara et al., 2010). Accumulated evidence suggest that PREs are involved in the regulation of plant growth and development as well as response to plant hormones and environmental stimuli such as temperature and light (Lee et al., 2006; Wang et al., 2009; Zhang et al., 2009; Mara et al., 2010; Bai et al., 2012; Castelain et al., 2012; Hao et al., 2012; Ikeda et al., 2012, 2013).
PRE1/BANQUO1(BNQ1)/BHLH136 is one of the first identified PRE transcription factors. It was identified by screening Arabidopsis activation-tagged transgenic lines with enhanced resistance to Paclobutrazol (Lee et al., 2006). Overexpression of PRE1 resulted in gibberellin related phenotypes such as longer hypocotyl, elongated petioles and early flowering (Lee et al., 2006). Overexpression of PRE2/BNQ2/BHLH134, PRE3/ATBS1/TMO7, PRE4/BNQ3/BHLH161 and PRE5 also resulted in a phenotype similar to that of the PRE1 transgenic plants (Lee et al., 2006), while the kidari/pre6-D mutant plants showed less rosette leaves and conditional longer hypocotyl under blue light (Hyun and Lee, 2006). Consistent with these observation, PRE1-RNAi, PRE2-RNAi and pre3 mutant plants flowered later than the Col wild type plants (Mara et al., 2010), suppressing the expression of PRE2, PRE5, and PRE6/KIDARI by artificial microRNA resulted in dwarfed phenotype and decreased sensitivity to gibberellin (Hyun and Lee, 2006; Bai et al., 2012; Oh et al., 2014), and knock down mutants of PRE1, PRE2, PRE5, and PRE6 by artificial micro-RNA resulted in smaller leaves and shorter hypocotyl in the transgenic plants (Bai et al., 2012; Oh et al., 2014).
PREs have been also shown to be involved in the regulation of brassinosteroid and light signaling. PRE6/KIDARI(KDR) was originally identified through a genetic screen of activation tagged mutants for long hypocotyl phenotype under blue and far-red light conditions (Hyun and Lee, 2006). PRE3 is able to suppress the phenotype of brassinosteroid receptor mutant bri1, and is involved in the regulation of light signaling (Wang et al., 2009; Castelain et al., 2012). PRE1 also plays a role in the regulation of brassinosteroid and light signaling (Zhang et al., 2009; Mara et al., 2010). On the other hand, the pre4 mutants have light signaling related phenotypes including pale-green sepals and carpels, decreased chlorophyll levels and late flowering (Mara et al., 2010), and suppression of PRE2, PRE5, and PRE6 led to decreased sensitivity to brassinosteroid, but increased sensitivity to light signaling (Hyun and Lee, 2006; Bai et al., 2012; Oh et al., 2014).
In addition to gibberellin and brassinosteroid related phenotypes, PRE3 overexpression plants showed auxin related phenotypes including longer primary root and decreased lateral root density, which can be restored by exogenously applied IAA treatment (Castelain et al., 2012). PRE6 has recently been reported to be involved in shade avoidance response, a phenomenon related to auxin signaling (Tian and Reed, 2001; Halliday et al., 2009; Sassi et al., 2013; Gommers et al., 2017). Most importantly, PRE3 has been identified as a target of ARF5/MONOPTEROS (MP) (Schlereth et al., 2010), and PRE1 has been reported to cooperate with ARF6 to regulate cell elongation in Arabidopsis (Oh et al., 2014), suggesting that PREs may be involved in the regulation of auxin signaling.
In this study, we report the identification of PRE6 as an auxin response gene, we show that PRE6 is a target of ARF5 and ARF8, and we provide evidence that PRE6 is a transcriptional repressor and it negatively regulates auxin responses in Arabidopsis.
Materials and Methods
Plant Materials and Growth Conditions
All the mutants and transgenic plants are in the Columbia-0 (Col) background, and Col Arabidopsis was used for plant transformation and protoplast isolation. The T-DNA insertion lines for PRE6/KDR (At1g26945), SALK_033495C, and SALK_048383C were obtained from the ABRC (Gommers et al., 2017), and designated as pre6-1 and pre6-2, respectively. The DR5:GUS transgenic plants have been described previously (Ulmasov et al., 1997b). The DR5:GUS/pre6 and DR5:GUS/35S:PRE6 plants were generated by crossing DR5:GUS transgenic plants with pre6-1 mutant or 35S:PRE6-1 transgenic plants, and selecting lines that were homozygous for both the DR5:GUS reporter gene and the pre6 mutant or the 35S:PRE6 transgenic plant, respectively.
For protoplast isolation and plant transformation, Col wild type Arabidopsis seeds were germinated and grown in soil pots as described previously (Wang et al., 2015).
For RNA isolation, Arabidopsis seeds were surface-sterilized and sown on 0.6% (w/v) phytoagar (PlantMedia) solidified 1/2 MS (Murashige and Skoog) medium with vitamins (PlantMedia) and 1% w/v sucrose as described previously (Wang et al., 2015; Dai et al., 2016).
For phenotypic and auxin response analyses in seedlings, surface-sterilized seeds were sown and grown on 1.5% (w/v) phytoagar solidified 1/2 MS medium.
All plants were grown in a growth chamber at 22°C, with a 16 h/8 h photoperiod and a photon density at ∼125 μmol m-2 s-1, or under darkness in the case of hypocotyl length analysis.
RNA Isolation, RT-PCR, and Quantitative RT-PCR (qRT-PCR)
To examine the expression of PREs and Aux/IAA genes in response to auxin, 14-day-old Col wild type, pre6 mutants and 35S:PRE6 transgenic seedlings were transferred into plates containing 10 μM IAA and kept in darkness for 4 h on a shaker, then total RNA was isolated using EasyPure Plant RNA Kit (TransGen Biotech) by following the manufacturer’s instructions. To examine the expression of PRE6 in the pre6 mutants and the 35S:PRE6 transgenic plants, total RNA was isolated from 7-day-old seedlings.
Two μg of total RNA was subjected to cDNA synthesis via Oligo(dT)-primed reverse transcription by using the EasyScript First-Strand cDNA Synthesis Super Mix (TransGene Biotech). Synthesized cDNA was used for PCR reactions, and expression of ACTIN2 (ACT2) was used as a control.
The primers used for qRT-PCR analysis of Aux/IAA genes have been described previously (Liu et al., 2015). The ACT2 primers used for qRT-PCR and the ACT7 primers used for ChIP assay have been described previously (Wang et al., 2015). All other primers used in this study for PCR, RT-PCR, and qRT-PCR are listed in Table 1.
Table 1.
Primers used in this study.
| Primers | Sequences |
|---|---|
| PRE1-NdeIF | CAACATATGTCGAACAGAAGATCAAGG |
| PRE1-SacIR | CAAGAGCTCTTACATGAGTAGGCTTCTAATAACG |
| PRE2-NdeIF | CAACATATGTCTTCTAGCAGAAGGTCG |
| PRE2-SacIR | CAAGAGCTCTTATCCATTAATCAAGCTCCTAATAAC |
| PRE3-NdeIF | CAACATATGTCGGGAAGAAGATCAC |
| PRE3-AflIIR | CAACTTAAGTTATTGGGTAAGTAAGCTTCTG |
| PRE4-NdeIF | CAACATATGTCTAGCAGAAAATCACGTTC |
| PRE4-SacIR | CAAGAGCTCCTACTGCATAAGCAAACTTCG |
| PRE5-NdeIF | CAACATATGTCTAACAGAAGATCAAGACAAAC |
| PRE5-SacIR | CAAGAGCTCTTACATGAGTAAGCTTCTAATCACG |
| PRE6-NdeIF | CAACATATGTCTAGCAGAAGATCATCACG |
| PRE6-SacIR | CAAGAGCTCTTAATAATTAAGCAAGCTCCTAATGATGG |
| PRE6p-PstIF | CAACTGCAGGTGGTTAGTGTAGAGTC |
| PRE6p-SacIR | CAAGAGCTCCTTCTTTCTTGATATATTATAAG |
| P1 | ATTATAAGTGTGTTTGTTTGGGTGT |
| P2 | TCAAGAAGTTGTTCTCGTGGGA |
| P3 | AGTCCGTATAATGTGCAGAGTC |
| P4 | ACGACTCGTATGAGACGATACA |
| P5 | ACATCAAAGGTCAAACATGGATG |
| P6 | GCCTATCTCTGCATCTACCACA |
Constructs
The effector constructs GD, CAT, LD-VP, ARF5, ARF6, ARF7, ARF8 and ARF19, and the reporter construct LexA-Gal4:GUS used for protoplast transient transfection assays have been described previously (Tiwari et al., 2001, 2004; Wang et al., 2005, 2007).
To generate HA-, GFP- or GD-tagged PRE6 constructs for plant transformation and protoplast transfection, full-length open reading frame (ORF) of PRE6 was amplified by RT-PCR using RNA isolated from Col wild type seedlings, and cloned in frame with an N-terminal HA, GFP or GD tag, respectively, into the pUC19 vector under the control of the double CaMV 35S promoter (Tiwari et al., 2001; Wang et al., 2005). The 35S:HA-PRE6 (referred as 35S: PRE6) and the 35S:GFP-PRE6 constructs in pUC19 was digested with NdeI and SacI and subcloned into the binary vector pPZP211 (Hajdukiewicz et al., 1994) for plant transformation.
To generate the PRE6p:GUS construct, a 3571bp DNA fragment immediately before the start codon of the PRE6 gene was PCR amplified using DNA isolated from Col wild type seedlings, and used to replace the OFP1 promoter in the OFP1p:GUS in pUC19 (Wang et al., 2007). The PRE6p:GUS construct in pUC19 was digested with PstI and SacI and subcloned into the binary vector pPZP211 for plant transformation.
Plant Transformation and Transgenic Plants Selection
About 5-week-old Col wild type plants with several mature flowers were transformed with the 35S:PRE6, 35S:GFP-PRE6 and PRE6p:GUS constructs in pPZP211, respectively, via Agrobacterium tumefaciens strain GV3101 by using the floral dip method (Clough and Bent, 1998). T1 transgenic plant seedlings were selected on 1/2 MS plates containing 50 μg/L kanamycin and 100 μg/L carbenicillin. A minimum of 15 independent transgenic lines with similar phenotypes were obtained for each construct, the expression status of the transformed genes was confirmed by RT-PCR or GUS staining, and two independent homozygous transgenic lines (T3 and/or T4) were used for further analysis.
Auxin Sensitivity Assays
In the hypocotyl elongation assay, sterilized seeds of the Col wild type, pre6 mutants and 35S:PRE6 transgenic lines were germinated and grown on vertically placed 1/2 MS plates supplied with different concentrations of IAA in a growth room under darkness. Photographs were taken from 5-day-old etiolated seedlings, and ImageJ software was used to measure the hypocotyl length.
In the primary root elongation and lateral root formation assays, 5-day-old seedlings of the Col wild type, pre6 mutants and 35S:PRE6 transgenic lines grown on a vertically placed 1/2 MS plates were transferred to new 1/2 MS plates supplemented with different concentrations of IAA, and grown vertically for another 5 days. The positions of the primary root tips were marked immediately and 5 days after the transfer. The new elongated primary roots between the two marks were measured, and the number of lateral roots was counted after 5 days of the transfer.
In all the assays, 25–35 seedlings for each genotype were used, and student t-test1 was used for statistical analysis.
Plasmid DNA Isolation, Protoplast Isolation and Transfection
Plasmid DNA of the reporter and effectors were isolated using the GoldHi Endo Free Plasmid Maxi Kit (CWBIO) by following the manufacturer’s protocol. Protoplasts were isolated from rosette leaves of 3–4-week-old Col wild type plants, and transfected as described previously (Tiwari et al., 2003; Wang et al., 2005, 2007, 2015; Tian et al., 2015; Dai et al., 2016; Zheng et al., 2016).
To examine the transcriptional activity of PRE6, plasmids of the reporter gene LexA-Gal4:GUS and the effector genes LD-VP and GD-PRE6 or GD were co-transfected into protoplasts. To examine the activation of PRE6 by ARF activators, plasmids of the reporter gene PRE6p:GUS and the ARF effector genes were co-transfected into protoplasts. To examine the association of ARF5 and ARF8 with the promoter region of PRE6, plasmids of ARF5 or ARF8 were transfected into protoplasts. Transfected protoplasts were incubated at room temperature in darkness for 20–22 h for GUS activity assays or ChIP assay.
GUS Staining and GUS Activity Assays
Glucuronidase (GUS) activities in transfected protoplasts were measured using a SynergyTM HT microplate reader.
To examine the auxin response of the PRE6p:GUS and effects of PRE6 on the expression of the DR5:GUS reporter gene, 7-day-old seedlings of PRE6p:GUS, DR5:GUS, DR5:GUS/pre6 and DR5:GUS/35S:PRE6 were treated with 10 μM IAA for 12 h, and then used for GUS activity assays either by staining or by quantitative measurement.
For GUS staining, Arabidopsis seedlings or different tissues or organs were incubated in solution containing X-Gluc (5-bromo-4-chloro-3-indolyl-β-D-glucuronide, Rose Scientific Ltd.) as described previously (Ulmasov et al., 1997b). For quantitative measurement, Arabidopsis seedlings were frozen in liquid nitrogen, then proteins were extracted and GUS activity was measured as described previously (Strader and Bartel, 2009).
Chromatin Immunoprecipitation (ChIP) Assay
Chromatin immunoprecipitation (ChIP) assay was performed by using the procedure described previously (Wang et al., 2015) with some modification. Briefly, after incubation, the transfected protoplasts were cross-linked in 1% formaldehyde for 20 min, the chromatin complex was then sheared by sonication and immunoprecipitated by HA-antibodies or rabbit pre-immune serum as control, and the Chromatin–antibody complexes were collected by using protein A-agarose beads (Millipore). After washing, the DNA-protein cross-links obtained were reversed at 65°C for 12 h, and DNA was purified using PCR Cleanup Kit (Axygen) for PCR reactions.
Microscopy
Photographs of the seedlings and the GUS stained tissues and organs were taken under a Motic K microscope equipped with an EOS 1100D digital camera. Photographs of the GFP fluorescence in the 35S:GFP-PRE6 transgenic seedlings were taken under an Olympus FV1000 confocal microscope.
Results
PRE6 Is an Auxin Response Gene
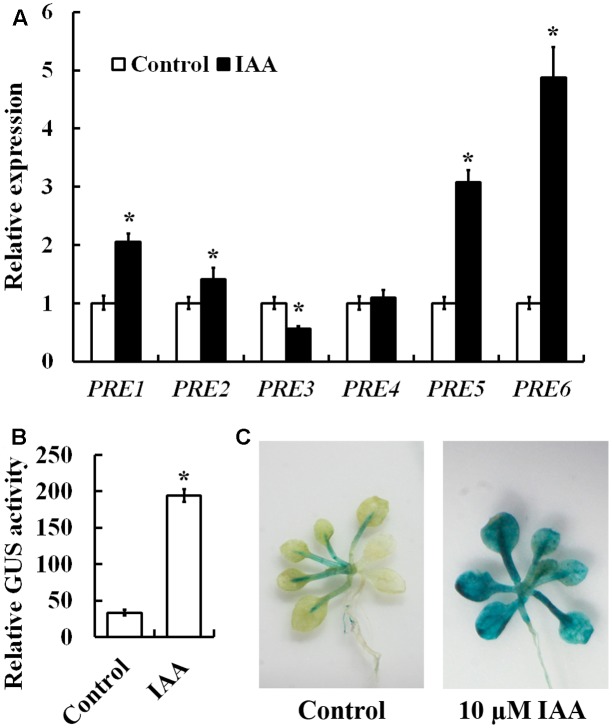
To examine whether PREs are involved in auxin signaling, we first examined the expression of PRE genes in response to auxin. Arabidopsis seedlings were treated with IAA, and qRT-PCR was used to examine the expression of PRE genes. As shown in Figure 1A, an ∼5-folds increase in response to auxin treatment was observed for PRE6, followed by ∼3, ∼2, and ∼1.5-folds for PRE5, PRE1, and PRE2, respectively. On the other hand, little increase was observed for PRE4 (Figure 1A). Unexpectedly, we found that expression of PRE3 was slightly suppressed by auxin treatment (Figure 1A).
FIGURE 1.
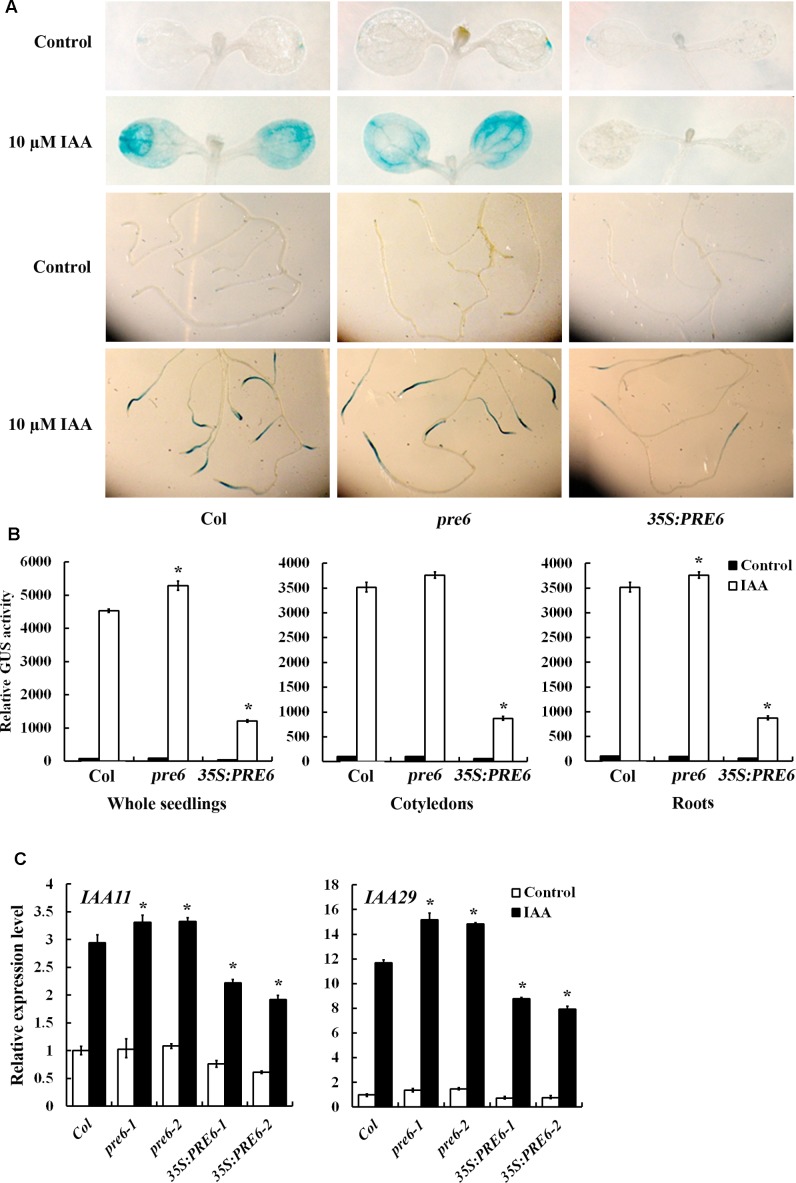
Induction of PRE genes by auxin and expression pattern of PRE6. (A) Expression of PREs in response to exogenously applied IAA. Fourteen-day-old seedlings were treated with 10 μM IAA for 4 h, then total RNA was isolated and subjected to quantitative RT-PCR analysis. ACT2 was used as a reference gene, and expression of each PRE gene in the absence of IAA was set as 1. Data represent the mean ± SD of three replicates. ∗Significantly different from control (p < 0.01). (B) Auxin response of the PRE6p:GUS in transfected protoplasts. Plasmids of PRE6p:GUS were transfected into protoplasts isolated from rosette leaves of 3 to 4-week-old Col wild type plants, and GUS activity was measured after the protoplasts were incubated at room temperature in darkness for 20–22 h. Data represent the mean ± SD of three replicates. ∗Significantly different from control (p < 0.01). (C) Auxin response of the PRE6p:GUS reporter gene in the transgenic seedlings. Fourteen-day-old seedlings of the PRE6p:GUS transgenic plants were treated with 10 μM IAA for 4 h, then GUS activity was stained 12 h after treatment by using X-Gluc. Left, control seedlings, right, seedlings treated with IAA.
PRE6 has been shown to be involved in light response and shade avoidance response (Hyun and Lee, 2006; Gommers et al., 2017). Because PRE6 is the most strongly induced PRE gene by auxin (Figure 1A), we thus wanted to investigate the potential role of PRE6 in auxin signaling in Arabidopsis. We first made a PRE6p:GUS reporter construct and examined its expression in response to exogenously applied IAA in transfected protoplasts, we found that GUS activities increased ∼5-folds in the presence of IAA (Figure 1B). We then generated PRE6p:GUS transgenic plants and assayed the auxin response of the reporter gene in plants. As shown in Figure 1C, GUS staining in the PRE6p:GUS transgenic seedlings was mainly observed in vascular system, and upon auxin treatment, GUS staining was enhanced in nearly all the tissues, indicating that the PRE6 promoter is functional. By using the PRE6p:GUS transgenic plants, we found that PRE6 was expressed in all the tissues and organs at seedling stage, but mainly expressed in the top of filament and style at maturity (Supplementary Figure S1).
Both Overexpression and Loss-of-Function of PRE6 in Arabidopsis Resulted in Auxin-Related Phenotypes
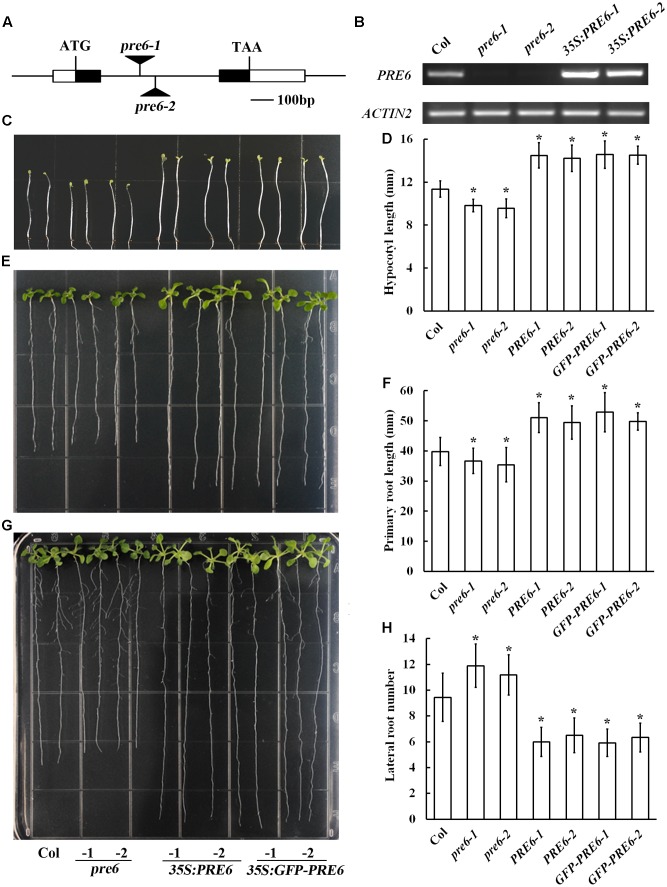
Previous studies indicated that overexpression of PRE6, as well as PRE1, PRE2, PRE3, and PRE4 increased hypocotyl length in the transgenic plants (Hyun and Lee, 2006; Lee et al., 2006; Mara et al., 2010; Hong et al., 2013; Gommers et al., 2017). To further examine whether Arabidopsis plants with altered expression level of PRE6 may exhibit auxin-related phenotypes, we generated Arabidopsis transgenic plants overexpressing PRE6, isolated two independent T-DNA insertion alleles of PRE6, pre6-1, and pre6-2 (Figure 2A), and compared their phenotypes by growing them side-by-side under same conditions. RT-PCR analysis showed that the PRE6 transcript was elevated in the 35S:PRE6 transgenic plants, confirming their overexpression status, and that the full-length transcript of PRE6 was undetectable in the pre6-1 and pre6-2 mutants, suggesting that they are likely representing loss-of-function alleles of PRE6 (Figure 2B).
FIGURE 2.
Phenotypes of the pre6 mutants and the PRE6 overexpression plants. (A) A diagram showing the T-DNA insertion sites in the pre6-1 and pre6-2 mutants. (B) Expression of PRE6 transcript in the pre6 mutants (pre6-1 and pre6-2) and transgenic plants overexpressing PRE6 (35S:PRE6-1 and 35S:PRE6-2). RNA was isolated from 14-day-old seedlings, and RT-PCR was used to examine the expression of PRE6. The expression of ACT2 was used as a control. (C) Photographs of the 5-day-old etiolated seedlings of the Col wild type, the pre6 mutants and the 35S:PRE6 transgenic plants. Sterilized seeds were grown on vertically placed 1/2 MS plates in darkness, and photographs were taken under a microscope. (D) Hypocotyl length of the 5-day-old etiolated seedlings of the Col wild type, the pre6 mutants and the 35S:PRE6 transgenic plants. Photographs were taken under a microscope, and ImageJ software was used to measure the hypocotyl length. Data represent the mean ± SD of 33–35 seedlings. ∗Significantly different from the Col wild type seedlings (p < 0.01). (E) Photographs of the 10-day-old light-grown seedlings of the Col wild type, the pre6 mutants and the 35S:PRE6 transgenic plants. Sterilized seeds were grown on vertically placed 1/2 MS plates, and photographs were taken under a microscope. (F) Primary root lengths of the 10-day-old light-grown seedlings of the Col wild type, the pre6 mutants and the 35S:PRE6 transgenic plants. Photographs were taken under a microscope, and ImageJ software was used to measure the primary root length. Data represent the mean ± SD of 33 seedlings. ∗Significantly different from the Col wild type seedlings (p < 0.01). (G) Photographs of the 14-day-old light-grown seedlings of the Col wild type, the pre6 mutants and the 35S:PRE6 transgenic plants. Sterilized seeds were grown on vertically placed 1/2 MS plates, and photographs were taken under a microscope. (H) Number of lateral roots in 14-day-old light-grown seedlings of the Col wild type, the pre6 mutants and the 35S:PRE6 transgenic plants. Data represent the mean ± SD of at least 25 seedlings. ∗Significantly different from the Col wild type seedlings (p < 0.01).
As expected, we found that dark-grown 35S:PRE6 transgenic seedlings had longer hypocotyls (Figures 2C,D). It has been reported recently that no difference on hypocotyl length is observed in light-grown pre6 mutants and Col wild type seedlings (Gommers et al., 2017). However, we found that the hypocotyl of pre6 mutants was slightly but statistically shorter than that of the Col wild type seedlings in dark-grown condition (Figures 2C,D). Similar to the results observed with hypocotyl length, we found that the primary roots were longer in the 35S:PRE6 transgenic seedlings, and shorter in the pre6 mutants (Figures 2E,F). On the other hand, we found that 35S:PRE6 transgenic plants produced fewer, but pre6 mutants produced more lateral roots than Col wild type seedlings (Figures 2G,H).
35S:PRE6 Transgenic Plants Are Hyposensitive Whereas pre6 Mutants Are Hypersensitive to Auxin
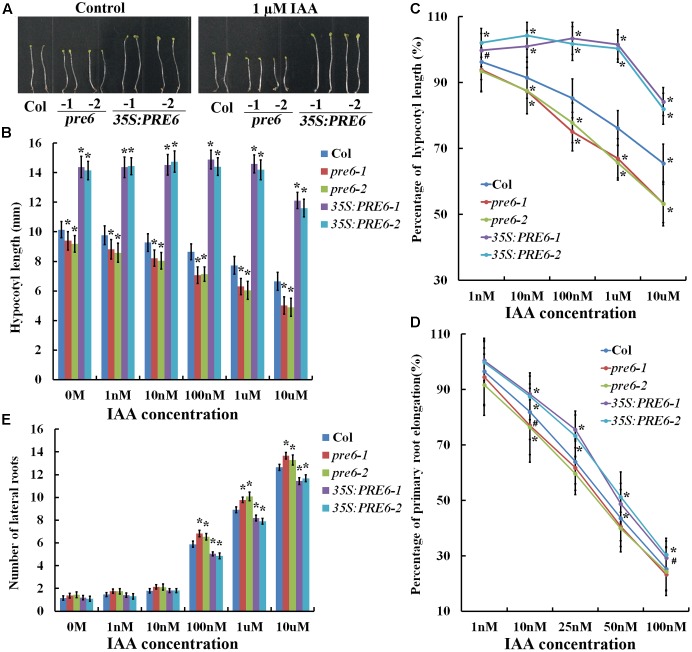
Having shown that Arabidopsis plants with altered expression level of PRE6 exhibited auxin-related phenotypes (Figure 2), we further examined the auxin response of the 35S:PRE6 transgenic plants and pre6 mutants in terms of hypocotyl elongation, primary root elongation and lateral root formation.
To examine hypocotyl elongation in response to auxin, seeds of Col wild type, pre6 mutants and 35S:PRE6 transgenic plants were sown directly on 1/2 MS plates containing different concentrations of IAA, and hypocotyl length was measured 5 days after the plates were kept in darkness. As shown in Figures 3A,B, auxin at all the concentrations tested inhibited hypocotyl elongation in both the Col wild type and the pre6 mutants, but showed little, if any effects in the 35S:PRE6 transgenic plants at lower concentrations (less than 1 μM). Analysis of percentage of hypocotyl elongation indicated that the pre6 mutants are more sensitive to auxin than the Col wild type plants (Figure 3C).
FIGURE 3.
Auxin sensitivities of the pre6 mutants and the PRE6 overexpression plants. (A) Hypocotyls in the 5-day-old etiolated seedlings of the Col wild type, the pre6 mutants and PRE6 transgenic plants in the absence and presence of 1 μM IAA. Sterilized seeds were grown on vertically placed 1/2 MS plates with or without 1 μM IAA in darkness, and photographs were taken under a microscope. (B) Hypocotyl length of the 5-day-old etiolated seedlings of the Col wild type, the pre6 mutants and PRE6 transgenic plants in the absence and presence of IAA. Photographs were taken under a microscope, and ImageJ software was used to measure the hypocotyl length. Data represent the mean ± SD of 33–35 seedlings. ∗Significantly different from the Col wild type seedlings (p < 0.01). (C) Percentage of hypocotyl length. The percentages of hypocotyl length were calculated by comparing the hypocotyl length of the seedlings at the presence of IAA to those of the control seedlings. Data represent the mean ± SD of 33–35 seedlings. ∗p < 0.01, #p < 0.05 significantly different from the Col wild type seedlings. (D) Percentage of primary root elongation. Five-day-old seedlings grown on a vertically placed 1/2 MS plates were transferred to new 1/2 MS plates supplemented with different concentrations of IAA, grown vertically for another 5 days, new elongated roots were measured, and the percentages of primary root elongation were calculated by comparing the primary root length of the seedlings at the presence of IAA to those of the control seedlings. Data represent the mean ± SD of 30 seedlings. ∗p < 0.01, #p < 0.05 significantly different from the Col wild type seedlings. (E) Lateral root formation. Five-day-old seedlings grown on a vertically placed 1/2 MS plates were transferred to new 1/2 MS plates supplemented with different concentrations of IAA, and number of lateral roots was counted 5 days after the transfer. Data represent the mean ± SD of 25–30 seedlings. ∗Significantly different from the Col wild type seedlings (p < 0.05).
To examine the effects of auxin on primary root elongation and lateral root formation, 5-day-old seedlings grown on vertically placed 1/2 MS plates were transferred to new plates containing different concentrations of IAA. Primary root elongation was measured, and number of lateral roots was counted 5 days after the transfer. We found that IAA at 1 nM has nearly no effects in the 35S:PRE6 transgenic plants, but inhibited primary root elongation in the Col wild type and pre6 mutants (Figure 3D). IAA at 10 nM and higher, however, inhibited primary root elongation in all the plants examined (Figure 3D). On the other hand, we found that the 35S:PRE6 transgenic plants produced less, whereas the pre6 mutants produced more lateral roots when compared with the Col wild type plants (Figure 3E). Taken together, these results suggest that the 35S:PRE6 transgenic plants were hyposensitive whereas pre6 mutants were hypersensitive to auxin, indicating that PRE6 may negatively regulate auxin signaling in Arabidopsis.
PRE6 Is a Transcriptional Repressor
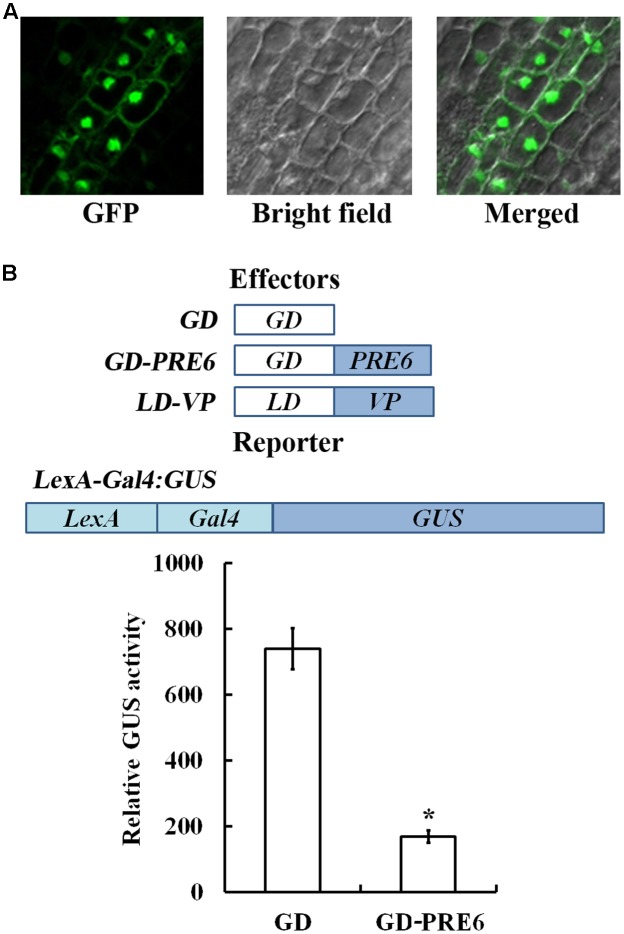
PRE proteins are atypical bHLH transcription factors that lack DNA binding basic motif in the bHLH domain, thus it was proposed that PRE proteins act as negative regulator of bHLH transcription factors via the formation of heterodimers (Hyun and Lee, 2006). Consistent with this, it has been shown that co-expression of PRE6 significantly reduced the transcriptional activation activities of HFR1 (Hong et al., 2013). Our results described above indicate that PRE6 is a negative regulator of auxin signaling in Arabidopsis. To further examine how PRE6 may regulate auxin signaling, we decided to examine whether PRE6 may regulate auxin response gene expression in Arabidopsis.
Previous results have only showed that PRE6 may not function as a transcriptional activator (Hong et al., 2013), thus we wanted to examine if PRE6 may function as a transcriptional repressor. Because previous protoplast transfection assays with PRE6-GFP plasmids indicated that PRE6 is localized in both nucleus and cytosol (Hong et al., 2013), we first wanted to examine if this is the case by generating transgenic plants expressing GFP-PRE6. As shown in Figure 2, the 35S:GFP-PRE6 transgenic seedlings showed a phenotype similar to that of the 35S:PRE6 transgenic seedlings, suggesting that GFP-PRE6 fusion proteins are functional. By examining GFP florescence in the roots of the 35S:GFP-PRE6 transgenic seedlings, we found that PRE6 is predominantly localized in the nucleus, but may also associate with membranes (Figure 4A). However, we could not rule out the possibility that the proteins were present in cytoplasm, but pushed to the cell periphery by the vacuole, which makes it looks like they were associated with membranes. We then examined the transcriptional activities of PRE6 by using Arabidopsis mesophyll protoplast transfection system. In this system, the transactivator LD-VP is recruited to the LexA promoter, whereas GD-PRE6 is recruited to Gal4 promoter of LexA-Gal4:GUS reporter gene. As shown in Figure 4B, co-transfection of the plasmids of the effector gene GD, LD-VP and the reporter gene LexA-Gal4:GUS resulted in the activation of the reporter gene, whereas co-transfection of the effector gene GD-PRE6, LD-VP and the reporter gene LaxA-Gal4:GUS resulted in the repression of the reporter gene activated by LD-VP, suggesting that PRE6 is a transcriptional repressor.
FIGURE 4.
PRE6 is a transcriptional repressor. (A) Subcellular localization of PRE6. GFP fluorescence in the root tips of 7-day-old 35S:GFP-PRE6 transgenic seedlings were observed and photographed under a fluorescence microscopy. Left, GFP channel; middle, bright field; right, merged image. (B) PRE6 is a transcriptional repressor. Plasmids of the effector LD-VP and GD-PRE6 or GD alone as a control, and the reporter LexA:Gal4:GUS (diagrammed on the top) plasmids were co-transfected into protoplasts isolated from rosette leaves of 3 to 4-week-old Col wild type plants, the protoplasts were incubated for 20–22 h at room temperature in darkness, and GUS activity was measured. Data represent the mean ± SD of three replicates. ∗Significantly different from the GD control (p < 0.01).
PRE6 Regulates Auxin Response Gene Expression
The results described above demonstrated that PRE6 is a transcriptional repressor and it is involved in the regulation of auxin response (Figures 3, 4). To determine how PRE6 may regulate auxin response in Arabidopsis, we examined the effects of PRE6 on the expression of auxin response genes. The transgenic plants with the integrated DR5:GUS auxin response reporter gene were crossed with 35S:PRE6 transgenic plants and the pre6 mutants, and the expression of the reporter genes in the absence and presence of auxin was examined by GUS staining. As shown in Figure 5A, in the absence of auxin, the GUS expression pattern in the 35S:PRE6 transgenic plants and the pre6 mutants was similar to that of the Col wild type. However, in the presence of auxin, GUS expression was dramatically increased in the Col wild type and the pre6 mutant seedlings, but not in the 35S:PRE6 transgenic seedlings (Figure 5A). Quantitative analysis showed that, in the presence of auxin, the GUS activity increased slightly in 35S:PRE6 transgenic seedlings, but decreased ∼4-folds as compared to the Col wild type and the pre6 mutant seedlings (Figure 5B).
FIGURE 5.
Expression of DR5: GUS reporter and endogenous Aux/IAA genes in the pre6 mutants and the PRE6 overexpression plants. (A) Expression of DR5:GUS reporter gene in the pre6 mutants and the PRE6 overexpression plants in response to exogenously applied IAA. Five-day-old seedlings with integrated DR5:GUS reporter gene were treated with 10 μM IAA for 12 h, then GUS activity was stained by using X-Gluc. Photographs were taken under a dissecting microscopy. (B) Quantitative analysis of GUS activities in the seedlings. After treated with 10 μM IAA for 12 h, seedlings were frozen in liquid nitrogen, proteins were extracted and GUS activity was measured. Data represent the mean ± SD of three replicates. ∗Significantly different from that of the control (p < 0.05). (C) Expression of IAA11 and IAA29 in response to exogenously applied IAA. Fourteen-day-old seedlings were treated with 10 μM IAA for 4 h, then total RNA was isolated and subjected to quantitative RT-PCR analysis. ACT2 was used as a reference gene, and expression of each IAA11 and IAA29 in Col wild type seedlings in the absence of IAA was set as 1. Data represent the mean ± SD of three replicates. ∗Significantly different from that of the control (p < 0.05).
We also examined the expression of endogenous Aux/IAA genes in response to exogenously applied IAA by using qRT-PCR. We found that the expression levels of IAA11 and IAA29 in response to IAA were increased in the pre6 mutants, but decreased in the 35S:PRE6 transgenic seedlings when compared to the Col wild type seedlings (Figure 5C).
ARF5 and ARF8 Directly Regulate the Expression of PRE6
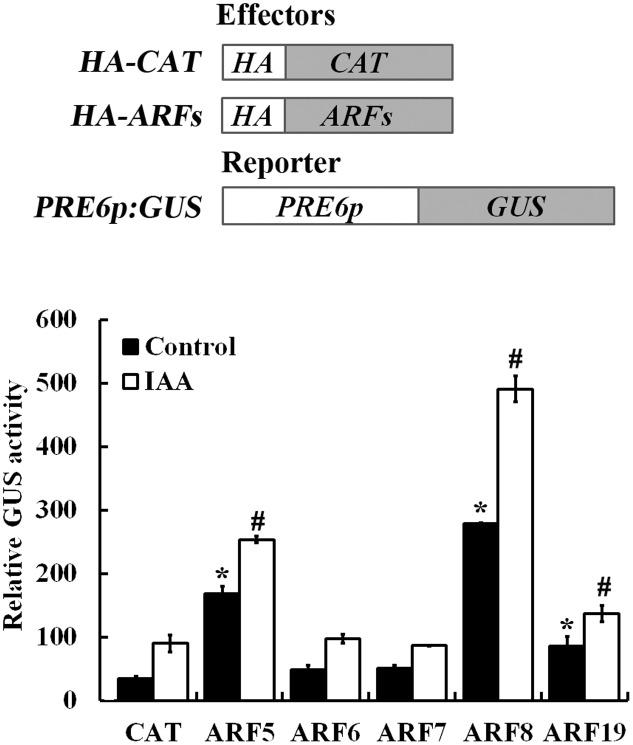
Auxin response genes are activated by ARF activators including ARF5, ARF6, ARF7, ARF8, and ARF19 (Wang et al., 2005). Because the expression of PRE6 was induced by auxin (Figure 1), we examined if any of the ARF activator may regulate the expression of PRE6 by using the Arabidopsis protoplast transient transfection assay. In this assay, plasmids of the ARF activator genes and the PRE6p:GUS reporter gene were co-transfected into the protoplasts, and GUS activity was measured after the transfected protoplasts were incubated overnight.
As shown in Figure 6, in the absence of auxin, ∼4 and ∼6-folds increased expression of the GUS gene was obtained when the reporter was co-transfected with ARF5 and ARF8, respectively, and GUS activity was further increased in the presence of auxin. The results also showed that co-transfection of ARF19 slightly induced the reporter gene expression, whereas co-transfection of ARF6 and ARF7 has little, if any effects on the expression of the reporter gene (Figure 6).
FIGURE 6.
ARF5 and ARF8 activate PRE6 expression in transfected protoplasts. Plasmids of the ARF effectors and the PRE6p:GUS reporter (diagrammed on the top) were co-transfected into protoplasts isolated from rosette leaves of 3 to 4-week-old Col wild type plants, the protoplasts were incubated for 20–22 h at room temperature in darkness, and GUS activity was measured. Co-transfection of CAT (chloramphenicol acetyltransferase) was used as a control. Data represent the mean ± SD of three replicates. ∗Significantly different from the CAT control in the absence of IAA (p < 0.05), #Significantly different from the CAT control in the presence of IAA (p < 0.05).
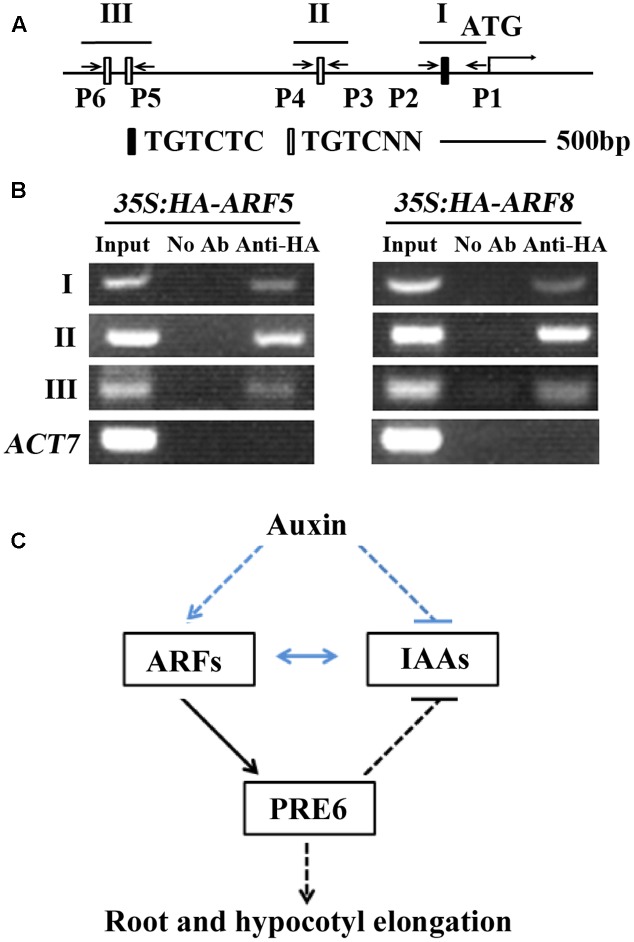
It is well-known that ARFs regulate auxin response gene expression via binding to the TGTCTC auxin response elements (Ulmasov et al., 1995, 1997a,b, 1999). Sequence analysis showed that PRE6 has one canonical TGTCTC element and three TGTC core elements within the 2 kb region upstream of its start codon (Figure 7A), indicating that ARF5 and ARF8 may regulate PRE6 expression via binding to its promoter. To test if this is the case, we performed ChIP assay to determine the association of ARF5 and ARF8 proteins to the promoter. Plasmids of ARF5 and ARF8 (with an HA tag) were transfected into Arabidopsis protoplasts, and anti-HA antibodies were used for immunoprecipitation. Immunoprecipitated DNA was amplified by using PCR with primers spanning the canonical TGTCTC element and the TGTC core elements (Figure 7A). Specific PCR products of the expected size were obtained for the three regions, but no bands were obtained when rabbit pre-immune serum was used for immunoprecipitation (Figure 7B). These results suggest that PRE6 is a target gene of ARF5 and ARF8.
FIGURE 7.
ARF5 and ARF8 are associated with the promoter region of PRE6. (A) ARF-binding sites in the 2 kb sequence upstream of the start codon of PRE6. I, II, and III indicate the regions for PCR amplification; P1 to P6 indicate the primers for PCR reactions. (B) Chromatin immunoprecipitation (ChIP) assay in transfected protoplasts. Plasmids of HA-ARF5 or HA-ARF8 were transfected into protoplasts isolated from rosette leaves of 3 to 4-week-old Col wild type plants, the transfected protoplasts were incubated for 20–22 h at room temperature in darkness, and ChIP assays were performed using anti-HA antibodies. Rabbit pre-immune serum was used as a mock control. Primer sets spanning the auxin response elements were used in PCR reactions. Amplification of ACT7 was used as a control. (C) A model illustrating the role of PRE6 in plant growth and development and auxin singling. ARFs directly activate the expression of PRE6, PRE6 regulates root and hypocotyls elongation, and it also serve as a feedback regulation loop in the regulation of auxin signaling via inhibiting the expression of Aux/IAA genes. Lines in blue indicate previously known results, and lines in black indicate results reported in this research.
Discussion
PREs have been reported to be involved in the regulation of gibberellin, brassinosteroid, temperature and light signaling in Arabidopsis (Lee et al., 2006; Wang et al., 2009; Zhang et al., 2009; Mara et al., 2010; Bai et al., 2012; Castelain et al., 2012; Oh et al., 2014). It has also been suggested that PRE1 is involved in the cross-talking between several different plant hormones including gibberellin, brassinosteroid and auxin (Oh et al., 2014), and the expression of PRE1 has been shown to be induced by several different plant hormones including gibberellin, auxin, and brassinosteroid (Lee et al., 2006; Zhang et al., 2009). In this study, we found that PRE6 is an auxin response gene, and it regulates auxin response in Arabidopsis.
PRE6 Is a Direct Target of ARF5 and ARF8
Among the six PRE genes, PRE3 was identified a target of ARF5 (Schlereth et al., 2010), and the expression of PRE1 and PRE5 has been shown to be induced by auxin (Zhang et al., 2009; Oh et al., 2014). Our qRT-PCR results showed that, in addition to PRE1, the expression of three other PRE genes, i.e., PRE2, PRE5, and PRE6 were also induced by auxin, with PRE6 being the highest responsive PRE gene to auxin (Figure 1A). Our results showed that the expression level of PRE1 increased only about twofold in response to auxin (Figure 1A), a result similar to the previous observation (Zhang et al., 2009). Unexpectedly, our results showed that the expression of PRE3 was slightly down regulated by auxin (Figure 1A), although PRE3 has been shown to be a target of ARF5 (Schlereth et al., 2010).
Auxin response of PRE6 was further confirmed by using PRE6p:GUS reporter gene in transfected protoplasts (Figure 1B), as well as in stable transformed plants (Figure 1C). The expression of auxin responsive genes are regulated by the interplay of ARFs activator and Aux/IAA proteins (Ulmasov et al., 1997a, 1999; Tiwari et al., 2003, 2004; Wang et al., 2005; Guilfoyle and Hagen, 2007; Chandler, 2016). By using protoplast transfection assays, we found that the expression of PRE6 was activated by ARF5 and ARF8, but not by AFR6 and ARF7, whereas ARF19 also slightly activated the expression of PRE6 (Figure 6). Sequencing analysis showed that there are four canonical or core auxin response elements in the promoter region of PRE6, and ChIP assay indicated that both ARF5 and ARF8 can be associated with those elements (Figure 7), indicating that PRE6 is a direct target of ARF5 and ARF8.
PRE6 Is Involved in the Regulation of Auxin Signaling in Arabidopsis
Alteration on hypocotyl and primary root length and lateral root numbers are observed in some well characterized auxin mutants such as iaa7, tir1-1, and iaa14 (Fukaki et al., 2002; Nakamura et al., 2006; Strader et al., 2008), and are considered as auxin-related phenotypes (Chapman and Estelle, 2009; Weijers and Wagner, 2016). Both the pre6 mutants and 35S:PRE6 transgenic seedlings showed auxin related phenotypes (Figure 2). Auxin response analysis showed that pre6 mutants are hypersensitive, whereas 35S:PRE6 transgenic plants are hyposensitive to auxin (Figure 3), indicating that PRE6 is a negative regulator of auxin signaling. Indeed, we found that the expression of the DR5:GUS reporter, as well as some of the endogenous Aux/IAA genes including IAA11 and IAA29 in response to exogenously applied IAA was reduced in the 35S:PRE6 transgenic plants (Figure 5). Consistent with this observation, transfection assays in protoplasts indicated that PRE6 function as a transcriptional repressor (Figure 4). However, considering that, expression of IAA11 and IAA29 was still highly induced by exogenously applied IAA (Figure 5), it is unlikely that PRE6 may directly regulate the expression of IAA11 and IAA29. It may be of great interest to find out how PRE6 may regulate the expression of Aux/IAA genes. Considering that stabilities of Aux/IAA proteins play an important role in auxin signaling, and PRE6 also function as a negative regulator of auxin signaling, it may also be of interest to examine whether PRE6 can be ubiquitinated like Aux/IAA proteins for degradation.
Auxin signaling is mainly controlled by the interplay of ARF activators and Aux/IAA protein. Our results showed that PRE6 is a direct target of ARF5 and ARF8 (Figures 6, 7), indicating that PRE6 functions downstream of ARF5 and ARF8 to regulate auxin signaling. Consistent with this, it has been reported that dark-grown arf6-2, arf8-3, and arf6-2 arf8-3 seedlings produced short hypocotyl (Nagpal et al., 2005; Oh et al., 2014), a phenotype similar to that of pre6 mutant seedlings (Figure 2). It should be noted that no difference in hypocotyl length was observed in light-grown pre6 mutants and Col wild type seedlings (Gommers et al., 2017). It is likely because light may affect auxin signaling, thus affecting plant growth and development.
PRE6 has been shown to regulate photomorphogenesis and light signaling via interacting with HFR1 (LONG HYPOCOTYL IN FAR-RED), a bHLH protein known to regulate photomorphogenesis in Arabidopsis, to interfere the interaction between HFR and PIF4 (PHYTOCHROMEINTERACTING FACTOR4), another bHLH protein known to regulate photomorphogenesis (Hyun and Lee, 2006; Hong et al., 2013). PIF4 has also been shown to interact with ARF6 to regulate hypocotyl cell elongation (Oh et al., 2014), thus it is likely that PRE6 may play a negative feedback role in auxin signaling by interfering the interaction of ARF activators and bHLH transcription factors, thus affecting the function of ARF activators. However, because PRE6 functioned as a transcriptional repressor in our protoplast transfection assays (Figure 4), and it suppressed the expression of the DR5:GUS reporter gene and some Aux/IAA genes in the transgenic plants, we could not rule out the possibility that PRE6 may be directly involved in the regulation of the expression of the auxin responsive genes.
PREs have been shown to function redundantly to regulate plant growth and development, as well as plant response to hormone such as gibberellins and brassinosteroid and light signaling (Hyun and Lee, 2006; Lee et al., 2006; Bai et al., 2012; Oh et al., 2014). PRE1 has been shown to cooperate with ARF6 to regulate hypocotyl cell elongation in Arabidopsis (Oh et al., 2014), and PRE3 has been shown to be a target of ARF5 (Schlereth et al., 2010). Considering that expression of PRE2 and PRE5 were also induced by exogenously applied IAA (Figure 1A), and PREs share high identities and similarities at the amino acid level (Mara et al., 2010), it is very likely that PREs may function redundantly to regulate auxin signaling in Arabidopsis.
We found in this study that PRE6 is an auxin response gene whose expression is regulated by ARF5 and ARF8, that PRE6 is a transcriptional repressor, and that PRE6 negatively regulates auxin signaling in Arabidopsis. Because Aux/IAA proteins can dimerize with ARFs to repress their functions, and inhibition of Aux/IAA genes by PRE6 may relieve this repression, it is very likely that ARF5 and ARF8 activated expression of PRE6 may function as positive feedback in auxin signaling (Figure 7C). Taken together, these results provide new insights into the function of PREs, and the regulation of auxin signaling in Arabidopsis.
Author Contributions
SW and J-GC conceived the study and designed the experiments. KZ, YW, NZ, QJ, XW, and CH performed the experiments. KZ and SW analyzed the data. KZ and SW drafted the manuscript. All the authors participated in the revision of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer DS and handling Editor declared their shared affiliation.
Acknowledgments
We thank Mr. Sajjad Ahmed for language editing of this manuscript.
Funding. This work was supported by the National Natural Science Foundation of China (31470297) and the National Key R&D Program of China (2016YFD0101902). KZ was supported by a visiting scholarship from the China Scholarship Council.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01813/full#supplementary-material
References
- Bai M. Y., Fan M., Oh E., Wang Z. Y. (2012). A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 24 4917–4929. 10.1105/tpc.112.105163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey P. C., Martin C., Toledo-Ortiz G., Quail P. H., Huq E., Heim M. A., et al. (2003). Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 15 2497–2502. 10.1105/tpc.151140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelain M., Le Hir R., Bellini C. (2012). The non-DNA-binding bHLH transcription factor PRE3/bHLH135/ATBS1/TMO7 is involved in the regulation of light signaling pathway in Arabidopsis. Physiol. Plant. 145 450–460. 10.1111/j.1399-3054.2012.01600.x [DOI] [PubMed] [Google Scholar]
- Chandler J. W. (2016). Auxin response factors. Plant Cell Environ. 39 1014–1028. 10.1111/pce.12662 [DOI] [PubMed] [Google Scholar]
- Chapman E. J., Estelle M. (2009). Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 43 265–285. 10.1146/annurev-genet-102108-134148 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Dai X., Zhou L., Zhang W., Cai L., Guo H., Tian H., et al. (2016). A single amino acid substitution in the R3 domain of GLABRA1 leads to inhibition of trichome formation in Arabidopsis without affecting its interaction with GLABRA3. Plant Cell Environ. 39 897–907. 10.1111/pce.12695 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. (2005a). The F-box protein TIR1 is an auxin receptor. Nature 435 441–445. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Weijers D., Lechner E., Yamada M., Hobbie L., et al. (2005b). Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9 109–119. [DOI] [PubMed] [Google Scholar]
- Farcot E., Lavedrine C., Vernoux T. (2015). A modular analysis of the auxin signaling network. PLOS ONE 10:e0122231. 10.1371/journal.pone.0122231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H., Tameda S., Masuda H., Tasaka M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29 153–168. 10.1046/j.0960-7412.2001.01201.x [DOI] [PubMed] [Google Scholar]
- Gommers C. M., Keuskamp D. H., Buti S., van Veen H., Koevoets I. T., Reinen E., et al. (2017). Molecular profiles of contrasting shade response strategies in wild plants: differential control of immunity and shoot elongation. Plant Cell 29 331–344. 10.1105/tpc.16.00790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle T. J., Hagen G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10 453–460. 10.1016/j.pbi.2007.08.014 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25 989–994. 10.1007/BF00014672 [DOI] [PubMed] [Google Scholar]
- Halliday K. J., Martínez-García J. F., Josse E. M. (2009). Integration of light and auxin signaling. Cold Spring Harb. Perspect. Biol. 1:a001586. 10.1101/cshperspect.a001586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Oh E., Choi G., Liang Z., Wang Z. Y. (2012). Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant 5 688–697. 10.1093/mp/sss011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S. Y., Seo P. J., Ryu J. Y., Cho S. H., Woo J. C., Park C. M. (2013). A competitive peptide inhibitor KIDARI negatively regulates HFR1 by forming nonfunctional heterodimers in Arabidopsis photomorphogenesis. Mol. Cells 35 25–31. 10.1007/s10059-013-2159-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y., Lee I. (2006). KIDARI, encoding a non-DNA binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol. Biol. 61 283–296. 10.1007/s11103-006-0010-2 [DOI] [PubMed] [Google Scholar]
- Ikeda M., Fujiwara S., Mitsuda N., Ohme-Takagi M. (2012). A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell 24 4483–4497. 10.1105/tpc.112.105023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Mitsuda N., Ohme-Takagi M. (2013). ATBS1 INTERACTING FACTORs negatively regulate Arabidopsis cell elongation in the triantagonistic bHLH system. Plant Signal. Behav. 8:e23448. 10.4161/psb.23448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S., Leyser O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435 446–451. 10.1038/nature03542 [DOI] [PubMed] [Google Scholar]
- Lee S., Lee S., Yang K. Y., Kim Y. M., Park S. Y., Kim S. Y., et al. (2006). Overexpression of PRE1 and its homologous genes activates gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 47 591–600. 10.1093/pcp/pcj026 [DOI] [PubMed] [Google Scholar]
- Li J. F., Bush J., Xiong Y., Li L., McCormack M. (2011). Large-scale protein–protein interaction analysis in Arabidopsis mesophyll protoplasts by split firefly luciferase complementation. PLOS ONE 6:e27364. 10.1371/journal.pone.0027364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Hu Q., Luo S., Li Q., Yang X., Wang X., et al. (2015). Expression of wild-type PtrIAA14.1, a poplar Aux/IAA gene causes morphological changes in Arabidopsis. Front. Plant Sci. 6:388. 10.3389/fpls.2015.00388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mara C. D., Huang T., Irish V. F. (2010). The Arabidopsis floral homeotic proteins APETALA3 and PISTILLATA negatively regulate the BANQUO genes implicated in light signaling. Plant Cell 22 690–702. 10.1105/tpc.109.065946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironova V., Teale W., Shahriari M., Dawson J., Palme K. (2017). The systems biology of auxin in developing embryos. Trends Plant Sci. 22 225–235. 10.1016/j.tplants.2016.11.010 [DOI] [PubMed] [Google Scholar]
- Mockaitis K., Estelle M. (2008). Auxin receptors and plant development: a new signaling paradigm. Annu. Rev. Cell Dev. Biol. 24 55–80. 10.1146/annurev.cellbio.23.090506.123214 [DOI] [PubMed] [Google Scholar]
- Nagpal P., Ellis C. M., Weber H., Ploense S. E., Barkawi L. S., Guilfoyle T. J., et al. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132 4107–4018. 10.1242/dev.01955 [DOI] [PubMed] [Google Scholar]
- Nakamura A., Nakajima N., Goda H., Shimada Y., Hayashi K., Nozaki H., et al. (2006). Arabidopsis Aux/IAA genes are involved in brassinosteroid-mediated growth responses in a manner dependent on organ type. Plant J. 45 193–205. 10.1111/j.1365-313X.2005.02582.x [DOI] [PubMed] [Google Scholar]
- Oh E., Zhu J. Y., Bai M. Y., Arenhart R. A., Sun Y., Wang Z. Y. (2014). Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3:e03031. 10.7554/eLife.03031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G., Calderon-Villalobos L. I., Prigge M., Peret B., Dharmasiri S., Itoh H., et al. (2009). Complex regulation of the TIR1/AFB family of auxin receptors. Proc. Natl. Acad. Sci. U.S.A. 106 22540–22545. 10.1073/pnas.0911967106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassi M., Wang J., Ruberti I., Vernoux T., Xu J. (2013). Shedding light on auxin movement: light-regulation of polar auxin transport in the photocontrol of plant development. Plant Signal. Behav. 8:e23355. 10.4161/psb.23355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth A., Möller B., Liu W., Kientz M., Flipse J., Rademacher E. H., et al. (2010). MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464 913–916. 10.1038/nature08836 [DOI] [PubMed] [Google Scholar]
- Strader L. C., Bartel B. (2009). The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid. Plant Cell 21 1992–2007. 10.1105/tpc.109.065821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L. C., Monroe-Augustus M., Bartel B. (2008). The IBR5 phosphatase promotes Arabidopsis auxin responses through a novel mechanism distinct from TIR1-mediated repressor degradation. BMC Plant Biol. 8:41. 10.1186/1471-2229-8-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale W. D., Paponov I. A., Palme K. (2006). Auxin in action: signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 7 847–859. 10.1038/nrm2020 [DOI] [PubMed] [Google Scholar]
- Tian H., Huo H., Dai X., Cheng Y., Zheng K., Wang X., et al. (2015). An ABA down-regulated bHLH transcription repressor gene, bHLH129 regulates root elongation and ABA response when overexpressed in Arabidopsis. Sci. Rep. 5:17587. 10.1038/srep17587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q., Reed J. W. (2001). Molecular links between light and auxin signaling pathways. J. Plant Growth Regul. 20 274–280. 10.1007/s003440010022 27597774 [DOI] [Google Scholar]
- Tiwari S. B., Hagen G., Guilfoyle T. J. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15 533–543. 10.1105/tpc.008417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S. B., Hagen G., Guilfoyle T. J. (2004). Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16 533–543. 10.1105/tpc.017384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S. B., Wang X.-J., Hagen G., Guilfoyle T. J. (2001). AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13 2809–2822. 10.1105/tpc.010289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T. J. (1997a). ARF1, a transcription factor that binds to auxin response elements. Science 276 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T. J. (1997b). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T. J. (1999). Dimerization and DNA binding of auxin response factors. Plant J. 19 309–319. 10.1046/j.1365-313X.1999.00538.x [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Liu Z. B., Hagen G., Guilfoyle T. J. (1995). Composite structure of auxin response elements. Plant Cell 7 1611–1623. 10.1105/tpc.7.10.1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T., Brunoud G., Farcot E., Morin V., Van den Daele H., Legrand J., et al. (2011). The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 7 508. 10.1038/msb.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhu Y., Fujioka S., Asami T., Li J., Li J. (2009). Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 21 3781–3791. 10.1105/tpc.109.072504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Chang Y., Guo J., Chen J. G. (2007). Arabidopsis Ovate Family Protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J. 50 858–872. 10.1111/j.1365-313X.2007.03096.x [DOI] [PubMed] [Google Scholar]
- Wang S., Tiwari S. B., Hagen G., Guilfoyle T. J. (2005). AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell 17 1979–1993. 10.1105/tpc.105.031096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang X., Hu Q., Dai X., Tian H., Zheng K., et al. (2015). Characterization of an activation-tagged mutant uncovers a role of GLABRA2 in anthocyanin biosynthesis in Arabidopsis. Plant J. 83 300–311. 10.1111/tpj.12887 [DOI] [PubMed] [Google Scholar]
- Weijers D., Wagner D. (2016). Transcriptional responses to the auxin hormone. Annu. Rev. Plant Biol. 67 539–574. 10.1146/annurev-arplant-043015-112122 [DOI] [PubMed] [Google Scholar]
- Zhang L. Y., Bai M. Y., Wu J., Zhu J. Y., Wang H., Zhang Z., et al. (2009). Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21 3767–3780. 10.1105/tpc.109.070441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Li X., Ma L. (2012). Basic helix-loop-helix transcription factors and epidermal cell fate determination in Arabidopsis. Plant Signal. Behav. 7 1556–1560. 10.4161/psb.22404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K., Tian H., Hu Q., Guo H., Yang L., Cai L., et al. (2016). Ectopic expression of R3 MYB transcription factor gene OsTCL1 in Arabidopsis, but not rice, affects trichome and root hair formation. Sci. Rep. 6:19254. 10.1038/srep19254 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.