Abstract
Corals that naturally thrive under extreme conditions are gaining increasing attention due to their importance as living models to understand the impact of global warming on world corals. Here, we present the first metagenomic study of viral communities in corals thriving in a thermally variable water body in which the temperature fluctuates between 11 and 39°C in different seasons. The viral assemblages of two of the most abundant massive (Porites harrisoni) and branching (Acropora downingi) corals in offshore and inshore reef systems in the northern Arabian Gulf were investigated. Samples were collected from five reef systems during summer, autumn and winter of 2011/2012. The two coral viromes contain 12 viral families, including 10 dsDNA viral families [Siphoviridae, Podoviridae, Myoviridae, Phycodnaviridae, Baculoviridae, Herpesviridae, Adenoviridae, Alloherpesviridae, Mimiviridae and one unclassified family], one-ssDNA viral family (Microviridae) and one RNA viral family (Retroviridae). Overall, sequences significantly similar to Podoviridae were the most abundant in the P. harrisoni and A. downingi viromes. Various morphological types of virus-like particles (VLPs) were confirmed in the healthy coral tissue by transmission electron microscopy, including large tailless VLPs and electron-dense core VLPs. Tailed bacteriophages were isolated from coral tissue using a plaque assay. Higher functional gene diversity was recorded in A. downingi than in P. harrisoni, and comparative metagenomics revealed that the Gulf viral assemblages are functionally distinct from Pacific Ocean coral viral communities.
Keywords: viral metagenomes, Acropora downingi, Porites harrisoni, extreme conditions, Arabian Gulf
Introduction
The field of coral virology remains in its infancy (Correa et al., 2016). Coral viruses remain the least-studied constituent of the coral holobiont, despite the fact that they are expected to infect and modulate all other members of the holobiont (for a review, see Vega Thurber and Correa, 2011). Viruses are able to influence living systems through both host mortality and horizontal gene transfer (Suttle, 2007) and are considered the largest reservoir of unexplored genetic diversity in the marine system (Monier et al., 2008). Presently, all available information about coral viruses is derived from studies performed on corals thriving in conditions of pristine warm water, where temperatures range between 16 and 32°C (Marhaver et al., 2008; Vega Thurber et al., 2008; Correa et al., 2013, 2016; Weynberg et al., 2014) and any increase or decrease in temperature outside the range causes coral to perish (Jokiel and Coles, 1990; Hoegh-Guldberg, 1999). No information is available about the viruses of corals that thrive naturally under extreme conditions, such as those dominating the Arabian Gulf.
The Arabian Gulf stands out among world seas because it is considered both one of the hottest seas and the most thermally varying coral-bearing water body in the world (Riegl and Purkis, 2012). The corals of the Arabian Gulf survive great fluctuations in water temperatures, which range from 39°C in summer to less than 11°C in winter (Spalding et al., 2001), making them a model system to study the impacts of rising global sea temperatures on corals (IPCC, 2007; Riegl and Purkis, 2012). In addition, as a semi-closed sea, the Gulf bears the direct impact of various human activities that increase the level of nutrients and pollutants in the system (Sheppard et al., 2010). It is well documented that coral reefs, which represent one of the world’s most productive marine ecosystems, are being rapidly depleted worldwide as a result of global and local disturbances that affect corals both directly and indirectly. The major global threats to coral reefs include climate change caused by global warming (Hoegh-Guldberg, 1999; Carpenter et al., 2008) and pollution (Hoegh-Guldberg, 2010). Recently, an important question has arisen regarding Arabian Gulf corals: how can Arabian Gulf corals survive fluctuations in water temperature better than corals elsewhere (see Riegl and Purkis, 2012)? Previous studies suggested that viruses might play a role in increasing the thermotolerance of their hosts similar to the case of Curvularia Thermal Tolerance Virus and its fungal host, Curvularia protuberata (Márquez et al., 2007). Also, Jacquet and Bratbak (2003) reported that phytoplankton Micromonas pusilla and Phaeocystis pouchetii became less sensitive to UVB when co-cultured with marine viruses. These findings support the idea that by transferring genetic material during viral infection, viruses can potentially enhance host metabolism, immunity, distribution, and evolution (Rohwer and Vega Thurber, 2009). Ongoing research to investigate the ability of corals to adapt to high temperature has focused either on the coral animal itself (Riegl and Purkis, 2012) or on the other members of the holobiont, especially Symbiodinium (Baker et al., 2004; Mostafavi et al., 2007; Shahhosseiny et al., 2011; Hume et al., 2013; Mahmoud and Al-Sarraf, 2016) and bacteria (Al-Dahash and Mahmoud, 2013; Mahmoud and Kalendar, 2016). Presently, no information is available about the viral assemblages of the Gulf corals.
The objective of this study was to determine the taxonomical and functional diversity of viruses in healthy Arabian Gulf corals. Therefore, we constructed and investigated two viral metagenomes to address major questions regarding the viral diversity associated with Acropora downingi and Porites harrisoni.
Materials and Methods
Sampling and Sample Sites
Coral reefs in the Gulf are categorized into inshore and offshore reef systems. Inshore reefs are in close proximity to land and are greatly influenced by adjacent human activities, including sewage discharge, oil production and seawater desalination activities, whereas offshore reefs are less affected by such activities (Carpenter et al., 1997; Al-Ghadban et al., 2007). A. downingi and P. harrisoni nubbins were collected by SCUBA from two inshore locations, including the Qit’at Benaya [28.6164° N, 48.4289° E] and Qit’at Alzor reefs [28.7607° N, 48.3912° E], and three offshore locations, including the Umm Al Maradim Island [28.6851° N, 48.6568° E], Kubbar Island [29.0754° N, 48.4926° E] and Qaro Island [28.8161° N, 48.7747° E] reef systems (Figure 1). The sites are located in the Arabian Gulf directly south of Kuwait and were selected based on the availability of previous data regarding coral-associated microbes at these sites (Al-Dahash and Mahmoud, 2013; Mahmoud and Al-Sarraf, 2016; Mahmoud and Kalendar, 2016).
FIGURE 1.
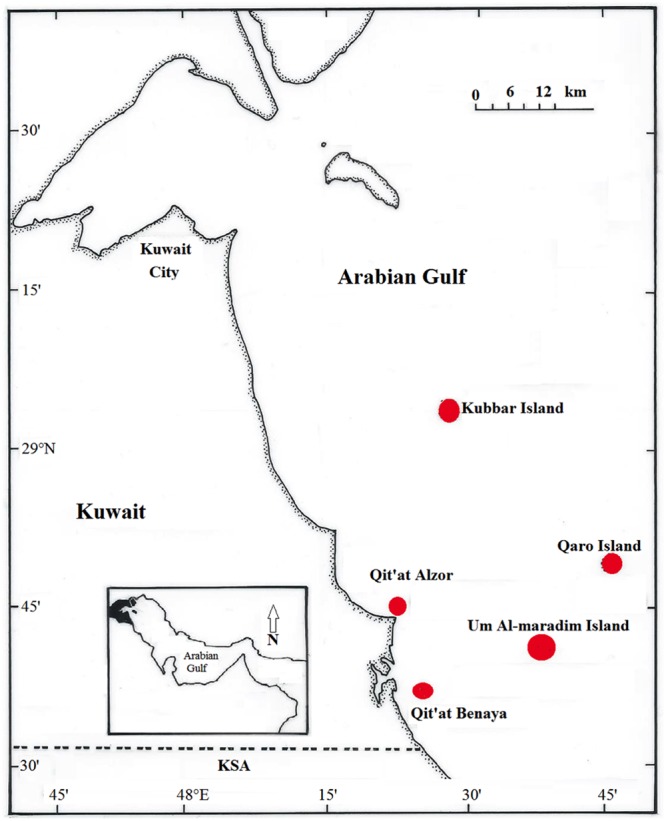
Kuwait map showing the three offshore reef systems (Kubbar Island, Qaro Island, and Umm Al Maradim Island) and the two inshore reef systems (Qit’at Alzor and Qit’at Benaya) sampled during the study.
Acropora downingi and P. harrisoni were chosen because they are ubiquitous in the Arabian Gulf and are categorized in the International Union of Conservation of Nature and Natural Resources (IUCN) red list of threatened species either as being of least concern or not having been assessed. A. downingi is more fragile (i.e., more susceptible to thermal stress) than P. harrisoni and recovers from diseases at a slower rate (Guest et al., 2016). As a consequence, Acroporidae-dominated reefs often become Poritidae-dominated reefs after incidents of massive bleaching (Coles and Riegl, 2013).
From each site, three coral colonies of each type were selected, tagged and repeatedly sampled in July and October 2011 and in January 2012. The water quality for each site at each sampling occasion is included in Supplementary Table S1. The sampled coral colonies remained healthy throughout the entire study period and exhibited no signs of stress or bleaching. Coral nubbins were collected by cutting 1-cm pieces of the three tagged colonies of both A. downingi and P. harrisoni from each site using a sterile hammer and chisel for massive coral and cutters for branching coral. Using different sets of sampling tools for different coral types reduced cross-contamination between colonies, and divers were wearing gloves throughout the sampling process. In addition, three subsamples of each colony were collected on each sampling date. All samples were placed underwater in sterile sampling bags, and they were packed on ice and returned to the laboratory for immediate processing.
Sample Processing
In the laboratory, the samples were divided into two sets. The first set was used for molecular analyses, whereas the second set was used for electron microscopy. The coral nubbins of the first set were washed with 0.02 μm-filtered sterile saline water (FSS, 3% NaCl) by placing coral nubbins in sterile stomacher bags and vigorously shaking them for 5 min. After shaking, samples were macerated in 10 ml of FSS using a sterile mortar and pestle (Vega Thurber et al., 2009b). The resulting slurry was liquefied with a syringe and used to extract viral DNA for metagenomic analysis. Coral nubbins of the second sample set were washed with FSS and immediately fixed in 2.5% v/v glutaraldehyde buffer (as described below).
Transmission Electron Microscopy (TEM)
The coral nubbins were processed according to the method described in Patten et al. (2008). The coral nubbins were fixed with 2.5% v/v glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4). Then they were decalcified using a solution made of 10% EDTA in 0.03 M NaOH and dehydrated in 30 to 100% of acetone series diluted in water (Wilson et al., 2005). Samples were kept in the dark at 4°C throughout the decalcification and fixation process. The decalcified samples were post-fixed in 1% osmium tetroxide in cacodylate buffer before being embedded in Epon 812 epoxy resin (TAAB laboratories, United Kingdom), sectioned and imaged by TEM (JEOL JEM-1200EX II; JEOL, Japan). 70 nm thick sections were used and they were positively stained using Uranly acetate and Lead citrate. Micrographs were obtained using a digital Erlangshen ES1000W CCD camera (Gatan, United States) and Gatan Digital Micrograph software (United States).
Isolation of Infectious Bacteriophages from Coral Using Plaque Assay (PA)
Bacteriophages associated with the coral nubbins in the current study were isolated using pure bacterial cultures. More than 100 different bacterial species were isolated from the two coral genera tissue and mucus from 2009 to 2012 (Al-Dahash and Mahmoud, 2013; Mahmoud and Kalendar, 2016). Coral nubbin homogenized slurry was filtered through sterile 0.22-μm Nalgene sterile bottom-top filter units (Sigma–Aldrich, United States), followed by a series of dilutions to bring the phage numbers to 20–200 viruses per aliquot as determined using SYBR green and epifluorescence microscopy (Noble and Fuhrman, 1998). The plaque assay was conducted following the protocol of Wilhelm and Poorvin (2001) (Supplementary Data), and bacteriophages were visualized using TEM following the protocol of Ackermann (2009).
Virus Purification and Concentration
To purify and concentrate viruses from coral samples, the liquefied coral-nubbin slurries were homogenized in a stomacher bag for 2 min. The resulting mixtures were filtered through 47 mm-diameter 8 μm-pore size nucleopore polycarbonate Whatman filters (Fisher Scientific, United States). The filtrate was centrifuged at 1000 × g for 15 min to remove the host cells and the coral skeleton. The supernatant was then sequentially filtered through 0.45- and 0.22-μm Nalgene sterile bottom-top filter units (Sigma–Aldrich, United States). The presence of viruses in the filtrate was confirmed using SYBR green and epifluorescence microscopy (Noble and Fuhrman, 1998). Viruses in the filtrate were concentrated using CsCl gradients according to the protocol described in Vega Thurber et al. (2009a). Briefly, CsCl concentrations of 1.7, 1.5, 1.35, and 1.15 g ml-1 were added to clear ultracentrifuge tubes. Next, identical volumes of the samples were added to the top of the created gradient. The tubes were centrifuged at 60,000 × g for 2 h at 4°C in a swinging-bucket rotor using a refrigerated Optima L-100 XP ultracentrifuge (Beckman Coulter, United States). The virions were collected using a sterile 18-gauge needle. The mouth of the needle was placed immediately below the 1.5 g ml-1 step, and viruses from the 1.3 to 1.5 g ml-1 fraction were collected. Samples were assessed with SYBR green and epifluorescence microscopy to confirm the presence of virions.
Viral DNA Extraction, Amplification and Sequencing
To remove naked DNA from each sample, 2.5 U of DNase I (Sigma–Aldrich, United States) was added per 100 μl of sample, and the mixture was incubated for 1 h at 37°C (Vega Thurber et al., 2009a). Both DNase I and RNA were heat-denatured by incubating the mixture at 70°C for 10 min. The viral DNA was then extracted using the QIAamp MinElute Virus Spin Kit (Qiagen, United States) by following the manufacturer’s protocol. The DNA concentration was determined, and DNA purity was confirmed using a Nanodrop nucleic acid analyser (Thermo Scientific, United States). The DNA of all the samples of a given coral host was pooled; this step facilitates the examination of a broader diversity of coral-associated viruses, as suggested by Marhaver et al. (2008).
The removal of bacterial and eukaryotic DNA from the extracted viral DNA was confirmed by screening the viral DNA samples for absence of amplification products with universal primers for 16S and 18S rDNA genes (Schafer and Muyzer, 2001).
A random-amplification PCR protocol described by Wang et al. (2003) was adapted for this work. A mixture containing 7 μl of genomic DNA (450 ng), 2 μl 5X Sequenase Buffer (13 units μl-1) (United States Biochemical, United States), and 1 μl Primer A (40 pmol μl-1) [GTT TCC CAG TCA CGA TCN NNN NNN NN] was incubated in a Gene Amp PCR system 9700 thermocycler (Applied Biosystems, United States) at 94°C for 2 min before being rapidly cooled to 10°C and held at 10°C for 5 min. Next, a sequenase mix (2.0 μl 5X Sequenase Buffer, 7.7 μl H2O, and 0.3 μl Sequenase) was prepared and added to the PCR tubes to give a total volume of 30 μl. The thermocycler temperature was ramped from 1 to 37°C over 8 min, at which point the temperature was maintained at 37°C for an additional 8 min. The temperature was then rapidly increased to 94°C and held for 2 min. The temperature was then decreased to 10°C and held for 5 min while 1.2 μl of diluted Sequenase (1:4 dilution) was added. The temperature was then increased from 10 to 37°C over 8 min and held for an additional 8 min before being increased to 94°C for an additional 8 min (Wang et al., 2003).
The resulting samples were amplified in 0.2-ml PCR tubes containing 6 μl of the produced DNA template, 4 μl of 50 mM MgCl2, 10 μl of 10X PCR Buffer, 1 μl of 25 mM dNTPs, 100 pmol μl-1 Primer B [GTTTCCCAGTCACGATC], 1 μl Taq polymerase (5 units μl-1) (hot start) and 77 μl water. The PCR program included 35 cycles of 30 s at 94°C, 30 s at 40°C, 30 s at 50°C and 1 min at 72°C. PCR amplicons were cleaned using a PCR clean-up spin kit (Qiagen, United States).
The samples were sequenced by the Yale Center for Genome Analysis facility (Connecticut, United States). Prior to sequencing, the samples were assessed for integrity on a bioanalyser and fragmented to ∼500 nt. The samples were sequenced on an Illumina HiSeq 2000 instrument using 76-nt paired-end sequencing.
Bioinformatics and Statistical Analysis
Prior to their assembly and annotation, reads were trimmed to remove primers using TagCleaner version 0.12 (Schmieder et al., 2010) and quality-trimmed for a minimum phred score of 20 using Trimmomatic version 0.36 (Bogler et al., 2014). The non-viral (non-target) sequence reads were identified and removed prior to assembly of contigs (Supplementary Data). All putative viral sequences were then assembled using Velvet version 1.2.10 (Zerbino and Birney, 2008) with k-mer lengths of 39 and 49. A final hybrid assembly was generated using Minimus II version 3.1.2 (Sommer et al., 2007) from Amos version 3.10 with a minimum overlap of 20 bp and 98% identity, and genes were predicted by FragGeneScan (Min et al., 2010). Viromes were annotated with a tBLASTx and BLASTp search (threshold E-value 10-5) against the GenBank viral non-redundant (nr and pep) and NCBI viral refseq databases (and protein database) and with MG-RAST version 3.2.45 (Meyer et al., 2008).
Variations in functional diversity between the Gulf coral viromes and Pacific coral viromes (Vega Thurber et al., 2009b) were determined using MG-RAST principal coordinates analysis (PCoA) and using XIPE-TOTEC software (Rodriguez-Brito et al., 2006). To produce MG-RAST PCoA for the compared viromes, the data were first normalized using DEseq (Anders and Huber, 2010) and then analyzed using Bray–Curtis distances using tools available through MG-RAST. Within XIPE-TOTEC, a non-parametric analysis of variance (ANOVA) was used to determine which metabolic subsystems in the MG-RAST data were significantly over- or underrepresented in the compared viromes. The Shannon index of diversity and sequence evenness scores for functional genes were calculated by following the protocol described in Dinsdale et al. (2008). The Gulf coral viral metagenome projects were submitted to GenBank under the following accession numbers: P. harrisoni (SRR5452095) and A. downingi (SRR5452096). Also the two viromes are submitted to MG-RAST under the following accession numbers: P. harrisoni (mgm4767658.3) and A. downingi (mgm4767672.3).
Results
Taxonomic Composition of the Two Gulf Coral Viromes
In total, 110,866,698 bp and 141,093,140 bp of raw read data were generated from A. downingi and Porites harrisoni, respectively, and an average of 21% of the reads were quality trimmed. The number of reads that remained from A. downingi and P. harrisoni after trimming was 81,703,324 and 118,925,078 bp, respectively. There was very little contamination with cellular gene markers, as demonstrated by the low number of matches to ribosomal proteins (0.3%). On average, 73% of the sequences in the two libraries did not match any nucleotide sequences in the NCBI database, based on tBLASTx analysis. Among the 17% of sequences in the two viromes that matched nucleotide sequences in the NCBI database, the majority of these sequences (97%) exhibited similarity to prokaryotic and eukaryotic genes rather than to viral genes, and only 2.3% were classified as viruses. The same results were confirmed using MG-RAST software.
The total number of viral reads from A. downingi that remained after removing the non-viral sequence reads was equal to 1,768,711 bp, which corresponded to 380 contiguous sequences with an average length of 204 bp and a maximum length of 3,257 bp; in P. harrisoni, the virome library was composed of 2,970,666 bp, with 321 contigs of an average length of 174 bp and a maximum length of 3,209 bp. The taxonomic compositions of the viral assemblages in the two viral metagenomes were determined (Figure 2). The tBLASTx results (Figures 2A,B) revealed that the two coral viromes contained 12 viral families, including ten dsDNA viral families [Caudovirales (Siphoviridae, Podoviridae, Myoviridae), Phycodnaviridae, Baculoviridae, Herpesviridae, Adenoviridae, Alloherpesviridae, Mimiviridae and one unclassified family], one ssDNA viral family (Microviridae) and one RNA viral family (Retroviridae).
FIGURE 2.
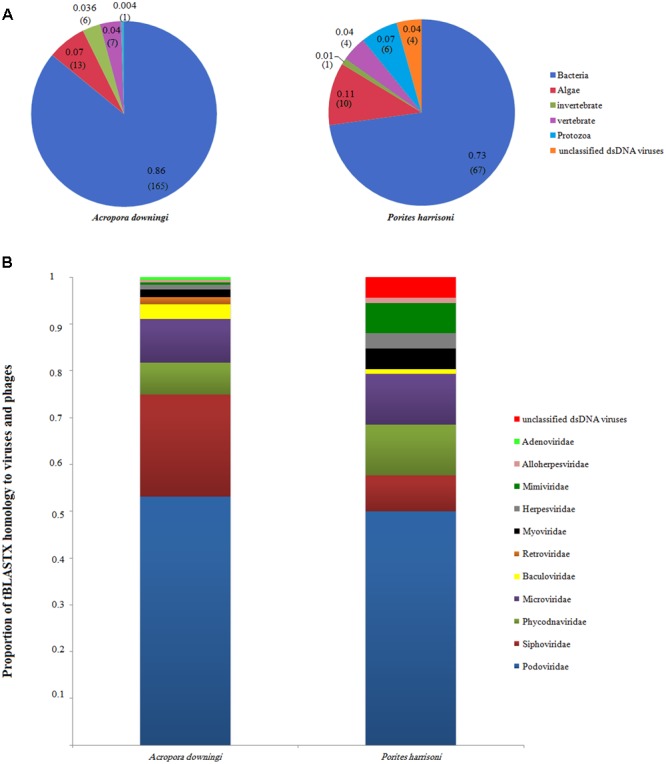
Annotated sequence classification according to tBLASTx hits in the GenBank non-redundant database (E-value 10-5). (A) Groups, percentage and proportion of viral annotated sequences after excluding eukaryote and prokaryote genes. The number of contigs for each group is also included between brackets. (B) The proportion of tBLASTX homology to virus and phage families in Acropora downingi and Porites harrisoni viromes.
The A. downingi viral assemblages were mainly dominated by viruses that target phages (86%, 165 contigs). Algal viruses (7%, 13 contigs) were the second most dominant group in this virome, followed by vertebrate (4%, 7 contigs), invertebrate (3%, 6 contigs) and protozoa (0.3%, 1 contig) viruses. On the other hand, phages (73%, 67 contigs) and algae (11%, 10 contigs), protozoa (7%, 6 contigs), vertebrate and unclassified (4%, 4 contigs), and invertebrate viruses (1%, 1 contig) dominated the P. harrisoni virome (Figure 2A). Figure 2B shows the variation within the detected viral families in A. downingi and P. harrisoni: Podoviridae (53.1%, 102 contigs), Siphoviridae (21.8%, 42 contigs), Microviridae (9.4%, 18 contigs) and Phycodnaviridae (6.8%, 13 contigs) dominated the A. downingi virome, while Podoviridae (52.3%, 46 contigs), Phycodnaviridae and Microviridae (11.4%, 10 contigs each) and Siphoviridae (6.8%, 7 contigs) dominated the P. harrisoni virome.
Metabolic Profiles of Gulf Coral Viromes and Comparison with Other Coral Viromes
The metabolic profiles of the two Gulf coral viromes were explored using MG-RAST, which assigns sequences to metabolic categories based on the best BLASTx hit against the subsystem database (E-value < 10-5). The most-represented categories in the A. downingi virome were related to respiration, then cluster-based subsystems, carbohydrate and phages, in that order. In P. harrisoni, the most-represented categories, in descending order, were respiration, phages and cluster-based subsystems (Figure 3).
FIGURE 3.
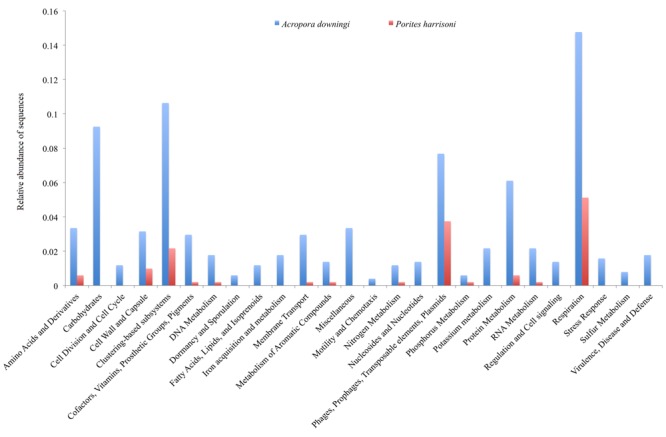
Relative functional gene sequences abundance in Arabian Gulf A. downingi and P. harrisoni viromes. Protein-encoding sequences were analyzed using BLASTX (E-value cut-off-10-5) to the MG-RAST subsystem database and grouped according to the highest hierarchal metabolic function or ‘subsystem.’
Thirteen categories, including carbohydrate, stress responses, virulence, disease and defense, regulation and cell signaling, motility and chemotaxis, cell division and cell cycles, dormancy and sporulation, and metabolisms (fatty acids, lipids, isoprenoids, iron, potassium, and nucleosides and nucleotides) were detected only in A. downingi and not in P. harrisoni (Figure 3). The carbohydrate category had a higher total abundance of sequences than the rest of the categories. Based on the functional genes obtained from MG-RAST, the virome of A. downingi (2.88) exhibited greater functional diversity (H′) than that of P. harrisoni (2.77), whereas the functional evenness of both coral viromes was 0.11.
A comparison in MG-RAST of the two Gulf coral viromes with 6 coral viromes that represent Pacific Ocean corals (Vega Thurber et al., 2009b) revealed clear segregation between the Gulf coral viromes and the other viromes (Figure 4). The coral viromes were compared based on the metabolic subsystems in MG-RAST; the resulting PCoA exhibited clear clustering of the Gulf coral viromes together and a clear distinction from the virome of P. compressa that inhabits the Pacific Ocean (Vega Thurber et al., 2009b) (Figure 4). A comparison of the P. harrisoni and the A. downingi viromes with the Pacific Ocean coral viromes using XIPE-TOTEC revealed significant overrepresentation of phage and prophage sequences in the Gulf coral viromes (P < 0.002) relative to those in the Pacific Ocean coral viromes. This comparison is clear in Supplementary Figure S1. Table 1 summarizes some of the sequences in the Gulf coral viromes with similarities to phage and prophages.
FIGURE 4.
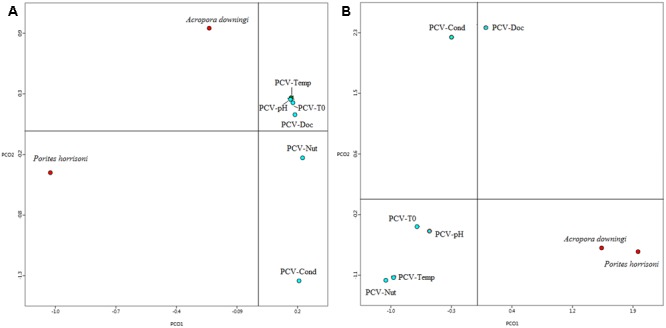
First two principle coordinates from the principal coordinate analysis of the viral communities from the Gulf coral viromes and Pacific coral virome (Vega Thurber et al., 2009b) as produced from the MG-RAST analysis for the (A) functional genes where protein-encoding sequences were analyzed using BLASTX (E-value < 10-5) to the subsystem database and grouped according to the highest hierarchal metabolic function or ‘subsystem. PCV1, pacific coral virome generated from stressed coral holobiont project; Temp, temperature; T0, control; Nut, nutrient; Cond, conductivity; DOC, dissolved organic carbon. (B) Best hit of viral classification.
Table 1.
Some of the gene annotations from the Arabian Gulf Porites harrisoni and Acropora downingi viromes showing those with similarities to phage and prophage sequences.
| Viral family (viridae) | Host | Accession # | Contig coverage | nt size | aa% Identity | E-value | Virus genome similarity | Gene annotation |
|---|---|---|---|---|---|---|---|---|
| Podo | Acropora | NC_001422.1 | 29038 | 3257 | 99 | 0 | Enterobacteria phage phiX174 sensu lato | DNA replication initiation protein gpA |
| Micro | Acropora | NC_004821.1 | 239 | 1494 | 88 | 0 | Bacillus prophage phBC6A52 | Hypothetical protein BC2597 |
| Micro | Acropora | NC_007817.1 | 12334 | 729 | 63 | 5e-96 | Enterobacteria phage ID2 Moscow/ID/2001 | Internal scaffolding protein |
| Sipho | Acropora | NC_008371.1 | 1131 | 511 | 93 | 1e-95 | Lactococcus phage jj50 | Hypothetical protein LPJV50_ORF31 |
| Sipho | Acropora | NC_017688.1 | 768 | 497 | 80 | 7e-60 | Lactococcus Phage ASCC191 | Hypothetical protein LLAPH_191_0037 |
| Sipho | Acropora | NC_001416.1 | 76 | 446 | 100 | 1e-91 | Enterobacteria phage lambda | Putative single-stranded DNA binding protein |
| Podo | Porites | NC_001422.1 | 41317 | 3209 | 100 | 0 | Enterobacteria phage phiX174 sensu lato | Minor spike protein |
| Micro | Porites | NC_004821.1 | 1472 | 1041 | 98 | 0 | Bacillus prophage phBC6A52 | Portal protein |
| Myo | Porites | YP_004323946.1 | 49 | 226 | 97 | 2e-39 | Synechococcus phage Syn19 | Hypothetical protein Syn19_113 |
| Sipho | Porites | YP_009277994.1 | 31 | 229 | 99 | 1e-44 | Propionibacterium phage QueenBey | Putative minor tail protein |
| Sipho | Porites | YP_009152377.1 | 25.72 | 230 | 99 | 7e-47 | Propionibacterium phage PHL092M00 | Putative protease |
Annotations are based on tBLASTx to the NCBI viral database (e-value threshold 10-5).
Electron Microscopic Analysis of Gulf Coral Tissue
Despite being the most dominant predicted viral group in the P. harrisoni and A. downingi virome, the presence of tailed bacteriophages was not detected inside coral tissue using electron microscopy. In contrast, most of the viruses that were detected in the coral tissue (Figure 5) and were identified as infecting prokaryotic and eukaryotic members of the holobionts were tailless viruses with or without envelope and with average sizes ranging from 75 to 300 nm. Various morphological types of virus-like particles (VLPs) were found in the healthy coral tissue by TEM and included large tailless VLPs and electron-dense core VLPs (Figure 5). The current work documents the presence of giant VLPs (>200 nm) in the tissue of P. harrisoni and A. downingi (Figures 5A–G). Figures 5C,D show what is known as the “virus factory,” which characterizes giant viruses known as the nucleocytoplasmic large viruses (NCLDV). In addition, giant VLPs within Symbiodinium cells are represented in Figures 5E–G, where the VLP size reached >300 nm. Electron-dense core VLPs with sizes ranging from 150 to 210 nm and with enveloped, icosahedral (non-tailed) capsids may be Herpes-like viral particles observed in the cell nuclei of both corals (Figures 5H,I). In contrast, tailed bacteriophages (Sipho-, Podo-, and Myo-) were not detected in the examined coral tissue by TEM. However, TEM images from A. downingi suggest that a tailless VLP with an icosahedral capsid 75 nm in diameter is infecting rod-shaped bacteria that inhabit the coral tissue; there is a possibility that this VLP could be a Podoviridae with a tail adsorbed to the host surface (Figures 5J,K). Tailed vibriophages were isolated from both A. downingi and P. harrisoni coral tissue using a phage plaque assay and cultures of a Vibrio sp. that were previously obtained from tissue of the same coral types (Al-Dahash and Mahmoud, 2013; Mahmoud and Kalendar, 2016). The tailed bacteriophage that were isolated from A. downingi tissue showed morphological features similar to members of the Siphoviridae family that have an isometric head and a long thin, flexible and non-contractile tail (Figure 6).
FIGURE 5.
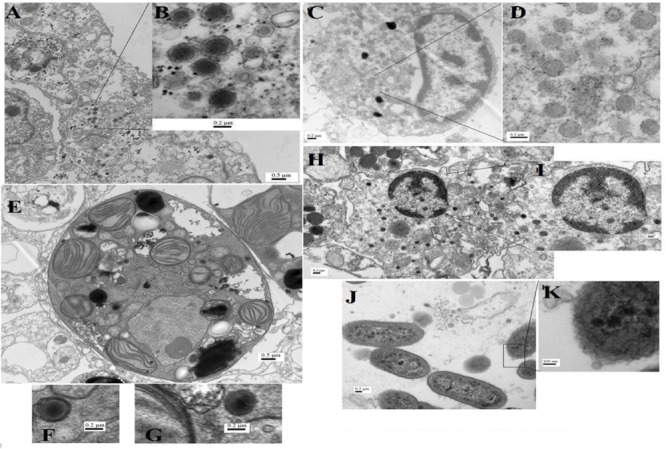
Transmission electron microscopy (TEM) images for virus-like particles (A–H) with sizes ranging from 75 to 200 nm and phages (J,K) from A. downingi and P. harrisoni tissue. (A,B) Large tailless nucleocytoplasmic VLP particles from epithelial cells of A. downingi. (C,D) Virus factory from epithelial cells. (E–G) Symbiodinium cell within the gastrodermis layer of A. downingi with large VLP (>300 nm). (H,I) VLP rupturing host nucleus and release of Herpes-like viral particles from P. harrisoni samples. (J,K) Tailless VLP attached to prokaryote cell within A. downingi tissue.
FIGURE 6.
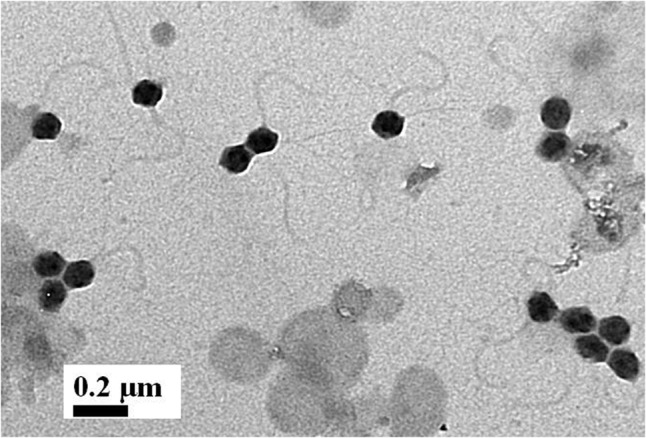
Electron micrographs showing vibriophages isolated from A. downingi using plaque assay, positively stained with 2% uranyl acetate solution and photographed at X60000 magnification.
Discussion
Few studies have focused on coral viromes (Marhaver et al., 2008; Vega Thurber et al., 2008, 2009b; Correa et al., 2013; Weynberg et al., 2014; Wood-Charlson et al., 2015; Correa et al., 2016), and they all concern corals from temperate and tropical regions that thrive in temperatures ranging from 16 to 32°C. We provide here the first analysis of viral communities in corals that naturally thrive in a thermally variable water body, where temperatures fluctuate between 11 and 39°C in different seasons.
Despite the stringent preparation protocols for the Gulf coral viromes and the low number of reads that were annotated as ribosomal RNA (an indicator of cellular contamination) in the prepared viromes, the two Gulf coral viromes were dominated by unknown sequences. As in studies of viromes that were obtained from different environments (Zhang et al., 2006; Fierer et al., 2007; Schoenfeld et al., 2008; Rosario et al., 2009; Parsley et al., 2010; Bibby et al., 2011; Park et al., 2011), including corals, the majority of the sequences had no homology to known genes of either viral or cellular origin and were classified as unknown (Mokili et al., 2012). This finding may suggest an insufficient representation of phages and viruses in the databases, implying that the majority of the viruses in these habitats remain uncharacterized (Culley, 2011). The majority of the known sequences (97%) were annotated to cellular organisms, including both prokaryotes and eukaryotes, due to host-derived genes, thus reflecting the “genetic mosaicism” of viruses (Vega Thurber et al., 2008, 2009b; Marhaver et al., 2008; Fancello et al., 2012).
The viral groups detected in Gulf corals were previously reported in corals elsewhere (Wood-Charlson et al., 2015). We caution that the taxonomic and functional analysis of viromes in this study is based on a small number of contigs. Keeping this in mind we find that the variation in the viral assemblages of the two most important coral types inhabiting the north Arabian Gulf (i.e., A. downingi and P. harrisoni) is evident in our study. The number of viral families that were identified in the A. downingi virome was greater than that found in P. harrisoni. The identified A. downingi and P. harrisoni viral sequences were dominated by sequences from phages, with the second most abundant sequences being from algal viruses, and the most-represented viral family in both corals was Podoviridae. This finding opposes the report of A. aspera that inhabit Heron Island, whose most dominant phage was Siphoviridae and least dominant was Podoviridae (Correa et al., 2016). However, we have to take into consideration that the Gulf viromes were produced by processing the entire coral nubbin, while in Correa et al. (2016), the viromes were produced by blasting only the coral tissue from the skeleton using an airbrush, which was followed by processing the blastate. This means that the Gulf coral viromes may include viruses that target other coral endolithic hosts. A previous study in P. astreoides that used full coral nubbins reported that the number of sequences with significant similarities to Podoviridae was the least abundant among dsDNA phage families in the produced metagenome (Wegley et al., 2007). On the other hand, Vega Thurber et al. (2008) showed that the Pacific Ocean P. compressa virome that was produced from whole coral nubbins was dominated by phages and that >60% of the viral sequences that were identified as eukaryotic viruses were similar to vertebrate viruses instead of algal viruses. Can the dominance of Podoviridae, which are phage with lytic lifestyles, in Gulf corals be related to coral strategies for surviving harsh conditions by controlling their symbiont density (Cunning and Baker, 2012)?
Only 12.7% of the identified viral sequences in the two viromes matched large viral families (i.e., Phycodnaviridae and Mimiviridae), despite the fact that the TEM images for the two coral tissues suggested that they are more abundant. This result may reflect the magnitude of discrimination against the genomes of these important viruses due to the method that was used. The viral isolation methods used in the present study succeeded in removing genomic contamination from cellular members of the holobionts, but the filtration step may have discriminated against large viruses. The importance of NCLVs to corals cannot be neglected. Martínez et al. (2014) stated that giant viruses are neither rare nor marginal players in marine ecosystems, whereas Correa et al. (2013) provided genomic evidence for the dominance of nucleocytoplasmic large DNA viruses (Mimiviridae and Phycodnaviridae) in Montastraea cavernosa and its algal endosymbiont. In addition, Correa et al. (2016) emphasized the importance of jointly applying electron microscopy and metagenomics when identifying dominant or core viral types, particularly NCLVs, within coral samples. In the current study, the TEM images of both A. downingi and P. harrisoni indicated the presence of giant VLPs (≥200 nm) in different parts of the healthy coral tissue and inside Symbiodinium cells (Figures 5E–G), raising questions about their role and importance in healthy Gulf corals. Our TEM and metagenomic data suggest the presence of giant VLPs, some of which are characterized by a Mimiviridae-like shape in the Gulf coral tissue (personal communication with Hans Ackermann; Correa et al., 2016). Claverie et al. (2009) reported that Mimiviridae are the second most abundant group in the marine system, exceeded only by bacteriophages. They suggested that ancestral Mimiviridae play a role in constructing octocorals’ mitochondrial genomes, as Mimiviridae exist in close proximity to corals and are able to infect coral tissue.
Our study also provides documentation for what is known as the virus factory (Figures 5C,D) within Gulf coral epidermal cells. These factories are created de novo in the viral host upon infection. Virus factories and large VLP particles that were in the cytoplasm of coral epithelial cells and associated with the Symbiodinium in the gastrodermal region were detected by TEM in the tested healthy Gulf corals. The images that we obtained (Figure 5) are similar to others that were previously published by Correa et al. (2016). Therefore, the role of NCLV in healthy Gulf coral tissue warrants further investigation.
Herpesviridae family sequences were found in the viromes of both Gulf corals. Furthermore, Herpes-like particles were observed in the cell nuclei of Gulf corals using TEM (Figures 5H,I). According to Vega Thurber et al. (2008), stressors induce production of Herpes-like viruses in Pacific Ocean corals; they found that the quantities of Herpes-like viral sequences increased up to six orders of magnitude in P. compressa that were exposed to high temperatures in comparison to their control counterparts. In addition, Correa et al. (2016) stated that Herpes-like annotations dominate dsDNA viral taxa in corals and that the most commonly identified VLPs in TEM images of both A. aspera and A. millepora tissue were particles with morphology highly reminiscent of herpesviruses. The viromes and the TEM images of Gulf corals do not support the previous findings.
Sequences significantly similar to Retroviridae members were found only in the A. downingi viral sequences. Retroviruses and RNA viruses with intermediate DNA stages are typically detected in DNA viral metagenome libraries (Correa et al., 2013, 2016; Weynberg et al., 2014; Wood-Charlson et al., 2015). Garcia et al. (2013) previously reported an abundance of retroviruses in the microbiomes of healthy and diseased Mussismilia braziliensis and A. millepora from the Atlantic Ocean. Additionally, Correa et al. (2013) revealed a high abundance of retroviruses in healthy and stressed Montastraea cavernosa, a common Caribbean reef-building coral. Recently, Correa et al. (2016) showed that retroviruses were the most dominant type of eukaryotic virus in the virome of A. aspera and that VLPs with physical shape similarities to retroviruses were detected by TEM in A. aspera tissue. Rohwer and Vega Thurber (2009) stated that retroviruses are potential vectors for horizontal gene transfer between animals and plants; in our case, such transfer might occur between coral cells and their algal symbiont. The presence of retroviruses in the A. downingi virome compared with the lack of retroviruses that were found in P. harrisoni in the current study may reflect physiological differences between the two corals (Gates and Ainsworth, 2011). However, the question of the role of retroviruses in corals remains unanswered.
The important roles of viruses as genomic manipulators of both prokaryotic and eukaryotic members of the coral holobiont are revealed by our deeper investigations into the functional diversity of annotated proteins within the Gulf coral viromes. A. downingi exhibited somewhat greater diversity in its functional genes than those of P. harrisoni did, and the two investigated viromes exhibited low evenness, reflecting the dominance of relatively few metabolic pathways in Gulf corals. Dinsdale et al. (2008) reported that coral-associated microbes generally exhibit low functional diversity because microbes associated with corals are taxonomically diverse.
Phage- and prophage-related genes were the second most dominant in P. harrisoni and the fourth most dominant in A. downingi. According to Bettarel et al. (2013), prophages protect their hosts from infection by a large number of related phages; this effect is considered vital for beneficial bacterial symbionts in coral holobionts. On the other hand, clustering-based subsystem-related genes were the second most dominant in the A. downingi and were the third most dominant in P. harrisoni. This finding may suggest the presence of functional coupling between certain genes, but without the knowledge of their precise functions, this point requires further investigation. The high occurrence of respiration-associated genes in the Gulf coral viromes may reflect the role of viruses in transferring this important group of genes that is responsible for host survival in the coral holobiont in environments in which the oxygenation diurnally fluctuates (Dinsdale et al., 2008) and, during summer, the increasing temperature decreases the oxygen solubility (Matear et al., 2000).
Finally, it was essential in the current study to compare the Gulf coral viromes with corresponding samples from corals inhabiting other, non-thermally fluctuating water bodies. When the two Gulf coral viromes were compared with six P. compressa viromes from the Pacific Ocean, they clustered separately based on functional gene and taxonomical diversity data obtained from MG-RAST. This finding supports the previous theory that viral assemblages differ in different coral species and in different geographical regions due to selective local pressures. However, we have to take into consideration the possibility that the separation noticed between the Gulf and Pacific coral viromes may be due to the differences in the algorithms applied by MG-RAST to analyze the compared viromes at the time of submission. In addition, Gulf coral viromes were produced by pooling samples collected from different seasons and sites together, which will add to the overall variability of these samples in comparison to the Pacific Ocean coral viromes, which were collected during Spring from one location and then exposed to different stressors under laboratory conditions. The XIPE-TOTEC results showed that Gulf corals exhibited over-representation of phage and prophage genes compared with the Pacific Ocean P. compressa virome, reflecting the importance of bacteriophages in the Gulf coral holobiont and their roles as top-down controllers of the diversity of coral-associated microbes (Wegley et al., 2007; Marhaver et al., 2008; Littman et al., 2011).
Conclusion
Our results confirm that healthy corals that thrive naturally in thermally fluctuating water bodies contain diverse groups of phages and viruses, including giant viruses, and that the presence of viruses is not necessarily related to the onset of infection and disease in corals. Further studies are needed to reveal the roles of viruses in maintaining the holobiont balance in the Gulf corals so that hypotheses can be formulated about how Gulf corals survive harsh environmental conditions.
Author Contributions
HM: initiator of the work, participate in all the practical involved in the work and run the metagenome practical fully, analyzed the metagenomes, prepared and wrote the manuscript. LJ: electron microscopic and phage work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful for Dr. Kyle Bibby (University of Pittsburgh, United States) and Prof. Jordan Peccia (Yale University) for all the help and guidance during the metagenome work. We are also grateful to Dr. Hans Ackermann (Laval University, Canada) for his valuable opinion in morphologically identifying viruses in some of the TEM images. Thanks to our diving team, Mr. Abdullah Al-Kandari and Mr. Raid Al-Kandari, for outstanding fieldwork. Many thanks to Ms. Ahlam Al-Kadi from the Nanoscopy Unit, Faculty of Science and Kuwait University, for assistance with the TEM work. We also acknowledge the Yale Center for Genome Analysis facility (Yale University) for sequencing the viromes using Illumina HiSeq 2000 facility, in addition we would like to acknowledge the general equipment and facilities provided by the Biotechnology Centre, Faculty of Science (Kuwait University), including the ABI 3130xI Genetic Analyzer (GS01/02), the epifluorescence microscope and the Nanodrop. We thank the core facility in the Faculty of Medicine (Kuwait University) for providing the ultracentrifuge facility (GM01/01 and GM01/05).
Footnotes
Funding. We thank Kuwait University, Research Administration grant number SL02/09, for supporting the study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02063/full#supplementary-material
References
- Ackermann H.-W. (2009). “Basic phage electron microscopy,” in Bacteriophages: Methods and Protocols: Isolation, Characterization, and Interactions Vol. 1 eds Clokie M. R. J., Kropinski A. M. (New York, NY: Humana Press; ), 113–126. 10.1007/978-1-60327-164-6 [DOI] [Google Scholar]
- Al-Dahash L. M., Mahmoud H. M. (2013). Harboring oil-degrading bacteria: A potential mechanism of adaptation and survival in corals inhabiting oil-contaminated reefs. Mar. Pollut. Bull. 72 364–374. 10.1016/j.marpolbul.2012.08.029 [DOI] [PubMed] [Google Scholar]
- Al-Ghadban A. N., Al-Yamani F., Al-Sammak A., Al-Hassam R., Behbehani M., Al-Hassan J., et al. (2007). Environmental stress of Kuwait’s coastal area due to the 1991 oil slick. IJOO 2 25–50. [Google Scholar]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Boil. 11:R106. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A. C., Starger C. J., McClanahan T. R., Glynn P. W. (2004). Coral reefs: corals’ adaptive response to climate change. Nature 430:741. 10.1038/430741a [DOI] [PubMed] [Google Scholar]
- Bettarel Y., Thuy N. T., Huy T. Q., Hoang P. K., Bouvier T. (2013). Observation of virus-like particles in this sections of the bleaching scleractinian coral Acropora cytherea. J. Mar. Biol. Assoc. 93 909–912. 10.1017/S0025315411002062 [DOI] [Google Scholar]
- Bibby K., Viau E., Peccia J. (2011). Viral metagenome analysis to guide human pathogen monitoring in environmental samples. Lett. Appl. Microbiol. 52 386–392. 10.1111/j.1472-765X.2011.03014.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogler A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter K. E., Abrar M., Aeby G., Aronson R. B., Banks S., Bruckner A., et al. (2008). One-third of reef-building corals face elevated extinction risk from climate change and local impacts. Science 321 560–563. 10.1126/science.1159196 [DOI] [PubMed] [Google Scholar]
- Carpenter K. E., Harrison P. L., Hodgson G., Alsaffar A. H., Alhazeem S. H. (1997). The Corals and Coral Reef Fishes of Kuwait. Kuwait: Kuwait Institute for Scientific Research. [Google Scholar]
- Claverie J.-M., Grzela R., Lartigue A., Bernadac A., Nitsche S., Vacelet J., et al. (2009). Mimivirus and Mimiviridae: Giant viruses with an increasing number of potential hosts, including corals and sponges. J. Invertebr. Pathol. 101 172–180. 10.1016/j.jip.2009.03.011 [DOI] [PubMed] [Google Scholar]
- Coles S. L., Riegl B. M. (2013). Thermal tolerances of reef corals in the Gulf: A review of the potential for increasing coral survival and adaptation to climate change through assisted translocation. Mar. Pollut. Bull. 72 323–332. 10.1016/j.marpolbul.2012.09.006 [DOI] [PubMed] [Google Scholar]
- Correa A. M. S., Ainsworth T. D., Rosales S. M., Thurber A. R., Butler C. R., Vega Thurber R. L. (2016). Viral outbreak in corals associated with an in situ bleaching event: atypical herpes-like viruses and a new megavirus infecting Symbiodinium. Front. Microbiol. 7:127. 10.3389/fmicb.2016.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa A. M. S., Welsh R. M., Vega Thurber R. L. (2013). Unique nucleocytoplasmic dsDNA and +ssRNA viruses are associated with the dinoflagellate endosymbionts of corals. ISME J. 7 13–27. 10.1038/ismej.2012.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley A. (2011). Virophages to viromes: a report from the frontier of viral oceanography. Curr. Opin. Virol. 1 52–57. 10.1016/j.coviro.2011.05.003 [DOI] [PubMed] [Google Scholar]
- Cunning R., Baker A. C. (2012). Excess algal symbionts increase the susceptibility of reef corals to bleaching. Nat. Clim. Change. 3 259–262. 10.1038/NCLIMATE1711 [DOI] [Google Scholar]
- Dinsdale E. A., Edwards R. A., Hall D., Angly F., Breitbart M., Brulc J. M., et al. (2008). Functional metagenomic profiling of nine biomes. Nature 452 629–632. 10.1038/nature06810 [DOI] [PubMed] [Google Scholar]
- Fancello L., Trape S., Robert C., Boyer M., Popgeorgiev N., Raoult D., et al. (2012). Viruses in the desert: a metagenomic survey of viral communities in four perennial ponds of the Mauritanian Sahara. ISME J. 7 359–369. 10.1038/ismej.2012.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N., Breitbart M., Nulton J., Salamon P., Lozupone C., Jones R., et al. (2007). Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl. Environ. Microbiol. 73 7059–7066. 10.1128/AEM.00358-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia G. D., Gregoracci G. B., Santos E. D. O., Meirelles P. M., Silva G. G. Z., Edwards R., et al. (2013). Metagenomic analysis of healthy and white plague-affected Mussismilia braziliensis corals. Microb. Ecol. 65 1076–1086. 10.1007/s00248-012-0161-4 [DOI] [PubMed] [Google Scholar]
- Gates R. D., Ainsworth T. D. (2011). The nature and taxonomic composition of coral symbiomes as drivers of performance limits in scleractinian corals. J. Exp. Mar. Biol. Ecol. 408 94–101. 10.1016/j.jembe.2011.07.029 [DOI] [Google Scholar]
- Guest J. R., Low J., Tun K., Wilson B., Ng C., Raingeard D., et al. (2016). Coral community response to bleaching on a highly disturbed reef. Sci. Rep. 6:20717. 10.1038/srep20717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegh-Guldberg O. (1999). Climate change, coral bleaching and the future of the world’s coral reefs. Mar. Freshwater Res. 50 839–866. 10.1071/MF99078 [DOI] [Google Scholar]
- Hoegh-Guldberg O. (2010). Dangerous shifts in ocean ecosystem function? ISME J. 4 1090–1092. 10.1038/ismej.2010.107 [DOI] [PubMed] [Google Scholar]
- Hume B., D’Angelo C., Burt J., Baker A. C., Riegl B., Wiedenmann J. (2013). Corals from the Persian/Arabian Gulf as models for thermotolerant reef-builders: prevalence of clade C3 Symbiodinium, host fluorescence and ex situ temperature tolerance. Mar. Pollut. Bull. 72 313–322. 10.1016/j.marpolbul.2012.11.032 [DOI] [PubMed] [Google Scholar]
- IPCC (2007). Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge: Cambridge University Press. [Google Scholar]
- Jacquet S., Bratbak G. (2003). Effects of ultraviolet radiation on marine virus-phytoplankton interactions. FEMS Microbiol. Ecol. 44 279–289. 10.1016/S0168-6496(03)00075-8 [DOI] [PubMed] [Google Scholar]
- Jokiel P. L., Coles S. L. (1990). Response of Hawaiian and other indo-pacific reef corals to elevated temperature. Coral Reefs 8 155–162. 10.1007/BF00265006 [DOI] [Google Scholar]
- Littman R., Willis B. L., Bourne D. G. (2011). Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef. Environ. Microbiol. Rep. 3 651–660. 10.1111/j.1758-2229.2010.00234.x [DOI] [PubMed] [Google Scholar]
- Mahmoud H. M., Al-Sarraf M. (2016). Molecular phylogeny and community fingerprinting of coral-associated Symbiodinium north of the Arabian Gulf. Mar. Pollut. Bull. 108 94–104. 10.1016/j.marpolbul.2016.04.053 [DOI] [PubMed] [Google Scholar]
- Mahmoud H. M., Kalendar A. A. (2016). Coral associated Actinobacteria: diversity, abundance and biotechnological potentials. Front. Microbiol. 7:204. 10.3389/fmicb.2016.00204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhaver K. L., Edwards R. A., Rohwer F. (2008). Viral communities associated with healthy and bleaching corals. Environ. Microbiol. 10 2277–2286. 10.1111/j.1462-2920.2008.01652.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez L. M., Redman R. S., Rodriguez R. J., Roossinck M. J. (2007). A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science 315 513–515. 10.1126/science.1136237 [DOI] [PubMed] [Google Scholar]
- Martínez J. M., Swan B. K., Wilson W. H. (2014). Marine viruses, a genetic reservoir revealed by targeted viromics. ISME J. 8 1079–1088. 10.1038/ismej.2013.214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matear R. J., Hirst A. C., McNeil B. I. (2000). Changes in dissolved oxygen in the Southern Ocean with climate change. Geochem. Geophys. Geosyst. 1 1–12. 10.1029/2000GC000086 [DOI] [Google Scholar]
- Meyer F., Paarmann D., D’Souza M., Olson R., Glass E. M., Kubal M., et al. (2008). The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. 10.1186/1471-2105-9-386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min R., Tang H., Ye Y. (2010). FragGeneScan: predicting genes in short and error-prone reads. Nucl. Acids Res. 38:e191. 10.1093/nar/gkq747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokili J. L., Rohwer F., Dutilh B. E. (2012). Metagenomics and future perspectives in virus discovery. Curr. Opin. Virol. 2 63–77. 10.1016/j.coviro.2011.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier A., Claverie J.-M., Ogata H. (2008). Taxonomic distribution of large DNA viruses in the sea. Genome Biol. 9:R106. 10.1186/gb-2008-9-7-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafavi P. G., Fatemi S. M., Shahhosseiny M. H., Hoegh-Guldberg O., Loh W. K. (2007). Predominance of clade D Symbiodinium in shallow-water reef-building corals off Kish and Larak Islands (Persian Gulf, Iran). Mar. Biol. 153 25–34. 10.1007/s00227-007-0796-8 [DOI] [Google Scholar]
- Noble R. T., Fuhrman J. A. (1998). Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14 113–118. 10.3354/ame014113 [DOI] [Google Scholar]
- Park E. J., Kim K.-H., Abell G. C. J., Kim M.-S., Roh S. W., Bae J.-W. (2011). Metagenomic analysis of the viral communities in fermented foods. Appl. Environ. Microbiol. 77 1284–1291. 10.1128/AEM.01859-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsley L. C., Consuegra E. J., Thomas S. J., Bahavsar J., Land A. M., Bhuiyan N. N., et al. (2010). Census of the viral metagenome within an activated sludge microbial assemblage. Appl. Environ. Microbiol. 76 2673–2677. 10.1128/AEM.02520-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten N. L., Harrison P. L., Mitchell J. G. (2008). Prevalence of virus-like particles within a staghorn scleractinian coral (Acropora muricata) from the Great Barrier Reef. Coral Reefs 27 569–580. 10.1007/s00338-008-0356-9 [DOI] [Google Scholar]
- Riegl B. M., Purkis S. J. (2012). “Coral reefs of the Gulf: adaptation to climatic extremes in the world’s hottest sea,” in Coral Reefs of the Gulf eds Riegl B. M., Purkis S. J. (Dordrecht: Springer; ) 1–4. 10.1007/978-94-007-3008-3 [DOI] [Google Scholar]
- Rodriguez-Brito B., Rohwer F., Edwards R. A. (2006). An application of statistics to comparative metagenomics. BMC Bioinformatics 7:162. 10.1186/1471-2105-7-162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer F., Vega Thurber R. (2009). Viruses manipulate the marine environment. Nature 459 207–212. 10.1038/nature08060 [DOI] [PubMed] [Google Scholar]
- Rosario K., Nilsson C., Lim Y. W., Ruan Y., Breitbart M. (2009). Metagenomic analysis of viruses in reclaimed water. Environ. Microbiol. 11 2806–2820. 10.1111/j.1462-2920.2009.01964.x [DOI] [PubMed] [Google Scholar]
- Schafer H., Muyzer G. (2001). “Denaturing gradient gel electrophoresis in marine microbial ecology,” in Marine Microbiology: Methods in Microbiology Vol. 30 ed. Paul J. H. (London: Academic Press; ) 425–468. [Google Scholar]
- Schmieder R., Lim Y., Rhower F., Edwards R. (2010). TagCleaner: Identification and removal of tag sequences from genomic and metagenomic datasets. BMC Bioinfomatics 11:341. 10.1186/1471-2105-11-341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfeld T., Patterson M., Wommack K. E., Young M., Mead D. (2008). Assembly of viral metagenomes from yellowstone hot springs. Appl. Environ. Microbiol. 74 4164–4174. 10.1128/AEM.02598-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahhosseiny M. H., Mostafavi P. G., Fatemi S. M. R., Karimi E. (2011). Clade identification of symbiotic zooxanthellae of dominant sclerectinian coral species of intertidal pools in Hengam Island. Afr. J. Biotechnol. 10 1502–1506. [Google Scholar]
- Sheppard C., Al-Husiani M., Al-Jamali F., Al-Yamani F., Baldwin R., Bishop J., et al. (2010). The Gulf: A young sea in decline. Mar. Pollut. Bull. 60 13–38. 10.1016/j.marpolbul.2009.10.017 [DOI] [PubMed] [Google Scholar]
- Sommer D., Delcher A., Salzberg S., Pop M. (2007). Minimus: a fast, lightweight genome assembler. BMC Bioinformatics 8:64. 10.1186/1471-2105-8-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding M. D., Ravilious C., Green E. P. (2001). World Atlas of Coral Reefs. Berkeley, CA: University of California Press. [Google Scholar]
- Suttle C. (2007). Marine viruses- major players in the global ecosystem. Nat. Rev. Microbiol. 5 801–812. 10.1038/nrmicro1750 [DOI] [PubMed] [Google Scholar]
- Vega Thurber R. L, Barott K. L., Hall D., Liu H., Rodriguez-Mueller B., Desnues C., et al. (2008). Metagenomic analysis indicates that stressors induce production of herpes-like viruses in the coral Porites compressa. Proc. Natl. Acad. Sci. U.S.A. 105 18413–18418. 10.1073/pnas.0808985105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega Thurber R., Hanes M., Breitbart M., Wegley L., Rohwer F. (2009a). Laboratory procedures to generate viral metagenomes. Nat. Protoc. 4 470–483. 10.1038/nprot.2009.10 [DOI] [PubMed] [Google Scholar]
- Vega Thurber R., Willner-Hall D., Rodriguez-Mueller B., Desnues C., Edwards R. A., Angly F., et al. (2009b). Metagenomic analysis of stressed coral holobionts. Environ. Microbiol. 11 2148–2163. 10.1111/j.1462-2920.2009.01935.x [DOI] [PubMed] [Google Scholar]
- Vega Thurber R. L., Correa A. M. S. (2011). Viruses of reef-building scleractinian corals. J. Exp. Mar. Biol. Ecol. 408 102–113. 10.1016/j.jembe.2011.07.030 28586690 [DOI] [Google Scholar]
- Wang D., Urisman A., Liu Y.-T., Springer M., Ksiazek T. G., Erdman D. D., et al. (2003). Viral discovery and sequence recovery using DNA microarrays. PLOS Biol. 1:E2. 10.1371/journal.pbio.0000002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegley L., Edwards R., Rodriguez-Brito B., Liu H., Rohwer F. (2007). Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 9 2707–2719. 10.1111/j.1462-2920.2007.01383.x [DOI] [PubMed] [Google Scholar]
- Weynberg K. D., Wood-Charlson E. M., Suttle C. A., van Oppen M. J. H. (2014). Generating viral metagenomes from the coral holobiont. Front. Microbiol. 5:206. 10.3389/fmicb.2014.00206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm S. W., Poorvin L. (2001). “Quantification of algal viruses in marine samples,” in Marine Microbiology: Methods in Microbiology Vol. 30 ed. Paul J. H. (London: Academic Press; ) 425–468. [Google Scholar]
- Wilson W. H., Dale A. L., Davy J. E., Davy S. K. (2005). An enemy within? Observations of virus-like particles in reef corals. Coral Reefs 24 145–148. 10.1007/s00338-004-0448-0 [DOI] [Google Scholar]
- Wood-Charlson E. M., Weynberg K. D., Suttle C. A., Roux S., van Oppen M. J. H. (2015). Metagenomic characterization of viral communities in corals: mining biological signal from methodological noise. Environ. Microbiol. 17 3440–3449. 10.1111/1462-2920.12803 [DOI] [PubMed] [Google Scholar]
- Zerbino D. R., Birney E. (2008). Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18 821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Breitbart M., Lee W. H., Run J.-Q., Wei C. L., Soh S. W. L., et al. (2006). RNA viral community in human feces: prevalence of plant pathogenic viruses. PLOS Biol. 4:e3. 10.1371/journal.pbio.0040003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


