Abstract
BACKGROUND
Conventional antidepressants usually require several weeks to achieve a full clinical response in patients with major depressive disorder, an illness associated with dysregulated circadian rhythms and a high incidence of suicidality. Two rapid-acting antidepressant strategies, low-dose ketamine (KT) and sleep deprivation (SD) therapies, dramatically reduce depressive symptoms within 24 hours in a subset of major depressive disorder patients. However, it is unknown whether they exert their actions through shared regulatory mechanisms. To address this question, we performed comparative transcriptomics analyses to identify candidate genes and relevant pathways common to KT and SD.
METHODS
We used the forced swim test, a standardized behavioral approach to measure antidepressant-like activity of KT and SD. We investigated gene expression changes using high-density microarrays and pathway analyses (Gene Ontology, Kyoto Encyclopedia of Genes and Genomes, Gene Set Enrichment Analysis) in KT- and SD-treated mice compared with saline-treated control male mice.
RESULTS
We show that KT and SD elicit common transcriptional responses implicating distinct elements of the circadian clock and processes involved in neuronal plasticity. There is an overlap of 64 genes whose expression is common in KT and SD. Specifically, there is downregulation of clock genes including Ciart, Per2, Npas4, Dbp, and Rorb in both KT- and SD-treated mice.
CONCLUSIONS
We demonstrate a potential involvement of the circadian clock in rapid antidepressant responses. These findings could open new research avenues to help design chronopharmacological strategies to treat major depressive disorder.
Keywords: Anterior cingulate cortex, Circadian clock, Depression, Ketamine, Sleep deprivation, Transcriptome
Major depressive disorder (MDD) is one of the most serious and common psychiatric disorders in the United States. According to the latest statistics, the National Institute of Mental Health estimated that 15.7 million adults over the age of 18 years in the United States had at least one major depressive episode, representing 6.7% of all U.S. adults (1). The two most rapid-acting antidepressant strategies, low-dose ketamine (KT) and sleep deprivation (SD) therapies, motivated a large number of studies into their mechanisms of action. In contrast to conventional antidepressants that can take weeks for full clinical response, 40% to 60% of patients with depression improve within hours of treatment (2,3). Importantly, both KT and SD decrease suicidality (4–10).
Circadian rhythms are intimately linked to the sleep-wake cycle (11). A subgroup of MDD patients has altered circadian processes including sleep, mood, temperature, and hormone secretions, all of which are regulated by circadian clock genes (12). Findings show a significant correlation between symptom severity and the degree of desynchronization. Moreover, many rhythms normalize as symptoms remit (13–16).
SD therapy usually involves keeping patients awake for approximately 36 hours. We proposed that by altering the sleep-wake cycle, the abnormal circadian clock genes that control rhythms could be reset (17,18). Although relapse can occur following recovery sleep, improvement can be sustained for weeks by circadian-related treatments. These include slowly advancing bedtimes (sleep-phase advance) and exposure to morning bright light (19).
Circadian studies of clock genes in mice show that in response to sleep deprivation, a subset of circadian clock genes (e.g., Per1, Per2) appear to behave as immediate early genes and are transcriptionally responsive within hours of treatment (20–23). It was also shown that depriving animals of sleep suppresses approximately 80% of rhythmic genes in the mouse (22,24).
Compared with SD, KT’s action on the circadian clock genes is less clear. In our earlier research in neuronal cell culture (NG108-15), we demonstrated KT’s effect in repressing circadian expression of a group of genes essential to maintaining circadian rhythmicity. We found dose-dependent reductions in the amplitude of circadian transcription including Bmal1, Per2, and Cry1 genes (25).
We propose that SD and KT may share common mechanisms of action that converge on circadian-related processes that act to accelerate antidepressant response (12). In this study, we performed comparative transcriptome analyses in SD- and KT-treated mice to identify candidate genes and pathways common to both treatments.
The first direct evidence for the dysregulation of the clock genes in the MDD brain comes from a microarray study. Control brains showed robust circadian rhythms across six brain areas that were dramatically altered in matched MDD patients. Of the brain regions studied, the anterior cingulate cortex (ACC) showed the most significant disruption in clock gene rhythms (26). The ACC is a major component of an extended neural network thought to regulate mood, and a growing body of data implicates the ACC as a key brain region associated with depression (27). Functional brain imaging studies show that increased activation of the ACC significantly correlates with improvement to a wide range of interventions including low-dose KT (28) and SD therapies (29).
Given the similarities in the rapid antidepressive effects elicited by KT and SD therapies, we hypothesized that these treatments may act through common molecular pathways in the ACC. To address this question, we analyzed the whole transcriptome in the ACC in groups of mice subjected to KT or SD treatment and compared them with a control group. We show that KT and SD antidepressant treatments activate common pathways and neuronal functions including synaptic plasticity, neurogenesis, and, notably, the circadian clock.
METHODS AND MATERIALS
Animals
Animals and protocols used in this study were reviewed and approved by the Institutional Animal Care and Use Committee of the University of California, Irvine. Six-month-old male mice from the C57BL6/J strain were obtained from Jackson Laboratories (Bar Harbor, ME).
Two groups of 4 mice each were habituated for 1 week in standard cages. A third group (n = 4) was habituated to a circular cage containing a rotatory bar that was stationary during habituation. All groups were housed with ad libitum food access and water at 24°C to 25°C room temperature. After habituation, mice were either sleep deprived (SD) or injected with ketamine (KT) or saline (control mice).
Sleep Deprivation
Mice housed in the circular cage were kept awake for 12 hours (zeitgeber time [ZT] 0 to ZT 12) by a slowly rotating bar (1.5 revolutions/min), suspended 1 cm above the floor and bedding within the mouse cage (30). During the SD period, ad libitum food and water were provided.
Ketamine
KT-treated mice were injected intraperitoneally with a KT solution of 3 mg/kg at ZT 5.
Control Mice
Control mice were injected intraperitoneally with saline solution in a volume of 100 µL/0.03 kg at ZT 5.
Forced Swim Test
We conducted the forced swim test (FST) 7 hours post-injection (ZT 12) of KT or saline. SD-treated mice were subjected to the FST at ZT 12 (following SD). FST was conducted according to a standardized protocol (31). Mice were placed in a cylindrical container (height 30 cm, diameter 20 cm) filled with tap water at a temperature of 25°C. Measurements were recorded in dark conditions using an infrared recording system for a period of 8 minutes. The immobility time was measured as 1-minute bins to identify subtle behavioral differences between the different experimental groups. At the end of the FST, the animals were gently dried and euthanized.
ACC Microdissection and RNA Isolation
Immediately after euthanization, brains were extracted, frozen in dry ice, and stored at −80°C. The microdissection of the ACC (bregma 1.34 to −0.5 mm) (Figure 1C) has been previously described (32). Total RNA was extracted from each sample using TRIzol (Invitrogen, Carlsbad, CA) following manufacturer's instructions and scaled 1:10. Total RNA was resuspended in 30 µL of RNase-free water and quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). Complementary DNA (cDNA) was synthesized from 50 ng of RNA using the cDNA Synthesis kit iScript (Bio-Rad, Hercules, CA). The obtained cDNA was then diluted 1:10 and 2 µL were used as the template for reverse transcriptase polymerase chain reaction (PCR) amplification using SYBR Green (Bio-Rad) as the fluorogenic intercalating dye and the CFX96 Real-Time System (Bio-Rad). The housekeeping gene β-actin was used as a control. The remaining RNA was used for microarray experiments. The primers used for reverse transcriptase PCR amplification are presented in Supplemental Table S3.
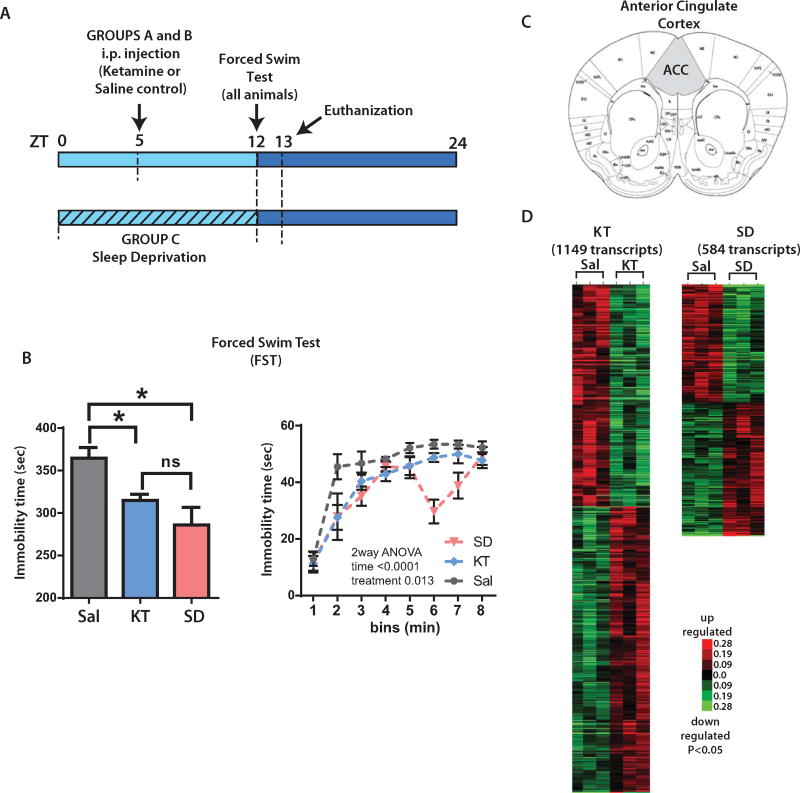
Figure 1.
Transcriptional profile of ketamine (KT) and sleep deprivation (SD) interventions. (A) Mice were divided into three groups and were administered either KT (3 mg/kg intraperitoneally [i.p.] at zeitgeber time [ZT] 5) (group A) or saline ([Sal], 100 µL/0.03 kg i.p.) at ZT 5 (group B, which is the control) or underwent SD (animals were kept awake from ZT 0 to ZT 12) (group C). All treatment groups were assessed for antidepressant activity on the forced swim test (FST) starting at ZT 12. At ZT 13, animals were euthanized and brains were dissected. (B) (Left) The bar graph depicts differences in immobility times in KT- and SD-treated mice versus saline (control mice) on the FST (*p < .05, t test). Differences between SD and KT immobility times were non-significant (ns). (Right) Graph indicates the expression of immobility in 1-minute bins, (two-way analysis of variance [ANOVA], followed by Bonferroni’s post hoc test, n = 4 per group). (C) Coronal section of the anterior cingulate cortex (ACC) (analogous to the human anterior cingulate cortex) was microdissected for gene expression analyses (shaded area). (D) Heat map plots showing transcriptionally altered genes in KT or SD mice depicting significant differences on gene expression (p < .05). Red indicates upregulated transcripts; green indicates downregulated transcripts (p < .05, t test, KT or SD compared with control mice; n = 3 per group).
Microarray Analysis
The remaining TRIzol-extracted RNA was diluted to 100 µL final volume of RNase-free water and cleaned using the Qiagen RNeasy Mini purification kit (Redwood, CA), following the manufacturer’s protocol. Eluted RNA was quantified with a NanoDrop spectrophotometer. The quality of the RNA was assessed on an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Probe synthesis and chip hybridization were performed at the Genomic High-Throughput Facility at the University of California, Irvine. Briefly, 100 ng of total RNA per sample was used as a template to obtain cDNA with the GeneChip cDNA synthesis Kit (Affymetrix, Santa Clara, CA). Mouse Gene ST arrays 2.0 (Affymetrix) were used to determine the transcriptome expression levels in the three groups of mice. The arrays were scanned in the GeneChip Scanner 3000 7G (Affymetrix), and the Command Console Software was used to produce the .CEL intensity file. The obtained data were analyzed in Affymetrix Expression Console software v1.1.1 using the PLIER algorithm to generate probe level summarization files (*.CHP). (Algorithm: PLIER version 2.0; quantification scale: linear; quantification type: signal and detection p value; background: GC composition-based background correction (PM-GCBG); normalization method: sketch-quantile). For each experimental condition, three microarrays were used.
Data Analysis
Each experimental group—SD or KT—was compared with the saline control. Because we analyzed simultaneously the expression profile in two experimental conditions, we selected genes using a moderate stringency (p < .05). This approach is commonly used to characterize brain function (33,34). For each experimental group, the obtained list of genes were hierarchically classified using the Gene Cluster program (35). To this end, the expression level of each gene was first log transformed and then median centered (relatively to its median expression across all samples), so that relative variations rather than absolute values were used for interpretation. The clustering method used here was an average linkage with the Pearson correlation coefficient as similarity metric. Results were displayed using the TreeView program (35). The list of genes were then classified according with its function using the Gene Ontology (GO) database and the functional classification tool Genecodis server (http://genecodis.cnb.csic.es/) (36). This latter program assigns genes to functional groups within various ontologies and calculates the statistical probability (hypergeometric p value corrected by false discovery rate method) of a particular functional group being overrepresented or underrepresented. We further classified the list of transcripts according to their functions with the Gene Set Enrichment Analysis (GSEA) using matrix, Kyoto Encyclopedia of Genes and Genomes (KEGG), GO molecular function, and biological properties. We used the following parameters: 1000 permutations; enrichment statistics as weighted; metric of ranking genes was signal to noise; minimal set of genes of 15. Then we merged the two sets of results from the SD and KT groups in a network using the Enrichment Map, version 2.01 (University of Toronto, Toronto, ON, Canada) for Cytoscape, version 3.2.1 (Agilent Technologies). We used a cutoff of p < .05. The data from the FST were analyzed by two-way analysis of variance as repeated measures to compare treatment and 1-minute bins as time, followed by Bonferroni’s post hoc test for multiple comparisons. The probability of gene overlap was calculated by hypergeometric test using R software, version 3.3.1 (R Foundation, Vienna, Austria). Unpaired Student’s t test was used to compare the total immobility time between groups. Data obtained from quantitative PCR was analyzed by unpaired Student’s t test. Correlation between gene expression from microarrays and quantitative PCR was analyzed using Pearson’s correlation coefficient using GraphPad Prism software, version 6.01 (La Jolla, CA). The Gene Expression Omnibus accession number for the microarray data set reported in this paper is GSE93041.
RESULTS
To explore possible common mechanisms of action between KT and SD therapies, we treated mice with KT (3 mg/kg), SD (12 hours), or saline (control). Immobility on the FST, a standardized test for antidepressant response, was used as a measure of efficacy (Figure 1A). When compared with control mice, we observed a decrease in immobility to both SD- and KT-treated mice (p < .05, t test) (Figure 1B, left) with no significant difference between treated groups. Our results are consistent with those of other studies (30,37–39). Interestingly, analyzing the activity in 1-minute bins, we observed that all animal groups gradually become immobile, with the saline group being the most immobile. However, the SD group shows a recovery within the sixth minute (Figure 1B, right).
We next analyzed the transcriptome in the mouse ACC (40), a region analogous to the ACC in humans (Figure 1C). Using a threshold of p < .05 (see Methods and Materials), we found changes in expression levels of 584 and 1149 genes in SD- and KT-treated animals, respectively (Figure 1D). We identified 64 transcripts common to the two groups (p = 6.107−8, hypergeometric test), (Figure 2A, Supplemental Table S1) associated with specific ontological categories related to the circadian clock and neuronal plasticity. These include entrainment of the circadian clock, regulation of dendrite morphogenesis, ribosome function, nucleic acid binding, and cellular metabolic processes (Figure 2B).
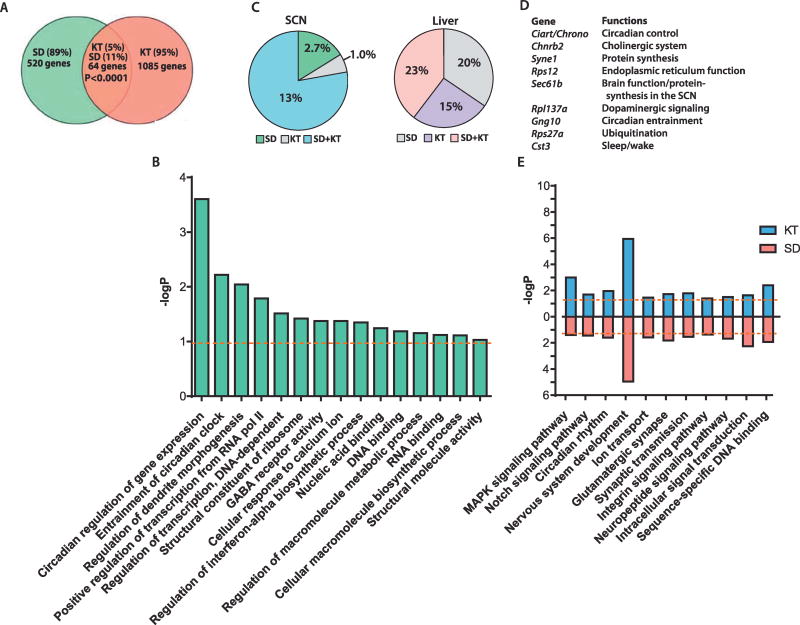
Figure 2.
Ketamine (KT) and sleep deprivation (SD) induce common molecular changes. (A) Venn diagram depicts shared transcripts between KT and SD. (B) Bar graph represents the Gene Ontology analysis from 64 common genes. (C) Pie charts illustrating percentage of known rhythmic genes in the suprachiasmatic nucleus (SCN) (left) and the liver (right). (D) Overlapping clock-controlled genes that are rhythmic in the SCN. (E) Bar graph represents the Gene Ontology analysis from SD or KT groups. Orange dotted lines denote significance threshold (p = .05, hypergeometric analysis corrected by false discovery rate method). GABA, gamma-aminobutyric acid; MAPK, mitogen-activated protein kinase.
Interestingly, we identified a higher proportion of overlapping rhythmic genes in the brain (suprachiasmatic nucleus) than in a peripheral clock organ (liver), implying that the actions of SD and KT genes predominantly affect the central nervous system (Figure 2C). Notably, we identified Ciart (circadian-associated repressor of transcription, also known as Chrono or Gm129), a gene whose product has been recently described to inhibit circadian transcription by direct binding to the CLOCK:BMAL1 activator complex (41,42) (Figure 2D). Moreover, on examination of the Expression Data Atlas, we found that this gene is highly expressed in the brain, specifically in the cerebral cortex and corpus striatum (Supplemental Figure S1) (43).
We used three systems to evaluate the overlap between KT and SD treatments in terms of biological pathways and processes: shared GO, KEGG, and GSEA. To circumvent the possible exclusion of potentially relevant genes, as noted, we grouped a restricted number of 64 common genes (representing a small portion of the differentially expressed genes), 5% (KT) and 11% (SD) (Figure 2A), and performed a gene-enrichment analysis with the full set of transcriptionally altered genes (p < .05, t test) from both experimental groups. Shared GO categories and KEGG pathways between SD and KT (p < .05, hypergeometric test corrected by false discovery rate method) include functional enrichment related to neuronal plasticity including nervous system development, synaptic transmission, and intracellular signal transduction, such as the mitogen-activated protein kinase (MAPK) signaling pathway (Figure 2E). Although we identified a relatively small number of overlapping transcripts, the sets of genes significantly responsive to either SD or KT treatments are relevant to specific biological pathways.
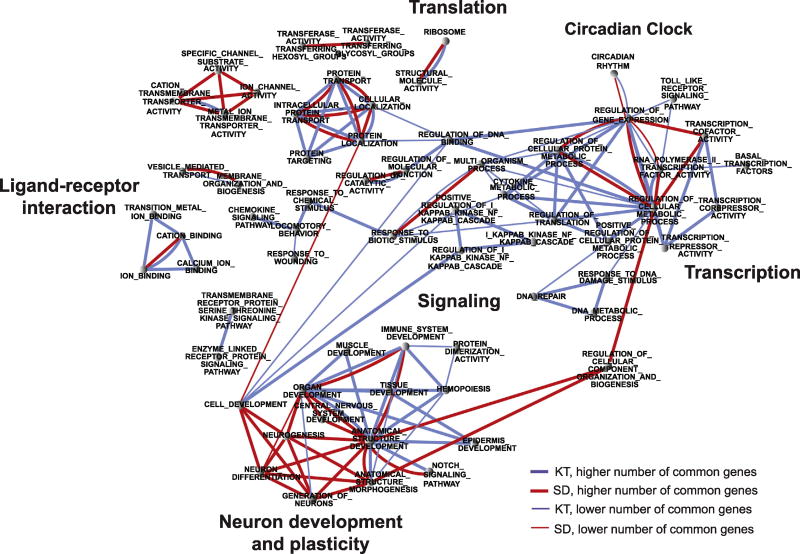
To further detect synchronized variations in the expression within groups of functionally correlated genes, we analyzed the data by GSEA using a cutoff value of p < .05 (see Methods and Materials). This analysis revealed that although some functions are exclusive for one or the other group (Figure 3), specific biological processes are shared between the two treatments, including ligand-receptor interactions, ion binding, neuron differentiation, anatomical structure development, protein transport, ribosome, cellular localization, and regulation of gene expression.
Figure 3.
Ketamine (KT) and sleep deprivation (SD) induce common biological processes. Functional annotation network showing the biological and molecular functions and Kyoto Encyclopedia of Genes and Genomes pathways that are altered by KT (blue lines) and SD (red lines) when compared with the control group (saline). Each node contains genes whose expression levels were altered by each treatment. Links between the nodes represent the proportion of genes that are common between gene sets. The edge thickness represents the degree of overlap between gene sets.
In support of a circadian hypothesis for the mode of action of KT and SD therapies, the GSEA identified a set of circadian rhythm genes (Figure 3). Distinct transcripts corresponding to core clock genes included Per2, Npas4, Rorb, Dbp, and Ciart that were downregulated by both treatments as revealed by quantitative PCR and microarray analyses (Figure 4A, B). We speculate that because the core clock controls an extensive array of neuronal functions (11,44), it might be centrally positioned to act as an effector on neuronal responses.
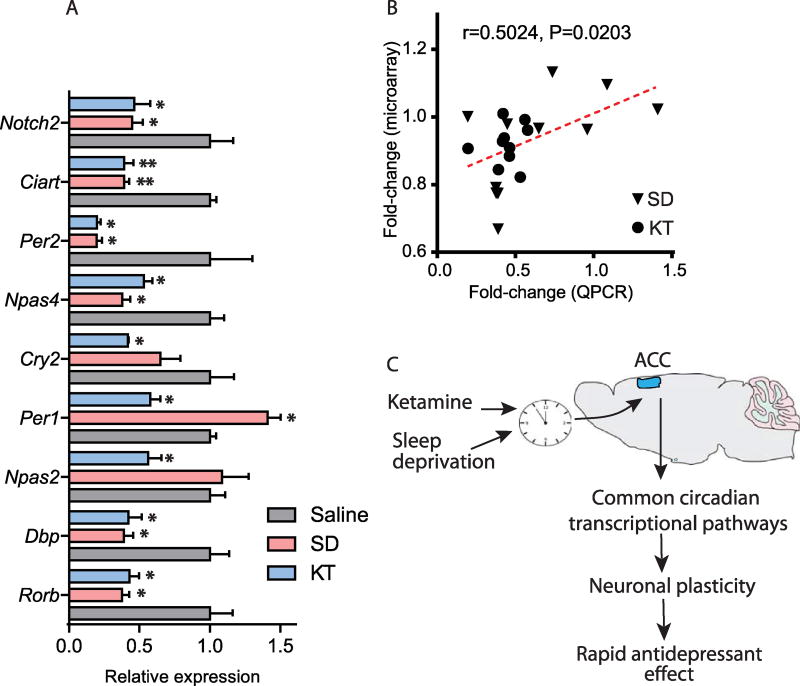
Figure 4.
Ketamine (KT) or sleep deprivation (SD) induce common neuronal responses. (A) Bar graph depicts the relative expression of known core clock genes and NOTCH pathway genes measured by quantitative polymerase chain reaction (qPCR). Saline is indicated by gray bars, SD by pink bars, and KT by blue bars (*p < .05, **p < .01, t test, n = 4 per group). (B) Correlation plot represents microarray and qPCR data. (C) Theoretical diagram illustrates the effects of KT or SD on the modulation of the limbic cortex or anterior cingulate cortex (ACC) circadian clock and pathways involved in neuronal plasticity. These neuronal adaptations could potentially provide a mechanism for the initiation of rapid antidepressant effects.
In addition, recent evidence demonstrates that the circadian clock controls some components of the NOTCH and MAPK signaling pathways (45–48). We found altered expression levels of transcripts belonging to the NOTCH and MAPK pathways with both KT and SD interventions (Figures 2 and 3, Supplemental Table S2).
We hypothesize that the fast-acting antidepressant effects of both KT and SD therapies could involve the reorganization of neuronal circuits controlled by the circadian clock (Figure 4C). The specific circadian signature(s) common to KT and SD therapies that are described here represent an important step toward a molecular understanding of how these antidepressants operate and establishes a valuable basis toward the development of pharmacological and therapeutic strategies to possibly reset abnormal circadian cycles.
DISCUSSION
To our knowledge, this is the first study that illustrates the presence of common transcriptional programs elicited in response to KT and SD therapies. While other studies have indicated the complex pharmacological actions of KT, only one (49) included transcriptome profiling and none, so far as we know, compared its transcriptional changes with SD.
We collected brain tissue at one time point (ZT 13) to avoid the confounding factor of time-dependent variability (50,51) and centered our analyses on the ACC to decrease regional variability (49). The ACC is a key region associated with depressive symptoms and is likely to play a role in the mechanism of action of antidepressants (12,27). Furthermore, circadian function in MDD patients, compared with nonpsychiatric control subjects, is profoundly disrupted in the ACC (12,26).
We administered the FST, a standardized behavioral measure used in rodents for predicting efficacy of antidepressants (31). Our results showed similar efficacy for SD and KT groups that was significantly different from the saline-treated control group. Antidepressant-like activity on the FST for KT (39,52,53) and SD (30,37) has been previously reported.
We applied three pathway analyses, GO, KEGG, and GSEA, to analyze our findings. GO identifies molecular function, cellular component, and biological processes (54). We found 64 genes common to both SD and KT. Of these, a greater number was responsive to SD (11%) compared with KT (5%). Shared GO and KEGG analysis (55) indicated an overrepresentation of genes relevant to neuronal plasticity, nervous system development, synaptic transmission, and intracellular signal transduction including the MAPK signaling pathway. We used the GSEA for the identification of biological functions. Importantly, we identified circadian rhythm genes, ligand-receptor interactions, ion binding, neuronal differentiation, anatomical structure development, and regulation of gene expression. Five circadian transcripts (Per2, Npas4, Rorb, Dbp, and Ciart) were downregulated by both treatments. In contrast, Per1 was differentially expressed in SD (upregulated) and KT (downregulated) treatments.
Our results are consistent with our earlier findings in neuronal cell culture (NG108-15) showing that KT downregulates Per2 via its inhibitory actions on the heterodimer, CLOCK/BMAL1, a key regulatory circadian component driving clock gene transcription (25). SD-induced transcriptional changes may involve similar mechanisms to repress DNA-binding of CLOCK/BMAL1 (50). This could potentially provide a mechanism by which resetting clock gene machinery could normalize dysregulated circadian rhythms and rapidly treat MDD.
KT is a noncompetitive high-affinity N-methyl-D-aspartate receptor antagonist, a glutamate receptor, with complex actions at multiple sites. These include the inhibition of intraneuronal N-methyl-D-aspartate receptors, the release of presynaptic glutamate, activation of postsynaptic α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptors, as well as activation of synaptogenic intracellular signaling pathways including brain-derived neurotrophic factor and mammalian target of rapamycin complex 1 (56). Gsk3β, a potent inhibitor of mammalian target of rapamycin complex 1 and a circadian-relevant gene, potentiates KT’s antidepressant effects in mice (57,58) and is thought to contribute to KT’s sustained actions beyond its relatively short half-life (2–3 hours) (59,60). These processes could facilitate synaptogenesis contributing to KT’s rapid antidepressant activity (56).
We showed that both KT and SD alter MAPK signaling (45–48,61). As seen in rodents, inhibition of MAPK blocks its antidepressant actions resulting in depressive-like behavior (62,63). Moreover, it has been observed that the MAPK pathway is able to control the circadian clock (47). The MAPK pathway responds to excitatory glutamatergic signaling controlling synaptic plasticity and higher brain processes such as learning and memory. Importantly, this pathway has been related to neuropathological processes including depression (64).
We also identified NOTCH as a common signaling pathway for SD and KT. The NOTCH signaling pathway has been reported in SD studies in the periphery in humans (65) and is associated with epigenetic mechanisms related to altered methylation following sleep loss, likely acting at the level of the CLOCK/BMAL1 complex to alter circadian clock gene expression (66). To our knowledge, there are no other studies showing KT’s effect on this pathway. The NOTCH pathway has been largely implicated in brain development (67,68). Moreover, recent discoveries revealed that this pathway has a prominent role in neuronal plasticity in adult brain (69). Also, it has been observed that the circadian clock controls some components of the NOTCH signaling pathway (45,46).
Both the MAPK and NOTCH pathways contribute to adult brain function by controlling processes such as cell migration, morphology, and nerve maturation (69,70). Bagot et al. (71) have used a model based on a social-interaction paradigm, subjecting the mice to a chronic social defeat stress to induce a depression-like state. This is likely to induce transcriptional responses that differ from our mice, because we used an experimental paradigm that does not include chronic stress (72). Notwithstanding, because both paradigms used KT as an antidepressant, we found important similarities in KT-related gene responses. Both studies identify activation of the MAPK pathway as transcriptionally altered by KT and overall underscore the effect of KT and SD on neuronal plasticity (Supplemental Table S2). Indeed, Bagot et al. (71) found genes encoding biological functions related to plasticity, including nerve maturation, cell maturation, and ion transport in the group of responders to KT. These were also found in our analysis. These findings imply that KT and SD may trigger similar mechanisms of neuronal reorganization, regulating central nervous system development, neuronal differentiation, and neuronal spine remodeling, which appear to be under circadian control (73–76). While the fact that Bagot et al. (71) did not find alterations in clock genes could be due to the timing of the collection of samples. It is important to stress that in our study the comparison is not between two pharmacological treatments, but rather between a pharmacological treatment and a nonpharmacological treatment.
Moreover, the ACC receives cholinergic afferents from the prefrontal cortex (40), which can be induced by environmental stressors, inducing depression and mood regulation (77). This observation coincides with our results showing a downregulation of the ACh receptor Chrnb2 (Supplemental Tables S1 and S2). Importantly, it has been postulated that rapid eye movement sleep disinhibition in depression is a consequence of a cholinergic neuronal overactivity (78). Therefore, our results support the notion that the effects of SD and KT as rapid antidepressants might involve a common mechanism on the sleep physiology and neuronal plasticity triggered by the circadian clock.
As noted previously, the ACC is an important pathophysiological structure in depression. Our findings that both KT and SD could modulate, in a similar manner, circadian clock genes in the ACC of mice reveal the presence of common mechanisms of action related to rapid improvement in depressive symptoms. The ACC has extensive projections to multiple brain regions including the hippocampus, amygdala, and striatum (27). Thus, disruption in clock gene expression in the ACC could account for profound changes associated with depression such as altered hormone secretion, temperature, mood, and sleep rhythms (12). Direct evidence from our microarray analyses of circadian clock genes in postmortem ACC of MDD patients, compared with matched control subjects, shows a significant disruption in gene expression (26). Thus, the common mode of actions of KT and SD on clock gene expression could help identify new targets for rapid and improved treatment.
The FST is a reliable measure of antidepressant efficacy in rodents (2,10,30,39,53,79), evoking despair-like behavior. Yet FST could be stressful and might trigger stress-related transcriptional responses. To minimize the effects of stress in our analysis, all groups of mice including the saline control were subjected to the FST. Moreover, depression is frequently accompanied by anxiety, and some antidepressants can also acts as anxiolytic agents (80). Also, anhedonia, the loss of interest in normally pleasurable rewarding activities, is a main symptom of depression (81). Thus, future behavioral studies will explore the possible role played by anxiety and anhedonia.
Although the response time to low-dose KT varies considerably in patients with MDD, this does not appear to be the case in laboratory mice. We have carefully evaluated the best possible experimental conditions for the comparison between KT and SD. We reasoned that the comparison of the effects elicited by both treatments should be carefully designed to avoid possible differential responses. Based on previous reports, we reasoned the following: 1) mice needed to be genetically identical and age matched; 2) an appropriate timing for both treatments needed to be applied to elicit very similar behavioral effects in mice (30,39,53); and 3) the molecular analysis needed to be timed at a period in which both treatments elicited similar effects (30,39,53). We considered that designing a clear and simple paradigm in a strain of mice already tested for both treatments was crucial to avoiding confounding factors and to identifying key molecular responses common in both treatments. Further insight could be obtained by exploring additional timings for treatments. These may reveal useful data in the possible design of chronotherapies. Future research that will apply multiple time points will reveal whether our findings are stable or change with time. Finally, future research could compare higher KT doses with doses of 3 mg/kg on circadian clock gene expression. A dose of 3 mg/kg has been previously reported to have an antidepressant effect on the FST starting 30 minutes postinjection in mice (39,53,82).
In conclusion, common actions of KT and SD on clock gene expression could help identify new targets for rapid and improved treatment. While further studies will allow the determination of the precise role of the circadian clock in the antidepressant actions of SD and KT, our findings reveal a previously unappreciated convergence of genes and pathways between SD and KT, opening new research avenues to design chronopharmacological strategies to treat MDD.
Supplementary Material
Acknowledgments
This study was supported by funds from the National Institutes of Health, the Novo Nordisk Foundation Challenge Grant, and the Institut National de la Sante et Recherche Medicale, France. RO-S was supported by a fellowship from the Government of Mexico (CONACYT) and by the Della Martin Foundation.
We thank all members of the Sassone-Corsi and Emiliana Borrelli laboratories for constructive comments and help.
Footnotes
DISCLOSURES
All authors report no biomedical financial interests or potential conflicts of interest.
Supplementary material cited in this article is available online at http://dx.doi.org/10.1016/j.biopsych.2017.02.1176.
References
- 1.NIMH. Major depression among adults. [Accessed September 18, 2016];2014 Available at: http://www.nimh.nih.gov/health/statistics/prevalence/major-depression-among-adults.shtml.
- 2.Berman RM, Cappiello A, Anand A, Oren DA, Heninger GR, Charney DS, Krystal JH. Antidepressant effects of ketamine in depressed patients. Biol Psychiatry. 2000;47:351–354. doi: 10.1016/s0006-3223(99)00230-9. [DOI] [PubMed] [Google Scholar]
- 3.Bunney BG, Bunney WE. Rapid-acting antidepressant strategies: mechanisms of action. Int J Neuropsychopharmacol. 2012;15:695–713. doi: 10.1017/S1461145711000927. [DOI] [PubMed] [Google Scholar]
- 4.DiazGranados N, Ibrahim LA, Brutsche NE, Ameli R, Henter ID, Luckenbaugh DA, et al. Rapid resolution of suicidal ideation after a single infusion of an N-methyl-D-aspartate antagonist in patients with treatment-resistant major depressive disorder. J Clin Psychiatry. 2010;71:1605–1611. doi: 10.4088/JCP.09m05327blu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larkin GL, Beautrais AL. A preliminary naturalistic study of low-dose ketamine for depression and suicide ideation in the emergency department. Int J Neuropsychopharmacol. 2011;14:1127–1131. doi: 10.1017/S1461145711000629. [DOI] [PubMed] [Google Scholar]
- 6.Price RB, Nock MK, Charney DS, Mathew SJ. Effects of intravenous ketamine on explicit and implicit measures of suicidality in treatment-resistant depression. Biol Psychiatry. 2009;66:522–526. doi: 10.1016/j.biopsych.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murrough JW, Soleimani L, DeWilde KE, Collins KA, Lapidus KA, Iacoviello BM, et al. Ketamine for rapid reduction of suicidal ideation: A randomized controlled trial. Psychol Med. 2015;45:3571–3580. doi: 10.1017/S0033291715001506. [DOI] [PubMed] [Google Scholar]
- 8.Zarate CA, Jr, Brutsche NE, Ibrahim L, Franco-Chaves J, Diazgranados N, Cravchik A, et al. Replication of ketamine's antidepressant efficacy in bipolar depression: A randomized controlled add-on trial. Biol Psychiatry. 2012;71:939–946. doi: 10.1016/j.biopsych.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Price RB, Iosifescu DV, Murrough JW, Chang LC, Al Jurdi RK, Iqbal SZ, et al. Effects of ketamine on explicit and implicit suicidal cognition: A randomized controlled trial in treatment-resistant depression. Depress Anxiety. 2014;31:335–343. doi: 10.1002/da.22253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dallaspezia S, Suzuki M, Benedetti F. Chronobiological therapy for mood disorders. Curr Psychiatry Rep. 2015;17:95. doi: 10.1007/s11920-015-0633-6. [DOI] [PubMed] [Google Scholar]
- 11.Masri S, Sassone-Corsi P. The circadian clock: A framework linking metabolism, epigenetics and neuronal function. Nat Rev Neurosci. 2013;14:69–75. doi: 10.1038/nrn3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bunney BG, Li JZ, Walsh DM, Stein R, Vawter MP, Cartagena P, et al. Circadian dysregulation of clock genes: Clues to rapid treatments in major depressive disorder. Mol Psychiatry. 2015;20:48–55. doi: 10.1038/mp.2014.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasler BP, Buysse DJ, Kupfer DJ, Germain A. Phase relationships between core body temperature, melatonin, and sleep are associated with depression severity: Further evidence for circadian misalignment in non-seasonal depression. Psychiatry Res. 2010;178:205–207. doi: 10.1016/j.psychres.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Troxel WM, Kupfer DJ, Reynolds CF, 3rd, Frank E, Thase ME, Miewald JM, et al. Insomnia and objectively measured sleep disturbances predict treatment outcome in depressed patients treated with psychotherapy or psychotherapy-pharmacotherapy combinations. J Clin Psychiatry. 2012;73:478–485. doi: 10.4088/JCP.11m07184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Avery DH, Shah SH, Eder DN, Wildschiodtz G. Nocturnal sweating and temperature in depression. Acta Psychiatr Scand. 1999;100:295–301. doi: 10.1111/j.1600-0447.1999.tb10864.x. [DOI] [PubMed] [Google Scholar]
- 16.Souetre E, Salvati E, Wehr TA, Sack DA, Krebs B, Darcourt G. Twenty-four-hour profiles of body temperature and plasma TSH in bipolar patients during depression and during remission and in normal control subjects. Am J Psychiatry. 1988;145:1133–1137. doi: 10.1176/ajp.145.9.1133. [DOI] [PubMed] [Google Scholar]
- 17.Bunney BG, Bunney WE. Mechanisms of rapid antidepressant effects of sleep deprivation therapy: Clock genes and circadian rhythms. Biol Psychiatry. 2013;73:1164–1171. doi: 10.1016/j.biopsych.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 18.Bunney WE, Bunney BG. Molecular clock genes in man and lower animals: Possible implications for circadian abnormalities in depression. Neuropsychopharmacology. 2000;22:335–345. doi: 10.1016/S0893-133X(99)00145-1. [DOI] [PubMed] [Google Scholar]
- 19.Wu JC, Kelsoe JR, Schachat C, Bunney BG, DeModena A, Golshan S, et al. Rapid and sustained antidepressant response with sleep deprivation and chronotherapy in bipolar disorder. Biol Psychiatry. 2009;66:298–301. doi: 10.1016/j.biopsych.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 20.Wisor JP, O'Hara BF, Terao A, Selby CP, Kilduff TS, Sancar A, et al. A role for cryptochromes in sleep regulation. BMC Neurosci. 2002;3:20. doi: 10.1186/1471-2202-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wisor JP, Pasumarthi RK, Gerashchenko D, Thompson CL, Pathak S, Sancar A, et al. Sleep deprivation effects on circadian clock gene expression in the cerebral cortex parallel electroencephalographic differences among mouse strains. J Neurosci. 2008;28:7193–7201. doi: 10.1523/JNEUROSCI.1150-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson CL, Wisor JP, Lee CK, Pathak SD, Gerashchenko D, Smith KA, et al. Molecular and anatomical signatures of sleep deprivation in the mouse brain. Front Neurosci. 2010;4:165. doi: 10.3389/fnins.2010.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 24.Maret S, Dorsaz S, Gurcel L, Pradervand S, Petit B, Pfister C, et al. Homer1a is a core brain molecular correlate of sleep loss. Proc Natl Acad Sci U S A. 2007;104:20090–20095. doi: 10.1073/pnas.0710131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bellet MM, Vawter MP, Bunney BG, Bunney WE, Sassone-Corsi P. Ketamine influences CLOCK:BMAL1 function leading to altered circadian gene expression. PLoS One. 2011;6:e23982. doi: 10.1371/journal.pone.0023982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li JZ, Bunney BG, Meng F, Hagenauer MH, Walsh DM, Vawter MP, et al. Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc Natl Acad Sci U S A. 2013;110:9950–9955. doi: 10.1073/pnas.1305814110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drevets WC, Savitz J, Trimble M. The subgenual anterior cingulate cortex in mood disorders. CNS Spectr. 2008;13:663–681. doi: 10.1017/s1092852900013754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salvadore G, Cornwell BR, Colon-Rosario V, Coppola R, Grillon C, Zarate CA, Jr, Manji HK. Increased anterior cingulate cortical activity in response to fearful faces: A neurophysiological biomarker that predicts rapid antidepressant response to ketamine. Biol Psychiatry. 2009;65:289–295. doi: 10.1016/j.biopsych.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mulert C, Juckel G, Brunnmeier M, Karch S, Leicht G, Mergl R, et al. Prediction of treatment response in major depression: Integration of concepts. J Affect Disord. 2007;98:215–225. doi: 10.1016/j.jad.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 30.Hines DJ, Schmitt LI, Hines RM, Moss SJ, Haydon PG. Antidepressant effects of sleep deprivation require astrocyte-dependent adenosine mediated signaling. Transl Psychiatry. 2013;3:e212. doi: 10.1038/tp.2012.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Can A, Dao DT, Arad M, Terrillion CE, Piantadosi SC, Gould TD. The mouse forced swim test. J Vis Exp. 2012;59:e3638. doi: 10.3791/3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orozco-Solis R, Ramadori G, Coppari R, Sassone-Corsi P. SIRT1 relays nutritional inputs to the circadian clock through the Sf1 neurons of the ventromedial hypothalamus. Endocrinology. 2015;156:2174–2184. doi: 10.1210/en.2014-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Surget A, Wang Y, Leman S, Ibarguen-Vargas Y, Edgar N, Griebel G, et al. Corticolimbic transcriptome changes are state-dependent and region-specific in a rodent model of depression and of antidepressant reversal. Neuropsychopharmacology. 2008;34:1363–1380. doi: 10.1038/npp.2008.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berton O, McClung CA, Dileone RJ, Krishnan V, Renthal W, Russo SJ, et al. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science. 2006;311:864–868. doi: 10.1126/science.1120972. [DOI] [PubMed] [Google Scholar]
- 35.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: A non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012;40:W478–W483. doi: 10.1093/nar/gks402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez-Rodriguez F, Kim J, Poland RE. Total sleep deprivation decreases immobility in the forced-swim test. Neuropsychopharmacology. 2004;29:1105–1111. doi: 10.1038/sj.npp.1300406. [DOI] [PubMed] [Google Scholar]
- 38.Scheuing L, Chiu CT, Liao HM, Chuang DM. Antidepressant mechanism of ketamine: Perspective from preclinical studies. Front Neurosci. 2015;9:249. doi: 10.3389/fnins.2015.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Autry AE, Adachi M, Nosyreva E, Na ES, Los MF, Cheng PF, et al. NMDA receptor blockade at rest triggers rapid behavioural antidepressant responses. Nature. 2011;475:91–95. doi: 10.1038/nature10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fillinger C, Yalcin I, Barrot M, Veinante P. Afferents to anterior cingulate areas 24a and 24b and midcingulate areas 24a' and 24b' in the mouse. Brain Struct Funct. 2017;222:1509–1532. doi: 10.1007/s00429-016-1290-1. [DOI] [PubMed] [Google Scholar]
- 41.Annayev Y, Adar S, Chiou YY, Lieb JD, Sancar A, Ye R. Gene model 129 (Gm129) encodes a novel transcriptional repressor that modulates circadian gene expression. J Biol Chem. 2014;289:5013–5024. doi: 10.1074/jbc.M113.534651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goriki A, Hatanaka F, Myung J, Kim JK, Yoritaka T, Tanoue S, et al. A novel protein, CHRONO, functions as a core component of the mammalian circadian clock. PLoS Biol. 2014;12:e1001839. doi: 10.1371/journal.pbio.1001839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petryszak R, Keays M, Tang YA, Fonseca NA, Barrera E, Burdett T, et al. Expression Atlas update—An integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 2016;44:D746–D752. doi: 10.1093/nar/gkv1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orozco-Solis R, Sassone-Corsi P. Epigenetic control and the circadian clock: Linking metabolism to neuronal responses. Neuroscience. 2014;264:76–87. doi: 10.1016/j.neuroscience.2014.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janich P, Pascual G, Merlos-Suárez A, Batlle E, Ripperger J, Albrecht U, et al. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature. 2011;480:209–214. doi: 10.1038/nature10649. [DOI] [PubMed] [Google Scholar]
- 46.Karpowicz P, Zhang Y, Hogenesch JB, Emery P, Perrimon N. The circadian clock gates the intestinal stem cell regenerative state. Cell Reports. 2013;3:996–1004. doi: 10.1016/j.celrep.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akashi M, Nishida E. Involvement of the MAP kinase cascade in resetting of the mammalian circadian clock. Genes Dev. 2000;14:645–649. [PMC free article] [PubMed] [Google Scholar]
- 48.Gerstner JR, Yin JCP. Circadian rhythms and memory formation. Nat Rev Neurosci. 2010;11:577–588. doi: 10.1038/nrn2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ficek J, Zygmunt M, Piechota M, Hoinkis D, Rodriguez Parkitna J, Przewlocki R, Korostynski M. Molecular profile of dissociative drug ketamine in relation to its rapid antidepressant action. BMC Genomics. 2016;17:362. doi: 10.1186/s12864-016-2713-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mongrain V, La Spada F, Curie T, Franken P. Sleep loss reduces the DNA-binding of BMAL1, CLOCK, and NPAS2 to specific clock genes in the mouse cerebral cortex. PLoS One. 2011;6:e26622. doi: 10.1371/journal.pone.0026622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Curie T, Mongrain V, Dorsaz S, Mang GM, Emmenegger Y, Franken P. Homeostatic and circadian contribution to EEG and molecular state variables of sleep regulation. Sleep. 2013;36:311–323. doi: 10.5665/sleep.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garcia LS, Comim CM, Valvassori SS, Reus GZ, Barbosa LM, Andreazza AC, et al. Acute administration of ketamine induces antidepressant-like effects in the forced swimming test and increases BDNF levels in the rat hippocampus. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32:140–144. doi: 10.1016/j.pnpbp.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 53.Weckmann K, Labermaier C, Asara JM, Muller MB, Turck CW. Time-dependent metabolomic profiling of Ketamine drug action reveals hippocampal pathway alterations and biomarker candidates. Transl Psychiatry. 2014;4:e481. doi: 10.1038/tp.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gene Ontology Consortium. Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abdallah CG, Adams TG, Kelmendi B, Esterlis I, Sanacora G, Krystal JH. Ketamine's mechanism of action: A path to rapid-acting antidepressants. Depress Anxiety. 2016;33:689–697. doi: 10.1002/da.22501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beurel E, Song L, Jope RS. Inhibition of glycogen synthase kinase-3 is necessary for the rapid antidepressant effect of ketamine in mice. Mol Psychiatry. 2011;16:1068–1070. [Google Scholar]
- 58.Liu RJ, Fuchikami M, Dwyer JM, Lepack AE, Duman RS, Aghajanian GK. GSK-3 inhibition potentiates the synaptogenic and antidepressant-like effects of subthreshold doses of ketamine. Neuropsychopharmacology. 2013;38:2268–2277. doi: 10.1038/npp.2013.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mathews DC, Zarate CA., Jr Current status of ketamine and related compounds for depression. J Clin Psychiatry. 2013;74:516–517. doi: 10.4088/JCP.13ac08382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kavalali ET, Monteggia LM. Synaptic mechanisms underlying rapid antidepressant action of ketamine. Am J Psychiatry. 2012;169:1150–1156. doi: 10.1176/appi.ajp.2012.12040531. [DOI] [PubMed] [Google Scholar]
- 61.Antle MC, Tse F, Koke SJ, Sterniczuk R, Hagel K. Non-photic phase shifting of the circadian clock: Role of the extracellular signal-responsive kinases I/II/mitogen-activated protein kinase pathway. Eur J Neurosci. 2008;28:2511–2518. doi: 10.1111/j.1460-9568.2008.06533.x. [DOI] [PubMed] [Google Scholar]
- 62.Duman CH, Schlesinger L, Kodama M, Russell DS, Duman RS. A role for MAP kinase signaling in behavioral models of depression and antidepressant treatment. Biol Psychiatry. 2007;61:661–670. doi: 10.1016/j.biopsych.2006.05.047. [DOI] [PubMed] [Google Scholar]
- 63.Reus GZ, Vieira FG, Abelaira HM, Michels M, Tomaz DB, dos Santos MA, et al. MAPK signaling correlates with the antidepressant effects of ketamine. J Psychiatr Res. 2014;55:15–21. doi: 10.1016/j.jpsychires.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 64.Thomas GM, Huganir RL. MAPK cascade signalling and synaptic plasticity. Nat Rev Neurosci. 2004;5:173–183. doi: 10.1038/nrn1346. [DOI] [PubMed] [Google Scholar]
- 65.Nilsson EK, Bostrom AE, Mwinyi J, Schioth HB. Epigenomics of total acute sleep deprivation in relation to genome-wide DNA methylation profiles and RNA expression. OMICS. 2016;20:334–342. doi: 10.1089/omi.2016.0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cedernaes J, Osler ME, Voisin S, Broman JE, Vogel H, Dickson SL, et al. Acute sleep loss induces tissue-specific epigenetic and transcriptional alterations to circadian clock genes in men. J Clin Endocrinol Metab. 2015;100:E1255–E1261. doi: 10.1210/JC.2015-2284. [DOI] [PubMed] [Google Scholar]
- 67.Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: Insights from mouse mutants. Nat Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- 68.Chiba S. Notch signaling in stem cell systems. Stem Cells. 2006;24:2437–2447. doi: 10.1634/stemcells.2005-0661. [DOI] [PubMed] [Google Scholar]
- 69.Ables JL, Breunig JJ, Eisch AJ, Rakic P. Not(ch) just development: Notch signalling in the adult brain. Nat Rev Neurosci. 2011;12:269–283. doi: 10.1038/nrn3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alberi L, Liu S, Wang Y, Badie R, Smith-Hicks C, Wu J, et al. Activity-induced Notch signaling in neurons requires Arc/Arg3.1 and is essential for synaptic plasticity in hippocampal networks. Neuron. 2011;69:437–444. doi: 10.1016/j.neuron.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bagot RC, Cates HM, Purushothaman I, Vialou V, Heller EA, Yieh L, et al. Ketamine and imipramine reverse transcriptional signatures of susceptibility and induce resilience-specific gene expression profiles. Biol Psychiatry. 2017;81:285–295. doi: 10.1016/j.biopsych.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Papadopoulou A, Siamatras T, Delgado-Morales R, Amin ND, Shukla V, Zheng YL, et al. Acute and chronic stress differentially regulate cyclin-dependent kinase 5 in mouse brain: Implications to glucocorticoid actions and major depression. Transl Psychiatry. 2015;5:e578. doi: 10.1038/tp.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Duman RS, Aghajanian GK. Synaptic dysfunction in depression: Potential therapeutic targets. Science. 2012;338:68–72. doi: 10.1126/science.1222939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mei L, Xiong WC. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci. 2008;9:437–452. doi: 10.1038/nrn2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Karatsoreos IN, Bhagat S, Bloss EB, Morrison JH, McEwen BS. Disruption of circadian clocks has ramifications for metabolism, brain, and behavior. Proc Natl Acad Sci U S A. 2011;108:1657–1662. doi: 10.1073/pnas.1018375108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maret S, Faraguna U, Nelson AB, Cirelli C, Tononi G. Sleep and waking modulate spine turnover in the adolescent mouse cortex. Nat Neurosci. 2011;14:1418–1420. doi: 10.1038/nn.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mineur YS, Obayemi A, Wigestrand MB, Fote GM, Calarco CA, Li AM, Picciotto MR. Cholinergic signaling in the hippocampus regulates social stress resilience and anxiety- and depression-like behavior. Proc Natl Acad Sci U S A. 2013;110:3573–3578. doi: 10.1073/pnas.1219731110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Berger M, Riemann D, Höchli D, Spiegel R. The cholinergic rapid eye movement sleep induction test with rs-86: State or trait marker of depression? Arch Gen Psychiatry. 1989;46:421–428. doi: 10.1001/archpsyc.1989.01810050035006. [DOI] [PubMed] [Google Scholar]
- 79.Benedetti F, Dallaspezia S, Lorenzi C, Pirovano A, Radaelli D, Locatelli C, et al. Gene-gene interaction of glycogen synthase kinase 3-β and serotonin transporter on human antidepressant response to sleep deprivation. J Affect Disord. 2012;136:514–519. doi: 10.1016/j.jad.2011.10.039. [DOI] [PubMed] [Google Scholar]
- 80.Ressler KJ, Mayberg HS. Targeting abnormal neural circuits in mood and anxiety disorders: From the laboratory to the clinic. Nat Neurosci. 2007;10:1116–1124. doi: 10.1038/nn1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nat Neurosci. 2010;13:1161–1169. doi: 10.1038/nn.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Serchov T, Clement HW, Schwarz MK, Iasevoli F, Tosh DK, Idzko M, et al. Increased signaling via adenosine A1 receptors, sleep deprivation, imipramine, and ketamine inhibit depressive-like behavior via induction of Homer1a. Neuron. 2015;87:549–562. doi: 10.1016/j.neuron.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.