Abstract
In evaluating pathological changes in drug efficacy and toxicity studies, morphometric analysis can be quite robust. In this experiment, we examined whether morphometric changes of major pathological findings in various tissue specimens stained with hematoxylin and eosin could be recognized and quantified using image processing software. Using Tissue Studio, hypertrophy of hepatocytes and adrenocortical cells could be quantified based on the method of a previous report, but the regions of red pulp, white pulp, and marginal zones in the spleen could not be recognized when using one setting condition. Using Image-Pro Plus, lipid-derived vacuoles in the liver and mucin-derived vacuoles in the intestinal mucosa could be quantified using two criteria (area and/or roundness). Vacuoles derived from phospholipid could not be quantified when small lipid deposition coexisted in the liver and adrenal cortex. Mononuclear inflammatory cell infiltration in the liver could be quantified to some extent, except for specimens with many clustered infiltrating cells. Adipocyte size and the mean linear intercept could be quantified easily and efficiently using morphological processing and the macro tool equipped in Image-Pro Plus. These methodologies are expected to form a base system that can recognize morphometric features and analyze quantitatively pathological findings through the use of information technology.
Keywords: morphometry, quantitative analysis, digital image processing, image processing software, digital pathology, hematoxylin and eosin
Pathological examination is the only way to directly confirm drug efficacy and toxicity through the observation of tissue specimens in both clinical and nonclinical situations1. Development of new drugs often fails due to drug toxicity, despite a potentially high efficacy. Conversely, when no abnormality on pathological investigation is observed, it is possible to approve a new drug with confidence.
Pathological evaluation of tissue specimens is of utmost importance, but it is usually performed qualitatively or semiquantitatively by pathologists2, 3. Bias in the evaluation can occur between individuals and between facilities, and a pathological evaluation requires a great deal of time. Furthermore, pathological and morphological analyses require special expertise, so researchers without pathological knowledge can have trouble interpreting the data. One solution to these problems is to quantify the morphological changes of pathological tissue automatically, leading to improvement of objectivity, reliability, and robustness. Automatic quantification will also lead to a reduction in time and increased data.
Quantification of histopathological changes using morphometry has been performed for some time. In the 1990’s, pathological images were digitized, and the histopathological changes were measured by image analysis software as an accurate numeric value4, 5. With the development of information technology, digital pathology has also developed, and whole slide images are easily acquired using a virtual slide scanner. Image processing software can analyze more complex tissue images. Recently, histopathological changes were analyzed in detail by image analysis software equipped with machine learning6 and a programmed tool7, 8. Automated diagnostic software for some cancer tissues is being developed for clinical applications.
We have been trying to recognize and quantify various types of histopathological findings and have pursued quantification of morphometric changes of pathological specimens. In this report, we introduce these methodologies on quantification of morphometric changes of histopathological images stained with hematoxylin and eosin (HE), which is a conventional staining allowing for comparisons with previous specimens. We address the possibility and the limitations of analysis using existing image processing software.
In this report, we used tissue specimens that were fixed in formalin, embedded in paraffin, and stained with HE in previous experiments. Whole digital slide images were obtained using virtual microscopy (Aperio AT2, Leica Microsystems, Wetzlar, Germany). Joint Photographic Expert Group (JPEG) images were extracted using image processing software (ImageScope, ver. 12.0.0.5039, Leica Microsystems). Tissue Studio (ver. 4.2.0, Definiens, Munich, Germany) and Image-Pro Plus (ver. 7.0.1.658, Media Cybernetics, Rockville, MD, USA) were used as image analysis software.
Table 1 shows a summary of the properties of quantification of various findings that were examined in this study using image processing software. The detailed methodology of each examination is described beginning in the following section.
Table 1. Summary of the Properties of Quantification of the Various Findings Using Image Processing Software.
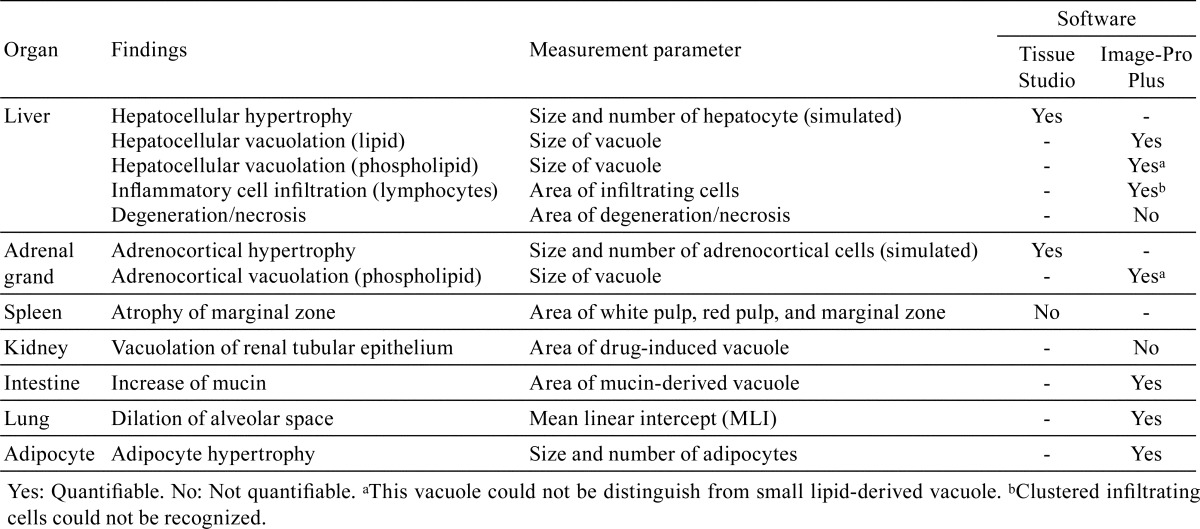
Hepatocellular hypertrophy was quantitatively analyzed based on the method of a previous report9 using Tissue Studio. Briefly, the nuclei of hepatocytes were detected (hematoxylin threshold, 0.08; typical nucleus size, 70 μm2), and the regions of the nuclei had expanded toward neighboring regions. Nuclei in sinusoids could be excluded by setting the conditions to an area <25, length/width >1.8, and hematoxylin intensity >0.4. The expanded regions were defined as simulated hepatocytes, and their areas were measured. Images colored based upon cell size are shown in Fig. 1A. The ratios of the numbers of larger cells were increased (Fig. 1B), as were the ratios of the areas occupied by the larger cells (Fig. 1C) in the hypertrophied hepatocytes compared with normal hepatocytes. The results of quantitative analysis of hypertrophy were usually consistent with the observations of a pathologist. Nuclei often do not appear in sections of hypertrophied hepatocytes, so not all hypertrophied hepatocytes could be simulated and evaluated. It is preferable to measure each actual hepatocyte region by detection of the cell membrane, but detection is difficult because it is difficult to recognize clearly the entire boundary of hepatocytes. A tool that can correctly recognize the boundary of the hepatocellular membrane or detect sinusoids is needed.
Fig. 1.
Analysis of cell size using Tissue Studio. (A) The hepatocellular region was simulated based upon the nucleus, and then these cells were colored based upon the cell size in normal (left image) and hypertrophied (right image) hepatocytes. Yellow cell, < 300 μm2; orange cell, 300–400 μm2; brown cell, >400 μm2. Uncolored hepatocytes were not simulated because their nuclei had not appeared. Bar = 100 μm. (B) The ratios of the numbers of hepatocytes per size. (C) The ratio of the total area of hepatocytes per size. (D) Adrenocortical cells were colored based upon the cell size in the normal (left image) and hypertrophied (right image) adrenal gland. Yellow cell, <200 μm2; orange cell, 200–300 μm2; brown cell, >300 μm2. Bar = 100 μm. (E) The ratios of the numbers of adrenocortical cells per size. (F) The ratio of the total area of the adrenocortical cells per size.
Hypertrophy of fascicular and reticular cells in the adrenal grand was analyzed using the same methods as used for hepatocytes. Images colored based upon cell size are shown in Fig. 1D. The ratios of the numbers of larger cells were increased (Fig. 1E), as were the ratios of the areas occupied by the larger cells (Fig. 1F) in the hypertrophied adrenocortical grand compared with in a normal adrenocortical gland.
Pathologically, the spleen has red pulp, white pulp, and a marginal zone, which is the border region between the red pulp and white pulp. These regions are sensitive to the immune response caused by drugs, and pathological toxicity is often detected in them. We attempted to recognize these regions using Tissue Studio for automatic analysis. Recognition was consistent with the observations of the pathologist in some specimens, but recognition was difficult in some specimens due to variable staining intensity between specimens. We were unable to determine criteria that could be used for recognition in all images. This problem may be solved by standardization of the staining intensity in digital images.
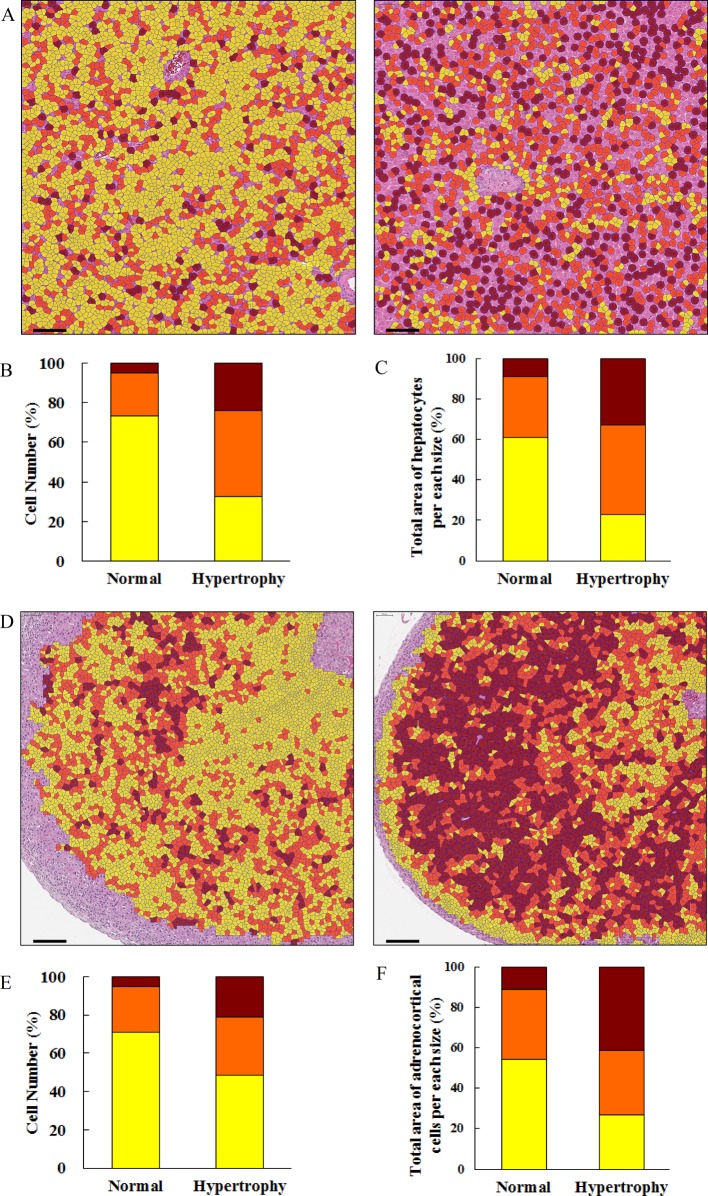
Quantification of hepatocellular lipid deposition in a histopathological image has been already reported. Unfortunately, though the ratio of the area occupied by lipid droplets in the evaluated region was demonstrated, the sizes of lipid droplets were not always evaluated10, 11. In this experiment, we used Image-Pro Plus to measure the size of each lipid droplet in JPEG images extracted from whole slide images of the liver. Lipid vacuoles in the histopathological image were defined as circular unstained objects. The size of the lipid vacuoles in hepatocytes was measured using two optimized criteria, an area of 1–500 μm2 and roundness of 1–1.5 units, in order to exclude non-lipid objects including large vessels, bile ducts, and sinusoidal spaces. The results of quantification were consistent with the observations of the pathologist (Fig. 2B). This method is very useful in the case of pathological conditions, including nonalcoholic steatohepatitis (NASH) that develops from microvesicular to macrovesicular steatosis. We have previously reported that the lipid droplet size became larger in a time-dependent manner in the NASH model12. In that report, the density of lipid droplets was high, and it was necessary to separate overlapping lipid droplets from one another, so several kinds of morphological image processing were performed. In this experiment, there were no overlapping lipid droplets, so advanced morphological image processing was not necessary. If the degree of overlapping lipid droplets is low, there is a method to separate them using the Watershed Sprit or Auto-Sprit command in Image-Pro Plus. The analysis method should be determined based upon the state of the vacuole, including its density. When morphological image processing is performed, it is necessary to check whether the original form of lipid droplets was unchanged. This method is applicable to other organs, unless there are unstained regions morphologically similar to the target vacuole.
Fig. 2.
Analysis of a cellular vacuole using Image-Pro Plus. (A) Analyzed image of the lipid-derived vacuole. Lipid droplets (green-colored objects) were recognized by the criteria of size and roundness, and the size was measured. Vessels, the bile duct, and sinusoidal spaces were excluded. Bar = 50 μm. (B) The total area of the lipid droplet per size in the hepatocyte with various grades of lipid deposition. (C) Representative images of tissue specimens with various grades (left, center, and right images: normal, −; very slight, ± ; and slight, +, respectively) of lipid deposition. Bar = 100 μm. (D) Quantification of the area of mucin in the intestinal mucosa. Left image: original image. Right image: analyzed image of mucin in the intestinal mucosa. The outline of the intestinal mucosa was drawn manually using the AOI setting (green line) in order to exclude nonspecific objects in the outside region, and then the inside region was analyzed. The mucin (light blue-colored objects) was recognized by the criteria of size. Bar = 200 μm.
In the intestinal mucosa, the area of mucin-derived vacuoles was measured using the same methods as used for lipid-derived vacuoles of the hepatocytes. Nonspecific objects in the outside region and internal space of the intestinal mucosa were excluded by setting the area of interest (AOI) of the outline of the mucosa and the size condition to 1–1,000 μm2. The results were consistent with the observations of the pathologist (Fig. 2D).
A vacuole derived from a phospholipid was observed in the case of drug-induced phospholipidosis. Unlike lipid-derived vacuoles, phospholipid-derived vacuoles are smaller and have a circular form and an unclear unstained region. The size of phospholipid-derived vacuoles was measured using an area of 1–40 μm2 as the criterion. The size of each droplet could be partially measured (data not shown). We could not distinguish between phospholipid-derived vacuoles and small lipid-derived vacuoles in the case of pathological conditions in the hepatocyte that typically accompanied microvesicular steatosis or in the case of the adrenal cortex with lipid droplets containing cholesterol.
We attempted to recognize and quantify a drug-induced vacuole in the renal tubules using an area of 1–1,000 μm2 as the criterion, but it was impossible to quantify only the vacuole specifically due to many unstained regions, including the renal tubular lumen, vessel lumen, and intercellular spaces. The kidney has many complex components, including the proximal tubule, distal tubule, renal pelvis, and glomerulus, and these regions are difficult to distinguish clearly. At present, the vacuole in each region cannot be precisely quantified.
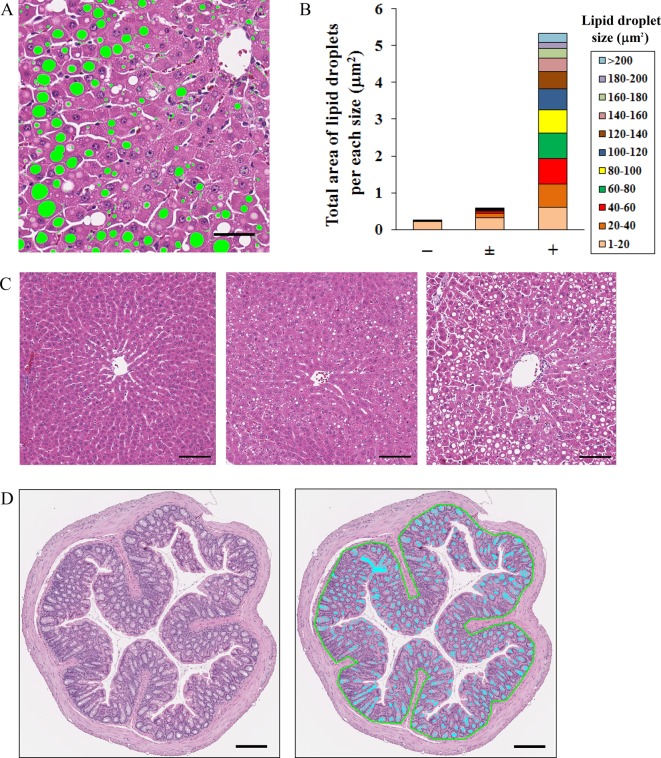
The histopathological characterization of infiltrating cells is basophilic and granular. In this experiment, using Image-Pro Plus, lymphocytes with mononuclear were analyzed quantitatively as inflammatory infiltrating cells in JPEG images of a 4 mm2 area extracted at 5 points from whole slide images of the liver. The nucleolus and contour of the nucleus of a hepatocyte were similar in form to a lymphocyte. These objects were impossible to distinguish by only setting the color and shape. By adding measurement conditions with fill holes, the area inside the contour of the nucleus was filled, and the nucleus was recognized as one object. Since the size of hepatocellular nucleus is larger than that of a lymphocyte, it was excluded by setting the size condition to 5–45 μm2. As a result, infiltrating cells could be analyzed quantitatively, and the results were consistent with the observations of the pathologist (Fig. 3B). However, using this size condition, clustered infiltrating cells could not be recognized, since the size of a cluster of lymphocytes is larger than 45 μm2. If there are many clustered infiltrating cells, the setting conditions must be changed. If neutrophils with lobulated nuclei and lymphocytes with mononuclear cells are mixed, it may be possible to distinguish them by setting the condition for roundness. In all cases, it is best to stain densely with hematoxylin.
Fig. 3.
Analysis of infiltrating cells (lymphocytes) using Image-Pro Plus. (A) Left image: original image of tissue specimen. Right image: analyzed image of infiltration cells. Green-colored objects were infiltrating cells, which were recognized using a size limitation of 5–45 μm2. The area of infiltrating cells was measured. Light blue-colored objects represent nuclei. By adding measurement conditions with fill holes, area inside the contour of a nucleus was filled, and the nucleus was recognized as one object, whose size was larger than 45 μm2. Clustered infiltrating cells (red arrows) had sizes larger than 45 μm2, so they were not recognized as infiltrating cells in this experiment. Bar = 20 μm. (B) Total area of infiltrating cells in the liver tissue with various grades of inflammatory cell infiltration. For analysis, JPEG images of 4 mm2 areas at 5 points were extracted from the whole slide image. (C) Representative images of various grades (left, center, and right images: normal, −; slight, +; and moderate, ++, respectively) of cell infiltration. Bar = 100 μm.
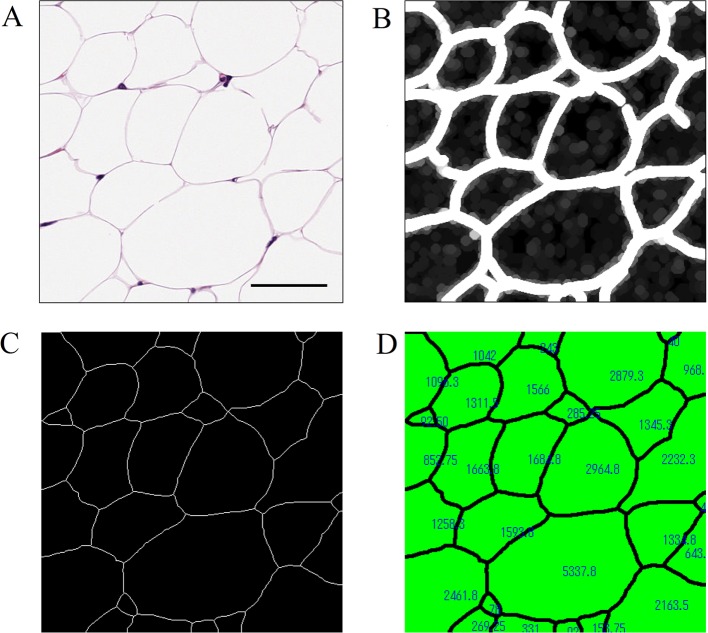
Adipocyte size could be quantitatively analyzed based upon a previous report13. Briefly, the image was converted by processing of an 8-bit gray scale and binarization. If needed, after manual adjustments with the erosion and paintbrush tools were performed, the adipocyte area was measured, with areas of less than 100 μm2 excluded from measurement. We previously reported the effect of drugs on adipocyte size using this method14, 15. In this experiment, we tried to avoid manual adjustments by utilizing the morphological filters equipped in Image-Pro Plus. In detail, the Well (7 × 7, pass 1, strength 10), Dilate (7 × 7 circle, pass 2), Close (7 × 7 circle, pass 1), and Pruning (threshold 50%) morphological filters were applied to the gray scale image (Fig. 4A–4C). The Dilate and Close morphological filters made it possible to thicken the contours of adipocytes, to connect slightly broken lines, and to eliminate small regions that were not adipocytes. Subsequently, the Pruning filter thinned the thickened contours of the adipocytes. Divided regions were defined as individual adipocytes, and their areas were measured (Fig. 4D). Although this method was more efficient compared with the previously reported one, it was necessary to verify that the images subjected to morphological processing were not extremely different from the original images.
Fig. 4.
An efficient method for measurement of adipocyte size using Image-Pro Plus. (A) A gray scale image of adipocytes. Bar = 50 μm. (B) Morphological processing with the Well, Dilate, and Close filters was performed. The contours of the adipocytes were thickened, slightly broken lines were connected, and small regions that were not adipocytes were eliminated. (C) Morphological processing with the Pruning filter was performed to thin the thickened contours. These divided regions were defined as adipocytes. (D) Each area was measured, and the process was almost automatic using the macro tool.
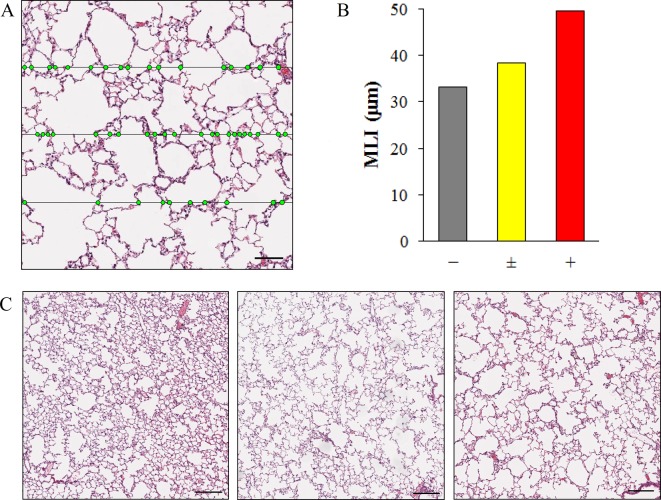
The mean linear intercept (MLI) in the lung is the mean distance between the alveolar walls, and it is an indicator of alveolar space. The MLI is calculated from the number of intersections between a straight line and the alveolar walls in a pathological tissue image16. The value of the MLI was analyzed easily and efficiently by using Image-Pro Plus. Specifically, the binarized image was superimposed on an image masked with grid lines (Process-Grid mask: Objects, LineOrth) (Fig. 5A), and only the intersections of both images remained (Process-Operation: OR). The number of intersections was measured. The MLI can be calculated as the ratio of the total length of the grid lines to the total number of intersections. In this experiment, each JPEG image of 0.63 mm2 areas extracted at 5 points from the left lobe and at each 2 points each from other lobes was analyzed. As a result, the MLI was larger in a lung exposed to cigarette smoke compared with a normal lung (Fig. 5B).
Fig. 5.
An efficient method for measurement of the MLI in the lung using Image-Pro Plus. (A) The lung tissue image was superimposed on grid lines (horizontal lines). The number of intersections (green points) between the alveolar walls and grid lines was measured almost automatically using the macro tool. The MLI was calculated from the ratio of the total length of the grid lines to the total number of intersections. Bar = 100 μm. (B) The MLI in the lung with various grades of alveolar space dilation. For analysis, each JPEG image of 0.63 mm2 areas extracted at 5 points from the left lobe and at 2 points each from the accessory, anterior, middle, and posterior lobe was analyzed. (C) Representative images of the various grades (left, center, and right images: normal, −; very slight, ± ; and slight, +, respectively) of alveolar space dilation. Bar = 200 μm.
We attempted to recognize a region of degeneration/necrosis in hepatocytes using Image-Pro Plus. Degeneration/necrosis of a hepatocyte has certain features, including an eosinophilic cytoplasm and an atrophied nucleus, but there are many regions with colors similar to those of a normal hepatocyte. In the present study, we were not able to automatically recognize a region of degeneration/necrosis. To quantify degeneration/necrosis using Image-Pro Plus, the AOI must be traced manually.
The convenience of automated quantification of a pathological tissue image is currently under discussion5, 17, 18. A means of automated clinical diagnosis of cancer has been developed19, 20. To date, there is no tool that can comprehensively analyze various types of pathological findings. Pathological tissues are very complex, and pathological changes have many variations in each organ. It may not be feasible to establish a general system that can automatically and quantitatively analyze the pathological changes of all tissue specimens.
In this report, we introduced several concrete examples of the quantitative analysis methods for pathological tissue findings examined using Tissue Studio and Image-Pro Plus as the image processing software. The demonstrated methods were just one example. The degree of pathological changes differs depending on the experiment, and morphologically similar findings of interest may occur in other experiments. Eventually, it will be necessary to set conditions appropriate for recognition and quantification of findings, but this study was designed as a starting point for future quantitative analysis. The establishment of a universal and efficient system for the recognition and quantification of histopathological changes will accelerate the creation of new drugs and serve as a potent support tool for pathologists.
Acknowledgments
The authors thank Satomi Nishikawa,Yuko Ono, Yuichi Murakami, Mao Mizukawa, Atsushi Fukunari, and Hiroyuki Utsumi for supporting the present study and helpful discussions.
Footnotes
Disclosure of Potential Conflict of InterestAll authors are employees of Mitsubishi Tanabe Pharma Corporation. The authors declare that they have no conflict of interest.
References
- 1.Potts SJ. Digital pathology in drug discovery and development: multisite integration. Drug Discov Today. 14: 935–941. 2009. [DOI] [PubMed] [Google Scholar]
- 2.Klopfleisch R. Multiparametric and semiquantitative scoring systems for the evaluation of mouse model histopathology--a systematic review. BMC Vet Res. 9: 123 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prasad K, and Prabhu GK. Image analysis tools for evaluation of microscopic views of immunohistochemically stained specimen in medical research-a review. J Med Syst. 36: 2621–2631. 2012. [DOI] [PubMed] [Google Scholar]
- 4.Isse K, Lesniak A, Grama K, Roysam B, Minervini MI, and Demetris AJ. Digital transplantation pathology: combining whole slide imaging, multiplex staining and automated image analysis. Am J Transplant. 12: 27–37. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, and Yener B. Histopathological image analysis: a review. IEEE Rev Biomed Eng. 2: 147–171. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kakimoto T, Kimata H, Iwasaki S, Fukunari A, and Utsumi H. Automated recognition and quantification of pancreatic islets in Zucker diabetic fatty rats treated with exendin-4. J Endocrinol. 216: 13–20. 2013. [DOI] [PubMed] [Google Scholar]
- 7.Kakimoto T, Okada K, Hirohashi Y, Relator R, Kawai M, Iguchi T, Fujitaka K, Nishio M, Kato T, Fukunari A, and Utsumi H. Automated image analysis of a glomerular injury marker desmin in spontaneously diabetic Torii rats treated with losartan. J Endocrinol. 222: 43–51. 2014. [DOI] [PubMed] [Google Scholar]
- 8.Kato T, Relator R, Ngouv H, Hirohashi Y, Takaki O, Kakimoto T, and Okada K. Segmental HOG: new descriptor for glomerulus detection in kidney microscopy image. BMC Bioinformatics. 16: 316 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asaoka Y, Togashi Y, Mutsuga M, Imura N, Miyoshi T, and Miyamoto Y. Histopathological image analysis of chemical-induced hepatocellular hypertrophy in mice. Exp Toxicol Pathol. 68: 233–239. 2016. [DOI] [PubMed] [Google Scholar]
- 10.Li M, Song J, Mirkov S, Xiao SY, Hart J, and Liu W. Comparing morphometric, biochemical, and visual measurements of macrovesicular steatosis of liver. Hum Pathol. 42: 356–360. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar D, Farrell GC, Fung C, and George J. Hepatitis C virus genotype 3 is cytopathic to hepatocytes: Reversal of hepatic steatosis after sustained therapeutic response. Hepatology. 36: 1266–1272. 2002. [DOI] [PubMed] [Google Scholar]
- 12.Horai Y, Utsumi H, Ono Y, Kishimoto T, Ono Y, and Fukunari A. Pathological characterization and morphometric analysis of hepatic lesions in SHRSP5/Dmcr, an experimental non-alcoholic steatohepatitis model, induced by high-fat and high-cholesterol diet. Int J Exp Pathol. 97: 75–85. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen HC, and Farese RV., Jr Determination of adipocyte size by computer image analysis. J Lipid Res. 43: 986–989. 2002. [PubMed] [Google Scholar]
- 14.Fukuda-Tsuru S, Kakimoto T, Utsumi H, Kiuchi S, and Ishii S. The novel dipeptidyl peptidase-4 inhibitor teneligliptin prevents high-fat diet-induced obesity accompanied with increased energy expenditure in mice. Eur J Pharmacol. 723: 207–215. 2014. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe Y, Nakayama K, Taniuchi N, Horai Y, Kuriyama C, Ueta K, Arakawa K, Senbonmatsu T, and Shiotani M. Beneficial effects of canagliflozin in combination with pioglitazone on insulin sensitivity in rodent models of obese type 2 diabetes. PLoS One. 10: e0116851 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunnill MS. Quantitative methods in the study of pulmonary pathology. Thorax. 17: 320–328. 1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aeffner F, Wilson K, Bolon B, Kanaly S, Mahrt CR, Rudmann D, Charles E, and Young GD. Commentary: Roles for pathologists in a high-throughput image analysis team. Toxicol Pathol. 44: 825–834. 2016. [DOI] [PubMed] [Google Scholar]
- 18.Garrido R, Zabka TS, Tao J, Fielden M, Fretland A, and Albassam M. Quantitative histological assessment of xenobiotic-induced liver enzyme induction and pituitary-thyroid axis stimulation in rats using whole-slide automated image analysis. J Histochem Cytochem. 61: 362–371. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwak JT, Hewitt SM, Kajdacsy-Balla AA, Sinha S, and Bhargava R. Automated prostate tissue referencing for cancer detection and diagnosis. BMC Bioinformatics. 17: 227 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atupelage C, Nagahashi H, Kimura F, Yamaguchi M, Tokiya A, Hashiguchi A, and Sakamoto M. Computational hepatocellular carcinoma tumor grading based on cell nuclei classification. J Med Imaging (Bellingham). 1: 034501 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]